Understanding how multiple micro RNAs (miRNAs) regulate their targets genome-wide holds promise for deciphering complex cellular regulatory networks. In this study, Hoefert et al. use CLEAR-CLIP to dissect miRNA-200 family targeting and reveal a novel role for these miRNAs in regulating cell adhesion and hair follicle morphogenesis.
Abstract
The microRNA (miRNA)-200 (miR-200) family is highly expressed in epithelial cells and frequently lost in metastatic cancer. Despite intensive studies into their roles in cancer, their targets and functions in normal epithelial tissues remain unclear. Importantly, it remains unclear how the two subfamilies of the five-miRNA family, distinguished by a single nucleotide within the seed region, regulate their targets. By directly ligating miRNAs to their targeted mRNA regions, we identify numerous miR-200 targets involved in the regulation of focal adhesion, actin cytoskeleton, cell cycle, and Hippo/Yap signaling. The two subfamilies bind to largely distinct target sites, but many genes are coordinately regulated by both subfamilies. Using inducible and knockout mouse models, we show that the miR-200 family regulates cell adhesion and orientation in the hair germ, contributing to precise cell fate specification and hair morphogenesis. Our findings demonstrate that combinatorial targeting of many genes is critical for miRNA function and provide new insights into miR-200’s functions.
Graphical Abstract
Introduction
miRNAs are a prominent class of small noncoding RNAs that regulate gene expression posttranscriptionally (Bartel, 2009; Ambros, 2011). In animals, miRNAs have been shown to recognize their targets by perfect base pairing between their 5′ end sequences, often called the seed region, and cognate mRNA sequences (Bartel, 2009). Studies of miRNA target recognition using molecular, computational, and structural approaches have independently demonstrated the importance of nucleotides 2–8 for miRNA target recognition (Lewis et al., 2003; Lim et al., 2005; Schirle et al., 2014). However, the prevalence of any given 7mer motif in mammalian genomes makes sequence-based miRNA target prediction challenging. Furthermore, nonperfectly matched miRNA–mRNA interactions have also been reported (Chi et al., 2012; Helwak et al., 2013; Moore et al., 2015). This further complicates efforts to reliably predict miRNA targets. In recent years, the development of techniques such as high-throughput sequencing of RNA (HITS) isolated by cross-linking immunoprecipitation (CLIP; HITS-CLIP) and photoactivatable ribonucleoside-enhanced (PAR)-CLIP, which directly purify mRNAs from the RNA-induced silencing complex (RISC) followed by next-generation sequencing, has established an experimental platform to identify RISC-bound mRNAs in a cellular context–specific manner (Chi et al., 2009; Hafner et al., 2010). To preserve the information for individual miRNA–mRNA binding events, several strategies have been devised to ligate miRNA to mRNA fragments when both are still bound by the same RISC (Helwak et al., 2013; Grosswendt et al., 2014; Moore et al., 2015).
Despite these technical advancements in identifying miRNA targets, however, functional studies of miRNAs remain challenging. It has become clear that a single miRNA–mRNA targeting event usually confers mild regulation of gene expression. In addition, many mRNAs are often bound by several different miRNAs. As a result, approaches focusing on a single miRNA are usually inadequate to resolve the redundancy embedded in the miRNA-regulated network. Indeed, large-scale knockout (KO) studies for individual miRNAs in Caenorhabditis elegans and mice indicate that many individual miRNAs are dispensable for animal development (Miska et al., 2007; Park et al., 2012). These observations are in contrast with the dire consequences reported in numerous tissue-specific KOs of Drosha, Dgcr8, and Dicer1, in which all miRNAs are ablated by blocking miRNA biogenesis. The differences in phenotype between loss of individual miRNAs and blocking all miRNAs could stem from the number of dysregulated targets and/or the degree of compromised regulation for the same target. Therefore, a different approach to investigate miRNA functions beyond considering only shared seed sequences should be explored.
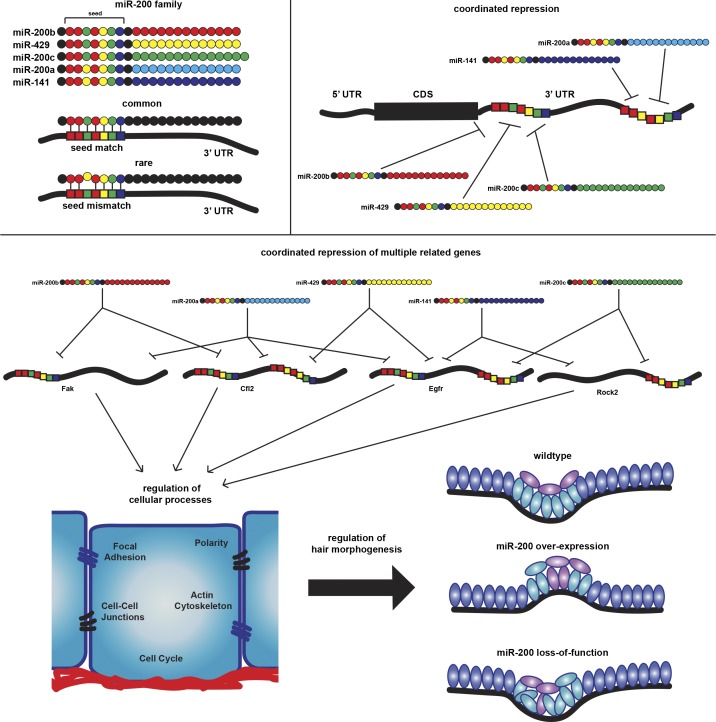
To begin to solve this problem, we focused on the miR-200 miRNA family, which consists of five miRNAs clustered in two genomic loci in both mouse and human. miR-200b/200a/429 are grouped into the miR-200b cluster, and miR-200c/141 are grouped into the miR-200c cluster (Fig. 1 A). Although classified as a single miRNA family, they share two seed sequences distinguished by a single-nucleotide difference within the seed region (Fig. 1 A). It is unclear whether the miR-200s sharing the same seed sequences recognize the same target sites and whether these two subfamilies regulate unique targets. At the functional level, miR-200s have been well documented for their tumor-suppressing roles in epithelial cancers, especially during metastasis. However, the tumor suppressor roles were largely demonstrated through gain-of-function approaches because miR-200s are frequently lost in metastatic epithelial cancer (Gregory et al., 2008; Korpal et al., 2008; Shimono et al., 2009). In addition, miR-200s have also been shown to promote metastasis by inhibiting individual targets such as Sec23a (Korpal et al., 2011). In contrast, the functions of miR-200s in normal epithelial tissues, where they are highly expressed, remain poorly understood.
Figure 1.
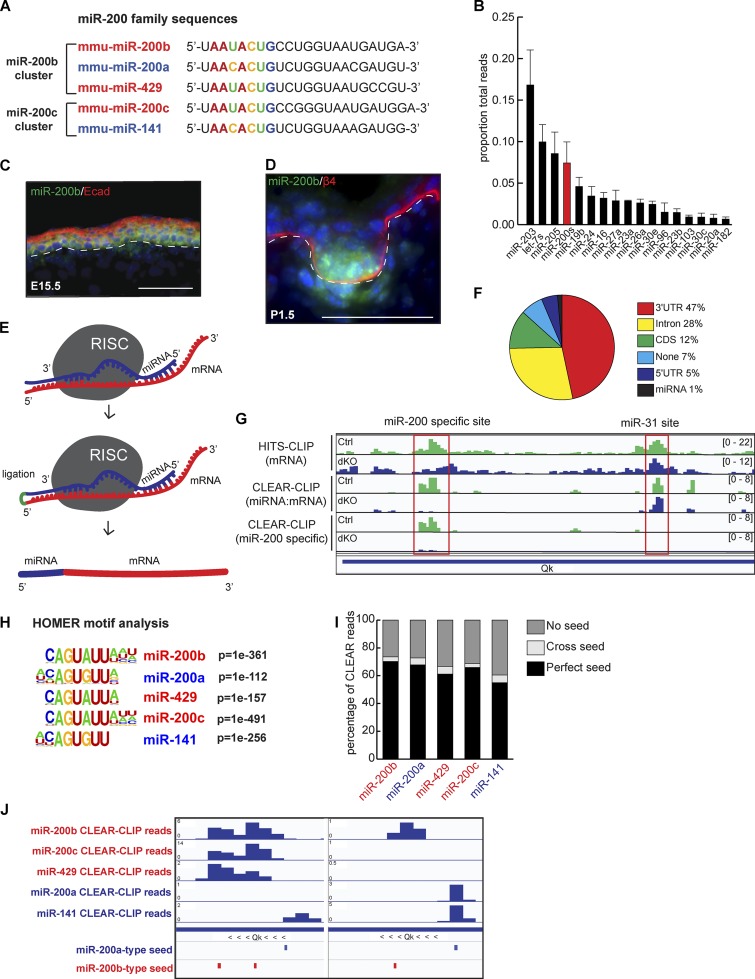
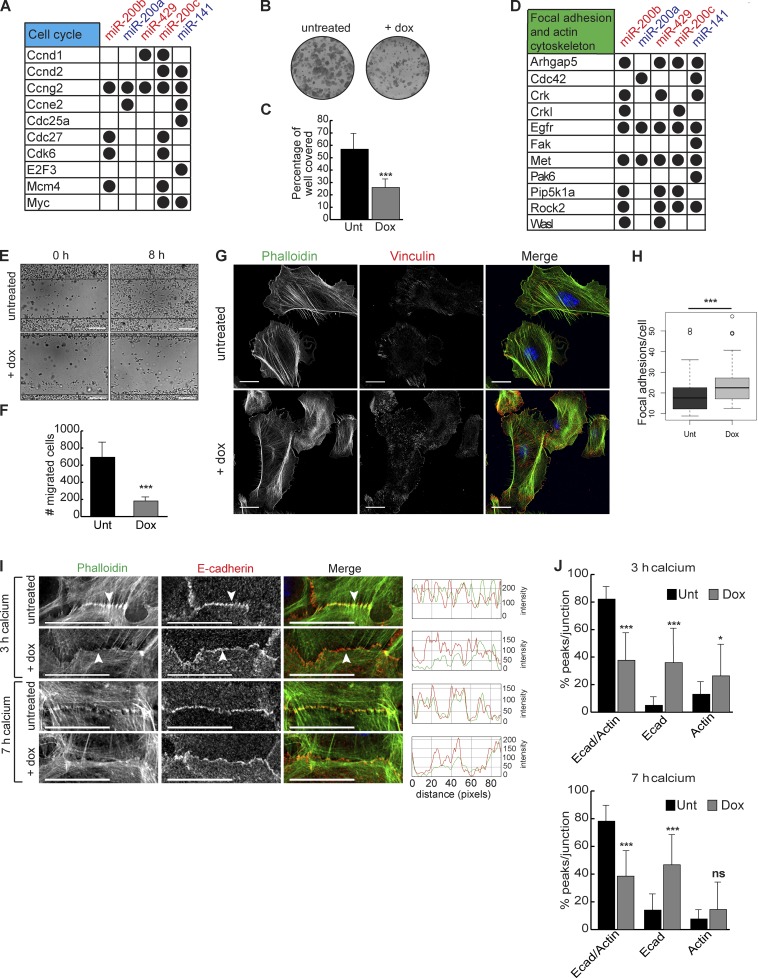
CLEAR-CLIP identifies targets for the miR-200 family. (A) The two miR-200 family clusters are shown by genomic cluster with the seed region in color. (B) miRNA-seq on whole epidermis at P4.5; n = 3; error bars are SD. (C and D) Fluorescence in situ hybridization of miR-200b in back skin at E15.5 (C) and P1.5 (D). Bars, 50 µm. (E) Schematic of CLEAR-CLIP technique. (F) Percentage of miR-200 family CLEAR-CLIP reads within each genomic region. CDS, coding sequence. (G) Integrative Genomics Viewer tracks showing reads from HITS-CLIP, CLEAR-CLIP, and miR-200–specific CLEAR-CLIP. (H) HOMER motif analysis of the mRNA fragment portion of CLEAR-CLIP reads from each miR-200 family member. (I) Percentage of CLEAR-CLIP reads from each family member containing a perfect seed, a cross seed, or no seed. (J) Integrative Genomics Viewer tracks for individual miR-200 member reads within a region of the Qk 3′ UTR.
Mammalian skin is an ideal system to study miRNA functions. When Dgcr8, Dicer1, or Argonaute (Ago) genes are conditionally ablated in developing skin, hair follicle morphogenesis is compromised, whereas epidermal lineages remain largely intact (Andl et al., 2006; Yi et al., 2006, 2008, 2009; Wang et al., 2012). The most prominent defects observed in hair morphogenesis are located within the hair germ (HG), a group of progenitor cells that undergo complex regulation to coordinate adhesion, migration, proliferation, and signaling events (St-Jacques et al., 1998; Andl et al., 2002; Jamora et al., 2003; Ahtiainen et al., 2014; Ouspenskaia et al., 2016). In this study, we apply the covalent ligation of endogenous Argonaute-bound RNAs (CLEAR)-CLIP technique for cell type–specific miRNA–mRNA target identification and, guided by these results, use gain- and loss-of-function mouse models to examine the functions of the miR-200 family in regulating cell adhesion and proliferation in vitro and in regulating cell adhesion, polarity, and cell fate specification during hair morphogenesis in vivo. We identify numerous novel targets enriched in the pathways of focal adhesion (FA), actin cytoskeleton, cell polarity, cell cycle, and Hippo signaling as miR-200’s targets. Importantly, many of these targets are coordinately regulated by multiple members of the miR-200 family. When the levels of miR-200s are genetically altered, cell adhesion and orientation in the HG are compromised, leading to imperfect cell fate specification. Our results provide new insights into the functions of the miR-200 family in skin development and establish an example of deciphering complex miRNA-regulated gene networks.
Results
Global identification of miR-200 family targets with CLEAR-CLIP
We performed quantitative miRNA sequencing (miR-seq; Zhang et al., 2013) and found that the miR-200 family is among the most abundantly expressed miRNAs in the neonatal skin (Fig. 1 B). Of note, all five members of the miR-200 family are expressed at similar levels judging from the miR-seq data (Fig. S1 A). We then visualized spatiotemporal expression of the family using a probe for miR-200b during embryonic skin development. miR-200b is broadly expressed in epidermal progenitors at embryonic day 15.5 (E15.5; Fig. 1 C) but is more strongly enriched in the HG by E17.5 (Fig. S1 B) and postnatal day 1.5 (P1.5; Fig. 1 D). For the miR-200 family, a single-nucleotide difference within the seed region of the subfamilies raises the uncertainty of assigning miRNA targets to individual miRNAs based on bioinformatic prediction alone. To gain the most comprehensive view of miR-200 family target regulation as well as to distinguish between targeting by different family members, we adapted a recently reported CLEAR-CLIP technique to capture miRNAs and their targets in mouse keratinocytes (Moore et al., 2015). This method directly ligates Ago2-bound miRNAs to their cognate mRNA fragments after cross-linked immunoprecipitation (Fig. 1 E). We have previously demonstrated that Ago2 is the most abundantly expressed (∼60%) of the Ago proteins in the skin and that its associated miRNA profile is generally similar to that of the other two more minor Ago proteins in the skin, Ago1 and Ago3 (Wang et al., 2012). Thus, Ago2 CLEAR-CLIP should represent global miRNA–mRNA interactions. To control for specificity, we generated miR-200 double KO (dKO) keratinocytes (see below for details) and used both WT and dKO cells for CLEAR-CLIP experiments.
Among all unique reads mapped to mRNAs, we identified 1,521,535 miRNA–mRNA hybrid reads and 80,154,975 mRNA-only reads (HITS-CLIP reads). Among miR-200 CLEAR-CLIP reads, 47% were mapped to the 3′UTR regions of mRNAs, comparable with previously published results (Fig. 1 F; Moore et al., 2015). Because of the prominence of 3′UTRs in mediating miRNA-dependent regulation (Bartel, 2009), we focused our remaining analyses on those regions. We used the HITS-CLIP reads to map Ago2-interacting mRNA regions and used the CLEAR-CLIP reads to determine miRNA-guided miRNA–mRNA interactions. Comparison of HITS-CLIP and CLEAR-CLIP reads recovered from the same libraries revealed that CLEAR-CLIP provides information for miRNAs with more specific coverage of mRNA sequences (Fig. 1 G). Importantly, CLEAR-CLIP allowed us to distinguish binding events between different miRNAs (Fig. S1 C). We observed loss of miR-200 family interactions within the 3′UTR of Quaking (Qk) in the dKO sample but no loss of signal for miR-31, another highly expressed miRNA associated with a distinct region of the same 3′UTR (Fig. 1 G). This indicates that CLEAR-CLIP can distinguish each miRNA–mRNA binding event.
The miR-200 family shares two nearly identical seed sequences (Fig. 1 A). We thus referred to the seed of miR-200a/141 as the a-type seed and the seed of miR-200b/429/200c as the b-type seed. The absence of miR-200–mRNA CLEAR-CLIP reads and large reduction of HITS-CLIP reads in the dKO suggested that CLEAR-CLIP can specifically identify miR-200–recognized mRNA regions (Fig. 1 G). We then performed hypergeometric optimization of motif enrichment (HOMER) analysis (Heinz et al., 2010) on mRNA fragments ligated to each of the miR-200 family members. The most highly represented motif for each family member was its own seed sequence (Fig. 1 H). The most invariable sequences were perfectly matched to nucleotides 2–7 of each of the miR-200s. The eighth nucleotide position also showed a strong preference toward matching (a G–C pair). For the a-type miRNAs (miR-200a/141), A or U was enriched for the ninth nucleotide position. These data indicate that cross-seed recognition between a- and b-type miR-200s is rare. Indeed, when we directly searched miR-200–interacting mRNAs for seed sequences, we found that the majority of reads contained the cognate seed; just a small percentage of chimeric reads appear to be generated from an a-type miR-200 family member interacting with a b-type target site, and vice versa (Fig. 1 I). Interestingly, for each miR-200 member, ∼30% of targeted mRNA fragments did not harbor seed sequences (Fig. 1 I), indicating that a considerable amount of miRNA–mRNA association may be independent of the canonical seed match (Helwak et al., 2013; Moore et al., 2015).
CLEAR-CLIP reads allowed for unique identification of each miRNA binding site in an miRNA-specific manner. Interestingly, we observed many instances of concurrent regulation by multiple miR-200 family members on the same 3′UTR. In some cases, the same binding sites were recognized by multiple members, indicating that the miR-200 family cooperates in target recognition (Figs. 1 J and S1 C). Overall, ∼50% of our identified miR-200 targets were bound by more than one miR-200 member. These data and analyses further confirmed that miRNAs sharing the same seed sequences could bind the same site and that cross-seed binding events are rare. Simply based on the detected interactions between miR-200s and their bound mRNAs, we observed strong enrichment of miR-200s’s targets in pathways in cancer, endocytosis, FA, and actin cytoskeleton, among others (Fig. S1 D). Altogether, our CLEAR-CLIP study with miR-200 WT and dKO samples has generated a comprehensive map of miR-200–recognized targets in an miRNA–mRNA pair-specific manner and revealed strong coordination of target recognition among the five miR-200 family members.
miR-200s regulate FA, actin cytoskeleton, cell cycle, and the Hippo signaling pathway
We developed an inducible transgenic (Tg) mouse model to probe the effect of the coordinated regulation. Because the miR-200b cluster harbors both a- and b-type seeds and contains three out of the five miR-200s, we created a pTRE2-200b-cluster mouse model. These mice were then bred to mice harboring a Krt14rtTA allele (Jackson et al., 2013) to generate progeny with doxycycline-inducible overexpression of miR-200s (referred to as Tg; Fig. 2, A and B; and Fig. S2, A–C).
Figure 2.
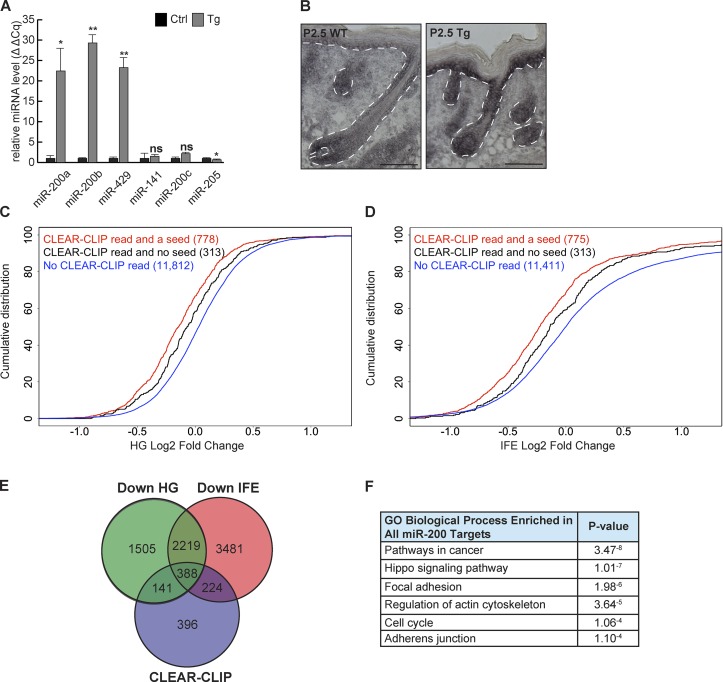
Identification of miR-200 family targets in the skin. (A and B) Validation of the induction of the miR-200b cluster by quantitative RT-PCR (A; n = 3; *, P < 0.05; **, P < 0.01; Student’s two-tailed t test; error bars are SD) and in situ hybridization (B; bars, 100 µm) from control and Tg whole epidermis. (C and D) Genes containing CLEAR-CLIP reads with a seed match are more strongly down-regulated than those without a seed match in Tg skin. Plots show HG-enriched population (C) and IFE-enriched population (D). (C) Red versus blue, P < 2.2−16; black versus blue, P = 3.26−5; red versus black, P = 0.005. (D) Red versus blue, P < 2.2−16; black versus blue, P = 0.0003; red versus black, P = 0.0009. Kolmogorov-Smirnov test. (E) Venn diagram of CLEAR-CLIP–identified targets and genes down-regulated in HG or IFE. (F) GO-term analysis on all miR-200 family targets.
Given the high expression of miR-200s within the HG, we examined whether miR-200s have specific targets in early stage hair follicles and the interfollicular epidermis (IFE). We purified HG and IFE populations using an established FACS strategy (Rhee et al., 2006) from control and miR-200 Tg skin at P0.5 (Fig. S2 D). We then performed RNA sequencing (RNA-seq) on each population and validated the enrichment of the HG population with gene set enrichment analysis (GSEA; Fig. S2 E; Subramanian et al., 2005).
The detected genes in the transcriptomes of cultured keratinocytes, HGs, and IFE largely overlapped, indicating that CLEAR-CLIP performed on keratinocytes would capture miR-200 targets in vivo (Fig. S2 F). Indeed, we observed that genes identified as miR-200 family targets through CLEAR-CLIP were robustly down-regulated with miR-200 overexpression in both HG and IFE (Fig. 2, C and D). Of note, CLEAR-CLIP–identified targets harboring the canonical seed match were more robustly down-regulated. However, genes with a CLEAR-CLIP read but no miR-200 seed match were still down-regulated, although to a more modest extent (Fig. 2, C and D). Thus, we defined genes with a CLEAR-CLIP read for any of the miR-200 family members and down-regulated >10% in our RNA-seq as miR-200 family targets (Fig. 2 E). Using these criteria, we identified 141 miR-200 family targets specific to HGs, 224 specific to IFE, and 388 shared between both populations, for a total of 753 miR-200 family targets in the skin.
The genome-wide CLEAR-CLIP and RNA-seq datasets allowed us to comprehensively identify most miR-200–regulated genes in the skin. For example, previously identified targets such as Sec23a (Korpal et al., 2011) and Cfl2 (Bracken et al., 2014) were recovered. However, we did not observe any miR-200–Zeb1/2 interaction because these mesenchymal genes are not expressed in normal epidermal cells, validating our cellular context–specific target identification. To gain insights into the miR-200–regulated pathways, we performed Gene Ontology (GO)-term analysis (Huang et al., 2009). We did not see strong enrichment for any distinct signatures between HG and IFE. This was not surprising given that HG and IFE populations are derived from the same lineage and share similar gene expression. We then considered all 753 targets identified for miR-200s from both populations and observed strong enrichment for gene categories such as pathways in cancer, the Hippo signaling pathway, FA, actin cytoskeleton, cell cycle, and adherens junctions (AJs; Fig. 2 F).
miR-200s coordinately repress many targets within a pathway
Although the regulation of some targets involved with the control of FA, actin cytoskeleton, and cell cycle has been previously reported in cancer cell lines (Bracken et al., 2014), the widespread and coordinated regulation of these pathways by the miR-200 family in normal skin cells is striking. In each of these regulated pathways, many, rather than a few, targets were identified as miR-200–regulated genes. In addition, we have identified many regulators of the Hippo signaling pathway as new miR-200 targets (Fig. 3 A). Furthermore, using the chimeric reads generated by CLEAR-CLIP, we were able to identify which miR-200 family member interacts with a specific target site. By examining genes involved with different processes, we observed that not only do miR-200s cooperate to repress the same targets, they further coordinate to repress multiple genes within these pathways (see below; Fig. 3 A).
Figure 3.
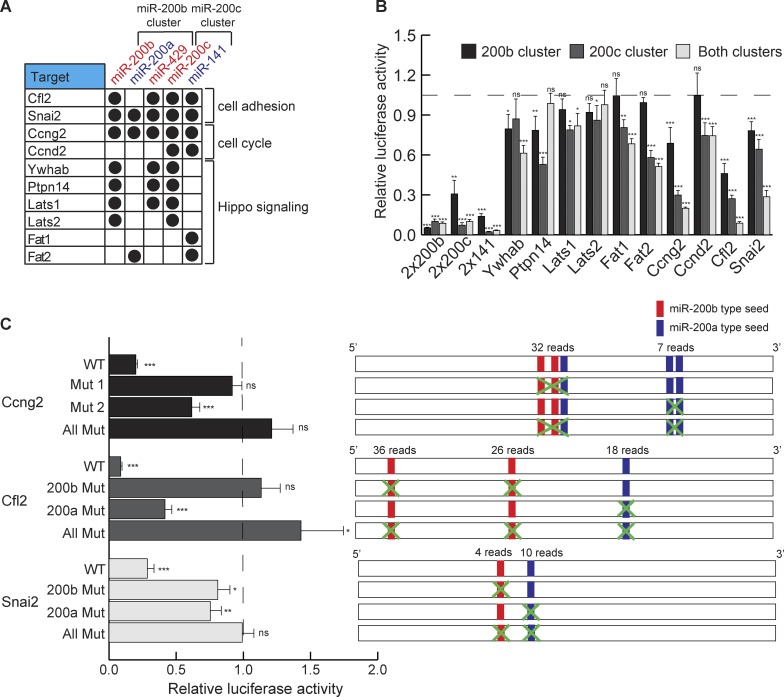
miR-200 members cooperate in repressing targets. (A) Genes identified as miR-200 family targets with specific miR-200 interactions. (B) Relative luciferase activity of 3′ UTRs from targets with expression of the miR-200b cluster, the miR-200c cluster, or both. The dashed line represents luciferase activity with no miR-200s; n = 6. (C) Relative luciferase activity of WT or mutated 3′ UTRs with expression of both miR-200 family clusters in dKO cells. Construct design for mutations is shown in the right panel. The dashed line represents luciferase activity with no miR-200s. n = 6. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s two-tailed t test; error bars are SD.
To validate these miR-200 targets and study coordinated regulation by the miR-200 family, we selected 10 new or previously validated targets for a heterologous luciferase assay (Fig. 3 B). All of the selected targets were confirmed by the luciferase assay, supporting our strategy to identify miR-200 targets by combining CLEAR-CLIP and RNA-seq. Of note, genes such as Ywhab, which was targeted by individual miR-200s from both clusters, were repressed more strongly in the presence of both clusters. In contrast, genes such as Fat1, which was recognized only by miR-141, were repressed only by the miR-200c but not the miR-200b cluster. Finally, the most potently repressed genes were Ccng2, Cfl2, and Snai2, all of which were coordinately targeted by both clusters. In the case of Ccng2 and Snai2, all five miR-200 members recognized their 3′UTRs, leading to strong regulation (Fig. 3 B). To further dissect how miR-200s coordinately regulate each target by binding to multiple sites in the 3′UTR, we mutated miR-200 target sites in the 3′UTRs of Ccng2, Cfl2, and Snai2 individually and combinatorially and then measured their response with the luciferase assay. We found that each site was effective in mediating repression and that combined deletion of all sites always led to the strongest derepression of luciferase activity (Fig. 3 C). Notably, sites with more CLEAR-CLIP reads generally showed stronger effects of miR-200–mediated repression, perhaps reflecting the quantitative nature of the CLEAR-CLIP for target capture (Fig. 3 C). Collectively, we have identified a large number of coordinately regulated targets of miR-200s in several important pathways of the skin.
Induction of the miR-200 family represses cell proliferation, migration, and AJs
To probe the cellular functions of miR-200s, we first analyzed the defects in inducible keratinocytes in culture. Among the strongest signatures we observed for miR-200 family targets were positive regulators of cell cycle (Fig. 4 A). Consistent with these findings, induction of the miR-200b cluster by doxycycline caused a marked reduction in colony formation ability (Fig. 4, B and C; and Fig. S3 A).
Figure 4.
miR-200 overexpression represses proliferation and migration and affects FA and AJ formation. (A) miR-200 targets involved with cell cycle. (B and C) Colony formation assay of Tg keratinocytes untreated (Unt) or treated with doxycycline (Dox). n = 6. (D) miR-200 targets involved with FA and actin cytoskeleton. (E) Scratch assay on Tg keratinocytes untreated or treated with doxycycline 0 or 8 h after scratching. (F) Number of cells migrated in E. n = 3. (G and H) Immunofluorescence and quantification of vinculin and phalloidin stain for actin on Tg keratinocytes untreated or treated with doxycycline. n = 76 untreated; n = 77 doxycycline treated. (I) Immunofluorescence of E-cadherin (Ecad) and phalloidin staining for actin on Tg keratinocytes untreated or treated with doxycycline and induced with calcium for 3 or 7 h. Arrowheads indicate actin localization to E-cadherin. Red-green-blue (RGB) line scans over 90 pixels across each cell junction. Representative images from n > 20 cell junctions imaged for each treatment. (J) Percentage of peaks per junction shared between E-cadherin and actin or exclusive to E-cadherin or actin treated as in I. 3-h untreated, n = 21; 3-h doxycycline, n = 23; 7-h untreated, n = 19; 7-h doxycycline, n = 26. *, P < 0.05; ***, P < 0.001; Student’s two-tailed t test; error bars are SD. Bars: (E) 100 µm; (G and I) 20 µm.
Another strong signature we observed for miR-200 family targets were regulators involved in cell migration, including FA and actin cytoskeleton (Figs. 4 D and S3, C and D). Induction of the miR-200b cluster inhibited keratinocyte migration in a scratch assay (Fig. 4, E and F; and Fig. S3 B). Because several key regulators of FA and actin such as Ptk2 (also known as FA kinase [Fak]), Cdc42, Crk, Rock2, and Wasl (also known as N-WASP) were identified as miR-200 targets, we visualized FA and actin cytoskeleton in cultured keratinocytes. Upon doxycycline treatment of Tg but not WT keratinocytes, we observed an increased number of FAs judging by staining for vinculin and actin (Fig. 4, G and H; and Fig. S3, E and F). The size of the FAs in the induced cells also appeared to be smaller than those observed in the control cells. This finding suggests that when many components involved with FA formation are repressed by miR-200s, it leads to smaller but more numerous FAs. Because actin cytoskeletal dynamics and the formation of AJs are intimately linked (Vasioukhin et al., 2000; Kanchanawong et al., 2010; Ratheesh and Yap, 2012; Case and Waterman, 2015) and because many genes involved with filopodia formation such as Rock2, Cdc42, Wasl, and Actn1 (also known as α-actinin) were targets of miR-200s (Figs. 4 D and S3, C and D), we examined the formation of AJs in control and miR-200b cluster–induced keratinocytes upon calcium treatment. As expected, the relocation of E-cadherin to the cell membrane as well as the attachment of actin stress fiber to the newly formed AJs were readily detectable by 3 h after calcium addition in control cells (Figs. 4 I and S3, G and H). Strikingly, although the relocation of E-cadherin was still visible in Tg cells, the attachment of actin stress fibers to E-cadherin was strongly repressed (Fig. 4, I and J). By 7 h after calcium addition, whereas the control cells had largely sealed cell–cell contacts, and the colocalization of E-cadherin and actin stress fibers was clearly visible on the cell cortex, the miR-200b cluster–induced cells still showed E-cadherin zippers, and actin stress fibers were generally not organized at the cell cortex, reflecting impaired AJ formation (Fig. 4, I and J; and Fig. S3 H).
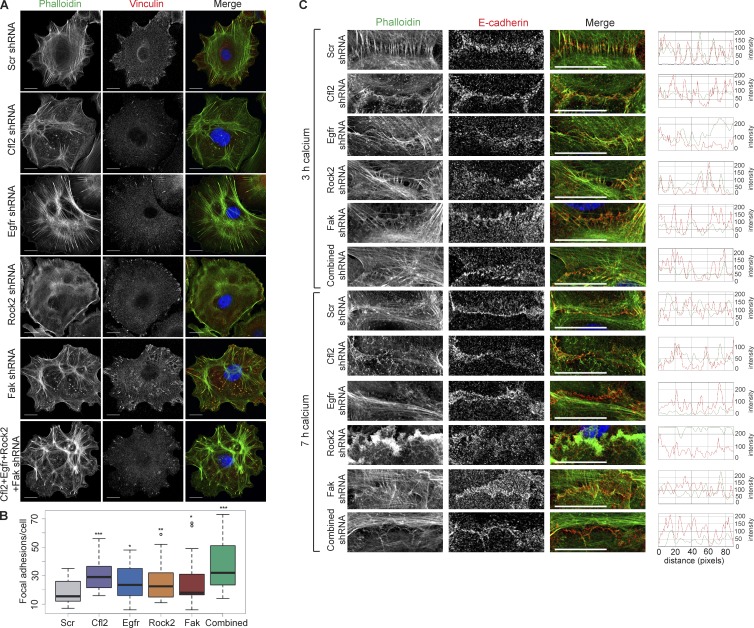
To determine how miR-200s regulate the formation of FA and AJs through the repression of multiple targets, we first knocked down (KD) Cfl2, Egfr, Rock2, and Fak individually and combinatorially (Fig. S3 I) and examined the effects on the FAs and AJs. We noted that individual KD experiments resulted in a substantial decrease in gene expression (∼80% down-regulation), whereas combinatorial KD experiments resulted in a mild decrease in gene expression (∼30% down-regulation for each target) that was more similar to miRNA-mediated gene repression. In WT keratinocytes, KD of each target resulted in an increase in the number of FAs, and KD of all four targets together resulted in the largest increase (Fig. 5, A and B). This suggests that coinhibition of multiple targets in the FA and actin cytoskeleton pathways mimics the effect of miR-200s expression. We next measured the AJ formation under the same KD conditions. KD of individual targets had variable effects on the attachment of actin stress fibers to E-cadherin, with KD of Cfl2 providing the strongest effect from an individual target. However, the combinatorial KD showed strong defects in actin attachment, similar to what was observed in miR-200b–induced cells (Fig. 5 C). Together, these data provide proof-of-principle evidence that corepression of multiple targets by miR-200s is critical for their role in regulating FAs and AJs.
Figure 5.
Multiple miR-200 targets are involved with FA and AJ formation. (A) Immunofluorescence of vinculin and phalloidin stain for actin on WT keratinocytes infected with shScr, shCfl2, shEgfr, shRock2, shFak, or a combination of all shRNAs. (B) Quantification of FAs per cell in A. n = 34 Scr, n = 19 shCfl2, n = 26 shEgfr, n = 35 shFak, n = 20 shRock2, and n = 24 combined. (C) Immunofluorescence of E-cadherin and phalloidin stain for actin on WT keratinocytes infected with shScr, shCfl2, shEgfr, shRock2, shFak, or a combination of all shRNAs and induced with calcium for 3 or 7 h. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s two-tailed t test. Bars, 20 µm.
Induction of the miR-200 family represses the Hippo pathway
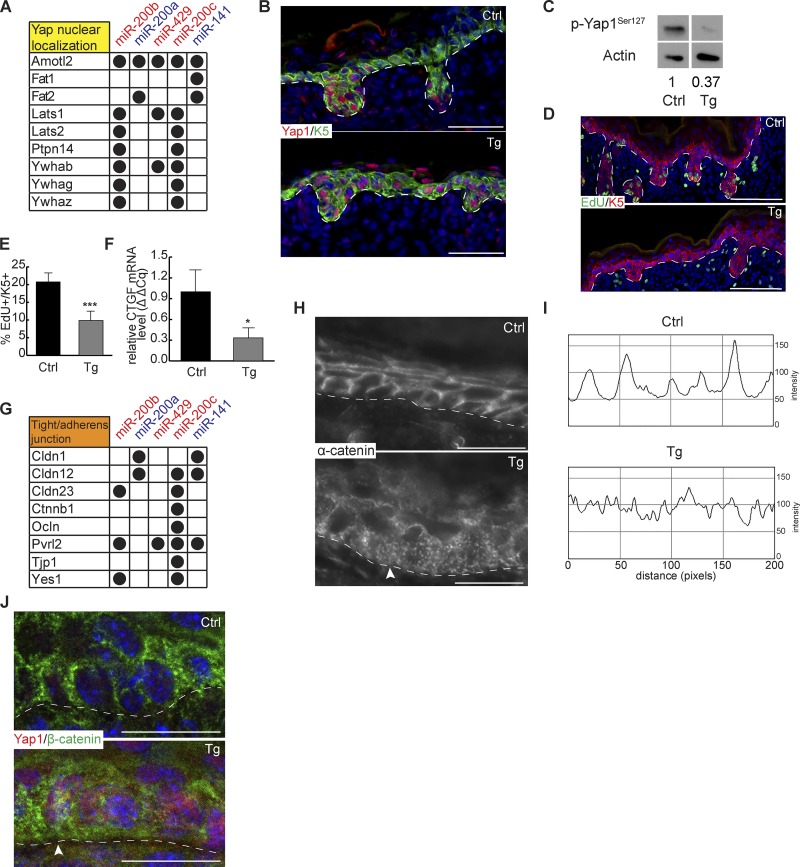
We next probed the effect of miR-200s on the Hippo signaling pathway. We detected many miR-200–targeted genes whose expression positively enhances Yap1 phosphorylation including Lats1, Lats2, Ptpn14, and others that promote cytoplasmic retention of phosphorylated Yap1 such as Ywhab (Fig. 6 A; Meng et al., 2016; Plouffe et al., 2016). Thus, we first determined the nuclear/cytoplasmic localization of Yap1 in induced skin. Consistent with the inhibition of these targets by miR-200s, we observed increased nuclear localization of Yap1 within the Tg IFE in vivo and within miR-200b cluster–induced keratinocytes in vitro (Figs. 6 B and S4, A and B). Western blotting of phosphorylated Yap1 further confirmed the reduced Yap1 phosphorylation in the Tg skin (Fig. 6 C). Nuclear accumulation of Yap1 is usually associated with increased cell growth and hyperproliferation (Meng et al., 2016). However, multiple positive cell cycle regulators were direct miR-200 targets (Fig. 4 A). Consistent with the decreased colony formation in induced keratinocytes in vitro (Fig. 4, B and C), we observed reduced cell proliferation in both IFEs and HGs of the Tg skin in vivo (Fig. 6, D and E). In addition, among miR-200–targeted genes, we noticed that direct targets of Yap1 such as Ctgf were repressed by miR-200s (Fig. 6 F). These data indicate that the progrowth effect of nuclear Yap1 accumulation is in turn dampened by miR-200s. Because cytoplasmic Yap1 has been shown to associate with cytoskeletal proteins such as α-catenin (Schlegelmilch et al., 2011) and many miR-200–regulated genes are key regulators of tight junctions and AJs (Figs. 6 G and S4 C), we next analyzed the cytoplasmic effect of miR-200 overexpression. We hypothesized that increased miR-200 expression may inhibit tight junction and AJ formation through combined down-regulation of their targets in these pathways. Indeed, when we visualized α-catenin expression in Tg skin, we observed more scattered and diffuse patterns of α-catenin in contrast with the strong membrane-localized expression in control skin (Fig. 6, H and I). Furthermore, puncta-like and diffuse α-catenin signals were seen near the basal surface, in contrast with the apical and lateral localization of α-catenin in the control (Fig. 6 H). We also noted that nuclear Yap1 appeared to correlate with cells having more puncta-like β-catenin expression on the membrane, whose diffuse patterns mirrored that of α-catenin (Fig. 6 J). These data reveal the functions of miR-200s in repressing cytoplasmic localization of Yap1 when overexpressed.
Figure 6.
miR-200s regulate Yap nuclear localization and cell junction formation. (A) miR-200 targets involved with Yap nuclear localization. (B) Immunofluorescence of Yap1 and Krt5 on control and Tg back skin at P0.5. Representative image from n = 6. (C) Immunoblot of p-Yap1Ser127 and actin from control and Tg whole epidermis at P0.5. n = 4. P = 0.073. Numbers underneath blots indicate relative expression level compared with control, which was set to 1. (D) Immunofluorescence of EdU and Krt5 on ctrl and Tg back skin at P0.5. (E) Quantification of EdU+/K5+ cells in control and Tg epidermis. n = 6. (F) Quantitative RT-PCR for Ctgf in control and Tg whole–back skin epidermis. n = 3. (G) miR-200 targets involved in tight junctions and AJs. (H) Immunofluorescence of α-catenin in control and Tg back skin at P0.5. The arrowhead indicates punctate localization of α-catenin in Tg skin. Representative image from n = 3. (I) α-catenin fluorescence line scan parallel to basement membrane across 200 pixels. (J) Immunofluorescence of β-catenin and Yap1 in control and Tg back skin at P0.5. The arrowhead indicates colocalization of nuclear Yap1 with disrupted β-catenin. Representative image from n = 6. *, P < 0.05; Student’s two-tailed T test; error bars are SD. Bars: (B and D) 100 µm; (H and J) 50 µm.
Induction of the miR-200 family compromises hair follicle development
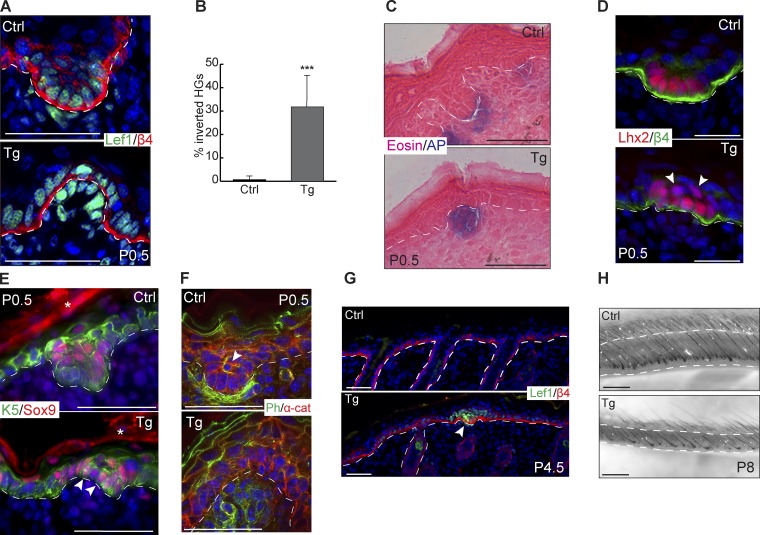
We next examined the phenotypes of Tg hair follicles. In addition to the compromised cell proliferation, the other prominent defect in Tg skin was evagination of HGs (Fig. 7 A). Overall, ∼30% of HGs in Tg skin showed abnormal evagination into the epidermis (Fig. 7 B). The evaginated HGs still had a close interaction between the epidermal cells and the dermal papillae cells as shown by both Lef1 (Fig. 7 A) and alkaline phosphatase staining (Fig. 7 C). During normal HG formation, the cells at the leading edge receive strong Wnt signaling and express a high level of Lhx2 (Rhee et al., 2006). In contrast, Sox9-expressing cells are usually located in the suprabasal layer of the HG, one cell layer away from the leading edge, as a result of asymmetric cell division and reduced Wnt activity in those cells (Ouspenskaia et al., 2016). In Tg skin, however, the demarcation between the Lhx2+ cells at the leading edge and the suprabasal Sox9+ cells was compromised. Instead, Lhx2+ cells were also seen to expand into the suprabasal layers, and Sox9+ cells were sometimes located at the basal layer (Fig. 7, D and E). Phalloidin staining further revealed reduced actin cytoskeleton signal in the HG cells. The strong actin bundle signals characteristic of the Sox9+ cells were largely lost, and dermal papillae cells also had reduced actin bundle signals (Fig. 7 F). By P4.5, the evaginated HGs appeared to be arrested, suggesting that evaginated HGs do not lead to productive hair follicles (Fig. 7 G). By P8, we observed a marked decrease in the ability of hair follicles to fully grow down into the dermis, a hallmark of hair follicle maturity at this stage (Fig. 7 H). These results were consistent with the inhibition of cell growth and migration, leading to the failure of hair follicle downward growth. Collectively, when miR-200s are induced in the skin, cell proliferation and hair morphogenesis are severely compromised, resulting in evaginated HG formation with compromised progenitor cell fate specification.
Figure 7.
miR-200 overexpression causes evaginated HGs. (A) Immunofluorescence of Lef1 and β4-integrin on control and Tg HGs at P0.5. (B) Percentage of control and Tg HGs with evaginated appearance. ***, P < 0.001; Student’s two-tailed t test; error bars are SD. (C) Alkaline phosphatase and eosin staining on control and Tg back skin at P0.5. (D) Immunofluorescence of Lhx2 and β4-integrin on control and Tg HGs at P0.5. Arrowheads indicate suprabasal Lhx2+ cells. (E) Immunofluorescence of Sox9 and Krt5 on control and Tg HGs at P0.5. Arrowheads indicate mislocalized Sox9+ cells, and asterisks indicate nonspecific secondary antibody staining. (F) Immunofluorescence of α-catenin with phalloidin stain for actin on control and Tg HGs at P0.5. The arrowhead indicates an actin bundle in central suprabasal cell. (G) Immunofluorescence of Lef1 and β4-integrin on control and Tg back skin at P4.5. The arrowhead indicates arrested HG. (H) Brightfield images of whole-mounted control and Tg back skin at P8. All images representative from n = 6 pairs of animals except for P4.5 (n = 3) and P8 (n = 1 because of neonatal lethality). Bars: (A and C–F) 50 µm; (G) 100 µm; (H) 500 µm.
Development of a miR-200 dKO mouse model
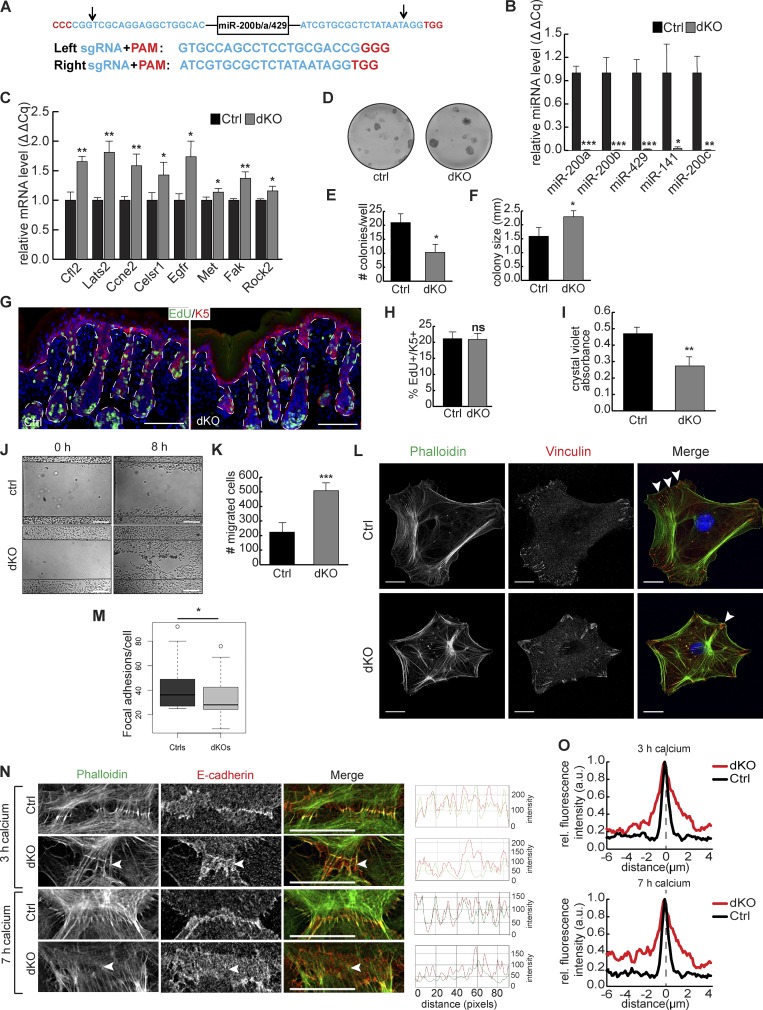
To study the requirement for miR-200s in skin development, we generated a dKO mouse model that ablates all five miR-200 family members. We first used CRISPR-Cas9–mediated genome editing to delete the entire miR-200b cluster by coinjecting Cas9 mRNA and two single-guide RNAs (sgRNAs) with predicted cleavage sites flanking the miR-200b cluster (Figs. 8 A and S4, D and E). We then bred mice heterozygous for the miR-200b/a/429 cluster (miR-200b/a/429+/−) with mice harboring an existing floxed allele for the miR-200c cluster (Park et al., 2012) and also harboring Krt14-cre. We confirmed by quantitative PCR (qPCR) that all five miR-200 members were deleted (Fig. 8 B), and several miR-200s–targeted genes were robustly derepressed in the dKO skin (Fig. 8 C).
Figure 8.
Ablation of all five miR-200 members enhances proliferation, cell migration, and cell adhesion and also affects FA and AJ formation. (A) Schematic of sequence targeted and guide RNAs used to delete miR-200b cluster. Arrows indicate predicted CRISPR cleavage sites. These sites were subsequently validated in Fig. S4 D. (B) Quantitative RT-PCR from whole epidermis of control and dKO animals at P0.5. n = 3. (C) Quantitative RT-PCR for miR-200 targets in control and dKO keratinocytes. n = 3. (D) Colony formation assay on control and dKO keratinocytes. (E and F) Number (E) and size (F) of colonies formed by control and dKO keratinocytes. n = 3. (G) Immunofluorescence for EdU and Krt5 in back skin of control and dKO animals at P0.5. (H) Percentage of EdU+/K5+ cells in control and dKO epidermis. n = 3. (I) Crystal violet absorbance of control and dKO keratinocytes plated in cell adhesion assay. n = 3. (J) Scratch assays on control and dKO keratinocytes 0 or 8 h after scratching. (K) Number of migrated cells from J. n = 3. (L) Immunofluorescence of vinculin and phalloidin stain for actin on control and dKO keratinocytes. Arrowheads indicate examples of FAs. (M) Number of FAs per cell in L. n = 43 control cells; n = 40 dKO cells. (N) Immunofluorescence of E-cadherin and phalloidin staining for actin on control and dKO keratinocytes induced with calcium for 3 or 7 h. Arrowheads indicate E-cadherin localization to cell cortex. RGB line scans over 90 pixels across each cell junction. Representative images from n > 20 cell junctions imaged for each treatment. Bars: (G) 100 µm; (L and N) 20 µm. (O) Mean fluorescence intensity of E-cadherin across cell junction. Distance of 0 and dashed line indicate center of junction. 3-h K14cre, n = 22; 3-h dKO, n = 22; 7-h K14cre, n = 21; 7-h dKO, n = 29; normalized to maximum intensity for each condition. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s two-tailed t test; error bars are SD.
Single KOs for either miR-200b/a/429 or miR-200c/141 were indistinguishable from control littermates and lacked any discernible skin defects. The lack of skin defects in single KOs was not unexpected, given the high degree of coordinated targeting of many genes by all five family members. At birth, loss of the entire miR-200 family in the skin produced dKO pups indistinguishable from littermate controls (Fig. S4 F). However, usually after the first week upon birth, dKO pups became significantly smaller than littermates, leading to unexpected neonatal lethality (Fig. S4 G). Although the exact cause was unknown, it was likely a result of malnutrition and competition with the control pups. We analyzed newborn skin to avoid complications from secondary defects associated with lethality. Immediately after birth, epidermal differentiation was normal, as judged by faithful expression of loricrin and normal skin morphology (Fig. S4, H and I), and hair follicle staging revealed no difference in either the stage or the number of hair follicles in dKO animals compared with controls at P0.5 (Fig. S4, J and K).
Loss of miR-200s increases proliferation and migration and compromises AJs in vitro
We first examined how loss of miR-200s affected proliferation and migration, given the strong alterations observed in the Tg model. Complete loss of all five miR-200s caused an increased colony-forming ability. Although fewer colonies formed from dKO keratinocytes, the colonies that formed were larger, indicating an enhanced proliferative ability of dKO cells (Fig. 8, D–F) However, when we quantified EdU incorporation in IFE and hair follicles in newborn skin, we did not detect discernible differences, indicating that miR-200s are not required to suppress cell cycle at this stage in vivo, when most cells are still highly proliferative (Fig. 8, G and H; and Fig. S4 L). We also observed a decreased ability of miR-200 dKO keratinocytes to adhere in a cell adhesion assay (Fig. 8 I). We next observed that loss of the entire family enhanced the ability of cells to migrate in a scratch assay, corroborating the compromised cell adhesion of the dKO cells (Fig. 8, J and K). Because many miR-200 family targets are involved with FA and actin cytoskeleton, we assessed FA formation and actin stress fibers in control and dKO keratinocytes. We observed fewer but much larger FAs in the dKO cells (Fig. 8, L and M). Actin stress fibers showed enhanced actin bundle signals toward FAs. Furthermore, radially organized actin bundles were often seen in dKO keratinocytes, in contrast with circumferentially organized actin bundles usually seen in control cells (Fig. 8 L). We further examined the formation of AJs in miR-200 dKO cells. In contrast with the inability of actin bundles to attach to E-cadherin with miR-200 overexpression, E-cadherin relocation to the cell cortex was disrupted with loss of miR-200s accompanied by disorganized actin stress fibers (Figs. 8 N and S5 A). Whereas control cells showed precise localization of E-cadherin along the cell junction, loss of miR-200s resulted in more diffuse localization (Fig. 8, N and O). When E-cadherin was properly localized to the membrane in dKO cells, actin polymerization and attachment was detected (Fig. 8 N). Together, these results suggest that loss of the entire miR-200 family enhanced cell proliferation, reduced cell adhesion, enlarged FAs, and compromised AJ formation in vitro.
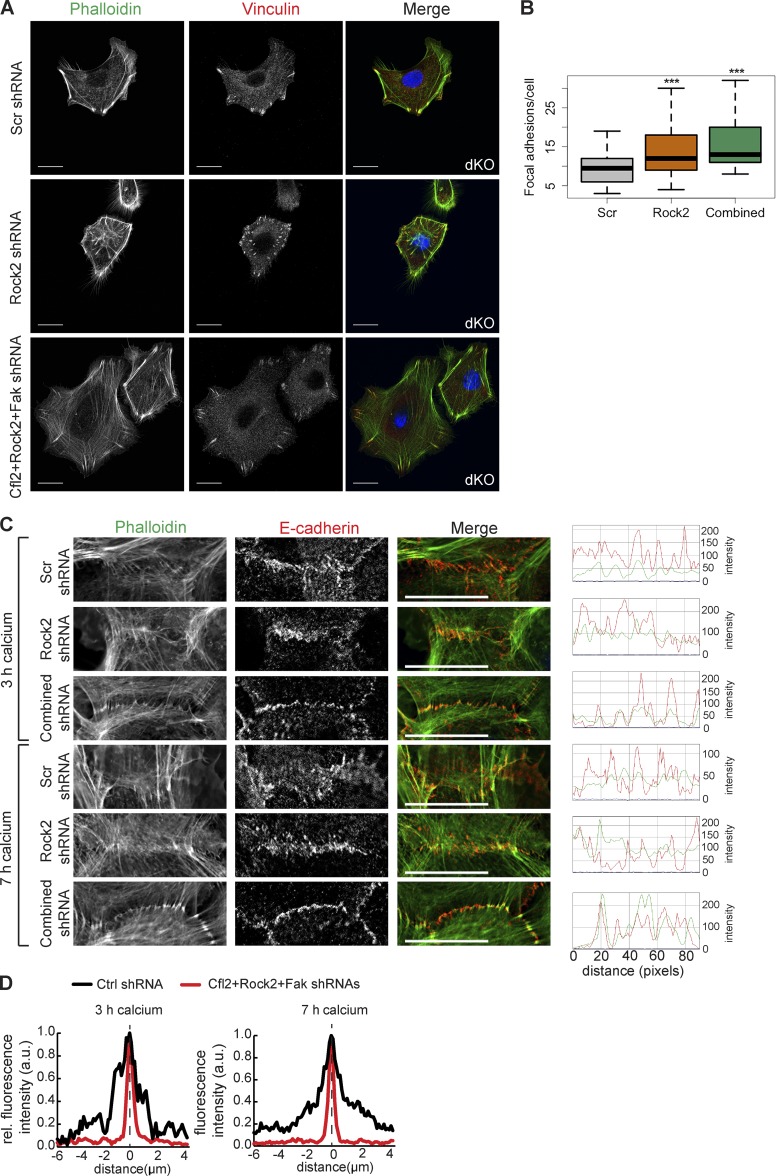
Next, we asked whether derepression of multiple miR-200 targets contributes to the FA and AJ defects. We performed the same KD experiments from Fig. 5 in miR-200 dKO keratinocytes. However, the KD of Cfl2, Egfr, and Fak individually as well as all four targets combinatorially led to widespread death of dKO cells, preventing us from analyzing the phenotypes. Instead, we were able to KD Rock2 individually and Cfl2, Rock2, and Fak combinatorially (Fig. S5 B). We indeed observed the rescue of FA numbers and, in the case of combinatorial KD, the size and number of FAs were both rescued to the level similar to WT control cells (Fig. 9, A and B). In a similar manner, the combinatorial KD of Cfl2, Rock2, and Fak in dKO cells largely rescued the inability of E-cadherin to precisely localize to the cell membrane and thus also rescued the ability of actin fibers to attach to the zipper during the AJ formation (Fig. 9, C and D). Altogether, these data further support the idea that derepression of multiple targets in the miR-200 dKO contributes to the regulation of FAs and AJs.
Figure 9.
Simultaneous KD of multiple miR-200 targets in dKO cells rescues FA and cell junction phenotypes. (A) Immunofluorescence of vinculin and phalloidin stain for actin on dKO keratinocytes infected with Scr, Rock2, or a combination of Rock2, Cfl2, and Fak shRNAs. (B) Number of FAs per cell in A. n = 42 Scr; n = 25 shRock2; n = 42 combined. (C) Immunofluorescence of E-cadherin and phalloidin staining for actin on dKO keratinocytes infected with Scr, shRock2, or a combination of Cfl2, Rock2, and Fak shRNAs and induced with calcium for 3 or 7 h; line scans over 90 pixels across each cell junction; representative images from n > 20 cell junctions imaged for each treatment. (D) Mean fluorescence intensity of E-cadherin across cell junction. Distance of 0 and dashed line indicate center of junction. 3-h Scr, n = 10; 3-h combined, n = 28; 7-h Scr, n = 18; 7-h combined, n = 38; normalized to maximum intensity for each condition. *, P < 0.05; Student’s two-tailed t test. Bars, 20 µm.
Loss of miR-200s compromises cell orientation and increases the variability of cell fate specification
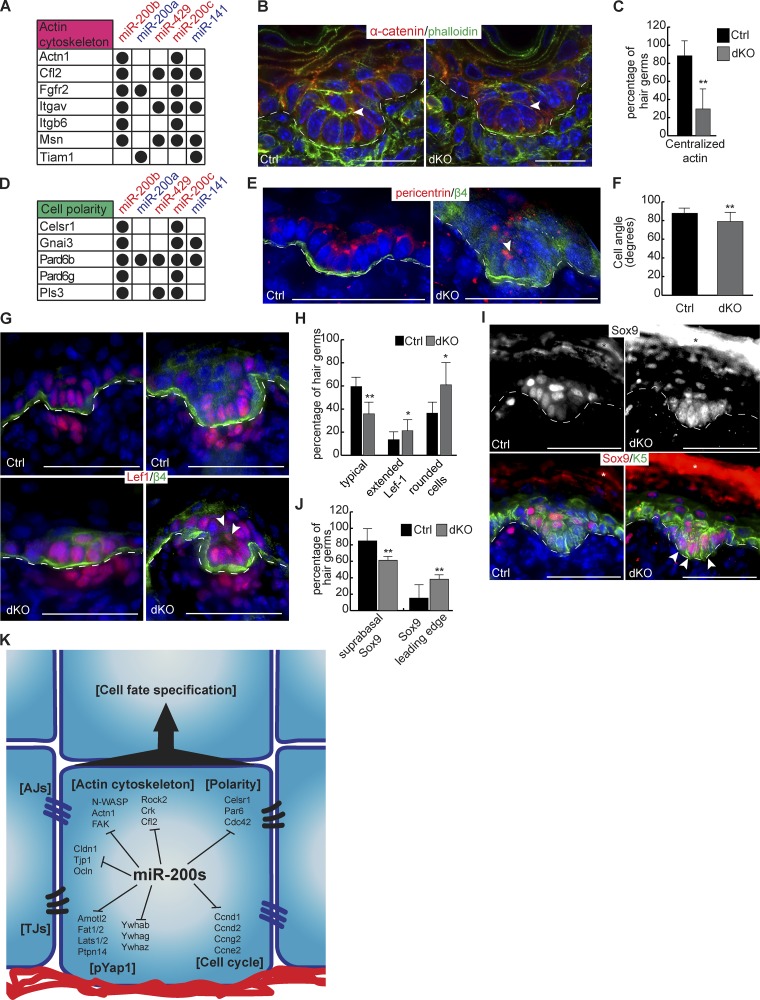
To further understand the requirement of the miR-200 family in vivo, we examined HG morphogenesis in miR-200 dKO skin. Because many miR-200 targets are regulators of actin cytoskeleton (Figs. 4 D and 10 A) and because actin bundles were defective in Tg and dKO keratinocytes, we first visualized the actin cytoskeleton in HGs. Consistent with asymmetric partitioning of different cell fates between the basal cells at the leading edge and the suprabasal cells of the HG, in which the leading edge cells express strong Lef1 and Lhx2 and the suprabasal cells express Sox9, phalloidin staining of actin bundles revealed that the suprabasal cells have distinctly strong actin signals (Fig. 10 B). Statistical analyses showed that nearly 90% of the HGs of the control skin had the characteristic signals of actin bundles (Fig. 10 C). In contrast, the actin signals were largely absent from the dKO, and only ∼30% of stage-matched dKO HGs were found to harbor such cells, and with weaker actin signals (Fig. 10, B and C). This result, combined with several miR-200 targets involved in cell polarity such as Par6 (also known as Pard6b) and Celsr1 (Fig. 10 D), prompted us to examine cell orientation of the basal and suprabasal cells of the HG. As expected, basal cells in control HGs occupied the basement membrane in a largely perpendicular orientation as determined by β4-integrin and pericentrin staining (Fig. 10, E and F; and Fig. S5, C and D). In contrast, basal cells of stage-matched dKO HGs had the compromised orientation, and nearly half of dKO basal cells were >15° off the perpendicular axis (Fig. 10, E and F; and Fig. S5, C and D).
Figure 10.
Loss of miR-200s in the skin leads to mild defects in hair follicle development. (A) miR-200 targets involved with actin cytoskeleton. (B) Immunofluorescence of α-catenin with phalloidin stain for actin on control and dKO HGs at P0.5. The arrowhead indicates the actin bundle. HFs shown are stage 2. (C) Percentage of control and dKO HGs at P0.5 with centralized actin or lack of clear signal. n = 6. HFs analyzed are stage 2. (D) miR-200 targets involved with cell polarity. (E) Immunofluorescence of Pericentrin and β4-integrin on control and dKO HGs at P0.5. The arrowhead indicates a misoriented cell. HFs shown are stage 2. (F) Angle of HG cells relative to the basement membrane from control and dKO animals at P0.5. HFs analyzed are matched at stages 1 or 2. n = 4 pairs of animals, n = 16 control cells, and n = 30 dKO cells. (G) Immunofluorescence of Lef1 and β4-integrin on control and dKO HGs at P0.5. Arrowheads indicate suprabasal Lef1+ cells. HFs in left panels are early stage 2, and HFs in right panels are late stage 2. (H) Percentage of control and dKO HGs at P0.5 with typical appearance, extended Lef1, and/or rounded cells at the leading edge; extended Lef1 and rounded cell phenotypes are not mutually exclusive, so HGs may be in both categories. HFs analyzed are stage 2. n = 6. (I) Immunofluorescence of Sox9 and Krt5 on control and dKO HGs at P0.5. Arrowheads indicate leading edge Sox9+ cells. HFs shown are early stage 2. (J) Percentage of HGs from control and dKO animals at P0.5 with typical Sox9 expression pattern or Sox9 at the leading edge. HFs analyzed are at stage 2. n = 6. (K) Model of miR-200 family function during hair follicle development. TJ, tight junction. *, P < 0.05; **, P < 0.01; Student’s two-tailed t test; error bars are SD. Bars: (B) 20 µm; (E, G, and I) 50 µm.
Because asymmetric cell division and perpendicular cell orientation are integral to cell fate specification within the HG (Ouspenskaia et al., 2016), we next examined whether cell fate is altered in dKO HGs. In contrast with the control placode and HG, Lef1 was often detected throughout the entire HG in the dKO (Fig. 10, G and H). Cell morphology was also mildly changed from columnar to a more rounded shape, perhaps reflecting compromised cell orientation. In contrast, Sox9 expression was expanded from the suprabasal cells to the basal cells at the leading edge of dKO HGs (Fig. 10, I and J). Although we did find a single miR-141–Sox9 CLEAR-CLIP read (Fig. S5 E), a luciferase assay failed to confirm the regulation of the Sox9 3′UTR by miR-200s (Fig. S5 F). We concluded that Sox9 is not a direct target of miR-200s, and its expanded expression may be caused by the altered actin cytoskeleton and cell orientation in dKO HGs. Collectively, our results show that loss of miR-200s in the skin has mild defects and leads to increased variation in cell fate specification during hair follicle development.
Discussion
In this study, we have determined, for the first time to our knowledge, miRNA-specific interactions with targeted mRNA sites for the miR-200 family in normal epithelial cells. Our results comprise a comprehensive, high-confidence set of targets for miR-200s and have revealed a highly coordinated gene expression regulatory network controlled by this miRNA family in the skin. Although the seed sequences that distinguish the two subfamilies differ by only a single nucleotide, the two subfamilies rarely recognize cross-seed targets. In contrast, the members that share a seed can recognize the same target site. These data underscore the importance of perfect seed matches in miRNA target recognition. However, for each miR-200, ∼30% of targets are recognized without seed matches. Thus, direct capture of miRNA–mRNA duplexes, rather than seed-based prediction, is critical for identifying all miRNA targets. The ability of each subfamily to recognize distinct target sites can both expand the range of miR-200–regulated genes as well as potentiate miR-200–mediated regulation if these different sites are located within the same 3′UTR. Indeed, we have identified many genes whose 3′UTRs are bound by both subfamilies. We further demonstrate that combinatorial target recognition by multiple miR-200s indeed confers stronger regulation of targets (Fig. 3, B and C). Thus, the miR-200 family can be more potent in target regulation than an individual miRNA or a single-seed miRNA family, and as a result, the miR-200–regulated target network is more complex.
Our genome-wide approach to identify miR-200 targets that combines CLEAR-CLIP for mapping physical interactions with RNA-seq for measuring the regulatory effect on mRNA levels has greatly facilitated functional studies for this miRNA family. Intriguingly, miR-200–targeted genes are highly enriched for key regulators of FA, actin cytoskeleton, AJs, cell polarity, cell cycle, and Hippo signaling (Fig. 10 K). Indeed, using our gain- and loss-of-function cells and mouse models, we have shown defects in these pathways when miR-200 levels are genetically altered. Although the inhibitory effects of miR-200s on cell cycle and migration have been previously shown during tumorigenesis (Gregory et al., 2008; Korpal et al., 2008; Shimono et al., 2009), the prominent defects we observed in the formation of AJs are novel and exciting. In miR-200–induced keratinocytes, whereas E-cadherin localization to the cell membrane upon calcium switching appears to be normal, the attachment of actin stress fibers to the newly formed AJs is severely reduced (Fig. 4, I and J). Consistent with such defects in vitro, the localizations of both α-catenin and β-catenin to the cell cortex are compromised when miR-200s are overexpressed in vivo (Fig. 6, H and J). However, when miR-200s are lost, a prominent defect we observed was compromised E-cadherin localization to the cell junction in vitro. The observed phenotypes suggest functions for miR-200 family targets in regulating the localization of both E-cadherin and actin to cell junctions. The disruptions observed with miR-200 overexpression are likely caused by the coordinated down-regulation of many genes involved with AJ formation, several of which may be important for proper actin attachment to nascent junctions. Indeed, this is supported by defects in actin attachment to AJs observed with KD of individual miR-200 family targets (Fig. 5 C). Even more intriguingly, combined KD of several targets together in WT keratinocytes showed robust defects (Fig. 5 C), and simultaneous KD of Cfl2, Rock2, and Fak in dKO keratinocytes resulted in a partial rescue of the AJ defects (Fig. 9, C and D). Therefore, miR-200–mediated regulation promoting proper levels of these genes is required for precise E-cadherin localization and overall junction formation. Indeed, such regulation of precise gene expression levels by miR-200s is likely to be important for many of the observed phenotypes both in vitro and in vivo.
In contrast with the direct regulation of adhesion and cytoskeleton, miR-200–mediated regulation of the Hippo pathway is enigmatic. Although we identified a strong signature for the inhibitory effect of miR-200s on phosphorylated Yap1 (pYap1) and indeed detected reduced pYap1 and increased nuclear Yap1 accumulation, we did not observe anticipated outcomes consistent with nuclear Yap1 activation such as downstream target activation and enhanced cell proliferation (Fig. 6). On one hand, it is likely that direct miR-200 targeting of the cell cycle machinery as well as Ctgf, one of the best-characterized Yap1 targets (Meng et al., 2016), may dampen the effect of nuclear Yap1. On the other hand, it could be caused by reduced pYap1 located in the cytoplasm, which may have unexplored functions. Notably, E-cadherin and α-catenin, two essential components of AJs, have both been shown to regulate Yap1 functions (Kim et al., 2011; Schlegelmilch et al., 2011). Thus, miR-200–mediated regulation of the Hippo pathway, especially the role of pYap1 in the cytoplasm, warrants further investigation.
In addition to showing specific defects in miR-200–regulated pathways, we have identified a novel role for these miRNAs in regulating hair follicle morphogenesis. As is detailed throughout this study, both miR-200 overexpression and loss of function result in disruptions to cell adhesion, junction formation, and proliferation. As these processes are critical for hair follicle development, it is not surprising that we observed faulty hair morphogenesis when miR-200 levels were manipulated. Intriguingly, we noticed defects in cell fate specification with both miR-200 overexpression (Fig. 7) and loss of function (Fig. 10), suggesting that precise regulation of cell adhesion and proliferation are required for cell fate determination. We note that the gain-of-function phenotypes are more severe than the phenotypes observed in the loss-of-function models, suggesting potential redundancy between miR-200s and other regulators.
In conclusion, we have demonstrated critical functions of the miR-200 family in regulating cell adhesion and proliferation during mammalian skin development. Our combinatorial approach using genome-wide miRNA target identification and genetic analyses of miRNA functions has provided a new example by which to dissect miRNA-mediated gene expression regulatory networks. The novel insights into miR-200 family targets and functions detailed in this study will help to further elucidate the roles of this important miRNA family in a wide range of conditions including epithelial development, regeneration, and cancer.
Materials and methods
Fluorescence in situ hybridization
In situ hybridization was performed as previously described (Yi et al., 2008) with minor modifications. A double-digitonin–labeled locked nucleic acid probe was used. Where shown, sections were incubated after TSA amplification with a β4-integrin primary antibody (1:100; clone 346-11A; 553745; BD) followed by an Alexa Fluor 594–conjugated secondary antibody (1:2,000; A11012 and A11007; Invitrogen/Molecular Probes).
Mice
The Tg mouse line was generated through standard Tg injection of the linearized pTRE2-200bcl DNA into an FVB background. Founders were bred with mice harboring a keratin14 reverse tetracycline trans-activator (rtTA) to produce mice with skin-specific doxycycline-inducible expression of the miR-200b cluster. Doxycycline chow used in experiments was 625 mg/kg (Teklad rodent diet TD-7012).
The miR-200b cluster KO allele was generated using two sgRNAs with the CRISPR-Cas9 system. In vitro–transcribed Cas9 mRNA and the two sgRNAs were injected into the cytoplasm of a fertilized mouse oocyte (FVB) and cultured overnight. Those that developed to the two-cell stage were subsequently transplanted into pseudopregnant females. Founders were bred with miR-200c/141fl/fl animals (Mirc13tm1Mtm/Mmjax; 013706; Jackson ImmunoResearch Laboratories, Inc.) and then to Krt14-Cre animals (a gift from E. Fuchs, The Rockefeller University, New York, NY). Sequences for miR-200bcl cloning, genotyping, and sgRNAs can be found in Table S1. Mice were housed and bred in a pathogen-free facility according to the guidelines of the Institutional Animal Care and Use Committee at the University of Colorado Boulder.
Primary keratinocyte harvesting and cell culture
Primary keratinocytes were isolated from animals between the ages of P0 and P4 as previously described, with some modifications (Yi et al., 2008). Mice were sacrificed, back and belly skin was removed, and excess fat was scraped from the tissue. The skin was then placed dermal-side down in 1× dispase for 30–60 min at 37°C. The epidermis was subsequently removed from the dermis with forceps and placed into 0.05% trypsin-EDTA for 10 min at 37°C. Trypsin was quenched with culture media, and cells were strained through a 40-µm filter and subsequently plated in E-low Ca2+ media onto dermal feeder cells as previously described (Yi et al., 2008).
CLEAR-CLIP
The CLEAR-CLIP protocol was adapted from the following citations: (Moore et al., 2014, 2015; Riemondy et al., 2015). Mouse keratinocytes of the designated genotype were maintained in E-low calcium medium. Tg cells were treated with 3 µg/ml final concentration doxycycline for 24 h before performing CLEAR-CLIP. One 15-cm dish of confluent cells was used per sample. RNA isolation from the nitrocellulose membrane was performed as previously described (Zarnegar et al., 2016). Libraries were mixed in equal amounts and sequenced on an Illumina HiSeq 4000 by the Microarray and Genomics Core at the University of Colorado Anschutz Medical Campus.
Samples for CLEAR-CLIP
CLEAR-CLIP was performed in two large experiments. The first experiment (set 1) consisted of six replicates of control cells (K14-Cre–only keratinocytes) and three replicates of miR-200 dKO keratinocytes. The second experiment (set 2) consisted of three replicates of control cells (Tg keratinocytes that were not induced with doxycycline), three replicates of Tg keratinocytes induced with 3 µg/ml doxycycline (final concentration) for 24 h, and three replicates of miR-200 dKO keratinocytes.
Assigning chimeric reads and genome annotation
Fastq files were obtained from the Microarray and Genomics Core and quality filtered using the FastX toolkit fastq_filter.pl requiring the first 25 bp to have a mean quality of 20. The 5′ and 3′ adapter sequences were then removed using CutAdapt. Next, the two base pairs at the 3′ end of the read that correspond with the NN of the 3′ adapter were trimmed, and only reads at least 43 bp long were kept (minimum of 19 bps for the miRNA, 20 for mapping the mRNA, and 4 bp for the barcode at the 5′ end). Next, the barcode was trimmed from the read but kept as a separate entry in the file using stripbarcode.pl, which is part of the cross-linking–induced mutation site (CIMS) package from the Darnell laboratory. Reads were then mapped using BLAST to a Fasta file containing the mouse miRNA database (BLAST settings: -word_size 11, -outfmt 6 –strand plus). BLAST results were filtered requiring at least 19 matching base pairs matching and E < 0.05. Only reads containing an miRNA that mapped to the 5′ end of the read were selected, and the resulting 3′ end sequence was then mapped to the mouse genome (mm10) using Novoalign. Reads between 20–24 bp were mapped using Novoalign, requiring an exact match to the genome. Reads of ≥25 bp were mapped using Novoalign and the –t 85 setting, which allows two deletions, two substitutions, or one deletion and one substitution. Novoalign files were parsed using the novoalign2bed.pl program from the Darnell laboratory CIMS package, and then mapped reads corresponding with 20–24 bp and ≥25 bp were combined into one file for each sample. Reads were annotated to regions of the genome using Bedtools intersect against annotated regions of the genome using annotation files downloaded from the University of California, Santa Cruz, table browser for the mouse mm10 genome. Similarly, reads were annotated to genes using Bedtools intersect against gene annotation files for the mouse mm10 genome.
Area selection for control-only analysis
Reads mapped to any miR-200 family member from all control samples were combined, and the genome coverage was computed using Bedtools GenomeCoverageBed. Areas where three or more reads overlapped were selected and merged into one area. Because merging can reduce the area significantly, making seed matching difficult, areas <50 bp were expanded symmetrically up to 50 bp. Areas of interest were then intersected with reads from individual miR-200 family members to determine which family members were interacting with the area. For each miR-200 family member, areas that mapped to 3′ UTRs were then used for motif analysis.
Area selection for combined analysis
Individual samples were combined by sample type into sets for each experiment (set 1 samples were controls [six replicates] and dKOs [three replicates], and set 2 samples included controls [three replicates], Tgs [three replicates], and dKOs [three replicates]). Reads mapped to any miR-200 family member were combined into one dataset per sample. For analysis of controls and Tgs, areas of interest were required to be found in two of the library sets between set 1 controls, set 2 controls, and set 2 Tgs. Overlapping areas were selected, and areas <40 bp were extended to 40 bp in the direction of the 3′ end of the mRNA (this increased the percentage of reads with seed matches). Areas of interest were then intersected with a database of mouse (mm10) gene 3′ UTRs obtained from the UCSC table browser.
Unbiased motif finding using HOMER
HOMER was downloaded from http://homer.ucsd.edu/homer/motif/. Motif finding was performed for each miR-200 family member individually only on reads found in controls. Areas corresponded with where that family member had a read of overlapping areas where three or more miR-200 family members overlapped (see the Area selection for control-only analysis section). The background for motif finding was all mouse mm10 3′ UTRs. Settings for motif finding were: -size given, -rna, -chopify, -len 4,5,6,7,8,9,10, -noweight, and –nlen 0. The top-ranked motif is shown for each miR-200 family member.
FACS and RNA-seq
Females harboring the pTRE2-200bcl allele and a K14RFP allele were bred with males harboring K14rtTA and fed doxycycline chow throughout pregnancy. Pups were collected at P0.5, and whole–back skin epidermis keratinocytes were isolated as described above except 10 mM EDTA was used instead of 0.05% Trypsin-EDTA to avoid degrading p-cadherin (see the Primary keratinocyte harvesting and cell culture section). After being filtered through a 40-µm filter, cells were spun down for 10 min at 300 g and resuspended in 1× PBS with 3% chelexed FBS on ice for 1 h with the following antibodies: p-cadherin (1:100; FAB761P; R&D Systems) and α6-integrin (1:75; clone NKI-GoH3; MCA699A647; AbD Serotec). Cells were again spun down for 10 min at 300 g and resuspended in 1× PBS with 3% chelexed FBS containing Hoechst 33342 (1:10,000; Invitrogen). Sorting for HG and IFE populations was performed on a MoFlow XDP cell sorter (Beckman Coulter). The HG population was gated as K14RFP+/p-cadhi/α6hi, and the IFE was gated as K14RFP+/p-cadlow/α6hi. Cells were sorted into 1× PBS with 3% chelexed FBS, subsequently spun down for 10 min at 300 g, and resuspended in 200 µl of TRIzol reagent.
Libraries for RNA-seq were assembled using the SMARTer stranded total RNA sample prep kit low input mammalian (634861; Takara Bio Inc.). RNA-seq raw reads were first trimmed to remove the 3′ adapter sequence (5′-GATCGGAAGAGCACACGTCTGAACTCCAGTC-3′; CutAdapt, default settings). Reads were then aligned to the mouse genome with TopHat with a supplied .gtf transcript annotation file (mm10; Illumina iGenomes; settings = –bowtie1–library-type fr-firststrand). Alignments of uniquely overlapping coding sequences were counted with HTSeq Count (settings = -s reverse –t CDS). Differential expression was calculated with DESeq using default settings. GO-term analysis was performed using DAVID Bioinformatics Resources (6.8; National Institute of Allergies and Infectious Diseases; National Institutes of Health).
Generation of cumulative distribution plots
Gene groups were intersected with RNA-seq log2 fold change, ranked, and plotted in R as a cumulative distribution. Statistical analysis was done using a Kolmogorov-Smirnov test.
Colony formation, scratch assays, and cell adhesion assays
Colony formation assays were performed by plating 2,000 low-passage keratinocytes (Tg, K14cre control, or dKO) per well of a six-well plate. Tg cells were treated with 3 µg/ml doxycycline or PBS every 24 h for the duration of the assay. 10 d after plating, cells were fixed for 10 min with 100% methanol and then stained for 2 h with 25% crystal violet. Wells were washed with water, and plates were imaged with a scanner. Each well was cropped to an independent image. For signal intensity measurements, images were converted to 8-bit grayscale in ImageJ (National Institutes of Health; Image>Type>8-bit) and inverted (Edit>Invert), and then the signal was measured (Analyze>Measure). Colony number and size were measured manually.
For scratch assays, low-passage keratinocytes were plated in a six-well plate and cultured until confluence. Tg cells were treated with 3 µg/ml doxycycline or PBS for 48 h before scratching. Once confluent, a scratch was made using a 200-µl pipette tip. Scratches were imaged every 2 h until the scratch was closed.
For cell adhesion assays, low-passage keratinocytes were plated on 24-well plates coated with 1 µg/ml fibronectin. Tg cells were treated with 3 µg/ml doxycycline or PBS for 24 h before plating. After 1 h, wells were washed three times with 1× PBS to remove unadhered cells and then were fixed with 4% PFA overnight. Wells were then stained with 0.25% crystal violet in 70% ETOH for 1 h and subsequently rinsed with deionized water. Wells were allowed to air dry for 30 min, and then 300 µl of 100% methanol was added to each well for 20 min to solubilize the crystal violet. 200 µl of the crystal violet mix from each well was transferred to a 96-well plate, and absorbance was measured at 595 nm on a plate reader.
FA and AJ assays
Tg, control, dKO, WT + shRNA, or dKO + shRNA keratinocytes were sparsely plated in on coverslips coated with 1 µg/ml fibronectin. Tg keratinocytes were treated with PBS or 3 µg/ml doxycycline for 24 h after plating. For cell–cell adhesion imaging, cells were treated with 2 mM calcium chloride for 3 or 7 h before imaging. Cells were fixed with 4% PFA, washed twice in 1× PBS, and permeabilized for 10 min in 1× PBS with 0.01% Triton X-100. Blocking was performed with 5% normal goat serum in 1× PBS for 1 h at room temperature. Coverslips were then incubated with primary antibody for 1 h at room temperature. Primary antibodies used were vinculin (mouse; 1:200; V9131; Sigma-Aldrich) or E-cadherin (rat; 1:200; gift from E. Fuchs). Coverslips were washed three times for 5 min in 1× PBS and then incubated with Alexa Fluor 594–, Alexa Fluor 488–, or Alexa Fluor 647–conjugated secondary antibodies (1:2,000; Invitrogen/Molecular Probes) and phalloidin (1:50; Alexa Fluor 488 conjugated; A12379; Invitrogen) for 1 h at room temperature. Nuclei were stained with Hoechst 33342 (1:5,000; Invitrogen). Coverslips were mounted onto slides with ProLong Gold antifade (P36930; Invitrogen) and left at room temperature overnight to cure. Cells were then imaged on an A1 laser-scanning confocal microscope (Nikon) in the University of Colorado Boulder Light Microscopy Core Facility. A custom FIJI macro was used for FA quantification (ImageJ; Bennett et al., 2016).
Cloning and 3′ UTR luciferase assays
3′ UTR fragments were generated by PCR amplification from cDNA. Primers used for cloning can be found in Table S1. These were then cloned into the pGL3 control vector (Promega). 2 ng renilla luciferase control, 20 ng of pGL3 reporter, and 380 ng of MSCV-IRES-GFP retroviral vector (MIGR), MIGR-200bcl, or K14-200ccl were cotransfected into miR-200 dKO keratinocytes in a 24-well plate using the LT-1 transfection reagent (Mirus Bio). For luciferase assays with both clusters, 190 ng each of MIGR-200bcl and K14-200ccl were transfected. Cell lysates were collected after 48 h, and renilla and firefly activity were measured using the Dual-Glo luciferase assay system (Promega).
Quantitative RT-PCR
cDNA was generated from 500–1,000 ng starting RNA using the miScript II RT kit (miRNA detection; 218160; QIAGEN) or the SuperScript III first-strand synthesis supermix for quantitative RT-PCR (mRNAs; 11752-050; Thermo Fisher Scientific). cDNA was diluted 1:10 for subsequent reactions. qPCR was performed using iQ SYBR green supermix (170-8880; Bio-Rad Laboratories) and quantified using the ΔΔC(t) method. miRNAs were normalized to sno25, and mRNAs were normalized to HPRT. Primers used can be found in Table S1.
shRNA infection
WT or dKO keratinocytes were infected with lentivirus containing MISSION shRNAs (Sigma-Aldrich) directed against mouse Scrambled (Scr; TRCN002), Cfl2 (TRCN00000071542), Egfr (TRCN0000055218), Ptk2 (TRCN0000023485), Rock2 (TRCN0000022922), or a combination (see text for details). Lentiviral supernatant was added to WT or dKO keratinocytes, spun for 45 min at 2,000 rpm, and then incubated for another 1 h at 37°C before changing media. Cells were then subjected to 2 µg/ml puromycin selection 24 h after infection for 72 h before quantitative RT-PCR to confirm KD and performance of assays. Lentiviral supernatant was acquired from the Functional Genomics Facility at the University of Colorado, Boulder.
Immunofluorescence, alkaline phosphatase, and H&E
Optimal cutting temperature–embedded tissues were sectioned to 10–14 µm and fixed with 4% PFA for 10 min at room temperature. Sections were permeabilized for 10 min at room temperature with 0.1% Triton X-100 in 1× PBS. When staining with mouse monoclonal antibodies, we used the mouse-on-mouse basic kit (BMK-2202; Vector Laboratories). Otherwise, blocking was performed in 5% normal serum of the same species the secondary antibody was raised in. Sections were incubated with primary antibody overnight at 4°C. The following antibodies and concentrations were used: cytokeratin 5 (chicken; 1:2,000; SIG-3475; Covance), β4-integrin (rat; 1:500; clone 346-11A), lef-1 (rabbit; 1:500; 2230; Cell Signaling Technology), p-cadherin (rat; 1:500; MAB761; R&D Systems), E-cadherin (rat; 1:200; gift from E. Fuchs), α-catenin (rabbit; 1:200; 3236S; Cell Signaling Technology), sox9 (rabbit; 1:500; AB5535; EMD Millipore), yap1 (rabbit; 1:500; clone D8H1X; 14074; Cell Signaling Technology), pericentrin (rabbit; 1:500; PRB-432C; Covance), β-catenin (mouse; 1:2,000; 610153; BD), and lhx2 (rabbit; 1:5,000; gift from E. Fuchs). After incubation with primary antibodies, sections were washed three times in 1× PBS and incubated for 1 h at room temperature with Alexa Fluor 594–, Alexa Fluor 488–, or Alexa Fluor 647–conjugated secondary antibodies (1:2,000; Invitrogen/Molecular Probes). EdU incorporation was detected using the Click-iT EdU Alexa Fluor 488 imaging kit (C10337; Thermo Fisher Scientific). Nuclei were stained with Hoechst 33342 (1:5,000; Invitrogen).
H&E staining was performed as previously described (Yi et al., 2008). Alkaline phosphatase staining was performed by incubating fixed and permeabilized sections with nitro blue tetrazolium (NBT)-5-bromo-4-chloro-3′-indolyphosphate (BCIP; 7.5 µl NBT and 5.6 µl BCIP in B3 solution [100 mM Tris, pH 7.5, 50 mM NaCl, and 25 mM MgCl2]) for 15–30 min at room temperature. Coverslips were mounted with 100% glycerol for H&E and alkaline phosphatase and with Fluoromount-G (SouthernBiotech) or ProLong Gold (Thermo Fisher Scientific) for fluorescence. Widefield imaging was performed on a DM5500B microscope (Leica Microsystems) using 20× 0.70 NA and 40× 0.75 NA objectives with an attached C10600-10B camera (Hamamatsu Photonics) and acquired with MetaMorph software (7.7; Molecular Devices). Confocal imaging was performed using an A1 laser-scanning confocal microscope with a 100× 1.49 NA objective lens and acquired with NIS Elements (Nikon) software in the University of Colorado, Boulder, Light Microscopy Core Facility. All imaging was performed at room temperature.
Statistical analysis
Statistical analyses were performed for all indicated data using the two-tailed Student’s t test with the exception of cumulative distribution functions, where analysis was done using a Kolmogorov-Smirnov test. Data distribution was assumed to be normal, but this was not formally tested. In figures, statistical significance is indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001) as is also shown in the figure legends. Sample sizes (n) of tested datasets are stated in their respective figure legends.
Data access
Sequencing data have been deposited into NCBI Gene Expression Omnibus with the accession numbers GSE102291 (RNA-seq) and GSE102716 (CLEAR-CLIP). A full list of targets identified for miR-200s can be found at http://yilab.colorado.edu/miR200_targets.xlsx.
Online supplemental material
Fig. S1 shows the relative expression level of each miR-200 family member in neonatal epidermis, in situ hybridization for miR-200b in the skin at E17.5, sequences of selected chimeric reads from CLEAR-CLIP, and KEGG pathways and GO biological processes enriched in miR-200 family targets identified with CLEAR-CLIP. Fig. S2 shows Tg and control mice at P4.5, H&E staining of Tg and control back skin at P0.5, staging of hair follicles from Tg and control animals at P0.5, FACS settings used to separate HGs from intermolecular epidermis, GSEA on isolated HG cells, and the overlap between the transcriptomes of cultured keratinocytes, HGs, and IFE. Fig. S3 shows colony formation assays, cell migration assays, FA assays, and cell junction formation assays performed on WT keratinocytes treated with doxycycline or untreated, additional miR-200 targets involved with FA and actin cytoskeleton, larger images of cell junctions shown in Fig. 4, and levels of KD achieved with shRNAs used in Fig. 5. Fig. S4 shows Yap1 nuclear/cytoplasmic localization in Tg keratinocytes treated with doxycycline or untreated, additional miR-200 targets involved with tight junctions and AJs, targeting/repair of CRISPR deletion of the miR-200b cluster, genotyping of the deletion, control, and dKO mice at P0.5 and P4.5, H&E staining of dKO and control back skin at P0.5, staging and number of hair follicles from dKO and control animals at P0.5, and percentage of EdU+ cells in dKO and control HGs at P0.5. Fig. S5 shows larger images of cell junctions shown in Fig. 8, level of KD achieved with shRNAs used in Fig. 9, the sequence of the single CLEAR-CLIP read identified for miR-141 and Sox9, and a luciferase assay assessing the ability of the Sox9 3′ UTR to be repressed by the miR-200 family clusters.
Supplementary Material
Acknowledgments
We thank M. Schober (New York University, New York, NY) and all members of the Yi laboratory for suggestions and helpful discussions. We thank E. Fuchs for Lhx2 antibody, B. Gao and K. Diener for sequencing, Y. Teng for CRISPR-Cas9/sgRNA injections, J. Orth for confocal microscopy, and Y. Han for FACS.
This work was supported by National Institutes of Health grants R01-AR059697, R01-AR059697-07S1, and R01-AR066703 and an American Cancer Society research scholar award (124718-RSG-13-197-01-DDC to R. Yi). J.E. Hoefert was supported by a National Institutes of Health Ruth L. Kirschstein predoctoral National Research Service Award (F31-AR066463). G.A. Bjerke was supported by an American Cancer Society postdoctoral fellowship (129540-PF-16-059-01-RMC).
The authors declare no competing financial interests.
Author contributions: J.E. Hoefert and R. Yi conceived of the study and wrote the manuscript. J.E. Hoefert performed most experiments and analysis, G.A. Bjerke performed CLEAR-CLIP and analyzed the data, and D. Wang generated the mouse strains for the miR-200b–inducible model and the miR-200b CRISPR-mediated KO model.
References
- Ahtiainen L., Lefebvre S., Lindfors P.H., Renvoisé E., Shirokova V., Vartiainen M.K., Thesleff I., and Mikkola M.L.. 2014. Directional cell migration, but not proliferation, drives hair placode morphogenesis. Dev. Cell. 28:588–602. 10.1016/j.devcel.2014.02.003 [DOI] [PubMed] [Google Scholar]
- Ambros V. 2011. MicroRNAs and developmental timing. Curr. Opin. Genet. Dev. 21:511–517. 10.1016/j.gde.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andl T., Reddy S.T., Gaddapara T., and Millar S.E.. 2002. WNT signals are required for the initiation of hair follicle development. Dev. Cell. 2:643–653. 10.1016/S1534-5807(02)00167-3 [DOI] [PubMed] [Google Scholar]
- Andl T., Murchison E.P., Liu F., Zhang Y., Yunta-Gonzalez M., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., and Millar S.E.. 2006. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr. Biol. 16:1041–1049. 10.1016/j.cub.2006.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136:215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.G., Riemondy K., Chapnick D.A., Bunker E., Liu X., Kuersten S., and Yi R.. 2016. Genome-wide analysis of Musashi-2 targets reveals novel functions in governing epithelial cell migration. Nucleic Acids Res. 44:3788–3800. 10.1093/nar/gkw207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken C.P., Li X., Wright J.A., Lawrence D.M., Pillman K.A., Salmanidis M., Anderson M.A., Dredge B.K., Gregory P.A., Tsykin A., et al. . 2014. Genome-wide identification of miR-200 targets reveals a regulatory network controlling cell invasion. EMBO J. 33:2040–2056. 10.15252/embj.201488641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case L.B., and Waterman C.M.. 2015. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 17:955–963. 10.1038/ncb3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi S.W., Zang J.B., Mele A., and Darnell R.B.. 2009. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 460:479–486. 10.1038/nature08170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi S.W., Hannon G.J., and Darnell R.B.. 2012. An alternative mode of microRNA target recognition. Nat. Struct. Mol. Biol. 19:321–327. 10.1038/nsmb.2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., and Goodall G.J.. 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10:593–601. 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- Grosswendt S., Filipchyk A., Manzano M., Klironomos F., Schilling M., Herzog M., Gottwein E., and Rajewsky N.. 2014. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol. Cell. 54:1042–1054. 10.1016/j.molcel.2014.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr., Jungkamp A.-C., Munschauer M., et al. . 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 141:129–141. 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A., Kudla G., Dudnakova T., and Tollervey D.. 2013. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 153:654–665. 10.1016/j.cell.2013.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., and Lempicki R.A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Jackson S.J., Zhang Z., Feng D., Flagg M., O’Loughlin E., Wang D., Stokes N., Fuchs E., and Yi R.. 2013. Rapid and widespread suppression of self-renewal by microRNA-203 during epidermal differentiation. Development. 140:1882–1891. 10.1242/dev.089649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamora C., DasGupta R., Kocieniewski P., and Fuchs E.. 2003. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 422:317–322. 10.1038/nature01458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P., Shtengel G., Pasapera A.M., Ramko E.B., Davidson M.W., Hess H.F., and Waterman C.M.. 2010. Nanoscale architecture of integrin-based cell adhesions. Nature. 468:580–584. 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.-G., Koh E., Chen X., and Gumbiner B.M.. 2011. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. USA. 108:11930–11935. 10.1073/pnas.1103345108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M., Lee E.S., Hu G., and Kang Y.. 2008. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283:14910–14914. 10.1074/jbc.C800074200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M., Ell B.J., Buffa F.M., Ibrahim T., Blanco M.A., Celià-Terrassa T., Mercatali L., Khan Z., Goodarzi H., Hua Y., et al. . 2011. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 17:1101–1108. 10.1038/nm.2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.-h., Jones-Rhoades M.W., Bartel D.P., and Burge C.B.. 2003. Prediction of mammalian microRNA targets. Cell. 115:787–798. 10.1016/S0092-8674(03)01018-3 [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., and Johnson J.M.. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 433:769–773. 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- Meng Z., Moroishi T., and Guan K.-L.. 2016. Mechanisms of Hippo pathway regulation. Genes Dev. 30:1–17. 10.1101/gad.274027.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska E.A., Alvarez-Saavedra E., Abbott A.L., Lau N.C., Hellman A.B., McGonagle S.M., Bartel D.P., Ambros V.R., and Horvitz H.R.. 2007. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 3:e215 10.1371/journal.pgen.0030215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Zhang C., Gantman E.C., Mele A., Darnell J.C., and Darnell R.B.. 2014. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat. Protoc. 9:263–293. 10.1038/nprot.2014.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Scheel T.K.H., Luna J.M., Park C.Y., Fak J.J., Nishiuchi E., Rice C.M., and Darnell R.B.. 2015. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 6:8864 10.1038/ncomms9864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouspenskaia T., Matos I., Mertz A.F., Fiore V.F., and Fuchs E.. 2016. WNT-SHH Antagonism Specifies and Expands Stem Cells prior to Niche Formation. Cell. 164:156–169. 10.1016/j.cell.2015.11.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.Y., Jeker L.T., Carver-Moore K., Oh A., Liu H.J., Cameron R., Richards H., Li Z., Adler D., Yoshinaga Y., et al. . 2012. A resource for the conditional ablation of microRNAs in the mouse. Cell Reports. 1:385–391. 10.1016/j.celrep.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouffe S.W., Meng Z., Lin K.C., Lin B., Hong A.W., Chun J.V., and Guan K.-L.. 2016. Characterization of Hippo Pathway Components by Gene Inactivation. Mol. Cell. 64:993–1008. 10.1016/j.molcel.2016.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratheesh A., and Yap A.S.. 2012. A bigger picture: classical cadherins and the dynamic actin cytoskeleton. Nat. Rev. Mol. Cell Biol. 13:673–679. 10.1038/nrm3431 [DOI] [PubMed] [Google Scholar]
- Rhee H., Polak L., and Fuchs E.. 2006. Lhx2 maintains stem cell character in hair follicles. Science. 312:1946–1949. 10.1126/science.1128004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemondy K., Wang X.J., Torchia E.C., Roop D.R., and Yi R.. 2015. MicroRNA-203 represses selection and expansion of oncogenic Hras transformed tumor initiating cells. eLife. 4 10.7554/eLife.07004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle N.T., Sheu-Gruttadauria J., and MacRae I.J.. 2014. Structural basis for microRNA targeting. Science. 346:608–613. 10.1126/science.1258040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelmilch K., Mohseni M., Kirak O., Pruszak J., Rodriguez J.R., Zhou D., Kreger B.T., Vasioukhin V., Avruch J., Brummelkamp T.R., and Camargo F.D.. 2011. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell. 144:782–795. 10.1016/j.cell.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono Y., Zabala M., Cho R.W., Lobo N., Dalerba P., Qian D., Diehn M., Liu H., Panula S.P., Chiao E., et al. . 2009. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 138:592–603. 10.1016/j.cell.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jacques B., Dassule H.R., Karavanova I., Botchkarev V.A., Li J., Danielian P.S., McMahon J.A., Lewis P.M., Paus R., and McMahon A.P.. 1998. Sonic hedgehog signaling is essential for hair development. Curr. Biol. 8:1058–1069. 10.1016/S0960-9822(98)70443-9 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V., Bauer C., Yin M., and Fuchs E.. 2000. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 100:209–219. 10.1016/S0092-8674(00)81559-7 [DOI] [PubMed] [Google Scholar]
- Wang D., Zhang Z., O’Loughlin E., Lee T., Houel S., O’Carroll D., Tarakhovsky A., Ahn N.G., and Yi R.. 2012. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev. 26:693–704. 10.1101/gad.182758.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R., O’Carroll D., Pasolli H.A., Zhang Z., Dietrich F.S., Tarakhovsky A., and Fuchs E.. 2006. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 38:356–362. 10.1038/ng1744 [DOI] [PubMed] [Google Scholar]
- Yi R., Poy M.N., Stoffel M., and Fuchs E.. 2008. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 452:225–229. 10.1038/nature06642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R., Pasolli H.A., Landthaler M., Hafner M., Ojo T., Sheridan R., Sander C., O’Carroll D., Stoffel M., Tuschl T., and Fuchs E.. 2009. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc. Natl. Acad. Sci. USA. 106:498–502. 10.1073/pnas.0810766105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar B.J., Flynn R.A., Shen Y., Do B.T., Chang H.Y., and Khavari P.A.. 2016. irCLIP platform for efficient characterization of protein-RNA interactions. Nat. Methods. 13:489–492. 10.1038/nmeth.3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Lee J.E., Riemondy K., Anderson E.M., and Yi R.. 2013. High-efficiency RNA cloning enables accurate quantification of miRNA expression by deep sequencing. Genome Biol. 14:R109 10.1186/gb-2013-14-10-r109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.