Arf6/ARF-6 is a crucial regulator of the endosomal PI(4,5)P2 pool in endocytic recycling. Chen et al. show that SAC-1 is important for epithelial cell recycling in C. elegans. SAC-1 acts as a novel interactor for ARF-6, curbing ARF-6 activity by limiting the access of ARF-6(GDP) to its GEF BRIS-1.
Abstract
Arf6/ARF-6 is a crucial regulator of the endosomal phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) pool in endocytic recycling. To further characterize ARF-6 regulation, we performed an ARF-6 interactor screen in Caenorhabditis elegans and identified SAC-1, the homologue of the phosphoinositide phosphatase Sac1p in yeast, as a novel ARF-6 partner. In the absence of ARF-6, basolateral endosomes show a loss of SAC-1 staining in epithelial cells. Steady-state cargo distribution assays revealed that loss of SAC-1 specifically affected apical secretory delivery and basolateral recycling. PI(4,5)P2 levels and the endosomal labeling of the ARF-6 effector UNC-16 were significantly elevated in sac-1 mutants, suggesting that SAC-1 functions as a negative regulator of ARF-6. Further analyses revealed an interaction between SAC-1 and the ARF-6-GEF BRIS-1. This interaction outcompeted ARF-6(guanosine diphosphate [GDP]) for binding to BRIS-1 in a concentration-dependent manner. Consequently, loss of SAC-1 promotes the intracellular overlap between ARF-6 and BRIS-1. BRIS-1 knockdown resulted in a significant reduction in PI(4,5)P2 levels in SAC-1-depleted cells. Interestingly, the action of SAC-1 in sequestering BRIS-1 is independent of SAC-1’s catalytic activity. Our results suggest that the interaction of SAC-1 with ARF-6 curbs ARF-6 activity by limiting the access of ARF-6(GDP) to its guanine nucleotide exchange factor, BRIS-1.
Introduction
Endocytosis is a fundamental biological process. In contrast to clathrin-dependent endocytosis, clathrin-independent endocytosis does not require dynamin (Grant and Donaldson, 2009). The cargo proteins that come into the cell via clathrin-independent endocytosis will enter early endosomes, where the cargo is sorted and delivered to different destinations. The small GTPase Arf6 is implicated in the recycling and trafficking of clathrin-independent cargo. Its functionality is closely associated with phosphoinositide phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) metabolism (Donaldson, 2003).
Arf6 activity is managed by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). In the human genome, 15 Arf-GEFs are encoded, of which eight are classified as Arf6-GEFs and belong to three families: cytohesin, EFA6, and BRAG (Casanova, 2007). Previous studies showed that different Arf6-GEFs could be activated by distinct mechanisms and thus participate in various functional processes (Hongu and Kanaho, 2014). The cytohesin family members Cytohesin1, ARNO, and Grp1 do not exclusively manage Arf6 (Casanova, 2007), whereas the EFA6 family proteins specifically catalyze the nucleotide exchange of Arf6 (Franco et al., 1999; Derrien et al., 2002; Matsuya et al., 2005; Sakagami et al., 2006). Studies on BRAG family members demonstrated that the GEF activity of GEP100 is specific to Arf6, and GEP100 partially colocalizes with Arf6 on endosomes (Someya et al., 2001; Dunphy et al., 2006; Hiroi et al., 2006).
PI(4,5)P2 mainly resides in the plasma membrane (De Matteis and Godi, 2004), and its synthetic precursor, phosphatidylinositol 4-phosphate (PI(4)P), is primarily derived from the Golgi apparatus and the plasma membrane (Dickson et al., 2016). PI(4,5)P2 contributes to endocytosis by recruiting proteins such as AP-2, epsin, and dynamin, and it is rapidly degraded after endocytosis (McMahon and Boucrot, 2011). Arf6 also localizes in the plasma membrane but does not seem to regulate clathrin-dependent endocytosis. In contrast, Arf6 inactivation is vital for cargo sorting after endocytosis (Donaldson, 2003). The constitutively activated Arf6 continuously activates phosphatidylinositol-4-phosphate 5 kinase (PIP5 kinase), producing unusually high amounts of endosomal PI(4,5)P2 and disrupting the sorting and subsequent recycling transport (Brown et al., 2001; Naslavsky et al., 2003). In Caenorhabditis elegans, RAB-10 can recruit CNT-1/ARF-6-GAP to endosomes, shutting down ARF-6 activity. In the absence of RAB-10, the level of PI(4,5)P2 was significantly increased, and the recycling of clathrin-independent cargo hTAC-GFP was severely affected (Shi et al., 2012). Arf6 is also involved in the regulation of actin cytoskeleton, and the overexpression of dominant-negative mutant Arf6(T27N) inhibited a series of actin-related processes, including cell spreading and wound healing (Song et al., 1998; Radhakrishna et al., 1999; Boshans et al., 2000; Palacios et al., 2001; Santy and Casanova, 2001). The regulatory effects of Arf6 on actin can also be mediated by PI(4,5)P2, which recruits actin regulators to influence actin dynamics (Yin and Janmey, 2003).
Phosphoinositide phosphatase Sac1p was first identified in the study of suppressors of actin mutant alleles in yeast (Novick et al., 1989). Accumulating evidence indicates that Sac1p homologues are associated with various biological processes, including affecting cell shape changes in the amnioserosa and JNK signaling during dorsal closure in Drosophila melanogaster (Wei et al., 2003). Sac1p contains an N-terminal SAC domain and two C-terminal transmembrane helices (Konrad et al., 2002; Hsu and Mao, 2015). The SAC domains of Sac1p and its homologues have the specificity of hydrolyzing PI(3)P, PI(4)P, and PI(3,5)P2 (Guo et al., 1999; Hughes et al., 2000b; Nemoto et al., 2000; Chung et al., 2015; Dickson et al., 2016; Vanhauwaert et al., 2017). The SAC domain contains seven conserved motifs, and the consensus sequence RXNCXDCLDRTN in the sixth motif is the phosphatase catalytic core (Hughes et al., 2000a). Mutations of the three amino acid sites (D394, A445, and R483) within the catalytic core efficiently disrupted the enzymatic activity (Kearns et al., 1997; Rivas et al., 1999; Liu et al., 2008). The SAC domain also contributes to protein interactions. A catalytically inactive mutant of hSAC1 is not capable of interacting with the COPI complex (Rohde et al., 2003). Likewise, in C. elegans, the SAC domain of UNC-26/synaptojanin is required for the targeting of UNC-26 to synapses (Dong et al., 2015). Sac1p/hSAC1 resides in the ER and Golgi and shuttles between ER and Golgi via COPI and COPII (Kochendörfer et al., 1999; Rohde et al., 2003; Blagoveshchenskaya et al., 2008; Bajaj Pahuja et al., 2015; Chung et al., 2015). The distribution of hSAC1 on the Golgi is directed by the first transmembrane helix and the N-terminal cytoplasmic region (Wang et al., 2013). It has also been reported that hSAC1 intracellular localization can be directed by additional proteins. For example, in mammalian neurons, palmitoylated JNK3 was found to recruit hSAC1 to the Golgi, thereby down-regulating the abundance of PI(4)P and post-Golgi trafficking (Yang et al., 2013).
The C. elegans genome encodes two yeast Sac1p homologues, F30A10.6/SAC-1 and W09C5.7/SAC-2, homologous to mammalian SAC1 and SAC2, respectively. To gain insight into the mechanism by which ARF-6 functions in recycling transport, we conducted an ARF-6 interactor screen and identified SAC-1 as a novel binding protein. SAC-1 is widely expressed in a variety of tissues, including the intestine, pharynx, pharyngeal neurons, and spermatheca (Kumar et al., 2012). In C. elegans intestinal cells, we observed that SAC-1 maintains significant colocalization with ARF-6. In the absence of ARF-6, the subcellular distribution of GFP-SAC-1 was significantly affected. In particular, we noted that SAC-1 acts to ensure proper apical secretory delivery and basolateral recycling transport. Further analysis revealed that in sac-1 mutants, PI(4,5)P2 levels significantly increased and the ARF-6 effector UNC-16 accumulated in the cytosol, suggesting that SAC-1 down-regulates ARF-6 activity. As expected, we found that SAC-1 competes with ARF-6(T27N) for binding to BRIS-1/ARF-6-GEF in a concentration-dependent manner. Depletion of SAC-1 led to the significant increase of the overlap between ARF-6-GFP and BRIS-1-mCherry. Our observations indicate that SAC-1, as a novel interactor of ARF-6, exerts an adverse effect on ARF-6 activation by interfering in the interaction between inactive ARF-6(GDP) and the GEF protein BRIS-1.
Results
SAC-1 interacts with both active and inactive ARF-6
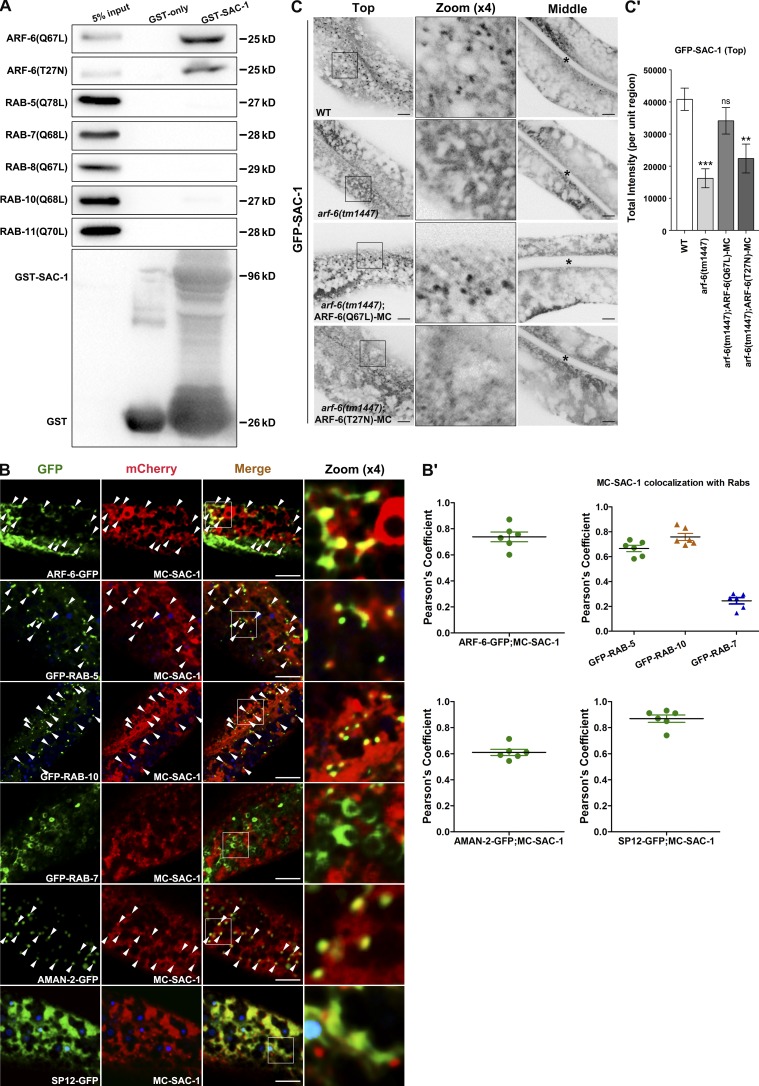
Arf6 is predominantly localized in the plasma membrane, and Arf6 needs to be inactivated after endocytosis for appropriate cargo sorting in endosomes (Donaldson, 2003). Overexpression of Arf6(Q67L), the constitutively active mutant form of Arf6, induces the persistent activity of PIP5 kinase. This leads to an unusually high amount of PI(4,5)P2 in the endosomal membrane, thereby impairing clathrin-independent cargo sorting and recycling (Brown et al., 2001; Naslavsky et al., 2003). Our previous study showed that rab-10 mutant is superficially normal in growth and development (Shi et al., 2010; Wang et al., 2016). In particular, rab-10 mutants overaccumulate hTAC-GFP in enlarged structures in the deep cytosol of intestinal epithelia (Chen et al., 2006). Thus, we performed a genome-wide RNAi screen using rab-10(RNAi) as the positive control to identify candidate genes whose expression knockdown lead to the hTAC-GFP overaccumulation in the intestine. After initial screen and the follow up validation, we noted that RNAi-mediated knockdown of 54 candidates resulted in intracellular hTAC-GFP aggregation, including SAC-1, ARX-7, GGTB-1, EXOC-7, HUM-2, and SEC-10. The previous study in mammal showed that HUM-2/myosin functions with Rab10 to mediate the delivery of GLUT4 storage vesicles to the plasma membrane (Chen et al., 2012). Recently, SEC-10 and EXOC-7 were found to be involved in the regulation of hTAC recycling in C. elegans intestine (Chen et al., 2014). We then conducted a small-scale screen for potential ARF-6(Q67L) interacting partner via GST pull-down assays. Out of 54 candidates, SAC-1 is the only one that exhibits a significant interaction with ARF-6(Q67L). Remarkably, SAC-1 interacts with ARF-6(Q67L) via its SAC domain with notable specificity (Fig. 1 A and Fig. S1, A and B). Also, we observed a moderate binding of SAC-1 to the GDP-bound mutant form ARF-6(T27N) (Fig. 1 A). We further performed coimmunoprecipitation experiments and validated the interaction of SAC-1 with ARF-6(Q67L) and ARF-6(T27N) (Fig. S1 C). Arf1 is predominantly an ER- and Golgi-associated small GTPase (Donaldson and Honda, 2005). In addition, Arf1 was reported to function at the plasma membrane to modulate the cortical actin (Caviston et al., 2014). To determine whether SAC-1 binds to ARF-1.2/Arf1, we examined the interaction of GST-SAC-1 with HA-ARF-1.2(Q71L) or HA-ARF-1.2(T31N). Nevertheless, both active and inactive mutant forms of ARF-1.2 failed to interact with SAC-1 (Fig. S1 D). We also selected the constitutively active mutant form of endocytic trafficking–related Rabs for the GST pull-down assay. Our results indicate that there was no significant binding between GST-SAC-1 and HA-tagged RAB-5(Q78L), RAB-7(Q68L), RAB-8(Q67L), RAB-10(Q68L), and RAB-11(Q70L), respectively (Fig. 1 A).
Figure 1.
SAC-1 interacts with the active and inactive mutant forms of ARF-6, and the residence of SAC-1 in basolateral puncta requires ARF-6. (A) Western blot showing GST pull-down with in vitro–translated HA-tagged proteins. Glutathione beads loaded with GST and GST-SAC-1 were incubated with in vitro–expressed HA-tagged RAB-5(Q78L), RAB-7(Q68L), RAB-8(Q67L), RAB-10(Q68L), RAB-11(Q70L), ARF-6(Q67L), and ARF-6(T27N). Eluted proteins were separated on the SDS-PAGE gel and analyzed by Western blotting using anti–HA antibody. Input lanes contain in vitro–expressed HA-tagged proteins in the binding assays (5%). (B and B′) SAC-1 colocalized well with ARF-6–, RAB-5–, or RAB-10–labeled endosomes. In addition, SAC-1 overlaps significantly with the TGN marker AMAN-2 and the ER marker SP12. Nevertheless, SAC-1 displayed little colocalization with the late endosome marker RAB-7. Arrowheads indicate positive overlap. Pearson’s correlation coefficients for GFP and mCherry signals were calculated (n = 6 animals). (C and C′) Compared with wild-type animals, the labeling of GFP-SAC-1 in basolateral puncta was significantly reduced in the absence of ARF-6 (top focal plane). Transgenic expression of ARF-6(Q67L)-mCherry rescued the puncta labeling GFP-SAC-1, and the transgenic expression of ARF-6(T27N)-mCherry failed to restore the endosomal localization of GFP-SAC-1. Asterisks in the panels indicate intestinal lumen. Error bars represent SEM (n = 18 each), and asterisks indicate the significant differences in the one-tailed Student’s t test (ns, no significance; **, P < 0.01; ***, P < 0.001). Bars, 10 µm.
To define the subcellular localization of SAC-1, we performed colocalization assays in intestinal cells. In wild-type animals, mCherry-SAC-1 labels ER-like structures in the deep cytosol and punctate structures near the basolateral plasma membrane, and GFP-tagged ARF-6 resides in basolateral tubules and puncta (Shi et al., 2012; Gleason et al., 2014). We observed that mCherry-SAC-1 and ARF-6-GFP had significant colocalization in basolateral puncta (Fig. 1, B and B′; Pearson’s correlation coefficient, ∼73.8%). RAB-5/Rab5 is a marker of early endosomes (Nielsen et al., 2000; Chen et al., 2006; Murray et al., 2016). We compared the localization of SAC-1 with RAB-5 and observed that mCherry-SAC-1 overlapped well with GFP-RAB-5 on endosomal structures (Fig. 1, B and B′; Pearson’s correlation coefficient, ∼67%). Previous studies showed that recycling regulator RAB-10 partially colocalizes with RAB-5 on early endosomes, where it appears to promote the maturation of early endosomes to recycling endosomes (Chen et al., 2006; Shi et al., 2010). In our study, SAC-1 and RAB-10 had significant overlap in cytosolic punctate structures (Fig. 1, B and B′; Pearson’s correlation coefficient, ∼75.8%). We also compared the localization of SAC-1 with other endosomal markers. SAC-1 failed to significantly overlap with the late endosome marker RAB-7 (Fig. 1, B and B′; Pearson’s correlation coefficient, ∼24.4%). Sac1p/hSAC1 has been reported to reside in the ER and Golgi, where it participates in the regulation of PI(4)P pool (Kochendörfer et al., 1999; Blagoveshchenskaya et al., 2008; Bajaj Pahuja et al., 2015; Chung et al., 2015). As expected, we observed a partial colocalization between SAC-1 and the TGN marker AMAN-2 (Fig. 1, B and B′; Pearson’s correlation coefficient, ∼62%). There was extensive overlap between the ER marker SP12-GFP and mCherry-SAC-1 in patch-like structures (Fig. 1, B and B′; Pearson’s correlation coefficient, ∼87%). It is also worth emphasizing that the mCherry-SAC-1–labeled basolateral puncta are independent of the SP12-GFP–positive structures.
Localization of SAC-1 in basolateral puncta requires the presence of ARF-6(GTP)
Small GTPases such as Arfs and Rabs usually recruit effectors to exert specific cellular effects (Hutagalung and Novick, 2011; Cherfils, 2014). In the absence of a specific small GTPase, the intracellular localization or distribution of its effector is often compromised. For example, in rab-10 mutants, the RAB-10 effectors CNT-1 and TBC-2 membrane association were impaired, showing cytoplasmic dispersion (Shi et al., 2012; Liu and Grant, 2015). To test whether the localization of SAC-1 is dependent on the presence of ARF-6, we prepared transgenic animals that specifically expressed GFP-SAC-1 in intestinal cells. In the wild-type background, GFP-SAC-1 mostly labels basolateral puncta and ER-like patch structures (Fig. 1 C). In arf-6(tm1447) mutants, the labeling of GFP-SAC-1 in basolateral puncta was significantly diminished (Fig. 1, C and C′; ∼60% intensity decrease). Instead, GFP-SAC-1–labeled patch-like structures in the deep cytosol were intact. We performed colocalization assays to dissect the observed localization variation of SAC-1. As expected, SAC-1 presented little overlap with RAB-10 in arf-6(RNAi) animals (Fig. 1, B and B′; and Fig. S1, E and E′). In contrast, the remaining SAC-1–positive puncta still colocalized with AMAN-2-GFP foci (Fig. 1, B and B′; and Fig. S1, E and E′), suggesting that ARF-6 is not involved in the Golgi positioning of SAC-1. Also, the overlap between mCherry-SAC-1 and ER marker SP12-GFP was not affected in the absence of ARF-6 (Fig. 1, B and B′; and Fig. S1, E and E′). These results suggest that the residence of SAC-1 in basolateral endosomes requires the presence of ARF-6, and the Golgi and ER positioning mechanisms of SAC-1 are distinct from that in endosomes.
SAC-1 is a transmembrane protein (Konrad et al., 2002; Hsu and Mao, 2015), and the potential explanation for our results is that ARF-6 is responsible for clustering SAC-1 from connected membranous organelles to the sorting endosomes. We then performed rescue experiments by coexpressing ARF-6(Q67L)-mCherry or ARF-6(T27N)-mCherry in arf-6(tm1447) animals (Fig. 1, C and C′). Interestingly, the transgenic expression of ARF-6(Q67L) significantly recuperated the puncta labeling GFP-SAC-1, whereas the expression of ARF-6(T27N) failed to restore the endosomal localization of GFP-SAC-1 (Fig. 1, C and C′).
To determine whether the endosomal localization of SAC-1 requires the presence of ARF-1.2, we examined the GFP-SAC-1 distribution in arf-1.2(RNAi) animals. GFP-SAC-1 appeared in the enlarged structures upon loss of ARF-1.2 (Fig. S1, F and F′). Moreover, there was substantial colocalization between Golgi marker AMAN-2-GFP and mCherry-SAC-1 in those enlarged structures (Fig. S1, G and G′). Together with the biochemical data (Fig. S1 D), these observations suggested that ARF-1.2, although not directly interacting with SAC-1, is required for the secretory delivery of SAC-1 in intestinal cells.
Clathrin-independent recycling cargo hTAC-GFP is trapped in early endosomes in sac-1 mutants
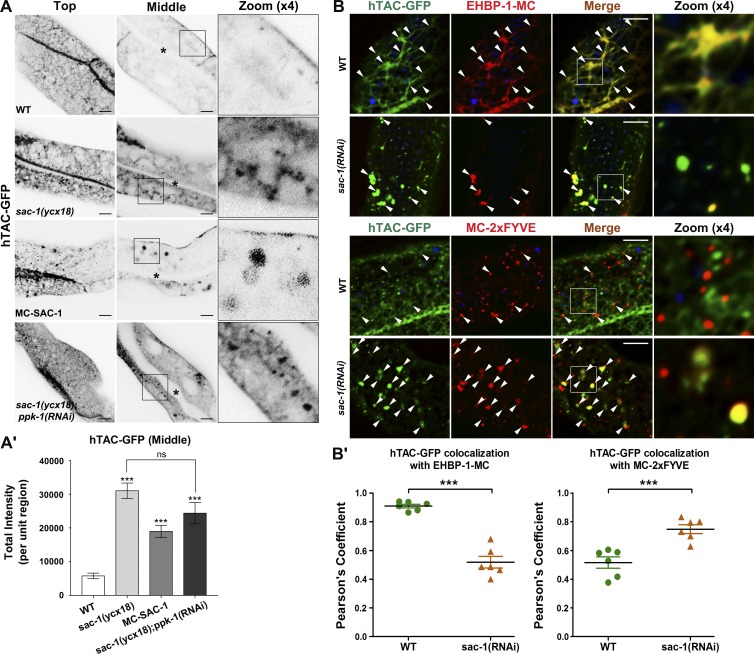
ARF-6/Arf6 is essential for the basolateral recycling flow of clathrin-independent cargo (Donaldson, 2003; Shi et al., 2012). Our finding that SAC-1 is an ARF-6 interactor prompted us to determine whether SAC-1 is required for ARF-6–mediated recycling transport in vivo. We sought to examine the steady-state distribution of the well-established clathrin-independent cargo hTAC-GFP (human IL-2 receptor α-chain) and clathrin-dependent cargo hTfR-GFP (human transferrin receptor; Chen et al., 2006; Shi et al., 2012). We developed a transgenic strain sac-1(ycx18), an intestine-specific CRISPR/Cas9 somatic mutant (Fig. 2 A and Table S2; Li et al., 2015). Live-cell imaging revealed that hTAC-GFP overaccumulated in intracellular aggregates upon loss of SAC-1 (Fig. 2, A and A′). Normally, hTAC-GFP predominantly coexists with recycling endosome marker EHBP-1, and to a lesser extent, early endosome marker EEA-1-2xFYVE (Wang et al., 2016; Fig. 2, B and B′). In the absence of SAC-1, the localization of hTAC-GFP in recycling endosomes was reduced, and the localization in 2xFYVE-labeled early endosomes was significantly enhanced, indicating a blockage of early endosome to recycling endosome transport (Fig. 2, B and B′). It is also worth noting that hTfR-GFP accumulated in intracellular structures in sac-1 mutants (Fig. S1, H and H'). A nonrecycling cargo, GFP-CD4-LL, showed a similar cytosolic distribution abnormality (Fig. S1, I and I′).
Figure 2.
The recycling transport of hTAC-GFP is impaired in sac-1 mutants. (A and A′) In the top focal plane, IL-2 receptor α chain hTAC-GFP–labeled tubular structures were severely perturbed in sac-1(ycx18) mutants. In the middle focal plane, hTAC-GFP overaccumulated on the cytosolic endosomal structures. An approximately 5.4-fold escalation of total hTAC-GFP intensity was observed in sac-1 animals. Also, in mCherry-SAC-1–overexpressing intestinal cells, hTAC-GFP accumulated on the enlarged structures. RNAi-mediated knockdown of PPK-1 failed to alleviate the overaccumulation phenotype of hTAC-GFP in sac-1(ycx18) mutants. Asterisks in the panels indicate intestinal lumen. Error bars represent SEM (n = 18 each), and asterisks indicate significant differences in the one-tailed Student’s t test (***, P < 0.001). (B and B′) The subcellular localization of the accumulated hTAC-GFP in sac-1(ycx18) mutants. In the absence of SAC-1, the overlap between hTAC-GFP and EHBP-1-mCherry was reduced, whereas the colocalization between hTAC-GFP and mCherry-2xFYVE was increased. Arrowheads indicate positive overlap. Pearson’s correlation coefficients for GFP and mCherry signals were calculated (n = 6 animals). Error bars represent SEM. ***, P < 0.001. Bars, 10 µm.
A recent study showed that the SAC domain protein SAC2 localizes at early endosomes and recycling endosomes in mouse N2A cells. Overexpression of the catalytically inactive mutant of SAC2 disrupts TfR (Transferrin receptor) recycling (Hsu et al., 2015). We then examined whether the recycling transport of hTAC-GFP was affected in sac-2(ok2743) mutant cells. In vivo imaging results showed that the absence of SAC-2 had no significant effect on the steady-state distribution of hTAC-GFP (Fig. S1, J and J′).
SAC-1 depletion exclusively impairs apical secretory transport in intestinal epithelia
Sac1p/hSAC1 is implicated in the secretory transport via modulating actin and lipid metabolism (Cleves et al., 1989; Whitters et al., 1993; Kearns et al., 1997; Hughes et al., 2000b; Schorr et al., 2001; Dickson et al., 2016). To determine whether SAC-1 acts to regulate secretory transport in intestinal epithelia, we used two different types of Golgi apparatus derived secretory cargo proteins: NPY-GFP (neuropeptide Y) and PGP-1-GFP (ATP-binding cassette transporter; Nagai et al., 2002; Sato et al., 2007). Neuropeptide Y fusion proteins have been proven to be secretory cargo in PC12 and HeLa cells (Nagai et al., 2002; Barg et al., 2010; Shibata et al., 2016). In C. elegans intestinal epithelia, we observed that NPY-GFP localizes mainly in the basolateral meshwork (top focal plane), lacking apparent apical membrane labeling (Fig. S2, A and A′). The absence of exocyst component SEC-6 resulted in NPY-GFP overaccumulation in cytosolic structures, and the labeling of NPY-GFP in basolateral tubules decreased, indicating a secretory defect (Fig. S2, A and A′). RAB-8/Rab8 is specifically required for the proper delivery of apical secretory cargo in C. elegans and mammalian intestinal epithelia (Sato et al., 2007; Shibata et al., 2016). In rab-8 mutants, we failed to detect the distribution abnormality of NPY-1-GFP, suggesting that NPY-GFP is delivered with basolateral directionality (Fig. S2, A and A′). There was no irregularity of NPY-GFP transport in SAC-1–depleted cells (Fig. S2, A and A′). However, we noticed that apical secretory cargo PGP-1-GFP significantly accumulated in intracellular aggregates in sac-1(ycx18) mutants (Fig. S2, B and B′), in a manner very similar to rab-8 mutants (Fig. S2, B and B′). Our results suggest that SAC-1 functions specifically to promote apical secretory transport without affecting the basolateral secretion in the intestinal epithelia of C. elegans.
Moreover, we observed that the labeling levels of TGN marker AMAN-2-GFP moderately decreased (Fig. S2, C–C″). This phenotypical change could be associated with the PGP-1-GFP secretion defects. There were no noticeable changes in the patterns of AMAN-2-GFP or SP12-GFP labeled ER upon loss of SAC-1, suggesting that the detrimental effect of SAC-1 depletion on apical secretion does not disrupt the morphology or distribution of the Golgi and ER (Fig. S2, C–D′).
Loss of SAC-1 leads to significantly elevated PI(4,5)P2 levels in endosomes
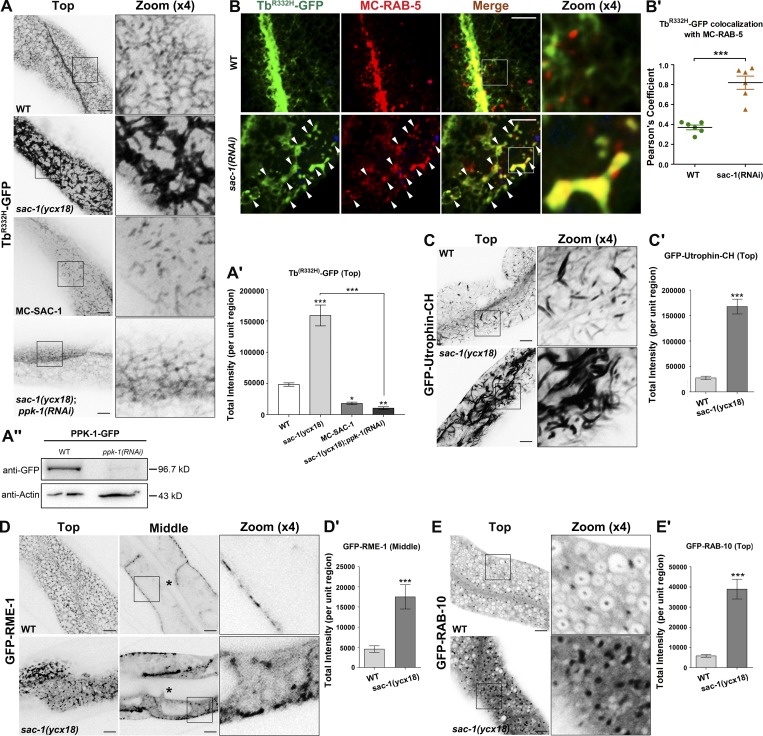
The SAC homologue domain has phosphoinositide phosphatase specificities to PI(3)P, PI(4)P, and PI(3,5)P2 (Guo et al., 1999; Hughes et al., 2000b; Nemoto et al., 2000; Vanhauwaert et al., 2017). Specifically, the SAC domain of yeast Sac1p and its homologues participate in the modulation of intracellular PI(4)P pool (Liu et al., 2008; Cheong et al., 2010; Dickson et al., 2016). Arf6/ARF-6 was reported to up-regulate intracellular PI(4,5)P2 levels by activating downstream PIP5 kinase (Brown et al., 2001; Naslavsky et al., 2003; Shi et al., 2012). To clarify whether SAC-1, as an ARF-6 interactor, manipulates endosomal P(4,5)P2 in intestinal cells, we used a newly developed PI(4,5)P2 biosensor, TbR332H-GFP, to examine the intracellular levels of PI(4,5)P2 in sac-1 mutant animals (Quinn et al., 2008; Hardie et al., 2015). As expected, in C. elegans intestinal cells, TbR332H-mCherry colocalizes extensively with the canonical PI(4,5)P2 biosensor PH-GFP (Fig. S3, A and A′; Pearson’s correlation coefficient, ∼92.7%). We also assayed the distribution of the PI(3)P marker GFP-2xFYVE, the PI(4)P marker GFP-P4M, and the PI(3,5)P2 marker GFP-2xML1N, respectively (Li et al., 2013; Hammond et al., 2014; Wang et al., 2016). In the absence of SAC-1, TbR332H-GFP overaccumulated on the intracellular aggregates and tubules (Fig. 3, A and A', ∼3.29-fold intensity increase). The PH-GFP–labeled structures also accumulated in sac-1(ycx18) mutant cells (Fig. S3, B and B′; ∼3.37-fold intensity increase). To determine the endosomal identity of TbR332H-GFP aggregates, we compared the overlap between the early endosome marker mCherry-RAB-5 and TbR332H-GFP. Although the colocalization level between the tubular TbR332H-GFP and the punctate mCherry-RAB-5 is apparently low in the wild-type background, TbR332H-GFP significantly overlapped with mCherry-RAB-5 in the enlarged endosomes upon loss of SAC-1 (Fig. 3, B and B′). In agreement with the perturbed localization of hTAC-GFP in sac-1(RNAi) animals, this finding indicates that TbR332H-GFP usually labels recycling endosomes (Fig. S3, C and C′), and the loss of SAC-1 affects phosphoinositides (PIPs) homeostasis, which in turn causes PI(4,5)P2 to accrue abnormally in early endosomes (Fig. 3, B and B′). Together, these results suggest that SAC-1 is a negative regulator of PI(4,5)P2 abundance in endosomes.
Figure 3.
Loss of SAC-1 leads to increased PI(4,5)P2 levels and overaccumulation of actin structures. (A and A′) Compared with wild-type animals, the labeling intensity of TbR332H-GFP in the basolateral endosomes was significantly increased (by ∼3.3-fold) in sac-1(ycx18) mutants. However, in mCherry-SAC-1–overexpressing intestinal cells, TbR332H-GFP labeling in puncta and tubules decreased remarkably. Furthermore, RNAi-mediated knockdown of PPK-1 (PIP5 kinase) significantly reversed the intensity of TbR332H-GFP in sac-1 mutants. Error bars represent SEM (n = 18 each), and asterisks indicate significant differences in the one-tailed Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (A″) Western blot showing the PPK-1-GFP level in animals before and after the RNAi-mediated PPK-1 knockdown. (B and B′) The colocalization level between TbR332H-GFP and mCherry-RAB-5 is relatively low in wild-type animals. TbR332H-GFP overlapped with mCherry-RAB-5 in the enlarged early endosomes in sac-1(RNAi) animals. Arrowheads indicate positive overlap. Pearson’s correlation coefficients for GFP and mCherry signals were calculated (n = 6 animals). Error bars represent SEM. ***, P < 0.001. (C and C′) In sac-1(ycx18) mutants, the intensity of GFP-Utrophin-CH-labeled actin structures was extensively up-regulated (by ∼6.1-fold). Error bars represent SEM (n = 18 each), and asterisks indicate significant differences in the one-tailed Student’s t test (***, P < 0.001). (D and D′) In wild-type animals, GFP-RME-1 labeled basolateral recycling endosomes, showing the tubular and punctate distribution (top focal plane). However, the basolateral meshwork labeled by GFP-RME-1 was greatly disrupted in sac-1(ycx18) mutants. GFP-RME-1–positive structures accumulated in the cytoplasmic puncta (middle focal plane). Asterisks in the panels indicate intestinal lumen. Error bars represent SEM (n = 18 each). Asterisks indicate significant differences in the one-tailed Student’s t test (***, P < 0.001). (E and E′) Compared with wild-type animals, loss of SAC-1 resulted in an accumulation of RAB-10–labeled puncta. Error bars represent SEM (n = 18 each), and asterisks indicate significant differences in the one-tailed Student’s t test (***, P < 0.001). Bars, 10 µm.
We also noted that the labeling intensity of GFP-2xFYVE–labeled puncta increased significantly (Fig. S3, D and D′). Consistent with the up-regulation of PI(3)P, GFP-RAB-5–labeled early endosomes accumulated in large quantities upon loss of SAC-1 (Fig. S4, C and C′). PI(3)P is required for cargo sorting and transport regulation in early endosomes (Norris et al., 1995; Shin et al., 2005; Posor et al., 2013; Hsu et al., 2015). The accumulation of hTfR-GFP correlates with the significant PI(3)P buildup in SAC-1–deficient cells, suggesting that this phenotype is independent of the PI(4,5)P2 hike in sac-1 mutants. Consistent with this hypothesis, the accumulation of hTfR-GFP was not mitigated in sac-1(ycx18);ppk-1(RNAi) animals (Fig. S1, H and H′). Various studies indicated that the SAC domain of Sac1p has the specificity of hydrolyzing PI(4)P and PI(3,5)P2 (Guo et al., 1999; Hughes et al., 2000b; Nemoto et al., 2000; Chung et al., 2015; Dickson et al., 2016; Vanhauwaert et al., 2017). However, the PI(4)P marker GFP-P4M or the PI(3,5)P2 marker GFP-2xML1N had no significant changes in distribution or intensity in the absence of SAC-1 (Fig. S3, E–F′). Consistently, although SAC-1 localizes in TGN, we did not observe any noteworthy abnormalities of AMAN-2-GFP subcellular distribution in sac-1 mutants (Fig. S2, C–C″). To clarify whether ER- and Golgi-located SAC-1 was efficiently depleted in sac-1(ycx18) mutants, we conducted colocalization experiment to determine the colabeling of the TGN marker AMAN-2 or the ER marker SP12 with SAC-1 in sac-1(ycx18) animals. The results showed that SAC-1 has a significant colocalization with AMAN-2 or SP12 in wild-type animals (Fig. 1, B and B′). However, in the absence of SAC-1, these significant overlaps were severely disrupted because of the lack of the CRISPR/Cas9 editing–sensitive mCherry-SAC-1 labeling (Fig. S3, G and G′). These findings suggest that, at least in C. elegans intestinal cells, Golgi- and ER-located SAC-1 is not the sole regulator of PI(4)P pool, and additional mechanisms are underlying the metabolism of PI(4)P in the Golgi and ER. Given these findings, we further examined the colocalization of mCherry-SAC-1 with TbR332H-GFP or GFP-2xFYVE. mCherry-SAC-1 and TbR332H-GFP exhibited a significant colocalization in intracellular puncta (Fig. S3, H and H′; Pearson’s correlation coefficient, ∼72%). mCherry-SAC-1 also partially overlapped with GFP-2xFYVE on punctate structures in the cytosol (Fig. S3, H and H′; Pearson’s correlation coefficient, ∼58%).
In agreement with the escalation of TbR332H-GFP labeling, the intensity of PI(4,5)P2 immunostaining significantly increased in sac-1 mutants (Fig. S3, I and I′). The absence of SAC-1–induced PI(4,5)P2 up-regulation appears to be correlated with the alteration in the total level of PIPs, which moderately increased upon loss of SAC-1 (Fig. S3, J and J′). PI(4,5)P2 synthetic precursor PI(4)P is mainly derived from the Golgi and plasma membrane (Dickson et al., 2016). However, the overall intensity of the PI(4)P marker GFP-P4M remains mostly unchanged in sac-1 mutant cells (Fig. S3, E and E′). These data suggest that the prominent increase of PI(4,5)P2 is unlikely at the expense of a loss of the PI(4)P pool, and the increase in PI(4,5)P2 levels could justify the up-regulation of the total cellular PIPs. We validated the specificity of the intestine-specific CRISPR/Cas9 sac-1(ycx18) mutants by coexpressing a CRISPR/Cas9 editing–resistant form of mCherry-SAC-1 in the intestine. We found that the CRISPR/Cas9 editing–resistant transgene fully rescued the TbR332H-GFP overaccumulation phenotype in sac-1(ycx18) mutants, where the TbR332H-GFP intensity was even lower than those of wild-type animals (Fig. S3, K and K′). These results also indicate that the requirement for SAC-1 in PI(4,5)P2 modulation is cell autonomous.
PI(4,5)P2 is a crucial actin cytoskeleton regulator, affecting actin organization via actin-binding proteins (Yin and Janmey, 2003; Wang et al., 2016). In wild-type animals, actin marker GFP-Utrophin-CH predominantly labels puncta- and strip-like structures within the cytoplasm. We observed that the overall intensity of GFP-Utrophin-CH-positive structures substantially increased in the sac-1 mutant background (Fig. 3, C and C′; ∼6.1-fold increase). In the absence of SAC-1, ACT-5-GFP–labeled puncta also overaccumulated and formed cytosolic aggregates (Fig. S4, A and A′).
The functionality decay of ARF-6 underlies irregular PI(4,5)P2 abundance and hTAC-GFP recycling defects in SAC-1–depleted cells
The substantial increase of PI(4,5)P2 levels in sac-1 mutants suggests that SAC-1 functions to attenuate ARF-6 activity. To test this possibility, we assessed the overall intensity of TbR332H-GFP in mCherry-SAC-1–overexpressing intestinal cells. The TbR332H-GFP labeling intensity in puncta and tubules decreased significantly (by ∼62.8%) compared with wild-type animals (Fig. 3, A and A′). Furthermore, RNAi-mediated knockdown of ARF-6 effector PPK-1/PIP5 kinase significantly alleviated the intensity boost of TbR332H-GFP in sac-1 mutants (Fig. 3, A and A′; intensity decreased by ∼93.5%).
Our previous studies indicated that the consistent inactivation of ARF-6 induced by the overexpression of ARF-6-GAP/CNT-1 led to a significant decrease in endosomal levels of PI(4,5)P2, which impaired the recycling of hTAC-GFP (Shi et al., 2012). In the intestinal cells overexpressing mCherry-SAC-1, we observed that the overaccumulation phenotype of hTAC-GFP resembled that of CNT-1 overexpression cells (Fig. 2, A and A′; Shi et al., 2012). The recycling defect of hTAC-GFP was not rescued in sac-1(ycx18);ppk-1(RNAi) animals (Fig. 2, A and A′). These experiments demonstrate that hTAC-GFP transport defects in SAC-1–depleted cells are closely associated with disturbed PI(4,5)P2 homeostasis (Shi et al., 2012).
The level of PI(4,5)P2 is positively correlated with the membrane recruitment of the PI(4,5)P2-binding protein RME-1 (Shi et al., 2012). As expected, in sac-1 mutants, the GFP-RME-1–labeled meshwork collapsed, and GFP-RME-1 mislocalized and accumulated in cytosolic aggregates (Fig. 3, D and D′). These aggregated GFP-RME-1 overlapped well with early endosome marker mCherry-RAB-5 in the enlarged endosomal structures (Fig. S4, B and B′). Of note, PI(4,5)P2 accumulation in early endosomes could account for the aggregation of GFP-RME-1 in the perturbed membranous compartments (Fig. 3, B and B′). Furthermore, the irregular deposits of recycling regulator RME-1 could explain the defects in hTAC-GFP recycling in sac-1 mutants. Previously, we reported that in arf-6 or cnt-1 mutants, the clathrin-independent cargo recycling regulator RAB-10 accumulated in intracellular endosomes, whereas distribution of the late endosomal marker RAB-7 was not affected (Shi et al., 2012). Likewise, in sac-1 mutants, RAB-10–labeled puncta overaccumulated in the cytoplasm (Fig. 3, E and E′), and the distribution of RAB-7–labeled structures was normal (Fig. S4, D and D′). Consistently, the RAB-10 effector EHBP-1–labeled tubular network was fully disrupted (Fig. S4, E and E′).
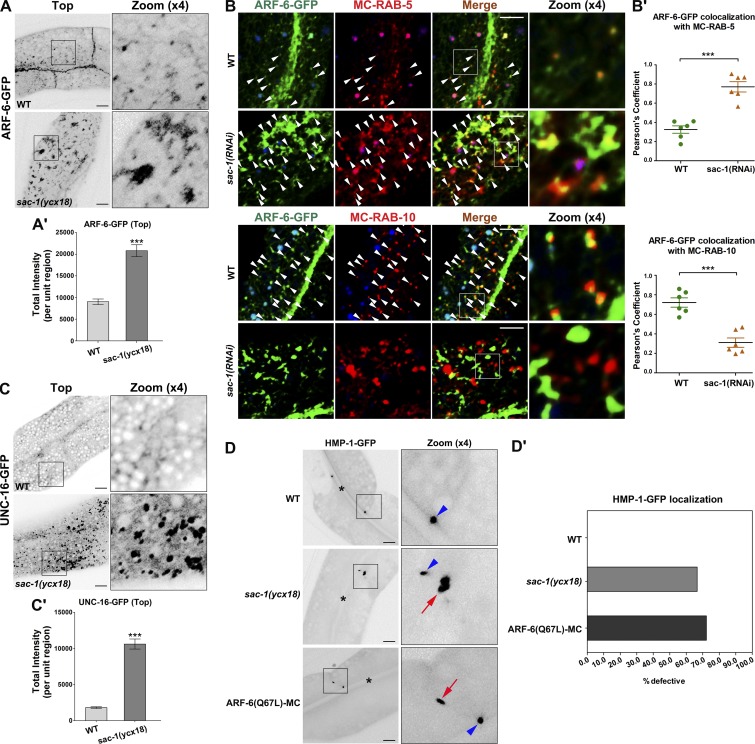
Loss of SAC-1 leads to intracellular accumulation of ARF-6 and UNC-16
The precise regulation of endosomal PI(4,5)P2 is required for the proper flow of recycling transport. Irregular increase or decrease of PI(4,5)P2 levels will impair the recycling process. To further characterize the implication of SAC-1 in the recycling process, we assayed the subcellular distribution of ARF-6-GFP in sac-1(ycx18) animals (Fig. 4, A and A′). Loss of SAC-1 resulted in the accumulation of ARF-6-GFP in endosomal structures, and sac-1(ycx18) mutants displayed significantly reduced meshwork labeling of ARF-6-GFP, as expected if recycling transport is disrupted (Shi et al., 2012). We conducted two sets of colocalization experiment to assess the labeling of the accumulated ARF-6. In wild-type animals, ARF-6 has a significant colocalization with the recycling endosome marker mCherry-RAB-10 in punctate structures and appears less localized in early endosome marker RAB-5–positive structures (Fig. 4, B and B′). SAC-1 depletion significantly increased the overlap between ARF-6 and RAB-5 in the enlarged endosomal structures, whereas the colocalization between ARF-6 and RAB-10 was significantly reduced (Fig. 4, B and B′).
Figure 4.
Loss of SAC-1 leads to intracellular accumulation of ARF-6 and UNC-16 and HMP-1 mislocalization. (A and A′) Depletion of SAC-1 resulted in the accumulation of ARF-6–labeled structures (by approximately twofold). Error bars represent SEM (n = 18 each). Asterisks indicate significant differences in the one-tailed Student’s t test (***, P < 0.001). (B and B′) In sac-1(RNAi) animals, the overlap between ARF-6-GFP and mCherry-RAB-5 was enhanced. In sac-1(RNAi) animals, the colocalization between ARF-6-GFP and mCherry-RAB-10 was decreased. Arrowheads indicate positive overlap. Pearson’s correlation coefficients for GFP and mCherry signals were calculated (n = 6 animals). Error bars represent SEM. ***, P < 0.001. (C and C′) Loss of SAC-1 resulted in overaccumulation of UNC-16–labeled puncta (by approximately fourfold). Error bars represent SEM (n = 18 each). Asterisks indicate the significant difference in the one-tailed Student’s t test (***, P < 0.001). (D and D′) In wild-type animals, HMP-1-GFP is localized in the subapical contact zone between cells. In sac-1(ycx18)– or ARF-6(Q67L)–overexpressing animals, HMP-1-GFP mislocalized in lateral plasma membranes. Arrowheads indicate subapical labeling of HMP-1-GFP. Arrows indicate the abnormal labeling of HMP-1-GFP in lateral membranes. Percentages of defective HMP-1 labeling in 18 intestinal cells (total of six animals per each genotype) were quantified and plotted. Bars, 10 µm.
The scaffold proteins JIP3 and JIP4, the mammalian homologues of C. elegans UNC-16, are considered to be Arf6 effectors (Isabet et al., 2009; Montagnac et al., 2009, 2011). Our data indicate that SAC-1 negatively regulates endosomal PI(4,5)P2 in intestinal epithelia. A plausible explanation is that the ARF-6 interactor SAC-1 regulates ARF-6 activity through a negative feedback loop. If so, then we would expect to observe the overaccumulation of the ARF-6 putative effector UNC-16 in sac-1 mutants. In wild-type animals, UNC-16-GFP labeled basolateral tubular and punctate structures in intestinal cells. As expected, in the absence of SAC-1, UNC-16-GFP concentrated in the cytosolic puncta (Fig. 4, C and C′), suggesting that ARF-6 activity is abnormally promoted.
Apical junctional protein HMP-1 mislocalizes in lateral membrane of sac-1 mutant cells
A recent study in sea urchin primary mesenchyme cells showed that constitutively active Arf6 induced mislocalization of junctional cadherin (Stepicheva et al., 2017). In C. elegans, apical junctions contain cadherin, HMP-1 (α-catenin), and HMP-2 (β-catenin; Cox and Hardin, 2004). To determine whether ARF-6 is also required for the proper localization of apical junctions in C. elegans epithelia, we examined the distribution of HMP-1 in vivo (Cox and Hardin, 2004; Segbert et al., 2004). Usually, HMP-1-GFP exclusively labels the subapical contact zone between cells (Liu et al., 2018). In the presence of constitutively active ARF-6(Q67L), HMP-1-GFP unexpectedly appeared in punctate structures in lateral plasma membranes (72.2% defective; Fig. 4, D and D′), consistent with a role of ARF-6/Arf6 in mediating the recycling of apical junction proteins. Also, a similar mislocalization of HMP-1-GFP was observed in sac-1(ycx18) mutant animals (66.6% defective; Fig. 4, D and D′). Our data further indicate that ARF-6 activity is abnormally elevated in SAC-1–depleted cells.
SAC-1 interacts with ARF-6 GEF protein BRIS-1
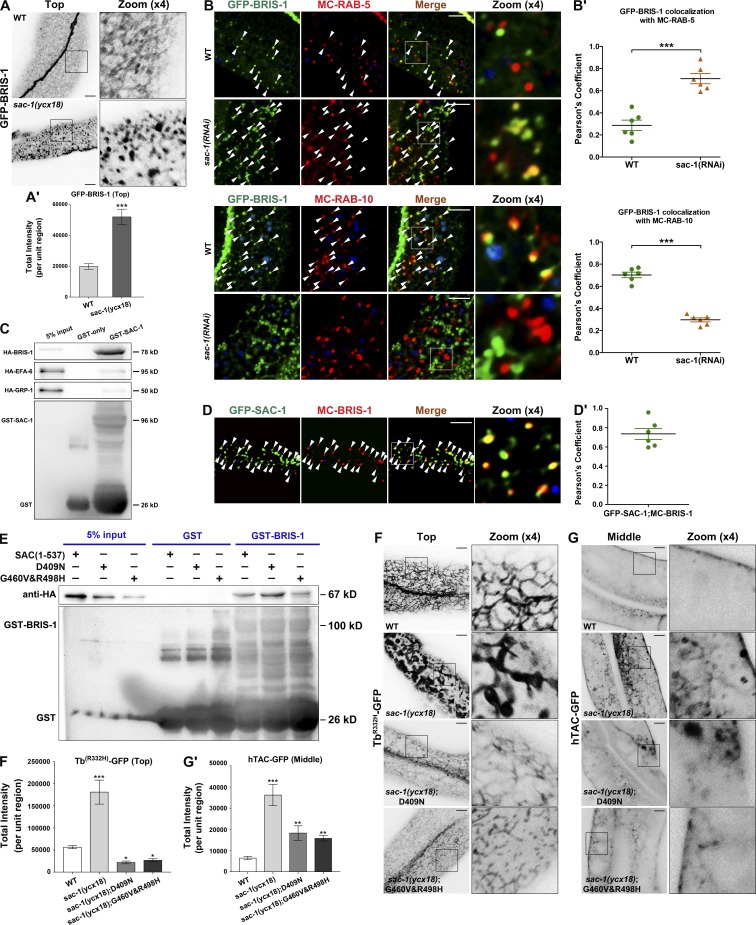
Thus far, our evidence suggests that SAC-1 plays a role in curbing ARF-6 activity in C. elegans intestines by a mechanism not yet determined. To explore the underlying mechanism, we first investigated whether CNT-1/ACAPs/ARF-6-GAP interacts with SAC-1 by GST pull-down. No significant interaction between GST-SAC-1 and HA-CNT-1 was recorded (Fig. S5 A). We then assayed the subcellular localization of previously reported Arf6/ARF-6 GEFs. Three ARF-6-GEF proteins are encoded in the genome of C. elegans, including cytohesin family protein GRP-1, EFA6 family protein EFA-6, and BRAG family protein BRIS-1 (Fig. S5 B). We developed transgenic animals that specifically express GFP fusion proteins of GRP-1, EFA-6, and BRIS-1 in intestinal cells. The subcellular localization of BRIS-1-GFP was affected by the loss of SAC-1. In particular, BRIS-1-GFP-labeled basolateral tubular meshwork was mostly absent, and GFP fluorescence signals appeared in overaccumulated puncta (Fig. 5, A and A′; ∼2.6-fold intensity increase). However, the distribution of GRP-1 within nuclei remained intact after depletion of SAC-1 (Fig. S5, C and C′). Also, SAC-1 deficiency only moderately disturbed the integrity of EFA-6-positive meshwork, whereas the overall intensity of EFA-6 labeling was not significantly affected (Fig. S5, D and D'). Next, we performed colocalization assays to examine the colabeling of BRIS-1 with the early endosome marker mCherry-RAB-5 and the recycling endosome marker mCherry-RAB-10. In agreement with the localization changes of ARF-6, a remarkable overlap between BRIS-1 and RAB-5 in enlarged endosomes was observed in SAC-1–depleted cells (Fig. 5, B and B′), whereas the colocalization of BRIS-1 with RAB-10 in punctate structures was significantly reduced (Fig. 5, B and B′).
Figure 5.
SAC domain mediates the interaction of SAC-1 with BRIS-1/ARF-6-GFE. (A and A′) Compared with wild-type animals, loss of SAC-1 led to the disruption of basolateral tubular network labeled by GFP-BRIS-1, and GFP-BRIS-1 appeared in aggregates. Error bars represent SEM (n = 18 each). Asterisks indicate significant differences in the one-tailed Student’s t test (***, P < 0.001). (B and B′) In sac-1(RNAi) animals, the overlap between GFP-BRIS-1 and mCherry-RAB-5 was enhanced. In sac-1(RNAi) animals, the colocalization between GFP-BRIS-1 and mCherry-RAB-10 was decreased. Arrowheads indicate positive overlap. Pearson’s correlation coefficients for GFP and mCherry signals were calculated (n = 6 animals). Error bars represent SEM. ***, P < 0.001. (C) Glutathione beads loaded with GST and GST-SAC-1 were incubated with HA-tagged BRIS-1, EFA-6, and GRP-1, respectively. Eluted proteins were separated on the SDS-PAGE gel and analyzed by Western blotting using anti-HA antibody. Input lanes contain in vitro expressed HA-tagged proteins in the binding assays (5%). (D and D′) GFP-SAC-1 overlapped with mCherry-BRIS-1 significantly in intracellular puncta (Pearson’s coefficient, ∼75.8%). Pearson’s correlation coefficients for GFP and mCherry signals were calculated (n = 6 animals). (E) The fragment (aa 1–537) containing the SAC domain can efficiently associate with BRIS-1. Similarly, the SAC domain with catalytic core mutations (D409N or G460V&R498H) maintains the capability of binding to BRIS-1. (F and F′) The labeling intensity of TbR332H-GFP in the basolateral endosomes was significantly promoted in sac-1(ycx18) mutants. In mCherry-SAC-1(D409N)–overexpressing intestinal cells, TbR332H-GFP labeling in puncta and tubules decreased. In mCherry-SAC-1(G460V&R498H)–overexpressing cells, TbR332H-GFP labeling in puncta and tubules was reduced. Error bars represent SEM (n = 18 each), and asterisks indicate the significant differences in the one-tailed Student’s t test (*, P < 0.05; ***, P < 0.001). (G and G′) The labeling intensity of hTAC-GFP was significantly increased in sac-1(ycx18) mutants. In mCherry-SAC-1(D409N)–overexpressing intestinal cells, hTAC-GFP labeling was also increased. In mCherry-SAC-1(G460V&R498H)–overexpressing cells, hTAC-GFP labeling was increased. Error bars represent SEM (n = 18 each), and asterisks indicate the significant differences in the one-tailed Student’s t test (**, P < 0.01; ***, P < 0.001). Bars, 10 µm.
To better assess the correlation of SAC-1 with BRIS-1, we performed GST pull-down assays to examine the interaction of GST-SAC-1 with HA-tagged GRP-1, EFA-6, and BRIS-1, respectively. As shown in Fig. 5 C, SAC-1 displayed a robust protein–protein interaction with BRIS-1, whereas no significant binding existed between SAC-1 and GRP-1 or EFA-6 (Fig. 5 C). Coimmunoprecipitation experiments were performed to validate the interaction between SAC-1 and BRIS-1 (Fig. S1 C). Consistently, there was substantial colocalization between GFP-SAC-1 and mCherry-BRIS-1 in intracellular puncta (Fig. 5, D and D′; Pearson’s correlation coefficient, ∼75.8%).
Catalytic activity of SAC domain is not implicated in the interaction with BRIS-1
Previous studies showed that SAC domains are involved in the regulation of protein interactions (Rohde et al., 2003). To determine whether the SAC domain mediates the binding of SAC-1 with BRIS-1, we selected the fragment (aa 1–537) containing the N-terminal SAC domain as bait to examine the interaction with BRIS-1. We observed that the SAC domain is sufficient to mediate the interaction with BRIS-1 (Fig. 5 E). Next, we used the SAC domain with catalytic core mutations for binding testing (Kearns et al., 1997; Liu et al., 2009). However, the association of BRIS-1 with the SAC domain was not affected by point mutations D409N or G460V&R498H (Fig. 5 E and Table S2). Therefore, our biochemical experiments described above indicate that the SAC domain serves as the interface for binding with BRIS-1, but phosphatase activity seems to be unrelated to this interaction.
To determine whether the PI(4,5)P2 and hTAC phenotypes are correlated with the catalytic activity of SAC-1, we examined the subcellular localization of the TbR332H-GFP and hTAC-GFP in SAC-1–depleted cells coexpressing the CRISPR/Cas9 editing–resistant forms of SAC-1 with catalytic core mutations (Fig. 5, F–G′). Notably, TbR332H-GFP labeling defects were significantly complemented by the transgenic expression of SAC-1(D409N) and SAC-1(G460V&R498H) (Fig. 5, F and F′). The consistent inactivation of ARF-6 leads to the decrease in PI(4,5)P2 levels and the overaccumulation of hTAC-GFP (Shi et al., 2012). Accordingly, hTAC-GFP cytosolic accumulation was observed in the animals overexpressing SAC-1(D409N) or SAC-1(G460V&R498H) (Fig. 5, G and G′). Together these results suggest that the functional role of SAC-1 in PI(4,5)P2 abundance modulation and the recycling transport is independent of the phosphoinositide phosphatase activity of SAC-1, thus rationalizing the interaction of BRIS-1 with the SAC domain harboring point mutations D409N or G460V&R498H.
SAC-1 competes with ARF-6(T27N) for interaction with BRIS-1/ARF-GEF
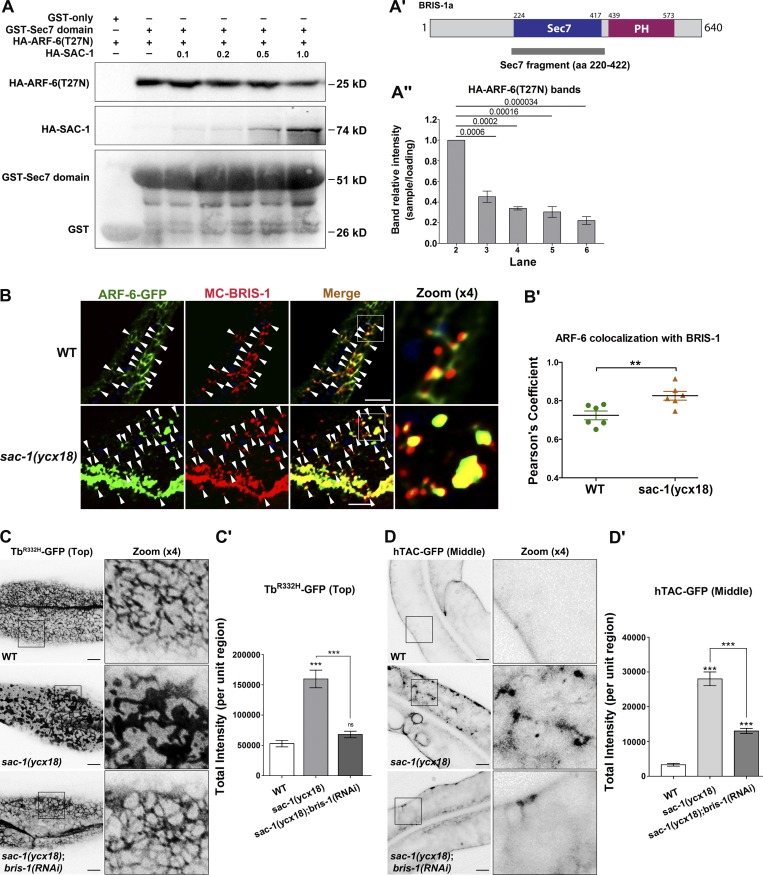
Thus far, our experiments demonstrated that SAC-1 interacts with ARF-6-GEF protein BRIS-1 and functions to restrain ARF-6 activity. Furthermore, we observed that the Sec7 domain of BRIS-1 mediates the interaction of BRIS-1 with SAC-1 (Fig. S5 E). It is worth noting that Arf small GTPases preferentially associate with the Sec7 domain of their GEF proteins, as previously described (Renault et al., 2003). This evidence raises the possibility that SAC-1 competes with ARF-6(GDP) for the Sec7 domain of BRIS-1, thereby interfering with the interaction of BRIS-1 and ARF-6(GDP). To test this proposed mechanism, we first examined whether the presence of SAC-1 affects the affinity of the BRIS-1-Sec7 domain to the GDP-bound mutant form ARF-6(T27N). In the absence of SAC-1, there was a certain level of binding between GST-Sec7 and HA-ARF-6(T27N) (Fig. 6, A and A′). However, the affinity between the Sec7 domain and ARF-6(T27N) was significantly reduced after adding HA-SAC-1 to the reaction (Fig. 6, A–A″). Specifically, HA-SAC-1 outcompeted HA-ARF-6(T27N) for binding to GST-Sec7 in a concentration-dependent manner. The presence of increasing concentrations of HA-SAC-1 gradually weakened the GST-Sec7:HA-ARF-6(T27N) interaction. Our results support the SAC-1:BRIS-1:ARF-6(GDP) competition model, which predicts that SAC-1 antagonizes BRIS-1 during the activation of ARF-6.
Figure 6.
SAC-1 competes with ARF-6(T27N) for binding to the Sec7 domain of BRIS-1. (A–A″) SAC-1 outcompeted ARF-6(T27N) for interaction with the Sec7 domain in a concentration-dependent manner, as determined by GST pull-down. Three independent experiments are represented in the histogram, error bars represent SEM (n = 3 each), and P values are indicated above. (B and B′) In wild-type animals, mCherry-BRIS-1 mainly labeled basolateral tubules and puncta and partially colocalized with ARF-6-GFP (Pearson’s coefficient, ∼72.4%). In sac-1(ycx18) mutants, mCherry-BRIS-1 and ARF-6-GFP were concentrated in the intracellular aggregates, and their colocalization level was substantially promoted (Pearson’s coefficient, ∼85.4%). Arrowheads indicate positive overlap. Pearson’s correlation coefficients for GFP and mCherry signals were calculated (n = 6 animals). Asterisks indicate significant differences in the one-tailed Student’s t test (**, P < 0.01). (C and C′) Compared with wild-type animals, the labeling intensity of TbR332H-GFP in the basolateral endosomes was grossly increased in sac-1(ycx18) mutants. Furthermore, RNAi-mediated knockdown of BRIS-1 significantly eased the intensity of TbR332H-GFP in sac-1 mutants. Error bars represent SEM (n = 18 each), and asterisks indicate significant differences in the one-tailed Student’s t test (***, P < 0.001). (D and D′) RNAi-mediated knockdown of BRIS-1 greatly reduced the accretion of hTAC-GFP in sac-1 mutants. Error bars represent SEM (n = 18 each), and asterisks indicate the significant differences in the one-tailed Student’s t test (***, P < 0.001). Bars, 10 µm.
mCherry-BRIS-1 predominantly labeled the basolateral tubular and punctate endosomes and colocalized well with ARF-6-GFP (Fig. 6, B and B′; Pearson’s correlation coefficient, ∼72.4%). Consistent with the competition model, mCherry-BRIS-1 and ARF-6-GFP were concentrated in enlarged structures, and their colocalization was substantially promoted in sac-1(ycx18) mutants (Fig. 6, B and B′; Pearson’s correlation coefficient, ∼85.4%). To investigate the functional conservation of SCAM1L, the mammalian homologue of C. elegans SAC-1, we examined the colabeling between Arf6 and its GEF protein, BRAG2, in HeLa cells. Arf6-mCherry and GFP-BRAG2 colocalize well in the intracellular puncta in control cells (Fig. S5, F–G'). In the absence of SCAM1L, the overlap between Arf6 and BRAG2 was remarkably boosted in the enlarged structures (Fig. S5, G and G′), suggesting that loss of SCAM1L led to an excessive association of BRAG2 with Arf6 in endosomes. Furthermore, after RNAi-mediated knockdown of BRIS-1, we observed a significant reversal of TbR332H-GFP intensity in SAC-1–depleted cells (Fig. 6, C and C′), suggesting that BRIS-1 mediates SAC-1–deficiency induced PI(4,5)P2 buildup. Consistently, the overaccumulation phenotype of hTAC-GFP was notably reduced in sac-1(ycx18);bris-1(RNAi) animals (Fig. 6, D and D′). Moreover, we observed the remarkable reduction of UNC-16-GFP accumulation in the cytosol (Fig. S5, J and J′). In summary, our experiments identify SAC-1 as a novel ARF-6 interactor that mediates negative feedback regulation of ARF-6 in C. elegans intestinal cells, likely with the involvement of the ARF-6 GEF protein BRIS-1.
Discussion
Arf6 is implicated in the recycling regulation of clathrin-independent cargo, and cargo sorting after endocytosis requires timely shutdown of Arf6 activity (Donaldson, 2003). However, Arf6 needs to be reactivated for proper endocytic recycling. Constitutive inactivation of ARF-6 leads to irregular hTAC-GFP recycling transport (Shi et al., 2012). The on-and-off activity of Arf6 is regulated by GEFs and GAPs. To better characterize the regulatory mechanism of ARF-6, we identified SAC-1 as a previously unrecognized ARF-6 interactor in C. elegans. In sac-1 mutant animals, ARF-6 activity was significantly boosted. The presence of SAC-1 restrained the interaction of ARF-6(T27N) with BRIS-1 in a concentration-dependent manner, suggesting that SAC-1 has an antagonistic effect on ARF-6(GTP) levels via a feedback loop.
Sac1p/hSAC1 contains an N-terminal SAC domain and two C-terminal transmembrane helices (Konrad et al., 2002; Hsu and Mao, 2015). Previous studies showed that Sac1p/hSAC1 localizes predominantly to the ER and Golgi (Kochendörfer et al., 1999; Rohde et al., 2003; Blagoveshchenskaya et al., 2008; Bajaj Pahuja et al., 2015; Chung et al., 2015). The distribution of hSAC1 in the Golgi is determined by the first transmembrane helix, and the N-terminal cytoplasmic region contributes to localization (Wang et al., 2013). The Golgi residence of Arabidopsis AtSAC1 also depends on the C-terminus, and the AtSAC1 truncation without the C-terminal segment results in abnormal distribution in the cytoplasm (Zhong et al., 2005). Consistently, C. elegans SAC-1 localizes to the trans-Golgi network and ER in our study. Furthermore, we found that SAC-1 appears on endosomes near the basolateral plasma membrane, and the endosomal residence of SAC-1 relies on ARF-6. Previous studies indicate that there is direct cargo flow between the Golgi and recycling endosomes (Bai and Grant, 2015), and the pool of PI(4)P in the Golgi contributes to the homeostasis of PI(4,5)P2 in the plasma membrane (Dickson et al., 2014). Similar to the shuttling of hSAC1 between the ER and plasma membrane via dynamic changes in ER-PM junctions (Dickson et al., 2016), SAC-1 intracellular localization in intestinal cells suggests that SAC-1 is probably also involved in the regulation of PIP homeostasis between the Golgi and sorting endosomes. Validating this possibility would require further experimentation.
Accumulating evidence indicates that Sac1p and its homologues could efficiently regulate intracellular PI(3)P, PI(4)P, and PI(3,5)P2 levels. Meanwhile, hSAC1 also manipulates PI(4,5)P2 levels indirectly by various mechanisms. For example, a recent study showed that hSAC1 localizes at discrete ER-PM junctions, where it limits the production of PI(4,5)P2 via limiting PI(4)P pool in the plasma membrane (Dickson et al., 2016). There is another mechanism by which hSAC1 regulates PI(4,5)P2 level indirectly in tsA-201 cells (Dickson et al., 2014). The Golgi PI(4)P pool is tightly correlated with the PI(4,5)P2 levels in the plasma membrane, and hSAC1 may indirectly influence PI(4,5)P2 in the plasma membrane by regulating PI(4)P in the Golgi. Our experiments demonstrate that SAC-1, as an ARF-6 interactor, can confine ARF-6 activity and influence the level of endosomal PI(4,5)P2. It is likely that these distinct PI(4,5)P2 regulatory mechanisms of hSAC1/SAC-1 have their characteristic implication in terminally differentiated cell types. However, without extensive experimental studies, we cannot rule out the possibility that these mechanisms coexist in the same cellular context. In addition to hSAC1, mammalian SAC2 was also found in recycling endosomes in the mouse neuroblastoma N2A cells (Hsu et al., 2015). SAC2 is a phosphoinositide 4-phosphatase that hydrolyzes PI(4)P. The overexpression of the catalytically inactive mutant of SAC2 disrupted the recycling transport of TfR (Hsu et al., 2015). However, we failed to observe abnormal hTAC-GFP distribution in sac-2 mutants. PI(4)P biosensor GFP-P4M–labeled structures were not significantly affected in sac-1 mutant cells. These findings suggest that the regulatory role of SAC-1 in basolateral recycling is not directly associated with PI(4)P modulation. It is worth noting that basolateral secretory cargo NPY-GFP–labeled meshwork was intact in sac-1 mutants. However, similar to rab-8 mutant animals, apical secretory cargo PGP-1-GFP severely overaccumulated in the cytoplasm of SAC-1–depleted cells. Our data suggest that SAC-1 is involved in promoting apical secretory delivery without affecting the basolateral counterpart in the intestinal epithelia of C. elegans. Surprisingly, the morphology or distribution of AMAN-2-GFP–labeled TGN or SP12-GFP–labeled ER was not affected by the loss of SAC-1. These results indicate that the apical and basolateral secretory transport has distinct regulatory mechanisms in intestinal epithelia, whereas SAC-1 is only involved in the apical secretory process. Additionally, the disturbance of the SAC-1–mediated secretory delivery is not directly associated with the morphological abnormality in the Golgi and ER, suggesting that the phenotype is caused by functionality damage. Sac1p was reported to play a crucial role in mediating ATP transport into the ER (Mayinger et al., 1995). Another plausible explanation is the regulatory redundancy in basolateral secretory transport. SAC domains in yeast Inp52p and Inp53, and mammalian and Drosophila synaptojanin also maintain hydrolyzing specificity toward PI(3)P, PI(4)P, and PI(3,5)P2 (Guo et al., 1999; Hughes et al., 2000b; Nemoto et al., 2000; Chung et al., 2015; Dickson et al., 2016; Vanhauwaert et al., 2017). C. elegans OCRL-1, the homologue of Inp52p and Inp53p, was found to be expressed in the intestinal cells (Spencer et al., 2011). Similarly, UNC-26/synaptojanin expression was found in the intestine (McKay et al., 2003; Hunt-Newbury et al., 2007). These SAC domain proteins may compensate for the lack of SAC-1 during basolateral secretory transport.
Arf6-GEFs can be regulated by distinct mechanisms in diverse cellular processes (Hongu and Kanaho, 2014). We found that the subcellular localization of BRIS-1 was disrupted in the absence of SAC-1, whereas the distribution of GRP-1 and EFA-6 remained virtually unchanged. Consistently, SAC-1 had no significant physical interaction with GRP-1 or EFA-6. In intestinal epithelia, the interaction between SAC-1 and BRIS-1 effectively governs the activity of ARF-6 and the endosomal PI(4,5)P2. Therefore, we speculate that the precise spatial and/or temporal control of Arf6-GEFs activity determines the working specificity of Arf6, and this functional commitment is likely to be established by the dedicated regulators of Arf6-GEFs in distinct biological processes. A recent study in human retinal microvascular endothelial cells demonstrated that various GEFs were sequentially used to activate Arf6 to coordinate VEGFR2 trafficking (Zhu et al., 2017). Arf6-GEF ARNO activates Arf6 and promotes VEGFR2 internalization, whereas Arf6-GEF GEP100 acts later to activate Arf6 to encourage VEGFR2 recycling. Although the exact mechanism of sequential usage of Arf6-GEFs in VEGFR2 trafficking is still unknown, the involvement of highly specific Arf6-GEF regulation is emerging as a common theme in the mechanisms of vesicle trafficking.
In the current study, we observed that the absence of ARF-6 resulted in the significantly reduced localization of GFP-SAC-1 in basolateral puncta. SAC-1 is a transmembrane protein, and presumably, there is no process of SAC-1 recruitment from the cytoplasm to the endosome. One explanation for our results is that ARF-6 is required for clustering SAC-1 from other connected membranous organelles to the sorting endosomes. To this end, we performed rescue experiments by coexpressing ARF-6(Q67L)-mCherry or ARF-6(T27N)-mCherry in arf-6(tm1447);GFP-SAC-1 animals, respectively. The results showed that the expression of ARF-6(Q67L) significantly recovered the puncta labeling GFP-SAC-1, whereas the expression of ARF-6(T27N) failed to restore the endosomal localization of GFP-SAC-1, suggesting that the interaction between SAC-1 and ARF-6(GTP) predominantly determines the endosomal positioning of SAC-1. Additionally, we found that the presence of HA-SAC-1 significantly reduced the binding of GST-BRIS-1(Sec7 domain) to HA-ARF-6(T27N), suggestive that SAC-1 could outcompete ARF-6(GDP) to occupy BRIS-1. It should be noted that unlike Arf1(GDP), Arf6(GDP) is mostly retained on membranes (Cavenagh et al., 1996; Song et al., 1998; Donaldson, 2003). Thus, we propose that the interaction of ARF-6(GDP) with SAC-1 on the membrane does not administrate the endosomal localization of SAC-1 (Fig. 1 C) but instead maintains the proximity of SAC-1 on sorting endosomes to facilitate the competition with ARF-6(GDP) itself. This speculation is corroborated by the additional experiments (Fig. S5, H and I). The addition of HA-SAC-1 failed to affect the interaction between ARF-6(T27N) and another ARF-6 GEF protein, EFA-6 (Fig. S5 H), suggesting that the interaction between SAC-1 and BRIS-1/GEF, rather than the interaction between SAC-1 and ARF-6(T27N), plays a prominent role in the competition for binding to BRIS-1. Moreover, the interaction between the ARF-6 GAP protein GST-CNT-1 and HA-ARF-6(Q67L) was not affected by the presence of HA-SAC-1 (Fig. S5 I), implying that the interaction between SAC-1 and ARF-6(Q67L) is not involved in the competition for BRIS-1. Taken together, our results suggested that ARF-6(GTP) is primarily responsible for clustering SAC-1 to sorting endosomes, whereas the interaction of ARF-6(GDP) with SAC-1 could be involved in the maintenance of SAC-1 endosomal proximity, allowing SAC-1 to compete with ARF-6(GDP) for the binding interface on the GEF protein BRIS-1.
Materials and methods
General methods and strains
The strains were originated from Bristol strain N2. Culturing and genetic manipulation of the animals were performed according to the standard procedure (Brenner, 1974). All strains were grown on NGM plates at 20°C, with Escherichia coli strain OP50 as the source of food. A complete list of strains can be found in Table S1.
RNAi
RNAi-mediated interference is based on standard feeding protocol (Timmons and Fire, 1998). The feeding constructs used in this study were either from the Ahringer library (Kamath and Ahringer, 2003) or prepared by subcloning cDNA fragments into the RNAi vector L4440 (Timmons and Fire, 1998). In most of our experiments, synchronized L1 stage animals were cultured for 72 h and scored as adults.
Antibodies
The antibodies used in the current study are listed below: rabbit anti–actin polyclonal antibody (sc-1616-R; Santa Cruz Biotechnologies), rabbit anti–HA monoclonal antibody (C29F4; Cell Signaling Technology), rabbit anti–GST monoclonal antibody (91G1; Cell Signaling Technology), rabbit anti–mCherry polyclonal antibody (ab167453; Abcam), rabbit anti–GFP polyclonal antibody–chromatin immunoprecipitation grade (ab290; Abcam), mouse anti–P(4,5)P2 monoclonal antibody (Z-P045; Echelon Biosciences), and mouse anti–PIPn monoclonal antibody (Z-P999; Echelon Biosciences).
Somatic CRISPR/Cas9 mutant strains
CRISPR/Cas9 plasmids were prepared following the previously described protocol (Shen et al., 2014; Li et al., 2015). The somatic CRISPR/Cas9 expression plasmids were prepared by replacing the eft-3 promoter in pDD162 (47549; Addgene) with the C. elegans intestine-specific promoter Pvha-6 via overlapping PCR using Phusion high-fidelity DNA polymerase (Thermo Scientific). CRISPR design tool (http://crispr.mit.edu) was deployed to identify the targets. Three sac-1 target sequences were chosen in this study (5′-AGCCAGCGAACGTTTCATATGG-3′, 5′-CAGCAAGCTGGTCATCTGTTGG-3′, and 5′-GAACAATTAAACTAGTCTCAGG-3′). CRISPR/Cas9 plasmids were transformed into DH5α competent cells. The clones were validated to include the target sequences via sequencing. CRISPR/Cas9 plasmids transgenic animals were developed by microinjection of plasmids at 50 ng/µl and a selection marker Podr-1::gfp into the N2 hermaphrodites germline (Zhou et al., 2016).
Cell cultures and transfection
HeLa cells were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, and 100 U/ml streptomycin. Exponentially growing cells were trypsinized. Solutions with 50,000 cells/ml were prepared in 90% medium (no phenol red or antibiotics) with 10% vol/vol serum. Cells were incubated at 37°C for 2 h before siRNA transfection. 12 µl X-tremeGENE siRNA Transfection Reagent (Sigma), 2 µg siRNA nontargeting control or siRNA plasmid, 2 µg GFP-BRAG2 plasmid, and 2 µg Arf6-mCherry plasmid were added into 100 µl OPTI-MEM I (Gibco). The transfection mix was incubated at room temperature for 5 min and then mixed for 20 min 24 h after transfection, and cells were fixed with 5% paraformaldehyde for 5 min at room temperature. Human SCAM1L siRNA oligonucleotide was synthesized with the sequence 5′-GAGATGGGATCGACTAAGT-3′ (Ribobio Inc.).
GST pull-down assay and Western blotting
rab-5(Q78L), rab-7(Q67L), rab-8(Q67L), rab-10(Q68L), rab-11.1(Q70L), arf-6(Q67L), arf-6(T27N), sac-1, sac-1(1-537aa), sac-1(1-537aa,D409N), sac-1(1-537aa,G460V&R498H), sac-1(Δ133-507aa), bris-1, bris-1(220-422aa), efa-6, grp-1, and cnt-1 cDNAs were transferred into a modified vector pcDNA3.1(+) (Invitrogen) with a 2xHA epitope tag and Gateway cassette (Invitrogen) for in vitro transcription and translation. sac-1, bris-1, and bris-1(220-422aa) PCR products were introduced into pGEX-2T vector (GE Healthcare Life Sciences) with a Gateway cassette. The N-terminally HA-tagged proteins were synthesized using the TNT-coupled transcription-translation system (Promega). GST-only, GST-SAC-1, GST-BRIS-1, and GST-BRIS-1(220–422 aa) fusion proteins were expressed in the ArcticExpress strain of E. coli (Stratagene). Bacterial pellets were lysed in 50 ml lysis buffer (50 mM Hepes, pH 7.5, 400 mM NaCl, 1 mM DTT, and 1 mM PMSF) with Complete Protease Inhibitor Mixture Tablets (Sigma). Extracts were cleared by centrifugation, and supernatants were collected and incubated with glutathione Sepharose 4B beads (GE Healthcare Life Sciences) at 4°C overnight. Beads were washed six times with cold STET buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, and 0.1% vol/vol Tween-20). In vitro–synthesized HA-tagged protein (15 µl TNT mix diluted in 500 µl STET) was added to the beads and allowed to bind at 4°C overnight. After six additional washes in STET, the proteins were eluted by boiling in 30 µl 2×SDS-PAGE sample buffers. Eluted proteins were separated on SDS-PAGE (12% wt/vol polyacrylamide) and blotted onto nitrocellulose. After blocking with 5% milk and washing with TBS-T buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.05% vol/vol Tween-20), the blot was probed with the anti–HA antibody and then stripped and reprobed with the anti–GST antibody.
Plasmid construction and transgenic strains
For sac-1(ycx18) mutant rescue assay, corresponding sgRNA resistant plasmids were prepared. We introduced silent mutations in the codon of each target sequence (5′-GGCTTCAGAGAGGTTTATCTGG-3′, 5′-CTGCCAATTGATCGTCAGTAGG-3′, and 5′-GTACTATCAAGTTGGTAAGTGG-3′). To construct GFP or mCherry fusion transgenes expressed in C. elegans intestine, previously described vha-6 promoter-driven vectors with a Gateway cassette inserted upstream or downstream of the GFP or mCherry coding region were used. The sequences of sac-1, bris-1, efa-6, grp-1, Tubby-PH(R332H) (Quinn et al., 2008; Hardie et al., 2015), 2xML1N (reporter of PI(3,5)P2), and P4M (reporter of PI4P) were cloned in-frame into entry vector pDONR221 by BP reaction, and then transferred into intestinal expression vectors by LR reaction to generate fusion plasmids (Chen et al., 2006). Low-copy integrated transgenic lines were obtained using the microparticle bombardment method (Praitis et al., 2001). Additional transgenic strains were obtained using standard microinjection techniques.
Whole-worm immunoprecipitation
Adult animals were collected and washed with M9 buffer (9-cm plates × 10). The worm pellet was lysed in ice-cold lysis buffer (25 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM PMSF, 1 mM Na3VO4, 1 µg/ml Pepstatin-A, and 10 mM NaF) containing protease inhibitor cocktail (Sigma). The lysates were incubated for 30 min at 4°C and centrifugated at 13,000 g for 30 min. The supernatant was incubated with 80 µl Protein A+G Agarose (Beyotime) for 1 h at 4°C to preclear nonspecific bead–protein interactions. 2 µl anti–GFP antibody (ab290) was added into precleared supernatant and incubated overnight at 4°C, followed by incubation with 80 µl Protein A+G Agarose for 4 h at 4°C. Precipitates were washed five times (10 min each) with lysis buffer and subjected to immunoblotting using anti–actin, anti–mCherry, and anti–GFP antibodies.
PIP immunostaining
Animal dissection was performed as previously described (Grant et al., 2001). Dissected intestines were fixed in 4% paraformaldehyde (Beyotime) in TBS for 20 min at room temperature, and washed three times (10 min each) in TBS, followed by treatment with 0.5% Saponin (S-7900; Sigma) in TBS for 15 min at room temperature. The specimens were then washed three times (10 min each) in TBS and blocked with 10% goat serum in TBS overnight at 4°C. Whole-mount staining using mouse anti-PIP antibody (Echelon Biosciences) diluted in TBS was performed for 1 h at 37°C. The specimens were then washed three times (10 min each) in TBS/goat serum 1%. Secondary antibodies conjugated to Alexa Fluor 488 (Thermo Fisher Scientific) were used at a dilution of 1:2,000 in TBS for 30 min at 37°C. Observations were performed with a 100×, 1.2-NA oil-immersion objective len at room temperature using a C2 laser scanning confocal microscope (Nikon).
Microscopy and imaging analysis
Live worms were mounted onto 2% agarose pads with 100 mM levamisole. Monofluorescence and multiwavelength fluorescence images were acquired at 20°C using a C2 laser scanning confocal microscope equipped with a 100× 1.2-NA oil-immersion objective and NIS-Elements AR 4.40.00 software. Z-series of optical sections were acquired using a 0.5 µm step size.
Monofluorescence images were analyzed by Metamorph software version 7.8.0.0 (Universal Imaging). The “Integrated Morphometry Analysis” function of Metamorph software was employed to measure the fluorescent intensity that is significantly brighter than the background (total intensity), fluorescence area (total area), and puncta number (structure count) within unit regions. From a total of six animals of each genotype, “total intensity,” “structure count,” and “total area” were sampled in three randomly selected unit regions of each animal defined by a 100 × 100 (pixel2) box positioned at random (n = 18 each). In the current study, total area was used to compare tubularity, as the endosomal tubule covers more area than when the tubular network collapses into puncta. Quantification of colocalization images was performed using the open source Fiji (ImageJ) software (Schindelin et al., 2012). Pearson’s correlation coefficients for GFP and mCherry signals were calculated, and six animals for each genotype were assessed.
Statistical analysis
Statistical analyses in the current study were performed using Prism software version 5.01 (GraphPad Software).
Online supplemental material
Fig. S1 demonstrates that the endosomal localization of mCherry-SAC-1 is impaired in arf-6(RNAi) animals. However, the Golgi and ER labeling of mCherry-SAC-1 is intact in arf-6(RNAi) animals. Fig. S2 shows that the transport of basolateral secretory cargo NPY-GFP is not affected in sac-1(ycx18) animals. In contrast, apical secretory cargo protein PGP-1-GFP accumulates in sac-1(ycx18) mutants. Fig. S3 shows that the level of PI(4,5)P2 in basolateral endosomes is promoted in sac-1(RNAi) animals. Also, the level of total PIPs increases moderately in sac-1(RNAi) animals. Fig. S4 shows that the recycling marker GFP-RME-1 accumulates in RAB-5–positive endosomes in SAC-1-depleted cells. Fig. S5 shows that loss of SCAM1L boosts the colocalization between GFP-BRAG2 and Arf6-mCherry. Moreover, SAC-1 fails to outcompete ARF-6(T27N) for interaction with EFA-6. Table S1 lists transgenic and mutant strains. Table S2 shows the alignment of C. elegans SAC-1 and the closely related homologues in human and yeast.
Supplementary Material
Acknowledgments
We thank Tian Xia (Huazhong University of Science and Technology, Huazhong, China) for support regarding this work. We thank Barth D. Grant (Rutgers University, New Brunswick, NJ) and Roger C. Hardie (University of Cambridge, Cambridge, England, UK) for important reagents. We also thank Xin Zhang, Hang Liu, Shimin Wang, Jia Zeng, Xin Fu, Jianli Guo, and Zhenrong Yang for technical assistance.
This work was supported by the Program for HUST Interdisciplinary Innovation Team and the Fundamental Research Funds for the Central University (grant 2016JCTD108), the National Natural Science Foundation of China (grants 31771570, 31571466, and 81371418), the Specialized Research Fund for the Doctoral Program of Higher Education (grant 20130142110071), the Program for New Century Excellent Talents in University (grant NCET-13-0234), and the Junior Thousand Talents Program of China (to A. Shi).
The authors declare no competing financial interests.
Author contributions: D. Chen and A. Shi designed the study; D. Chen, C. Yang, S. Liu, and W. Hang performed the experiments; X. Wang contributed the reagents; D. Chen, C. Yang, S. Liu, W. Hang, J. Chen, and A. Shi analyzed the data; and D. Chen and A. Shi wrote the paper with input and final approval from all authors.
References
- Bai Z., and Grant B.D.. 2015. A TOCA/CDC-42/PAR/WAVE functional module required for retrograde endocytic recycling. Proc. Natl. Acad. Sci. USA. 112:E1443–E1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj Pahuja K., Wang J., Blagoveshchenskaya A., Lim L., Madhusudhan M.S., Mayinger P., and Schekman R.. 2015. Phosphoregulatory protein 14-3-3 facilitates SAC1 transport from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 112:E3199–E3206. 10.1073/pnas.1509119112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barg S., Knowles M.K., Chen X., Midorikawa M., and Almers W.. 2010. Syntaxin clusters assemble reversibly at sites of secretory granules in live cells. Proc. Natl. Acad. Sci. USA. 107:20804–20809. 10.1073/pnas.1014823107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoveshchenskaya A., Cheong F.Y., Rohde H.M., Glover G., Knödler A., Nicolson T., Boehmelt G., and Mayinger P.. 2008. Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. J. Cell Biol. 180:803–812. 10.1083/jcb.200708109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshans R.L., Szanto S., van Aelst L., and D’Souza-Schorey C.. 2000. ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol. Cell. Biol. 20:3685–3694. 10.1128/MCB.20.10.3685-3694.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown F.D., Rozelle A.L., Yin H.L., Balla T., and Donaldson J.G.. 2001. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 154:1007–1017. 10.1083/jcb.200103107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.E. 2007. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 8:1476–1485. 10.1111/j.1600-0854.2007.00634.x [DOI] [PubMed] [Google Scholar]
- Cavenagh M.M., Whitney J.A., Carroll K., Zhang C., Boman A.L., Rosenwald A.G., Mellman I., and Kahn R.A.. 1996. Intracellular distribution of Arf proteins in mammalian cells. Arf6 is uniquely localized to the plasma membrane. J. Biol. Chem. 271:21767–21774. 10.1074/jbc.271.36.21767 [DOI] [PubMed] [Google Scholar]
- Caviston J.P., Cohen L.A., and Donaldson J.G.. 2014. Arf1 and Arf6 promote ventral actin structures formed by acute activation of protein kinase C and Src. Cytoskeleton (Hoboken). 71:380–394. 10.1002/cm.21181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.C., Schweinsberg P.J., Vashist S., Mareiniss D.P., Lambie E.J., and Grant B.D.. 2006. RAB-10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Mol. Biol. Cell. 17:1286–1297. 10.1091/mbc.E05-08-0787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Li L., Li J., Liu B., Zhu X., Zheng L., Zhang R., and Xu T.. 2014. SEC-10 and RAB-10 coordinate basolateral recycling of clathrin-independent cargo through endosomal tubules in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 111:15432–15437. 10.1073/pnas.1408327111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang Y., Zhang J., Deng Y., Jiang L., Song E., Wu X.S., Hammer J.A., Xu T., and Lippincott-Schwartz J.. 2012. Rab10 and myosin-Va mediate insulin-stimulated GLUT4 storage vesicle translocation in adipocytes. J. Cell Biol. 198:545–560. 10.1083/jcb.201111091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong F.Y., Sharma V., Blagoveshchenskaya A., Oorschot V.M., Brankatschk B., Klumperman J., Freeze H.H., and Mayinger P.. 2010. Spatial regulation of Golgi phosphatidylinositol-4-phosphate is required for enzyme localization and glycosylation fidelity. Traffic. 11:1180–1190. 10.1111/j.1600-0854.2010.01092.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J. 2014. Arf GTPases and their effectors: assembling multivalent membrane-binding platforms. Curr. Opin. Struct. Biol. 29:67–76. 10.1016/j.sbi.2014.09.007 [DOI] [PubMed] [Google Scholar]
- Chung J., Torta F., Masai K., Lucast L., Czapla H., Tanner L.B., Narayanaswamy P., Wenk M.R., Nakatsu F., and De Camilli P.. 2015. INTRACELLULAR TRANSPORT. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science. 349:428–432. 10.1126/science.aab1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves A.E., Novick P.J., and Bankaitis V.A.. 1989. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J. Cell Biol. 109:2939–2950. 10.1083/jcb.109.6.2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E.A., and Hardin J.. 2004. Sticky worms: adhesion complexes in C. elegans. J. Cell Sci. 117:1885–1897. 10.1242/jcs.01176 [DOI] [PubMed] [Google Scholar]
- De Matteis M.A., and Godi A.. 2004. PI-loting membrane traffic. Nat. Cell Biol. 6:487–492. 10.1038/ncb0604-487 [DOI] [PubMed] [Google Scholar]
- Derrien V., Couillault C., Franco M., Martineau S., Montcourrier P., Houlgatte R., and Chavrier P.. 2002. A conserved C-terminal domain of EFA6-family ARF6-guanine nucleotide exchange factors induces lengthening of microvilli-like membrane protrusions. J. Cell Sci. 115:2867–2879. [DOI] [PubMed] [Google Scholar]
- Dickson E.J., Jensen J.B., and Hille B.. 2014. Golgi and plasma membrane pools of PI(4)P contribute to plasma membrane PI(4,5)P2 and maintenance of KCNQ2/3 ion channel current. Proc. Natl. Acad. Sci. USA. 111:E2281–E2290. 10.1073/pnas.1407133111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson E.J., Jensen J.B., Vivas O., Kruse M., Traynor-Kaplan A.E., and Hille B.. 2016. Dynamic formation of ER-PM junctions presents a lipid phosphatase to regulate phosphoinositides. J. Cell Biol. 213:33–48. 10.1083/jcb.201508106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J.G. 2003. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J. Biol. Chem. 278:41573–41576. 10.1074/jbc.R300026200 [DOI] [PubMed] [Google Scholar]
- Donaldson J.G., and Honda A.. 2005. Localization and function of Arf family GTPases. Biochem. Soc. Trans. 33:639–642. 10.1042/BST0330639 [DOI] [PubMed] [Google Scholar]
- Dong Y., Gou Y., Li Y., Liu Y., and Bai J.. 2015. Synaptojanin cooperates in vivo with endophilin through an unexpected mechanism. eLife. 4:4 10.7554/eLife.05660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy J.L., Moravec R., Ly K., Lasell T.K., Melancon P., and Casanova J.E.. 2006. The Arf6 GEF GEP100/BRAG2 regulates cell adhesion by controlling endocytosis of beta1 integrins. Curr. Biol. 16:315–320. 10.1016/j.cub.2005.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco M., Peters P.J., Boretto J., van Donselaar E., Neri A., D’Souza-Schorey C., and Chavrier P.. 1999. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 18:1480–1491. 10.1093/emboj/18.6.1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason R.J., Akintobi A.M., Grant B.D., and Padgett R.W.. 2014. BMP signaling requires retromer-dependent recycling of the type I receptor. Proc. Natl. Acad. Sci. USA. 111:2578–2583. 10.1073/pnas.1319947111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B.D., and Donaldson J.G.. 2009. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10:597–608. 10.1038/nrm2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B., Zhang Y., Paupard M.C., Lin S.X., Hall D.H., and Hirsh D.. 2001. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat. Cell Biol. 3:573–579. 10.1038/35078549 [DOI] [PubMed] [Google Scholar]
- Guo S., Stolz L.E., Lemrow S.M., and York J.D.. 1999. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 274:12990–12995. 10.1074/jbc.274.19.12990 [DOI] [PubMed] [Google Scholar]
- Hammond G.R., Machner M.P., and Balla T.. 2014. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol. 205:113–126. 10.1083/jcb.201312072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie R.C., Liu C.H., Randall A.S., and Sengupta S.. 2015. In vivo tracking of phosphoinositides in Drosophila photoreceptors. J. Cell Sci. 128:4328–4340. 10.1242/jcs.180364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi T., Someya A., Thompson W., Moss J., and Vaughan M.. 2006. GEP100/BRAG2: activator of ADP-ribosylation factor 6 for regulation of cell adhesion and actin cytoskeleton via E-cadherin and alpha-catenin. Proc. Natl. Acad. Sci. USA. 103:10672–10677. 10.1073/pnas.0604091103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongu T., and Kanaho Y.. 2014. Activation machinery of the small GTPase Arf6. Adv. Biol. Regul. 54:59–66. 10.1016/j.jbior.2013.09.014 [DOI] [PubMed] [Google Scholar]
- Hsu F., and Mao Y.. 2015. The structure of phosphoinositide phosphatases: Insights into substrate specificity and catalysis. Biochim. Biophys. Acta. 1851:698–710. 10.1016/j.bbalip.2014.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F., Hu F., and Mao Y.. 2015. Spatiotemporal control of phosphatidylinositol 4-phosphate by Sac2 regulates endocytic recycling. J. Cell Biol. 209:97–110. 10.1083/jcb.201408027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes W.E., Cooke F.T., and Parker P.J.. 2000a Sac phosphatase domain proteins. Biochem. J. 350:337–352. 10.1042/bj3500337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes W.E., Woscholski R., Cooke F.T., Patrick R.S., Dove S.K., McDonald N.Q., and Parker P.J.. 2000b SAC1 encodes a regulated lipid phosphoinositide phosphatase, defects in which can be suppressed by the homologous Inp52p and Inp53p phosphatases. J. Biol. Chem. 275:801–808. 10.1074/jbc.275.2.801 [DOI] [PubMed] [Google Scholar]
- Hunt-Newbury R., Viveiros R., Johnsen R., Mah A., Anastas D., Fang L., Halfnight E., Lee D., Lin J., Lorch A., et al. 2007. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 5:e237 10.1371/journal.pbio.0050237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung A.H., and Novick P.J.. 2011. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91:119–149. 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabet T., Montagnac G., Regazzoni K., Raynal B., El Khadali F., England P., Franco M., Chavrier P., Houdusse A., and Ménétrey J.. 2009. The structural basis of Arf effector specificity: the crystal structure of ARF6 in a complex with JIP4. EMBO J. 28:2835–2845. 10.1038/emboj.2009.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., and Ahringer J.. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 30:313–321. 10.1016/S1046-2023(03)00050-1 [DOI] [PubMed] [Google Scholar]
- Kearns B.G., McGee T.P., Mayinger P., Gedvilaite A., Phillips S.E., Kagiwada S., and Bankaitis V.A.. 1997. Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature. 387:101–105. 10.1038/387101a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochendörfer K.U., Then A.R., Kearns B.G., Bankaitis V.A., and Mayinger P.. 1999. Sac1p plays a crucial role in microsomal ATP transport, which is distinct from its function in Golgi phospholipid metabolism. EMBO J. 18:1506–1515. 10.1093/emboj/18.6.1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad G., Schlecker T., Faulhammer F., and Mayinger P.. 2002. Retention of the yeast Sac1p phosphatase in the endoplasmic reticulum causes distinct changes in cellular phosphoinositide levels and stimulates microsomal ATP transport. J. Biol. Chem. 277:10547–10554. 10.1074/jbc.M200090200 [DOI] [PubMed] [Google Scholar]
- Kumar S., Koutsovoulos G., Kaur G., and Blaxter M.. 2012. Toward 959 nematode genomes. Worm. 1:42–50. 10.4161/worm.19046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Yi P., and Ou G.. 2015. Somatic CRISPR-Cas9-induced mutations reveal roles of embryonically essential dynein chains in Caenorhabditis elegans cilia. J. Cell Biol. 208:683–692. 10.1083/jcb.201411041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang X., Zhang X., Zhao M., Tsang W.L., Zhang Y., Yau R.G., Weisman L.S., and Xu H.. 2013. Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc. Natl. Acad. Sci. USA. 110:21165–21170. 10.1073/pnas.1311864110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu O., and Grant B.D.. 2015. Basolateral Endocytic Recycling Requires RAB-10 and AMPH-1 Mediated Recruitment of RAB-5 GAP TBC-2 to Endosomes. PLoS Genet. 11:e1005514 10.1371/journal.pgen.1005514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang S., Hang W., Gao J., Zhang W., Cheng Z., Yang C., He J., Zhou J., Chen J., and Shi A.. 2018. LET-413/Erbin acts as a RAB-5 effector to promote RAB-10 activation during endocytic recycling. J. Cell Biol. 217:299–314. 10.1083/jcb.201705136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Boukhelifa M., Tribble E., Morin-Kensicki E., Uetrecht A., Bear J.E., and Bankaitis V.A.. 2008. The Sac1 phosphoinositide phosphatase regulates Golgi membrane morphology and mitotic spindle organization in mammals. Mol. Biol. Cell. 19:3080–3096. 10.1091/mbc.E07-12-1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Boukhelifa M., Tribble E., and Bankaitis V.A.. 2009. Functional studies of the mammalian Sac1 phosphoinositide phosphatase. Adv. Enzyme Regul. 49:75–86. 10.1016/j.advenzreg.2009.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuya S., Sakagami H., Tohgo A., Owada Y., Shin H.W., Takeshima H., Nakayama K., Kokubun S., and Kondo H.. 2005. Cellular and subcellular localization of EFA6C, a third member of the EFA6 family, in adult mouse Purkinje cells. J. Neurochem. 93:674–685. 10.1111/j.1471-4159.2005.03072.x [DOI] [PubMed] [Google Scholar]
- Mayinger P., Bankaitis V.A., and Meyer D.I.. 1995. Sac1p mediates the adenosine triphosphate transport into yeast endoplasmic reticulum that is required for protein translocation. J. Cell Biol. 131:1377–1386. 10.1083/jcb.131.6.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay S.J., Johnsen R., Khattra J., Asano J., Baillie D.L., Chan S., Dube N., Fang L., Goszczynski B., Ha E., et al. 2003. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 68:159–169. 10.1101/sqb.2003.68.159 [DOI] [PubMed] [Google Scholar]
- McMahon H.T., and Boucrot E.. 2011. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12:517–533. 10.1038/nrm3151 [DOI] [PubMed] [Google Scholar]
- Montagnac G., Sibarita J.B., Loubéry S., Daviet L., Romao M., Raposo G., and Chavrier P.. 2009. ARF6 Interacts with JIP4 to control a motor switch mechanism regulating endosome traffic in cytokinesis. Curr. Biol. 19:184–195. 10.1016/j.cub.2008.12.043 [DOI] [PubMed] [Google Scholar]
- Montagnac G., de Forges H., Smythe E., Gueudry C., Romao M., Salamero J., and Chavrier P.. 2011. Decoupling of activation and effector binding underlies ARF6 priming of fast endocytic recycling. Curr. Biol. 21:574–579. 10.1016/j.cub.2011.02.034 [DOI] [PubMed] [Google Scholar]
- Murray D.H., Jahnel M., Lauer J., Avellaneda M.J., Brouilly N., Cezanne A., Morales-Navarrete H., Perini E.D., Ferguson C., Lupas A.N., et al. 2016. An endosomal tether undergoes an entropic collapse to bring vesicles together. Nature. 537:107–111. 10.1038/nature19326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., and Miyawaki A.. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87–90. 10.1038/nbt0102-87 [DOI] [PubMed] [Google Scholar]
- Naslavsky N., Weigert R., and Donaldson J.G.. 2003. Convergence of non-clathrin- and clathrin-derived endosomes involves Arf6 inactivation and changes in phosphoinositides. Mol. Biol. Cell. 14:417–431. 10.1091/mbc.02-04-0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y., Kearns B.G., Wenk M.R., Chen H., Mori K., Alb J.G. Jr., De Camilli P., and Bankaitis V.A.. 2000. Functional characterization of a mammalian Sac1 and mutants exhibiting substrate-specific defects in phosphoinositide phosphatase activity. J. Biol. Chem. 275:34293–34305. 10.1074/jbc.M003923200 [DOI] [PubMed] [Google Scholar]
- Nielsen E., Christoforidis S., Uttenweiler-Joseph S., Miaczynska M., Dewitte F., Wilm M., Hoflack B., and Zerial M.. 2000. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 151:601–612. 10.1083/jcb.151.3.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris F.A., Auethavekiat V., and Majerus P.W.. 1995. The isolation and characterization of cDNA encoding human and rat brain inositol polyphosphate 4-phosphatase. J. Biol. Chem. 270:16128–16133. 10.1074/jbc.270.27.16128 [DOI] [PubMed] [Google Scholar]
- Novick P., Osmond B.C., and Botstein D.. 1989. Suppressors of yeast actin mutations. Genetics. 121:659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios F., Price L., Schweitzer J., Collard J.G., and D’Souza-Schorey C.. 2001. An essential role for ARF6-regulated membrane traffic in adherens junction turnover and epithelial cell migration. EMBO J. 20:4973–4986. 10.1093/emboj/20.17.4973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posor Y., Eichhorn-Gruenig M., Puchkov D., Schöneberg J., Ullrich A., Lampe A., Müller R., Zarbakhsh S., Gulluni F., Hirsch E., et al. 2013. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 499:233–237. 10.1038/nature12360 [DOI] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., and Austin J.. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 157:1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn K.V., Behe P., and Tinker A.. 2008. Monitoring changes in membrane phosphatidylinositol 4,5-bisphosphate in living cells using a domain from the transcription factor tubby. J. Physiol. 586:2855–2871. 10.1113/jphysiol.2008.153791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna H., Al-Awar O., Khachikian Z., and Donaldson J.G.. 1999. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J. Cell Sci. 112:855–866. [DOI] [PubMed] [Google Scholar]
- Renault L., Guibert B., and Cherfils J.. 2003. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature. 426:525–530. 10.1038/nature02197 [DOI] [PubMed] [Google Scholar]
- Rivas M.P., Kearns B.G., Xie Z., Guo S., Sekar M.C., Hosaka K., Kagiwada S., York J.D., and Bankaitis V.A.. 1999. Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to “bypass Sec14p” and inositol auxotrophy. Mol. Biol. Cell. 10:2235–2250. 10.1091/mbc.10.7.2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde H.M., Cheong F.Y., Konrad G., Paiha K., Mayinger P., and Boehmelt G.. 2003. The human phosphatidylinositol phosphatase SAC1 interacts with the coatomer I complex. J. Biol. Chem. 278:52689–52699. 10.1074/jbc.M307983200 [DOI] [PubMed] [Google Scholar]
- Sakagami H., Suzuki H., Kamata A., Owada Y., Fukunaga K., Mayanagi H., and Kondo H.. 2006. Distinct spatiotemporal expression of EFA6D, a guanine nucleotide exchange factor for ARF6, among the EFA6 family in mouse brain. Brain Res. 1093:1–11. 10.1016/j.brainres.2006.02.058 [DOI] [PubMed] [Google Scholar]
- Santy L.C., and Casanova J.E.. 2001. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J. Cell Biol. 154:599–610. 10.1083/jcb.200104019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Mushiake S., Kato Y., Sato K., Sato M., Takeda N., Ozono K., Miki K., Kubo Y., Tsuji A., et al. 2007. The Rab8 GTPase regulates apical protein localization in intestinal cells. Nature. 448:366–369. 10.1038/nature05929 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorr M., Then A., Tahirovic S., Hug N., and Mayinger P.. 2001. The phosphoinositide phosphatase Sac1p controls trafficking of the yeast Chs3p chitin synthase. Curr. Biol. 11:1421–1426. 10.1016/S0960-9822(01)00449-3 [DOI] [PubMed] [Google Scholar]
- Segbert C., Johnson K., Theres C., van Fürden D., and Bossinger O.. 2004. Molecular and functional analysis of apical junction formation in the gut epithelium of Caenorhabditis elegans. Dev. Biol. 266:17–26. 10.1016/j.ydbio.2003.10.019 [DOI] [PubMed] [Google Scholar]
- Shen Z., Zhang X., Chai Y., Zhu Z., Yi P., Feng G., Li W., and Ou G.. 2014. Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev. Cell. 30:625–636. 10.1016/j.devcel.2014.07.017 [DOI] [PubMed] [Google Scholar]
- Shi A., Chen C.C., Banerjee R., Glodowski D., Audhya A., Rongo C., and Grant B.D.. 2010. EHBP-1 functions with RAB-10 during endocytic recycling in Caenorhabditis elegans. Mol. Biol. Cell. 21:2930–2943. 10.1091/mbc.E10-02-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A., Liu O., Koenig S., Banerjee R., Chen C.C., Eimer S., and Grant B.D.. 2012. RAB-10-GTPase-mediated regulation of endosomal phosphatidylinositol-4,5-bisphosphate. Proc. Natl. Acad. Sci. USA. 109:E2306–E2315. 10.1073/pnas.1205278109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S., Kawanai T., Hara T., Yamamoto A., Chaya T., Tokuhara Y., Tsuji C., Sakai M., Tachibana T., and Inagaki S.. 2016. ARHGEF10 directs the localization of Rab8 to Rab6-positive executive vesicles. J. Cell Sci. 129:3620–3634. 10.1242/jcs.186817 [DOI] [PubMed] [Google Scholar]
- Shin H.W., Hayashi M., Christoforidis S., Lacas-Gervais S., Hoepfner S., Wenk M.R., Modregger J., Uttenweiler-Joseph S., Wilm M., Nystuen A., et al. 2005. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 170:607–618. 10.1083/jcb.200505128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya A., Sata M., Takeda K., Pacheco-Rodriguez G., Ferrans V.J., Moss J., and Vaughan M.. 2001. ARF-GEP(100), a guanine nucleotide-exchange protein for ADP-ribosylation factor 6. Proc. Natl. Acad. Sci. USA. 98:2413–2418. 10.1073/pnas.051634798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Khachikian Z., Radhakrishna H., and Donaldson J.G.. 1998. Localization of endogenous ARF6 to sites of cortical actin rearrangement and involvement of ARF6 in cell spreading. J. Cell Sci. 111:2257–2267. [DOI] [PubMed] [Google Scholar]
- Spencer W.C., Zeller G., Watson J.D., Henz S.R., Watkins K.L., McWhirter R.D., Petersen S., Sreedharan V.T., Widmer C., Jo J., et al. 2011. A spatial and temporal map of C. elegans gene expression. Genome Res. 21:325–341. 10.1101/gr.114595.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepicheva N.A., Dumas M., Kobi P., Donaldson J.G., and Song J.L.. 2017. The small GTPase Arf6 regulates sea urchin morphogenesis. Differentiation. 95:31–43. 10.1016/j.diff.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., and Fire A.. 1998. Specific interference by ingested dsRNA. Nature. 395:854 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Vanhauwaert R., Kuenen S., Masius R., Bademosi A., Manetsberger J., Schoovaerts N., Bounti L., Gontcharenko S., Swerts J., Vilain S., et al. 2017. The SAC1 domain in synaptojanin is required for autophagosome maturation at presynaptic terminals. EMBO J. 36:1392–1411. 10.15252/embj.201695773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Chen J., Enns C.A., and Mayinger P.. 2013. The first transmembrane domain of lipid phosphatase SAC1 promotes Golgi localization. PLoS One. 8:e71112 10.1371/journal.pone.0071112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Liu H., Wang Y., Liu O., Zhang J., Gleason A., Yang Z., Wang H., Shi A., and Grant B.D.. 2016. RAB-10 Promotes EHBP-1 Bridging of Filamentous Actin and Tubular Recycling Endosomes. PLoS Genet. 12:e1006093 10.1371/journal.pgen.1006093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H.C., Sanny J., Shu H., Baillie D.L., Brill J.A., Price J.V., and Harden N.. 2003. The Sac1 lipid phosphatase regulates cell shape change and the JNK cascade during dorsal closure in Drosophila. Curr. Biol. 13:1882–1887. 10.1016/j.cub.2003.09.056 [DOI] [PubMed] [Google Scholar]
- Whitters E.A., Cleves A.E., McGee T.P., Skinner H.B., and Bankaitis V.A.. 1993. SAC1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J. Cell Biol. 122:79–94. 10.1083/jcb.122.1.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Zhou X., Zhu J., Liu R., Zhang S., Coquinco A., Chen Y., Wen Y., Kojic L., Jia W., and Cynader M.S.. 2013. JNK3 couples the neuronal stress response to inhibition of secretory trafficking. Sci. Signal. 6:ra57 10.1126/scisignal.2003727 [DOI] [PubMed] [Google Scholar]
- Yin H.L., and Janmey P.A.. 2003. Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 65:761–789. 10.1146/annurev.physiol.65.092101.142517 [DOI] [PubMed] [Google Scholar]
- Zhong R., Burk D.H., Nairn C.J., Wood-Jones A., Morrison W.H. III, and Ye Z.H.. 2005. Mutation of SAC1, an Arabidopsis SAC domain phosphoinositide phosphatase, causes alterations in cell morphogenesis, cell wall synthesis, and actin organization. Plant Cell. 17:1449–1466. 10.1105/tpc.105.031377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Zeng J., Ouyang C., Luo Q., Yu M., Yang Z., Wang H., Shen K., and Shi A.. 2016. A novel bipartite UNC-101/AP-1 μ1 binding signal mediates KVS-4/Kv2.1 somatodendritic distribution in Caenorhabditis elegans. FEBS Lett. 590:76–92. 10.1002/1873-3468.12043 [DOI] [PubMed] [Google Scholar]
- Zhu W., Shi D.S., Winter J.M., Rich B.E., Tong Z., Sorensen L.K., Zhao H., Huang Y., Tai Z., Mleynek T.M., et al. 2017. Small GTPase ARF6 controls VEGFR2 trafficking and signaling in diabetic retinopathy. J. Clin. Invest. 127:4569–4582. 10.1172/JCI91770 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.