Cao et al. show that UGT8 promotes BLBC progression through activating sulfatide–αVβ5 axis. ZA is identified as a direct inhibitor of UGT8 and suppresses BLBC progression, suggesting that inhibition of UGT8 offers a promising opportunity for treating BLBC.
Abstract
Basal-like breast cancer (BLBC) is associated with a poor clinical outcome as a result of the few treatment options and poor therapeutic response. Here, we report that elevated expression of urine diphosphate–galactose ceramide galactosyltransferase (UGT8) specifically occurs in BLBC and predicts poor prognosis in breast cancer patients. UGT8 expression is transcriptionally up-regulated by Sox10, triggering the sulfatide biosynthetic pathway; increased sulfatide activates integrin αVβ5-mediated signaling that contributes to BLBC progression. UGT8 expression promotes, whereas UGT8 knockdown suppresses tumorigenicity and metastasis. Importantly, we identify that zoledronic acid (ZA), a marketed drug for treating osteoporosis and bone metastasis, is a direct inhibitor of UGT8, which blocks the sulfatide biosynthetic pathway. Significantly, a clinically achievable dosage of ZA exhibits apparent inhibitory effect on migration, invasion, and lung metastasis of BLBC cells. Together, our study suggests that UGT8 is a potential prognostic indicator and druggable target of BLBC and that pharmacologic inhibition of UGT8 by ZA offers a promising opportunity for treating this challenging disease.
Introduction
Breast cancer is a heterogeneous disease comprised of four major molecular subtypes, including luminal A, luminal B, HER2/ERBB2, and basal-like (Vargo-Gogola and Rosen, 2007). Basal-like breast cancer (BLBC) is an especially aggressive subtype that typically afflicts younger and premenopausal women and possesses the worst prognosis of any breast cancer subtypes (Kreike et al., 2007; Rakha et al., 2008). BLBC tends to be negative for the expression of estrogen receptor (ER), progesterone receptor (PR), and epidermal growth factor receptor 2 (HER2; i.e., triple-negative), a situation that lacks effective targeted therapies, such as endocrine and anti-HER2 therapies (Fadare and Tavassoli, 2008; Korsching et al., 2008; Rakha et al., 2008). <30% patients with metastatic BLBC have a 5-yr survival rate despite adjuvant chemotherapy that remains the mainstay of BLBC treatment (Dent et al., 2007). The highly aggressive nature and the absence of effective therapeutics make it a high priority to elucidate its determinants of aggressiveness and identify its potential therapeutic targets.
Metabolic alterations contribute to rewire metabolic and oncogenic signaling pathways to meet the demands of cancer cell survival and metastasis (Hanahan and Weinberg, 2011). Sulfatide is a sphingolipid commonly found on the surface of most of eukaryotic cells (Xiao et al., 2013). Besides its structural role for the cell membrane, sulfatide is also involved in cell adhesion and aggregation, protein trafficking, axon–myelin interactions, learning and memory, modulation of sodium and potassium channels, and neural plasticity (Xiao et al., 2013). Abnormal metabolism of sulfatide is correlated with development of many diseases, including metachromatic leukodystrophy, diabetes, autoimmune diseases, and cancers (Takahashi and Suzuki, 2012). Sulfatide biosynthesis is associated with a simple two-step pathway and two enzymes. UGT8 is the first key enzyme that catalyzes the transfer of galactose to ceramide for the synthesis of galactosylceramide (GalCer; Bosio et al., 1996a), which GalCer sulfotransferase (GAL3ST1), the second enzyme in this pathway, converts into sulfatide by sulfonation reaction (Honke et al., 2001). There is growing evidence for the involvement of sulfatide in the regulation of proliferation, differentiation, apoptosis, and senescence of cancer cells (Takahashi and Suzuki, 2012; Xiao et al., 2013). High expression of sulfatide is observed in several human cancer tissues, including the colon, kidney, lung, liver, and ovary (Takahashi and Suzuki, 2012; Xiao et al., 2013). Significantly elevated expression of sulfatide in ovarian carcinomas and colorectal carcinomas predicts poor prognosis (Morichika et al., 1996; Makhlouf et al., 2004), suggesting a critical role for sulfatide in cancer progression. Thus, a better understanding of the biological functions and mechanisms of the sulfatide biosynthetic pathway will improve our ability to define its contribution to tumor progression and treatment options.
In this study, we report that UGT8 expression is dramatically up-regulated in BLBC and predicts poor prognosis in breast cancer patients. UGT8 expression provides tumorigenic and metastatic advantages in BLBC through activating sulfatide–αVβ5 axis. Our study provides an understanding of how UGT8 contributes to BLBC aggressiveness, suggesting a potential prognostic indicator and druggable target of BLBC.
Results
UGT8 expression is up-regulated in BLBC subtype
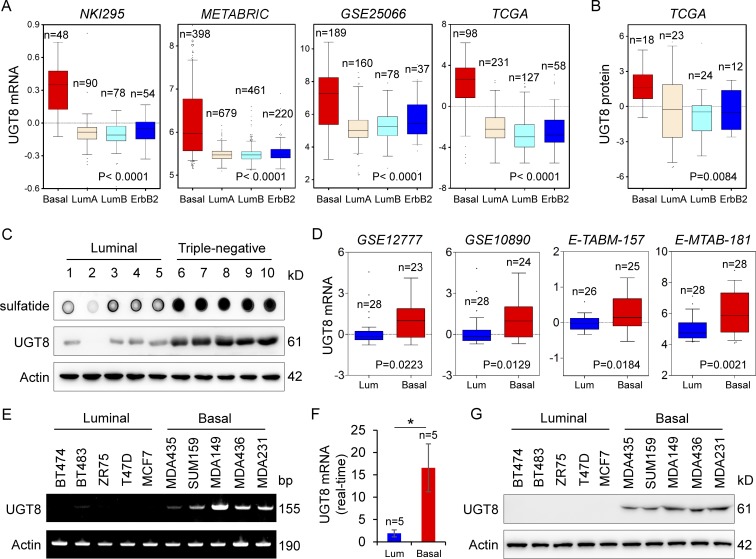
We recently reported two metabolic enzymes, aldo-keto reductase 1 member B1 (AKR1B1) and fructose-1, 6-biphosphatase (FBP1), that were tightly associated with BLBC aggressiveness (Dong et al., 2013; Wu et al., 2017). To further acquire other clinically relevant metabolic determinants required for BLBC, we systematically analyzed multiple publicly available gene expression datasets (NKI295, METABRIC, GSE25066, The Cancer Genome Atlas [TCGA], GSE1456, GSE7390, GSE2034, and GSE22358), which contain >5,000 breast cancer patients (van de Vijver et al., 2002; Pawitan et al., 2005; Wang et al., 2005; Desmedt et al., 2007; Hatzis et al., 2011; Curtis et al., 2012; Glück et al., 2012). In addition to some previously identified genes, such as lactate dehydrogenase B (LDHB), AKR1B1, and FBP1, we noticed that UGT8 mRNA expression that involved in controlling the sulfatide biosynthetic pathway was remarkably up-regulated in BLBC (Fig. 1 A and Fig. S1 A). In line with this observation, UGT8 protein expression also was significantly elevated in BLBC by proteogenomic analysis of TCGA dataset that contains 105 breast tumor samples (Fig. 1 B; Mertins et al., 2016). To confirm this observation, we examined the UGT8 level in fresh frozen breast tumor tissues. Consistently, the expressions of UGT8 and its downstream metabolite sulfatide were up-regulated in triple-negative breast cancer (TNBC) that is mostly also BLBC and were dramatically down-regulated in luminal subtype of breast cancers (Fig. 1 C). To further explore the association of UGT8 with basal subtype, we also analyzed UGT8 expression in four gene expression datasets (GSE12777, GSE10890, E-TABM-157, and E-MTAB-181), which contain 51, 52, 51, and 56 breast cancer cell lines, respectively (Neve et al., 2006; Hoeflich et al., 2009; Heiser et al., 2012). Consistently, UGT8 expression was significantly high in BLBC cell lines (Fig. 1 D). We confirmed this observation by either semiquantitative RT-PCR or quantitative RT-PCR in a panel of breast cancer cell lines that contained five luminal and five basal subtype cell lines, showing that UGT8 mRNA expression was much higher in BLBC cells than in luminal cells (Fig. 1, E and F). We further tested UGT8 protein expression in these cell lines. Strikingly, elevated UGT8 protein level was observed in BLBC cell lines (Fig. 1 G). Consistently, UGT8 activity was much higher in BLBC cells than in luminal cells (Fig. S1 B), supporting that UGT8 activity positively correlates with its expression. Together, our data suggest that UGT8 overexpression is primarily restricted to BLBC.
Figure 1.
Elevated UGT8 expression highly correlates with BLBC. (A) Box plots indicated UGT8 mRNA expression in different subtypes of breast cancer from four gene expression datasets (NKI295, METABRIC, GSE25066, and TCGA). Comparisons were analyzed by one-way ANOVA. (B) Box plot indicated UGT8 protein expression in different subtypes of breast cancer from TCGA dataset. Comparisons were analyzed by one-way ANOVA. (C) Expression of UGT8 and sulfatide was examined in tumor samples from five cases of luminal and five cases of TNBC. (D) Box plots indicated UGT8 mRNA expression in luminal and BLBC cell lines from four gene expression datasets (GSE12777, GSE10890, E-TABM-157, and E-MTAB-181). Comparisons were made using the two-tailed Student’s t test. (E and F) Expression of UGT8 mRNA was analyzed by either semi-quantitative RT-PCR (E) or quantitative RT-PCR (F) in a representative panel of breast cancer cell lines. Data are shown as mean ± SD based on three independent experiments. *, P < 0.05 by Student’s t test. (G) Expression of UGT8 in cells from E was examined by Western blotting.
UGT8 positively correlates with Sox10 and is a direct target of Sox10
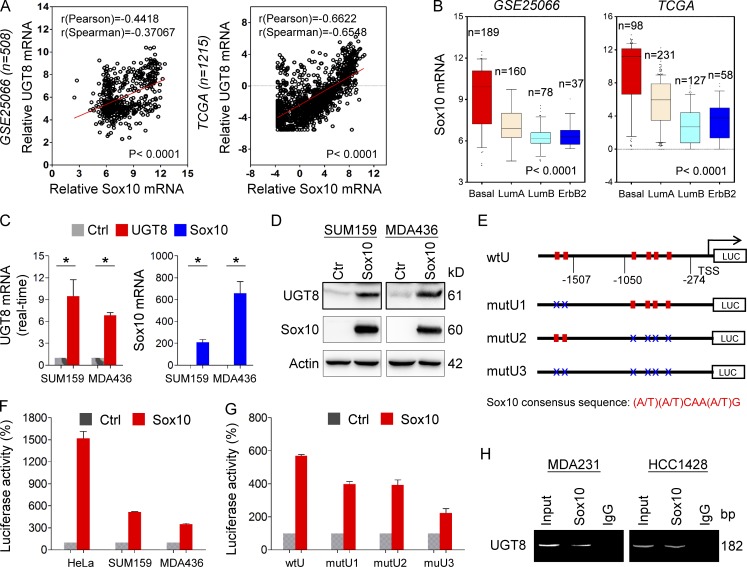
Given the intimate link between UGT8 and BLBC, we next determined how UGT8 was up-regulated in BLBC. Coexpression analysis of UGT8 with other genes in two large gene expression datasets (GSE25066 and TCGA) showed that UGT8 expression positively correlated with Sox10 expression (Fig. 2 A). We also analyzed Sox10 expression in different subtypes of breast cancer, showing that similar to UGT8, Sox10 was dramatically up-regulated in BLBC in multiple gene expression datasets (Fig. 2 B and Fig. S2 A). To investigate the causal relationship between UGT8 and Sox10, we expressed Sox10 in SUM159 and MDA-MB436 cells. Strikingly, Sox10 up-regulated UGT8 expression in mRNA and protein levels in all these cell lines (Fig. 2, C and D). Next, SUM149 and HCC1428 cells with endogenous Sox10 expression were transfected with empty vector or shSox10 vector, showing that knockdown of Sox10 expression down-regulated UGT8 expression (Fig. S2 B). These results indicate that Sox10, as a transcriptional activator, may induce UGT8 expression through transcriptional regulation.
Figure 2.
UGT8 positively correlates with Sox10 and is a direct transcriptional target of Sox10. (A) Analysis of GSE25066 and TCGA datasets for the expression of UGT8 and Sox10. The relative level of UGT8 was plotted against that of Sox10. Correlations were analyzed using Pearson’s correlation method and Spearman’s rank correlation test. (B) Box plots indicated Sox10 mRNA expression in different subtypes of breast cancer from GSE25066 and TCGA datasets. Comparisons were analyzed by one-way ANOVA. (C) Expression of UGT8 and Sox10 was analyzed by quantitative RT-PCR in SUM159 and MDA-MB436 cells infected with empty vector or Sox10-expressing vector. Data are shown as mean ± SD based on three independent experiments. *, P < 0.01 by Student's t test. (D) Expression of UGT8 and Sox10 was examined by Western blotting in SUM159 and MDA-MB436 cells infected with empty vector or Sox10-expressing vector. (E) Schematic diagram showing positions of potential Sox10-binding motifs in UGT8 promoter. UGT8 promoter luciferase construct and mutated derivatives were also shown. Sox10 consensus sequence: (A/T)(A/T)CAA(A/T)G. (F) UGT8 promoter luciferase construct (wtU) was coexpressed with empty vector or Sox10-expressing vector in HeLa, SUM159, and MDA-MB436 cells, respectively. After 48 h, luciferase activities were analyzed (mean ± SD in three separate experiments). (G) UGT8 promoter luciferase construct (wtU) as well as its mutants (mutU1, mutU2, and mutU3) were coexpressed with empty vector or Sox10-expressing vector in HEK-293T cells. Luciferase activities were analyzed as in F. Data are shown as mean ± SD based on three independent experiments. (H) ChIP analysis for binding of Sox10 to the UGT8 promoter in MDA-MB231 and HCC1428 cells.
Having identified their tight association and immediate induction of UGT8 expression by Sox10, we next determined whether UGT8 expression was regulated directly by Sox10. We noticed that UGT8 promoter contained 10 putative consensus Sox10-binding motifs (A/T)(A/T)CAA(A/T)G from −2,211 bp to transcription start site (TSS; Fig. S2 C). To investigate which motifs are critical for Sox10-mediated gene transcription, we cloned the human UGT8 promoter and created several deletion mutants of promoter-luciferase constructs based on the location of these motifs, including wtU, wtU1 and wtU2 (Fig. S2 C). By expressing the wtU in HeLa, SUM159, and MDA-MB436 cells, an approximately 3- to 15-fold increase in UGT8 promoter luciferase activity was observed in cells undergoing Sox10 overexpression (Fig. 2 F). The wtU1 without the region between −2,211 and −1,507 bp partially lost the reporter activity (wtU1 vs. wtU), whereas wtU2 without the region between −2,211 and −1,050 bp did not further reduce the reporter activity to respond to Sox10 expression (wtU2 vs. wtU1), indicating that the regions between −2,211 and −1,507 bp and between −1,050 bp and −274 bp are important for Sox10-mediated UGT8 activation (Fig. S2 C). To further evaluate the binding motifs inside the UGT8 promoter, several constructs with point mutants were generated in the UGT8-binding motifs (mutU1, mutU2, and mutU3; Fig. 2 E). Either the mutU1 or the mutU2 significantly reduced, whereas the mutU3 containing all mutations of both mutU1and mutU2 almost completely lost, the reporter activity induced by Sox10 (Fig. 2 G), suggesting that Sox10 activates the UGT8 promoter in a Sox10 motif–dependent fashion and that the motifs in the regions between −2,211 and −1,507 bp and between −1,050 bp and −274 bp are required for Sox10-mediated transcriptional activation. To further investigate whether Sox10 directly bound to the UTG8 promoter, we performed chromatin immunoprecipitation (ChIP) assays in MDA-MB231 and HCC1428 cells with endogenous Sox10 expression. A dramatic enrichment of Sox10 in the UGT8 promoter was observed in these cells (Fig. 2 H). These data suggest that UGT8 is a direct target of Sox10.
UTG8 expression activates the sulfatide biosynthetic pathway and enhances breast cancer cell migration and invasion
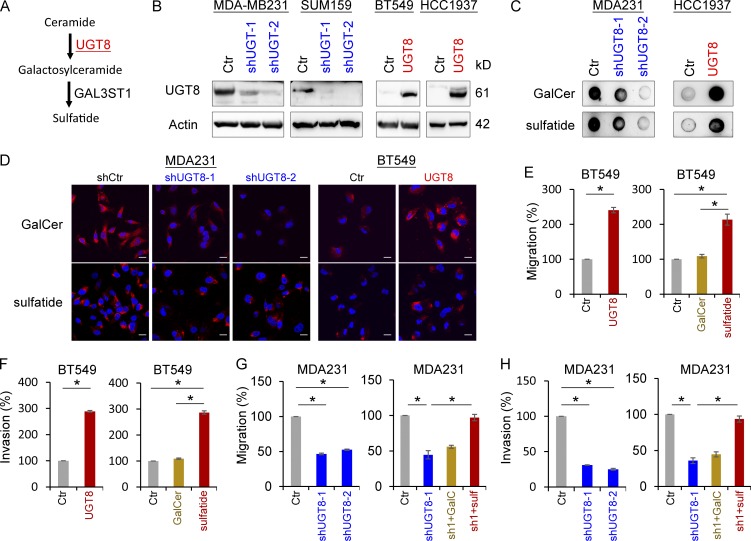
To investigate the molecular function and mechanism of UGT8, we generated stable transfectants with empty vector or knockdown of UTG8 expression in MDA-MB231 and SUM159 cells and also created stable clones with empty vector or UGT8 expression in BT549 and HCC1937 cells (Fig. 3 B). We first examined the production of GalCer and sulfatide, two downstream metabolites of UGT8 in the sulfatide biosynthetic pathway (Fig. 3 A). Immunoblotting data showed that knockdown of UGT8 expression caused a remarkable decrease, whereas exogenous UGT8 expression resulted in a dramatic increase in both GalCer and sulfatide levels (Fig. 3 C). Similar results were obtained in these cells by immunostaining-confocal analysis (Fig. 3 D and Fig. S3 A). These data suggest that UGT8 is required for increased GalCer and sulfatide production in breast cancer cells. To further understand the metabolic consequence of UGT8 expression, we examined the effect of UGT8 expression and both metabolites on breast cancer cell migration and invasion. Either UGT8 expression or sulfatide, but not GalCer, markedly induced the migration and invasion of BT549 cells in vitro (Fig. 3, E and F). Consistently, knockdown of UGT8 expression dramatically repressed the migration and invasion of MDA-MB231 and SUM159 cells in vitro, whereas sulfatide, but not GalCer, significantly restored the decreased migration and invasion of MDA-MB231 and SUM159 cells with stable knockdown of UGT8 expression (Fig. 3, G and H; and Fig. S3, B and C). These data indicate an important role of sulfatide in UGT8-mediated acquisition of migratory and invasive ability in breast cancer cells.
Figure 3.
UGT8 activates the sulfatide biosynthetic pathway and enhances breast cancer cell migration and invasion. (A) Sulfatide biosynthetic pathway. (B) Stable transfectants with empty vector or knockdown of UGT8 expression were established in MDA-MB231 and SUM159 cells, and stable clones with empty vector or UGT8 expression were also generated in BT549 and HCC1937 cells. (C) Expression of GalCer and sulfatide was examined by immunoblotting in MDA-MB231 cells with stable empty vector or knockdown of UGT8 expression, as well as HCC1937 cells with stable empty vector or UGT8 expression. (D) Expression of GalCer and sulfatide was measured by immunofluorescent staining in MDA-MB231 cells with stable empty vector or knockdown of UGT8 expression as well as BT549 cells with stable empty vector or UGT8 expression. Nuclei were visualized with DAPI (blue). Bars, 20 µm. (E and F) Migratory ability (E) and invasiveness (F) of BT549 cells with stable empty vector or UGT8 expression as well as BT549 cells treated with or without GalCer (2 µM) or sulfatide (2 µM) were analyzed. The percentage of migratory and invasive cells was shown in the bar graph (mean ± SD in three separate experiments). (G and H) Migratory ability (G) and invasiveness (H) of MDA-MB231 cells with stable empty vector or knockdown of UGT8 expression, as well as shUGT8-expressing MDA-MB231 cells treated with or without GalCer (2 µM) or sulfatide (2 µM) were analyzed. The percentage of migratory and invasive cells was analyzed as in E and F. *, P < 0.01 by Student’s t test. Data are shown as mean ± SD based on three independent experiments.
Zoledronic acid (ZA) is a direct inhibitor of UGT8 and suppresses the sulfatide biosynthetic pathway and breast cancer cell migration and invasion
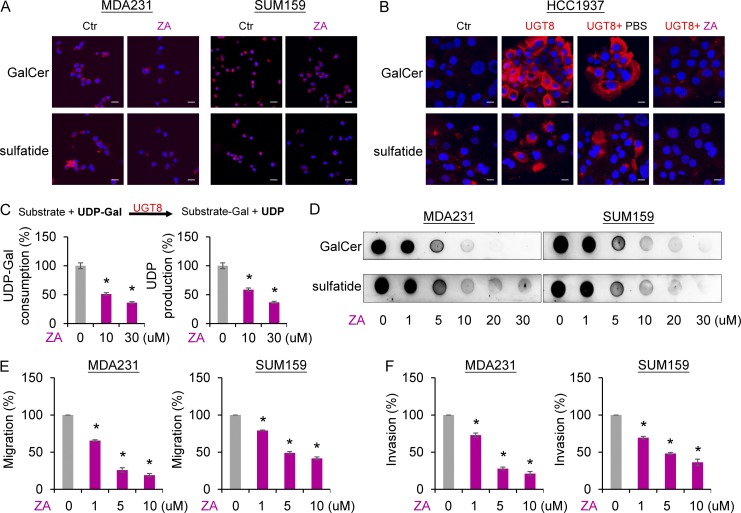
It has been speculated that ZA might be a potential inhibitor of UGT8 through computational modeling (Pannuzzo et al., 2016); however, until now this possibility has not been verified by further experiments. To further determine the relationship between ZA and UGT8, we first examined the effect of ZA on two downstream metabolites of UGT8 in the sulfatide biosynthetic pathway. Immunostaining-confocal analysis showed that ZA dramatically decreased the expression GalCer and sulfatide in endogenously UGT8-expressing MDA-MB231 and SUM159 cells and ectopically UGT8-expressing HCC1937 cells (Fig. 4, A and B). Next, we examined whether ZA could directly inhibit the enzymatic activity of UGT8. An in vitro galactosidation assay showed that UGT8 efficiently induced urine diphosphate (UDP)-galactose consumption and UDP production, whereas ZA strongly blocked this process, indicating that ZA functions as a direct inhibitor of UGT8 (Fig. 4 C). To further confirm the inhibitory effect of ZA on UGT8, we tested the effect of different concentrations of ZA on GalCer and sulfatide production in breast cancer cells by immunoblotting. We found that ZA efficiently blocked the expression of GalCer and sulfatide in a concentration dependent manner in MDA-MB231 and SUM159 cells (Fig. 4 D). These data demonstrate a potent inhibitory efficacy of ZA against GalCer and sulfatide production in breast cancer cells. We then evaluated the effect of ZA on breast cancer cell migration and invasion. As anticipated, ZA remarkably inhibited the migration and invasion of MDA-MB231 and SUM159 cells in a concentration-dependent manner (Fig. 4, E and F). These data indicate that ZA inhibits migratory and invasive ability of breast cancer cells by a similar mechanism with UGT8 knockdown.
Figure 4.
ZA is a direct inhibitor of UGT8 and inhibits breast cancer cell migration and invasion. (A and B) Expression of GalCer and sulfatide was measured by immunofluorescent staining in MDA-MB231 and SUM159 cells treated with or without ZA (20 µM; A), HCC1937 cells with empty vector, or stable UGT8 expression as well as UGT8-expressing HCC1937 cells treated with or without ZA (20 µM; B). Nuclei were visualized with DAPI (blue). Bars, 20 µm. (C) In vitro activity assay of UGT8 was performed by mixing UDP-galactose, substrate, and lysate of MDA-MB231 cells. After treatment of the indicated concentration of ZA, UDP-galactose consumption and UDP production were tested by HPLC system. The percentage of UDP-galactose consumption and UDP production was shown in the bar graph (mean ± SD in three separate experiments). (D) Expression of GalCer and sulfatide was examined by immunoblotting in MDA-MB231 and SUM159 cells treated with the indicated concentration of ZA. (E and F) Migratory ability (E) and invasiveness (F) of MDA-MB231 and SUM159 cells treated with the indicated concentration of ZA. The percentage of migratory and invasive cells was shown in the bar graph (mean ± SD in three separate experiments). *, P < 0.05 by Student’s t test.
UGT8 activates αVβ5 signaling via up-regulating sulfatide
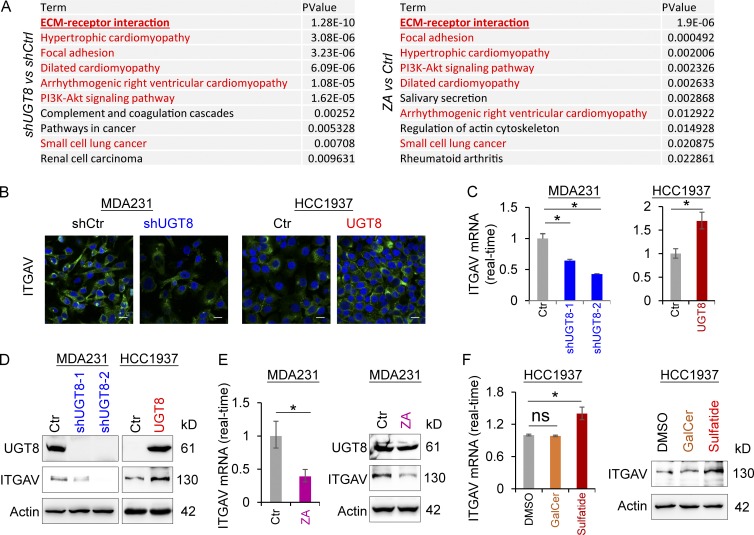
To understand the molecular mechanisms underlying UGT8-mediated metabolic changes in BLBC, we performed expression profiling analysis of MDA-MB231 cells undergoing knockdown of UGT8 expression or ZA treatment. We first compared genes that were transcriptionally down-regulated in MDA-MB231 cells undergoing knockdown of UGT8 expression (GSE112900) to that in MDA-MB231 cells treated with ZA studied previously (GSE33552; Vintonenko et al., 2012). KEGG pathway analysis of down-regulated genes showed that 7 of the top 10 most significant KEGG pathway terms were completely consistent between UGT8 knockdown and ZA treatment (Fig. 5 A), indicating that UGT8 knockdown and ZA may share similar mechanisms to mediate metabolic and functional alterations.
Figure 5.
UGT8 knockdown and ZA treatment have similar gene expression profiles and UGT8-mediated sulfatide induces ITGAV expression. (A) KEGG pathway analysis of down-regulated genes after UGT8 knockdown (fold change less than −2; left panel) and ZA treatment (30 µM; fold change less than −1.5; right panel) in MDA-MB231 cells. The top 10 most significant KEGG pathway terms were listed. (B) Expression of ITGAV was measured by immunofluorescent staining in MDA-MB231 cells with stable empty vector or knockdown of UGT8 expression as well as HCC1937 cells with stable empty vector or UGT8 expression. Nuclei were visualized with DAPI (blue). Bars, 20 µm. (C) Expression of ITGAV mRNA was analyzed by quantitative RT-PCR in MDA-MB231 cells with stable empty vector or knockdown of UGT8 expression as well as HCC1937 cells with stable empty vector or UGT8 expression. (D) Expression of ITGAV was analyzed by Western blotting in MDA-MB231 cells with stable empty vector or knockdown of UGT8 expression as well as HCC1937 cells with stable empty vector or UGT8 expression. (E) Expression of ITGAV was analyzed by quantitative RT-PCR (left) or Western blotting (right) in MDA-MB231 cells treated with or without ZA (10 µM). (F) Expression of ITGAV was analyzed by quantitative RT-PCR (left) or by Western blotting (right) in HCC1937 cells treated with or without GalCer (2 µM) or sulfatide (2 µM). Data are shown as mean ± SD based on three independent experiments. *, P < 0.01 by Student’s t test.
After KEGG pathway analysis, we noticed that the most significant pathway was related to the ECM-receptor interaction (Fig. 5 A). Among the genes of this pathway, ITGAV was noted to encode integrin α chain V, which is involved in multiple signaling pathways by combining different integrin β chains (Desgrosellier and Cheresh, 2010). Up-regulation of ITGAV is correlated with an aggressive phenotype in a variety of cancers (Desgrosellier and Cheresh, 2010). It has been reported that sulfatide up-regulates ITGAV expression (Wu et al., 2013). Consistently, UGT8 knockdown or ZA treatment caused a remarkable decrease, whereas UGT8 expression led to an obvious increase of ITGAV expression in mRNA and protein levels (Fig. 5, B–E). Intriguingly, sulfatide but not GalCer significantly elevated the expression of ITGAV mRNA and protein (Fig. 5 F), further supporting that UGT8 acts mainly via sulfatide-mediated cellular program.
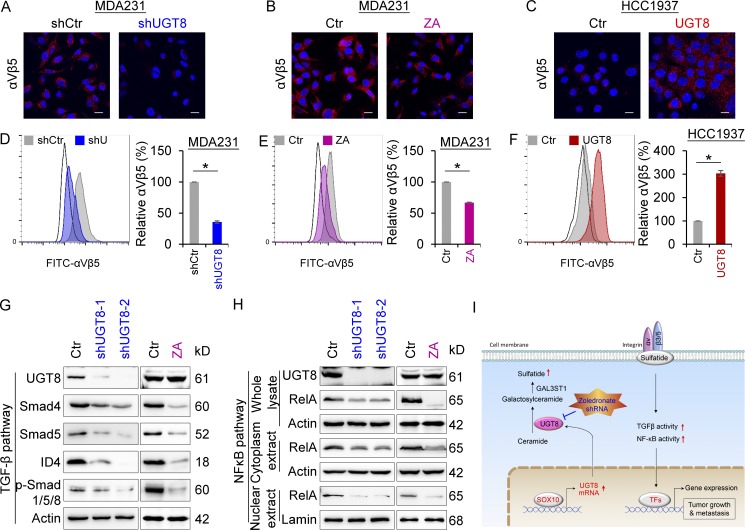
Integrins αVβ3 and αVβ5 are important malignant drivers that are the most thoroughly studied in the αV subfamily (Desgrosellier and Cheresh, 2010). To understand the association of UGT8 with αVβ3 and αVβ5, we determined the effect of UGT8 on αVβ3 and αVβ5 clustering. Our results showed that knockdown of UGT8 expression or ZA treatment resulted in a significant decrease (Fig. 6, A and B), whereas UGT8 expression led to a dramatic increase in αVβ5 clustering by immunostaining-confocal analysis (Fig. 6 C). Similar results were obtained by flow cytometry analysis (Fig. 6, D–F). Unexpectedly, knockdown of UGT8 expression or UGT8 expression only caused a slight change of αVβ3 clustering (Fig. S3, D and E), indicating that UGT8 functions mainly via αVβ5-mediated signaling. The αVβ3 and αVβ5 enhances cell survival through multiple mechanisms, including p53 inactivation, increased BCL2 expression, and activation of TGF-β signaling, NFκB, or PI3K pathway (Desgrosellier and Cheresh, 2010). Given the tight association of TGF-β signaling and NFκB pathways with BLBC aggressiveness (Desgrosellier and Cheresh, 2010; Dong et al., 2012; Wu et al., 2017), we chose them as examples to characterize the regulatory mechanism of UGT8 in BLBC. As expected, knockdown of UGT8 expression or ZA treatment caused a dramatic decrease of Smad4, Smad5, p-Smad1/5/8, and ID4 levels (Fig. 6 G), and also led to a remarkable reduction of RelA levels in both nucleus and cytoplasm (Fig. 6 H), supporting the involvement of UGT8 in TGF-β signaling and NFκB pathway.
Figure 6.
UGT8 activates αVβ5 signaling. (A–C) Expression of αVβ5 was measured by immunofluorescent staining in MDA-MB231 cells with stable empty vector or knockdown of UGT8 expression (A), MDA-MB231 cells treated with or without ZA (20 µM; B) as well as HCC1937 cells with stable empty vector or UGT8 expression (C). Nuclei were visualized with DAPI (blue). Bars, 20 µm. (D–F) The level of αVβ5 was analyzed by flow cytometry in MDA-MB231 cells with stable empty vector or knockdown of UGT8 expression (D), MDA-MB231 cells treated with or without ZA (10 µM; E) as well as HCC1937 cells with stable empty vector or UGT8 expression (F). Representative images were shown (left). Isotype controls are used to determine the staining specificity (unfilled). The level of αVβ5 in cells with knockdown of UGT8 expression or UGT8 expression as well as cells treated with ZA was shown as a percentage of the control (mean ± SD in three separate experiments). *, P < 0.01 by Student’s t test. (G) Expression of UGT8, Smad4, Smad5, p-Smad1/5/8, and ID4 was analyzed by Western blotting in MDA-MB231 cells with stable empty vector or knockdown of UGT8 expression (left), as well as MDA-MB231 cells treated with or without ZA (10 µM; right). (H) Expression of UGT8 and RelA was analyzed by Western blotting in MDA-MB231 cells with stable empty vector or knockdown of UGT8 expression (left) as well as MDA-MB231 cells treated with or without ZA (10 µM; right). (I) A proposed model to illustrate the transcription activation of UGT8 by Sox10, which activates sulfatide–αVβ5 signaling axis in BLBC (see Discussion). TFs, transcription factors.
UGT8 promotes tumorigenicity of breast cancer
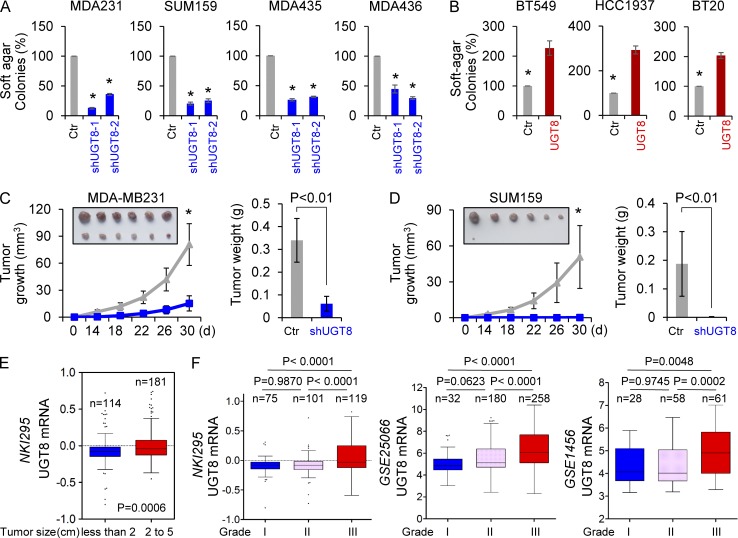
Having identified the critical association of UGT8-mediated metabolic alteration with oncogenic signaling in BLBC, we sought to evaluate the functional role of UGT8 in vitro and in vivo. We first tested the in vitro tumorigenicity using soft agar assay. Knockdown of UGT8 expression caused a remarkable decrease of colony-formation in MDA-MB231, SUM159, MDA-MB435, and MDA-MB436 cells (Fig. 7 A), whereas ectopic expression of UGT8 led to a significant increase of colonies in BT549, HCC1937, and BT20 cells (Fig. 7 B). We then examined the tumorigenicity in vivo by tumor xenograft experiments in which female SCID mice were injected with MDA-MB231 and SUM159 cells with stable empty vector or knockdown of UGT8 expression. As shown in Fig. 7 (C and D), MDA-MB231 and SUM159 cells with stable knockdown of UGT8 expression resulted in significantly decreased tumor growth compared with their corresponding vector control cells. To investigate the clinical implications of UGT8 expression for breast cancer progression, we extended our observations to clinicopathologically relevant parameters. We first assessed the UGT8 expression and its association with tumor size of breast cancer patients in NKI295 dataset. We segregated patients into two groups according to primary tumor size, showing that high UGT8 expression was associated with a larger tumor size of breast cancer patients (Fig. 7 E). We then evaluated the correlation between UGT8 expression and histological grades of the tumors in NKI295, GSE25066, and GSE1456 datasets in which tumors had been scored for tumor grade. Patients were separated into three groups according to histological grades of tumors. Our results showed that UGT8 expression was present predominantly in Grade 3 tumors, but less commonly in Grade 1 and Grade 2 tumors (Fig. 7 F). These data suggest that UGT8 functions as a critical mediator of BLBC aggressiveness.
Figure 7.
Knockdown of UGT8 expression inhibits tumorigenicity in vitro and in vivo. (A) Soft-agar assay was performed using MDA-MB231, SUM159, MDA-MB435, and MDA-MB436 cells with stable empty vector or knockdown of UGT8 expression (A), as well as BT549, HCC1937, and BT20 cells with stable empty vector or UGT8 expression (B). Data are presented as a percentage of empty vector cell lines (mean ± SD in three separate experiments). (C and D) MDA-MB231 cells (C) and SUM159 cells (D) with stable empty vector or knockdown of UGT8 expression were injected into the mammary fat pad of SCID mice. The growth of tumors was examined every 2 d. Tumor size and weight were recorded. Data are shown as mean ± SEM from six mice. *, P < 0.001. (E) Box plots indicated UGT8 expression in different tumor size of breast cancer from NKI295 dataset. Comparisons were made using the two-tailed Student’s t test. (F) Box plots indicated UGT8 expression in different histological grades of breast cancer from multiple datasets (NKI295, GSE25066, and GSE1456). Comparisons between two groups were made using the two-tailed Student’s t test.
Inhibition of UGT8 by shRNA or ZA suppresses metastasis of breast cancer
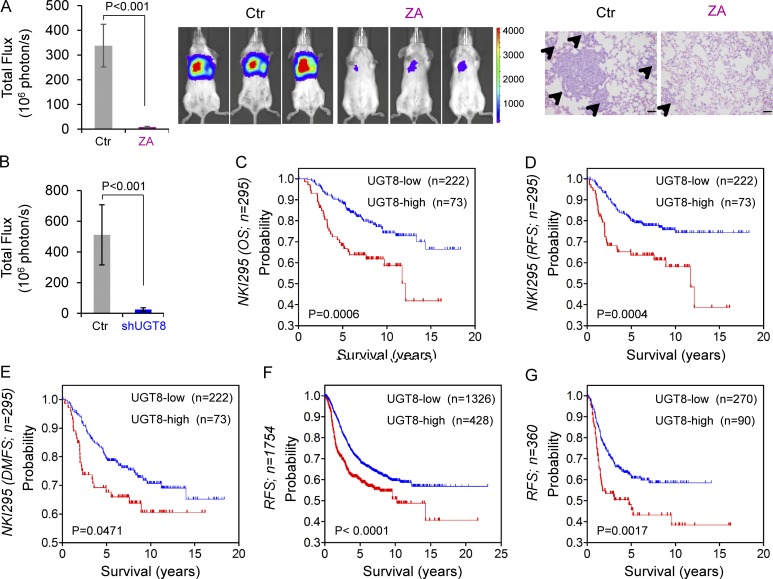
Because UGT8-mediated signaling was associated with cell migration and invasion, we reasoned that UGT8 might be critical for breast cancer metastasis in vivo. To test this notion, we assessed whether inhibition of UGT8 affected tumor metastasis in a xenograft metastasis model in which MDA-MB231 cells were injected via tail vein to generate pulmonary metastases. Remarkably, knockdown of UGT8 expression or ZA treatment suppressed lung metastasis in vivo (Fig. 8, A and B; and Fig. S4 A). We also examined the expressions of sulfatide and UGT8 in metastatic nodules from two group mice in Fig. S4 A. As expected, knockdown of UGT8 expression significantly caused a decrease in sulfatide expression (Fig. S4 B). These data suggest that UGT8 is critical for metastasis of BLBC cells and that pharmacologic inhibition of UGT8 may prevent metastasis of BLBC cells in vivo.
Figure 8.
Inhibition of UGT8 suppresses metastasis in vivo and elevated UGT8 predicts poor survival. (A and B) MDA-MB231 cells (A) and MDA-MB231 cells with stable empty vector or knockdown of UGT8 expression (B) were injected into SCID mice via the tail vein. For evaluation of ZA, the mice received ZA (0.0186 mg/kg/d) or sterile PBS subcutaneously. After 4 wk, the development of lung metastases was monitored using bioluminescence imaging and quantified by measuring photon flux (mean of six animals + SEM; left). Three representative mice from each group were shown (middle). Lung metastatic nodules were examined in paraffin-embedded sections stained with hematoxylin and eosin. The arrowheads indicate lung metastases. Bar, 100 µm (A, right). (C–E) Kaplan-Meier survival analysis for overall survival (OS), RFS, and distant metastasis-free survival (DMFS) of patients in the NKI295 dataset according to UGT8 expression status. The p-value was determined using the log-rank test. (F and G) Kaplan-Meier survival analysis for RFS of patients with various subtypes (F) or BLBC (G) in an aggregate breast cancer dataset according to UGT8 expression status. The p-value was determined using the log-rank test.
Given the critical function of UGT8 expression in breast cancer, we then evaluated whether UGT8 expression was correlated with patient survival in NKI295 dataset (van de Vijver et al., 2002). Patients were ranked based on UGT8 expression, and the top quartile and the other three quartiles were compared. Kaplan-Meier survival analysis showed that high UGT8 expression had shorter overall survival, relapse-free survival (RFS), and distant metastasis-free survival (Fig. 8, C–E). To extend this observation, we used an aggregate breast cancer dataset with 1,754 breast cancer samples to determine its clinical relevance (Györffy et al., 2010). Survival analysis demonstrated that tumors with an elevated UGT8 expression exhibited shorter RFS (Fig. 8 F). Similar result was observed by analyzing BLBC patient samples of this dataset (Fig. 8 G). These data support the critical roles of UGT8 in breast cancer aggressiveness.
Discussion
In this study, we report that inhibition of UGT8 suppresses the tumorigenic and metastatic capacity of BLBC cells by attenuating sulfatide–αVβ5 axis. Our study reveals several mechanistic and therapeutic insights into the crucial roles of UGT8 in BLBC progression.
UGT8 represents a potential prognostic indicator for breast cancer patients and is a major downstream target of Sox10 in BLBC
Our results showed that UGT8 expression increased tumorigenicity, whereas knockdown of UGT8 expression in BLBC cells suppressed tumorigenicity and metastasis in vitro and in vivo. Given the tight association of UGT8 with the aggressive properties of breast cancer, it may be beneficial to evaluate the possibility of UGT8 as a prognostic indicator for the assessment of the prognosis and the design of treatment of breast cancer patients. We analyzed several factors that indicate patients who are at risk of tumor progression, including (1) breast cancer subtypes: UGT8 overexpression occurs specifically in BLBC; (2) tumor grade: a significantly higher UGT8 expression is associated with higher tumor grade; (3) tumor size: UGT8 overexpression is correlated with larger tumor size; (4) survival rate: UGT8 overexpression has poor survival. These results strongly suggest the potential use of UGT8 in prognostic stratification of breast cancer patients.
Analysis of copy number alterations across cancer genomes reveals that less than 0.5% of primary tumors have UGT8 amplifications in TCGA dataset that contain 1,098 breast cancer patients (unpublished data), indicating the involvement of other genetic or epigenetic mediators in UGT8 overexpression in BLBC. Sox10, a transcriptional activator that in humans is encoded by the Sox10 gene, is also highly expressed in BLBC (Dravis et al., 2015). This protein as a nucleocytoplasmic shuttle protein is important for neural crest and peripheral nervous system development and tumor progression (Sarkar and Hochedlinger, 2013; Dravis et al., 2015). Sox10 has been identified to regulate several genes that are involved in the process of myelination, and Sox10-deficient mice exhibit defects in several neural crest-derived cell types as well as a failure of oligodendroglia to terminally differentiate and produce myelin (Stolt et al., 2002). Consistently, UGT8-mediated synthesis of GalCer and sulfatide is involved in myelin sheath development and function (Bosio et al., 1998), suggesting a possible link between Sox10 and UGT8. Indeed, our data identified Sox10 as a transcription inducer of UGT8 and demonstrated a positive correlation between the expression of Sox10 and UGT8 in two large breast cancer gene expression datasets, revealing that Sox10 is a direct transcriptional activator responsible for high UGT8 expression in BLBC.
UGT8 facilitates BLBC aggressiveness by activating sulfatide-αVβ5 axis
UGT8 is an endoplasmic reticulum–localized enzyme responsible for synthesis of GalCer from ceramide (Bosio et al., 1996c). Ceramide, as a substrate of UGT8, is intimately involved in the regulation of cancer cell growth, differentiation, senescence, and apoptosis (Maceyka and Spiegel, 2014). Because ceramide can be formed from multiple metabolic pathways, it is difficult to change its expression levels just by changing one single enzyme (Maceyka and Spiegel, 2014). We thus investigated two downstream metabolites, GalCer and sulfatide. Consistent with previous studies that mice lacking either UGT8 or GAL3ST1 gene caused an absence of sulfatide (Bosio et al., 1996b; Honke et al., 2002), our data showed that knockdown of UGT8 expression led to a dramatic decrease of both GalCer and sulfatide. However, only sulfatide, but not GalCer, enhanced the migratory and invasive ability of tumor cells, indicating the critical role of sulfatide in UGT8-mediated cellular program. Aberrant expression of sulfatide is associated with a variety of cancers (Takahashi and Suzuki, 2012; Xiao et al., 2013); thus, elucidating the biological functions of sulfatide will reveal mechanisms underlying the development of these diseases. Emerging evidence has demonstrated that sulfatide induces ITGAV expression by enhancing its promoter activity (Wu et al., 2013). Our data showed that sulfatide, but not GalCer, promoted ITGAV expression and induced integrin αVβ5 formation. Elevated expression of integrin αVβ5 is associated with tumor cell proliferation, angiogenesis, tumor migration, and metastasis (Desgrosellier and Cheresh, 2010). Thus, sulfatide functions as a key signaling molecule to allow crosstalk between metabolic and oncogenic pathways.
Pathological examination shows that malignant cells have often detached from the tumor mass at the periphery of or at the invading front of tumor. Interestingly, integrin αVβ5 expression often confines to invasion front in many cancers (Hood and Cheresh, 2002). It is well documented that integrin αVβ5 can regulate the TGF-β and NFκB pathways that are required for BLBC progression and metastasis (Desgrosellier and Cheresh, 2010; Dong et al., 2012; Wu et al., 2017). Consistently, our study revealed that knockdown of UGT8 expression significantly suppressed TGF-β and NFκB pathways in BLBC cells, supporting the crucial roles of UGT8-sulfatide–αVβ5 axis in BLBC.
Together, UGT8 expression is transcriptionally up-regulated by Sox10 in BLBC cells, resulting in activation of the sulfatide biosynthetic pathway; increased sulfatide induces integrin αVβ5 formation, triggering TGF-β and NFκB pathways that associate with BLBC aggressiveness. These findings provide a link between UGT8-mediated metabolic flux and oncogenic signaling pathways, which contributes to BLBC progression (Fig. 6 I).
UGT8 is a potential druggable target for treating BLBC
Treatment of BLBC represents an unmet medical need. Despite extensive study, few effective drug targets have been identified for BLBC. Thus, there is an urgent need to identify therapeutic targets for this subtype of breast cancer. Given the critical role of UGT8 in controlling the sulfatide biosynthetic pathway and BLBC progression, UGT8 overexpression represents an oncogenic event that associated with BLBC aggressiveness; thus, blocking of UGT8 by small molecules or antibodies may provide an attractive new approach for the clinical treatment of BLBC. ZA is a marketed drug that has been licensed for the treatment of osteoporosis or bone metastasis (Zekri et al., 2014). Accumulating evidence indicates that ZA induces apoptosis of cancer cells and suppresses migratory and invasive ability of cancer cells (Desgrosellier and Cheresh, 2010). Recent clinical trial data showed that adjuvant use of ZA could significantly improve disease-free survival and reduce the risk of locoregional and distant recurrence in postmenopausal breast cancer patients with low levels of circulating female hormones (Coleman et al., 2011, 2014). However, the exact mechanism underlying the anticancer property of ZA remains unknown. Here, we identified that ZA is a direct inhibitor of UGT8, which can efficiently inhibit the enzymatic activity of UGT8 and block sulfatide production. Our detailed microarray analysis demonstrates that both ZA treatment and knockdown of UGT8 expression have highly similar gene expression profiles, providing strong support for the notion that ZA is an inhibitor of UGT8. This finding is especially significant because ZA may become a potentially valuable target drug to suppress BLBC progression. Indeed, ZA remarkably suppressed cancer cell migration and invasion in vitro, inhibited lung metastasis of BLBC cells in mice models, exhibiting apparent efficacy against BLBC.
Unexpectedly, ZA did not result in an apparent effect on tumor growth in our xenograft experiments (unpublished data), which is different from the UGT8 knockdown that caused dramatically reduced tumor growth in vivo. There are two possible reasons for this discrepancy. First, after injection of a single dose of ZA, its circulating plasma levels are short lasting (Brown et al., 2007). Such dosage may not suppress tumor growth, but is enough to inhibit tumor metastasis. Indeed, our results showed that ZA at relatively low concentration displayed significant inhibition of cell migration and invasion. Second, ZA inhibits the enzymatic activity of UGT8, but doesn’t affect the expression of UGT8. Thus, we cannot exclude the possibility that UGT8 has other functions besides its enzymatic activity. Further study is required to unveil this question.
Drug repositioning has been used as an efficient strategy to identify and develop new uses for existing drugs because marketed drugs have established safety profiles and pharmacokinetic data (Mizushima, 2011). ZA is generally safe and well-tolerated in clinical trials at a dose of 4 mg every 3–4 wk or 1 yr for the treatment of osteoporosis or bone complications of cancer (Brown et al., 2007; Coleman et al., 2011, 2014). Because it can be rapidly cleared to the bones or kidneys within hours of intravenous treatment (Chen et al., 2002), repeated pulses of ZA may be required for the maintenance of plasma ZA concentration when the efficacy of ZA on tumor growth and lung metastasis is evaluated in mice model. Here, the mice were injected subcutaneously with ZA at a dose of 0.0186 mg/kg/d for a consecutive 28 d, and the cumulative dosage of ZA in mice is approximately equivalent to a single dose of 4 mg in humans, according to the conversion of animal doses to human equivalent doses based on body surface area. Significantly, this clinically achievable dosage of ZA remarkably suppressed lung metastasis of BLBC cells, displaying potent inhibitory effect and indicating that the optimal dose and frequency of ZA should be considered in future clinical trials. More than 40 clinical trials have been performed to investigate the potential anticancer activity of ZA (Coleman et al., 2011, 2014; Early Breast Cancer Trialists’ Collaborative Group, 2015); however, few clinical trials mainly focus on TNBC or BLBC. Recently, two retrospective studies from randomized trials of ZA plus chemotherapy versus chemotherapy alone demonstrated that a trend favoring ZA treatment was observed in TNBC that is mostly also BLBC despite relatively small sample size of TNBC patients in both studies (including 34 and 103 cases, respectively; Hasegawa et al., 2015; Kroep et al., 2016). Our study and these retrospective findings strongly support the translational value of ZA as a direct inhibitor of UGT8, and TNBC or BLBC might be the most promising subtype to be effectively treated with additional ZA. Together, the adoption of ZA is worthy of further exploration because in contrast to commonly used chemotherapy and adjuvant therapies; it has relatively lower toxicity and may be more acceptable for cancer treatment. Our study provides a proof of principle that UGT8 is a potentially valuable therapeutic target against BLBC and that pharmacological inhibition of UGT8 by ZA offers a promising opportunity for the clinical treatment of this challenging disease.
Materials and methods
Plasmids, shRNA, sulfatide, GalCer, and antibodies
UTG8 shRNA was purchased from MISSION shRNA at Sigma-Aldrich. Human UGT8 and Sox10 were amplified from a MDA-MB231 cDNA library and subcloned into pLVX-Puro.
Sulfatide and GalCer were purchased from Avanti Polar Lipids and Abcam, respectively. Antibody against UGT8 was from Protein Tech Group. Antibodies against Sox10, Smad4, Smad5, Integrin αV, and Integrin αVβ5 were from Abcam. Antibodies against RelA and Phospho-Smad1 (Ser463/465)/Smad5 (Ser463/465)/Smad9 (Ser465/467) were from Santa Cruz and Cell Signaling Technology, respectively. Antibodies for galactocerebroside, sulfatide, and integrin αVβ3 were purchased from Millipore Sigma. Antibodies for ID4 and β-actin were from BioCheck and Sigma-Aldrich, respectively.
Cell culture
MDA-MB231, SUM159, MDA-MB435, and MDA-MB436 cells were grown in DMEM/F12 supplemented with 10% FBS. HCC1937 and BT549 cells were grown in RPMI1640 plus 10% FBS. BT20 cells were cultured in DMEM/F12 supplemented with 10% FBS and insulin (5 µg/ml). For establishing stable transfectants with UGT8 expression, BLBC cells were transfected with pLVX-UGT8; stable clones were selected with puromycin (300 ng/ml) for 4 wk.
Immunostaining
Experiments were performed as described previously (Dong et al., 2013). Cells were grown on chamber slides, fixed with 4% paraformaldehyde, and incubated with primary antibodies. Secondary antibodies used were Texas red–conjugated goat anti–mouse or FITC-conjugated goat anti–rabbit (Molecular Probe).
Quantitative RT-PCR
Total RNA was isolated using RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. Specific quantitative RT-PCR experiments were performed using SYBR green Power Master Mix following manufacturer’s protocol (Applied Biosystems).
Luciferase reporter assay
Experiments were performed as described previously (Lin et al., 2010; Dong et al., 2012). All experiments were performed three times in triplicate.
Chromatin ChIP
ChIP assays were performed as described previously (Lin et al., 2010; Dong et al., 2012). The primers used for ChIP assays were 5′-CTGAATGGGAGCTTGAAGGATAC-3′ and 5′-GAAATCAGTGAGGTTCATTTCAC-3′ for the UGT8 promoter. The cells were prepared to perform ChIP assay with the Imprint ChIP kit (Sigma-Aldrich) according to the manufacturer’s instructions as we described recently (Dong et al., 2012).
Lipid extraction
Lipids from 100 mg of cells were extracted in 2 ml of chloroform, 4 ml of methanol, and 1.6 ml of water. The extraction was set in a constant temperature shaker (40°C) overnight. After the cell residues were separated by centrifugation, the solvents were evaporated using a stream of nitrogen. The pooled extracts then were resolved in 2 ml of chloroform/methanol (1:1; vol/vol). Phospholipids were decomposed by mild alkaline hydrolysis for 2 h at 40°C using 200 µl of 1 M KOH. After cooling, the samples were neutralized using glacial acidic acid. Changing the solvent ratio to methanol/chloroform/water (2:2:1.8; vol/vol/vol) caused a phase separation, with chloroform in the lower phase. After the chloroform phase was evaporated, the residue was solubilized in chloroform/methanol (1:1; vol/vol), and then was submitted for immunoblotting analysis.
Immunoblotting of tumor samples
The tumor samples were collected from resected breast tumors from patients with informed consent. The experiments were performed according to the approved guidelines established by the institutional review board at the Zhejiang University (Hangzhou, China). The sample was homogenized using 20 strokes of a Dounce homogenizer in 1 ml of homogenizing buffer. After centrifugation, the pellet was collected. For the detection of proteins, the resuspended pellet in Laemmli buffer was boiled and analyzed by SDS-PAGE, and then proteins were transferred onto PVDF membranes (Thermo Fisher Scientific). For the detection of sulfatide and GalCer, the extracted lipids from the pellet were spotted onto PVDF membranes. Immunoreactive blots were visualized by chemiluminescence.
Flow cytometry
Cells were washed twice and suspended with 1 ml PBS and then incubated with 5 µl LM609-αVβ3 antibody or P1F6-αVβ5 antibody at 4°C for 30 min. After two washes with PBS, the cells were collected by centrifugation (350 RCF, 5 min) and incubated with 1 ml Alexa Fluor 488 anti–mouse IgG mAb at 4°C for 30 min in the dark. After two washes with PBS, the stained cells were suspended in 500 ml PBS, and detected by Cytomic FC 500MCL.
Enzyme assays
In vitro activity assay of UGT8 was performed in a final volume of 120 µl with 25 mM Tris, pH 7.5, 5 mM calcium chloride, 10 mM manganese chloride, lysate from MDA-MB231 cells, 0.6 mM aglycone substrate, and 1.2 mM UDP sugar. The reactions were performed with the addition of cell lysates at 37°C for 1 h and then were terminated with the addition of 200 µl ethanol. After filtering, supernatants were subjected to HPLC system, consisting of C18 reversed-phase column (5 µm, 4.6 × 250 mm) and a UV detector. The chromatography was performed with 10.5% acetonitrile and 89.5% 0.02 M sodium phosphate buffer containing 10 mM tetrabutylammonium bromide as ion pair reagent. The flow rate was 1 ml/min and the UV detection was operated at 260 nm.
Colony formation assay
Colony formation assay was performed using double-layer soft agar in 24-well plates with a top layer of 0.35% agar and a bottom layer of 0.7% agar. Cells were seeded into 24-well plates in desired medium and cultured at 37°C for 15–20 d, and the colonies were stained and counted.
Migration and invasion assays
Migration and invasion assays were performed as described previously (Lin et al., 2010; Dong et al., 2012). All experiments were performed at least twice in triplicate. Statistical analysis was performed using the Student’s t test; a p-value of <0.05 was considered significant.
Tumorigenesis assay and lung metastasis model
Animal experiments were performed according to procedures approved by the Institutional Animal Care and Use Committee at the Zhejiang University. To examine the effect of UGT8 on tumorigenesis, female SCID mice (5–8 wk old) were injected with 106 exogenous UGT8 knockdown cells on the left flank and vector control cells on the right flank. For evaluation of the drug, mice were injected with MDA-MB231 cells (106 cells/mouse) on the left flank of every mouse; ZA (0.0186 mg/kg/d) was administered subcutaneously and sterile PBS was used as vehicle control. Tumor formation was monitored every 2 to 4 d for 30 d. Tumors size and weight were measured. To test the effect of UGT8 on tumor metastasis, SCID mice were injected with MDA-MB231 cells (106 cells/mouse) with stable empty vector or knockdown of UGT8 expression via tail vein (six mice/group). To evaluate the drug efficacy, mice were injected with MDA-MB231 cells (106 cells/mouse) via tail vein (six mice/group), and received ZA (0.0186 mg/kg/day) or sterile PBS as vehicle control subcutaneously. After 4 wk, lung metastasis was analyzed by an IVIS-100 imagining system (Xenogen). After mice were sacrificed, lung metastatic nodules were detected in paraffin-embedded sections stained with hematoxylin and eosin. Data were analyzed using the Student’s t test; a p-value <0.05 was considered significant.
Statistical analysis
Results are expressed as mean ± SD or SEM as indicated. Comparisons were made by the two-tailed Student’s t test or one-way ANOVA. Correlations between UGT8 and Sox10 were analyzed by Pearson’s correlation method and Spearman’s rank correlation test. Survival curves were performed using the Kaplan-Meier method, and differences were analyzed by the log-rank test. In all statistical tests, P < 0.05 was considered statistically significant.
Accession numbers
Microarray data of MDA-MB231 cells with UGT8 knockdown were deposited at the Gene Expression Omnibus with the accession number GSE112900.
Online supplemental material
Fig. S1 shows expression and activity of UGT8 in BLBC. Fig. S2 shows the correlation between UGT8 and Sox10. Fig. S3 shows the effect of UGT8 on GalCer and sulfatide production, and migration and invasion of breast cancer cells. Fig. S4 shows the effect of knockdown of UGT8 expression on metastasis of breast cancer cells.
Supplementary Material
Acknowledgments
This work was supported by grants from Natural Science Foundation of China (nos. 81772801 and 81472455 to C. Dong), Key Program of Zhejiang Province Natural Science Foundation (no. LZ17H160002 to C. Dong), National Key R&D Program of China (no. 2016YFC1303200 to C. Dong), Fundamental Research Funds for Central Universities of China (to C. Dong), and the Thousand Young Talents Plan of China (to C. Dong). Information about TCGA, the investigators, and institutions that constitute TCGA research network can be found at http://cancergenome.nih.gov/.
The authors declare no competing financial interests.
Author contributions: C. Dong raised conceptions and participated in the design and coordination of this research. Q. Cao and X. Chen designed and performed most of experiments. X. Wu and R. Liao performed the semiquantitative RT-PCR, quantitative RT-PCR, and colony formation assay. P. Huang and Y. Tan generated most of DNA constructs. L. Wang, G. Ren, and J. Huang analyzed UGT8 and sulfatide expression in breast tumor tissues. C. Dong supervised the work and wrote the manuscript.
References
- Bosio A., Binczek E., Le Beau M.M., Fernald A.A., and Stoffel W.. 1996a The human gene CGT encoding the UDP-galactose ceramide galactosyl transferase (cerebroside synthase): cloning, characterization, and assignment to human chromosome 4, band q26. Genomics. 34:69–75. 10.1006/geno.1996.0242 [DOI] [PubMed] [Google Scholar]
- Bosio A., Binczek E., and Stoffel W.. 1996b Functional breakdown of the lipid bilayer of the myelin membrane in central and peripheral nervous system by disrupted galactocerebroside synthesis. Proc. Natl. Acad. Sci. USA. 93:13280–13285. 10.1073/pnas.93.23.13280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio A., Binczek E., and Stoffel W.. 1996c Molecular cloning and characterization of the mouse CGT gene encoding UDP-galactose ceramide-galactosyltransferase (cerebroside synthetase). Genomics. 35:223–226. 10.1006/geno.1996.0342 [DOI] [PubMed] [Google Scholar]
- Bosio A., Binczek E., Haupt W.F., and Stoffel W.. 1998. Composition and biophysical properties of myelin lipid define the neurological defects in galactocerebroside- and sulfatide-deficient mice. J. Neurochem. 70:308–315. 10.1046/j.1471-4159.1998.70010308.x [DOI] [PubMed] [Google Scholar]
- Brown J.E., Ellis S.P., Lester J.E., Gutcher S., Khanna T., Purohit O.P., McCloskey E., and Coleman R.E.. 2007. Prolonged efficacy of a single dose of the bisphosphonate zoledronic acid. Clin. Cancer Res. 13:5406–5410. 10.1158/1078-0432.CCR-07-0247 [DOI] [PubMed] [Google Scholar]
- Chen T., Berenson J., Vescio R., Swift R., Gilchick A., Goodin S., LoRusso P., Ma P., Ravera C., Deckert F., et al. . 2002. Pharmacokinetics and pharmacodynamics of zoledronic acid in cancer patients with bone metastases. J. Clin. Pharmacol. 42:1228–1236. 10.1177/009127002762491316 [DOI] [PubMed] [Google Scholar]
- Coleman R.E., Marshall H., Cameron D., Dodwell D., Burkinshaw R., Keane M., Gil M., Houston S.J., Grieve R.J., Barrett-Lee P.J., et al. AZURE Investigators . 2011. Breast-cancer adjuvant therapy with zoledronic acid. N. Engl. J. Med. 365:1396–1405. 10.1056/NEJMoa1105195 [DOI] [PubMed] [Google Scholar]
- Coleman R., Cameron D., Dodwell D., Bell R., Wilson C., Rathbone E., Keane M., Gil M., Burkinshaw R., Grieve R., et al. AZURE investigators . 2014. Adjuvant zoledronic acid in patients with early breast cancer: final efficacy analysis of the AZURE (BIG 01/04) randomised open-label phase 3 trial. Lancet Oncol. 15:997–1006. 10.1016/S1470-2045(14)70302-X [DOI] [PubMed] [Google Scholar]
- Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y., et al. . 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 486:346–352. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent R., Trudeau M., Pritchard K.I., Hanna W.M., Kahn H.K., Sawka C.A., Lickley L.A., Rawlinson E., Sun P., and Narod S.A.. 2007. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 13:4429–4434. 10.1158/1078-0432.CCR-06-3045 [DOI] [PubMed] [Google Scholar]
- Desgrosellier J.S., and Cheresh D.A.. 2010. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 10:9–22. 10.1038/nrc2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmedt C., Piette F., Loi S., Wang Y., Lallemand F., Haibe-Kains B., Viale G., Delorenzi M., Zhang Y., d’Assignies M.S., et al. TRANSBIG Consortium . 2007. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin. Cancer Res. 13:3207–3214. 10.1158/1078-0432.CCR-06-2765 [DOI] [PubMed] [Google Scholar]
- Dong C., Wu Y., Yao J., Wang Y., Yu Y., Rychahou P.G., Evers B.M., and Zhou B.P.. 2012. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J. Clin. Invest. 122:1469–1486. 10.1172/JCI57349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Yuan T., Wu Y., Wang Y., Fan T.W., Miriyala S., Lin Y., Yao J., Shi J., Kang T., et al. . 2013. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 23:316–331. 10.1016/j.ccr.2013.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravis C., Spike B.T., Harrell J.C., Johns C., Trejo C.L., Southard-Smith E.M., Perou C.M., and Wahl G.M.. 2015. Sox10 Regulates Stem/Progenitor and Mesenchymal Cell States in Mammary Epithelial Cells. Cell Reports. 12:2035–2048. 10.1016/j.celrep.2015.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists’ Collaborative Group 2015. Adjuvant bisphosphonate treatment in early breast cancer: meta-analyses of individual patient data from randomised trials. Lancet. 386:1353–1361. 10.1016/S0140-6736(15)60908-4 [DOI] [PubMed] [Google Scholar]
- Fadare O., and Tavassoli F.A.. 2008. Clinical and pathologic aspects of basal-like breast cancers. Nat. Clin. Pract. Oncol. 5:149–159. 10.1038/ncponc1038 [DOI] [PubMed] [Google Scholar]
- Glück S., Ross J.S., Royce M., McKenna E.F. Jr., Perou C.M., Avisar E., and Wu L.. 2012. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine ± trastuzumab. Breast Cancer Res. Treat. 132:781–791. 10.1007/s10549-011-1412-7 [DOI] [PubMed] [Google Scholar]
- Györffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., and Szallasi Z.. 2010. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123:725–731. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- Hanahan D., and Weinberg R.A.. 2011. Hallmarks of cancer: the next generation. Cell. 144:646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hasegawa Y., Tanino H., Horiguchi J., Miura D., Ishikawa T., Hayashi M., Takao S., Kim S.J., Yamagami K., Miyashita M., et al. JONIE Study Group . 2015. Randomized Controlled Trial of Zoledronic Acid plus Chemotherapy versus Chemotherapy Alone as Neoadjuvant Treatment of HER2-Negative Primary Breast Cancer (JONIE Study). PLoS One. 10:e0143643 10.1371/journal.pone.0143643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzis C., Pusztai L., Valero V., Booser D.J., Esserman L., Lluch A., Vidaurre T., Holmes F., Souchon E., Wang H., et al. . 2011. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 305:1873–1881. 10.1001/jama.2011.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser L.M., Sadanandam A., Kuo W.L., Benz S.C., Goldstein T.C., Ng S., Gibb W.J., Wang N.J., Ziyad S., Tong F., et al. . 2012. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl. Acad. Sci. USA. 109:2724–2729. 10.1073/pnas.1018854108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich K.P., O’Brien C., Boyd Z., Cavet G., Guerrero S., Jung K., Januario T., Savage H., Punnoose E., Truong T., et al. . 2009. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin. Cancer Res. 15:4649–4664. 10.1158/1078-0432.CCR-09-0317 [DOI] [PubMed] [Google Scholar]
- Honke K., Tsuda M., Koyota S., Wada Y., Iida-Tanaka N., Ishizuka I., Nakayama J., and Taniguchi N.. 2001. Molecular cloning and characterization of a human beta-Gal-3′-sulfotransferase that acts on both type 1 and type 2 (Gal beta 1-3/1-4GlcNAc-R) oligosaccharides. J. Biol. Chem. 276:267–274. 10.1074/jbc.M005666200 [DOI] [PubMed] [Google Scholar]
- Honke K., Hirahara Y., Dupree J., Suzuki K., Popko B., Fukushima K., Fukushima J., Nagasawa T., Yoshida N., Wada Y., and Taniguchi N.. 2002. Paranodal junction formation and spermatogenesis require sulfoglycolipids. Proc. Natl. Acad. Sci. USA. 99:4227–4232. 10.1073/pnas.032068299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood J.D., and Cheresh D.A.. 2002. Role of integrins in cell invasion and migration. Nat. Rev. Cancer. 2:91–100. 10.1038/nrc727 [DOI] [PubMed] [Google Scholar]
- Korsching E., Jeffrey S.S., Meinerz W., Decker T., Boecker W., and Buerger H.. 2008. Basal carcinoma of the breast revisited: an old entity with new interpretations. J. Clin. Pathol. 61:553–560. 10.1136/jcp.2008.055475 [DOI] [PubMed] [Google Scholar]
- Kreike B., van Kouwenhove M., Horlings H., Weigelt B., Peterse H., Bartelink H., and van de Vijver M.J.. 2007. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 9:R65 10.1186/bcr1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroep J.R., Charehbili A., Coleman R.E., Aft R.L., Hasegawa Y., Winter M.C., Weilbaecher K., Akazawa K., Hinsley S., Putter H., et al. . 2016. Effects of neoadjuvant chemotherapy with or without zoledronic acid on pathological response: A meta-analysis of randomised trials. Eur. J. Cancer. 54:57–63. 10.1016/j.ejca.2015.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Wu Y., Li J., Dong C., Ye X., Chi Y.I., Evers B.M., and Zhou B.P.. 2010. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 29:1803–1816. 10.1038/emboj.2010.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceyka M., and Spiegel S.. 2014. Sphingolipid metabolites in inflammatory disease. Nature. 510:58–67. 10.1038/nature13475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhlouf A.M., Fathalla M.M., Zakhary M.A., and Makarem M.H.. 2004. Sulfatides in ovarian tumors: clinicopathological correlates. Int. J. Gynecol. Cancer. 14:89–93. 10.1111/j.1048-891x.2004.014223.x-1 [DOI] [PubMed] [Google Scholar]
- Mertins P., Mani D.R., Ruggles K.V., Gillette M.A., Clauser K.R., Wang P., Wang X., Qiao J.W., Cao S., Petralia F., et al. NCI CPTAC . 2016. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 534:55–62. 10.1038/nature18003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima T. 2011. Drug discovery and development focusing on existing medicines: drug re-profiling strategy. J. Biochem. 149:499–505. 10.1093/jb/mvr032 [DOI] [PubMed] [Google Scholar]
- Morichika H., Hamanaka Y., Tai T., and Ishizuka I.. 1996. Sulfatides as a predictive factor of lymph node metastasis in patients with colorectal adenocarcinoma. Cancer. 78:43–47. [DOI] [PubMed] [Google Scholar]
- Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F., et al. . 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 10:515–527. 10.1016/j.ccr.2006.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannuzzo G., Graziano A.C., Pannuzzo M., Masman M.F., Avola R., and Cardile V.. 2016. Zoledronate derivatives as potential inhibitors of uridine diphosphate-galactose ceramide galactosyltransferase 8: A combined molecular docking and dynamic study. J. Neurosci. Res. 94:1318–1326. 10.1002/jnr.23761 [DOI] [PubMed] [Google Scholar]
- Pawitan Y., Bjöhle J., Amler L., Borg A.L., Egyhazi S., Hall P., Han X., Holmberg L., Huang F., Klaar S., et al. . 2005. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 7:R953–R964. 10.1186/bcr1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakha E.A., Reis-Filho J.S., and Ellis I.O.. 2008. Basal-like breast cancer: a critical review. J. Clin. Oncol. 26:2568–2581. 10.1200/JCO.2007.13.1748 [DOI] [PubMed] [Google Scholar]
- Sarkar A., and Hochedlinger K.. 2013. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 12:15–30. 10.1016/j.stem.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt C.C., Rehberg S., Ader M., Lommes P., Riethmacher D., Schachner M., Bartsch U., and Wegner M.. 2002. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 16:165–170. 10.1101/gad.215802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., and Suzuki T.. 2012. Role of sulfatide in normal and pathological cells and tissues. J. Lipid Res. 53:1437–1450. 10.1194/jlr.R026682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver M.J., He Y.D., van’t Veer L.J., Dai H., Hart A.A., Voskuil D.W., Schreiber G.J., Peterse J.L., Roberts C., Marton M.J., et al. . 2002. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347:1999–2009. 10.1056/NEJMoa021967 [DOI] [PubMed] [Google Scholar]
- Vargo-Gogola T., and Rosen J.M.. 2007. Modelling breast cancer: one size does not fit all. Nat. Rev. Cancer. 7:659–672. 10.1038/nrc2193 [DOI] [PubMed] [Google Scholar]
- Vintonenko N., Jais J.P., Kassis N., Abdelkarim M., Perret G.Y., Lecouvey M., Crepin M., and Di Benedetto M.. 2012. Transcriptome analysis and in vivo activity of fluvastatin versus zoledronic acid in a murine breast cancer metastasis model. Mol. Pharmacol. 82:521–528. 10.1124/mol.111.077248 [DOI] [PubMed] [Google Scholar]
- Wang Y., Klijn J.G., Zhang Y., Sieuwerts A.M., Look M.P., Yang F., Talantov D., Timmermans M., Meijer-van Gelder M.E., Yu J., et al. . 2005. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 365:671–679. 10.1016/S0140-6736(05)70933-8 [DOI] [PubMed] [Google Scholar]
- Wu W., Dong Y.W., Shi P.C., Yu M., Fu D., Zhang C.Y., Cai Q.Q., Zhao Q.L., Peng M., Wu L.H., and Wu X.Z.. 2013. Regulation of integrin αV subunit expression by sulfatide in hepatocellular carcinoma cells. J. Lipid Res. 54:936–952. 10.1194/jlr.M031450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Li X., Fu Q., Cao Q., Chen X., Wang M., Yu J., Long J., Yao J., Liu H., et al. . 2017. AKR1B1 promotes basal-like breast cancer progression by a positive feedback loop that activates the EMT program. J. Exp. Med. 214:1065–1079. 10.1084/jem.20160903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Finkielstein C.V., and Capelluto D.G.. 2013. The enigmatic role of sulfatides: new insights into cellular functions and mechanisms of protein recognition. Adv. Exp. Med. Biol. 991:27–40. 10.1007/978-94-007-6331-9_3 [DOI] [PubMed] [Google Scholar]
- Zekri J., Mansour M., and Karim S.M.. 2014. The anti-tumour effects of zoledronic acid. J. Bone Oncol. 3:25–35. 10.1016/j.jbo.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.