Abstract
We further investigated the role of the Arabidopsis CBF regulatory genes in cold acclimation, the process whereby certain plants increase in freezing tolerance upon exposure to low temperature. The CBF genes, which are rapidly induced in response to low temperature, encode transcriptional activators that control the expression of genes containing the C-repeat/dehydration responsive element DNA regulatory element in their promoters. Constitutive expression of either CBF1 or CBF3 (also known as DREB1b and DREB1a, respectively) in transgenic Arabidopsis plants has been shown to induce the expression of target COR (cold-regulated) genes and to enhance freezing tolerance in nonacclimated plants. Here we demonstrate that overexpression of CBF3 in Arabidopsis also increases the freezing tolerance of cold-acclimated plants. Moreover, we show that it results in multiple biochemical changes associated with cold acclimation: CBF3-expressing plants had elevated levels of proline (Pro) and total soluble sugars, including sucrose, raffinose, glucose, and fructose. Plants overexpressing CBF3 also had elevated P5CS transcript levels suggesting that the increase in Pro levels resulted, at least in part, from increased expression of the key Pro biosynthetic enzyme Δ1-pyrroline-5-carboxylate synthase. These results lead us to propose that CBF3 integrates the activation of multiple components of the cold acclimation response.
Understanding the mechanisms that plants have evolved to survive freezing is of basic scientific interest and has the potential to offer new strategies to improve the freezing tolerance of agronomic plants. Toward this end, many investigators have focused on studying the phenomenon of cold acclimation, the process whereby certain plants increase in freezing tolerance in response to low temperatures (Hughes and Dunn, 1996; Thomashow, 1999). One line of investigation has been to identify genes that are induced at low temperature and to determine whether they have roles in freezing tolerance. This has led to the identification of scores of cold-induced genes, many of which encode either LEA (late embryogenesis abundant) or novel polypeptides (Thomashow, 1998). Recent results obtained with Arabidopsis indicate that at least one COR (cold-regulated) gene has a role in freezing tolerance. Overexpression of COR15a, which encodes a novel polypeptide that is targeted to the chloroplasts, has been shown to increase the freezing tolerance of chloroplasts in vivo and protoplasts in vitro (Artus et al., 1996). This increase in freezing tolerance results from the COR15a-encoded protein stabilizing membranes against freezing injury (Artus et al., 1996; Steponkus et al., 1998). There is evidence that the mature COR15a polypeptide, COR15am, acts directly as a cryoprotective protein that decreases the propensity of lipid bilayers to form deleterious hexagonal II phase lipids (Steponkus et al., 1998), a major type of freeze-induced membrane lesion that occurs in nonacclimated plants (Steponkus et al., 1993).
Whereas overexpression of COR15a has a detectable effect on freezing tolerance of chloroplasts and protoplasts, the effect is small (1°C–2°C; Artus et al., 1996; Steponkus et al., 1998). Moreover, it has little if any effect on whole-plant freezing survival (Jaglo-Ottosen et al., 1998). Expression of the entire battery of COR genes, which includes the COR6.6, COR47, and COR78 gene pairs (in addition to the COR15 gene pair), however, does (Jaglo-Ottosen et al., 1998). The ability to express all of the COR genes in concert at warm temperature was made possible by the discovery of the CBF family of transcriptional activators (Stockinger et al., 1997; Gilmour et al., 1998), also known as DREB1 proteins (Liu et al., 1998; Shinwari et al., 1998). COR6.6, COR15a, COR47, and COR78, and presumably other yet to be discovered CRT(C-repeat)/DRE(dehydrationresponsive element)-regulated COR genes, contain in their promoters a cold- and dehydration-responsive DNA regulatory element known as the CRT/DRE (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994). The first CRT/DRE binding factor to be discovered, CBF1 (CRT/DRE binding factor 1), was initially shown to activate expression of reporter genes in yeast that carried the CRT/DRE element (Stockinger et al., 1997). This indicated that the protein, which has an AP2/EREBP DNA binding motif (Riechmann and Meyerowitz, 1998), was a transcriptional activator. Overexpression of CBF1 in Arabidopsis was subsequently shown to activate expression of the entire battery of known CRT/DRE-regulated COR genes and to enhance whole plant freezing survival without a low temperature stimulus (Jaglo-Ottosen et al., 1998).
Additional studies have shown that CBF1 is a member of a small gene family encoding nearly identical proteins (Gilmour et al., 1998; Shinwari et al., 1998). The genes, CBF1, CBF2, and CBF3 (also known as DREB1b, DREB1c, and DREB1a, respectively), are located in tandem on chromosome 4 (Gilmour et al., 1998; Shinwari et al., 1998). Overexpression of CBF3 in Arabidopsis, like overexpression of CBF1, activates COR gene expression and enhances freezing tolerance at warm nonacclimating temperatures (Liu et al., 1998; Kasuga et al., 1999). All three CBF genes are cold-induced; CBF transcript levels increase within 15 min of transferring plants to low temperature followed at approximately 2 h by accumulation of transcripts for the target CRT/DRE-regulated COR genes (Gilmour et al., 1998; Shinwari et al., 1998). The mechanism whereby the CBF genes are activated by low temperature is not known but does not appear to involve autoregulation (Gilmour et al., 1998).
Many biochemical changes occur in plants during cold acclimation in addition to COR gene expression and are likely to have roles in freezing tolerance. It is well documented that lipid composition changes during cold acclimation in a wide range of plants and there are compelling data to indicate that this contributes to greater freezing tolerance (Steponkus et al., 1993). In a similar manner, the levels of Pro and Suc increase in Arabidopsis (McKown et al., 1996; Wanner and Junttila, 1999) and other plants (Guy et al., 1992; Koster and Lynch, 1992) during cold acclimation and are likely to have roles in freezing tolerance. There is evidence that Pro can protect both membranes and proteins against freeze-induced damage in vitro (Rudolph and Crowe, 1985; Carpenter and Crowe, 1988) and direct evidence that increased levels of Pro enhances whole plant freezing tolerance (Nanjo et al., 1999). Suc and other simple sugars have also been shown to be effective cryoprotectants in vitro (Strauss and Hauser, 1986; Carpenter and Crowe, 1988) and there is correlative evidence indicating a role in freezing tolerance in cold-acclimated plants (Guy et al., 1992; Koster and Lynch, 1992; Wanner and Junttila, 1999). A question thus raised is whether the CBF transcription factors are limited to activating the expression of COR genes encoding cryoprotective polypeptides, or alternatively, have a role in activating multiple components of the cold acclimation response. The results presented here support the latter model.
RESULTS
Identification of Transgenic Arabidopsis Plants That Overexpress CBF3
Transgenic Arabidopsis plants that overexpress CBF3 at normal growth temperature were created by placing the CBF3 coding sequence under control of the cauliflower mosaic virus (CaMV) 35S promoter and transforming the construct into Arabidopsis plants using the floral dip transformation procedure. Twenty-two independent lines were identified in which the T2 plants segregated 3:1 for kanamycin resistance (the selectable marker carried on the transformation vector). These lines presumably carried a single active T-DNA locus. The kanamycin resistant T2 plants were then screened by western analysis for constitutive expression of COR15a, a target gene of the CBF transcriptional activators. Eight lines were identified that constitutively produced the COR15am protein at levels that were equal to or greater than that which occurred in cold-acclimated wild-type plants. However, further analysis indicated that in five of these lines, COR15a expression was not uniform among the plants; i.e. although all of the plants were kanamycin resistant, not all produced the COR15am polypeptide. This variation may have resulted from gene silencing events. Regardless, these lines were not used further for this study.
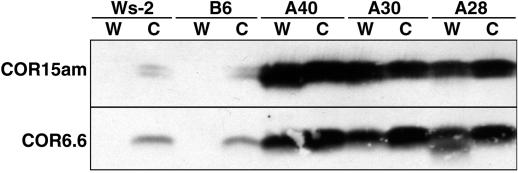
Three independent transgenic lines (A28, A30, and A40) were identified that produced the COR15am polypeptide at high levels uniformly among plants grown at normal temperatures. Northern analysis (Fig. 1) indicated that the transcript levels for CBF3 were about equal in nonacclimated and cold-treated A28, A30, and A40 plants and were much greater than those observed in either nonacclimated or cold-treated control plants (i.e. non-transformed plants or transgenic plants carrying the transformation vector without an insert). The transcript levels for two target COR genes, COR15a and COR6.6, were also nearly equal in nonacclimated and cold-treated A28, A30, and A40 plants and approximated the levels observed in cold-acclimated control plants (Fig. 1). Western analysis (Fig. 2) indicated that the proteins encoded by COR15a and COR6.6 were present in both nonacclimated and cold-acclimated A28, A30, and A40 plants at 3- to 5-fold higher levels than those found in cold-acclimated control plants.
Figure 1.
Transcript levels of CBF3 and target COR genes in transgenic plants overexpressing CBF3. Northern analysis of total RNA (20 μg for CBF3; 5 μg for the other genes) prepared from control Arabidopsis Ws-2 and B6 plants and from CBF3-expressing A40, A28, and A30 plants. Plants were either grown at 20°C (W) or at 20°C and then cold treated at 5°C for 7 d (C). eIF4a (eukaryotic initiation factor 4a) is a constitutively expressed gene used as a loading control (Metz et al., 1992). Upon long exposure, CBF3 transcripts can be detected in the total RNA samples prepared from cold-treated Ws-2 and B6 plants (not shown).
Figure 2.
Protein levels of COR15am and COR6.6 in transgenic plants overexpressing CBF3. Western analysis of total soluble protein (50 μg) prepared from control Arabidopsis Ws-2 and B6 plants and from CBF3-expressing A40, A30, and A28 plants. Plants were either grown at 20°C (W), or grown at 20°C and then cold treated at 5°C for 7 d (C). Protein transfers were treated with antiserum made to recombinant COR15am and COR6.6 polypeptides (Gilmour et al., 1996).
Effects of CBF3 Overexpression on Vegetative Growth, Time to Flowering, and Freezing Tolerance
Liu et al. (1998) have reported that transgenic Arabidopsis plants overexpressing CBF3 (DREB1a) have a “dwarf” phenotype. This was also true of the A28, A30, and A40 transgenic plants. After the same number of days of vegetative growth at normal temperature, the size of the leaves and overall dimensions of the CBF3-expressing plants were considerably less than those of the control plants (Fig. 3, A and B). Additional effects on growth that were not previously noted were also evident. One was that CBF3-expressing plants had a pronounced prostrate growth habit; whereas the leaves of the control plants generally had an upright stature, those of the transgenic plants laid flat to the soil (Fig. 3C). The CBF3-expressing plants also had much shorter petioles when compared with those of the control plants (Fig. 3D) and the leaves had a slight bluish-green tint (Fig. 3, A and B). Also, there was a substantial difference in time to flowering between the control and CBF3-expressing plants; i.e. control plants bolted and formed flowers well before the CBF3-expressing plants did (Fig. 3E). In one experiment, for instance, the control plants began to bolt at 17 d, whereas the A40, A30, and A28 plants took 21, 26, and 28 d, respectively, to initiate bolting (Table I). The CBF3-expressing plants went on to form flowers and set seed, although as noted by Liu et al. (1998), the final plant mass and seed yield were considerably less than that obtained with control plants (Fig. 3F). The lower yield of seed was due at least in part to the CBF3-expressing plants producing fewer axillary shoots. The delay in flowering observed in the CBF3-expressing plants, significantly, did not “simply” involve a slower overall growth rate, but appeared to involve a developmental delay in flowering. In one experiment, for instance, the control plants produced an average of 4.5 and 4.6 leaves per rosette, whereas the A40, A30, and A28 transgenic plants produced 6.0, 9.7, and 12.5 leaves per rosette, respectively (Table I).
Figure 3.
Growth characteristics of CBF3-expressing transgenic plants. A, Non-transformed Ws-2 plants and transgenic CBF3-expressing A30 plants after 2 weeks growth at 20°C. B, Non-transformed Ws-2 and transgenic B6 “control” plants and transgenic CBF3-expressing A40, A30, and A28 plants after 15 d growth at 20°C. C and D, Non-transformed Ws-2 plants and transgenic CBF3-expressing A28 plants after 16 d growth at 20°C. E, Non-transformed Ws-2 plants and CBF3-expressing A30 and A28 plants after 5 weeks growth at 20°C. F, As in E except after 9 weeks of growth at 20°C.
Table I.
Effects of CBF3 expression on time to flowering and rosette leaf number
| Plants | Time to Flowering (d)a | Rosette Leaves Per Plant (n) |
|---|---|---|
| Ws-2 | 17 | 4.5 (8) |
| B6 | 17 | 4.6 (8) |
| A40 | 21 | 6.0 (7) |
| A30 | 26 | 9.7 (15) |
| A28 | 28 | 12.5 (8) |
Period of time between planting seeds in soil and appearance of first flower buds in a population of “n” plants.
Overexpression of CBF3 (DREB1a) (Liu et al., 1998; Kasuga et al., 1999), like overexpression of CBF1 (Jaglo-Ottosen et al., 1998), has been reported to increase the freezing tolerance of nonacclimated plants. Similarly, we found that nonacclimated control plants were killed by freezing at −6°C for 24 h whereas nonacclimated CBF3-expressing plants were not; results for Arabidopsis (L.) Heynh. ecotype Wassilewskija (Ws)-2 and A30 plants are shown in Figure 4A. To quantify the increase in freezing tolerance we carried out electrolyte leakage tests. The results indicated that the freezing tolerance of nonacclimated CBF3-expressing plants was greater than that of the nonacclimated control plants; nonacclimated control plants had an EL50 (temperature that caused a 50% leakage of electrolytes) of approximately −4.5°C, whereas the three CBF3-expressing lines had EL50 values of approximately −8°C (Fig. 4B). The freezing tolerance of cold-acclimated CBF3-expressing plants was significantly greater than that of both nonacclimated CBF3-expressing plants and cold-acclimated control plants. CBF3-expressing plants that had been cold acclimated for 7 d had EL50 values of −11°C and lower (Fig. 4, C and D). In these particular experiments, the cold-acclimated control plants had EL50 values of approximately −6°C (Fig. 4, C and D). In other experiments, we have obtained EL50 values as low as −8°C, but have never observed cold-acclimated control Ws-2 plants with EL50 values as low as those obtained with the cold-acclimated CBF3-expressing plants.
Figure 4.
Effect of CBF3 overexpression on plant freezing tolerance. A, Seedlings of control Arabidopsis Ws-2 plants and CBF3-expressing A30 plants were grown at 20°C on solid medium and then frozen at −2°C for 24 h followed by 24 h at −6°C. B, Control Arabidopsis Ws-2 plants and CBF3-expressing A40, A30, and A28 plants were grown at 20°C, and the freezing tolerance of leaves was measured using the electrolyte leakage test. C and D, Same as B except that plants were grown at 20°C followed by 7-d-cold acclimation at 5°C.
Overexpression of CBF3 Affects Pro Metabolism
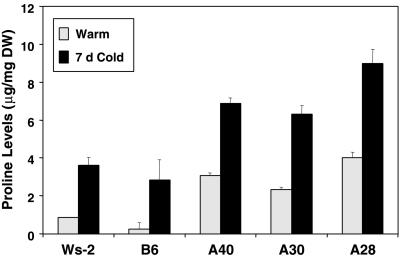
Pro levels increase during cold acclimation in Arabidopsis and other plants (Koster and Lynch, 1992; Alberdi et al., 1993; McKown et al., 1996; Wanner and Junttila, 1999). We reasoned that if the increase in Pro that occurs in Arabidopsis with cold acclimation was brought about by genes that were regulated by the CBF activators, then overexpression of CBF3 might result in elevated levels of Pro in nonacclimated plants. This was the case. Under nonacclimating growth conditions, the free Pro levels in the CBF3-expressing plants were approximately 5-fold higher than they were in the control plants, levels which were about the same as those found in cold-acclimated control plants (Fig. 5). The Pro levels in the CBF3-expressing plants increased further (approximately 2-fold) upon cold acclimation and were 2- to 3-fold higher than those found in the cold-acclimated control plants (Fig. 5).
Figure 5.
Effect of CBF3 expression on Pro levels. Free Pro levels were determined in leaf tissue from control Arabidopsis Ws-2 and B6 plants and CBF3-expressing A40, A30, and A28 plants grown at 20°C (warm), or plants grown at 20°C and cold-treated at 5°C for 7 d (7-d cold).
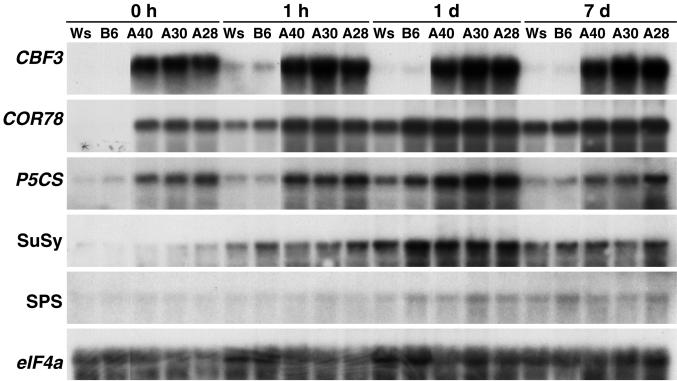
The Pro biosynthetic enzyme Δ1-pyrroline-5-carboxylate synthase (P5CS) has a key role in determining Pro levels in plants (Yoshiba et al., 1997). Because of this, and that P5CS transcript levels have been shown to increase in Arabidopsis in response to low temperature (Xin and Browse, 1998), it was of interest to determine whether P5CS transcript levels were elevated in the CBF3-expressing plants. Northern analysis indicated that they were; P5CS transcript levels were approximately 4-fold higher in nonacclimated CBF3-expressing plants than they were in nonacclimated control plants and were about equal to those found in the control plants that had been cold-treated for 1d (Fig. 6). The P5CS transcript levels in 7-d cold-acclimated CBF3-expressing plants were 2- to 3-fold higher than in cold-acclimated control plants (Fig. 6), a finding that was consistent with the 2- to 3-fold higher levels of Pro found in the cold-acclimated CBF3-expressing plants (Fig. 5).
Figure 6.
Effect of CBF3 expression on transcript levels of genes involved in Pro and sugar metabolism. Northern analysis of total RNA (20 μg for CBF3; 5 μg for other genes) isolated from control Arabidopsis Ws-2 and B6 plants and from CBF3-expressing A40, A30, and A28 plants. Plants were grown at 20°C then cold-treated at 5°C for the times indicated. The blots were hybridized with probes for CBF3, COR78, P5CS2, Suc synthase (SuSy), Suc-phosphate synthase (SPS), and eIF4a, a constitutively expressed gene used as a loading control (Metz et al., 1992).
Overexpression of CBF3 Affects Sugar Metabolism
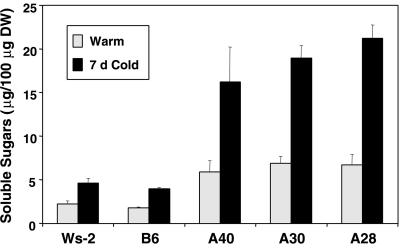
The accumulation of simple sugars is another commonly observed biochemical change that occurs with cold acclimation in Arabidopsis and other plants (Guy et al., 1992; Koster and Lynch, 1992; McKown et al., 1996; Wanner and Junttila, 1999). Thus, it was of interest to determine whether CBF3 expression affected the sugar levels in plants. This was tested by determining the amount of total soluble sugars in control and CBF3-expressing plants at both nonacclimating and cold-acclimating temperatures. The results indicated (Fig. 7) that the levels of total sugars in nonacclimated CBF3-expressing plants were approximately 3-fold higher than those in nonacclimated control plants. Upon cold acclimation, sugar levels went up in both the control and CBF3-expressing plants approximately 2-fold; thus, sugar levels in the CBF3-expressing plants remained approximately 3-fold higher in the control plants. Total soluble sugars notably accounted for as much as 20% of the total dry weight of plant material in the cold-acclimated CBF3-expressing plants.
Figure 7.
Effect of CBF3 expression on levels of total soluble sugars. Total soluble sugars were determined for leaf tissue from control Ws-2 and B6 plants and CBF3-expressing A40, A30, and A28 plants grown at 20°C (warm) or plants grown at 20°C and cold treated at 5°C for 7 d (7-d cold).
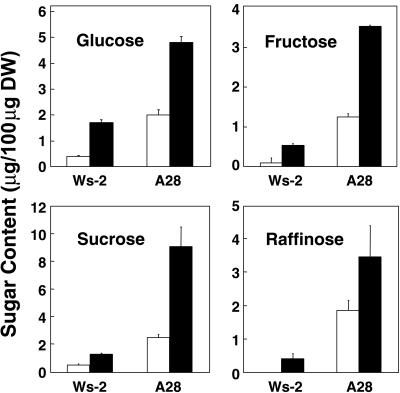
Analysis of the total soluble sugars by gas chromatography was performed on samples prepared from Ws-2 and B6 control plants and A28, A30, and A40 CBF3-expressing plants. The results indicated that CBF3 expression brought about increased levels of several sugars, namely Glc, Fru, Suc, and raffinose. A representative experiment comparing nonacclimated and cold-acclimated Ws-2 and A28 plants is presented in Figure 8. The results indicate that the levels of Glc, Fru, Suc, and raffinose increased in the Ws-2 control plants upon cold acclimation by approximately 2- to 5-fold depending on the particular sugar. In nonacclimated A28 plants, the levels of the four sugars were equal to or higher than they were in the cold-acclimated Ws-2 plants and increased further upon cold acclimation. In the cold-acclimated A28 plants, Suc plus raffinose accounted for approximately 12% of the total dry weight and Glc plus Fru accounted for approximately 8%.
Figure 8.
Effect of CBF3 expression on the levels of specific soluble sugars. Total sugars were prepared from leaf tissue of control Ws-2 plants and CBF3-expressing A28 plants grown at 20°C (white bars) or at 20°C followed by 7 d at 5°C (black bars) and the levels of individual sugars determined by gas chromatography.
Two enzymes that have key roles in determining the levels of Suc in plant cells are Suc-phosphate-synthase (SPS) and Suc synthase (SuSy; Winter and Huber, 2000). Transcript levels for genes encoding both SPS (Strand et al., 1997) and SuSy (Déjardin et al., 1999) have been shown to increase in Arabidopsis in response to low temperature. Thus, it was of interest to determine whether CBF3 had an effect on the expression of these genes. As previously reported, we found that the transcript levels for both the SPS and SuSy genes increased in response to low temperature (Fig. 6). However, these increases did not appear to involve the CBF transcriptional activators as there was little if any difference in the transcript levels for the SPS and SuSy encoding genes in the CBF3-expressing and control plants (Fig. 6). Thus, the effects of CBF3 on sugar levels do not appear to be mediated by altering transcription of the SPS or SuSy encoding genes.
DISCUSSION
The increase in freezing tolerance that occurs with cold acclimation is thought to involve the activation of multiple freezing tolerance mechanisms. Here we show that overexpression of the CBF3 transcriptional activator results in multiple biochemical changes that are commonly observed to occur in plants during cold acclimation. We show specifically that CBF3 overexpression in Arabidopsis results in the accumulation of COR polypeptides, the accumulation of Pro, and the accumulation of soluble sugars including Suc, raffinose, Glc, and Fru. There is evidence to indicate that each of these “classes” of biochemical alterations—COR polypeptides (Thomashow, 1998), Pro (Rudolph and Crowe, 1985; Carpenter and Crowe, 1988; Nanjo et al., 1999), and sugars (Strauss and Hauser, 1986; Carpenter and Crowe, 1988; Koster and Lynch, 1992; Wanner and Junttila, 1999)—contribute to an enhancement of freezing tolerance. It is interesting that we also found that Arabidopsis plants overexpressing CBF3 displayed a prostrate growth habit, a phenotype that has been associated with cold acclimation, and increased freezing tolerance in other plants (Omran et al., 1968; Roberts, 1990). Taken together, these findings lead us to propose that Arabidopsis CBF3 is a key regulatory gene that acts to integrate the activation of multiple mechanisms that work in concert to enhance freezing tolerance. Presumably CBF1 and CBF2 have overlapping, if not identical, roles to those of CBF3.
How does CBF3 bring about the biochemical changes that we observed? In the case of COR gene induction, the answer is known; CBF3 binds to CRT/DRE regulatory elements located in the promoters of these genes and activates their expression (Gilmour et al., 1998). Whether CBF3 also binds to the promoters of genes directly involved in sugar and Pro synthesis remains to be determined. However, our results indicate that CBF3 overexpression causes an increase in the transcript levels for at least one of the two known P5CS Pro biosynthetic genes (Strizhov et al., 1997). A CBF3-induced increase in P5CS gene expression thus seems likely to contribute to the increase in Pro levels observed in the CBF3-expressing plants. An examination of the promoter region of the P5CS2 gene (Strizhov et al., 1997; GenBank accession no. X86788) reveals that the core conserved sequence of the CRT/DRE regulatory element, CCGAC (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994), is present twice within 350 nucleotides upstream of the ATG start codon. Whether CBF3 binds to these sequences and activates expression of the P5CS2 gene, however, remains to be determined. It should be noted that Kasuga et al. (1999) did not observe any effect of CBF3 (DREB1A) expression on the levels of P5CS transcripts, nor did they observe an increase in P5CS transcript levels in response to low temperature, as we observed here (Fig. 6) and as Xin and Browse (1998) previously reported. The reason for this difference is not immediately obvious, but may be due, in part, to the P5CS probes used; we used a cDNA for the P5CS2 gene whereas Kasuga et al. (1999) used a probe for P5CS1.
Xin and Browse (1998) have identified an Arabidopsis gene, ESKIMO1 (ESK1), that affects the levels of Pro and sugars and has a major effect on freezing tolerance. Whereas wild-type plants grown at normal temperatures had a median lethal temperature (LT)50 of −2.8°C in a whole plant freeze test, plants carrying an esk1 mutation had a median lethal temperature of −10.6°C. The concentrations of free Pro and total sugars were elevated in the esk1 mutant plants about 30-fold and 2-fold, respectively. In addition, P5CS transcript levels in nonacclimated esk1 plants were approximately 8-fold higher than in nonacclimated wild-type plants. The esk1 mutation, significantly, did not affect expression of the COR genes; the transcript levels for COR6.6, COR15a, COR47, and COR78 remained low under normal growth conditions in the esk1 plants and were highly induced in response to low temperature. Therefore, Xin and Browse (1998) reasonably argued that it is not appropriate to consider cold acclimation as a simple, linear signaling pathway activating the full set of processes responsible for increasing freezing tolerance. Instead, they proposed a model in which parallel or branched signaling pathways activate “distinct suites” of cold acclimation responses. As envisioned, activation of one pathway would be able to result in considerable freezing tolerance without support from other components. Because esk1 plants did not overexpress the COR genes, Xin and Browse (1998) proposed that esk1 defines a signaling pathway of cold acclimation that is distinct from that which mediates expression of the COR genes.
Our findings with the CBF3 overexpressing plants do not contradict the general cold acclimation model of Xin and Browse (1998), but suggest a more integrated circuitry for cold acclimation signaling. In particular, our results provide evidence that the synthesis of COR proteins, Pro and sugars are coordinately regulated and that CBF3 has a key role in this regulation. The fact that the esk1 mutation affects the levels of P5CS expression, but not COR gene expression, can be explained in ways that do not necessitate the existence of distinct signaling pathways for Pro and COR protein synthesis. The two available esk1 alleles are recessive and, as mentioned above, cause an 8-fold increase in P5CS transcript levels. Thus, ESK1 appears to be a negative regulator of P5CS transcription at warm temperatures. One simple possibility is that ESK1 is a transcriptional repressor that binds to the promoter of one or both of the Arabidospis P5CS genes (Strizhov et al., 1997) at warm temperature and keeps transcription at a relatively low level. At low temperature, CBF3 could either directly bind to the P5CS promoter(s) and overcome repression by ESK1, or induce the expression of some other protein that inactivates ESK1. In these scenarios, the esk1 mutation would not affect expression of the COR genes because the COR genes would not have the ESK1 binding sequence. Of course, more complex models could also explain the current findings. The fundamental point is that the new information provided by this study argues for an integrated control of cold acclimation with CBF3, and presumably the other CBF proteins as well, having a key role in inducing multiple biochemical and physiological changes that act to increase plant freezing tolerance.
The finding that CBF3 overexpression results in accumulation of COR polypeptides, Pro, and sugars suggests that a considerable portion of the cold acclimation response falls under control of the CBF genes. However, our results also indicate that the freezing tolerance of the CBF3 overexpressing plants increased further in response to low temperature (Fig. 4). Thus, overexpression of CBF3 at warm temperature did not result in maximum freezing tolerance even though expression of its target genes, as judged by the levels of COR proteins (Fig. 2), was somewhat greater than that which occurred in cold-acclimated control plants. These findings could signify that CBF3 controls only a subset of low temperature-induced freezing tolerance genes and that additional non-CBF3-controlled genes might be induced in response to low temperature and activate additional freezing tolerance mechanisms. This would be akin to the parallel pathway model proposed by Xin and Browse (1998). However, it remains possible that CBF3 actually regulates all of the genes that are important in cold acclimation, but that the activities of the proteins encoded by these genes might be greater at low temperature; the proteins might be activated in response to phosphorylation catalyzed by a temperature-regulated protein kinase, for instance, or their action affected by changes in metabolite pool levels brought about by slower plant growth at low temperature. There is also the more trivial explanation that expression of CBF3 under control of the CaMV 35S promoter might not completely mimic the activation of CBF3 by low temperature.
The effect that CBF3 overexpression has on flowering is curious. Time to flowering is controlled by multiple physiological and environmental factors (Koornneef et al., 1998; Levy and Dean, 1998). In some plants, including Arabidopsis, the transition to flowering is responsive to vernalization, a long period (weeks) of low temperature treatment. In “facultative” plants such as Arabidopsis, the effect of vernalization is to shorten the time to flowering (the magnitude of the effect varies greatly among Arabidopsis ecotypes). If CBF3 expression at warm temperature was fully mimicking exposure to low temperature, then one might think that if it had any effect on flowering, that it would decrease the transition time and cause a corresponding decrease in the number of rosette leaves per plant. Our results, however, indicate that CBF3 expression had the opposite effect; it increased the time to flowering and increased the number of rosette leaves per plant. A mechanism to explain this effect is not immediately obvious but would seem to be consistent with the notion that CBF3 expression does not control all low temperature responses in Arabidopsis.
We previously reported that CBF1 overexpression induced COR gene expression and increased plant freezing tolerance without a low temperature stimulus (Jaglo-Ottosen et al., 1998). A significant difference in that study and the results presented here is that we did not note any obvious effect of CBF1 overexpression on plant growth. A likely explanation for this difference is suggested by the results of Liu et al. (1998). These investigators found a positive correlation between the level of CBF3 (DREB1a) expression, the level of expression of the target gene COR78 (RD29a), and the degree to which the plants were stunted in growth. In our previous experiments (Jaglo-Ottosen et al., 1998), the level of COR15am protein in nonacclimated CBF1-expressing plants was approximately equal to the levels of COR15am in control plants that had been cold-acclimated for 7 d. Here, the COR15am levels in the nonacclimated CBF3-expressing plants was greater than in control plants that had been cold-acclimated for 7 d (Fig. 2). Thus, it seems likely that the lack of effect on growth and development observed with the CBF1-expressing plants was due to lower levels of CBF-induced target gene expression. It should be kept in mind, however, that it has not yet been established that all three CBF genes control the same sets of genes. A detailed comparison between the levels of CBF1, CBF2, and CBF3 expression in transgenic Arabidopsis plants, the levels of target gene expression assessed on a genomic scale through microarray analysis, and a detailed analysis of the growth and development of the plants should help resolve this issue.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (L.) Heynh. ecotype Wassilewskija (Ws)-2 and transgenic plants in the Ws-2 background were grown in controlled environment chambers at 20°C under constant illumination from cool-white fluorescent lights (100–150 μmol m−2 s−1) in Baccto planting mix (Michigan Peat, Houston). Pots were subirrigated with deionized water as necessary. All seeds were cold-treated (5°C) for 4 d immediately after planting to ensure uniform germination. Plants were cold acclimated by placing pots at 5°C under continuous light (20–60 μmol m−2 s−1) as described (Gilmour et al., 1988).
Constructs and Plant Transformation
A 910-bp BamHI/HindIII fragment from a cDNA clone containing the whole coding region of CBF3 (Gilmour et al., 1998) was inserted into the BglII and HindIII sites of the binary transformation vector pGA643 (An et al., 1988). The resulting plasmid, pMPS13, which contains the CBF3 coding sequence under control of the CaMV 35S promoter, was transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. Arabidopsis plants were transformed with plasmid pMPS13 or the transformation vector pGA643 using the floral dip method (Clough and Bent, 1998). Transformed plants were selected on the basis of kanamycin resistance. Homozygous T3 or T4 plants were used in all experiments.
Western Analysis
Total soluble protein was obtained essentially as described (Gilmour et al., 1996) by grinding leaf material (approximately 100 mg) in 0.4 mL of extraction buffer (50 mm PIPES (1,4-piperazinediethanesulfonic acid), pH 7.0, 25 mm EDTA) containing 2.5% (w/v) polyvinyl-polypyrrolidone and removing insoluble material by centrifugation (16,000g × 20 min). Protein concentration in the supernatant was measured using the dye-binding method of Bradford (1976) with bovine serum albumin as the standard. Protein samples (50 μg of total protein) were fractionated by Tricine (N-[tris(hydroxymethyl)methyl]glycine) SDS/PAGE (Schägger and von Jagow, 1987) and transferred to 0.2 micron nitrocellulose membranes by electroblotting (Towbin et al., 1979). COR15am and COR6.6 were detected using the enhanced chemiluminescence kit (Amersham, Buckinghamshire, UK) with antiserum raised to recombinant COR15am and COR6.6 (Gilmour et al., 1996).
RNA Hybridization and cDNA Probes
Total RNA was extracted from Arabidopsis plants as described previously (Gilmour et al., 1988). Northern transfers were prepared and hybridized as described (Hajela et al., 1990) using high stringency wash conditions (Stockinger et al., 1997). 32P-labeled probes were prepared by random priming (Feinberg and Vogelstein, 1983). A gene-specific probe to CBF3 was made to the 3′ end of the cDNA clone by PCR as described previously (Gilmour et al., 1998). Arabidopsis cDNA clones encoding Arabidopsis Suc synthase (182C20T7), corresponding to the SUS1 gene (Martin et al., 1993), and Δ1-pyrroline-5-carboxylate synthase (125M17T7), corresponding to the P5CS2 gene (Strizhov et al., 1997), were obtained from the Arabidopsis Biological Resource Center at Ohio State University (Columbus). Probes made against the P5CS2 coding sequence would be expected to cross-hybridize with transcripts from the highly similar P5CS1 gene (Strizhov et al., 1997) and thus serve as a measure of total P5CS transcripts. A cDNA encoding a putative Suc-phosphate synthase gene from Arabidopsis was obtained from James Zhang (Mendel Biotechnology, Hayward, CA).
Pro Analysis
Lyophilized leaf material (30 mg) was extracted with 3 mL of deionized water at 80°C for 15 min. The samples were shaken for approximately 1 h at room temperature and then allowed to stand overnight at 4°C. The extracts were filtered through glass wool and analyzed for Pro content using the acid ninhydrin method (Troll and Lindsley, 1955). Briefly, 500 μL of the aqueous extract was mixed with 500 μL of glacial acetic acid and 500 μL of acid ninhydrin reagent (12.5 mg of ninhydrin, 0.3 mL of glacial acetic acid, 0.2 mL of 18 n orthophosphoric acid) and heated at 100°C for 45 min. After cooling, 500 μL of the reaction mix was partitioned against toluene (2 mL) and the absorbance of the organic phase was determined at 515 nm. The resulting values were compared with a standard curve constructed using known amounts of Pro (Sigma, St. Louis). Pro levels in certain samples were confirmed by amino acid analysis using an amino acid analyzer at the Macromolecular Structure Facility in the Biochemistry Department at Michigan State University.
Sugar Analysis
Total soluble sugars were extracted from lyophilized leaf material (20 mg) in 80% (v/v) ethanol (2 mL) at 80°C for 15 min. The samples were shaken for approximately 1 h at room temperature and allowed to stand overnight at 4°C. Extracts were filtered through glass wool and chlorophyll removed by shaking samples (0.4 mL) with water (0.4 mL) and chloroform (0.4 mL). The aqueous extract was assayed for sugar content using the phenol-sulfuric acid assay (Dubois et al., 1956). Certain samples were analyzed using a miniaturized version of the gas chromatography method described in Everard et al. (1994) with the following modifications. Vacuum dried carbohydrates were solubilized overnight in 0.5 mL of dry pyridine containing 30 mg mL−1 hydroxylamine-HCl and 0.1 mg mL−1 phenyl β-d-glucoside (derivatization efficiency internal standard). After heating at 75°C for 1 h, samples were cooled to room temperature and derivatized by the addition of 0.5 mL of hexamethyldisilazane and 0.05 mL of trifluoroacetic acid. Precipitates were allowed to settle for 1 h at room temperature prior to transferring the liquid phase to automatic sampler vials (0.3 mL). Samples were analyzed on a Hewlett-Packard 6890 gas chromatograph fitted with a 0.25-μm film, 30 m × 320 μm DB17 capillary column (J&W Scientific, Folsom, CA), and a flame ionization detector. The chromatography details were as follows: 1-μL injections with a 1:50 split ratio; injector and detector temperatures 305°C; initial column temperature 150°C with a ramp of 3°C min−1 to 200°C, followed immediately by a 20°C min−1 ramp to 290°C with a 10-min hold at this final temperature. The carrier gas was hydrogen at a linear velocity of 46 cm s−1.
Whole Plant Freeze Test
Ws-2 and A30 seedlings were grown (13 and 20 d, respectively) on Gamborg's B-5 medium containing 0.2% (w/v) Suc under sterile conditions in Petri dishes. The plants were tested for freezing tolerance by first placing the plates at −2°C in the dark for 3 h followed by ice nucleation with sterile ice chips. The plates were incubated an additional 21 h at −2°C, then the temperature of the freezer was turned down to −6°C, and the plates were left at this temperature for an additional 24 h. The plates were taken from the freezer and placed at 4°C in the dark for 18 h, followed by 2 d at 22°C under cool-white fluorescent lights (40–50 μmol m−2 s−1) with an 18-h photoperiod. The plates were then scored for freezing damage.
Electrolyte Leakage Freeze Test
Electrolyte leakage freeze tests were performed essentially as described (Uemura et al., 1995) with minor modifications. Tubes (16 × 12 mm) containing 3 to 4 leaves were placed in a low temperature bath set at −2°C in a randomized design. The randomization was performed with the aid of the SAS system (SAS Institute, Cary, NC). Ice chips were added to each tube after 1 h of incubation. Each tube was capped with foam plugs and incubated a further 1 h at −2°C. The bath temperature was then lowered 1°C every 20 min. Tubes were removed at each temperature and incubated an additional hour at that same temperature in a separate bath. Tubes were placed on ice after removal from the bath until all tubes had been removed. The samples were then thawed overnight at 2.5°C, and electrolyte leakage was measured as described (Gilmour et al., 1988).
ACKNOWLEDGMENTS
We gratefully acknowledge the assistance of Daniel Cook for the whole plant freeze test and the Macromolecular Structure Facility in the Biochemistry Department at Michigan State University for performing amino acid analysis. We also thank James Zhang for providing a clone of Suc-phosphate synthase and the Arabidopsis Biological Resource Center at Ohio State University for providing cDNA clones for P5CS2 and Suc synthase.
Footnotes
This work was supported in part by a subcontract (no. 593–0219–06) under the U.S. Department of Agriculture/Cooperative State Research, Education and Extension Service Cooperative Agreement (no. 96–34340–2711) North Central Biotechnology Initiative and by funds from Mendel Biotechnology, Inc., and the Michigan Agricultural Experiment Station.
LITERATURE CITED
- Alberdi M, Corcuers LJ, Maldonado C, Barrientos M, Fernandez J, Henriquez O. Cold acclimation in cultivars of Avena sativa. Phytochemistry. 1993;33:57–60. [Google Scholar]
- An G, Ebert PR, Mitra A, Ha SB. Binary vectors. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. p. A3. , 1–19. [Google Scholar]
- Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin CT, Thomashow MF. Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA. 1996;93:13404–13409. doi: 10.1073/pnas.93.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol. 1994;24:701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye dining. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carpenter JF, Crowe JH. The mechanism of cryoprotection of proteins by solutes. Cryobiology. 1988;25:244–255. doi: 10.1016/0011-2240(88)90032-6. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Déjardin A, Sokolov LN, Kleczkowski LA. Sugar-osmoticum levels modulate differential abscisic acid-independent expression of two stress-responsive sucrose synthase genes in Arabidopsis. Biochem J. 1999;344:503–509. [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- Everard JD, Gucci R, Kann SC, Flore JA, Loescher WH. Gas exchange and carbon partitioning in the leaves of celery (Apium graveolens L.) at various levels of root zone salinity. Plant Physiol. 1994;106:281–292. doi: 10.1104/pp.106.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabelling restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Hajela RK, Thomashow MF. Cold acclimation in Arabidopsis thaliana. Plant Physiol. 1988;87:745–750. doi: 10.1104/pp.87.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Lin C, Thomashow MF. Purification and properties of Arabidopsis thaliana COR (cold-regulated) gene polypeptides COR15am and COR6.6 expressed in Escherichia coli. Plant Physiol. 1996;111:293–299. doi: 10.1104/pp.111.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- Guy CL, Huber JLA, Huber SC. Sucrose phosphate synthase and sucrose accumulation at low temperature. Plant Physiol. 1992;100:502–508. doi: 10.1104/pp.100.1.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajela RK, Horvath DP, Gilmour SJ, Thomashow MF. Molecular cloning and expression of cor (cold-regulated) genes in Arabidopsis thaliana. Plant Physiol. 1990;93:1246–1252. doi: 10.1104/pp.93.3.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MA, Dunn MA. The molecular biology of plant acclimation to low temperature. J Exp Bot. 1996;47:291–305. [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotech. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Peeters AJM, Soppe W. Genetic control of flowering time in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:345–370. doi: 10.1146/annurev.arplant.49.1.345. [DOI] [PubMed] [Google Scholar]
- Koster KK, Lynch DV. Solute accumulation and compartmentation during the cold acclimation of puma rye. Plant Physiol. 1992;98:108–113. doi: 10.1104/pp.98.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Dean C. The transition to flowering. Plant Cell. 1998;10:1973–1989. doi: 10.1105/tpc.10.12.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T, Frommer WB, Salanoubat M, Willmitzer L. Expression of an Arabidopsis sucrose synthase gene indicates a role in metabolism of sucrose both during phloem loading and in sink organs. Plant J. 1993;4:367–377. doi: 10.1046/j.1365-313x.1993.04020367.x. [DOI] [PubMed] [Google Scholar]
- McKown R, Kuroki G, Warren G. Cold responses of Arabidopsis mutants impaired in freezing tolerance. J Exp Bot. 1996;47:1919–1925. [Google Scholar]
- Metz AM, Timmer RT, Browning KS. Sequences for two cDNAs encoding Arabidopsis thaliana eukaryotic protein synthesis initiation factor 4A. Gene. 1992;120:313–314. doi: 10.1016/0378-1119(92)90112-3. [DOI] [PubMed] [Google Scholar]
- Nanjo T, Kobayashi M, Yoshiba Y, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Lett. 1999;461:205–210. doi: 10.1016/s0014-5793(99)01451-9. [DOI] [PubMed] [Google Scholar]
- Omran AO, Atkins IM, Gilmour ECJ. Heritability of cold hardiness in flax. Crop Sci. 1968;8:716–719. [Google Scholar]
- Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biol Chem. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- Roberts DWA. Identification of loci on chromosome 5A of wheat involved in control of cold hardiness, vernalization, leaf length, rosette growth habit, and height of hardened plants. Genome. 1990;33:247–259. [Google Scholar]
- Rudolph AS, Crowe JH. Membrane stabilization during freezing: the role of two natural cryoprotectants, trehalose and proline. Cryobiology. 1985;22:367–377. doi: 10.1016/0011-2240(85)90184-1. [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K. An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem Biophys Res Commun. 1998;250:161–170. doi: 10.1006/bbrc.1998.9267. [DOI] [PubMed] [Google Scholar]
- Steponkus PL, Uemura M, Joseph RA, Gilmour SJ, Thomashow MF. Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1998;95:14570–14575. doi: 10.1073/pnas.95.24.14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steponkus PL, Uemura M, Webb MS. A contrast of the cryostability of the plasma membrane of winter rye and spring oat-two species that widely differ in their freezing tolerance and plasma membrane lipid composition. In: Steponkus PL, editor. Advances in Low-Temperature Biology. Vol. 2. London: JAI Press; 1993. pp. 211–312. [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand A, Hurry V, Gustafsson P, Gardström P. Development of Arabidopsis thaliana leaves at low temperatures releases the suppression of photosynthesis and photosynthetic gene expression despite the accumulation of soluble carbohydrates. Plant J. 1997;12:605–614. doi: 10.1046/j.1365-313x.1997.00605.x. [DOI] [PubMed] [Google Scholar]
- Strauss G, Hauser H. Stabilization of lipid bilayer vesicles by sucrose during freezing. Proc Natl Acad Sci USA. 1986;83:2422–2426. doi: 10.1073/pnas.83.8.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strizhov N, Ábrahám E, Ökrész L, Blicking S, Zilberstein A, Schell J, Koncz C, Szabados L. Differential expression of two P5CS genes controlling proline accumulation during salt-stress required ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. 1997;12:557–569. doi: 10.1046/j.1365-313x.1997.00557.x. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 1998;118:1–7. doi: 10.1104/pp.118.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troll W, Lindsley J. A photometric method for the determination of proline. J Biol Chem. 1955;215:655–660. [PubMed] [Google Scholar]
- Uemura M, Joseph RA, Steponkus PL. Cold acclimation of Arabidopsis thaliana: effect on plasma membrane lipid composition and freeze-induced lesions. Plant Physiol. 1995;109:15–30. doi: 10.1104/pp.109.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner LA, Junttila O. Cold-induced freezing tolerance in Arabidopsis. Plant Physiol. 1999;120:391–400. doi: 10.1104/pp.120.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter H, Huber SC. Regulation of sucrose metabolism higher plants: localization and regulation of activity of key enzymes. Crit Rev Plant Sci. 2000;19:31–67. doi: 10.1080/10409230008984165. [DOI] [PubMed] [Google Scholar]
- Xin Z, Browse J. eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc Natl Acad Sci USA. 1998;95:7799–7804. doi: 10.1073/pnas.95.13.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol. 1997;38:1095–1102. doi: 10.1093/oxfordjournals.pcp.a029093. [DOI] [PubMed] [Google Scholar]