Abstract
Background
Since the description of the normal human gut microbiome in healthy individuals using broad-range polymerase chain reaction, there has been great advancement in the techniques used to conduct microbiome research and applications of this research across health, gastrointestinal diseases, and nongastrointestinal diseases.
Summary and Key Messages
In inflammatory bowel disease (IBD), studies have reported gut dysbiosis meaning that the microbial composition, diversity, and richness are altered. Persons with IBD harbor on average 25% fewer microbial genes than healthy persons. Reduced diversity has been reported in both the fecal and mucosal microbiome of IBD. Crohn disease and ulcerative colitis are systemic diseases with immunological alterations in both intestinal and circulating compartments. As knowledge about the impact of the gut microbiome on the intestinal and systemic immune response has grown, researchers have begun exploring how the gut microbiome may impact on other systemic conditions. Considering the role of microbes especially on regulatory T cells, it was plausible that the gut microbiome may have a role in other chronic immune-mediated inflammatory diseases. In this review, studies of the gut microbiome in other chronic immune-mediated inflammatory diseases are discussed including how the information can inform our understanding of the gut microbiome in IBD.
Keywords: Animal models, Chronic immune-mediated inflammatory diseases, Dysbiosis, Gut microbiome, Inflammatory bowel disease
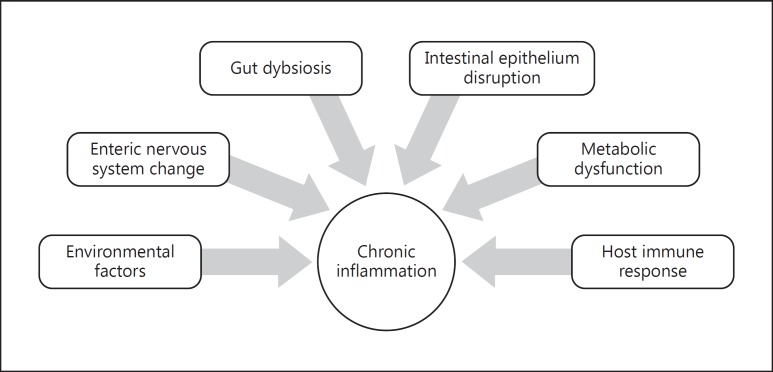
Since the description of the normal human gut microbiome in healthy individuals using broad-range polymerase chain reaction, there has been great advancement in the techniques used to conduct microbiome research and applications of this research across health, gastrointestinal diseases, and nongastrointestinal diseases [1]. The seminal study by Eckburg et al. [1] reported that the mucosal microbiome was quite similar within an individual in different segments of the colon from the cecum to the rectum but was quite different than the fecal microbiome from the same individual. The mucosal microbiome, which is in intimate contact with the gut epithelium, may play a more active role in orchestrating the intestinal immune response (Fig. 1). The luminal microbiome (which ultimately ends up in the stool) reflects the shed debris plus remnants of food metabolism and hence is comprised of a different microbial content than the mucosal microbiota. Some bacterial species present in the intestinal lumen may not access the mucus layer and epithelial crypts [2]. These luminal (fecal) microbes might be more essential for energy and metabolic interactions. While some fecal microbes may be similar to those predominantly residing at the mucosal interface, it is not clear to what extent the fecal microbiome, while conveniently accessible, reflects the key findings of the mucosal microbiome.
Fig. 1.
Contributing factors in the development of immune-mediated inflammatory disease.
Studies of both gut mucosal biopsies and stool have rapidly proliferated, exploring changes in various gastrointestinal diseases including inflammatory bowel disease (IBD). IBD includes Crohn disease (CD) and ulcerative colitis (UC), i.e., diseases thought to emerge primarily because of changes in the gut microbiome that drive aberrant intestinal immune responses which ultimately lead to the clinical manifestations of these diseases. Some common themes have emerged from studies in IBD; however, by no means has a definitive uniform pattern been defined for the varied phenotypes of either CD or UC [3]. Typically, studies have reported gut dysbiosis meaning that the microbial composition, diversity, and richness are altered in IBD. Persons with IBD harbor on average 25% fewer microbial genes than healthy persons [4]. Reduced diversity has been reported in both the fecal and mucosal microbiome of IBD. Firmicutes, one of the two predominant phyla in the healthy human gut, are typically reduced 5- to 10-fold and different studies have reported a reduction of particular species such as Faecalibacterium prausnitzii. Moreover, ileal and colonic CD have distinct microbiota patterns. In a study of mucosal biopsies from persons with CD and UC in different states of disease activity and from healthy controls, shifts in microbe abundance were evident comparing the inflamed mucosa between CD and UC. However, these shifts were more dramatic in the noninflamed mucosa between CD and UC [5]. It is problematic to discern whether microbiota changes reported in IBD are a cause or effect; thus, finding changes in the noninflamed mucosa may be particularly informative.
As noted, CD and UC are systemic diseases with immunological alterations in both intestinal and circulating compartments. While CD is associated with greater changes in Th1 pathways and mediators and UC is associated with greater Th2 changes, the Th17 pathway has proved to have an important role, particularly in CD. Intestinal regulatory T cells can be stimulated by the interface of certain microbes at the intestinal mucosa. The importance of the gut microbiota in the development of both the intestinal mucosal and systemic immune systems can be readily appreciated from studies of germ-free animals. Germ-free animals contain abnormal numbers of several immune cell types and immune cell products, and they have deficits in local and systemic lymphoid structures [6, 7]. Secreted levels of IgA and IgG are reduced and there are notable changes in circulating cytokine levels [7, 8]. Germ-free animals are devoid of microbes but there is evidence that different microbes can induce different immunological responses. The introduction of germ-free mice with Bacteroides fragilis, for example, induces proliferation of CD4+ T cells [9]. Other species of the Bacteroidetes phylum are similarly thought to be important for the differentiation of Th17 cells [10]. In addition, Lactobacilli species have been shown to differentially regulate dendritic cells [11].
As knowledge about the impact of the gut microbiome on the intestinal and systemic immune response has grown, researchers have begun exploring how the gut microbiome may impact on other systemic conditions. Considering the role of microbes especially on regulatory T cells, it was plausible that the gut microbiome may have a role in other chronic immune-mediated inflammatory diseases. There are a number of lines of evidence as to why there is rationale to consider common etiological themes for various chronic immune-mediated inflammatory diseases. First, there is epidemiological co-occurrence of these diseases. While they have been widely prevalent in developed countries over the past century, they have also been simultaneously emerging in developing countries. Second, there are some environmental factors that seem to similarly adversely affect these diseases such as smoking and a lack of vitamin D. Many of these diseases also share comorbidities. While rheumatoid arthritis (RA) is not typically comorbid with IBD, persons with IBD quite commonly get inflammatory arthropathies. Persons with IBD, however, are statistically more likely to get multiple sclerosis (MS) than the general population [12]. Fourth, some of these immune-mediated inflammatory diseases (like IBD and RA) share responses to biological agents that may have specific immunological effects. Yet, in other instances, there may be paradoxical effects such as the possibility that antibodies to tumor necrosis factor (TNF) could induce psoriasis even though they can be very effective at treating psoriasis, and they could also induce demyelinating disease (but have no proven benefit in treating MS). Finally, these immune-mediated inflammatory diseases have considerable overlap of genetic susceptibility loci. Overlap can comprise a shared locus for which the same single nucleotide polymorphism confers an increased risk for more than one disease or a shared locus for which the same polymorphism increases the risk for one disease but is protective of another, or finally a shared locus for which different polymorphisms are implicated [13]. It is not just that the genotype drives the immune response (many of the genetic single nucleotide polymorphisms common to immune-mediated inflammatory diseases are immune response loci) but the genotype can also impact on the gut microbiome. In a large study of 416 twin pairs, the greatest similarities in the gut microbiome were found in monozygotic compared to dizygotic twins (both twin types share a household and diet [14]).
Multiple Sclerosis
Activated T cells migrate to, adhere to and penetrate through the blood-brain barrier. In the central nervous system, T cells are reactivated by antigens presented on major histocompatibility complex sites predominantly by microglial cells. The reactivated T cells secrete pro-inflammatory cytokines, such as interferon-γ or interleukin-2 (IL-2 [15]). While an immunoinflammatory response is undeniably occurring in the central nervous system of persons with MS, the ongoing mystery is identifying the primary antigenic trigger that activates the initial T cell response. There is evidence that this may occur at the level of the gut microbiome. In this context, research investigating the gut microbiota is in fact framed by our understanding of MS immunopathogenesis.
There is a growing body of research exploring the interactions between the host, the brain, and the microbiome. Increased gut permeability may enable microbes or microbial metabolites to enter the bloodstream (reviewed in Tremlett et al. [16]). Further, gut microbes produce neuromodulating metabolites (e.g., short chain fatty acids) and can induce the production of neurotransmitters such as serotonin and neuromodulating hormones such as peptide YY. Interaction between the central nervous system and the gut may be bidirectional and enteric innervation by the vagus nerve, which has anti-inflammatory effects, may play a key role. In addition, stress has been shown to alter the gut microbiome, directly or indirectly, through intestinal immune responses [17]. In turn, the gut microbiome may mediate depression and anxiety, a nascent field of study [18, 19].
In therelapsing/remitting experimental autoimmune encephalomyelitis (EAE) mouse model, Ochoa-Repáraz et al. [20] showed that animals that received oral antibiotics or antibiotics present in drinking water had significantly reduced clinical activity scores for 3 weeks after initiation compared to animals that received no antibiotics or that received intraperitoneal antibiotics. This was accompanied by a reduction in interferon-γ and TNF production by lymphocytes and an increase in IL-10 and IL-13. It was also accompanied by a reduction in interferon-γ-producing and IL-17-producing T lymphocytes with an increase in IL-10- and IL-13-producing T lymphocytes.
Berer et al. [21] studied relapsing/remitting EAE in a germ-free mouse model. EAE was evident within 10 weeks in the specific pathogen-free mice but never emerged in germ-free mice; with microbial reconstitution of germ-free mice, EAE emerged. The differential roles of T and B cells in this relapsing/remitting EAE mouse model were also studied. The model involved injection ofmyelin oligodendrocyte glycoprotein in Freund's adjuvant into mice and transgenic CD4+ T cells infiltrated the central nervous system. Activation of myelin oligodendrocyte glycoprotein-specific T cells in the gut-associated lymphoid tissue was necessary (but not sufficient) for the emergence of relapsing/remitting EAE. Full relapsing/remitting EAE required recruitment of myelin oligodendrocyte glycoprotein-reactive B cells. In germ-free mice, there was a deficit of Th17 cells in Peyer's patches and in the lamina propria in addition to a deficit of myelin oligodendrocyte glycoprotein antibodies, all of which was reversed with gut microbiota reconstitution.
A number of studies have reported on the gut microbiome in adult MS [22, 23, 24]. In one stool microbiota study [23], untreated persons with MS had a significantly increased relative abundance of the archaeal phylum Euryarchaeota (particularly, Methanobrevibacter, which is a predominant colonic Archaeal genus) and bacteria Verrucomicrobia (Akkermansia) compared to healthy controls. A reduced abundance of Bacteroidetes (Prevotella and Butyricimonas) and Actinobacteria (Collinsella and Slackia) was also reported. Increased Methanobrevibacter and Akkermansia had negative correlations with some known anti-inflammatory cytokines in autoimmune demyelination such as TNFP1 and other key T cell and monocyte pathways often implicated in MS pathogenesis. Tremlett et al. [25] similarly identified an enrichment of Methanobrevibacter among early pediatric MS patients, whereas others have reported a depleted abundance of Archaea [26]. The potential inflammatory role of Euryarchaeota has previously been recognized. Considering their known ability to cause human dendritic cell activation (particularly Methanosphaera stadtmanae and Methanobrevibacter smithii[27]) and the increased abundance of M. stadtmanae often reported in IBD, further investigation is warranted. Further, others have also reported increased abundances of Akkermansia [28]. Interestingly, while a link might exist in MS and a pro-inflammatory T cell response, reduced Akkermansia muciniphila has been reported in IBD [29].
Treated persons with MS had increases in the bacterial genera Prevotella and Sutterella, suggesting that immune therapy may normalize some of the MS-related changes in the microbiota. There were no significant differences in the microbiota among those treated with interferon compared with those treated with glatiramer acetate. A lower abundance of Butyricimonas, a group of butyrate-producing bacteria, in MS patients was also found.
Others have also found increased stool Prevotella in MS [24]. In a Japanese study, there was no difference in stool diversity or richness between persons with MS and controls [22]. However, species belonging to Bacteroides, Faecalibacterium, Prevotella and Anaerostipes were less abundant in the gut microbiota of persons with MS than in healthy controls. The reduction in Prevotella was consistent with data noted above on untreated persons with MS; however, in this Japanese study, two thirds of subjects were receiving treatment. Of the 19 reduced species in MS samples, 14 belonged to Clostridia clusters XIVa and IV of which F. prausnitzii was reduced.
Tremlett et al. [30] studied the gut microbiota in pediatric patients with relapsing/remitting MS and found that relative to controls, MS cases had a significant enrichment in the relative abundance for members of the Desulfovibrionaceae family and depletion in Lachnospiraceae and Ruminococcaceae (all p < 0.000005). They further showed that microbiota richness correlated positively with Th17 for cases but not controls, whereas Bacteroidetes inversely correlated with Th17 for cases but not controls. Fusobacteria correlated with regulatory cells in controls but not in cases [31]. This group also studied the risk of relapse in pediatric MS adjusting for age and immune drug exposure status [32]. A shorter time to relapse was associated with a significant increase in Firmicutes, a significant reduction in Fusobacteria and the absence of the Archaea Euryarchaeota. The authors also reported quite different fecal microbiotas in users of distinct immune drugs, which is an important reminder for those conducting studies in humans with chronic immune diseases using different immune-altering drugs.
In a study assessing the interaction of vitamin D supplementation in persons with MS using glatiramer acetate, investigators found changes in the fecal microbiotas of persons with MS compared to healthy controls [33]. Vitamin D supplementation increased the abundance of Akkermansia, Ruminococcus, and Fecalibacterium to similar levels seen in healthy controls; however, this was only evident in persons on glatiramer acetate, underscoring how different interventions may be additive in their beneficial effects on the gut microbiome.
Hence, from MS data there are conflicting findings between particular microbes and different (or the same) diseases. For example, MS studies have reported either increases or decreases in Prevotella and increases in Akkermansia and reductions in Collinsella reported in MS differ from results reported in other chronic immune-mediated inflammatory diseases (including IBD). In contrast, consistent with studies of other chronic immune-mediated inflammatory diseases, such as CD, MS data often report reductions of F. prausnitzii. Studies of the MS gut microbiome, however, are limited in their sample size. Further studies are clearly needed to allow for more defined trends to be evaluated.
Rheumatoid Arthritis
Ironically, in one of the earliest animal model studies manipulating the gut microbiota, germ-free mice had worse adjuvant arthritis than conventional or specific pathogen-free mice [34]. Even in the absence of accompanying microbiome studies, O'Dell et al. [35] showed that oral minocycline was effective in treating RA compared with placebo. More recently, Chen et al. [36] have shown reduced fecal microbial diversity in persons with RA compared to controls, a typical trend observed in IBD. Of note, in this study, methotrexate and hydroxychloroquine, two longstanding RA treatments, improved gut microbial diversity. Hence, either these drugs have a direct or indirect effect on the gut microbiome or alternatively by improving the RA course, the gut microbiota normalized suggesting that alterations in the gut microbiota are effect rather than cause. As noted with MS, further research in the gut microbiome and RA is needed to account for concurrent immune-modulating therapy. Using principle coordinate analysis based on the Bray Curtis distance matrix changes in Actinobacteria were most striking between RA and controls but a decrease in Faecalibacterium was also noted. Thus, current findings suggest that Faecalibacterium plays an important role in many diseases and further research should be aimed at elucidating the bacteria's plausible function in immune-mediated inflammatory diseases. Sokol et al. [37] initially reported reduced F. prausnitzii as potentially important in predicting relapse of CD. Evidence of reduction across different immune-mediated inflammatory diseases may suggest that reduction of this microbe is truly associated with enhanced immune activation and inflammatory disease. IBD studies can be confounded by active gut inflammation but MS, RA, and other immune-mediated inflammatory disease studies are not necessarily confounded by localized presence of disease. Chen et al. [36] recently reported an increase in the abundance of Eggerthella and Actinomyces of the Actinobacteria and Turicibacter and Streptococcus of the Firmicutes. Moreover, of the Actinobacteria, Collinsella abundance correlated with increased gut permeability in Caco-2 cells, andwas also correlated with high levels of α-aminoadipic acid, asparagine, and IL-17A production. Kugasathan et al. [38] recently presented data on a prospective pediatric IBD cohort drawn from 28 referral centers across the US. They reported that the presence of increased Collinsella in stool samples was associated with a fistulizing phenotype of CD. At present, relatively little is known regarding what role, if any, Turicibacter may have on IBD. One study has reported an increase in Turicibacter in mice with depleted CD8+ T cells [39], another suggested its presence might be related to TNF expression [40] and the bacterium has also been isolated from the serum of an acutely ill patient [41]. These findings suggest that the function of this bacterium should be further evaluated for its plausible role in disease.
A few candidate microorganisms have been associated with RA; Prevotella, for example, has shown interesting trends. Scher et al. [42] showed elevated levels of Prevotella copri in treatment-naïve new-onset RA patients; however, levels were paradoxically decreased in chronic RA, psoriatic arthritis patients and even healthy controls, suggesting that this microbe may play a more important role in disease onset (vs. disease perpetuation). Further investigation revealed that mice colonized with P. copri presented with exacerbated colitis when treated with dextran sulfate sodium. The role of Lactobacillus has also been extensively studied with respect to RA. Liu et al. [43] showed increased Lactobacillus and corresponding diversity levels in the RA gut. This proposes that an increase in these levels may be involved in disease onset and/or progression. Other RA data also show an association between increased Lactobacillus populations and disease [44]. Moreover, increased abundances of both Lactobacillus and Bifidobacterium populations in active IBD have also been shown [45]. This is relevant, as particular Lactobacillus strains are utilized as supplementary treatment in chronic immune-mediated inflammatory; these microorganisms are considered probiotic, with the potential to confer a health benefit to the host, and have been evaluated in a clinical context for probiotic treatment of such diseases. To date, no Lactobacillus treatment studies have proven effective in IBD [46].
Psoriasis and Psoriatic Arthritis
Increasing evidence suggests that the microbiome plays an important role in psoriatic disease, including psoriasis and psoriatic arthritis, the latter of which represents another form of chronic arthritis. Though studies of the skin microbiome, mycobiome, and virome in psoriatic disease are numerous (reviewed in Yan et al. [47]), few studies have specifically focused on the gut microbes. Scher et al. [48] compared the gut community composition in 16 psoriatic arthritis patients, 15 patients with psoriasis of the skin and 17 healthy matched controls. Both psoriatic disease cohorts demonstrated reduced diversity compared to that observed in healthy controls. The relative abundance of Coprococcus species was reduced in both disease groups; the gut of psoriatic arthritis patients was characterized by a reduction in Akkermansia, Ruminococcus, and Pseudobutyrivibrio, which was positively correlated with heptanoate and hexanoate (medium chain fatty acids). The genus Akkermansia was also inversely correlated with soluble IgA and short chain fatty acids such as acetate and butyrate. Akkermansia and Ruminococcus are consistently reported to be reduced in IBD, particularly CD. In CD, it is difficult to discern whether changes in specific stool microbes are integral to or are secondary to the inflammatory process. Perhaps we can learn from other immune-mediated inflammatory diseases, like psoriasis, about the relevance of microbes like Akkermansia and Ruminococcus for the systemic immune response. Of note, in the context of the local gut immunological response, Scher et al. [48] also reported the fecal supernatant of psoriatic arthritis patients to show an increase in secretory IgA and a corresponding decrease in RANKL. More recently, Masallat et al. [49] found a reduction of the Actinobacteria phylum in psoriasis patients, which was negatively correlated with disease severity (PASI score). An increased Firmicutes/Bacteroidetes ratio in psoriasis patients was similarly reported, and was positively correlated with disease severity. Furthermore, F. prausnitzii has also been shown to be depleted in psoriasis as well as in many other chronic immunoinflammatory diseases [50].
Conclusions
As discussed above, several gut microbiota dysbiosis trends are common between many chronic immune-mediated inflammatory diseases, but also, findings that are not shared between particular diseases are also evident. Accordingly, our group led by Gary Van Domselaar has recently conducted a study to determine the gut microbiome in several immune-mediated inflammatory diseases including 19 CD patients, 19 UC patients, 18 MS patients and 21 RA patients relative to 32 healthy controls. The goal of this study was to determine if there were microbes consistently or uniquely disproportionate among several chronic inflammatory diseases. We observed the gut microbiota of CD to be consistently different in terms of richness and diversity compared to other chronic immune-mediated inflammatory diseases and healthy controls, and that the UC, MS and RA microbiota demonstrated similar diversity and richness. In terms of taxonomic distribution, while several taxa were uniquely increased or decreased in each chronic disease, the greatest number was found in CD. Moreover, we identified Eggerthella and Clostridium XIVa to be increased in all disease cohorts, whereas Coriobacteriaceae were decreased in all diseases relative to healthy controls. It would be particularly useful for further studies to include several additional chronic immune-mediated inflammatory diseases that preferentially affect distinct organ systems to elucidate commonalities or distinctions of the gut microbiome. Figure 2 shows a list of candidate microorganisms that are highly interesting in the context of immune-mediated inflammatory diseases.
Fig. 2.

Microorganisms of interest in chronic immune-mediated inflammatory diseases.
Animal models have provided some common themes across immune disease research involving the gut microbiome. Germ-free animal models of disease have reduced or absent development of organ-specific inflammation. Reconstitution with specific microbes can lead to worsened organ-specific inflammation. This adds to the rationale for studying the gut microbiome in human chronic immune-mediated inflammatory diseases. There has been considerable enthusiasm for the notion that an alteration of the gut microbiome is important at directing the immune dysregulation of IBD. However, as described here, for non-gut immunoinflammatory diseases, there is also increasing evidence for a role of the gut microbiota. Gut microbiome studies in these other chronic immune-mediated inflammatory diseases can inform the research and the analysis of studies in IBD. If microbe excess or deficiency is important across several chronic immune diseases then it adds to the likelihood that the connection between those microbes and the intestinal and subsequently systemic immune response is important, and makes the changes in IBD less likely to be secondary to the intestinal inflammation. Further, gut microbial changes in IBD that are not evident in other chronic immune-mediated inflammatory diseases are more likely to be IBD-specific.
Disclosure Statement
Charles Bernstein has consulted to Abbvie Canada, Ferring Canada, Janssen Canada, Napo Pharmaceuticals, Pfizer Canada, Shire Canada, Takeda Canada, and Mylan Pharmaceuticals. He has received unrestricted educational grants from Abbvie Canada, Janssen Canada, Shire Canada, and Takeda Canada. He has been on speaker's bureau of Abbvie Canada, Ferring Canada and Shire Canada.
References
- 1.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swidsinski A, Loening-baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol. 2005;11:1131–1140. doi: 10.3748/wjg.v11.i8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forbes JD, Van Domselaar G, Bernstein CN. The gut microbiota in immune-mediated inflammatory diseases. Front Microbiol. 2016;7:1081. doi: 10.3389/fmicb.2016.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin J, Li R, Raes J, Arumugam M, Burgdorf S, Manichanh C, Nielsen T, Pons N, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto J, Hansen T, Paslier D Le, Linneberg A, Nielsen HB, Pelletier E, Renault P, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H. A human gut microbial gene catalog established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forbes JD, Van Domselaar G, Bernstein CN. Microbiome survey of the inflamed and noninflamed gut at different compartments within the gastrointestinal tract of inflammatory bowel disease patients. Inflamm Bowel Dis. 2016;22:817–825. doi: 10.1097/MIB.0000000000000684. [DOI] [PubMed] [Google Scholar]
- 6.Sekirov I, Russell S, Antunes L. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 7.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 8.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72:4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Ivanov II, Frutos R de L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen HR, Pestka JJ, Alerts E. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J Immunol. 2017;168:171–178. doi: 10.4049/jimmunol.168.1.171. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein CN, Wajda A, Blanchard JF. The clustering of other chronic inflammatory diseases in inflammatory bowel disease: a population-based study. Gastroenterology. 2005;129:827–836. doi: 10.1053/j.gastro.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 13.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 14.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Treuren W Van, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinha S, Boyden AW, Itani FR, Crawford MP, Karandikar NJ. CD8+ T-cells as immune regulators of multiple sclerosis. Front Immunol. 2015;6:619. doi: 10.3389/fimmu.2015.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tremlett H, Bauer KC, Appel-Cresswell S, Finlay BB, Waubant E. The gut microbiome in human neurological disease: a review. Ann Neurol. 2017;81:369–382. doi: 10.1002/ana.24901. [DOI] [PubMed] [Google Scholar]
- 17.Mahony SMO, Marchesi JR, Scully P, Codling C, Ceolho A, Quigley EMM, Cryan JF, Dinan TG. Microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry. 2009;65:263–267. doi: 10.1016/j.biopsych.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 18.Collins SM, Bercik P. The relationship between intestinal microbiota and the central nervous system in normal gastrointestinal function and disease. Gastroenterology. 2009;136:2003–2014. doi: 10.1053/j.gastro.2009.01.075. [DOI] [PubMed] [Google Scholar]
- 19.Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Invest. 2015;125:926–938. doi: 10.1172/JCI76304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochoa-Repáraz J, Mielcarz DW, Ditrio LE, Burroughs AR, Foureau DM, Haque-Begum S, Kasper LH. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J Immunol. 2009;183:6041–6050. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- 21.Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, Wekerle H, Krishnamoorthy G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 22.Miyake S, Kim S, Suda W, Oshima K, Nakamura M, Matsuoka T, Chihara N, Tomita A, Sato W, Kim S, Morita H, Hattori M, Yamamura T. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to clostridia XIVa and IV clusters. PLoS One. 2015;10:1–16. doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, Patel B, Mazzola MA, Liu S, Glanz BL, Cook S, Tankou S, Stuart F, Melo K, Nejad P, Smith K, Topçuolu BD, Holden J, Kivisäkk P, Chitnis T, De Jager PL, Quintana FJ, Gerber GK, Bry L, Weiner HL. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Soldan MMP, Luckey DH, Marietta E V, Jeraldo PR, Chen X, Weinshenker BG, Rodriguez M, Kantarci OH, Nelson H, Murray JA, Mangalam AK. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep. 2016;6:28484. doi: 10.1038/srep28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tremlett H, Fadrosh D, Lynch S, Hart K, Graves J, Lulu S, Aaen G, Belman A, Benson L, Casper C, Chitnis T, Gorman M, Krupp L, Lotze T, Ness J, Roalst S, Weinstock-Guttman B, Waubant E. Gut microbiome in early pediatric multiple sclerosis: a case-control study (P4.027) Neurology. 2015:84. [Google Scholar]
- 26.Castillo Alvarez F, Perez Matute P, Colina Lizuain S. Intestinal microbiota in multiple sclerosis: influence of treatment with interferon β-1b. https://onlinelibrary.ectrims-congress.eu/ectrims/2016/32nd/146290/federico.castillo.lvarez.intestinal.microbiota.in.multiple.sclerosis.influence.html?f=m3
- 27.Bang C, Weidenbach K, Gutsmann T, Heine H, Schmitz RA. The intestinal archaea Methanosphaera stadtmanae and Methanobrevibacter smithii activate human dendritic cells. PLoS One. 2014;9:e99411. doi: 10.1371/journal.pone.0099411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cekanaviciute E, Debelius J, Singh S, Runia T, Nelson C, Yoo B, Kanner R, Crabtree-Hartman E, Mazmanian S, Knight R, Katz Sand I, Casaccia P, Cree B, Baranzini S. Gut dysbiosis is a feature of MS and it is characterized by bacteria able to regulate lymphocyte differentiation in vitro. Mult Scler J. 2016;22:58–59. [Google Scholar]
- 29.Png CW, Linden SK, Gilshenan KS, Zoetendal EG, Mcsweeney CS, Sly LI, McGuckin MA, Florin THJ. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 30.Tremlett H, Fadrosh DW, Faruqi AA, Zhu F, Hart J, Roalstad S, Graves J, Lynch S, Waubant E, Aaen G, Belman A, Benson L, Charles Casper T, Chitnis T, Gorman M, Harris Y, Krupp L, Lotze TE, Ness J, Olsen C, Rodriguez M, Rose J, Simmons TC, Tillema JM, Weinstock-Guttman B. Gut microbiota in early pediatric multiple sclerosis: a case control study. Eur J Neurol. 2016;23:1308–1321. doi: 10.1111/ene.13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tremlett H, Fadrosh DW, Faruqi AA, Hart J, Roalstad S, Graves J, Spencer CM, Lynch SV, Zamvil SS, Waubant E, US Network of Pediatric MS Centers Associations between the gut microbiota and host immune markers in pediatric multiple sclerosis and controls. BMC Neurol. 2016;16:182. doi: 10.1186/s12883-016-0703-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tremlett H, Fadrosh DW, Faruqi AA, Hart J, Roalstad S, Graves J, Lynch S, Waubant E. Gut microbiota composition and relapse risk in pediatric MS: a pilot study. J Neurol Sci. 2016;363:153–157. doi: 10.1016/j.jns.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cantarel BL, Waubant E, Chehoud C, Kuczynski J, DeSantis TZ, Warrington J, Venkatesan A, Fraser CM, Mowry EM. Gut microbiota in multiple sclerosis. J Investig Med. 2015;63:729–734. doi: 10.1097/JIM.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohashi O, Kuwata J, Umehara K, Uemura F, Takahashi T, Ozawa A. Susceptibility to adjuvant-induced arthritis among germfree, specific-pathogen-free, and conventional rats. Infect Immun. 1979;26:791–794. doi: 10.1128/iai.26.3.791-794.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Dell JR, Haire CE, Palmer W, Drymalski W, Wees S, Blakely K, Churchill M, Eckhoff PJ, Weaver A, Doud D, Erikson N, Dietz F, Olson R, Maloley P, Klassen LW, Moore GF. Treatment of early rheumatoid arthritis with minocycline or placebo: results of a randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 1997;40:842–848. doi: 10.1002/art.1780400510. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, Nelson H, Matteson EL, Taneja V. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8:43. doi: 10.1186/s13073-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, Blugeon S, Bridonneau C, Furet J-P, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kugathasan S, Denson LA, Walters TD, Kim M-O, Marigorta U, Schirmer M, Mondal K, Liu C, Griffiths AM, Noe JD, Crandall WV, Snapper SB, Rabizadeh S, Rosh JR, Shapiro JM, Guthery S, Mack DR, Kellermayer R, Kappelman M, Steiner S, Moulton DE, Keljo DJ, Cohen S, Oliva-Hemker M, Heyman MB. Early anti-TNF is effective in preventing internal penetrating but not stricturing disease complications in children newly diagnosed with Crohn's disease: a prospective risk prediction model for disease behavior study. Gastroenterology. 2017;152:S64. [Google Scholar]
- 39.Presley LL, Wei B, Braun J, Borneman J. Bacteria associated with immunoregulatory cells in mice. Appl Environ Microbiol. 2010;76:936–941. doi: 10.1128/AEM.01561-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones-hall YL, Kozik A, Nakatsu C. Ablation of tumor necrosis factor is associated with decreased inflammation and alterations of the microbiota in a mouse model of inflammatory bowel disease. PLoS One. 2015;10:e0119441. doi: 10.1371/journal.pone.0119441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bosshard PP, Zbinden R, Altwegg M. Turicibacter sanguinis gen. nov., sp. nov., a novel anaerobic, Gram-positive bacterium. Int J Syst Evol Microbiol. 2002;52:1263–1266. doi: 10.1099/00207713-52-4-1263. [DOI] [PubMed] [Google Scholar]
- 42.Scher J, Sczesnak A, Longman R, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer E, Abramson S, Huttenhower C, Littman D. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Zou Q, Zeng B, Fang Y, Wei H. Analysis of fecal lactobacillus community structure in patients with early rheumatoid arthritis. Curr Microbiol. 2013;67:170–176. doi: 10.1007/s00284-013-0338-1. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, Lan Z, Chen B, Li Y, Zhong H, Xie H, Jie Z, Chen W, Tang S, Xu X, Wang X, Cai X, Liu S, Xia Y, Li J, Qiao X, Al-Aama JY, Chen H, Wang L, Wu Q, Zhang F, Zheng W, Li Y, Zhang M, Luo G, Xue W, Xiao L, Li J, Chen W, Xu X, Yin Y, Yang H, Wang J, Kristiansen K, Liu L, Li T, Huang Q, Li Y, Wang J. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 45.Wang W, Chen L, Zhou R, Wang X, Song L, Huang S, Wang G, Xia B. Increased proportions of bifidobacterium and the lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel. J Clin Microbiol. 2014;52:398–406. doi: 10.1128/JCM.01500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernstein CN. Treatment of IBD: where we are and where we are going. Am J Gastroenterol. 2015;110:114–126. doi: 10.1038/ajg.2014.357. [DOI] [PubMed] [Google Scholar]
- 47.Yan D, Issa N, Afifi L, Jeon C, Chang H, Liao W. The role of the skin and gut microbiome in psoriatic disease. Curr Derm Rep. 2017;6:94–103. doi: 10.1007/s13671-017-0178-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, Marmon S, Neimann A, Brusca S, Patel T, Manasson J, Pamer EG, Littman DR, Abramson SB. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67:128–139. doi: 10.1002/art.38892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masallat D, Moemen D, State A. Gut bacterial microbiota in psoriasis: a case control study. African J Microbiol Res. 2016;10:1337–1343. [Google Scholar]
- 50.Eppinga H, Weiland CJS, Bing Thio H, van der Woude CJ, Nijsten TEC, Peppelenbosch MP, Konstantinov SR. Similar depletion of protective Faecalibacterium prausnitzii in psoriasis and inflammatory bowel disease, but not in Hidradenitis suppurativa. J Crohns Colitis. 2016;10:1067–1075. doi: 10.1093/ecco-jcc/jjw070. [DOI] [PubMed] [Google Scholar]