Abstract
To meet the ever-growing demand for effective, safe, and affordable protein therapeutics, decades of intense efforts have aimed to maximize the quantity and quality of recombinant proteins produced in CHO cells. Bioprocessing innovations and cell engineering efforts have improved product titer; however, uncharacterized cellular processes and gene regulatory mechanisms still hinder cell growth, specific productivity, and protein quality. Herein we summarize recent advances in systems biology and data-driven approaches aiming to unravel how molecular pathways, cellular processes, and extrinsic factors (e.g. media supplementation) influence recombinant protein production. In particular, as the available omics data for CHO cells continue to grow, predictive models and screens will be increasingly used to unravel the biological drivers of protein production, which can be used with emerging genome editing technologies to rationally engineer cells to further control the quantity, quality and affordability of many biologic drugs.
Graphical abstract
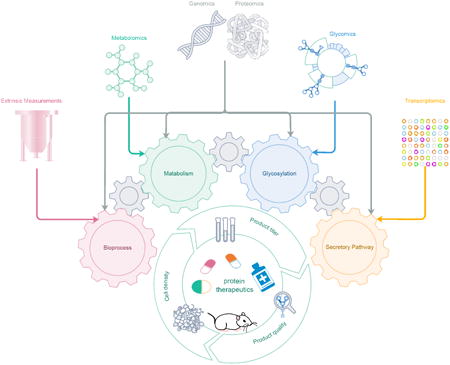
Introduction
Over the past few decades, Chinese hamster ovary (CHO) cells have emerged as the primary host for the biopharmaceutical industry. CHO cell lines were derived from the same hamster in 1957, and variants of the cell line have (e.g., CHO-K1, CHO-S and DG44) been further developed to meet different production requirements (see [1,2] for detailed genetic and phenotypic differences across the common CHO cell lines). These cell lines have been adopted by industry for various reasons, including the development of DHFR-deficient CHO cell line including which enable efficient transgene transfection and amplification. They also exhibit excellent capabilities to perform human-compatible post-translational modifications (PTMs), and they are highly adaptable to suspension-growth culture conditions in chemically-defined media. They also exhibit favorable safety profiles, e.g., being less prone to virus infection [3–5]. Over the past several decades, extensive efforts have aimed to increase the productivity of these cells to reduce the costs associated with culturing the cells and purifying product. Thus, innovations have effectively increased the yield of recombinant proteins (e.g., monoclonal antibodies) by three orders of magnitude, from 10-50mg/L in the 1980s to >10g/L in the 2010s [6,7].
Historically, at least three waves of innovations have offered additional toolboxes to further enhance biotherapeutic production (Figure 1) . The first wave significantly improved the volumetric yield, and leveraged innovations in bioprocessing techniques, media optimization [8] and tools that improve production by engineering the transgene and vectors (e.g., to optimize mRNA copy number and codon usage). The second wave involves targeted engineering of the host cells to enhance productivity and per-cell yield [9]. The third wave is beginning to use systems-level engineering to boost protein productivity by modulating cellular pathways to optimize cellular processes (e.g., metabolism [10]). It is being enabled by systems biology models [11,12], large-scale omics datasets [13,14]), and combinatorial genome editing [15,16], which are discovering and leveraging more comprehensive knowledge about the cell pathways influencing protein quantity and quality. Each wave continues to contribute novel innovations, and are resulting in improved protein production (Figure 2).
Fig 1. Three waves of different technologies have enabled continued improvement of recombinant protein production in CHO cells.
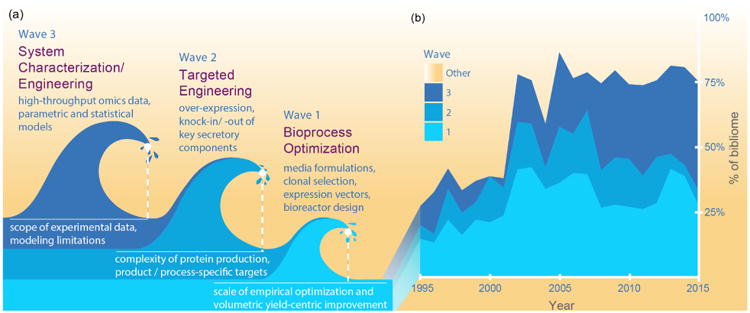
(a) Recombinant protein production has steadily improved over the past few decades thanks to innovations in bioprocessing, targeted genetic manipulation of cells, and systems biology approaches. Together, novel technologies, approaches and discoveries in each field have been of great importance. (b) A comprehensive survey of the CHO bioprocessing literature [60] highlights the historical development of the field in CHO cell research. The first wave-bioprocess development has been driving most of the earlier studies, while the targeted genetic manipulation, omics studies, and modeling efforts have become increasingly important after the mid-2000s with the increased prevalence of genomic resources, genome editing technologies, and development of novel computational models and algorithms.
Fig. 2. Published specific productivity, cell density and total product titer has improved steadily over the years.
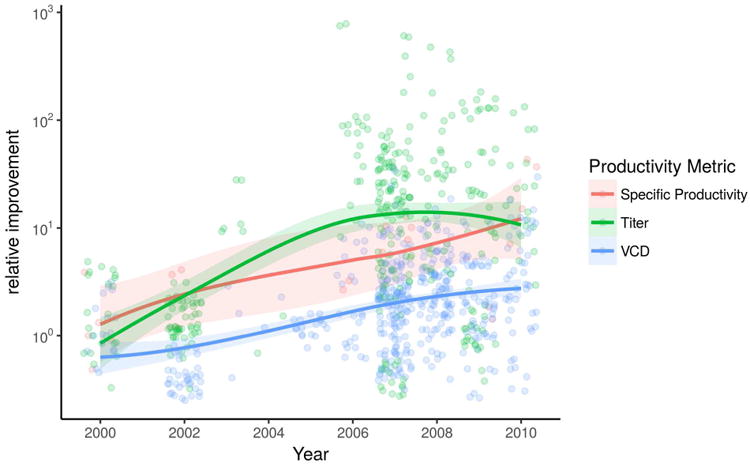
The trend for three major productivity metrics reported by literature from 2000- 2010 [60]. As a result of development in bioprocessing and feeding strategies, the volumetric yield has been greatly improved. The introduction of cell engineering to CHO has further improved the per-cell productivity since the mid-2000s.
In this review, we mention a few important innovations in each wave, and focus primarily on emerging efforts in systems biology and data-driven approaches that can advance our understanding of the cellular mechanisms contributing to recombinant protein production in CHO cells. These techniques are starting to guide efforts to engineer the cellular pathways and improve the product quality and protein productivity. These emerging efforts are ushering in an era of rational cell factory design in mammalian cell bioprocessing.
Wave 1: Bioprocess and transgene expression optimization
Bioprocess and transgene expression optimization has improved recombinant protein titer in CHO cells by ∼100-fold over the past few decades. This increase in volumetric yield has been primarily achieved through media optimization [17], clonal selection processes [18], expression vectors [19], genetic elements [20], bioprocess controls [21], and bioreactor design [22]. Recent innovations further enhance production through high-throughput assays to test everything from genetic elements to media conditions [23], leveraging tools from robotics to microfluidics [24].
Wave 2: Targeted engineering of CHO cells
Optimizing these extrinsic factors has improved titers, often by achieving higher cell densities; therefore, opportunities remain to further enhance the per-cell yield by directly engineering the cells [25]. Several cellular processes are associated with protein production, such as metabolism [26] and the secretory pathway [27]. Thus, following the success from bioprocess optimization came the second wave of strategies to engineer host cell lines. The advent of targeted genetic modification technologies, including knock-in strategies, have enabled the study of genes that improve protein production [28]. For example, overexpression of secretory pathway elements has been used to locate the faulty step in protein secretion while comparing the expression of easy- and difficult-to-express proteins [29]. Additional tools, such as ZFNs, TALENs [30] and CRISPR/Cas9 [15] enable efforts to edit individual host cell genes to fine-tune cell physiology, and precisely control product quality, such as glycosylation [31,32]. Further improvements to protein production could be achieved as additional emerging technologies are applied to CHO to activate or repress host cell genes with the CRISPRa/i system [33] and targeted epigenetic changes [34].
Wave 3: Characterizing and Engineering the CHO Protein Secretion System
Genome-wide analysis of protein secretion through omics technologies
The advances in the first two waves have provided powerful tools to enhance protein production. However, the synthesis and secretion of a single protein depends the concerted function of hundreds or thousands of other proteins. Thus, truly effective engineering strategies may require multiple genetic changes to the host cell. To achieve this, efforts have been made to comprehensively study the molecular changes that occur to enable high rates of protein secretion, thus shedding light on molecular and physiological factors making certain cells high producers.
Omics data have been used extensively to study productive clones. For example, a differential proteomic analysis identified the up-regulation of glutathione biosynthesis and the down-regulation of DNA replication to be characteristic of high-producing CHO cells [35]. Likewise, transcriptomic profiling of various CHO cell lines indicated that certain favorable metabolic and glycosylation patterns are associated with differential expression of key genes [36]. Ribosome profiling and polysome profiling have also been used to quantify translation of recombinant proteins and the endogenous mRNA in antibody-producing CHO cells [37,38].
Metabolite profiling of CHO can improve production by measuring metabolite accumulation and nutrient consumption. Indeed, several studies have profiled both extracellular and intracellular metabolites in CHO cell cultures with different growth media to connect cell culture media, productivity and growth rate [39–41]. Metabolomics has also successfully identified novel apoptosis-inducing metabolites that accumulate in the culture media [42].
These and many additional studies, show that omics data have emerged as valuable assays that provide insights into which genes, proteins, metabolites are associated with desired traits in protein production in CHO cells. Furthermore, they are helping to identify potential targets for cell engineering and bioprocess optimization for enhanced protein production.
Mapping out the CHO secretory pathway
The aforementioned high-throughput omics experiments often provide many differentially expressed genes, and it can be unclear which genes are most responsible for the improvements in production. Since bottlenecks in the secretory pathway frequently limit recombinant protein production [43], analysis of omics data in the context of this pathway can be informative. Recent progress in high-throughput omics technologies now allow researchers to systematically map out and dissect portions of the secretory pathway, such as protein synthesis, the unfolded protein response, glycosylation, and metabolism.
Various omics technologies are helping identify components of the secretory machinery. For example, a systematic discovery of genes involved in protein folding was carried out in yeast with synthetic genetic arrays [44]. More recently, a similar screen conducted at the single-cell level with combinatorial CRISPR interference revealed the bifurcation of unfolded protein response in unprecedented detail [45].
Such studies are fueling efforts to connect the known secretory machinery components. A network reconstruction of the CHO secretory pathway characterized the functional roles and localizations of the secretory machinery components, allowing better integration of omics data in the context of the secretory pathway [46]. Similarly, the machinery required for protein synthesis, post-translational modification, and secretion of individual recombinant proteins has been mapped out for mammals, enabling insights into product-specific needs [47].
Another component of the CHO secretory pathway required for most biotherapeutics is human-compatible glycosylation [48]. Recent advances in glycomics have enabled the profiling of glycan structures under various glycosyltransferase genes knockouts [31] and lectin binding preference [49] in CHO.
Developing predictive models for elevating cell productivity and product quality
Efforts to map out the protein secretion pathway are enabling more informative analyses of omic datasets. Such resources provide a platform for systems biology and machine learning algorithms to understand cell mechanisms for the production of recombinant proteins in CHO cells. Modeling efforts centered around the mechanisms in CHO protein production usually fall into one of the two frameworks: knowledge-based parametric models, and data-driven statistical models.
The knowledge-based modeling paradigm links the genotype to phenotype on a mechanistic basis. With careful curation, the models could help distill biological causation from observed data correlation. Genome scale models (GEMs) directly couple cellular functions such as cell growth and protein synthesis to enzyme activities [50]. The most comprehensive genome scale metabolic reconstruction in CHO [12] has provided recent insights into changes in lipid metabolism in antibody-producing CHO cells [51]. Apart from the stoichiometrically motivated GEMs, kinetic models characterize the dynamics of the cellular processes. These models have provided valuable insights in smaller-scale systems such as glycolysis and the pentose phosphate pathways [52]. N- and O-linked glycosylation profiles can also be modelled [53] through rule-based kinetic [54] and Markov models [55,56]. In addition, specific productivity was found to influence mAb glycosylation through an integrated model that couples glycosylation with cellular metabolism and secretory capacity [57].
Data-driven models do not rely on labor-intensive human curation, and they make fewer assumptions about the host cells. Therefore, these methods are particularly valuable in poorly characterized systems. Such models have been deployed to study the productivity of recombinant proteins and antibodies using CHO gene expression [58], product sequence features [59] and measurements of various bioprocess parameters [60]. Other bioprocess variables such as lactate consumption can also be accurately predicted [61]. One dilemma facing data-driven models is the shortage of high-coverage experimental data, used as training sets [62]. As biological data can be difficult or expensive to obtain, having a community-driven repository for various types of omics data can be one way to mitigate the shortage of training data [63,64].
Both of these powerful modeling frameworks are enabling the simulation and analysis of cellular responses influencing recombinant protein production in CHO cells. Furthermore, they are facilitating detailed analysis and integration of multiple omics data types. With the rather recent introduction of systems biology and machine learning methods to recombinant production in CHO cells, we expect to see a more widespread adoption of these tools for guiding rational design of CHO cell factories.
Conclusion
While innovations have driven a 1000-fold increase in protein titer in CHO cells, many challenges remain surrounding the production of many therapeutic proteins at high specific cellular productivity and high quality. Thus, further innovations in bioprocess optimization are needed to optimize expression conditions. Similarly, to speed up screening efforts, we need higher efficiency in genome editing strategies and high expression targeted integration sites for transgenes. Finally, omics studies and model-guided approaches will continue to map out the cellular pathways influencing the quantity and quality of secreted proteins. Fundamentally, a better general understanding of CHO cells is needed. For example, clonal variation and genomic instability in CHO lead to variable protein production over time. A recent multi-omics study showed that ∼40% of differentially expressed genes in a producer cell line contained different copy number variations, suggesting CNVs as a driver of transcriptional activation as opposed to epigenetic or regulatory changes [51]. Thus, to unravel which genetic and epigenetic changes underlie desired protein production traits, large scale genetic screens coupled with multi-omics data and computational models [45,65,66] will be invaluable to understand and engineer desired characteristics such as specific productivity, viability, morphology, and growth rate for large-scale bioprocesses. We anticipate that such data and novel computational tools will be increasingly valuable to therapeutic protein production.
Supplementary Material
Supplementary Table 1: The classification of the CHO bibliome into the 3 different waves according to the types of phenotypic and bioprocess data contained therein.
Highlights.
3 waves of innovation have enhanced protein production in CHO cells.
Systems-level methods are now unravelling drivers of protein production
Predictive models will facilitate rational cell engineering for protein production
Article Highlight.
*The art of CHO cell engineering:
A comprehensive overview of the targeted engineering efforts in CHO cells.
*What can mathematical modelling say…
A review of the development of the field of modelling CHO metabolism and protein glycosylation.
**A consensus genome-scale…
The first genome-scale reconstruction of the CHO metabolic pathways.
*high-throughput screening and selection of …
A summary of the cell line selection methods that isolate high-producing clones with a particular focus on the recent development of high-throughput essay techniques.
**Engineered CHO cells for production of diverse…
19 glycosyltransferases were knocked out (singly and combinatorially) in CHO cells stably expressing EPO. The resulting glycosylation patterns were subsequently profiled.
*ribosome profiling-guided depletion…
Ribosome profiling provided a genome-wide view of protein translation in an IgG-producing CHO cell line.
*A multiplexed single-cell CRISPR
Key genes regulating the unfolded protein response are identified using a genome-scale CRISPR interference screen.
**Network reconstruction of the mouse secretory pathway…
This secretory pathway network reconstruction for CHO facilitates the interpretation of omics data related to protein secretion, and identifies targets for engineering improved growth and IgG production.
*A markov chain model for N-linked protein glycosylation
A non-kinetic low-parameter Markov model for N-glycosylation used glycosyltransferase reaction rules to predict glycoprofiles following in glycoengineering efforts.
**Mammalian Systems Biotechnology Reveals Global Cellular Adaptations in a Recombinant CHO Cell Line
The authors proposed a workflow combining multi-omics data and genome scale models to study the genotypic and phenotypic differences between a wild-type and recombinant antibody-producing Chinese hamster ovary (CHO) cell line.
*Quantitative feature extraction from the Chinese hamster ovary bioprocess bibliome using a novel meta-analysis workflow
The authors compiled the CHO bibliome: a repository covering all published CHO cell studies from 1995 to 2015, and demonstrated that data can be extracted for further analysis.
Acknowledgments
This work was conducted with support from the Novo Nordisk Foundation provided to the Center for Biosustainability at the Technical University of Denmark (NNF10CC1016517) and from NIGMS (R35 GM119850). Support was also provided from a fellowship from the Government of Mexico (CONACYT) and the University of California Institute for Mexico and the United States (UC-MEXUS), and the Taiwan Ministry of Education.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lewis NE, Liu X, Li Y, Nagarajan H, Yerganian G, O'Brien E, Bordbar A, Roth AM, Rosenbloom J, Bian C, et al. Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome. Nat Biotechnol. 2013;31:759–765. doi: 10.1038/nbt.2624. [DOI] [PubMed] [Google Scholar]
- 2.Wurm FM, Hacker D. First CHO genome. Nat Biotechnol. 2011;29:718–720. doi: 10.1038/nbt.1943. [DOI] [PubMed] [Google Scholar]
- 3.Kantardjieff A, Jacob NM, Yee JC, Epstein E, Kok YJ, Philp R, Betenbaugh M, Hu WS. Transcriptome and proteome analysis of Chinese hamster ovary cells under low temperature and butyrate treatment. J Biotechnol. 2010;145:143–159. doi: 10.1016/j.jbiotec.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Rita Costa A, Elisa Rodrigues M, Henriques M, Azeredo J, Oliveira R. Guidelines to cell engineering for monoclonal antibody production. Eur J Pharm Biopharm. 2010;74:127–138. doi: 10.1016/j.ejpb.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Nishimiya D. Proteins improving recombinant antibody production in mammalian cells. Appl Microbiol Biotechnol. 2014;98:1031–1042. doi: 10.1007/s00253-013-5427-3. [DOI] [PubMed] [Google Scholar]
- 6.Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol. 2004;22:1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- 7.Huang YM, Hu W, Rustandi E, Chang K, Yusuf-Makagiansar H, Ryll T. Maximizing productivity of CHO cell-based fed-batch culture using chemically defined media conditions and typical manufacturing equipment. Biotechnol Prog. 2010;26:1400–1410. doi: 10.1002/btpr.436. [DOI] [PubMed] [Google Scholar]
- 8.Hacker DL, De Jesus M, Wurm FM. 25 years of recombinant proteins from reactor-grown cells - where do we go from here? Biotechnol Adv. 2009;27:1023–1027. doi: 10.1016/j.biotechadv.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Fischer S, Handrick R, Otte K. The art of CHO cell engineering: A comprehensive retrospect and future perspectives. Biotechnol Adv. 2015;33:1878–1896. doi: 10.1016/j.biotechadv.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 10.Seth G, Hossler P, Yee JC, Hu WS. Engineering Cells for Cell Culture Bioprocessing – Physiological Fundamentals. In Advances in Biochemical Engineering/Biotechnology. 2006:119–164. doi: 10.1007/10_017. [DOI] [PubMed] [Google Scholar]
- 11.Galleguillos SN, Ruckerbauer D, Gerstl MP, Borth N, Hanscho M, Zanghellini J. What can mathematical modelling say about CHO metabolism and protein glycosylation? Comput Struct Biotechnol J. 2017;15:212–221. doi: 10.1016/j.csbj.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hefzi H, Ang KS, Hanscho M, Bordbar A, Ruckerbauer D, Lakshmanan M, Orellana CA, Baycin-Hizal D, Huang Y, Ley D, et al. A Consensus Genome-scale Reconstruction of Chinese Hamster Ovary Cell Metabolism. Cell Syst. 2016;3:434–443.e8. doi: 10.1016/j.cels.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis AM, Abu-Absi NR, Borys MC, Li ZJ. The use of 'Omics technology to rationally improve industrial mammalian cell line performance. Biotechnol Bioeng. 2016;113:26–38. doi: 10.1002/bit.25673. [DOI] [PubMed] [Google Scholar]
- 14.Kildegaard HF, Baycin-Hizal D, Lewis NE, Betenbaugh MJ. The emerging CHO systems biology era: harnessing the 'omics revolution for biotechnology. Curr Opin Biotechnol. 2013;24:1102–1107. doi: 10.1016/j.copbio.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Lee JS, Grav LM, Lewis NE, Faustrup Kildegaard H. CRISPR/Cas9-mediated genome engineering of CHO cell factories: Application and perspectives. Biotechnol J. 2015;10:979–994. doi: 10.1002/biot.201500082. [DOI] [PubMed] [Google Scholar]
- 16.Grav LM, Lee JS, Gerling S, Kallehauge TB, Hansen AH, Kol S, Lee GM, Pedersen LE, Kildegaard HF. One-step generation of triple knockout CHO cell lines using CRISPR/Cas9 and fluorescent enrichment. Biotechnol J. 2015;10:1446–1456. doi: 10.1002/biot.201500027. [DOI] [PubMed] [Google Scholar]
- 17.Reinhart D, Damjanovic L, Kaisermayer C, Kunert R. Benchmarking of commercially available CHO cell culture media for antibody production. Appl Microbiol Biotechnol. 2015;99:4645–4657. doi: 10.1007/s00253-015-6514-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SY, Kim HU. Systems strategies for developing industrial microbial strains. Nat Biotechnol. 2015;33:1061–1072. doi: 10.1038/nbt.3365. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Menzel C, Meier D, Zhang C, Dübel S, Jostock T. A comparative study of different vector designs for the mammalian expression of recombinant IgG antibodies. J Immunol Methods. 2007;318:113–124. doi: 10.1016/j.jim.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Kober L, Zehe C, Bode J. Optimized signal peptides for the development of high expressing CHO cell lines. Biotechnol Bioeng. 2013;110:1164–1173. doi: 10.1002/bit.24776. [DOI] [PubMed] [Google Scholar]
- 21.Hacker DL, De Jesus M, Wurm FM. 25 years of recombinant proteins from reactor-grown cells - where do we go from here? Biotechnol Adv. 2009;27:1023–1027. doi: 10.1016/j.biotechadv.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Porter AJ, Dickson AJ, Racher AJ. Strategies for selecting recombinant CHO cell lines for cGMP manufacturing: realizing the potential in bioreactors. Biotechnol Prog. 2010;26:1446–1454. doi: 10.1002/btpr.442. [DOI] [PubMed] [Google Scholar]
- 23.Priola JJ, Calzadilla N, Baumann M, Borth N, Tate CG, Betenbaugh MJ. High-throughput screening and selection of mammalian cells for enhanced protein production. Biotechnol J. 2016;11:853–865. doi: 10.1002/biot.201500579. [DOI] [PubMed] [Google Scholar]
- 24.Droz X, Harraghy N, Lançon E, Le Fourn V, Calabrese D, Colombet T, Liechti P, Rida A, Girod PA, Mermod N. Automated microfluidic sorting of mammalian cells labeled with magnetic microparticles for those that efficiently express and secrete a protein of interest. Biotechnol Bioeng. 2017;114:1791–1802. doi: 10.1002/bit.26270. [DOI] [PubMed] [Google Scholar]
- 25.Carinhas N, Oliveira R, Alves PM, Carrondo MJT, Teixeira AP. Systems biotechnology of animal cells: the road to prediction. Trends Biotechnol. 2012;30:377–385. doi: 10.1016/j.tibtech.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Richelle A, Lewis NE. Improvements in protein production in mammalian cells from targeted metabolic engineering. Current Opinion in Systems Biology. 2017;6:1–6. doi: 10.1016/j.coisb.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jossé L, Smales CM, Tuite MF. Engineering the chaperone network of CHO cells for optimal recombinant protein production and authenticity. Methods Mol Biol. 2012;824:595–608. doi: 10.1007/978-1-61779-433-9_32. [DOI] [PubMed] [Google Scholar]
- 28.Hansen HG, Pristovšek N, Kildegaard HF, Lee GM. Improving the secretory capacity of Chinese hamster ovary cells by ectopic expression of effector genes: Lessons learned and future directions. Biotechnol Adv. 2017;35:64–76. doi: 10.1016/j.biotechadv.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Le Fourn V, Girod PA, Buceta M, Regamey A, Mermod N. CHO cell engineering to prevent polypeptide aggregation and improve therapeutic protein secretion. Metab Eng. 2014;21:91–102. doi: 10.1016/j.ymben.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 31.Yang Z, Wang S, Halim A, Schulz MA, Frodin M, Rahman SH, Vester-Christensen MB, Behrens C, Kristensen C, Vakhrushev SY, et al. Engineered CHO cells for production of diverse, homogeneous glycoproteins. Nat Biotechnol. 2015;33:842–844. doi: 10.1038/nbt.3280. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Yin B, Chung CY, Betenbaugh MJ. Glycoengineering of CHO Cells to Improve Product Quality. Methods Mol Biol. 2017;1603:25–44. doi: 10.1007/978-1-4939-6972-2_2. [DOI] [PubMed] [Google Scholar]
- 33.Chavez A, Tuttle M, Pruitt BW, Ewen-Campen B, Chari R, Ter-Ovanesyan D, Haque SJ, Cecchi RJ, Kowal EJK, Buchthal J, et al. Comparison of Cas9 activators in multiple species. Nat Methods. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orellana CA, Marcellin E, Schulz BL, Nouwens AS, Gray PP, Nielsen LK. High-antibody-producing Chinese hamster ovary cells up-regulate intracellular protein transport and glutathione synthesis. J Proteome Res. 2015;14:609–618. doi: 10.1021/pr501027c. [DOI] [PubMed] [Google Scholar]
- 36.Vishwanathan N, Yongky A, Johnson KC, Fu HY, Jacob NM, Le H, Yusufi FNK, Lee DY, Hu WS. Global insights into the Chinese hamster and CHO cell transcriptomes. Biotechnol Bioeng. 2015;112:965–976. doi: 10.1002/bit.25513. [DOI] [PubMed] [Google Scholar]
- 37.Kallehauge TB, Li S, Pedersen LE, Ha TK, Ley D, Andersen MR, Kildegaard HF, Lee GM, Lewis NE. Ribosome profiling-guided depletion of an mRNA increases cell growth rate and protein secretion. Sci Rep. 2017;(7):40388. doi: 10.1038/srep40388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Godfrey CL, Mead EJ, Daramola O, Dunn S, Hatton D, Field R, Pettman G, Smales CM. Polysome profiling of mAb producing CHO cell lines links translational control of cell proliferation and recombinant mRNA loading onto ribosomes with global and recombinant protein synthesis. Biotechnol J. 2017;12 doi: 10.1002/biot.201700177. [DOI] [PubMed] [Google Scholar]
- 39.Mohmad-Saberi SE, Hashim YZHY, Mel M, Amid A, Ahmad-Raus R, Packeer-Mohamed V. Metabolomics profiling of extracellular metabolites in CHO-K1 cells cultured in different types of growth media. Cytotechnology. 2013;65:577–586. doi: 10.1007/s10616-012-9508-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sellick CA, Croxford AS, Maqsood AR, Stephens GM, Westerhoff HV, Goodacre R, Dickson AJ. Metabolite profiling of CHO cells: Molecular reflections of bioprocessing effectiveness. Biotechnol J. 2015;10:1434–1445. doi: 10.1002/biot.201400664. [DOI] [PubMed] [Google Scholar]
- 41.Dietmair S, Hodson MP, Quek LE, Timmins NE, Chrysanthopoulos P, Jacob SS, Gray P, Nielsen LK. Metabolite profiling of CHO cells with different growth characteristics. Biotechnol Bioeng. 2012;109:1404–1414. doi: 10.1002/bit.24496. [DOI] [PubMed] [Google Scholar]
- 42.Mulukutla BC, Kale J, Kalomeris T, Jacobs M, Hiller GW. Identification and control of novel growth inhibitors in fed-batch cultures of Chinese hamster ovary cells. Biotechnol Bioeng. 2017;114:1779–1790. doi: 10.1002/bit.26313. [DOI] [PubMed] [Google Scholar]
- 43.Dinnis DM, James DC. Engineering mammalian cell factories for improved recombinant monoclonal antibody production: lessons from nature? Biotechnol Bioeng. 2005;91:180–189. doi: 10.1002/bit.20499. [DOI] [PubMed] [Google Scholar]
- 44.Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016;167:1867–1882.e21. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lund AM, Kaas CS, Brandl J, Pedersen LE, Kildegaard HF, Kristensen C, Andersen MR. Network reconstruction of the mouse secretory pathway applied on CHO cell transcriptome data. BMC Syst Biol. 2017;(11):37. doi: 10.1186/s12918-017-0414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feizi A, Gatto F, Uhlen M, Nielsen J. Human protein secretory pathway genes are expressed in a tissue-specific pattern to match processing demands of the secretome. NPJ Syst Biol Appl. 2017;(3):22. doi: 10.1038/s41540-017-0021-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiang AW, Li S, Spahn PN, Richelle A, Kuo CC, Samoudi M, Lewis NE. Modulating carbohydrate-protein interactions through glycoengineering of monoclonal antibodies to impact cancer physiology. Curr Opin Struct Biol. 2016;40:104–111. doi: 10.1016/j.sbi.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L, Luo S, Zhang B. The use of lectin microarray for assessing glycosylation of therapeutic proteins. MAbs. 2016;8:524–535. doi: 10.1080/19420862.2016.1149662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Brien EJ, Monk JM, Palsson BO. Using Genome-scale Models to Predict Biological Capabilities. Cell. 2015;161:971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yusufi FNK, Lakshmanan M, Ho YS, Loo BLW, Ariyaratne P, Yang Y, Ng SK, Tan TRM, Yeo HC, Lim HL, et al. Mammalian Systems Biotechnology Reveals Global Cellular Adaptations in a Recombinant CHO Cell Line. Cell Syst. 2017;4:530–542.e6. doi: 10.1016/j.cels.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 52.Almquist J, Cvijovic M, Hatzimanikatis V, Nielsen J, Jirstrand M. Kinetic models in industrial biotechnology – Improving cell factory performance. Metab Eng. 2014;24:38–60. doi: 10.1016/j.ymben.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 53.Spahn PN, Lewis NE. Systems glycobiology for glycoengineering. Curr Opin Biotechnol. 2014;30:218–224. doi: 10.1016/j.copbio.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 54.Krambeck FJ, Bennun SV, Andersen MR, Betenbaugh MJ. Model-based analysis of N-glycosylation in Chinese hamster ovary cells. PLoS One. 2017;12:e0175376. doi: 10.1371/journal.pone.0175376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spahn PN, Hansen AH, Hansen HG, Arnsdorf J, Kildegaard HF, Lewis NE. A Markov chain model for N-linked protein glycosylation--towards a low-parameter tool for model-driven glycoengineering. Metab Eng. 2016;33:52–66. doi: 10.1016/j.ymben.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spahn PN, Hansen AH, Kol S, Voldborg BG, Lewis NE. Predictive glycoengineering of biosimilars using a Markov chain glycosylation model. Biotechnol J. 2017;12 doi: 10.1002/biot.201600489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jimenez Del Val I, Fan Y, Weilguny D. Dynamics of immature mAb glycoform secretion during CHO cell culture: An integrated modelling framework. Biotechnol J. 2016;11:610–623. doi: 10.1002/biot.201400663. [DOI] [PubMed] [Google Scholar]
- 58.Clarke C, Doolan P, Barron N, Meleady P, O'Sullivan F, Gammell P, Melville M, Leonard M, Clynes M. Predicting cell-specific productivity from CHO gene expression. J Biotechnol. 2011;151:159–165. doi: 10.1016/j.jbiotec.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 59.Pybus LP, James DC, Dean G, Slidel T, Hardman C, Smith A, Daramola O, Field R. Predicting the expression of recombinant monoclonal antibodies in Chinese hamster ovary cells based on sequence features of the CDR3 domain. Biotechnol Prog. 2014;30:188–197. doi: 10.1002/btpr.1839. [DOI] [PubMed] [Google Scholar]
- 60.Golabgir A, Gutierrez JM, Hefzi H, Li S, Palsson BO, Herwig C, Lewis NE. Quantitative feature extraction from the Chinese hamster ovary bioprocess bibliome using a novel meta-analysis workflow. Biotechnol Adv. 2016;34:621–633. doi: 10.1016/j.biotechadv.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 61.Le H, Kabbur S, Pollastrini L, Sun Z, Mills K, Johnson K, Karypis G, Hu WS. Multivariate analysis of cell culture bioprocess data--lactate consumption as process indicator. J Biotechnol. 2012;162:210–223. doi: 10.1016/j.jbiotec.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 62.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning Edited by Hastie T, Tibshirani R, Friedman J. Springer; New York: 2009. High-Dimensional Problems: p N; pp. 649–698. [Google Scholar]
- 63.Gerstl MP, Hanscho M, Ruckerbauer DE, Zanghellini J, Borth N. CHOmine: an integrated data warehouse for CHO systems biology and modeling. Database. 2017;2017 doi: 10.1093/database/bax034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kremkow BG, Baik JY, MacDonald ML, Lee KH. CHOgenome.org 2.0: Genome resources and website updates. Biotechnol J. 2015;10:931–938. doi: 10.1002/biot.201400646. [DOI] [PubMed] [Google Scholar]
- 65.Opdam S, Richelle A, Kellman B, Li S, Zielinski DC, Lewis NE. A Systematic Evaluation of Methods for Tailoring Genome-Scale Metabolic Models. Cell Syst. 2017;4:318–329.e6. doi: 10.1016/j.cels.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon K, Klepper K, Pekin D, Beckett AN, et al. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat Methods. 2017;14:573–576. doi: 10.1038/nmeth.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: The classification of the CHO bibliome into the 3 different waves according to the types of phenotypic and bioprocess data contained therein.


