Abstract
In medical sciences, a disease condition is typically associated with multiple risk and protective factors. Although many studies report results of multiple factors, nearly all meta-analyses separately synthesize the association between each factor and the disease condition of interest. The collected studies usually report different subsets of factors, and the results from separate analyses on multiple factors may not be comparable because each analysis may use different subpopulation. This may impact on selecting most important factors to design a multi-factor intervention program. This article proposes a new concept, multivariate meta-analysis of multiple factors (MVMA-MF), to synthesize all available factors simultaneously. By borrowing information across factors, MVMA-MF can improve statistical efficiency and reduce biases compared with separate analyses when factors were missing not at random. As within-study correlations between factors are commonly unavailable from published articles, we use a Bayesian hybrid model to perform MVMA-MF, which effectively accounts for both within- and between-study correlations. The performance of MVMA-MF and the conventional methods are compared using simulations and an application to a pterygium dataset consisting of 29 studies on 8 risk factors.
Keywords: Bayesian hybrid model, Missing data, Multiple factors, Multivariate meta-analysis, Within-study correlation
1. Introduction
As a disease condition is typically associated with many risk and protective factors in medical sciences, many randomized controlled trials and observational studies considered multiple factors.1–4 Reliable summary of association between each factor and the disease condition is crucial for the design of a multi-factor intervention program. The growth of interest in evidence-based medicine has led to a dramatic increase in attention paid to systematic reviews and meta-analyses. In prevention studies, it has become increasingly popular to perform meta-analyses on multiple risk and protective factors to summarize existing evidence; however, currently, nearly all meta-analyses are performed on each factor separately.5–7 Different studies usually focus on different subsets of all risk and protective factors, and may only selectively report some significant factors in peer-reviewed articles; some factors may be reported by only a few studies. Hence, if we organize the collected data in a matrix with rows and columns indexing studies and factors respectively, then the data matrix is expected to contain many missing entries; see the example in Table 1. The conventional meta-analysis separately estimates each factor’s association with the disease condition, so it cannot borrow information across the factors. Moreover, results from separate meta-analyses may not be directly comparable because they may be based on different subpopulations. This limits medical investigators to select most important factors for the design of a multi-factor intervention program.
Table 1.
The pterygium data containing 29 studies with 8 risk factors. The effect size is log odds ratio with within-study standard error in parentheses. The blank entries indicate that the risk factors are unavailable from the corresponding studies.
| Risk factor | ||||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Study | (1) | (2) | (3) | (4) | (5) | (6) | (7) | (8) |
| 1 | −0.08 (0.34) | |||||||
| 2 | 1.54 (0.10) | |||||||
| 3 | 0.28 (0.17) | 0.53 (0.10) | 0.53 (0.13) | |||||
| 4 | 0.45 (0.11) | 0.41 (0.10) | 0.97 (0.23) | |||||
| 5 | 0.30 (0.40) | |||||||
| 6 | 0.12 (0.40) | 0.48 (0.70) | ||||||
| 7 | 1.40 (0.23) | |||||||
| 8 | 0.39 (0.13) | 0.05 (0.13) | ||||||
| 9 | 0.55 (0.22) | −0.04 (0.26) | ||||||
| 10 | 3.04 (1.03) | |||||||
| 11 | 1.95 (0.40) | 1.21 (0.42) | 0.67 (0.36) | −1.34 (0.34) | 0.12 (0.31) | |||
| 12 | 1.10 (0.30) | |||||||
| 13 | 2.03 (0.39) | −0.69 (0.21) | −1.14 (0.38) | −1.64 (0.21) | 2.99 (0.74) | |||
| 14 | 0.83 (0.09) | 0.11 (0.09) | 0.91 (0.09) | −0.58 (0.12) | 1.14 (0.13) | |||
| 15 | 0.41 (0.22) | |||||||
| 16 | −0.20 (0.24) | 1.73 (0.18) | ||||||
| 17 | 0.42 (0.15) | −0.11 (0.25) | 0.38 (0.21) | 0.22 (0.15) | −0.64 (0.15) | −0.52 (0.37) | ||
| 18 | 0.63 (0.12) | 0.03 (0.05) | 1.24 (0.24) | |||||
| 19 | 0.89 (0.08) | −0.08 (0.07) | 1.70 (0.08) | −0.17 (0.08) | −0.06 (0.14) | |||
| 20 | 0.39 (0.22) | −0.13 (0.30) | ||||||
| 21 | 0.90 (0.30) | 0.85 (0.49) | ||||||
| 22 | −0.48 (0.25) | 0.31 (0.25) | 0.87 (0.20) | |||||
| 23 | 0.66 (0.26) | |||||||
| 24 | −0.46 (0.72) | −0.73 (0.52) | ||||||
| 25 | 0.76 (0.34) | −0.48 (0.55) | 1.05 (0.33) | |||||
| 26 | 0.03 (0.11) | 0.19 (0.09) | ||||||
| 27 | 1.16 (0.36) | 0.53 (0.23) | 0.83 (0.25) | |||||
| 28 | 0.12 (0.03) | 0.14 (0.20) | 1.43 (0.28) | 1.43 (0.22) | ||||
| 29 | 0.41 (0.09) | −0.26 (0.10) | 0.46 (0.09) | |||||
Risk factors: (1) occupation type; (2) smoking status; (3) education attainment; (4) use of hat; (5) use of spectacles; (6) area of residence; (7) use of sunglasses; and (8) latitude of residence.
Recently, Serghiou et al.8 introduced field-wide systematic review and meta-analysis to report and assess the entire field of putative factors for a disease condition. Based on this concept, researchers can learn the selective availability and different adjustments of factors and the patterns of modeling. Although multiple factors were collected, the authors pooled the results for each factor separately; this is not efficient to analyze the multivariate data from a field-wide systematic review.
This article proposes multivariate meta-analysis of multiple factors (MVMA-MF) to jointly synthesize all risk and protective factors in the field-wide systematic review. Compared with separate meta-analyses that use different subpopulations, the results produced by jointly modeling of multiple factors have better interpretation and generalizability because they are based on the whole population in the multivariate meta-analysis. Multivariate meta-analysis methods have gained much attention in the recent literature9–12, and they improve effect estimates by borrowing information from the correlations between multiple endpoints.13–15 The multivariate meta-analysis models have been applied to several areas, such as meta-analysis of diagnostic tests16–18, meta-analysis of multiple outcomes19,20, and network meta-analysis of mixed treatment comparisons.21–26 Mixed treatment comparisons use both direct and indirect evidence of treatment contrasts to synthesize the comparisons between multiple treatments; its focus differs from MVMA-MF, because MVMA-MF is interested in estimating the effect of multiple factors, not the contrasts between them.
Multivariate random-effects model generally requires correlations within each collected study. In some situations, within-study correlations are known to be zero. For example, in meta-analysis of diagnostic tests, the study-specific sensitivity and specificity are uncorrelated within studies because they are calculated from the true negative and true positive patients respectively.10 However, in MVMA-MF, the factors can be correlated within each study because they may be measured on same patients. Such within-study correlations are unknown unless individual patient data are available. Ignoring within-study correlations in the standard multivariate random-effects model may have great impact on the estimated overall effect sizes.27
To deal with unknown within-study correlations, this article considers an alternative Bayesian model for MVMA-MF. The conventional multivariate model partitions the overall variance-covariance matrix into two parts: the within-study level that is due to sampling error and the between-study level that is due to heterogeneity between the collected studies. Instead of partitioning the overall correlations into the two levels, the alternative model directly specifies one single overall correlation matrix; hence, it may be viewed as a hybrid approach. This model is the Bayesian version of the model introduced by Riley et al.28 Currently, Riley’s model is implemented in a frequentist way, such as using the restricted maximum likelihood method.29 However, the data of MVMA-MF are usually fairly sparse (Table 1), and our simulations in Section 5 show that the frequentist method generated poor 95% confidence intervals for sparse data; also, the algorithm for maximizing the (restricted) likelihood did not converge for many simulated data. Instead of using the frequentist method, we present a fully Bayesian method to perform MVMA-MF. Although Bayesian methods have been introduced to deal with multivariate meta-analyses by Lu and Ades30 and Wei and Higgins31, the former paper considered mixed treatment comparisons which focused on treatment contrasts, and the latter assumed known within-study covariance matrices. Hence, both Bayesian methods may not be directly applied to MVMA-MF. The Bayesian version of Riley’s model will be detailed in Section 4. We will use simulations and reanalyze the real data in Serghiou et al.8 to demonstrate the benefit of the joint Bayesian model for MVMA-MF, and we will conclude this article with brief discussion.
2. The motivating pterygium data
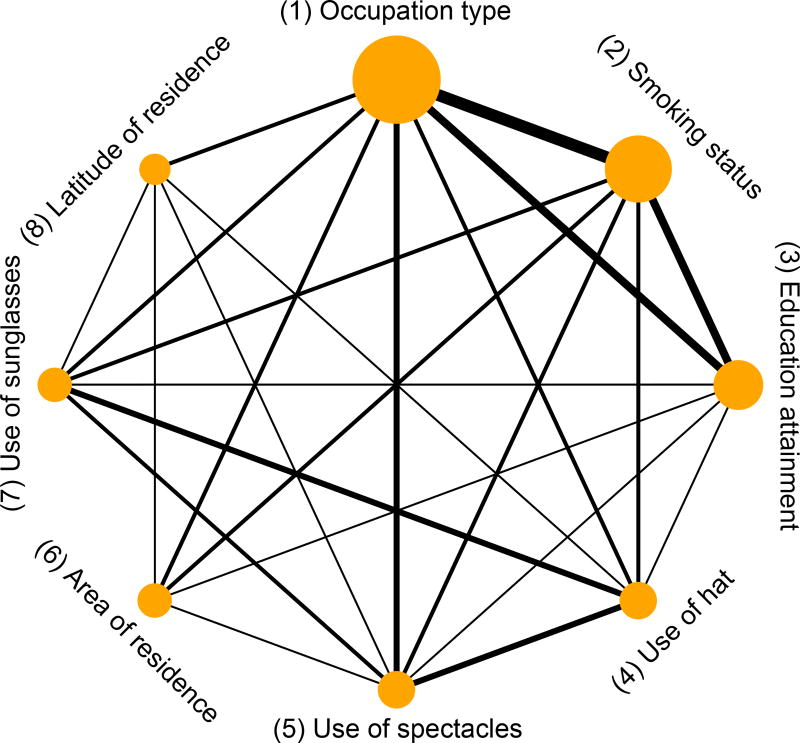
Pterygium is one of the most common eye conditions, and it may be related to many risk factors.32 Instead of reporting only one risk factor at a time, Serghiou et al.8 collected the odds ratios of all putative risk factors for pterygium. Specifically, they identified 60 eligible studies reporting on a total of 65 risk factors. Since most risk factors were only reported in less than 3 studies, we focus on the following 8 risk factors, each of which was reported in at least 4 studies: (1) occupation type (outdoor vs. indoor); (2) smoking status (yes vs. no); (3) education attainment (low vs. high); (4) use of hat (yes vs. no); (5) use of spectacles (yes vs. no); (6) area of residence (rural vs. urban); (7) use of sunglasses (yes vs. no); and (8) latitude of residence (low vs. high). These risk factors are sorted according to their frequencies reported in the collected studies (from high to low). Also, we cleaned the data by removing the log odds ratios that were obtained using multivariate regression model, because they were adjusted for different risk factors in different studies. Table 1 presents the cleaned data and Figure 1 shows the network plot of the 8 risk factors. The network indicates that most pairs of risk factors are simultaneously reported in some studies, but several pairs, such as area of residence and use of hat, are not. From Table 1, the risk factor latitude of residence was reported in only 4 studies, while 23 studies reported occupation type. Most studies reported different subsets of the 8 risk factors, and many entries in Table 1 are missing. The estimated overall effect sizes produced by univariate models may be poor because some risk factors have few samples. Also, many factors (e.g., area of residence and education attainment) are expected to be correlated, so a multivariate model may be more appropriate for this dataset than univariate models.
Figure 1.
Network plot of the pterygium data. The nodes represent the risk factors, and the edge between two nodes indicate that these nodes are simultaneously reported in common studies. The node size is proportional to the number of studies that report the corresponding risk factor, and the edge thickness is proportional to the number of studies that simultaneously report the corresponding two risk factors.
3. Conventional meta-analysis models
This section reviews some existing models for general multivariate meta-analysis. Suppose that n independent studies are collected; each study reports a p-dimensional vector of effect sizes, denoted as yi = (yi1, …, yip)T, and denote its within-study variance-covariance matrix as Si (i = 1, …, n). The conventional univariate meta-analysis pools the results for each j = 1, …, p separately; a fixed- or random-effects model is applied to the data , where vij is the within-study variance, i.e., the jth diagonal element in Si.33,34 Since most studies are conducted by different research teams in different places using different methods, the studies are usually expected to be heterogeneous.35 Also, the random-effects model may produce more conservative results than the fixed-effects model36,37, so this article focuses on the random-effects setting that accounts for the heterogeneity between studies. We denote the univariate model as Model U, which ignores both within- and between-study correlations.
Multivariate meta-analysis recently gains much attention to simultaneously synthesize the p-dimensional effect sizes.9–11 Given that the within-study variance-covariance matrices Si are known, the commonly used random-effects model is specified as follows to analyze the multivariate data :
where μi represents study i’s underlying true effect sizes, μ = (μ1, …, μp)T contains the overall effect sizes, and T = (τij) is the p × p between-study variance-covariance matrix. We denote this multivariate model as Model M. Note that within-study variances are routinely reported in published articles, but within-study correlations are usually unavailable. Let Di = diag(Si) be the diagonal matrix consisting of the within-study variances. To analyze the data when the within-study correlations are unknown, a naïve multivariate method is to simply ignore these correlations by setting them to zero but still account for the between-study correlations; we denote this model as Model M0. Nevertheless, ignoring within-study correlations could lead to poor estimated effect sizes, especially when the within-study correlations are comparable to or greater than the between-study correlations.27 The following section introduces an alternative model that can incorporate both within- and between-study correlations for MVMA-MF.
4. Multivariate meta-analysis of multiple factors
4.1 Multivariate hybrid meta-analysis model
Model M may be deemed ideal to perform MVMA-MF: it uses the within-study correlations between multiple factors that are usually unknown, and it provides a benchmark for the performance of other potential models. Note that study i’s marginal variance-covariance matrix in Model M is Mi = Si + T, and it can be written as Mi = (Di + Δ)1/2Ri(Di + Δ)1/2, where contains between-study variances (i.e., diagonal elements in T), and Di is the diagonal matrix containing within-study variances (i.e., diagonal elements in Si). The marginal correlation matrix of study i, Ri, is determined by both Si and T, and thus it needs to be estimated if the within-study correlations are unknown. It may be inefficient to use the data from merely n studies to simultaneously estimate all Ri’s, which involve too many parameters.
Alternatively, extending the bivariate model in Riley et al.28, we consider a multivariate model that does not require within-study correlations to perform MVMA-MF:
where is a diagonal matrix that consists of additional variances beyond sampling error due to between-study heterogeneity for the p effect sizes. In this model, all collected studies are assumed to share a common marginal correlation matrix R. This assumption effectively reduces the number of parameters to be estimated and accounts for both within- and between-study correlations. Like Model M, the alternative model partitions the marginal variances of yi, Di + Ψ, into the within- and between-study levels; however, it directly uses the matrix R to model the overall correlations, instead of partitioning the correlations into the previous two levels. Therefore, the alternative model may be deemed hybrid, and we denote it as Model H.
4.2 Missing data
So far, the models for complete data have been discussed. In MVMA-MF, each collected study often reports a small subset of the complete factors, and many factors are missing as in Table 1. It is straightforward to extend the four methods (i.e., Models U, M, M0, and H) to deal with missing data under the assumption of missing at random. Model H for missing data will be detailed here. Suppose that ỹi = (ỹi1, …, ỹip)T contains the complete p factors in study i; however, we only observe ti factors and their within-study variances (1 ≤ ti ≤ p). Denote the effect sizes of the ti factors as yi = (yi1, …, yiti)T, which is a ti-dimensional sub-vector of ỹi, and let Di be the ti × ti diagonal matrix containing the within-study variances. We write yi = Xiỹi, where Xi = (ei1, …, eiti)T is a ti × p matrix indicating missingness.31 Specifically, for each j = 1, …, ti, eij = (eij1, …, eijp)T with eijk = 1 if the observed yij is the effect size of factor k and eijk = 0 otherwise. For example, for study 3 in the pterygium data (Table 1),
Recall that μ, Ψ, and R represent the overall mean effect sizes, the between-study variances, and the marginal correlation matrix for the complete p factors, respectively. Hence, for the observed ti-dimensional vector yi, its overall mean is Xi μ, its marginal variances form the diagonal matrix , and its marginal correlation matrix is . Consequently, the hybrid model for missing data can be specified as
Although the derivation here is based on the assumption of missing at random38, the simulations in Section 5 will compare the performance of Model H with Models M, M0, and U when some factors are missing under various mechanisms (including missing not at random).
4.3 Bayesian hybrid model
Currently the existing statistical software, such as the Stata command “mvmeta”, can only implement Model H in a frequentist way.28,29 However, when the dimension of factors p is large compared with the number of collected studies, the estimated variance-covariance matrix using the frequentist method may be inconsistent39, leading to poor interval estimates. Indeed, the simulations in Section 5 show that the frequentist method produced poor 95% confidence intervals when the data of MVMA-MF were sparse, and the algorithm for maximizing the (restricted) likelihood did not converge for many simulated data. Therefore, we use a fully Bayesian approach to estimating the overall multivariate effect size μ and its variance-covariance matrix that are of interest in Model H. Vague priors are assigned to both the mean and variance-covariance structures. The Supporting Information provides the details of implementing the Bayesian MVMA-MF and its R and JAGS code. The simulations in Section 5 indicate that the 95% CrIs obtained using the Bayesian method generally had higher coverage probabilities than those obtained using the frequentist method for sparse data, which are common in MVMA-MF.
5. Simulations
We conducted simulations under various settings to compare the performance of the hybrid model (Model H) with the ideal model that uses within-study correlations (Model M), the model that ignores within-study correlations but accounts for between-study correlations (Model M0), and the univariate model that ignores both types of correlations (Model U). Bias and root mean squared error (RMSE) of point estimate and 95% CI/CrI coverage probability were used to evaluate the models’ performance. We set the number of studies in each simulated MVMA-MF dataset to 30 and considered 5 factors in total. The true overall effect sizes of the 5 factors were zero, i.e., μ = (0, 0, 0, 0, 0)T. Also, the between-study standard deviation τ was 1 for each factor; the within-study standard deviation of each factor σ was set to 0.5, 1, or 2. These choices for σ represented different extents of heterogeneity between studies. Moreover, we considered the exchangeable correlation structure for both the between- and within-study correlation matrices, RB = (rBij) and RW = (rWij), which were determined by the correlation parameters ρB and ρW, respectively; that is, rBij = ρB and rWij = ρW for 1 ≤ i ≠ j ≤ 5. The between-study correlations ρB were 0.2, 0.5, or 0.8, and the within-study correlations ρW were drawn from U(0, 0.3), U(0.3, 0.6), or U(0.6, 0.9). For each setting, 1,000 MVMA-MF datasets were simulated using the ideal Model M with Si = σ2RW and T = τ2RB. Three missingness scenarios were considered: (I) all 5 factors were observed in all studies, i.e., the data were complete; (II) the data of factors 1, 3, and 5 in 10 studies were missing completely at random; and (III) the smallest 10 effect sizes of factors 1, 3, and 5 were missing. The missingness that was not at random in scenario (III) may be viewed as the effect of reporting bias. We also considered a missingness scenario that was similar to (III) but contained more missing data: (III′) the smallest 25 effect sizes of factors 1, 3, and 5 were missing; in this case, the three factors were only available from 5 studies, so the simulated MVMA-MF data were much sparser than the previous scenarios. Both the REML and Bayesian methods were applied to implement the four models. For the Bayesian analyses, we used the priors specified in Section S1.2 of the Supporting Information. The Markov chain Monte Carlo (MCMC) algorithm was used for estimation with one chain, having a run of 10,000 updates after a 10,000-run burn-in period.
Table 2 presents the results when ρB was 0.5, and Tables S1 and S2 in the Supporting Information present those when ρB was 0.2 and 0.8, respectively. First, since the data of factors 2 and 4 were complete under all scenarios, their corresponding results produced by the four models were almost the same; all models led to nearly unbiased estimates and proper 95% CI/CrI coverage probabilities for these two factors. Second, the results of factors 1, 3, and 5 produced by the four models also differed little when the data were complete or missing completely at random. This is expected from the perspective of missing data analysis.38 Third, if the missingness was not at random, the results produced by the four models were noticeably different. The univariate model led to the largest bias and RMSE and the lowest 95% CI/CrI coverage probability. Since Model M0 still accounted for the between-study correlations, its performance was similar to Model H when ρW was small compared to ρB. However, Model H outperformed Model M0 when ρW was larger than ρB, because the within-study level dominated the estimation of the overall effect sizes in this situation, but Model M0 ignored correlations at this level. Finally, note that Model M was ideal as it used the within-study correlations that are usually unavailable in real world. Although Model H did not use the within-study correlations, the biases and RMSEs produced by Model H were fairly close to the ideal Model M under various settings. Also, the 95% CI/CrI coverage probability produced by Model H was generally higher than those produced by Models M0 and U.
Table 2.
The simulation results produced by Models M, H, M0, and U in various settings of within-study variances and correlations under different missingness scenarios. The results outside parentheses were obtained using the Bayesian method; those inside parentheses were obtained using the restricted maximum likelihood method.
| Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 5 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|||||||||||
| Model | Bias | RMSE | CP (%) | Bias | RMSE | CP (%) | Bias | RMSE | CP (%) | Bias | RMSE | CP (%) | Bias | RMSE | CP (%) |
| τ = 1, σ = 1, ρB = 0.5, ρW ~ U(0.6, 0.9): | |||||||||||||||
| No missing data (I) | |||||||||||||||
| M | 0.00 (0.00) | 0.26 (0.26) | 94 (94) | 0.00 (0.00) | 0.26 (0.26) | 92 (93) | 0.00 (0.00) | 0.25 (0.25) | 94 (95) | 0.01 (0.01) | 0.26 (0.26) | 93 (93) | 0.01 (0.01) | 0.25 (0.25) | 94 (95) |
| H | 0.00 (0.00) | 0.26 (0.26) | 95 (94) | 0.00 (0.00) | 0.26 (0.26) | 95 (93) | 0.00 (0.00) | 0.25 (0.25) | 96 (95) | 0.01 (0.01) | 0.26 (0.26) | 95 (93) | 0.01 (0.01) | 0.25 (0.25) | 96 (94) |
| M0 | 0.00 (0.00) | 0.26 (0.26) | 96 (95) | 0.00 (0.00) | 0.26 (0.26) | 96 (94) | 0.00 (0.00) | 0.25 (0.25) | 97 (96) | 0.01 (0.01) | 0.26 (0.26) | 96 (95) | 0.01 (0.01) | 0.25 (0.25) | 97 (96) |
| U | 0.00 (0.00) | 0.26 (0.26) | 95 (94) | 0.00 (0.00) | 0.26 (0.26) | 94 (94) | 0.00 (0.00) | 0.25 (0.25) | 96 (95) | 0.01 (0.01) | 0.26 (0.26) | 95 (94) | 0.01 (0.01) | 0.25 (0.25) | 96 (95) |
| Factors 1, 3, and 5 in 10 studies were missing completely at random (II) | |||||||||||||||
| M | 0.00 (0.00) | 0.29 (0.30) | 95 (93) | 0.00 (0.00) | 0.26 (0.26) | 93 (94) | 0.01 (0.00) | 0.28 (0.30) | 95 (93) | 0.01 (0.01) | 0.26 (0.26) | 94 (93) | 0.01 (0.01) | 0.28 (0.29) | 94 (93) |
| H | 0.00 (0.00) | 0.29 (0.31) | 96 (92) | 0.00 (0.00) | 0.26 (0.26) | 94 (93) | 0.01 (0.00) | 0.29 (0.30) | 96 (92) | 0.01 (0.01) | 0.26 (0.26) | 95 (93) | 0.00 (0.01) | 0.28 (0.30) | 96 (92) |
| M0 | 0.00 (0.00) | 0.29 (0.30) | 97 (96) | 0.00 (0.00) | 0.26 (0.26) | 95 (94) | 0.01 (0.00) | 0.28 (0.29) | 98 (95) | 0.01 (0.01) | 0.26 (0.26) | 96 (94) | 0.01 (0.01) | 0.28 (0.29) | 97 (96) |
| U | 0.00 (−0.01) | 0.32 (0.32) | 95 (94) | 0.00 (0.00) | 0.26 (0.26) | 94 (94) | 0.02 (−0.01) | 0.31 (0.32) | 96 (94) | 0.01 (0.01) | 0.26 (0.26) | 95 (94) | 0.01 (0.01) | 0.31 (0.32) | 95 (93) |
| Factors 1, 3, and 5 in 10 studies were missing not at random (III) | |||||||||||||||
| M | 0.48 (0.46) | 0.56 (0.54) | 54 (51) | 0.00 (0.00) | 0.26 (0.26) | 94 (94) | 0.48 (0.46) | 0.55 (0.54) | 52 (48) | 0.01 (0.01) | 0.26 (0.26) | 94 (94) | 0.48 (0.47) | 0.56 (0.55) | 51 (47) |
| H | 0.51 (0.49) | 0.58 (0.57) | 54 (43) | 0.00 (0.00) | 0.26 (0.26) | 96 (94) | 0.52 (0.48) | 0.59 (0.56) | 49 (41) | 0.01 (0.01) | 0.26 (0.26) | 95 (94) | 0.53 (0.50) | 0.60 (0.57) | 48 (41) |
| M0 | 0.58 (0.56) | 0.64 (0.62) | 53 (44) | 0.00 (0.00) | 0.26 (0.26) | 95 (94) | 0.61 (0.55) | 0.67 (0.62) | 46 (41) | 0.01 (0.01) | 0.26 (0.26) | 95 (94) | 0.62 (0.56) | 0.67 (0.63) | 46 (41) |
| U | 0.75 (0.75) | 0.80 (0.80) | 19 (16) | 0.00 (0.00) | 0.26 (0.26) | 94 (94) | 0.75 (0.75) | 0.80 (0.80) | 18 (14) | 0.01 (0.01) | 0.26 (0.26) | 94 (94) | 0.75 (0.75) | 0.80 (0.80) | 17 (13) |
|
| |||||||||||||||
| τ = 1, σ = 1, ρB = 0.5, ρW ~ U(0, 0.3): | |||||||||||||||
| Factors 1, 3, and 5 in 10 studies were missing not at random (III) | |||||||||||||||
| M | 0.69 (0.68) | 0.74 (0.74) | 26 (23) | 0.01 (0.01) | 0.26 (0.26) | 95 (94) | 0.70 (0.68) | 0.75 (0.73) | 23 (22) | 0.02 (0.02) | 0.26 (0.26) | 94 (93) | 0.70 (0.68) | 0.75 (0.74) | 25 (22) |
| H | 0.68 (0.67) | 0.74 (0.73) | 30 (21) | 0.01 (0.01) | 0.26 (0.26) | 96 (94) | 0.69 (0.67) | 0.74 (0.73) | 27 (21) | 0.02 (0.02) | 0.26 (0.26) | 95 (93) | 0.69 (0.67) | 0.74 (0.73) | 30 (21) |
| M0 | 0.71 (0.69) | 0.76 (0.75) | 26 (22) | 0.01 (0.01) | 0.26 (0.26) | 95 (94) | 0.72 (0.70) | 0.77 (0.75) | 24 (21) | 0.02 (0.02) | 0.26 (0.26) | 94 (93) | 0.72 (0.70) | 0.77 (0.75) | 25 (22) |
| U | 0.75 (0.75) | 0.80 (0.80) | 19 (16) | 0.01 (0.01) | 0.26 (0.26) | 95 (94) | 0.75 (0.75) | 0.80 (0.80) | 16 (12) | 0.02 (0.02) | 0.26 (0.26) | 94 (93) | 0.75 (0.75) | 0.80 (0.80) | 18 (14) |
|
| |||||||||||||||
| τ = 1, σ = 0.5, ρB = 0.5, ρW ~ U(0.6, 0.9): | |||||||||||||||
| Factors 1, 3, and 5 in 10 studies were missing not at random (III) | |||||||||||||||
| M | 0.47 (0.44) | 0.52 (0.50) | 36 (31) | 0.00 (0.00) | 0.21 (0.21) | 95 (93) | 0.46 (0.43) | 0.51 (0.49) | 35 (30) | 0.01 (0.01) | 0.21 (0.21) | 95 (93) | 0.47 (0.45) | 0.52 (0.50) | 33 (28) |
| H | 0.46 (0.45) | 0.51 (0.50) | 43 (30) | 0.00 (0.00) | 0.21 (0.21) | 96 (93) | 0.46 (0.44) | 0.51 (0.49) | 39 (29) | 0.01 (0.01) | 0.21 (0.21) | 96 (93) | 0.47 (0.45) | 0.52 (0.50) | 37 (26) |
| M0 | 0.45 (0.45) | 0.51 (0.51) | 39 (31) | 0.00 (0.00) | 0.21 (0.21) | 95 (93) | 0.46 (0.44) | 0.51 (0.49) | 37 (30) | 0.01 (0.01) | 0.21 (0.21) | 95 (93) | 0.47 (0.46) | 0.52 (0.51) | 35 (27) |
| U | 0.60 (0.59) | 0.64 (0.64) | 15 (13) | 0.00 (0.00) | 0.21 (0.21) | 95 (93) | 0.59 (0.59) | 0.63 (0.63) | 14 (12) | 0.01 (0.01) | 0.21 (0.21) | 94 (93) | 0.60 (0.60) | 0.64 (0.64) | 14 (12) |
|
| |||||||||||||||
| τ = 1, σ = 2, ρB = 0.5, ρW ~ U(0.6, 0.9): | |||||||||||||||
| Factors 1, 3, and 5 in 10 studies were missing not at random (III) | |||||||||||||||
| M | 0.57 (0.58) | 0.73 (0.74) | 74 (70) | 0.01 (0.01) | 0.41 (0.42) | 96 (96) | 0.59 (0.59) | 0.74 (0.74) | 73 (70) | 0.02 (0.02) | 0.41 (0.41) | 95 (96) | 0.59 (0.59) | 0.74 (0.74) | 73 (70) |
| H | 0.63 (0.60) | 0.78 (0.76) | 74 (66) | 0.01 (0.01) | 0.42 (0.42) | 97 (96) | 0.65 (0.61) | 0.80 (0.76) | 72 (65) | 0.02 (0.02) | 0.41 (0.41) | 97 (97) | 0.67 (0.62) | 0.81 (0.77) | 71 (64) |
| M0 | 0.96 (0.90) | 1.05 (1.00) | 62 (56) | 0.01 (0.01) | 0.42 (0.42) | 97 (96) | 1.01 (0.90) | 1.10 (1.00) | 56 (55) | 0.02 (0.02) | 0.41 (0.41) | 97 (97) | 1.02 (0.91) | 1.11 (1.01) | 56 (54) |
| U | 1.18 (1.18) | 1.26 (1.26) | 29 (25) | 0.01 (0.01) | 0.42 (0.42) | 96 (94) | 1.19 (1.19) | 1.27 (1.27) | 28 (24) | 0.02 (0.02) | 0.41 (0.41) | 96 (94) | 1.19 (1.19) | 1.27 (1.27) | 28 (24) |
| Factors 1, 3, and 5 in 25 studies were missing not at random (III′) | |||||||||||||||
| M | 1.57 (1.60) | 1.72 (1.76) | 76 (32) | 0.01 (−0.01) | 0.42 (0.41) | 95 (95) | 1.58 (1.60) | 1.73 (1.77) | 75 (31) | 0.02 (0.02) | 0.41 (0.42) | 96 (94) | 1.60 (1.59) | 1.74 (1.76) | 75 (34) |
| H | 2.36 (1.46) | 2.75 (1.64) | 65 (26) | 0.01 (−0.02) | 0.42 (0.43) | 96 (93) | 2.45 (1.47) | 2.73 (1.67) | 65 (27) | 0.02 (0.01) | 0.41 (0.43) | 96 (93) | 2.50 (1.48) | 2.79 (1.66) | 67 (28) |
| M0 | 3.05 (2.92) | 3.11 (2.99) | 73 (4) | 0.01 (−0.01) | 0.42 (0.41) | 96 (96) | 3.14 (2.95) | 3.20 (3.02) | 62 (3) | 0.02 (0.02) | 0.41 (0.42) | 97 (96) | 3.11 (2.93) | 3.17 (3.00) | 60 (3) |
| U | 3.21 (3.19) | 3.28 (3.26) | 11 (1) | 0.01 (−0.01) | 0.42 (0.41) | 95 (94) | 3.23 (3.21) | 3.30 (3.27) | 9 (1) | 0.02 (0.02) | 0.41 (0.42) | 96 (94) | 3.21 (3.18) | 3.28 (3.24) | 9 (1) |
RMSE: root mean square error; CP: 95% CI/CrI coverage probability.
In most situations, the biases and RMSEs of point estimates obtained using the REML method were close to those obtained using the Bayesian method. However, under the missingness scenarios (III) and (III′), the 95% CrI coverage probabilities for factors 1, 3, and 5 obtained using the Bayesian method were generally higher than those obtained using the REML method. As noted in Section 4.3, this may be due to that the estimated variance-covariance matrix was inconsistent when many observations were missing and the dimension was close to the sample size (only five observations for each of factors 1, 3, and 5). Also, under scenario (III′), the biases and RMSEs produced by Model H using the REML method were noticeably different from those using the Bayesian method for factors 1, 3, and 5; again, these differences may be caused by the sparsity of the simulated data. In addition, when Model H was used under scenario (III′), the optimization algorithm for the REML estimates did not converge for many simulated replicates. We ran around 2,500 iterations to obtain 1,000 datasets that produced converged REML estimates, and these 1,000 datasets were used to produce the results for scenario (III′). Hence, the Bayesian method is preferred to implement the multivariate hybrid model when the dimension of factors is high but the observations are limited.
Moreover, we conducted an additional simulation study driven by the pterygium data. The conclusions are similar to those presented here. For more details, see Section S2 in the Supporting Information.
6. Real data analysis
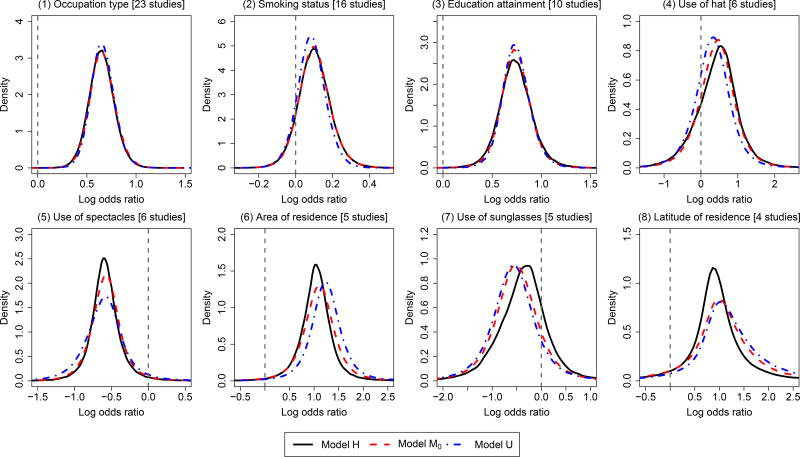
The pterygium dataset has been introduced in details as our motivating example. Since the within-study correlations were unknown, we used Models H, M0, and U but not Model M to estimate the overall log odds ratios of the 8 risk factors. Due to the sparsity of the dataset, the models were implemented using Bayesian analyses. The estimates were based on three MCMC chains, each containing a run of 100,000 updates after a 100,000-run burn-in period. The convergence of the chains was checked using their trace plots. Table 3 presents the median overall log odds ratios with 95% CrIs. Figure 2 shows the posterior density plots of the 8 risk factors.
Table 3.
The estimated overall log odds ratios (95% CrI) of the 8 risk factors in the pterygium data obtained by Model H (accounting for both between- and within-study correlations), Model M0 (only accounting for between-study correlations), and Model U (univariate model ignoring both between- and within-study correlations) using the Bayesian method.
| Risk factor | No. of studies |
Estimated overall log odds ratio | ||
|---|---|---|---|---|
|
| ||||
| Model H | Model M0 | Model U | ||
| (1) Occupation type | 23 | 0.65 (0.40, 0.92) | 0.65 (0.41, 0.93) | 0.66 (0.42, 0.91) |
| (2) Smoking status | 16 | 0.10 (−0.07, 0.29) | 0.10 (−0.07, 0.29) | 0.08 (−0.07, 0.25) |
| (3) Education attainment | 10 | 0.73 (0.40, 1.10) | 0.74 (0.42, 1.07) | 0.74 (0.46, 1.06) |
| (4) Use of hat | 6 | 0.47 (−0.87, 1.62) | 0.44 (−0.77, 1.55) | 0.32 (−0.84, 1.41) |
| (5) Use of spectacles | 6 | −0.59 (−1.02, −0.09) | −0.58 (−1.06, −0.06) | −0.60 (−1.26, 0.05) |
| (6) Area of residence | 5 | 1.04 (0.29, 1.78) | 1.10 (0.22, 1.97) | 1.24 (0.39, 2.11) |
| (7) Use of sunglasses | 5 | −0.36 (−1.53, 0.74) | −0.51 (−1.64, 0.62) | −0.57 (−1.76, 0.63) |
| (8) Latitude of residence | 4 | 0.92 (−0.74, 2.72) | 1.04 (−0.98, 3.10) | 1.14 (−0.70, 3.37) |
Figure 2.
Posterior density plots produced by Model H (accounting for both between- and within-study correlations), Model M0 (only accounting for between-study correlations), and Model U (univariate model ignoring both between- and within-study correlations) for the overall log odds ratios of the 8 risk factors in the pterygium data.
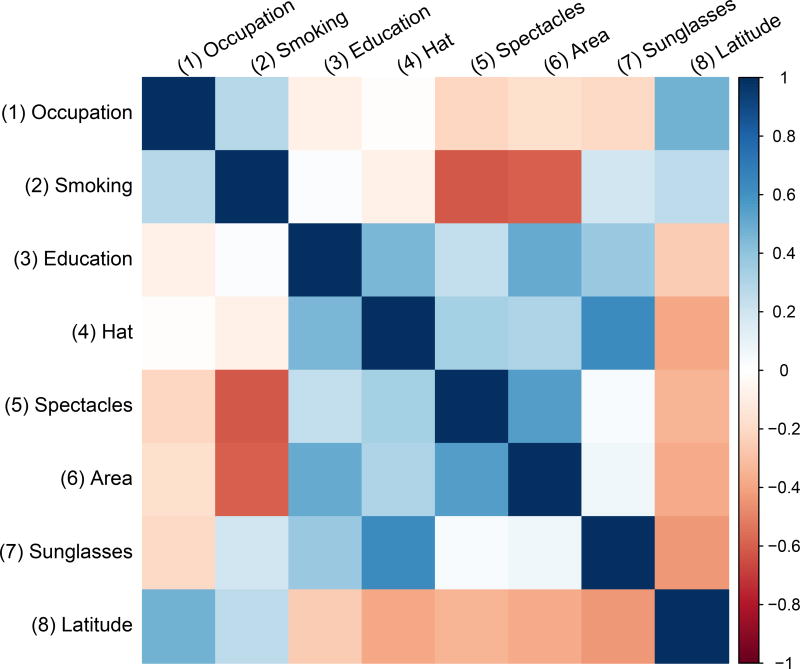
For risk factors that were reported in a relatively large number of studies (e.g., occupation type, smoking status, and education attainment), the three models yielded similar estimated overall log odds ratios; their density curves were also fairly similar. For risk factors that were only reported in a few studies (e.g., use of spectacles, area of residence, and latitude of residence), the peaks of the density curves produced by Model H were narrower and higher compared with those produced by Models M0 and U, indicating that Model H produced narrower 95% CrIs. Also, for the risk factor use of sunglasses, the location of its density curve produced by Model H was noticeably different from those produced by the other two models. Figure 3 depicts the estimated overall correlations between the 8 risk factors produced by Model H. It shows that many factors were correlated and some correlations were fairly high. Hence, ignoring the within-study correlations could lead to fairly different estimated overall log odds ratios; the unknown within-study correlations need to be carefully considered in MVMA-MF.
Figure 3.
Plot of the estimated overall correlations between the 8 risk factors produced by Model H in the pterygium data. Darker color implies higher correlation.
The priors in the above real data analyses were specified in Section S1.2 of the Supporting Information; specifically, uniform priors U(0, 10) were assigned for the between-study standard deviations. We examined the results’ sensitivity to prior specification by changing U(0, 10) priors to U(0, 3), U(0, 100), and U(0, 1000). In addition, we altered the order of the 8 risk factors in the data analysis; this basically changed the priors assigned to the correlation matrices in Models H and M0 (see Section S1 in the Supporting Information). Table S3 in the Supporting Information presents the results. In sum, changing priors had little impact on the risk factors occupation type, smoking status, and education attainment, which were reported in at least ten studies. When the priors for the between-study standard deviations varied from U(0, 3) to U(0, 100), the point estimates of the overall log odds ratios of all 8 risk factors remained nearly unchanged. The CrIs for risk factors use of sunglasses and latitude of residence noticeably became wider. Compared with other risk factors, since the data for these two factors were limited, the priors had larger influence on their posteriors. When the priors for the between-study standard deviations varied from U(0, 100) to U(0, 1000), most results did not change. Changing the priors for the correlation matrices in Models H and M0 had some impact on the CrIs for the risk factors that were reported in no more than six studies. Nevertheless, the CrIs produced by Model H were still narrower than those produced by Models M0 and U. For more details, see Section S3 in the Supporting Information.
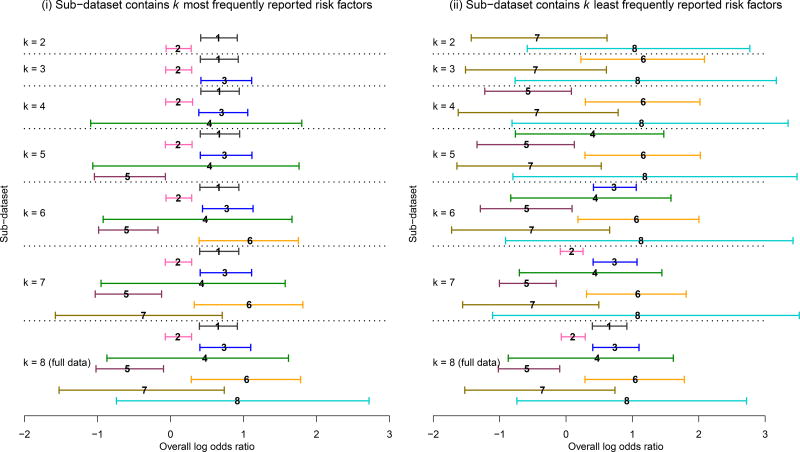
We also performed additional sensitivity analyses to investigate the impact of risk factor selection on the estimated overall log odds ratios. We considered two scenarios for the set of risk factors to be included in MVMA-MF: (i) sub-dataset consisted of the k most frequently reported risk factors; and (ii) sub-dataset consisted of the k least frequently reported risk factors (k = 2, …, 8). For example, when k = 2, the sub-dataset under scenario (i) contained the risk factors (1) occupation type and (2) smoking status; the sub-dataset under scenario (ii) contained the risk factors (7) use of sunglasses and (8) latitude of residence. Model H was implemented to analyze these sub-datasets.
Figure 4 shows the 95% CrIs of the overall log odds ratios under both scenarios; the labels of risk factors used in this figure can be found in Table 3’s first column. In scenario (i), starting from the two most frequently reported risk factors, infrequently reported risk factors were iteratively added to the MVMA-MF. Figure 4(i) shows that the estimated overall log odds ratios of risk factors 4 and 5 had some changes as new factors were added to the sub-datasets. The 95% CrIs of the three most frequently reported risk factors 1–3 changed little. This might be explained by two reasons. First, the correlations between these three factors were weak (Figure 3), so the addition of risk factor 3 had little impact on estimating the effect sizes of factors 1 and 2. Second, the later added factors 4–8 provided much fewer information (no more than six studies) compared with factors 1–3, which were reported in more than ten studies, so the correlations contributed little to the effect size estimation for factors 1–3.
Figure 4.
Bayesian estimates of overall log odds ratios produced by Model H based on subsets of the pterygium data. Each horizontal solid line represents 95% CrI of log odds ratio. The number placed at the median log odds ratio within each 95% CrI represents the corresponding risk factor’s label. The results of the sub-datasets that contain k = 2, …, 8 risk factors are accordingly listed from upper to lower, separated by the dotted lines.
Compared with scenario (i), Figure 4(ii) shows larger changes of estimated overall log odds ratios in scenario (ii). Under this scenario, starting from the two least frequently reported risk factors, more frequently reported factors were iteratively added to the MVMA-MF. The 95% CrIs of infrequently reported risk factors, such as 5 and 6, became narrower as more reported risk factors were included in the MVMA-MF. This illustrates the benefit of jointly modeling multiple factors: the inference on infrequently reported factors can be strengthened by borrowing information from frequently reported factors through the correlations between them.
7. Discussion
This article has introduced MVMA-MF with application to the pterygium data. In contrast to the tradition of meta-analyzing each single factor separately, this article encourages researchers to collect all possible factors and analyze them jointly to enhance the estimation of overall effect sizes. A Bayesian multivariate hybrid model has been introduced to implement MVMA-MF in which within-study correlations are usually unknown. The simulations have shown that, when many factors are missing not at random, Bayesian MVMA-MF can use the information across multiple factors to impute missing data and thus can produce better estimates than separate meta-analyses. Our results are consistent with those produced by Kirkham et al.14 using frequentist bivariate and univariate meta-analyses; that is, “borrowing of strength” from the correlations between factors can reduce bias under the setting of missing not at random. For example, consider two highly correlated factors with a correlation coefficient close to one, and the results for factor 1 are fully observed. Even if the results for factor 2 are missing not at random, the missing results can be largely recovered through the high correlation between the two factors, so the bias in the estimate of factor 2 may be greatly reduced.
The hybrid model effectively reduces model complexity by specifying a common marginal correlation matrix for all studies. This model could be a proper tradeoff between goodness-of-fit and model complexity in some situations; however, if the collected studies’ marginal correlation matrices differ a lot, the hybrid model may fit the data poorly. Future work is needed to generalize the hybrid model by allowing more degrees of freedom and select the best model using deviance information criterion.40
An important issue of MVMA-MF is to incorporate the effect size of a certain factor that has been adjusted for other factors. For example, in the original pterygium data presented in Serghiou et al.8, many collected studies only reported log odds ratios that were obtained using multivariate regression after adjusting different factors (e.g., age and gender), while log odds ratios without any adjustments were unavailable from these studies. We did not include such data in Table 1 due to the inconsistent adjustments. How to incorporate such data with different adjustments is of great interest to enrich the MVMA-MF and enhance the robustness and precision of the results.41,42 We leave this to future studies.
Another interesting but challenging problem is to identify the factors’ missingness mechanism and impute the missing factors more properly when the missingness is not at random. The factors could be missing completely at random, missing at random, or missing not at random.43 If the missingness of a factor does not depend on any observed or unobserved data, then this factor is missing completely at random. It is also possible that this factor is missing because of some observed data in some earlier studies; this is the case of missing at random. Bayesian inference under these two missingness mechanisms is valid when restricted to the observed data.38 However, the missingness not at random closely relates to publication and reporting bias, when some studies or factors were intended to be not published or reported due to their negative results.44,45 Although our simulations showed that the proposed hybrid model performed better than Models M0 and U when missingness was not at random, both its bias and 95% CrI coverage probability were expected to be further improved. Approaches to correcting publication or reporting bias have been introduced and widely used in univariate meta-analysis46; similar methods are highly needed for multivariate meta-analysis.
Supplementary Material
Acknowledgments
Funding: This work was supported by the National Institute of Allergy and Infectious Diseases [R21AI103012], the National Institute of Dental and Craniofacial Research [R03DE024750], the National Library of Medicine [R21LM012197], National Cancer Institute [P30CA077598], the National Institute on Minority Health and Health Disparities [U54MD008620], and the National Institute of Diabetes and Digestive and Kidney Diseases [U01DK106786].
Footnotes
Conflicts of interest: none.
Additional supporting information is available at the publisher’s web site.
References
- 1.Multiple Risk Factor Intervention Trial Research Group. Multiple risk factor intervention trial. JAMA. 1982;248(12):1465–1477. [PubMed] [Google Scholar]
- 2.Stamler J, Vaccaro O, Neaton JD, Wentworth D Multiple Risk Factor Intervention Trial Research Group. Diabetes, other risk factors, and 12-yr cardiovascular mortality for men screened in the Multiple Risk Factor Intervention Trial. Diabetes Care. 1993;16(2):434–444. doi: 10.2337/diacare.16.2.434. [DOI] [PubMed] [Google Scholar]
- 3.Ezzati M, Lopez AD, Rodgers A, Vander Hoorn S, Murray CJL. Selected major risk factors and global and regional burden of disease. Lancet. 2002;360(9343):1347–1360. doi: 10.1016/S0140-6736(02)11403-6. [DOI] [PubMed] [Google Scholar]
- 4.The Risk and Prevention Study Collaborative Group. n-3 fatty acids in patients with multiple cardiovascular risk factors. N Engl J Med. 2013;368(19):1800–1808. doi: 10.1056/NEJMoa1205409. [DOI] [PubMed] [Google Scholar]
- 5.Ebrahim S, Davey Smith G. Systematic review of randomised controlled trials of multiple risk factor interventions for preventing coronary heart disease. BMJ. 1997;314(7095):1666–1674. doi: 10.1136/bmj.314.7095.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole MG, Dendukuri N. Risk factors for depression among elderly community subjects: a systematic review and meta-analysis. Am J Psychiatry. 2003;160(6):1147–1156. doi: 10.1176/appi.ajp.160.6.1147. [DOI] [PubMed] [Google Scholar]
- 7.Flenady V, Koopmans L, Middleton P, et al. Major risk factors for stillbirth in high-income countries: a systematic review and meta-analysis. Lancet. 2011;377(9774):1331–1340. doi: 10.1016/S0140-6736(10)62233-7. [DOI] [PubMed] [Google Scholar]
- 8.Serghiou S, Patel CJ, Tan YY, Koay P, Ioannidis JPA. Field-wide meta-analyses of observational associations can map selective availability of risk factors and the impact of model specifications. J Clin Epidemiol. 2016;71:58–67. doi: 10.1016/j.jclinepi.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson D, White IR, Thompson SG. Extending DerSimonian and Laird's methodology to perform multivariate random effects meta-analyses. Stat Med. 2010;29(12):1282–1297. doi: 10.1002/sim.3602. [DOI] [PubMed] [Google Scholar]
- 10.Jackson D, Riley R, White IR. Multivariate meta-analysis: potential and promise. Stat Med. 2011;30(20):2481–2498. doi: 10.1002/sim.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H, Manning AK, Dupuis J. A method of moments estimator for random effect multivariate meta-analysis. Biometrics. 2012;68(4):1278–1284. doi: 10.1111/j.1541-0420.2012.01761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu D, Liu RY, Xie M. Multivariate meta-analysis of heterogeneous studies using only summary statistics: efficiency and robustness. J Am Stat Assoc. 2015;110(509):326–340. doi: 10.1080/01621459.2014.899235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riley RD, Abrams KR, Lambert PC, Sutton AJ, Thompson JR. An evaluation of bivariate random-effects meta-analysis for the joint synthesis of two correlated outcomes. Stat Med. 2007;26(1):78–97. doi: 10.1002/sim.2524. [DOI] [PubMed] [Google Scholar]
- 14.Kirkham JJ, Riley RD, Williamson PR. A multivariate meta-analysis approach for reducing the impact of outcome reporting bias in systematic reviews. Stat Med. 2012;31(20):2179–2195. doi: 10.1002/sim.5356. [DOI] [PubMed] [Google Scholar]
- 15.Riley RD, Jackson D, Salanti G, et al. Multivariate and network meta-analysis of multiple outcomes and multiple treatments: rationale, concepts, and examples. BMJ. 2017;358:j3932. doi: 10.1136/bmj.j3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reitsma JB, Glas AS, Rutjes AWS, Scholten RJPM, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58(10):982–990. doi: 10.1016/j.jclinepi.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 17.Chu H, Cole SR. Bivariate meta-analysis of sensitivity and specificity with sparse data: a generalized linear mixed model approach. J Clin Epidemiol. 2006;59(12):1331–1332. doi: 10.1016/j.jclinepi.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Ma X, Nie L, Cole SR, Chu H. Statistical methods for multivariate meta-analysis of diagnostic tests: an overview and tutorial. Stat Methods Med Res. 2013;25(4):1596–1619. doi: 10.1177/0962280213492588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Houwelingen HC, Zwinderman KH, Stijnen T. A bivariate approach to meta-analysis. Stat Med. 1993;12(24):2273–2284. doi: 10.1002/sim.4780122405. [DOI] [PubMed] [Google Scholar]
- 20.Berkey CS, Anderson JJ, Hoaglin DC. Multiple-outcome meta-analysis of clinical trials. Stat Med. 1996;15(5):537–557. doi: 10.1002/(SICI)1097-0258(19960315)15:5<537::AID-SIM176>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 21.Lu G, Ades AE. Combination of direct and indirect evidence in mixed treatment comparisons. Stat Med. 2004;23(20):3105–3124. doi: 10.1002/sim.1875. [DOI] [PubMed] [Google Scholar]
- 22.Salanti G, Higgins JPT, Ades AE, Ioannidis JPA. Evaluation of networks of randomized trials. Stat Methods Med Res. 2008;17(3):279–301. doi: 10.1177/0962280207080643. [DOI] [PubMed] [Google Scholar]
- 23.Salanti G. Indirect and mixed-treatment comparison, network, or multiple-treatments meta-analysis: many names, many benefits, many concerns for the next generation evidence synthesis tool. Res Synth Methods. 2012;3(2):80–97. doi: 10.1002/jrsm.1037. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Carlin BP, Neaton JD, et al. Network meta-analysis of randomized clinical trials: reporting the proper summaries. Clin Trials. 2014;11(2):246–262. doi: 10.1177/1740774513498322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin L, Zhang J, Hodges JS, Chu H. Performing arm-based network meta-analysis in R with the pcnetmeta package. J Stat Softw. 2017;80(5):1–25. doi: 10.18637/jss.v080.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin L, Chu H, Hodges JS. Sensitivity to excluding treatments in network meta-analysis. Epidemiology. 2016;27(4):562–569. doi: 10.1097/EDE.0000000000000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riley RD. Multivariate meta-analysis: the effect of ignoring within-study correlation. J R Stat Soc Ser A Stat Soc. 2009;172(4):789–811. [Google Scholar]
- 28.Riley RD, Thompson JR, Abrams KR. An alternative model for bivariate random-effects meta-analysis when the within-study correlations are unknown. Biostatistics. 2008;9(1):172–186. doi: 10.1093/biostatistics/kxm023. [DOI] [PubMed] [Google Scholar]
- 29.White IR. Multivariate random-effects meta-regression: updates to mvmeta. Stata J. 2011;11(2):255–270. [Google Scholar]
- 30.Lu G, Ades AE. Modeling between-trial variance structure in mixed treatment comparisons. Biostatistics. 2009;10(4):792–805. doi: 10.1093/biostatistics/kxp032. [DOI] [PubMed] [Google Scholar]
- 31.Wei Y, Higgins JPT. Bayesian multivariate meta-analysis with multiple outcomes. Stat Med. 2013;32(17):2911–2934. doi: 10.1002/sim.5745. [DOI] [PubMed] [Google Scholar]
- 32.Saw S-M, Tan D. Pterygium: prevalence, demography and risk factors. Ophthalmic Epidemiol. 1999;6(3):219–228. doi: 10.1076/opep.6.3.219.1504. [DOI] [PubMed] [Google Scholar]
- 33.Borenstein M, Hedges LV, Higgins JPT, Rothstein HR. A basic introduction to fixed-effect and random-effects models for meta-analysis. Res Synth Methods. 2010;1(2):97–111. doi: 10.1002/jrsm.12. [DOI] [PubMed] [Google Scholar]
- 34.Riley RD, Higgins JPT, Deeks JJ. Interpretation of random effects meta-analyses. BMJ. 2011;342:d549. doi: 10.1136/bmj.d549. [DOI] [PubMed] [Google Scholar]
- 35.Higgins JPT. Commentary: heterogeneity in meta-analysis should be expected and appropriately quantified. Int J Epidemiol. 2008;37(5):1158–1160. doi: 10.1093/ije/dyn204. [DOI] [PubMed] [Google Scholar]
- 36.Berlin JA, Laird NM, Sacks HS, Chalmers TC. A comparison of statistical methods for combining event rates from clinical trials. Stat Med. 1989;8(2):141–151. doi: 10.1002/sim.4780080202. [DOI] [PubMed] [Google Scholar]
- 37.Petitti DB. Meta-Analysis, Decision Analysis, and Cost-Effectiveness Analysis: Methods for Quantitative Synthesis in Medicine. New York, NY: Oxford University Press; 2000. [Google Scholar]
- 38.Seaman S, Galati J, Jackson D, Carlin J. What is meant by "missing at random"? Stat Sci. 2013;28(2):257–268. [Google Scholar]
- 39.Bickel PJ, Levina E. Regularized estimation of large covariance matrices. Ann Stat. 2008;36(1):199–227. [Google Scholar]
- 40.Spiegelhalter DJ, Best NG, Carlin BP, Van Der Linde A. Bayesian measures of model complexity and fit. J R Stat Soc Series B Stat Methodol. 2002;64(4):583–639. [Google Scholar]
- 41.Becker BJ, Wu M-J. The synthesis of regression slopes in meta-analysis. Stat Sci. 2007;22(3):414–429. [Google Scholar]
- 42.Wu MJ, Becker BJ. Synthesizing regression results: a factored likelihood method. Res Synth Methods. 2013;4(2):127–143. doi: 10.1002/jrsm.1063. [DOI] [PubMed] [Google Scholar]
- 43.Higgins JPT, White IR, Wood AM. Imputation methods for missing outcome data in meta-analysis of clinical trials. Clin Trials. 2008;5(3):225–239. doi: 10.1177/1740774508091600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothstein HR, Sutton AJ, Borenstein M. Publication Bias in Meta-Analysis: Prevention, Assessment and Adjustments. Chichester, UK: John Wiley & Sons; 2005. [Google Scholar]
- 45.Schmid CH. Outcome reporting bias: a pervasive problem in published meta-analyses. Am J Kidney Dis. 2017;69(2):172–174. doi: 10.1053/j.ajkd.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 46.Duval S, Tweedie R. A nonparametric "trim and fill" method of accounting for publication bias in meta-analysis. J Am Stat Assoc. 2000;95(449):89–98. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.