Abstract
Fluorescence imaging is a powerful technique for the real-time noninvasive monitoring of protein dynamics. Recently, fluorogen activating proteins (FAPs)/fluorogen probes for protein imaging were developed. Unlike the traditional fluorescent proteins (FPs), FAPs do not fluoresce unless bound to their specific small-molecule fluorogens. When using FAPs/fluorogen probes, a washing step is not required for the removal of free probes from the cells, thus allowing rapid and specific detection of proteins in living cells with high signal-to-noise ratio. Furthermore, with different fluorogens, living cell multi-color proteins labeling system was developed. In this review, we describe about the discovery of FAPs, the design strategy of FAP fluorogens, the application of the FAP technology and the advances of FAP technology in protein labeling systems.
KEY WORDS: Fluorogen activating proteins, Fluorogens, Genetically encoded sensors, Fluorescence imaging, Molecular imaging
Graphical abstract
Fluorescence imaging is a powerful technique for the real-time noninvasive monitoring of protein dynamics. Recently, fluorogen activating proteins (FAPs)/fluorogen probes for protein imaging were developed. In this review, we describe about the discovery of FAPs, the design strategy of FAP fluorogens, the application of the FAP technology and the advances of FAP technology in protein labeling systems.
1. Introduction
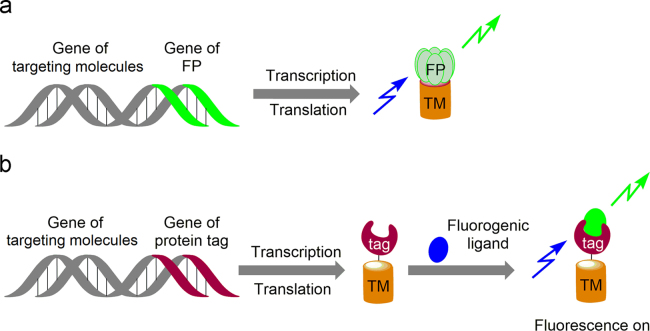
Fluorescence imaging is one of the most powerful techniques to observe biomolecules in real-time with high spatial and temporal resolution, which reveals the fundamental insights into the production, localization, trafficking, and biological functions of biomolecules in living systems1, 2, 3, 4. As the biological objects are poorly fluorescent, fluorescent probes including fluorescent proteins and organic fluorescent dyes are essential molecular tools for bio-imaging5, 6. Among them, a diverse set of genetically encodable fluorescent biosensors have been designed to probe dynamic cellular events. These sensors that generally involve the incorporation of a fluorescent tag into a protein or a selected protein domain, have enabled researchers to track various components of intracellular signaling networks in real time within the native cellular environment. In the past several decades, two approaches have been developed to construct genetically encodable biosensors for live cell studies (Fig. 1): 1) fluorescent protein-based reporters: chimeric genetic fusions of fluorescent proteins (e.g., GFP and its variants) with a protein (or RNA) domain7; 2) fluorogen-based reporters: a genetically encodable tag binds a fluorogenic ligand (endogenously present or exogenously applied) and activates its fluorescence. As the fluorogenic chromophore is non-fluorescent by its own and becomes strongly fluorescent only upon binding its target, unspecific fluorescence background in cells remains minimal even in the presence of an excess of dye, thus ensuring high imaging contrast8. Labeling with fluorogenic probes can be covalent, relying on chemical or enzymatic reaction, or non-covalent, relying on binding equilibrium. In the past 20 years, great efforts have been dedicated to exploring covalence-based self-labeling tags, such as the commercially available SNAP-tag9, 10, 11, CLIP-tag12 and Halo Tag13, 14, 15. Parallel to the development of covalent fluorogenic protein labeling strategies, methods based on the non-covalent interaction between a protein tag and a fluorogenic dye have emerged16, 17. Unlike the covalent labeling strategies, non-covalent labeling can be very fast since no covalent bond has to be created. Moreover, systems based on reversible non-covalent binding could provide an additional degree of control as fluorescence could also be switched off by washing away the fluorogenic ligand, given that the off-rate is fast enough. In this review, we describe the discovery of one of the non-covalence-based fluorogenic probes based on fluorogen activating proteins (FAPs), the design strategy of FAP fluorogens, the application of the FAP technology and the advances of FAP technology in protein labeling systems.
Figure 1.
(a) Fluorescent protein-based reporters and (b) fluorogen-based reporters for fluorescence imaging, TM: targeting molecules.
1.1. The discovery of the FAPs
FAP technology was first introduced in 2008 by Szent-Gyorgyi et al.18 by using single-chain antibodies as genetically encodable FAP. FAPs binding modified thiazole orange (TO) and malachite green (MG) were first generated by screening a yeast surface-displayed library of human single-chain antibodies (scFvs) using fluorescence-activating cell sorting (FACS). Eight unique FAPs were isolated from the library, among which six proteins specifically activate modified MG. scFvs are engineered proteins composed of Immunoglobulin (IgG) variable heavy (VH) and variable light (VL) domains tethered together via a short flexible peptide linker, which retain the wide range of antigen recognition capabilities of the full-length antibodies, and are also conformable to be used as recombinant tags in diverse fusion proteins19.
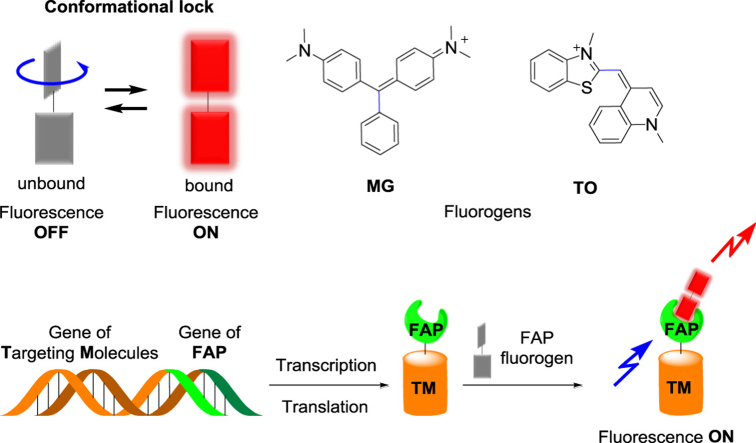
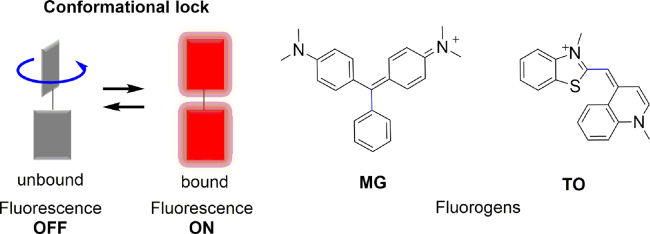
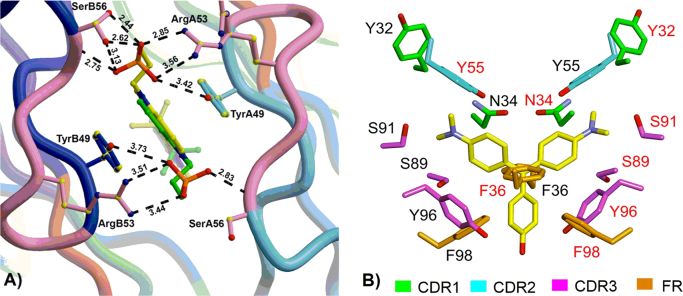
The FAP technology is a fluorogenic tagging approach that utilizes molecular recognition to directly activate the fluorescence of otherwise nonfluorescent small-molecule dyes (fluorogens). Selected FAPs bind TO and MG with nanomolar affinity and increase their respective green and red fluorescence by as much as thousands of fold. The fluorescence enhancement results from FAPs constraining the rapid rotation around a single bond within the chromophore (Fig. 2)6. The non-covalent interactions between the fluorogens and FAPs are like those of ligands and their receptors, mainly including van der Waals forces, π-effects and hydrogen bonds. Molecular recognition capabilities are largely determined by these loops of FAPs, termed complementarity determining regions (CDRs), which undergo somatic hypermutation during the immune response to generate specific high affinity binding to the antigen (Fig. 3)20, 21. FAPs represent a new class of fluorogen-based reporters, which provide a fluorescent tool for imaging fusion protein's location and abundance in time and space. FAP-fluorogen imaging system offers a number of distinct advantages in bio-applications: 1) unbound dye remains nonfluorescent in solution, allowing for the simple addition of dyes to the cellular media without any need for fixation or washout, a property that will enable imaging in more complicated tissue environments and live-cell imaging; 2) fluorogen binding to most FAPs occurs within seconds of addition, and can be carried out in a near physiological buffer or medium of choice. The interaction between the fluorogen and FAP is highly specific, with some FAP clones exhibiting subnanomolar affinity; 3) since FAPs are small in size (<30 kDa), they are easy to genetically engineer. The FAP technology thus allows specific fluorescent labeling of fusion proteins of interest in both living or chemically fixed cells; 4) the possibility offered to completely control the concentration of fluorogens paves the way for on-demand applications wherein fluorescence is desired only at a specific time or at a given density as exemplified with the FAPs; 5) fluorescence visualization can be spatially controlled by the appropriate choice of the membrane permeable and impermeable fluorogens, enabling one to selectively observe FAP fusion proteins inside cells, on the cell surface, or within trafficking vesicles; 6) variations of the fluorogens have been shown to produce a variety of distinct spectral and sensing properties for a given FAP, which is very useful in a variety of multicolor experiments. To sum up, the FAP-fluorogen system is a versatile, effective fluorogenic labeling strategy.
Figure 2.
Concept and examples of the fluorogen-based reporters operating by intramolecular rotation. Adapted with permission from Ref. 6. Copyright (2017) American Chemical Society.
Figure 3.
(A) The interactions between the DIR sulfonate and M8VL show the two orientations of the bound DIR in the crystal structure (green and yellow), and the A (light blue) and B (blue) VL chains. CDR2 is colored pink. SerB56 and ArgA53 also sample two alternate conformations. One sulfonate (top) has more interactions with the protein than the alternate (bottom) sulfonate. Adapted with permission from Ref. 20. Copyright (2012) American Chemical Society. (B) MG interactions with L5*. Spatial distribution of amino acid side chains that contact MG. Shown are contacting side chains from both VL A (black letters) and VL B (red letters) that comprise a set of pairwise symmetrical locations and orientations. Adapted with permission from Ref. 21.
1.2. The variations of the FAPs
For many of the scFvs, both VH and VL domains are essential for dye binding and fluorescence, however, the analysis of other scFvs revealed that either VH or the VL domain alone is sufficient to cause the fluorogenic dye activation19, 22 (Table 1). The existence of FAPs comprised of only of VH or VL domains that activate MG, such as L5, H6 and H8 FAPs, have already been demonstrated by Szent-Gyorgyi et al.18. In order to discover more desirable FAPs with appropriate and superior properties, such as high quantum yield and binding affinity, scFvs-based FAPs have been reconstructed and researched. For instance, the L5* FAP is a VL domain that binds MG to activate intense fluorescence, which is a leucine-to-serine point mutant (Ser89) obtained by directed evolution of L521. L5* binds to MG to form a bright fluorescent complex that improves on the quantum yield of the original L5 FAP by about 5-fold (quantum yield = 0.24). Another improved version of L5, the dL5** FAP (quantum yield = 0.20), is a synthetic dimer of a light chain with a disulfide forming pair of cysteines in each monomer. When expressed as a monomer putatively assembled into ternary complex with MG dye, the dL5** FAP confers tighter binding while maintaining increased brightness. The VH domain alone FAP, dH6.2 FAP, is a synthetic dimer derived from a heavy chain with the second cysteine changed to alanine in each monomer23. This kind of FAP was normally bright but considerably less photostable across multiple compartments, which has proven useful for super-resolution imaging22.
Table 1.
Properties of FAPs and fluorogens. Fluorogens are derivatives of MG or TO, FAPs are comprised of hypervariable heavy (H) and light (L) chains joined by a flexible linker. All information is taken from Refs. 18, 21.
| FAP | Fluorogen | Size (kDa) | Excitation λmax (nm) | Emission λmax (nm) | Quantum yield | Fluorescence enhancement |
|---|---|---|---|---|---|---|
| L5-MG | MG-2p | 11.5 | 640 | 668 | 0.048 | 4100 |
| H6-MG | MG-2p | 14.4 | 635 | 656 | 0.25 | 18,000 |
| H8-MG | MG-2p | 13.6 | 626 | 646 | ||
| HL4-MG | MG-2p | 26.1 | 629 | 649 | 0.16 | 15,700 |
| HL1.0.1-TO1 | TO1-2p | 25.9 | 509 | 530 | 0.47 | 2600 |
| L5*-MG | MG-2p | 14.2 | 634 | 667 | 0.24 |
In addition, these FAPs contained internal disulfide bonds, which restricted their use to non-reducing environments such as the cell surface and secretory apparatus, since these FAPs may not fold properly in the reducing environment of the cytosol. The engineering of disulfide-free FAPs, like p13-CW FAP, a classic heavy-light scFv (HL4) with the second cysteine in each domain changed to an alanine, improved labeling in the cytoplasm and various other reducing subcellular compartments24, 25. Furthermore, selection of scFvs against other fluorogens successfully extended the chromatic palette of FAPs26, 27. Of particular interest, some scFv promiscuously activate various dimethylindole red (DIR) analogs, providing access to wavelengths ranging from the blue to the near infrared (NIR, 650—900 nm)26. There are also promiscuous FAPs that can bind more than one fluorogen, with alternate excitation and emission wavelengths and varying affinity constants for fluorogen binding21. Further improvements in brightness would result in better sensitivity and lower phototoxicity under typical imaging conditions.
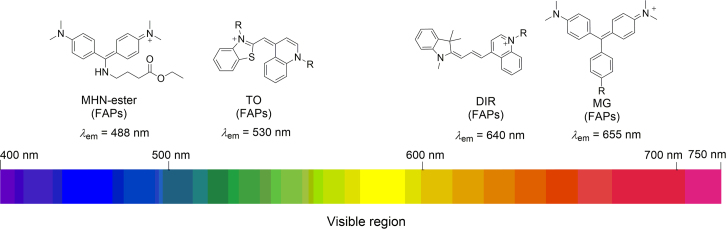
2. The fluorogens of the FAP technology
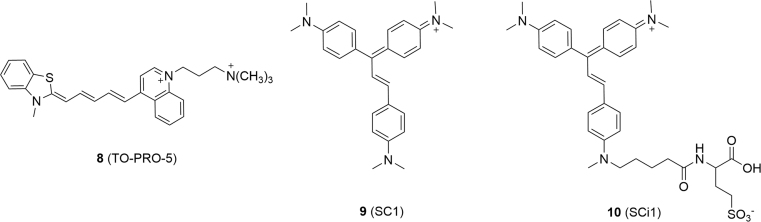
MG, TO and DIR are classic FAP fluorogens (Fig. 4). In aqueous buffer these fluorogens exhibit strong absorbance maxima at 607 nm (MG), 504 nm (TO) and 610 nm (DIR), but exhibit extremely low levels of fluorescence. When bound to their cognate FAPs, the fluorogens exhibit bathochromic excitation maxima that are well matched to lasers (MG and DIR, 633 nm; TO1, 488 or 514 nm) generally used in flow cytometry and microscopy.
Figure 4.
Fluorogens utilized for the development of FAP labeling methods. The maximal emission wavelengths of the fluorogens bound to their cognate FAPs are given.
Along with the progress of genetically encodable FAPs to specifically label a protein-of-interest within the cellular milieu, various fluorogens of FAPs based on MG, TO or DIR were explored for tracking cellular proteins and other signaling molecules within their endogenous environment, providing unprecedented insights into the dynamic regulation of signaling networks in living cells.
2.1. Cell membrane permeable and impermeable fluorogens
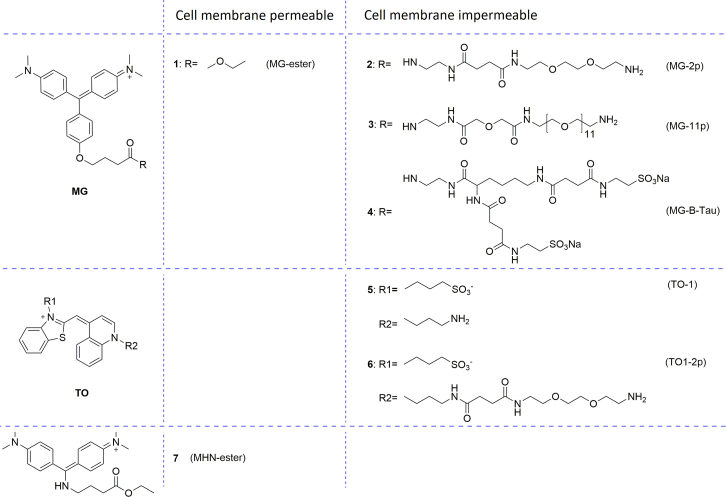
Cell surface proteins tagged with FAP are readily detected with cell membrane impermeable fluorogens, whereas proteins within the secretory compartments in the cell only could be stained with membrane permeable fluorogens. In order to increase or decrease cell membrane permeability, fluorogen 1—6 were developed based on MG or TO (Fig. 5). With the intention of shorting the emission wavelength to blue, fluorogen 7 (MHN-ester) was explored28. MG-ester18, 29 and MHN-ester (fluorogen 1 and 7) showed good cell permeability. On the other hand, sulfonated groups, polyethelyene glycol groups and amino groups were used to reduce the cell membrane permeability, resulting in cell impermeable sulfonated fluorogens TO1, TO1–2p, polyethelyene glycol modified fluorogens MG-2p, MG-11p and sulfonated fluorogens MG-B-Tau30, respectively.
Figure 5.
Structures of cell membrane permeable and impermeable fluorogens.
2.2. Fluorogens with large pseudo-Stokes shifts
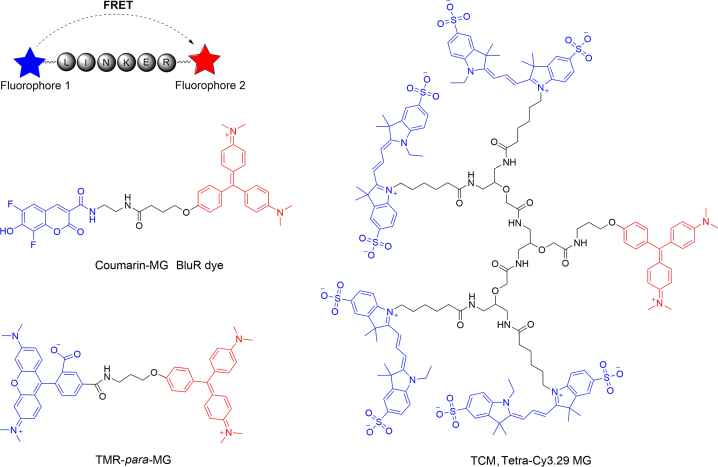
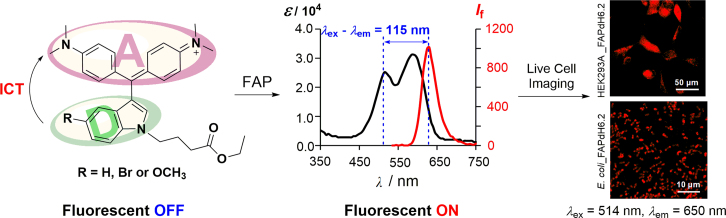
Large Stokes shift is desirable in fluorescent labeling applications of dyes, as it reduces self-quenching effects and interference from excitation source31. Usually, fluorogen MG exhibited small Stokes shifts (Δλ = 20 nm), great efforts have been dedicated to increase the wavelength length difference between the excitation and emission and three series of Förster resonance energy transfer (FRET)-based MG fluorogens with large pseudo-Stokes shifts have been developed (Fig. 6)32, 33, 34. Very recently, we have rationally designed and synthesized a series of novel 3-indole-Malachite Green-based FAP fluorogens35. The important features of this class of FAP fluorogens are the efficient internal charge transfer resulting in significant fluorescence enhancements, remarkable large “pseudo-Stokes” shifts, low toxicity to cells, as well as very fast onset in response to FAP in both live mammalian cells and bacterial cells (Fig. 7). They have the potential to be an alternative to FRET-based MG fluorogens with large pseudo-Stokes shifts in multiplexing applications with FAP imaging.
Figure 6.
Structures of FRET based MG fluorogens with large pseudo-Stokes shifts. Donor groups are depicted in blue, and acceptor groups in red.
Figure 7.
3-Indole Malachite Green based FAP fluorogens and their applications in live mammalian cells and bacterial cells imaging. Adapted with permission from Ref. 35. Copyright (2017) American Chemical Society.
2.3. In vivo FAP fluorogens
Cellular and tissue imaging in the near-infrared (NIR) wavelengths between 650 and 900 nm is advantageous for in vivo because of the low absorption of biological molecules in this region36, 37. In the past decade, significant advances have been made in the design of molecular probes for in vivo imaging. Two series of NIR FAP fluoromodules have been developed by modification of TO26 or MG38 via conjugating methine groups (Fig. 8), among which, MG modified fluorogens have been successfully used for the detection of protein–protein interactions in vivo.
Figure 8.
Structures of NIR FAP fluorogens.
3. The application of the FAP technology
3.1. Protein locations visualization
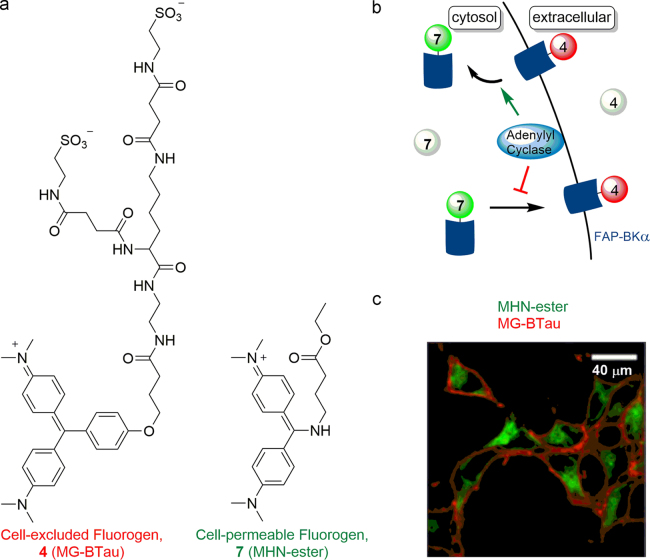
FAP-based fluoromodules have been successfully and widely utilized in selectively visualization of protein location, internalization and trafficking in mammalian and yeast cells since 2008. As discussed above, FAP technology has multiple advantages in biological imaging: 1) experimental flexibility—fluorescence is generated only upon addition of the fluorogen; 2) fast responses—FAPs can be visualized few seconds or minutes after addition of the fluorogen; 3) high spatial labeling discrimination—by playing with the cell-permeability of the fluorogen, fusion proteins in a given cell location can be selectively observed. For instance, comprehensive studies of plasma membrane G-protein coupled receptors (GPCRs), especially for β2-adrenergic receptor (β2-AR), expression, location, trafficking and quantification, as well as other membrane proteins, have been investigated by genetically encoding FAP tags in targeting molecules and imaging with modified TO or MG fluorogens29, 39, 40, 41, 42. Meanwhile, FAP-fluorogen technology was shown to be also suitable for labeling intracellular/cytosolic targets with the evolution of fluorogens. Based on FAPs and membrane permeable fluorogen MG-ester, Bruchez et al.25 presented a new labeling technology for cytoplasmic compartments, which is no-wash, far-red, highly fluorogenic, photostable, and nonphototoxic and functions in all organelles. Later, a two-color, Green-Inside Red-Outside (GIRO) (Fig. 9), compartment selective FAP-based approach that generates distinct signals from surface and internal proteins in live cells for simultaneous detection is also demonstrated by Bruchez's group28. Lately, the same group established a three-color labeling approach, allowing excitation-dependent visualization of extracellular, intracellular, and total protein pools in the same cells by using one fluorogenic tag that combines with distinct dyes to affect different spectral characteristics34.
Figure 9.
An example for membrane protein and intracellular protein visualization with cell permeable and impermeable fluorogens. (a) The chemical structure of the cell excluded MG-BTau (4), and cell permeable MHN-ester (7). (b) Schematized paradigm for GIRO labeling. (c) Live cells labeled with MG-BTau (red) and MHN-Ester (green) to label surface and internal protein in FAP-BKα expressing live cells. Adapted with permission from Ref. 28. Copyright (2015) American Chemical Society.
3.2. Drug discovery platform
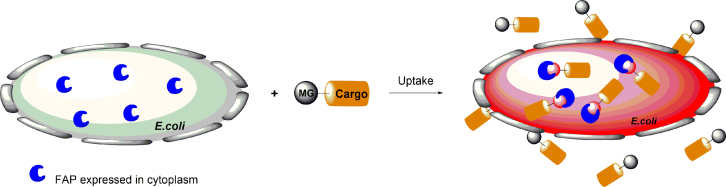
The FAP-based fluorescence detection and quantification approach also provides a platform for high-throughput screening of receptor proteins29. The most successful application is the discovery of novel cystic fibrosis transmembrane conductance regulator (CFTR) F508del correctors, using FAP tagging method in the trafficking studies of CFTR43, 44, 45. In addition, FAPs have an enormous potential for use in flow cytometry cell surface-based assays because fluorescence can be limited to proteins that are or have recently been resident on the surface membrane. Wu et al.46 developed a platform combining FAP technology with high-throughput flow cytometry to detect real-time protein trafficking to and from the plasma membrane in living cells. The hybrid platform allows drug discovery for trafficking receptors such as GPCRs, and has been validated using the β2-AR system. They later expanded the hybrid system to a new type of biosensor, which provides the opportunity to study multiple trafficking proteins in the same cell47. Recently, Jarvik et al.48 developed a novel approach based on FAPs and tethered fluorogen for visualizing regions of close apposition between the surfaces of living cells, which has the potential to provide a real-time readout of the proximity status of the membranes of the two cells. More recently, we and Brönstrup group first applied MG/FAP to study translocation efficiencies of molecular scaffolds designed to transport cargos in bacteria, which provided a general method for investigating the translocation capability of compounds across the membrane of bacterial cells (Fig. 10)49.
Figure 10.
Principle illustrating the application of the FAP system for uptake visualization of MG–carrier conjugates in E. coli. Adapted with permission from Ref. 49. Copyright (2017) Wiley-VCH.
3.3. Super-resolution imaging and single molecule tracking
In 2014, the Nobel Prize in Chemistry recognized super resolution imaging, a breakthrough in optical microscopy that enables researchers to visualize and investigate biological processes at the individual molecule level inside living cells. Fluorogen-based reporters were furthermore shown to open great prospects for super-resolution microscopy and single molecule tracking. Highly photostable far-red MG-based FAPs fluoromodules were reported to be well suitable for live cell imaging with stimulated emission depletion (STED) nanoscopy in mammalian cells and bacteria23, 50. The ability to control the labeling density of the FAPs by adjusting the fluorogen concentration in the milieu was used to obtain sparse labeling distribution of densely genetically tagged proteins for single molecule localization imaging in living cells, further demonstrating the interest of such probes for super-resolution microscopy22. This system is advantageous over traditional approaches51, 52, because it does not require special imaging buffers or photo-activation or photo-switching with a second laser line. The ability to label a subset of proteins independently of the expression level also enabled to track individual receptors in living cells53, confirming the usefulness of FAPs for single molecule studies.
3.4. Functional biomaterials and biosensors
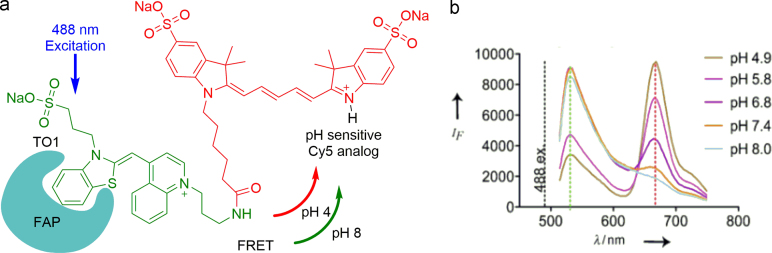
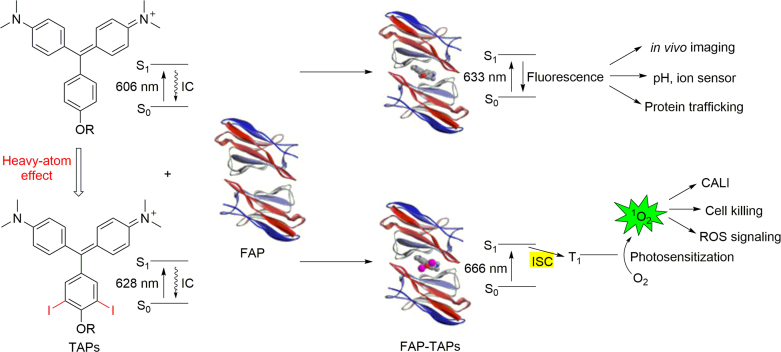
FAPs proved to be beneficial for the design of functional biomaterials and biosensors, as well. For instance, combined with FAP, a genetically encoded pH sensor was developed by coupling modified TO with a pH sensitive Cy5 analog. It was successfully used to track surface proteins through endocytosis, which undergoes a significant change in FRET efficiency in response to environmental pH change (Fig. 11)54. Saunders etal.55 developed a membrane materials in which dL5 FAP and AEAEAKAK, an amphiphilic peptide, are combined to form a solid-phase fluorescence detection platform. It is envisioned that dL5 FAP membranes can be established in diseased locales to monitor infiltration and migration of inflammatory cells marked with antibodies conjugated to MG. In combination with polymeric materials, targeted biosensor have been developed to distinct subcellular structures within living cells56. More recently, Meng group57 reported an injectable film by which antibodies can be localized in vivo. Their system builds upon a bifunctional polypeptide consisting of a FAP and a β-fibrillizing peptide (βFP). The FAP domain generates fluorescence that reflects IgG binding sites conferred by protein A/G (pAG) conjugated with the fluorogen MG. When injected into the subcutaneous space of mouse footpads, film-embedded IgG were retained locally, with distribution through the lymphatics impeded. Further, a targetable near-infrared photosensitizer (TAP)-FAP that has been described by Bruchez et al.58, which allows researchers to study protein inactivation, targeted-damage introduction and cellular ablation with unprecedented precision (Fig. 12).
Figure 11.
Illustration of the FRET-based pH biosensor. (a) The chemical structure of the FRET-based dye. A derivative of TO is linked to the pH-dependent Cy5 analogue. (b) The fluorescence emission spectra of the FAP-bound dye in citrate/phosphate buffer at various pH values. Upon excitation at λ=488 nm, energy transfer from the donor (TO) to the acceptor (Cy5 analogue) depends on the pH value. Adapted with permission from Ref. 54. Copyright (2012) Wiley-VCH.
Figure 12.
Mechanism of ROS generation by the targetable NIR photosensitizer, FAP-TAPs. IC, internal conversion by molecule's free rotation; ISC, intersystem crossing. Reprinted with permission from Ref. 58. Copyright (2016) Nature Publishing Group.
3.5. In vivo imaging
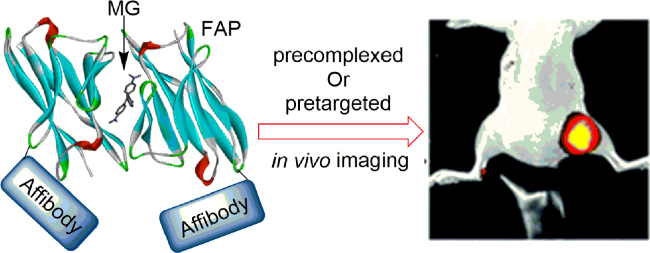
With the development and maturity of FAP technology, its applications have begun to expand into imaging of living animals. Pena et al.59 first described the combined use of novel genetically targeted probes and high resolution optical imaging technologies to explore mitochondrial metabolism, ROS generation and function/dysfunction in the context of the living zebrafish. Remarkably, Bruchez and co-workers60 developed a new tumor-targeting probe, affiFAP, containing a protein that specifically binds EGFR (affibody) and dL5** FAP. Fluorescence activation was achieved through either systemic (affiFAPs were pre-complexed with fluorogens prior to injection) or topical (affiFAPs were pre-targeted to the tumor site) administration of fluorogen. The latter approach was expected to minimize any undesired non-specific probe fluorescence and to demonstrate a possible no-wash platform for surgical guidance. They extended the application of affiFAP to in vivo imaging of a xenografted human EGFR-enriched tumor model in mice (Fig. 13), and establish its utility as a pretargeted fluorogen activating reagent, which is promising to be used in clinical settings to molecularly define tumor margins.
Figure 13.
Affibody-fused FAP for targeted in vivo tumor imaging. Affibody is a protein that specifically binds EGFR. Adapted with permission from Ref. 60. Copyright (2017) Royal Society of Chemistry.
4. Conclusion and perspectives
As summarized in this review, fluorogenic labeling is a general concept for imaging biomolecules with high contrast in living systems, with great potential for pushing the limit of biological imaging. In this review, we have discussed in detail about the discovery, development and application of the FAP technology as a novel effective fluorogenic labeling method. Clearly, the FAP-fluorogen system displays a few unprecedented attributes, such as a small size, no-wash, high signal-to-noise ratio, and tunable spectral properties, which make them interesting alternatives to classical fluorescent proteins and open great prospects for advanced imaging, such as super-resolution microscopies. In this two-component system, both the FAP and the fluorogen can be tuned in order to obtain the desired properties. For example, fluorogens can be designed to display improved spectral properties, brightness, and photostability. Meanwhile, through random mutagenesis and directed evolution, FAPs can be selected to accommodate a large repertoire of fluorogens. With further efforts, we expect that the FAP-fluorogen system will be useful for addressing exciting unexplored biological questions and will greatly advance our understanding of human disease and normal cell function. However, in all cases introduced above, the FAP was expressed from a recombinant gene that encoded a protein fusion between the FAP and the protein of interest. This approach results in two significant setbacks61: (1) time and labor regarding quality control and generation of each recombinant protein, and (2) artificial protein expression from a nonnative promoter, typically altering protein regulation and abundance in the cell. Therefore, careful upstream studies of their toxicity and their influence on cellular processes is still required. At present, our group is conducting an in-depth study on this issue. Moreover, whether the FAP approach is feasible with smaller antibody fragments such as nanobodies remains to be investigated.
Acknowledgments
The work of the Hu's lab is funded by the National Natural Science Foundation of China (NSFC) projects (Grant Nos. 21778077 and 21502236), Sino-German research project (GZ 1271), Beijing Nova Program (Z16111000490000) and CAMS Innovation Fund for Medical Sciences (CIFMS, 2017-12M-2-004).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
References
- 1.Lichtman J.W., Conchello J.A. Fluorescence microscopy. Nat Methods. 2005;2:910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 2.Xu W., Zeng Z., Jiang J.H., Chang Y.T., Yuan L. Discerning the chemistry in individual organelles with small-molecule fluorescent probes. Angew Chem Int Ed Engl. 2016;55:13658–13699. doi: 10.1002/anie.201510721. [DOI] [PubMed] [Google Scholar]
- 3.Tong H., Lou K., Wang W. Near-infrared fluorescent probes for imaging of amyloid plaques in Alzheimer's disease. Acta Pharm Sin B. 2015;5:25–33. doi: 10.1016/j.apsb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Zhang L, Nazare M, Yao Q, Hu HY. A novel nitroreductase-enhanced MRI contrast agent and its potential application in bacterial imaging. Acta Pharm Sin B 2018, Available from: <10.1016/j.apsb.2017.11.001> [DOI] [PMC free article] [PubMed]
- 5.Li C., Tebo A.G., Gautier A. Fluorogenic labeling strategies for biological imaging. Int J Mol Sci. 2017;18:1473–1483. doi: 10.3390/ijms18071473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klymchenko A.S. Solvatochromic and fluorogenic dyes as environment-sensitive probes: design and biological applications. Acc Chem Res. 2017;50:366–375. doi: 10.1021/acs.accounts.6b00517. [DOI] [PubMed] [Google Scholar]
- 7.Kong J., Shi Y., Wang Z., Pan Y. Interactions among SARS-CoV accessory proteins revealed by bimolecular fluorescence complementation assay. Acta Pharm Sin B. 2015;5:487–492. doi: 10.1016/j.apsb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jullien L., Gautier A. Fluorogen-based reporters for fluorescence imaging: a review. Methods Appl Fluoresc. 2015;3:042007. doi: 10.1088/2050-6120/3/4/042007. [DOI] [PubMed] [Google Scholar]
- 9.Keppler A., Gendreizig S., Gronemeyer T., Pick H., Vogel H., Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 10.Srikun D., Albers A.E., Nam C.I., Iavarone A.T., Chang C.J. Organelle-targetable fluorescent probes for imaging hydrogen peroxide in living cells via SNAP-Tag protein labeling. J Am Chem Soc. 2010;132:4455–4465. doi: 10.1021/ja100117u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leng S., Qiao Q.L., Gao Y., Miao L., Deng W.G., Xu Z.C. SNAP-tag fluorogenic probes for wash free protein labeling. Chin Chem Lett. 2017;28:1911–1915. [Google Scholar]
- 12.Gautier A., Juillerat A., Heinis C., Corrêa I.R., Jr, Kindermann M., Beaufils F. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Martin B.R., Giepmans B.N., Adams S.R., Tsien R.Y. Mammalian cell–based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 14.Los G.V., Encell L.P., McDougall M.G., Hartzell D.D., Karassina N., Zimprich C. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 15.Li D., Liu L., Li W.H. Genetic targeting of a small fluorescent zinc indicator to cell surface for monitoring zinc secretion. ACS Chem Biol. 2015;10:1054–1063. doi: 10.1021/cb5007536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan Q., Bruchez M.P. Advances in chemical labeling of proteins in living cells. Cell Tissue Res. 2015;360:179–194. doi: 10.1007/s00441-015-2145-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Specht E.A., Braselmann E., Palmer A.E. A critical and comparative review of fluorescent tools for live-cell imaging. Annu Rev Physiol. 2017;79:93–117. doi: 10.1146/annurev-physiol-022516-034055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szent-Gyorgyi C., Schmidt B.F., Creeger Y., Fisher G.W., Zakel K.L., Adler S. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat Biotechnol. 2008;26:235–240. doi: 10.1038/nbt1368. [DOI] [PubMed] [Google Scholar]
- 19.Falco C.N., Dykstra K.M., Yates B.P., Berget P.B. scFv-based fluorogen activating proteins and variable domain inhibitors as fluorescent biosensor platforms. Biotechnol J. 2009;4:1328–1336. doi: 10.1002/biot.200900075. [DOI] [PubMed] [Google Scholar]
- 20.Senutovitch N., Stanfield R.L., Bhattacharyya S., Rule G.S., Wilson I.A., Armitage B.A. A variable light domain fluorogen activating protein homodimerizes to activate dimethylindole red. Biochemistry. 2012;51:2471–2485. doi: 10.1021/bi201422g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szent-Gyorgyi C., Stanfield R.L., Andreko S., Dempsey A., Ahmed M., Capek S. Malachite green mediates homodimerization of antibody VL domains to form a fluorescent ternary complex with singular symmetric interfaces. J Mol Biol. 2013;425:4595–4613. doi: 10.1016/j.jmb.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan Q., Schwartz S.L., Maji S., Huang F., Szent-Gyorgyi C., Lidke D.S. Localization microscopy using noncovalent fluorogen activation by genetically encoded fluorogen-activating proteins. Chemphyschem. 2014;15:687–695. doi: 10.1002/cphc.201300757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitzpatrick J.A., Yan Q., Sieber J.J., Dyba M., Schwarz U., Szent-Gyorgyi C. STED nanoscopy in living cells using fluorogen activating proteins. Bioconjugate Chem. 2009;20:1843–1847. doi: 10.1021/bc900249e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yates B.P., Peck M.A., Berget P.B. Directed evolution of a fluorogen-activating single chain antibody for function and enhanced brightness in the cytoplasm. Mol Biotechnol. 2013;54:829–841. doi: 10.1007/s12033-012-9631-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Telmer C.A., Verma R., Teng H., Andreko S., Law L., Bruchez M.P. Rapid, specific, no-wash, far-red fluorogen activation in subcellular compartments by targeted fluorogen activating proteins. ACS Chem Biol. 2015;10:1239–1246. doi: 10.1021/cb500957k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Özhalici-Ünal H., Pow C.L., Marks S.A., Jesper L.D., Silva G.L., Shank N.I. A rainbow of fluoromodules: a promiscuous scFv protein binds to and activates a diverse set of fluorogenic cyanine dyes. J Am Chem Soc. 2008;130:12620–12621. doi: 10.1021/ja805042p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zanotti K.J., Silva G.L., Creeger Y., Robertson K.L., Waggoner A.S., Berget P.B. Blue fluorescent dye-protein complexes based on fluorogenic cyanine dyes and single chain antibody fragments. Org Biomol Chem. 2011;9:1012–1020. doi: 10.1039/c0ob00444h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pratt C.P., He J., Wang Y., Barth A.L., Bruchez M.P. Fluorogenic Green-Inside Red-Outside (GIRO) labeling approach reveals adenylyl cyclase-dependent control of BKα surface expression. Bioconjugate Chem. 2015;26:1963–1971. doi: 10.1021/acs.bioconjchem.5b00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holleran J., Brown D., Fuhrman M.H., Adler S.A., Fisher G.W., Jarvik J.W. Fluorogen-activating proteins as biosensors of cell-surface proteins in living cells. Cytometry A. 2010;77:776–782. doi: 10.1002/cyto.a.20925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan Q., Schmidt B.F., Perkins L.A., Naganbabu M., Saurabh S., Andreko S.K. Near-instant surface-selective fluorogenic protein quantification using sulfonated triarylmethane dyes and fluorogen activating proteins. Org Biomol Chem. 2015;13:2078–2086. doi: 10.1039/c4ob02309a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Araneda J.F., Piers W.E., Heyne B., Parvez M., McDonald R. High stokes shift anilido-pyridine boron difluoride dyes. Angew Chem Int Ed Engl. 2011;50:12214–12217. doi: 10.1002/anie.201105228. [DOI] [PubMed] [Google Scholar]
- 32.Szent-Gyorgyi C., Schmidt B.F., Fitzpatrick J.A., Bruchez M.P. Fluorogenic dendrons with multiple donor chromophores as bright genetically targeted and activated probes. J Am Chem Soc. 2010;132:11103–11109. doi: 10.1021/ja9099328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yushchenko D.A., Zhang M., Yan Q., Waggoner A.S., Bruchez M.P. Genetically targetable and color-switching fluorescent probe. Chembiochem. 2012;13:1564–1568. doi: 10.1002/cbic.201200334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naganbabu M., Perkins L.A., Wang Y., Kurish J., Schmidt B.F., Bruchez M.P. Multiexcitation fluorogenic labeling of surface, intracellular, and total protein pools in living cells. Bioconjugate Chem. 2016;27:1525–1531. doi: 10.1021/acs.bioconjchem.6b00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q., Wang Q., Sun Y., Zuo L., Fetz V., Hu H.Y. Superior fluorogen-activating protein probes based on 3-indole-malachite green. Org Lett. 2017;19:4496–4499. doi: 10.1021/acs.orglett.7b02055. [DOI] [PubMed] [Google Scholar]
- 36.Frangioni J.V. In vivo near-infrared fluorescence imaging. Curr Opin Chem Biol. 2003;7:626–634. doi: 10.1016/j.cbpa.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Kiyose K., Kojima H., Nagano T. Functional near-infrared fluorescent probes. Chem Asian J. 2008;3:506–515. [Google Scholar]
- 38.Zhang M., Chakraborty S.K., Sampath P., Rojas J.J., Hou W., Saurabh S. Fluoromodule-based reporter/probes designed for in vivo fluorescence imaging. J Clin Invest. 2015;125:3915–3927. doi: 10.1172/JCI81086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fisher G.W., Adler S.A., Fuhrman M.H., Waggoner A.S., Bruchez M.P., Jarvik J.W. Detection and quantification of β2AR internalization in living cells using FAP-based biosensor technology. J Biomol Screen. 2010;15:703–709. doi: 10.1177/1087057110370892. [DOI] [PubMed] [Google Scholar]
- 40.Shruti S., Urban-Ciecko J., Fitzpatrick J.A., Brenner R., Bruchez M.P., Barth A.L. The brain-specific Beta4 subunit downregulates BK channel cell surface expression. PLoS One. 2012;7:e33429. doi: 10.1371/journal.pone.0033429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fisher G.W., Fuhrman M.H., Adler S.A., Szent-Gyorgyi C., Waggoner A.S., Jarvik J.W. Self-checking cell-based assays for GPCR desensitization and resensitization. J Biomol Screen. 2014;19:1220–1226. doi: 10.1177/1087057114534299. [DOI] [PubMed] [Google Scholar]
- 42.Boeck J.M., Spencer J.V. Effect of human cytomegalovirus (HCMV) US27 on CXCR4 receptor internalization measured by fluorogen-activating protein (FAP) biosensors. PLoS One. 2017;12:e0172042. doi: 10.1371/journal.pone.0172042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holleran J.P., Glover M.L., Peters K.W., Bertrand C.A., Watkins S.C., Jarvik J.W. Pharmacological rescue of the mutant cystic fibrosis transmembrane conductance regulator (CFTR) detected by use of a novel fluorescence platform. Mol Med. 2012;18:685–696. doi: 10.2119/molmed.2012.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holleran J.P., Zeng J., Frizzell R.A., Watkins S.C. Regulated recycling of mutant CFTR is partially restored by pharmacological treatment. J Cell Sci. 2013;126:2692–2703. doi: 10.1242/jcs.120196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsen M.B., Hu J., Frizzell R.A., Watkins S.C. Simple image-based no-wash method for quantitative detection of surface expressed CFTR. Methods. 2016;96:40–45. doi: 10.1016/j.ymeth.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Y., Tapia P.H., Fisher G.W., Simons P.C., Strouse J.J., Foutz T. Discovery of regulators of receptor internalization with high-throughput flow cytometry. Mol Pharmacol. 2012;82:645–657. doi: 10.1124/mol.112.079897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y., Tapia P.H., Fisher G.W., Waggoner A.S., Jarvik J., Sklar L.A. High-throughput flow cytometry compatible biosensor based on fluorogen activating protein technology. Cytometry A. 2013;83:220–226. doi: 10.1002/cyto.a.22242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ackerman D.S., Vasilev K.V., Schmidt B.F., Cohen L.B., Jarvik J.W. Tethered fluorogen assay to visualize membrane apposition in living cells. Bioconjugate Chem. 2017;28:1356–1362. doi: 10.1021/acs.bioconjchem.7b00047. [DOI] [PubMed] [Google Scholar]
- 49.Ferreira K., Hu H.Y., Fetz V., Prochnow H., Rais B., Müller P.P. Multivalent siderophore-DOTAM conjugates as theranostics for imaging and treatment of bacterial infections. Angew Chem Int Ed Engl. 2017;56:8272–8276. doi: 10.1002/anie.201701358. [DOI] [PubMed] [Google Scholar]
- 50.Saurabh S., Perez A.M., Comerci C.J., Shapiro L., Moerner W.E. Super-resolution imaging of live bacteria cells using a genetically directed, highly photostable fluoromodule. J Am Chem Soc. 2016;138:10398–10401. doi: 10.1021/jacs.6b05943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 53.Schwartz S.L., Yan Q., Telmer C.A., Lidke K.A., Bruchez M.P., Lidke D.S. Fluorogen-activating proteins provide tunable labeling densities for tracking FcεRI independent of IgE. ACS Chem Biol. 2015;10:539–546. doi: 10.1021/cb5005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grover A., Schmidt B.F., Salter R.D., Watkins S.C., Waggoner A.S., Bruchez M.P. Genetically encoded pH sensor for tracking surface proteins through endocytosis. Angew Chem Int Ed Engl. 2012;51:4838–4842. doi: 10.1002/anie.201108107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saunders M.J., Liu W., Szent-Gyorgyi C., Wen Y., Drennen Z., Waggoner A.S. Engineering fluorogen activating proteins into self-assembling materials. Bioconjugate Chem. 2013;24:803–810. doi: 10.1021/bc300613h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Magenau A.J., Saurabh S., Andreko S.K., Telmer C.A., Schmidt B.F., Waggoner A.S. Genetically targeted fluorogenic macromolecules for subcellular imaging and cellular perturbation. Biomaterials. 2015;66:1–8. doi: 10.1016/j.biomaterials.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu W., Saunders M.J., Bagia C., Freeman E.C., Fan Y., Gawalt E.S. Local retention of antibodies in vivo with an injectable film embedded with a fluorogen-activating protein. J Control Release. 2016;230:1–12. doi: 10.1016/j.jconrel.2016.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He J., Wang Y., Missinato M.A., Onuoha E., Perkins L.A., Watkins S.C. A genetically targetable near-infrared photosensitizer. Nat Methods. 2016;13:263–268. doi: 10.1038/nmeth.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pena K., Larsen M., Calderon M., Tsang M., Watkins S.C., Bruchez M.P. Combining novel probes and high resolution imaging to dissect mitochondrial function in living systems. Microsc Microanal. 2017;23:1170–1171. [Google Scholar]
- 60.Wang Y., Ballou B., Schmidt B.F., Andreko S., St, Croix C.M., Watkins S.C. Affibody-targeted fluorogen activating protein for in vivo tumor imaging. Chem Commun. 2017;53:2001–2004. doi: 10.1039/c6cc09137g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gallo E., Vasilev K.V., Jarvik J. Fluorogen-activating-proteins as universal affinity biosensors for immunodetection. Biotechnol Bioeng. 2014;111:475–484. doi: 10.1002/bit.25127. [DOI] [PMC free article] [PubMed] [Google Scholar]