Abstract
Panax notoginseng is famous for its important therapeutic effects. Saponins are bioactive compounds found in different parts and developmental stages of P. notoginseng plants. Thus, it is urgently to study saponins distribution in different parts and growth ages of P. notoginseng plants. In this study, potential biomarkers were found, and their chemical characteristic differences were revealed through metabolomic analysis. High-performance liquid chromatography data indicated the higher content of saponins (i.e., Rg1, Re, Rd, and Rb1) in the underground parts than that in the aerial parts. 20(S)-Protopanaxadiol saponins were mainly distributed in the aerial parts. Additionally, the total saponin content in the 3-year-old P. notoginseng plant (188.0 mg/g) was 1.4-fold higher than that in 2-year-old plant (130.5 mg/g). The transcriptomic analysis indicated the tissue-specific transcription expression of genes, namely, PnFPS, PnSS, PnSE1, PnSE2, and PnDS, which encoded critical synthases in saponin biosyntheses. These genes showed similar expression patterns among the parts of P. notoginseng plants. The expression levels of these genes in the flowers and leaves were 5.2fold higher than that in the roots and fibrils. These results suggested that saponins might be actively synthesized in the aerial parts and transformed to the underground parts. This study provides insights into the chemical and genetic characteristics of P. notoginseng to facilitate the synthesis of its secondary metabolites and a scientific basis for appropriate collection and rational use of this plant.
Abbreviations: AACT, acetoacetyl-CoA acyltransferase; DS, dammarenediol-II synthase; DXPS, 1-deoxy-d-xylulose 5-phosphate synthase; DXPR, 1-deoxy-d-xylulose 5-phosphate reductoisomerase; FPS, farnesyl pyrophosphate synthase; FPP, farnesyl diphosphate; GDPS, gerenyl diphosphatesynthase; HDS, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; HPLC—UV, high-performance liquid chromatography-ultraviolet detection; IPP, isoprenyl diphosphate; IPPI, isopentenyl pyrophosphate isomerase; ISPD, 2-C-methylerythritol 4-phosphatecytidyl transferase; ISPE, 4-(cytidine-5′-diphospho)-2-C-methylerythritol kinase; ISPH, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase; MECPS, 2-C-methylerythritol-2,4-cyclophosphate synthase; MEP, 2-C-methyl-d-erythritol-4-phosphate; MVA, mevalonate; MVDD, mevalonate diphosphate decarboxylase; MVK, mevalonate kinase; OPLS-DA, orthogonal partial least-squares discrimination analysis; P450, P450-monooxygenase; PCA, principal component analysis; PDS, 20(S)-protopanaxadiol saponins; PMK, phosphomevalonate kinase; PTS, 20(S)-protopanaxatriol saponins; SE, squalene epoxidase; SS, squalene synthase; UGTs, UDP-glycosyltransferases; UPLC—MS, ultrahigh-performance liquid chromatography–mass spectrometry; VIP, variable importance in projection
KEY WORDS: Panax notoginseng, Saponin, Growth years, Metabolomic analyses, Gene expression
Graphical abstract
This study elaborates the distribution of saponins and the expression patterns of related genes in different parts and developmental stages of P. notoginseng plants.
1. Introduction
Panax notoginseng (Burk) F.H. Chen is a perennial plant famous for its important therapeutic effects, such as antidepressant, estrogen-like activity, anti-tumor, anti-oxidant, hepatoprotective, and anti-inflammation1, 2, 3, 4. P. notoginseng is commonly used as a single or major ingredient in capsules, tablets, pills, and medicinal liquors. P. notoginseng is widely consumed in the health food market5. The major active components of this plant are protopanaxadriol and protopanaxatriol saponins, whose pharmacological effects are related to the different plant parts and developmental stages6, 7. Saponins from the leaves and flowers of P. notoginseng exert anti-tumor and hepatoprotective effects8, 9. Additionally, the saponin contents of P. notoginseng can be easily influenced by region of origin, resource10, harvest time11, and processing12. Mature P. notoginseng are generally more expensive than the younger ones because of their higher quality5. 3-Year-old P. notoginseng plants mainly serve as medicinal materials in the market, and 2-year-old samples are also sold in the market. The parts and developmental stages of P. notoginseng plants can influence the safe use of medicinal materials. There are fewer studies to clarify the chemical characteristics in different parts and growth stages of P. notoginseng plants. Metabolomics is a scientific study that reveals metabolite differences for authentication of species13, 14, 15, 16, 17 differentiation of cultivation regions18, 19, age discrimination20, 21, and discrimination of raw and processed samples22. Thus, the chemical characteristics of saponins in the aerial and underground parts of 2- and 3-year-old P. notoginseng plants could be analyzed.
Study has reported the triterpene saponins synthesized through 2-C-methyl-d-erythritol-4-phosphate and mevalonate pathways23. Putative cytochrome P450-dependent monoxygenases and UDP-glycosyltransferases involved in triterpene saponin biosynthesis were discovered24. In this process, the main enzymes involved are farnesyl pyrophosphate synthase (FPS), squalene synthase (SS), squalene epoxidase (SE), and dammarenediol-II synthase (DS). Niu et al.25 cloned genes (i.e., PnFPS, PnSS, PnSEs, and PnDS) that encoded these enzymes and analyzed their tissue-specific expression pattern by using mature 4-year-old P. notoginseng plants. However, such expression patterns have not been reported in different parts of 2- and 3-year-old P. notoginseng plants. Further analysis of the expression patterns of these genes could contribute to reveal saponin distributions within P. notoginseng.
In this study, we investigated the characteristic chemical markers of saponins in different parts of 2- and 3-year-old P. notoginseng plants through ultrahigh-performance liquid chromatography–mass spectrometry (UPLC—MS) analysis. Saponin content was further determined through high-performance liquid chromatography (HPLC—UV) analysis to define their distribution. The tissue-specific expression patterns of genes related to saponin synthesis were analyzed to reveal their molecular characteristics. Our findings provide a scientific basis for appropriate collection and application of P. notoginseng, and help for resolving the issue of drug safety usage.
2. Materials and methods
2.1. Materials and chemicals
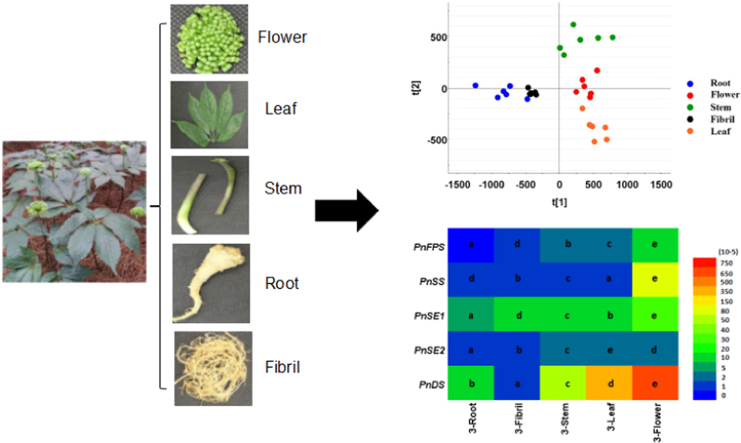
Six batches of whole 2- and 3-year-old P. notoginseng plants in their flowering stage were collected from Wenshan, Yunnan Province, China (N 23.5° and E 104°). Each plant was separated into five different parts including the root, fibril, stem, leaf, and flower (Fig. 1). All voucher specimens were carefully washed, cut into small pieces, and immediately deposited at −80 °C for further processing.
Figure 1.
Plant traits of P. notoginseng. (A) Stem; (B) root; (C) fibril; (D) flower; and (E) leaf.
Six standards of saponins (i.e., Rg1, Re, Rd, Rb1, Rb2, and Rc) and a standard of notoginsenoside R1 with 98.0% purity were purchased from Shanghai Tauto Biotech Company (Shanghai, China). The standard stock solutions were dissolved in methanol (Fair Lawn, NJ, USA) and stored at −20 °C prior to use. Other chemicals and solvents were of analytical grade.
2.2. HPLC—MS conditions
All samples were crushed, and 0.1 g of the powdered sample was weighed and mixed with 1.0 mL of pure methanol containing 0.1% formic acid under vortex for 10 s. The mixture was sonicated for 10 min, frozen at −20 °C for 1 h, and centrifuged at 10,000 rpm (Cetrifuge, Sigma 3-30KS, Germany) for 10 min. The upper layer was collected, filtered through 0.22 µm filter, and transferred to a sample vial. The vial was injected into the column for UPLC–quadruple time-of-flight (QTOF)/MS analysis.
The UPLC—MS analysis was performed using a UPLC system (Waters, UK) coupled to an electrospray ionization-QTOF/MS (Waters, UK). A C18 reversed phase column (50 mm × 2.1 mm, i.d. 1.7 µm, Acquity HPLC BEH, Waters, UK) was used for UPLC separation, and the sample injection volume was 10 µL. The column temperature was kept at 35 °C, and the flow rate was kept at 0.4 mL/min. The gradient was composed of water containing 0.1% formic acid (A) and acetonitrile containing 0.1% formic acid (B). The linear gradient was set as follows: 0–2 min for 1% B to 20% B, 2–3 min for 20% B to 50% B, 3–7 min for 50% B to 80% B, 7–7.5 min for 80% B to 99% B, 7.5–9 min for 99% B, 9–9.1 min for 99% B to 1% B, and 9.1–10 min for 1% B.
High-accuracy MS data were recorded using MassLynx 4.1 software (Waters, UK). The capillary voltage was set at 3.0 kV for positive and negative modes, and the cone voltage was set at 40 V for both modes. The source temperature was set at 120 °C with a cone gas flow of 50 L/h. The desolvation temperature was set at 400 °C with desolvation gas flow of 800 L/h. MSE data were acquired in continuous mode by using ramp collision energy in two scan functions. The conditions for low-energy mode are scan range of 50–2000 Da, scan time of 0.2 s, and collision energy of 6 V. The conditions for high-energy mode are scan range of 50–2000 Da, scan time of 0.2 s, and collision energy ramp of 10–40 V.
2.3. HPLC–UV conditions
Agilent HPLC 1260 series system (Agilent, USA) equipped with a quaternary pump, an autosampler, a column compartment, and a VWD was used for HPLC analysis. A C18 reversed phase column (250 mm × 4.6 mm, i.d. 5 µm, Eclipse XDB; Agilent, USA) was used for separation, and the sample injection volume was set as 10 µL. The column temperature was kept at 25 °C, the flow rate at 1.0 mL/min, and wavelength at 203 nm. The Gradient was composed of water (A) and acetonitrile (B), and the linear gradient was set as follows: 0–12 min for 19% B and 12–60 min for 19% B to 36% B.
2.4. Gene expression conditions
Total RNA was isolated from the root, fibril, stem, leaf, and flower by using a plant RNA Isolation Mini Kit (BioTeke, Beijing, China). Concentration and quality of the extracted RNA were tested using NanoDrop 2000 spectrophotometer and 1% agarose gel electrophoresis before cDNA synthesis.
Approximately 800 ng of DNase I-treated total RNA was converted into single-stranded cDNA by using FastQuant RT Kit (With gDNase) (TIANGEN, Beijing, China). The cDNA products were diluted 10fold with deionized water and used as a template for real-time PCR. All primers used in this study were synthesized based on the description of Niu et al.25. The quantitative reaction was conducted using an SYBR®Green Realtime PCR Master Mix (Toyobo, Japan). The reaction mixture contained 10 µL of SYBR Green qPCR Master Mix, 0.8 µL of 10 µmol/L each of the forward and reverse primers, and 2 µL of the cDNA template and added with sterile water to obtain a final volume of 30 µL. Each reaction was repeated three times. PCR amplification was conducted under the following conditions: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 55 °C for 15 s, and 72 °C for 15 s. The expression levels of the key genes were calculated by comparing the cycle threshold value (Ct) for each gene to the Ct of the reference gene 18S rRNA24.
2.5. Data analysis
LC—MS raw data were transformed into MassLynx 4.1 Software (Waters, USA) to obtain the molecular features of the samples for non-targeted metabolic analysis of the extracted features. Multivariate data analysis was conducted with SIMCA-P software. Principal component analysis (PCA) was performed to obtain an overview of the sample distribution and observe possible outliers. Orthogonal partial least-squares discrimination analysis (OPLS-DA) was performed to identify the metabolites that significantly contributed to clustering and discrimination. S-plot and variable importance in projection (VIP) were used to evaluate the variable contribution. Variables with VIP >1 were deemed as potential biomarkers. HPLC and gene expression data were managed by SPSS 17.0 software for analysis of variance. Mean values were set as significant or non-significant by paired t-tests (P<0.05).
3. Results
3.1. Analysis of metabolic profiling in P. notoginseng plants
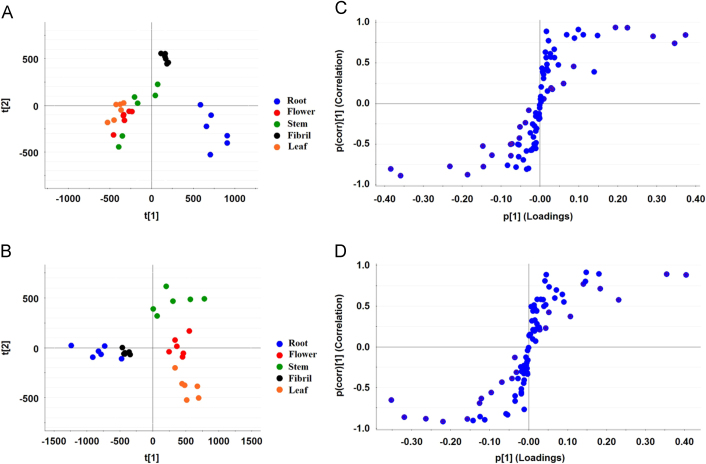
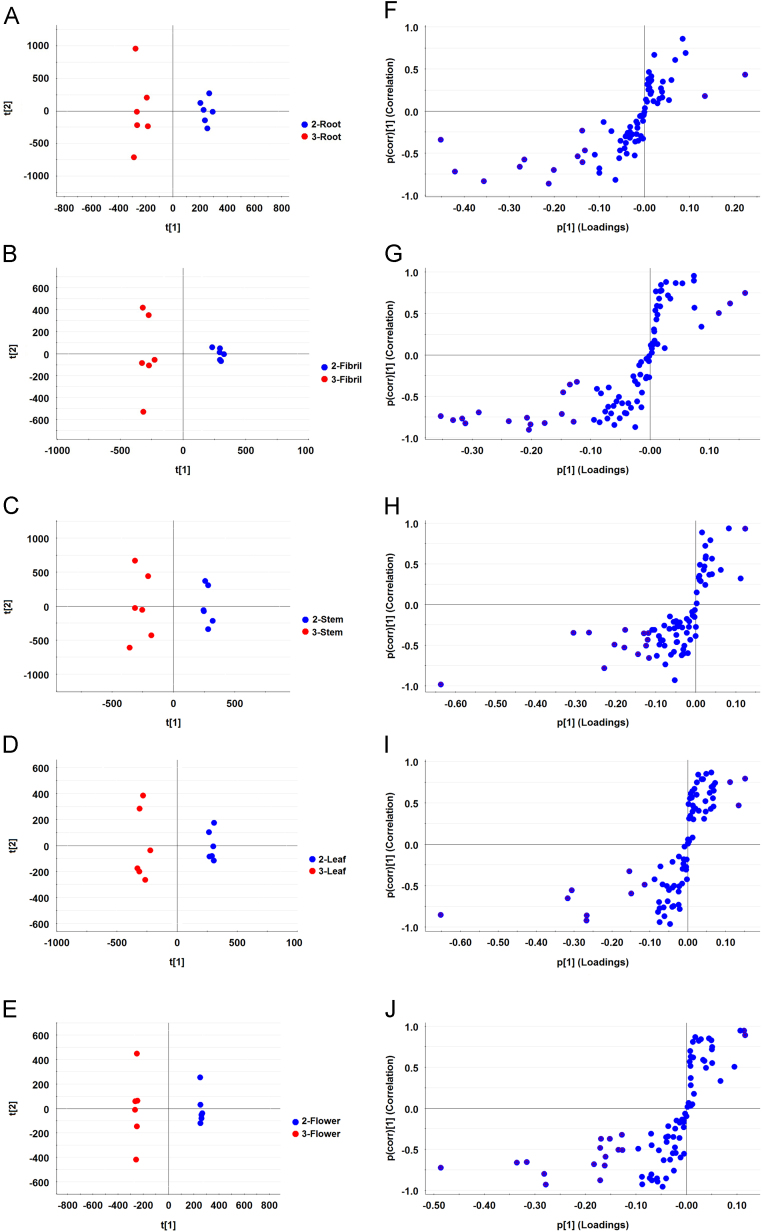
The chemical components in different parts and developmental stages of P. notoginseng plants were detected through UPLC—MS analysis (Supplementary Information Figs. S1 and S2). Saponin content was also detected in different parts and developmental stages of P. notoginseng (Fig. 2). In 2-year-old P. notoginseng plants, the root and fibril were clearly separated; however, the aerial parts (stem, leaf, and flower) overlapped each other and were not perfectly separated (Fig. 2A). The S-plot derived by OPLS-DA presents “S-form,” and the upper-right and lower-left variables were available ions that contributed to the differences (Fig. 2C). The 24 potential biomarkers with VIP values > 1 were presented in Supplementary Information Fig. S3A. The PCA results of 3-year-old P. notoginseng plant indicated that the five parts were separated from one another. PC1 revealed the differences between the underground parts (i.e., root and fibril) and aerial parts (i.e., stem, leaf, and flower) (Fig. 2B). Twenty-one potential biomarkers were identified based on the results of S-plot and VIP values (Fig. 2D and Supplementary Information Fig. S3B). The metabolic characteristics of saponin were compared in the same parts of 2- and 3-year-old P. notoginseng plants. The PCA results revealed that saponins differed in the same parts of 2- and 3-year P. notoginseng plants (Fig. 3A–E). OPLS—DA analysis showed potential markers in different growth stages of P. notoginseng plants. Ions that contributed to the separation were selected from the S-plot and regarded as potential markers (Fig. 3F–J). Potential markers that decided the differences were also identified by VIP values. A total of 13, 18, 14, 11, and 17 potential biomarkers (VIP>1) were identified in the roots, fibrils, stems, leaves, and flowers, respectively (Supplementary Information Fig. S4). These results indicated that metabolites exhibited different distribution patterns in different parts and growth stages of P. notoginseng plants.
Figure 2.
Metabolic analysis of saponins in different parts of P. notoginseng. 2- and 3-year-old P. notoginseng samples were used in saponin metabolite analysis by UPLC—MS. (A, C) PCA score plots and S-plot of the five parts in 2-year-old P. notoginseng; and (B, D) PCA score plots and S-plot of the five parts in 3-year-old P. notoginseng.
Figure 3.
Metabolic analysis of saponins in different developmental stages. 2- and 3-Year-old of P. notoginseng samples were used in the saponin metabolite analysis by UPLC—MS. (A—E) PCA score plots of the roots, fibrils, stems, leaves, and flowers at two developmental stages, respectively; (F—J) S-plot of the roots, fibrils, stems, leaves, and flowers at two developmental stages, respectively.
3.2. Analysis of saponin content in P. notoginseng
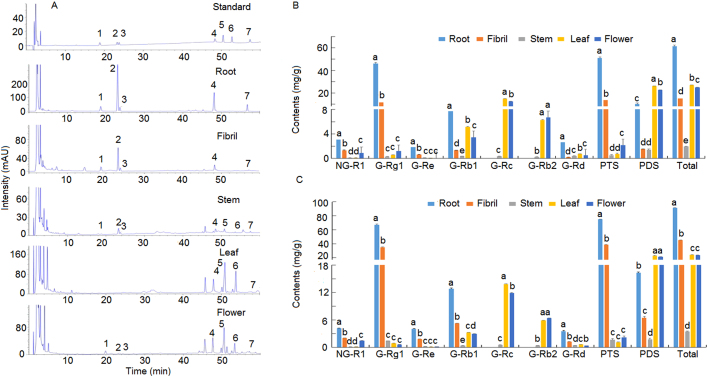
Saponin content was detected in different plant parts and developmental stages through HPLC analysis. The analysis method and the HPLC chromatogram of each compound were provided in Supplementary Information Table S1 and Fig. 4A. The linearity of all calibration curves were R2>0.999. The precision, repeatability, and stability variations were 0.03%–1.12%, 0.19%–2.68%, and 1.21%–2.99%, respectively. The recovery ranged from 95.84% to 102.37%, with variations of 0.56%–2.87%. These results showed that the established HPLC method could be used for accurate and sensitive quantitative analyses of notoginsenoside R1 and six major saponins (i.e., Rg1, Re, Rc, Rd, Rb1, and Rb2). The contents of main saponins significantly differed among different plant parts and developmental stages (Fig. 4B and C). In 2-year-old P. notoginseng plant, the highest levels of R1, Rg1, and Re were found in the roots (3.1, 46.1, and 1.8 mg/g, respectively), followed by fibrils (Fig. 4B). Moreover, Rb1 and Rd exhibited high levels in the roots and leaves. Rc and Rb2 were not detected in the underground parts (roots and fibrils) but were found at high levels in the leaves and flowers. In 3-year-old P. notoginseng plants, high levels of R1, Rg1, Re, Rb1, and Rd (4.1, 67.1, 4.0, 12.8, and 3.5 mg/g, respectively) were found in the roots. The distribution patterns of Rc and Rb2 were similar to those in 2-year-old P. notoginseng plant (Fig. 4C). In both 2- and 3-year old P. notoginseng plants, the contents of 20(S)-protopanaxatriol saponins (PTS), which included R1, Rg1, and Re, were high in the underground parts (i.e., roots and fibrils); meanwhile, the contents of 20(S)-protopanaxadiol saponins (PDS), which included Rb1, Rc, Rb2, and Rd, were high in the aerial parts (i.e., leaves and flowers). The total saponin content in 3-year-old P. notoginseng (188.0 mg/g) was 1.4-fold higher than that in 2-year-old one (130.5 mg/g). These results indicated the high saponin contents (R1, Rg1, Re, Rb1, and Rd) were in the roots, while high Rb2 and Rc contents in the leaves and flowers. Hence, the contents and types of saponins were unenvenly distributed in P. notoginseng.
Figure 4.
HPLC chromatograms and contents of saponins in P. notoginseng. (A) HPLC chromatogram profiles of P. notoginseng plants. 1. Notoginseng R1; 2. Ginsenoside Rg1; 3. Ginsenoside Re; 4. Ginsenoside Rb1; 5. Ginsenoside Rc; 6. Ginsenoside Rb2; 7. Ginsenoside Rd. (B) Contents of the seven saponins in 2-year-old P. notoginseng. (C) Contents of the seven saponins in 3-year-old P. notoginseng. NG: notoginsenoside; G: ginsenoside; PTS: total amount of 20(S)-protopanaxatriol saponins (PDS) including notoginsenoside R1 and ginsenosides Rg1 and Re; PDS: total amount of PDS including ginsenoside Rb1, Rc, Rb2, and Rd; Total: total amount of PTS and PDS including notoginsenoside R1 and ginsenosides Rg1, Re, Rb1, Rc, Rb2, and Rd. Different letters represent significant difference among the five parts at P<0.05.
3.3. Gene expression profiling
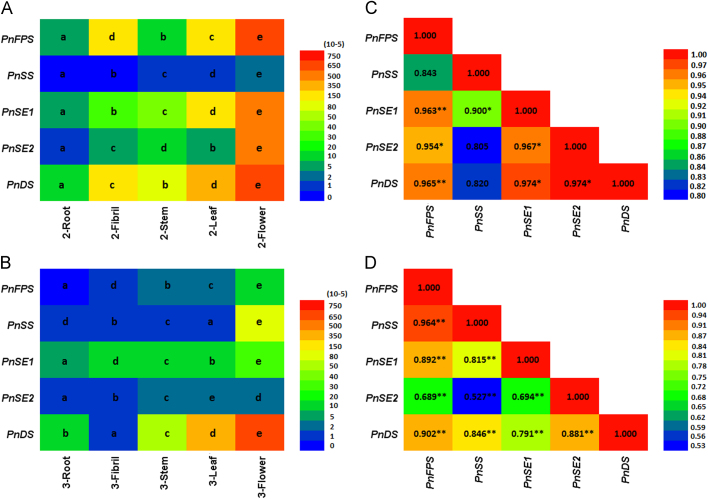
The tissue-specific expression patterns of five genes involved in triterpene saponin biosynthesis were evaluated through reverse transcriptional quantitative PCR. The relative expression of genes (i.e., PnFPS, PnSS, PnSE1, PnSE2, and PnDS) demonstrated significantly similar expression patterns among the five parts (i.e., roots, fibrils, stems, flowers, and leaves) at two developmental stages (Fig. 5). In 2-year-old P. notoginseng, the transcript levels of these genes were higher in the flowers and leaves than those in the roots and fibrils (Fig. 5A). The expression levels of PnFPS, PnSS, PnSE1, PnSE2, and PnDS in the flowers were 85.0-, 120.7-, 111.4-, 528.3-, and 49.0-fold higher than that in the roots, respectively. Compared with the 2-year-old P. notoginseng plant, the 3-year-old P. notoginseng plant showed similar gene expression profiling but slightly lower expression levels (Fig. 5B). Four genes (i.e., PnFPS, PnSS, PnSE1, and PnDS) were highly expressed in the flowers, with 100.3-, 124.0-, 5.2-, and 42.7-fold higher expression level than those in the roots. The expression of PnSE1 was the highest in the leaves and 7.1-fold higher than that in the roots. Additionally, the expression of the five genes exhibited significant correlations among the five parts (P<0.05, Fig. 5C and D). Hence, the expression levels of PnFPS, PnSS, PnSE1, PnSE2, and PnDS in the flowers were significantly higher than that in the roots.
Figure 5.
Expression profiling and Pearson correlation analysis of the five genes in P. notoginseng. Different letters represent significant differences. (A) Expression levels of the five genes in 2-year-old P. notoginseng. (B) Expression levels of the five genes in 3-year-old P. notoginseng. (C) Pearson correlation analysis of the five genes in 2-year-old P. notoginseng. (D) Pearson correlation analysis of the five genes in 3-year-old P. notoginseng. Different letters represent significant differences at P<0.05. The symbol * represents a significant correlation among the five genes at P<0.05, and the symbol ** represents a highly significant correlation between the five genes at P<0.01.
4. Discussion
In this study, integrated metabolomic and transcriptomic analysis were used to determine the saponin distribution and the expression patterns of genes encoding key enzymes involved in saponin biosynthesis in P. notoginseng plants. Potential biomarkers were found in the aerial and underground parts of 2- and 3-year-old P. notoginseng plants. PTS and PDS showed tissue-specific distribution. The total saponin content was markedly higher in the 3-year-old P. notoginseng plant than that in the 2-year-old plant. Five genes (i.e., PnFPS, PnSS, PnSE1, PnSE2, and PnDS) showed higher expression in the flowers and leaves than that in the underground parts.
Metabolite profiles have been applied to evaluate chemical characteristics in different root parts (i.e., main roots, rhizomes, branch roots, and fibrous roots) of P. notoginseng26. In the present study, the PCA results revealed that saponins differed between the underground and aerial parts of P. notoginseng plants. The results of S-plot and VIP analyses indicated that these biomarkers were mainly PTS/PDS-type saponins, and metabolic differences in different plant parts and growth stages.
Triterpenoids possess important pharmacological effects, such as hypoglycemic activity (Rg1)27, antioxidant activity (Re)28, neuroprotection activity (Rb1)29, improvement in learning and memory abilities (Rd)30, immunity activity (Rb2)31, and anti-inflammatory activity (Rc)32. Notoginsenoside R1 and ginsenoside (i.e., Rg1, Re, Rb1, and Re) constitute up to 90% of the total P. notoginseng saponins33. Re and Rb1 are potential biomarkers in different parts of 3-year-old P. notoginseng plants. Thus, these saponins are the main bioactive components, and their distribution was further analyzed. The HPLC data showed that the contents of notoginsenoside R1 and saponins (i.e., Rg1, Re, Rd, and Rb1) were the highest in the roots, followed by the fibrils. This finding is consistent with the Chinese Pharmacopoeia (2015)34. PTS was detected in the whole plant, and PDS was mainly distributed in the aerial parts; these results are similar to those reported in a previous study35. Hence, the aerial parts contained certain saponins and could be used to extract saponins for raw materials or health care products. The total saponin content in 3-year-old P. notoginseng (188.0 mg/g) was 1.4-fold higher than that in the 2-year-old plant (130.5 mg/g). This work provides references for rational use of different P. notoginseng parts.
Saponin biosynthesis is a complicated process that involves in various rate-limiting enzymes36. The biosynthesis of isoprenoid through both pathways is shown in Supplementary Information Fig. S5. Mevalonate-5-pyrophosphate decarboxylase, which is the last enzyme of the mevalonic pathway, produces isoprenyl diphosphate (IPP). Farnesyl diphosphate (FPP) is derived from IPP through the catalytic reaction of farnesyl diphosphate synthase (FPS)37. The two FPP molecules are placed into a C30 isopreniod squalene by SS38. The first oxygenation step in triterpenoid biosynthesis is performed by SE, which catalyzes the epoxidation of the double bond of squalene to form 2,3-oxidosqualene23. DS catalyzes the cyclization of 2,3-oxidosqualene to form various secondary metabolites39. The five genes (i.e., PnFPS, PnSS, PnSE1, PnSE2, and PnDS) were confirmed to encode FPS, SS, SE, and DS in saponin biosyntheses25. In the present study, the transcript levels of PnFPS, PnSS, PnSE1, PnSE2, and PnDS were higher in the flowers and leaves than that those in the roots. The saponin contents (i.e., Rg1, Re, Rd, and Rb1) were significantly higher in the underground parts than that in the aerial parts. Hence, saponins might be actively synthesized in the aerial parts and transformed to the underground parts; this finding is similar to previous reports40. In a previous study, the results of 13C-labeling assays indicated that biosynthesized ginsenosides could be transported from ginseng leaves into roots41. Schramek et al.42 reported that the products of photosynthetic metabolites (glucose and fructose) contributed to ginsenoside synthesis or movement into ginseng roots. In the present study, the high gene expression profiles in the flowers and leaves confirmed that saponins might be actively synthesized in the aerial parts and transformed to the underground parts.
This study elaborates the distribution of saponins and the expression patterns of related genes in different parts and developmental stages of P. notoginseng plants. Our data provide information for understanding the biochemical diversity of secondary metabolism and molecular characteristics of saponins. These results can guide the appropriate collection and rational use of P. notoginseng plants.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (No. 81603238).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.apsb.2017.12.010.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Ng T. Pharmacological activity of sanchi ginseng (Panaxnotoginseng) J Pharm Pharmacol. 2006;58:1007–1019. doi: 10.1211/jpp.58.8.0001. [DOI] [PubMed] [Google Scholar]
- 2.Chan P., Thomas G.N., Tomlinson B. Protective effects of trilinolein extracted from Panax notoginseng against cardiovascular disease. Acta Pharmacol Sin. 2002;23:1157–1162. [PubMed] [Google Scholar]
- 3.Xiang H., Liu Y., Zhang B., Huang J., Li Y., Yang B. The antidepressant effects and mechanism of action of total saponins from the caudexes and leaves of Panax notoginseng in animal models of depression. Phytomedicine. 2011;18:731–738. doi: 10.1016/j.phymed.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 4.Sun K., Fan J., Han J. Ameliorating effects of traditional Chinese medicine preparation, Chinese materia medica and active compounds on ischemia/reperfusion-induced cerebral microcirculatory disturbances and neuron damage. Acta Pharm Sin B. 2015;1:8–24. doi: 10.1016/j.apsb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang T., Guo R., Zhou G., Zhou X., Kou Z., Sui F. Traditional uses, botany, phytochemistry, pharmacology and toxicology of Panax notoginseng (Burk.) F.H. Chen: a review. J Ethnopharmacol. 2016;188:234–258. doi: 10.1016/j.jep.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Du Q.Z., Jerz G., Waibel R., Winterhalter P. Isolation of dammarane saponins from Panax notoginseng by high-speed countercurrent chromatography. J Chromatogr A. 2003;1008:173–180. doi: 10.1016/s0021-9673(03)00988-9. [DOI] [PubMed] [Google Scholar]
- 7.Sun H., Yang Z., Ye Y. Structure and biological activity of protopanaxatriol-type saponins from the roots of Panax notoginseng. Int Immunopharmacol. 2006;6:14–25. doi: 10.1016/j.intimp.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y., Wang W., Han L., Rayburn E., Hill D., Wang H. Isolation, structural determination, and evaluation of the biological activity of 20(S)-25-methoxyl-dammarane-3beta, 12beta, 20-triol [20(S)-25-OCH3-PPD], a novel natural product from Panax notoginseng. Med Chem. 2007;3:51–60. doi: 10.2174/157340607779317508. [DOI] [PubMed] [Google Scholar]
- 9.Yoshikawa M., Morikawa T., Kashima Y., Ninomiya K., Matsuda H. Structures of new dammarane-type triterpene saponins from the flower buds of Panax notoginseng and hepatoprotective effects of principal ginseng saponins. J Nat Prod. 2003;66:922–927. doi: 10.1021/np030015l. [DOI] [PubMed] [Google Scholar]
- 10.Guan J., Lai C., Li S. A rapid method for the simultaneous determination of 11 saponins in Panax notoginseng using ultra performance liquid chromatography. J Pharm Biomed Anal. 2007;44:996–1000. doi: 10.1016/j.jpba.2007.03.032. [DOI] [PubMed] [Google Scholar]
- 11.Dong T., Cui X., Song Z., Zhao K., Ji Z., Lo C. Chemical assessment of roots of Panax notoginseng in China: regional and seasonal variations in its active constituents. J Agric Food Chem. 2003;51:4617–4623. doi: 10.1021/jf034229k. [DOI] [PubMed] [Google Scholar]
- 12.Yu H., Zhang L., Song X., Liu Y., Zhang J., Cao M. Chemical constituents from processed rhizomes of Panax notoginseng. Chin J Chin Mater Med. 2013;38:3910–3917. [PubMed] [Google Scholar]
- 13.Chen Y., Zhao Z., Chen H., Yi T., Qin M., Liang Z. Chemical differentiation and quality evaluation of commercial Asian and American ginsengs based on a UHPLC—QTOF/MS/MS metabolomics approach. Phytochem Anal. 2015;26:145–160. doi: 10.1002/pca.2546. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen H., Lee D., Lee W., Lee G., Yoon S., Shin B. UPLC—QTOFMS based metabolomics followed by stepwise partial least square-discriminant analysis (PLS-DA) explore the possible relation between the variations in secondary metabolites and the phylogenetic divergences of the genus Panax. J Chromatogr B Anal Technol Biomed Life Sci. 2016;1012:61–68. doi: 10.1016/j.jchromb.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Chan T., But P., Cheng S., Kwok I., Lau F., Xu H. Differentiation and authentication of Panaxginseng, Panax quinquefolius, and ginseng products by using HPLC/MS. Anal Chem. 2000;72:1281–1287. doi: 10.1021/ac990819z. [DOI] [PubMed] [Google Scholar]
- 16.Yang W., Qiao X., Li K., Fan J., Bo T., Guo D. Identification and differentiation of Panaxginseng, Panaxquinquefolium, and Panax notoginseng by monitoring multiple diagnostic chemical markers. Acta Pharm Sin B. 2016;6:568–575. doi: 10.1016/j.apsb.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou P., Song Y., Lei W., Li J., Tu P., Jiang Y. Application of 1H NMR-based metabolomics for discrimination of different parts and development of a new processing workflow for Cistanche deserticola. Acta Pharm Sin B. 2017;7:647–656. doi: 10.1016/j.apsb.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee D., Kim J., Shrestha S., Seo K., Lee Y., Noh H. Quality evaluation of Panax ginseng roots using a rapid resolution LC—QTOF/MS-based metabolomics approach. Molecules. 2013;18:14849–14861. doi: 10.3390/molecules181214849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song H., Kim D., Woo S., Lee H., Oh S. An approach for simultaneous determination for geographical origins of Korean Panaxginseng by UPLC—QTOF/MS coupled with OPLS-DA models. J Ginseng Res. 2013;37:341–348. doi: 10.5142/jgr.2013.37.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim N., Kim K., Choi B., Lee D., Shin Y., Bang K. Metabolomic approach for age discrimination of Panax ginseng using UPLC—Q-Tof MS. J Agric Food Chem. 2011;59:10435–10441. doi: 10.1021/jf201718r. [DOI] [PubMed] [Google Scholar]
- 21.Kim N., Kim K., Lee D., Shin Y., Bang K., Cha S. Nontargeted metabolomics approach for age differentiation and structure interpretation of age-dependent key constituents in hairy roots of Panaxginseng. J Nat Prod. 2012;75:1777–1784. doi: 10.1021/np300499p. [DOI] [PubMed] [Google Scholar]
- 22.Park H., In G., Kim J., Cho B., Han G., Chang I. Metabolomic approach for discrimination of processed ginseng genus (Panaxginseng and Panax quinquefolius) using UPLC—QTOF MS. J Ginseng Res. 2014;38:59–65. doi: 10.1016/j.jgr.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichtenthaler H., Rohmer M., Schwender J. Two independent biochemical pathway for isopentenyl diphosphate and isopreniod biosynthesis in higher plants. Physiol Plant. 1997;101:643–652. [Google Scholar]
- 24.Luo H., Sun C., Sun Y., Wu Q., Li Y., Song J. Analysis of the transcriptome of Panax notoginseng root uncovers putative triterpene saponin-biosynthetic genes and genetic markers. BMC Genom. 2011;12:S5. doi: 10.1186/1471-2164-12-S5-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niu Y., Luo H., Sun C., Yang T., Dong L., Huang L. Expression profiling of the triterpene saponin biosynthesis genes FPS, SS, SE, and DS in the medicinal plant Panax notoginseng. Gene. 2014;533:295–303. doi: 10.1016/j.gene.2013.09.045. [DOI] [PubMed] [Google Scholar]
- 26.Wang J., Yau L., Gao W., Liu Y., Yick P., Liu L. Quantitative comparison and metabolite profiling of saponins in different parts of the root of Panax notoginseng. J Agric Food Chem. 2014;62:9024–9034. doi: 10.1021/jf502214x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Y., Chu S., Zhang Z., Chen N. Hepataprotective effects of ginsenoside Rg1— a review. J Ethnopharmacol. 2017;206:178–183. doi: 10.1016/j.jep.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Cho W., Chung W., Lee S., Leung A., Chen C., Yue K. Ginsenoside Re of Panax ginseng possesses significant antioxidant and antihyperlipidemic efficacies in streptozotocin-induced diabetic rats. Eur J Pharmacol. 2006;550:173–179. doi: 10.1016/j.ejphar.2006.08.056. [DOI] [PubMed] [Google Scholar]
- 29.Li Y., Tang J., Khatibi N., Zhu M., Chen D., Tu L. Treatment with ginsenoside Rb1, a component of Panax ginseng, provides neuroprotection in rats subjected to subarachnoid hemorrhage-induced brain injury. Acta Neurochir. 2011;110:75–79. doi: 10.1007/978-3-7091-0356-2_14. [DOI] [PubMed] [Google Scholar]
- 30.Hou J., Xue J., Lee M., Sung C. Ginsenoside Rd as a potential neuroprotective agent prevents trimethyltin injury. Biomed Rep. 2017;6:435–440. doi: 10.3892/br.2017.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Q., Gao B., Jie Q., Wei B., Fan J., Zhang H. Ginsenoside-Rb2 displays anti-osteoporosis effects through reducing oxidative damage and bone-resorbing cytokines during osteogenesis. Bone. 2014;66:306–314. doi: 10.1016/j.bone.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Yu T., Yang Y., Kwak Y., Song G., Kim M., Rhee M. Ginsenoside Rc from Panax ginseng exerts anti-inflammatory activity by targeting TANK-binding kinase1/interferon regulatory factor-3 and p38/ATF-2. J Ginseng Res. 2017;41:127–133. doi: 10.1016/j.jgr.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan J., Yang F., Li S., Wang Y., Cui X. Chemical characteristics for different parts of Panax notoginseng using pressurized liquid extraction and HPLC—ELSD. J Pharm Biomed Anal. 2006;41:1596–1601. doi: 10.1016/j.jpba.2006.01.058. [DOI] [PubMed] [Google Scholar]
- 34.Chinese Pharmacopoeia Commission. Pharmacopoeia of the People's Republic of China. Vol I. Beijing: China Medical Science Press; 2015.
- 35.Wang D., Li H., Chen K., Zhang Y., Yang C. HPLC analysis of ginsenoside saponins in different underground parts of Panaxnotoginseng. Acta Bot Yunnanica. 2005;7:685–690. [Google Scholar]
- 36.Laden B., Tang Y., Cloning Porter T. heterologous expression, and enzymological characterization of human squalene monooxygenase. Arch Biochem Biophys. 2000;374:381–388. doi: 10.1006/abbi.1999.1629. [DOI] [PubMed] [Google Scholar]
- 37.Augustin J., Kuzina V., Andersen S., Bak S. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry. 2011;72:435–457. doi: 10.1016/j.phytochem.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 38.Pan J., Bugni T., Poulter C. Recombinant squalene synthase, synthesis of cyclopentyl non-head-to-tail triterpenes. J Org Chem. 2009;74:7562–7565. doi: 10.1021/jo9014547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim Y., Kim Y., Uddin M., Lee S., Kim S., Park S. Enhanced triterpene accumulation in Panax ginseng hairy roots overexpressing mevalonate-5-pyrophosphate decarboxylase and farnesyl pyrophosphate synthase. ACS Synth Biol. 2014;3:773–779. doi: 10.1021/sb400194g. [DOI] [PubMed] [Google Scholar]
- 40.Zhang D., Li W., Xia E., Zhang Q., Liu Y., Zhang Y. The medicinal herb Panax notoginseng genome provides insights into ginsenoside biosynthesis and genome evolution. Mol Plant. 2017;10:903–907. doi: 10.1016/j.molp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 41.Kim Y., Zhang D., Yang D. Biosynthesis and biotechnological production of ginsenosides. Biotechnol Adv. 2015;33:717–735. doi: 10.1016/j.biotechadv.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 42.Schramek N., Huber C., Schmidt S., Dvorski S., Kmispel N., Ostrozhenkova E. Biosynthesis of ginsenosides in field-grown Panaxginseng. JSM Biotechnol Bioeng. 2014;2:1033. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material