Abstract
Uterine leiomyosarcoma (u‐LMS) and endometrial stromal sarcoma (ESS) are among the most frequent soft tissue sarcomas, which, in adults, lead to fatal lung metastases and patients have an extremely poor prognosis. Due to their rarity and heterogeneity, there are no suitable biomarkers for diagnosis and prognosis, although some biomarker candidates have appeared. In 2017, The Cancer Genome Atlas (TCGA) Research Network's work on u‐LMS has confirmed mutations and deletions in RB1,TP53 and PTEN. In addition, whole‐exome sequencing of u‐LMS has confirmed and demonstrated frequent alterations in TP53,RB1, α‐thalassemia/mental retardation syndrome X‐linked (ATRX) and mediator complex subunit 12 (MED12). MED12 is a useful biomarker to diagnose uterine‐derived LMS and tumors arising from (LM) with a relatively favorable prognosis. TP53 and ATRX mutations can be important mechanisms in the pathogenesis of u‐LMS and are correlated with a poor prognosis. In an update based on the 2014 WHO classification, low‐grade ESS is often associated with gene rearrangement bringing about the JAZF 1‐SUZ12 (formerly JAZF1‐JJAZ1) fusion gene, whereas high‐grade ESS is associated with the YWHAE‐NUTM fusion gene. Low‐grade ESS with JAZF1 rearrangement may correlate with metastasis. However, high‐grade ESS with metastasis with YWHAE rearrangement shows a relatively favorable prognosis. The genetic/molecular genetic aberrations in u‐LMS and ESS are reviewed, focusing on molecular biomarkers for these primary and metastatic tumors.
Keywords: endometrial stromal sarcoma, genetic aberrations, The Cancer Genome Atlas project, uterine leiomyosarcoma
1. INTRODUCTION
Malignant soft tissue tumors (soft tissue sarcomas [STS]) are rare and diverse. Although they are rare in adults, accounting for ≤1% of adult malignant tumors, they are known to account for 15% of pediatric malignant tumors.1 These tumors originate in the bone and soft tissues (Clinically relevant molecular subtypes in Leiomyosarcoma muscle, fat and nerves) throughout the body, and ≥75 histological types have been identified. Soft tissue tumors in adults lead to fatal lung metastases in 30% of cases, and they have an extremely poor prognosis (mean survival, 15 months). Due to the rarity and heterogeneity of these tumors, there are no suitable biomarkers and treatment methods, and the past few decades have not seen marked improvements in survival.2
Uterine leiomyosarcoma (u‐LMS) and endometrial stromal sarcoma (ESS) are among the most frequent STS in adult women, accounting for 1%‐2% of uterine malignant tumors, and their incidence tends to increase. LMS occurs frequently in 50‐55‐year‐old women, while 15% of STS occur in women under the age of 40 years. ESS is classified into low‐grade (LG‐ESS) and high‐grade (HG‐ESS). LG‐ESS frequently occurs before menopause. The prognoses of LMS and HG‐ESS are extremely poor.3
With the recent developments in genomic analysis techniques, the pathogenesis of these tumors has been becoming clearer, and some biomarker candidates have appeared. In the present article, the development and pathological states of uterine sarcoma (u‐LMS and ESS) are described based on findings from genomic analysis, and some molecular biomarker candidates for diagnosis and prognosis are discussed.
2. UTERINE LEIOMYOSARCOMA
Soft tissue sarcomas have been traditionally classified by their location of occurrence. However, based on the development of genomic analysis in the last few years, a new concept has arisen that STS can be divided into 2 groups. One, which accounts for one‐third of STS, is a relatively simple tumor and has a diploid karyotype with several chromosome abnormalities. The other, which accounts for two‐thirds of STS, has a complicated karyotype, with instability of many genes and mutation of the TP53 gene that encodes p53 in many cases;4 u‐LMS is considered to belong to the latter group. Moreover, u‐LMS demonstrates complicated tumor development and pathology that differs from LMS originating from other sites due to the involvement of estrogen.5
In 2017, The Cancer Genome Atlas (TCGA) Research Network performed a multiplatform molecular characterization of adult STS, including 53 soft tissue LMS and 27 u‐LMS cases. They reported that sarcomas had low mutational burdens compared with other tumors in the TCGA projects. Moreover, in integrated and individual platform analyses, u‐LMS and soft tissue LMS were generally more similar to each other than to other sarcomas. The analysis confirmed mutations and deletions in RB1, which encodes RB, TP53 and PTEN (Figure 1).4 In addition, whole‐exome sequencing of soft tissue LMS and u‐LMS has confirmed and demonstrated frequent alterations in TP53, RB1, α‐thalassemia/mental retardation syndrome X‐linked (ATRX) and mediator complex subunit 12 (MED12).6, 7 Other shared features of LMS were elevated microRNA (miRNA)‐143 and miRNA‐145 expressions, low miRNA expressions of inflammatory response genes, and low leukocyte fraction on methylation analysis. In contrast, u‐LMS and soft tissue LMS had significantly different methylation and miRNA expression signatures, with u‐LMS showing a higher DNA damage response score and hypomethylation of estrogen receptor 1 (ESR1) target genes, while soft tissue LMS had a more prominent hypoxia inducible factor (HIF) 1α signaling signature, suggesting that the use of different management approaches should be considered for u‐LMS and soft tissue LMS due to the predicted differences in hormonal responsiveness and stress responses.4
Figure 1.
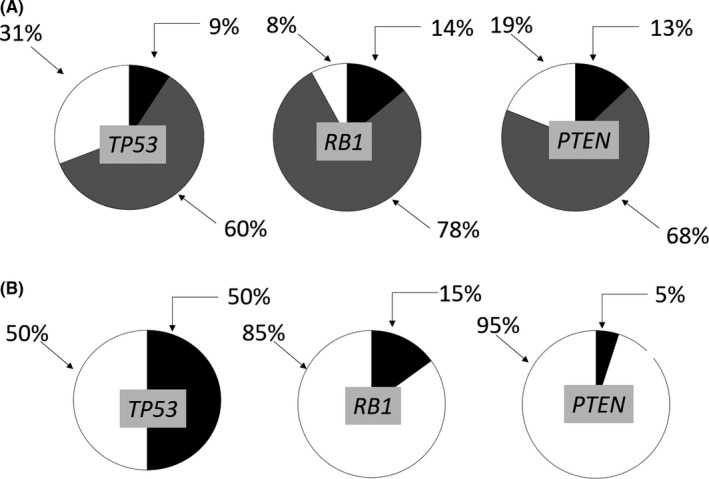
In the recurrent focal copy‐number alterations, the ratios of deletions of the tumor suppressors (A) and mutations (B) of TP53,RB1 and PTEN to each whole gene in leiomyosarcoma. The deletions of the tumor suppressors TP53 (9% deep and 60% shallow deletions), RB1 (14% deep, 78% shallow) and PTEN (13% deep, 68% shallow) (false discovery rate [FDR] is 0.25) are shown. Mutations of TP53 in 50%, RB1 in 15% and PTEN in 5% of samples are shown (FDR values computed by MuSic2 are <1e‐22, 2e‐15 and 0.49, respectively). A, Black indicates deep, a striped pattern indicates shallow deletions and white indicates no alterations. B, Black indicates mutations, and white indicates no mutations. Data from The Cancer Genome Atlas Research Network that performed a multiplatform molecular characterization of adult soft tissue sarcoma4
2.1. Whole molecular mechanisms of tumor development, metastasis and chemoresistance in leiomyosarcoma
To elucidate the molecular mechanisms of tumor development and metastasis, numerous experimental animal models have been created. Hernando et al found a defect in the PI3K‐AKT signal transduction pathway in many LMS. They succeeded in creating smooth muscle cell‐specific Pten knockout animals (Tagln‐Cre; Ptenflox/flox). This mouse model showed smooth muscle cell hyperplasia and abdominal LMS. However, development of LMS was not observed in the uterus and in lung metastases.8 Xing et al introduced Cre recombinase into the anti‐Mullerian hormone type II receptor (Amhr 2) locus in mice to conditionally inactivate p53 in the reproductive tract and reported that u‐LMS developed in 50% of the mice within 13 months. Furthermore, when the tumor suppressor gene BRCA1 was also inactivated, the frequency of tumor development increased to 82%.9 Strizzi et al reported that Cripto‐1 (CR‐1) protein, an active growth factor in the Wnt signaling pathway, is present in ≥70% of u‐LMS, and they created a CR‐1 overexpression transgenic mouse model. Wnt signaling and c‐Src and AKT signaling pathways were activated in these mice, and u‐LMS were detected in approximately 20% of the mice. These findings suggested that crosstalk between the Wnt signaling pathway and the Src/AKT pathway may play a significant role in the development of u‐LMS. Interestingly, p53 mutation was not involved in the development of u‐LMS in these animals.10 Kawabe et al developed a uterine sarcoma tissue‐derived orthotopic and metastatic mouse model using a green fluorescent protein stably expressed uterine sarcoma cell. They also identified the differential expression of genes related to cell proliferation and migration (TNNT1, COL1A2 and ZIC1) between orthotopic tumors with high and low metastatic potential.11 Animal models are indispensable tools for the study of u‐LMS because a real clinical sample makes large‐scale analysis of human samples challenging.
Doxorubicin is one of the key drugs in the treatment of u‐LMS, although resistance is the major hurdle, with a response rate of only 19% due to drug resistance.12 Overcoming resistance to chemotherapy and investigating molecular targeted therapies are challenges. Currently, multi drug resistant 1 (MDR1) is frequently associated with the overexpression of membrane‐embedded drug efflux transporters, such as ATP‐binding cassette transporters, known as P‐glycoprotein 1 (P‐gp) (also named ABCB1 ([TP‐binding Cassette Sub‐family B Member 1]), leading to the reduced accumulation of chemotherapeutic drugs and chemoresistance in LMS cells.13 Hung et al reported that the overexpression of frizzled 1 (FZD1) encoded by FZD1 gene, which is a G‐protein‐coupled receptor and involved in the Wnt/β‐catenin pathway, was observed in LMS cells and contributed to doxorubicin‐resistance via the activation of the protein kinase C δ/AP‐1 signaling transduction pathway, resulting in enhanced ABCB1 expression.14 Lin et al reported on the important role of progesterone receptor membrane component 1 (PGRMC1) encoded by PGRMC1, which is an adapter protein mediating cholesterol synthesis, steroid signaling and cytochrome p450 activation. PGRMC1 promoted cell proliferation and cell cycle progression to the S phase by mediating ERK activation, leading to doxorubicin‐resistance in LMS cells.15 In the last decade, several anticancer drugs or multityrosine kinase inhibitors have demonstrated efficacy with respect to progression‐free survival and overall survival, particularly the tyrosine kinase inhibitor (TKI) pazopanib in the treatment of u‐LMS.16 However, the biological mechanisms or optimal predictive biomarkers for these therapies are still being explored. The further development of in vitro or in vivo models using advanced genetic techniques will continue to increase our understanding of LMS biology.
2.2. Molecular biomarker candidates for diagnosis and prognosis of primary uterine leiomyosarcoma
2.2.1. Tumor suppressor gene
Molecular genetic studies have demonstrated that, in LMS (site of formation in the uterine or extrauterine tissues), frequent chromosomal losses are detected in the long arm of chromosome 10 (10q) and 13 (13q), the regions in which tumor suppressor PTEN and RB1 genes are present. In particular, losses at 10q21.3 and 13q14.2‐q14.3 were the most frequent, with frequencies of 75% at each of the loci. According to Yang et al,17 chromosomal gains were observed in 1q21, 5p14‐pter, 8q, 12q13‐15, 13q31, 17p11, 19p13 and 20q13.
Genetic deficiency of 10 q means inactivation of tumor suppressor gene PTEN, leading to activation of the PI3K/AKT pathway and the downstream mTOR pathway. Hernando et al reported abnormalities of the PI3K‐AKT signaling pathway in many LMS. They genetically inactivated Pten in the smooth muscle cell lineage by cross‐breeding Pten (loxP/loxP) mice with Tagln‐cre mice. The mice developed widespread smooth muscle cell hyperplasia and abdominal LMS in the peritoneal cavity. In addition, mTOR inhibitor suppressed tumor growth.17 Thus, the PTEN‐PI3K/AKT‐mTOR pathway can play important roles in not only the generation of LMS but also in the treatment and prediction of the prognosis of patients with LMS.18, 19 However, this pathway has not been well investigated in u‐LMS.
Loss at the 13q region leads to inactivation of the tumor suppressor RB1 gene. The RB1 gene is involved in the cell cycle, specifically at the G1‐S phase checkpoint. Regulatory aberration at this region is known to induce cells to divide indefinitely. Dei‐Tos et al20 observed defects in RB‐cyclin D1 signaling (RB1, CDKN2A which encodes p16, CCND1 which encodes cyclin D1, and CCND3 which encodes cyclin D3) in ≥90% of u‐LMS patients. It has also been reported that an abnormality in this pathway is an obvious factor related to poor prognosis.17
Many studies have shown that the frequencies of TP53 mutation in uterine leiomyoma (u‐LM), smooth muscle tumor of uncertain malignant potential (STUMP) and u‐LMS are 0%, 6%‐29% and 24%‐30%, respectively. It has also been reported that the frequencies of PTEN mutation are 5%, 33% and 42%‐58%, respectively.21 However, u‐LMS has shown a lower frequency of TP53 mutation, higher expression of MDM2, and a higher TP53/MDM2 ratio than the other sarcomas.
2.2.2. Genes related to the cell cycle
In general, it is difficult to distinguish between u‐LM and u‐LMS with protein expressions related to the cell cycle (p16, p21, p27, p53, Ki‐67 and PHH3).22 In 2004, Quade et al investigated 4 normal uterine myometrium samples, 7 u‐LM and 9 u‐LMS using microarrays of oligonucleotides representing approximately 7000 unique probe sets. They reported that, while there was a difference in CDKN1A, which encodes p21, which is involved in cell proliferation and the cell cycle, this difference was not significant. Moreover, they reported that there were no clear differences in cellular gene expressions between LMS that developed in the uterus vs other tissues.23 Miyata et al reported that overexpression of cell cycle‐related genes and frequent hypermethylation at the polycomb group target genes and protocadherin genes were observed in u‐LMS in single nucleotide polymorphism arrays, gene expression array analysis, and analysis involving comprehensive detection of methylated DNA.24 Furthermore, Aurora‐A kinase, which is a serine/threonine kinase and is involved in the regulation of the M phase in the cell cycle and division, has recently been reported to be overexpressed in u‐LMS. Matsumura et al demonstrate the important role of acrogranin, which is also known as PCDGF, progranulin or proepithelin. Acrogranin, an 88‐kDa glycoprotein, is one of the pluripotent growth factors that mediate cell cycle progression and cell motility. It activates the extracellular regulated kinases and PI3K signal cascades, increases expressions of cyclins D and B, and is overexpressed in u‐LMS.25
2.2.3. Genes related to oncogenesis of uterine leiomyosarcoma derived from uterine leiomyoma
There are currently many studies focused on the mediator complex subunit 12 (MED12) gene, which was discovered in genomic analysis of u‐LM. Makinen et al observed a mutation in MED12 that resides on the long arm of the X chromosome (Xq13.1.) in ≥70% of u‐LM and postulated that this mutation contributes greatly to the formation of u‐LM. Mediator is a complex that is composed of approximately 30 subunits that acts as a bridge between transcription factors and RNA polymerase II in the nucleus. These molecules consequently augment or suppress gene expression in a cell‐dependent manner. MED12 is known to be involved in p53 and Wnt/β‐catenin pathways.26 Interestingly, in vitro study of u‐LM has shown that the u‐LM cells with MED12 mutation grow poorly in culture and cannot be maintained through passage.27
In 2012, the same research group demonstrated that, albeit at low frequencies, MED12 mutations are observed in u‐LMS, as well as u‐LM, while no mutations were found in other sarcomas or in gastrointestinal stromal tumors (GIST), suggesting that a subgroup of u‐LMS may develop from an LM precursor. Furthermore, they report that such u‐LMS do not have greater malignancy than those from the group without mutations.28 In 2012, Pérot et al compared MED12 protein expression between benign and malignant smooth muscle tumors. They postulated that MED12 could be a tumor suppressor gene, and that MED12 protein expression inhibits LMS oncogenesis.29 Schwetye et al compared the frequencies of MED12 gene mutations among normal myometrium adjacent to LM, pelvic LM and extrauterine LM. MED12 mutations were detected in 54% of u‐LM, 15% of cases in myometrium adjacent to LM, and 0% of extrauterine LM. Moreover, MED12 mutations were also detected in 30% of u‐LMS compared with 4% of extrauterine LMS, suggesting that smooth muscle tumors in pelvic/retroperitoneal sites are subject to the same mutational changes as those of uterine myometrium.30 Recently, Zhang et al showed the frequencies of MED12, TP53 or PTEN mutations in LM, mitotically active LM, cellular LM, atypical LM, STUMP and u‐LMS. STUMP and atypical LM were more similar to LMS, with high frequencies of TP53 and PTEN mutations and low frequencies of MED12 mutation. However, atypical LM shared more molecular alterations, including the selected microRNA, oncogenes and tumor suppressors that are highly relevant to uterine smooth muscle tumors with u‐LMS, suggesting that atypical LM may be a precursor lesion of u‐LMS or have similar genetic changes during its early stage (Table 1).21
Table 1.
Mutation analysis of MED12, TP53 and PTEN and comparison with the literature for 6 types of uterine smooth muscle tumors
| Mutations | LMS | STUMP | Atypical LM | Cellular LM | Mitotically active LM | LM | Number of references |
|---|---|---|---|---|---|---|---|
| MED12 | 15% (21/141) | 10% (4/40) | 12% (8/69) | 12% (11/95) | 48% (16/33) | 67% (636/948) | 13 |
| TP53 | 28% (50/176) | 17% (6/35) | 10% (5/50) | 0% (0/26) | 0% (0/7) | 0% (0/59) | 7 |
| PTEN | 54% (76/142) | 33% (6/18) | 26% (11/42) | 5% (1/22) | 0% (0/7) | 5% (1/20) | 6 |
LM, leiomyoma; LMS, leiomyosarcoma.
Bertsch et al31 showed other genetic abnormalities in u‐LM. The overexpression of high‐mobility group AT‐hook 2 (HMGA 2), which is an oncogene, occurred in u‐LM (40%) and u‐LMS (25%) with no MED12 mutation, suggesting that they may act differently in u‐LM tumorigenesis. Because u‐LM is related to familial hereditary leiomyomatosis and renal cell cancer (HLRCC) syndrome, heterozygous germ‐line mutations in fumarate hydratase (FH), which is a tumor suppressor and encodes fumarase of the tricarboxylic acid cycle, has been reported to be related to the development of u‐LM.32 Recent studies have shown that a kind of u‐LMS can be derived from the precursor lesion of u‐LM based on the examination of 3 driver genes, MED12, HMGA 2 and FH.33
Based on several studies, MED12 mutation is also observed in u‐LMS, while it is more frequent in u‐LM than in u‐LMS. MED12 mutations are rarely observed in other sarcomas or in GIST, as well as extrauterine LMS,29, 30 suggesting that MED12 can be a useful biomarker to diagnose uterine‐derived LMS, although it is difficult to distinguish between u‐LM and u‐LMS only by examination of MED12 mutation.
2.3. Other biomarkers for primary uterine leiomyosarcoma
2.3.1. Stathmin 1
Stathmin 1 encoded by STMN1 is involved in the regulation of the cell cycle as the protein that destabilizes microtubules. It is also known as an activator of the PI3K‐AKT‐mTOR pathway. In 2017, the Gynecologic Oncology Group (GOG) and NRG showed that STMN1 expression was significantly associated with shorter progression‐free survival and overall survival for all analyzed uterine endometrial cancer cases in both GOG‐177 and TCGA.34 STMN1 expression was observed in all u‐LMS cases (100%), while it was observed in approximately 40% of benign u‐LM cases.35
2.3.2. Insulin‐like growth factor (IGF‐II) mRNA binding protein 3 (IMP3)
IMP3 encoded by IMP3 is an RNA‐binding carcinoembryonic protein consisting of 580 amino acids that is observed only in advanced tumor tissues, but not in normal tissues.36 Strong expression of IMP3 is observed in the cytoplasm in more than 50% of u‐LMS cases, while it is not observed in typical u‐LM.36 Yasutake et al37 reported a multivariate analysis that showed that advanced stage and IMP3 are independent predictors of a poor prognosis in u‐LMS.
2.3.3. Telomere‐maintenance mechanism by alternative lengthening of telomeres
It has been reported that the correlation of alternative lengthening of telomeres (ALT) activation and loss of α‐thalassemia/mental retardation syndrome X‐linked (ATRX) encoded by ATRX expression can suppress cell death in sarcoma.38 Further studies using next‐generation sequencing have shown that loss of function of p53 can be correlated with ATRX pathways involved in ALT. Although ATRX mutations were not correlated with ATRX protein expression, all ATRX‐mutated LMS cases showed the ALT phenotype. TP53 and ATRX mutations can be important mechanisms in the pathogenesis of u‐LMS and are correlated with a poor prognosis.39
2.3.4. MicroRNA (miRNA)‐181b
The Cancer Genome Atlas indicated that high miR‐181b was more common in u‐LMS and was an independent predictor of recurrence‐free survival in a multivariate model, including tumor size. MiR‐181b expression has been reported to promote proliferation and migration of vascular smooth muscle via the PI3K pathway.4
2.3.5. KIT expression
Raspollini et al40 showed a high frequency of positive immunostaining for KIT in u‐LMS. However, imatinib is not theoretically applicable for u‐LMS or ESS because of the lack of KIT hotspot mutations.41 Pazopanib inhibits VEGF, PDGFR, FGFR and c‐KIT. After a recent successful placebo‐controlled phase III trial, the PALETTE study, it was approved by the FDA (April 2012) for use in STS, including LMS. On subgroup analysis, u‐LMS patients had a partial response (11%), and median PFS was 3 months (95% CI 2.5‐4.7 months), with median OS of 17.5 months (95% CI 11.1‐19.6 months).42
To date, several anticancer drugs, multityrosine kinase inhibitors or small molecule inhibitors, have demonstrated efficacy with respect to progression‐free survival and overall survival. However, optimal predictive biomarkers for these therapies are still being explored. Some novel check point inhibitors, such as nivolumab, pembrolizumab and ipilimumab, have also been explored as potential therapies, but their efficacy is still limited. In contrast, in LMS as a whole, aberrant PI3K‐AKT‐mTOR signaling may be crucial, given recurrent deletion/mutation of PTEN. Indeed, mTOR inhibitors such as everolimus and temsirolimus have shown some clinical efficacy in LMS, although the use of definitive predictive biomarkers is still challenging. Thus, further investigations are needed to evaluate the predictors for these therapies and to provide the optimal precision medicine.
2.4. Molecular biomarker candidates for diagnosis and prognosis of metastases of uterine leiomyosarcoma
Recently, some researchers reported extremely interesting studies.11, 43, 44 They investigated the gene expression patterns of primary and metastatic lesions in patients and in vivo with distant metastatic u‐LMS, and they identified genes that were overexpressed in each type of lesion. Several genes, which are listed along with their functions in Table 2, are overexpressed at primary lesion sites, and the following genes were overexpressed or down underexpressed in metastatic lesions.
Table 2.
Gene expression patterns of primary and metastatic lesions in patients and in vivo with distant metastatic u‐LMS
| Functions | References | |
|---|---|---|
| Overexpressed gene at primary lesion site | ||
| Osteocrin (OSTN) | Muscle metabolism | Davidson et al43 |
| Neuroligin4X (NLGN4X) and Neuroligin1 (NLGN1) | Neural development, angiogenesis and tumor growth | Davidson et al43 |
| SLITRK4 encodes the Slitrk protein |
An integral membrane protein similar to Trk tyrosine kinase receptors Slitrk is primarily involved in nervous system development. |
Davidson et al43 |
| TSPAN7 (CD231) | The expression of tetraspanins, a family of transmembrane adhesion molecules including integrins, claudin‐1 and EGFR | Davidson et al43 |
| Overexpressed genes in metastatic lesions | ||
| TSPAN10 (metastatic site) | The expression of tetraspanins, a family of transmembrane adhesion molecules, including integrins, claudin‐1 and EGFR | Davidson et al43 |
| FOLR3 and SHMT1 | Folate metabolism | Davidson et al43 |
| TDO2 | The expression of tryptophan conversion enzyme | Davidson et al43 |
| TNNT1 | The expression of skeletal muscle protein |
Davidson et al43
Kawabe et al11 |
| Early growth response 2 (EGR2) | Neural development and has been reported to be involved in the prognosis of Ewing's sarcoma, | Davidson et al43 |
| SGK1 | A factor involved in the activation of ion channels for the transport of K, Na and cellular signal transduction of serine/threonine kinases. | Davidson et al43 |
| Gas 6 and Tyro 3 | Gas 6 is a gene product expressed when cell growth arrest occurs. It is involved in various biological activities, such as the promotion of cell proliferation, chemotaxis or cell adhesion factors as a common ligand of Tyro 3 receptor protein tyrosine kinase, Axl and Mer. | el Sayadi et al44 |
| COL1A2 | A genetic maker involved in aggressive malignancy | Kawabe et al11 |
| Low expression gene in highly metastatic lesions | ||
| ZIC1 | A transcription factor gene that induces cell‐cycle arrest/apoptosis and inhibits cell migration/invasion by blockade of PI3K/Akt and MAPK pathways | Kawabe et al11 |
u‐LMS, uterine leiomyosarcoma.
Increasing evidence of intratumor genetic heterogeneity (ITH) is emerging, both within individual tumor biopsies and spatially separated between biopsies of the same tumor. Furthermore, sequential analysis of tumors has also provided evidence that ITH evolves during the course of the disease.45 However, these genes may be useful in elucidating the mechanism of metastasis from u‐LMS.
3. ENDOMETRIAL STROMAL SARCOMA
Endometrial stromal sarcoma (ESS) is composed of cells similar to the endometrial stroma at the proliferative phase. Basically, it is diagnosed by histopathological criteria. The growing evidence provided by molecular genetic analysis has been increasingly used to diagnose ESS (Table 3) (Figure 2).
Table 3.
Classification of ESS by molecular genetic examinations
| Karyotype | Fusion transcript | Functions | Material and methods |
|---|---|---|---|
| LG‐ESS | |||
| t(7;17)(p15;q21) | JAZF1‐SUZ12 | JAZF1; zinc finger gene, one of the largest family of transcriptional repressor | FISH, PCR |
| SUZ12; a member of the polycomb group protein family involved in transcriptional repression | |||
| t(6;7)(p21;p15) | JAZF1‐PHF1 | JAZF1; zinc finger gene, one of the largest family of transcriptional repressors | FISH |
| PHF1; a member of the polycomb group protein family involved in transcriptional repression. | |||
| t(6;10)(p21;p11) | EPC1‐PHF1 | EPC1; part of the nucleosome acetyltransferase of histone H4 complex | FISH |
| PHF1; a member of the polycomb group protein family involved in transcriptional repression. | |||
| t(1;6)(p34;p21) | MEAF6‐PHF1 | MEAF6; part of histone acetyltransferase multi‐subunit complexes | FISH |
| PHF1; a member of the polycomb group protein family involved in transcriptional repression | |||
| t(X;17)(p11;q21) | CXorf67‐MBTD1 | CXorf6; chromosome X open reading frame 67 | FISH, PCR |
| MBTD1; a member of the polycomb group protein family involved in transcriptional repression | |||
| t(X;22)(p11;q13) | ZC3H7B‐BCOR | ZC3H7B; it is involved in protein‐nucleic acid interactions | FISH, PCR |
| BCOR; it interacts with polycomb group proteins | |||
| HG‐ESS | |||
| t(10;17)(q21;p13) | YWHAE‐NUTM | YWHAE; mediator of signal transduction by binding to phosphoserine‐containing proteins | FISH, PCR |
ESS, endometrial stromal sarcoma; HG‐ESS, high‐grade ESS; LG‐ESS, low‐grade ESS; PCR, polymerase chain reaction.
Figure 2.
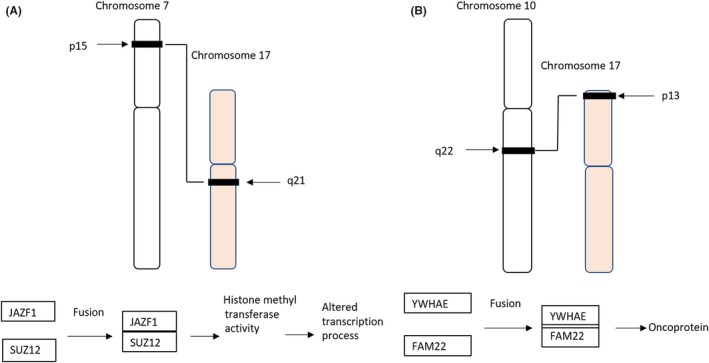
Schematic of chromosomal translocations present in low‐grade endometrial stromal sarcoma (LG‐ESS) (A) and high‐grade endometrial stromal sarcoma (HG‐ESS) (B). A, The most common translocation involves the short arm of chromosome 7 and long arm of chromosome 17(7;17) (p15;q21), leading to fusion of 2 zinc finger genes, JAZF1 and SUZ12, and production of JAZF1/SUZ12 gene fusion protein. B, Schematic of t(10;17) (q22;p13) chromosomal translocation present in HG‐ESS results in the fusion of the YWHAE and NUTM genes, leading to production of YWHAE/NUTM gene fusion proteins, which contributes to oncoprotein
3.1. Whole molecular mechanisms of tumor development, metastasis and chemoresistance in endometrial stromal sarcoma
Based on 2014 WHO classification, LG‐ESS is often associated with gene rearrangement bringing about the JAZF 1‐SUZ12 (formerly JAZF1‐JJAZ1) fusion gene, whereas HG‐ESS is associated with the YWHAE‐NUTM fusion gene. LG‐ESS with JAZF1 rearrangement may correlate with metastasis. On the other hand, HG‐ESS with metastasis with YWHAE rearrangement shows a relatively favorable prognosis.46
Low‐grade endometrial stromal sarcoma usually expresses ER/PR receptors and is an indolent tumor with a favorable prognosis. However, LG‐ESS is characterized by late recurrences even in patients with stage I disease, suggesting the need for long‐term follow‐up.2 In an in vitro study, Quan et al47 found that the histone deacetylase (HDAC) inhibitor suberanilohydroxamic acid (SAHA) combined with inhibition of PI3K (LY294002, LY) and mammalian target of rapamycin (mTOR) led to the strongest growth inhibition and slowest growth recovery in an ESS cell line. Lee et al report that ESS with YWHAE/NUTM (formerly YWHAE/FAM22A/B) fusion which gives rise to a 14‐3‐3 oncoprotein displays higher‐grade histology and more aggressive clinical course.48
3.2. Molecular biomarker candidates for diagnosis and prognosis for primaries of low‐grade endometrial stromal sarcoma
In 1988, Dal Cin et al49 first reported a chromosomal abnormality in LG‐ESS. Based on the growing evidence, LG‐ESS is classified into 3 categories based on molecular genetic abnormalities: translocation t(7;17) (p15;q21); 6p21‐rearrangements; and X;22 or 17‐rearrangements.
Chromosomes 7 and 17 are recombined in the first genetic hallmark to be discovered in ESS; namely, the translocation t(7;17) (p15; q21). Two zinc finger genes, the JAZF1 gene from chromosomal band 7p15 and SUZ12 (formerly JJAZ1) from 17q11, are fused by this translocation.50 Subsequent studies have shown that the fusion of JAZF1 and SUZ12 is recognized in almost all ESS cases and is more frequently expressed in LG‐ESS than in HG‐ESS.51 Micci et al52 show that the PHD finger protein 1 (PHF1) gene from chromosomal band 6p21 recombined with JAZF1 as the partner gene of the JAZF1 gene in LG‐ESS. Further studies have shown that the PHF1 gene from chromosomal band 6p21 recombined with JAZF1, EPC1, MEAF6 or BRD8, which encode the proteins involved in regulation of protein acetylation and/or histone acetyltransferase activity.53 All these fusions in ESS combined genes are involved in transcriptional regulation; that is, polycomb group complex‐mediated aberration methylation/acetylation; hence, their presumed oncogenic effects may be involved in the pathogenesis of ESS.52
Currently, ESS is considered to be a tumor with molecular genetic heterogeneity in gene rearrangement. Representative fusion genes such as JAZF1‐SUZ12, PHF1‐JAZF1, EPC1‐PHF1, MEAF6‐PHF1 and ZC3H7‐BCOR are observed in more than 50% of ESS cases.54, 55
3.3. Molecular biomarker candidates for diagnosis and prognosis for primaries of high‐grade endometrial stromal sarcoma
In 2012, Lee et al48 showed that a specific gene rearrangement, t(10;17) (q22;p13), brought about YWHAE‐NUTM (previously known as YWHAE‐FAM22) fusion, which is related to malignant potential. In an update based on 2014 WHO classification, LG‐ESS is often associated with gene rearrangement bringing about the JAZF 1‐SUZ12 fusion gene, whereas HG‐ESS is associated with the YWHAE‐NUTM fusion gene, suggesting that LG‐ESS and HG‐ESS are independent units.46 Clinically, HG‐ESS shows more malignant potential than LG‐ESS. Hoang et al (2017) show that ESS with ZC3H7B‐BCOR fusion morphologically closely resembles mucinous LMS. They also suggest that tumors with ZC3H7B‐BCOR fusion should be classified as ESS even if they are morphologically LMS.56
3.4. Other biomarkers for primary endometrial stromal sarcoma
3.4.1. Cyclin D1 expression
More than 70% of HG‐ESS cases with the YWHAE‐NUTM2A/B fusion gene show diffusely moderate to high staining of cyclin D1. In contrast, LG‐ESS with the JAZF1 fusion gene shows only 5% focal staining. Lee et al indicate that cyclin D1 is a sensitive and specific diagnostic immunomarker for YWHAE‐NUTM ESS. When evaluating HG‐ESS, cyclin D1 can be included in the immunohistochemical panel as an indicator of YWHAE‐NUTM ESS.57
3.4.2. KIT expression
It has been reported that overexpression of KIT, PDGFRA or EGFR is observed in a few cases, while KIT expression is observed in HG‐ESS with the YWHAE‐NUTM2A/B fusion gene, which frequently spreads beyond the uterus. If the extrauterine disease presents with a pelvic/abdominal mass, particularly in situations where its uterine origin is not definitive, an epithelioid GIST should be considered in the differential diagnosis because of the high frequency of KIT immunopositivity. Lee et al indicate that the high‐grade round cell component of YWHAE‐NUTM2A/B ESS consistently expresses KIT, but lacks KIT hotspot mutations, while GIST frequently contains activating gene mutations in KIT. Moreover, the expression of ANO1 (also known as DOG1), which is another widely used diagnostic immunomarker for GIST, is observed in approximately 90% of GIST, while ANO1 is not expressed by YWHAE‐NUTM2A/B ESS, suggesting that the evaluation of only Kit expression may be a potential diagnostic pitfall in the evaluation of pelvic/abdominal masses in female patients. Thus, the additional diagnostic strategy including the immunoprofiling of another antibody such as ANO1 or cyclin D1 or a mutation screen of KIT is needed.58 Likewise, expression of tyrosine kinase receptor was observed, but gene abnormalities were found in few cases.59
3.4.3. Murine double minute 2 amplification
Murine double minute 2 (MDM2) is an oncogene that can promote tumor occurrence and development by regulating the cell cycle and promoting cell proliferation. The evaluation of MDM2 protein expression may not be helpful in the differential diagnosis because it is often observed in various neoplasms, including ESS or other sarcomas. In contrast, the assessment of MDM2 gene amplification by FISH has become a routine ancillary tool in distinguishing well‐differentiated and dedifferentiated liposarcoma from classic lipoma, while MDM2 amplification is not common in ESS. Thus, examination of MDM2 amplification in ESS, particularly with metastatic lesions with myxoid morphology, can be useful for differentiation from liposarcoma.60
3.4.4. Interferon‐induced transmembrane protein 1
CD10 is a well‐established marker for endometrial stromal differentiation, although it has limited specificity. IFITM1 has been demonstrated as a potential marker based on the bioinformatics approach to identify more specific markers of endometrial stromal differentiation by searching a public database of protein expression profiles, the Human Protein Atlas. Immunohistochemical analysis of IFITM1 has shown high sensitivity and specificity in LG‐ESS and HG‐ESS.61
3.4.5. Molecular biomarkers for metastases of endometrial stromal sarcoma
In 2013, it was first reported that an LG‐ESS case with a PHF1‐JAZF1 fusion gene in the metastatic lesion, as well as the primary lesion, had invasive progression and fatal lung metastasis.62 It has also been reported that an LG‐ESS case with JAZF1 rearrangement and MDM2 amplification had lethal lung metastasis.60 On the other hand, it has been reported that a case with the JAZF1‐SUZ12 fusion gene in the primary and metastatic lesion had metastatic but not invasive disease.63
It has been reported that an HG‐ESS case with metastasis with YWHAE rearrangement responded to anthracycline‐based therapy and showed relatively long‐term survival.64 These facts indicate that further study to elucidate the relationship between the pattern of fusion genes and the prognosis or therapeutic effects in ESS is needed in the future.
4. CONCLUSIONS
Further advances in genome analysis are expected to elucidate not only the pathogenic genes but also the molecular basis of various abnormal traits, such as metastasis, for uterine sarcoma, which is characterized by its rarity and diversity. The development of molecular imaging methods or molecular targeted drugs based on these findings is also expected in the future.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
Tsuyoshi H, Yoshida Y. Molecular biomarkers for uterine leiomyosarcoma and endometrial stromal sarcoma. Cancer Sci. 2018;109:1743–1752. https://doi.org/10.1111/cas.13613
Funding information
This work was partially funded by a Grant‐in‐Aid for Scientific Research (15H04981) and Grant‐in‐Aid for Young Scientists (17K16840) from the Japan Society for the Promotion of Science.
REFERENCES
- 1. Toro JR, Travis LB, Wu HJ, Zhu K, Fletcher CD, Devesa SS. Incidence patterns of soft tissue sarcomas, regardless of primary site, in the surveillance, epidemiology and end results program, 1978‐2001: an analysis of 26,758 cases. Int J Cancer. 2006;119:2922‐2930. [DOI] [PubMed] [Google Scholar]
- 2. D'Angelo E, Prat J. Uterine sarcomas: a review. Gynecol Oncol. 2010;116:131‐139. [DOI] [PubMed] [Google Scholar]
- 3. Davidson B, Micci F. Molecular characteristics of uterine sarcomas. Expert Rev Mol Diagn. 2017;17:515‐522. [DOI] [PubMed] [Google Scholar]
- 4. Cancer Genome Atlas Research Network . Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171:950‐965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guo X, Jo VY, Mills AM, et al. Clinically relevant molecular subtypes in leiomyosarcoma. Clin Cancer Res. 2015;21:3501‐3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Agaram NP, Zhang L, LeLoarer F, et al. Targeted exome sequencing profiles genetic alterations in leiomyosarcoma. Genes Chromosom Cancer. 2016;55:124‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mäkinen N, Aavikko M, Heikkinen T, et al. Exome sequencing of uterine leiomyosarcomas identifies frequent mutations in TP53, ATRX, and MED12. PLoS Genet. 2016;12:e1005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hernando E, Charytonowicz E, Dudas ME, et al. The AKT‐mTOR pathway plays a critical role in the development of leiomyosarcomas. Nat Med. 2007;13:748‐753. [DOI] [PubMed] [Google Scholar]
- 9. Xing D, Scangas G, Nitta M, et al. A role for BRCA1 in uterine leiomyosarcoma. Cancer Res. 2009;69:8231‐8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Strizzi L, Bianco C, Hirota M, et al. Development of leiomyosarcoma of the uterus in MMTV‐CR‐1 transgenic mice. J Pathol. 2007;211:36‐44. [DOI] [PubMed] [Google Scholar]
- 11. Kawabe S, Mizutani T, Ishikane S, et al. Establishment and characterization of a novel orthotopic mouse model for human uterine sarcoma with different metastatic potentials. Cancer Lett. 2015;366:182‐190. [DOI] [PubMed] [Google Scholar]
- 12. Wen KC, Horng HC, Wang PH, et al. Uterine sarcoma Part I‐Uterine leiomyosarcoma: the Topic Advisory Group systematic review. Taiwan J Obstet Gynecol. 2016;55:463‐471. [DOI] [PubMed] [Google Scholar]
- 13. Hua J1, Mutch DG, Herzog TJ. Stable suppression of MDR‐1 gene using siRNA expression vector to reverse drug resistance in a human uterine sarcoma cell line. Gynecol Oncol. 2005;98:31‐38. [DOI] [PubMed] [Google Scholar]
- 14. Hung TH, Chen CM, Tseng CP, et al. FZD1 activates protein kinase C delta‐mediated drug‐resistance in multidrug‐resistant MES‐SA/Dx5 cancer cells. Int J Biochem Cell Biol. 2014;53:55‐65. [DOI] [PubMed] [Google Scholar]
- 15. Lin ST1, May EW, Chang JF, Hu RY, Wang LH, Chan HL. PGRMC1 contributes to doxorubicin‐induced chemoresistance in MES‐SA uterine sarcoma. Cell Mol Life Sci. 2015;72:2395‐2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferrero S, Leone Roberti Maggiore U, Aiello N, et al. Pharmacokinetic drug evaluation of pazopanib for the treatment of uterine leiomyosarcomas. Expert Opin Drug Metab Toxicol. 2017;13:881‐889. [DOI] [PubMed] [Google Scholar]
- 17. Yang J, Du X, Chen K, et al. Genetic aberrations in soft tissue leiomyosarcoma. Cancer Lett. 2009;275:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Setsu N, Yamamoto H, Kohashi K, et al. The Akt/mammalian target of rapamycin pathway is activated and associated with adverse prognosis in soft tissue leiomyosarcomas. Cancer. 2012;118:1637‐1648. [DOI] [PubMed] [Google Scholar]
- 19. Wong TF, Takeda T, Li B, et al. Curcumin disrupts uterine leiomyosarcoma cells through AKT‐mTOR pathway inhibition. Gynecol Oncol. 2011;122:141‐148. [DOI] [PubMed] [Google Scholar]
- 20. Dei Tos AP, Maestro R, Doglioni C, et al. Tumor suppressor genes and related molecules in leiomyosarcoma. Am J Pathol. 1996;148:1037‐1045. [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Q, Ubago J, Li L, et al. Molecular analyses of 6 different types of uterine smooth muscle tumors: emphasis in atypical leiomyoma. Cancer. 2014;120:3165‐3177. [DOI] [PubMed] [Google Scholar]
- 22. Mills AM, Ly A, Balzer BL, et al. Cell cycle regulatory markers in uterine atypical leiomyoma and leiomyosarcoma: immunohistochemical study of 68 cases with clinical follow‐up. Am J Surg Pathol. 2013;37:634‐642. [DOI] [PubMed] [Google Scholar]
- 23. Quade BJ, Wang TY, Sornberger K, Dal Cin P, Mutter GL, Morton CC. Molecular pathogenesis of uterine smooth muscle tumors from transcriptional profiling. Genes Chromosom Cancer. 2004;40:97‐108. [DOI] [PubMed] [Google Scholar]
- 24. Miyata T, Sonoda K, Tomikawa J, et al. Genomic, epigenomic, and transcriptomic profiling towards identifying omics features and specific biomarkers that distinguish uterine leiomyosarcoma and leiomyoma at molecular levels. Sarcoma. 2015;2015:412068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsumura N, Mandai M, Miyanishi M, et al. Oncogenic property of acrogranin in human uterine leiomyosarcoma: direct evidence of genetic contribution in in vivo tumorigenesis. Clin Cancer Res. 2006;12:1402‐1411. [DOI] [PubMed] [Google Scholar]
- 26. Mäkinen N, Mehine M, Tolvanen J, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334:252‐255. [DOI] [PubMed] [Google Scholar]
- 27. Nadine Markowski D, Tadayyon M, Bartnitzke S, Belge G, Maria Helmke B, Bullerdiek J. Cell cultures in uterine leiomyomas: rapid disappearance of cells carrying MED12 mutations. Genes Chromosom Cancer. 2014;53:317‐323. [DOI] [PubMed] [Google Scholar]
- 28. Kämpjärvi K, Mäkinen N, Kilpivaara O, et al. Somatic MED12 mutations in uterine leiomyosarcoma and colorectal cancer. Br J Cancer. 2012;107:1761‐1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pérot G, Croce S, Ribeiro A, et al. MED12 alterations in both human benign and malignant uterine soft tissue tumors. PLoS ONE. 2012;7:e40015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schwetye KE, Pfeifer JD, Duncavage EJ. MED12 exon 2 mutations in uterine and extrauterine smooth muscle tumors. Hum Pathol. 2014;45:65‐70. [DOI] [PubMed] [Google Scholar]
- 31. Bertsch E, Qiang W, Zhang Q, et al. MED12 and HMGA2 mutations: two independent genetic events in uterine leiomyoma and leiomyosarcoma. Mod Pathol. 2014;27:1144‐1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kämpjärvi K, Mäkinen N, Mehine M, et al. MED12 mutations and FH inactivation are mutually exclusive in uterine leiomyomas. Br J Cancer. 2016;114:1405‐1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mehine M, Kaasinen E, Heinonen HR, et al. Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers. Proc Natl Acad Sci USA. 2016;113:1315‐1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reyes HD, Miecznikowski J, Gonzalez‐Bosquet J, et al. High stathmin expression is a marker for poor clinical outcome in endometrial cancer: an NRG oncology group/gynecologic oncology group study. Gynecol Oncol. 2017;146:247‐253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Allen MM, Douds JJ, Liang SX, Desouki MM, Parkash V, Fadare O. An immunohistochemical analysis of stathmin 1 expression in uterine smooth muscle tumors: differential expression in leiomyosarcomas and leiomyomas. Int J Clin Exp Pathol. 2015;8:2795‐2801. [PMC free article] [PubMed] [Google Scholar]
- 36. Cornejo K, Shi M, Jiang Z. Oncofetal protein IMP3: a useful diagnostic biomarker for leiomyosarcoma. Hum Pathol. 2012;43:1567‐1572. [DOI] [PubMed] [Google Scholar]
- 37. Yasutake N, Ohishi Y, Taguchi K, et al. Insulin‐like growth factor II messenger RNA‐binding protein‐3 is an independent prognostic factor in uterine leiomyosarcoma. Histopathology. 2017;72:739‐748. [DOI] [PubMed] [Google Scholar]
- 38. Liau JY, Lee JC, Tsai JH, et al. Comprehensive screening of alternative lengthening of telomeres phenotype and loss of ATRX expression in sarcomas. Mod Pathol. 2015;28:1545‐1554. [DOI] [PubMed] [Google Scholar]
- 39. Yang CY, Liau JY, Huang WJ, et al. Targeted next‐generation sequencing of cancer genes identified frequent TP53 and ATRX mutations in leiomyosarcoma. Am J Transl Res. 2015;7:2072‐2081. [PMC free article] [PubMed] [Google Scholar]
- 40. Raspollini MR, Amunni G, Villanucci A, et al. c‐Kit expression in patients with uterine leiomyosarcomas: a potential alternative therapeutic treatment. Clin Cancer Res. 2004;10:3500‐3503. [DOI] [PubMed] [Google Scholar]
- 41. Serrano C, Mackintosh C, Herrero D, et al. Imatinib is not a potential alternative treatment for uterine leiomyosarcoma. Clin Cancer Res. 2005;11:4977‐4979. [DOI] [PubMed] [Google Scholar]
- 42. van der Graaf WT, Blay JY, Chawla SP, et al. Pazopanib for metastatic soft‐tissue sarcoma (PALETTE): a randomised, double‐blind, placebo‐controlled phase 3 trial. Lancet. 2012;379:1879‐1886. [DOI] [PubMed] [Google Scholar]
- 43. Davidson B, Abeler VM, Førsund M, et al. Gene expression signatures of primary and metastatic uterine leiomyosarcoma. Hum Pathol. 2014;45:691‐700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. el Sayadi H, Pissaloux D, Alberti L, et al. Autocrine role for Gas6 with Tyro3 and Axl in leiomyosarcomas. Target Oncol. 2013;8:261‐269. [DOI] [PubMed] [Google Scholar]
- 45. Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 2012;72:4875‐4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Conklin CM, Longacre TA. Endometrial stromal tumors: the new WHO classification. Adv Anat Pathol. 2014;21:383‐393. [DOI] [PubMed] [Google Scholar]
- 47. Quan P, Moinfar F, Kufferath I, et al. Effects of targeting endometrial stromal sarcoma cells via histone deacetylase and PI3K/AKT/mTOR signaling. Anticancer Res. 2014;34:2883‐2897. [PubMed] [Google Scholar]
- 48. Lee CH, Ou WB, Mariño‐Enriquez A, et al. 14‐3‐3 fusion oncogenes in high‐grade endometrial stromal sarcoma. Proc Natl Acad Sci USA. 2012;109:929‐934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dal Cin P, Talcott J, Abrams J, Li FP, Sandberg AA. Ins(10;19) in an endometrial stromal sarcoma. Cancer Genet Cytogenet. 1988;36:1‐5. [DOI] [PubMed] [Google Scholar]
- 50. Koontz JI, Soreng AL, Nucci M, et al. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc Natl Acad Sci USA. 2001;98:6348‐6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hrzenjak A, Moinfar F, Tavassoli FA, et al. JAZF1/JJAZ1 gene fusion in endometrial stromal sarcomas: molecular analysis by reverse transcriptase‐polymerase chain reaction optimized for paraffin‐embedded tissue. J Mol Diagn. 2005;7:388‐395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Micci F, Gorunova L, Agostini A, et al. Cytogenetic and molecular profile of endometrial stromal sarcoma. Genes Chromosom Cancer. 2016;55:834‐846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Micci F, Brunetti M, Dal Cin P, et al. Fusion of the genes BRD8 and PHF1 in endometrial stromal sarcoma. Genes Chromosom Cancer. 2017;56:841‐845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Panagopoulos I, Mertens F, Griffin CA. An endometrial stromal sarcoma cell line with the JAZF1/PHF1 chimera. Cancer Genet Cytogenet. 2008;185:74‐77. [DOI] [PubMed] [Google Scholar]
- 55. Panagopoulos I, Thorsen J, Gorunova L, et al. Fusion of the ZC3H7B and BCOR genes in endometrial stromal sarcomas carrying an X;22‐translocation. Genes Chromosom Cancer. 2013;52:610‐618. [DOI] [PubMed] [Google Scholar]
- 56. Hoang LN, Aneja A, Conlon N, et al. Novel high‐grade endometrial stromal sarcoma: a morphologic mimicker of myxoid leiomyosarcoma. Am J Surg Pathol. 2017;41:12‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee CH, Ali RH, Rouzbahman M, et al. Cyclin D1 as a diagnostic immunomarker for endometrial stromal sarcoma with YWHAE‐FAM22 rearrangement. Am J Surg Pathol. 2012;36:1562‐1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee CH, Hoang LN, Yip S, et al. Frequent expression of KIT in endometrial stromal sarcoma with YWHAE genetic rearrangement. Mod Pathol. 2014;27:751‐757. [DOI] [PubMed] [Google Scholar]
- 59. Cossu‐Rocca P, Contini M, Uras MG, et al. Tyrosine kinase receptor status in endometrial stromal sarcoma: an immunohistochemical and genetic‐molecular analysis. Int J Gynecol Pathol. 2012;31:570‐579. [DOI] [PubMed] [Google Scholar]
- 60. Schoolmeester JK, Sciallis AP, Greipp PT, et al. Analysis of MDM2 amplification in 43 endometrial stromal tumors: a potential diagnostic pitfall. Int J Gynecol Pathol. 2015;34:576‐583. [DOI] [PubMed] [Google Scholar]
- 61. Parra‐Herran CE, Yuan L, Nucci MR, Quade BJ. Targeted development of specific biomarkers of endometrial stromal cell differentiation using bioinformatics: the IFITM1 model. Mod Pathol. 2014;27:569‐579. [DOI] [PubMed] [Google Scholar]
- 62. Schoolmeester JK, Sukov WR, Maleszewski JJ, Bedroske PP, Folpe AL, Hodge JC. JAZF1 rearrangement in a mesenchymal tumor of nonendometrial stromal origin: report of an unusual ossifying sarcoma of the heart demonstrating JAZF1/PHF1 fusion. Am J Surg Pathol. 2013;37:938‐942. [DOI] [PubMed] [Google Scholar]
- 63. Tokinaga A, Furuya M, Niino H, et al. Colonic low‐grade endometrial stromal sarcoma and orthotopic endometrial stromal tumor with limited infiltration sharing the JAZF1‐SUZ12 gene fusion. Pathol Int. 2014;64:178‐182. [DOI] [PubMed] [Google Scholar]
- 64. Hemming ML, Wagner AJ, Nucci MR, et al. YWHAE‐rearranged high‐grade endometrial stromal sarcoma: two‐center case series and response to chemotherapy. Gynecol Oncol. 2017;145:531‐535. [DOI] [PMC free article] [PubMed] [Google Scholar]


