Abstract
Genome-wide association studies have detected many loci underlying susceptibility to disease, but most of the genetic factors that contribute to disease susceptibility remain unknown. Here we provide evidence that part of the missing heritability can be explained by an overestimation of heritability. We estimated the heritability of twelve complex human diseases using family history of disease in 1,555,906 white European individuals from the UK Biobank. Estimates using simple family-based statistical models were inflated on average by ~47% comparing with those from Structural Equation Models (SEM) that specifically accounted for shared familial environmental factors. In addition, heritabilities using SNP data explained an average of 44.2% of the simple family-based estimates across diseases and an average of 57.3% of SEM estimated heritability and accounted for almost all of the SEM heritability for hypertension. Our results show that both genetics and familial environment make substantial contributions to familial clustering of disease.
Introduction
The causation of most common human diseases is complex, being influenced by a combination of genetic and environmental factors1. The development of genome-wide association studies (GWAS) has allowed the detection of many genetic variants associated with these diseases. However, these variants only explain a fraction of the heritability estimated in previous family-based studies and hence there is a “missing heritability” that remains unidentified2. One possible explanation for this missing heritability is that previous heritability estimates could be inflated because family environmental effects were not specified in the model or because they could not be estimated due to the study design3. Furthermore, comparisons of heritability explained by SNPs identified through GWAS or the hidden heritability estimated from genome-wide arrays (that is, the SNP heritability which captures the contribution of common variants including those not yet detected as genome-wide significant due to lack of power) with published estimates of heritability possess some important challenges. For instance, the populations from which family-based heritability estimates were obtained may differ from those used in the GWAS studies in definition or prevalence of disease or genetic background. These, and other factors3, make assessments of heritability estimates for disease from familial and GWAS studies difficult and in some instances inappropriate.
The objective of the current study was to estimate the heritability of twelve complex human diseases using self-reported personal and family history of disease in 1,555,906 white European participants and relatives from the UK Biobank, which comprise over 2% of the UK population.
Results
Data overview and Relative Risks
The UK Biobank contains disease and trait data, as well as biological samples collected from around 500,000 participants and has as its main objective to identify ways of improving the prevention, diagnosis and treatment of complex diseases4. UK Biobank participants were measured for multiple traits and questioned about their lifestyle, environmental risk factors and medical history and gave their informed consent following strict protocols5. Here we use information from the family disease history reported by participants to estimate the heritability and the environmental contributions to the liability of twelve broadly defined complex diseases: heart disease, stroke, chronic bronchitis, hypertension, diabetes, Alzheimer's disease, Parkinson's disease, severe depression and lung, bowel, prostate and breast cancers (Supplementary Table 1). Accuracy of self-reported health status was assessed and is discussed in the supplementary information (Supplementary note and Supplementary Tables 2 and 3).
Disease prevalence was higher among men than among women for all diseases except for severe depression, which was more prevalent among women (Supplementary Table 4). Generally, disease prevalence was higher among the parents of the participants than among the participants and their siblings, suggesting an age-related increase in disease liability. The relative risks (RR) of parents (RRPO) and siblings (RRSIB) of ill individuals participating in UK Biobank were estimated for each disease. In addition, the relative risk for partners of affected individuals was estimated using information from the parents of the participants (RRPAR). All the relative risks were significantly larger than one (Supplementary Figure 1). Overall, the relative risks for the estimates of RRPO and RRSIB that combined information from blood and adopted relatives were higher than those for RRPAR, except for hypertension and lung cancer. These estimates of relative risks suggest that combinations of both genetics and shared environmental risk factors contribute to the causation of these diseases (Supplementary Figure 1).
Heritability Estimates using Falconer’s Method
We estimated heritability values (h2) from either the correlations or regression coefficients (b) of the first-degree family pairs: parents-offspring (participants), siblings-participants and parents-siblings of participants (to provide h2PO, h2SIB, and h2PSIB, respectively) following Falconer’s Method (Methods). Correlations or regression coefficients using information of adoptive parent-offspring (bAPO) and adoptive sibling (bASIB) pairs, and parents of participants (partners, bPAR) were also calculated. Estimates among concordant and discordant gender pairs were calculated using a method that takes into account differences between sexes1, then these estimates were combined using a weighted mean of b across all gender pairs. Across generation differences in disease prevalence were taken into account using a control population of the same age for comparison1. Genetic correlations between genders were close to one, but tended to be lower than one (Supplementary Table 5).
All heritability estimates from first-degree family pairs were significantly different from zero (Table 1). The highest h2PO value was noted for depression (0.491±0.007) whereas the highest h2SIB was observed for prostate cancer (0.707±0.062). Estimates of h2PO were significantly lower than those of h2SIB for heart disease, stroke, hypertension, diabetes, and prostate and breast cancers, suggesting the existence of non-additive genetic effects or a greater environmental similarity between siblings than between parents and their children. The highest value of the regression from adoptive parent-offspring pairs, bAPO, was observed for severe depression (0.250±0.036) suggesting an important influence of shared family environmental effects on this disease. The adoptive parent-offspring regression, although much smaller than for depression, was also significantly greater than zero for heart disease, bronchitis and breast cancer. Hypertension had a high value for the correlation between partners, bPAR, (0.203±0.002) and a low value for bAPO (0.035±0.021) indicating the importance of environmental effects shared by partners but that are not shared between parents and their offspring, and/or positive assortative mating for hypertension or a trait or combination of traits highly correlated with hypertension.
Table 1. Family-based heritability estimates not accounting for shared environmental effects calculated by Falconer’s method and regression coefficients derived from different relative pairs.
| Disease | h2PO (SE) | h2SIB (SE) | h2PSIB (SE) | bAPO (SE) | bASIB (SE) | bPAR (SE) |
|---|---|---|---|---|---|---|
| Heart Disease | 0.368 (0.005) | 0.557 (0.018) | 0.514 (0.010) | 0.114 (0.026) | 0.145 (0.108) | 0.151 (0.003) |
| Stroke | 0.162 (0.010) | 0.305 (0.044) | 0.260 (0.017) | -0.057 (0.054) | - | 0.038 (0.004) |
| Bronchitis | 0.420 (0.009) | 0.501 (0.034) | 0.567 (0.017) | 0.169 (0.039) | 0.338 (0.138) | 0.108 (0.005) |
| Hypertension | 0.366 (0.009) | 0.691 (0.010) | 0.477 (0.008) | 0.035 (0.021) | 0.190 (0.056) | 0.203 (0.002) |
| Diabetes | 0.474 (0.007) | 0.692 (0.019) | 0.485 (0.012) | 0.067 (0.037) | 0.185 (0.098) | 0.109 (0.004) |
| Alzheimer’s | 0.238 (0.061) | - | 0.349 (0.036) | - | - | 0.060 (0.005) |
| Parkinson’s | 0.247 (0.038) | - | 0.214 (0.053) | - | - | 0.028 (0.013) |
| Depression | 0.491 (0.007) | 0.443 (0.019) | 0.642 (0.013) | 0.250 (0.036) | 0.184 (0.083) | 0.162 (0.005) |
| Lung cancer | 0.117 (0.038) | - | 0.314 (0.025) | - | - | 0.119 (0.005) |
| Bowel cancer | 0.260 (0.017) | 0.387 (0.057) | 0.300 (0.023) | 0.171 (0.120) | - | 0.032 (0.005) |
| Prostate cancer | 0.361 (0.022)¥ | 0.707 (0.062)¥ | 0.321 (0.036) ¥ | -0.053 (0.183) | - | - |
| Breast cancer | 0.287 (0.014)Ψ | 0.393 (0.039)Ψ | 0.301 (0.025)Ψ | 0.144 (0.070) | - | - |
h2PO: heritability estimates using data of parents and offspring; (SE): Standard errors between brackets; h2SIB: heritability estimates using data of siblings; h2PSIB: heritability estimates using data of parents and siblings of participants; bAPO: regression coefficient of parents on adopted offspring; bASIB: regression coefficient of adoptive siblings; bPAR regression coefficient of parents of participants (partners); -: Effect was not estimated as there was less than one pair with both members affected
Only male-male pairs
Only female-female pairs.
Significant positive regression or correlation coefficients from adopted pairs and partners (e.g. parents of participants) suggest the potential existence of various environmental effects shared by family members. Hence estimates of heritability obtained using only blood relatives or from models that do not account for the full complexity of shared environmental effects may be inflated (Supplementary Table 6)6–13.
Heritability Estimates using Structural Equation Modelling
Heritabilities estimated from SEM were in general lower than those estimated using Falconer’s method, with significant family environmental effects detected for all the diseases except for Parkinson’s disease (Table 2, Supplementary Table 7). Although for most diseases, genetic effects were the major attributable contribution to disease liability, for hypertension the sum of the effects due to shared familial environment was more important than genetic effects (A = 0.28 and C+S+P = 0.33). The estimated partner effect (P = 0.13) for hypertension and the common family effect (C = 0.15) for depression were high. High values of P inform about shared environment among partners or perhaps the presence of assortative mating. The physiological nature of hypertension mitigates against the possibility of assortative mating and it seems more likely that the high estimate for P is due to environmental factors shared by partners such as diet. However we cannot conclusively differentiate among these possibilities without more information such as the length of cohabitation14. The relatively large estimate of the common family effect for depression (C=0.15) would account for approximately half the correlation in the liability for depression between first degree relatives (as the expected correlation = A/2 + C) and would be important to consider in future studies of depression. Similarly our estimates suggest that at least a half of the correlation in disease liability between siblings is due to the combined effects of common family (C) and sibling (S) environment for heart disease, hypertension and lung cancer.
Table 2. Genetic and environmental effects estimated using the parsimonious reduced SEM model.
| Disease | Model | A (CI±0.95) | C (CI±0.95) | S (CI±0.95) | P (CI±0.95) | E (CI±0.95) |
|---|---|---|---|---|---|---|
| Heart Disease | ACSPE | 0.27 (0.24-0.27) | 0.08 (0.07-0.12) | 0.08 (0.07-0.08) | 0.06 (0.06-0.07) | 0.51 (0.49-0.57) |
| Stroke | APE | 0.23 (0.21-0.25) | - | - | 0.04 (0.03-0.04) | 0.73 (0.71-0.76) |
| Bronchitis | ACE | 0.29 (0.25-0.33) | 0.10 (0.10-0.11) | - | - | 0.61 (0.60-0.64) |
| Hypertension | ACSPE | 0.28 (0.28-0.29) | 0.06 (0.06-0.06) | 0.14 (0.14-0.14) | 0.13 (0.12-0.13) | 0.39 (0.38-0.39) |
| Diabetes | ASPE | 0.50 (0.49-0.52) | - | 0.11 (0.09-0.13) | 0.07 (0.06-0.08) | 0.32 (0.29-0.34) |
| Alzheimer’s | ACE | 0.25 (0.17-0.33) | 0.05 (0.03-0.06) | - | - | 0.70 (0.63-0.78) |
| Parkinson’s | AE | 0.26 (0.20-0.34) | - | - | - | 0.74 (0.72-0.81) |
| Depression | ACE | 0.25 (0.21-0.29) | 0.15 (0.15-0.15) | - | - | 0.60 (0.58-0.63) |
| Lung cancer | ACE | 0.09 (0.02-0.14) | 0.11 (0.09-0.13) | - | - | 0.81 (0.75-0.86) |
| Bowel cancer | ACSE | 0.24 (0.21-0.26) | 0.03 (0.01-0.03) | 0.06 (0.03-0.12) | - | 0.67 (0.65-0.71) |
| Prostate cancer | ASE | 0.38 (0.32-0.44) | - | 0.19 (0.11-0.26) | - | 0.43 (0.36-0.51) |
| Breast cancer | ASE | 0.29 (0.26-0.33) | - | 0.06 (0.01-0.10) | - | 0.65 (0.60-0.69) |
A: Additive genetic effects; C: Environmental effects common to the whole family; S: Sibling environmental effects; P: Partner environmental effects; E: Residual environmental effect; Confidence Interval at 95% between brackets. -: Parameter dropped from parsimonious reduced model.
Simulations
To test the performance of our analytical methods we simulated data for the twelve different diseases using the genetic and environmental contributions to liability estimated under the full model for each disease and the corresponding values of prevalence for fathers, mothers, participants and siblings (Supplementary Table 8). We analyzed ten replicated simulations for each disease to estimate the liability components. The means of liability components of the ten replicates were similar to those used to perform the simulations (Supplementary Tables 8 and 9). Performing model comparison within each replicate (Methods), recovered the model used to simulate data in more than 50% of the replicates for 4 of the 12 diseases (heart disease, hypertension, severe depression and prostate cancer) (Supplementary Table 10). However, even for the instances where the true generating model was not recovered, the means of genetic parameters across replicates were similar to those used to simulate data (Supplementary Table 11). Fitting an AE model ignoring familial environment to the simulated data yielded an overestimation of the heritability for all diseases (Supplementary Table 12).
Heritability using SNPs
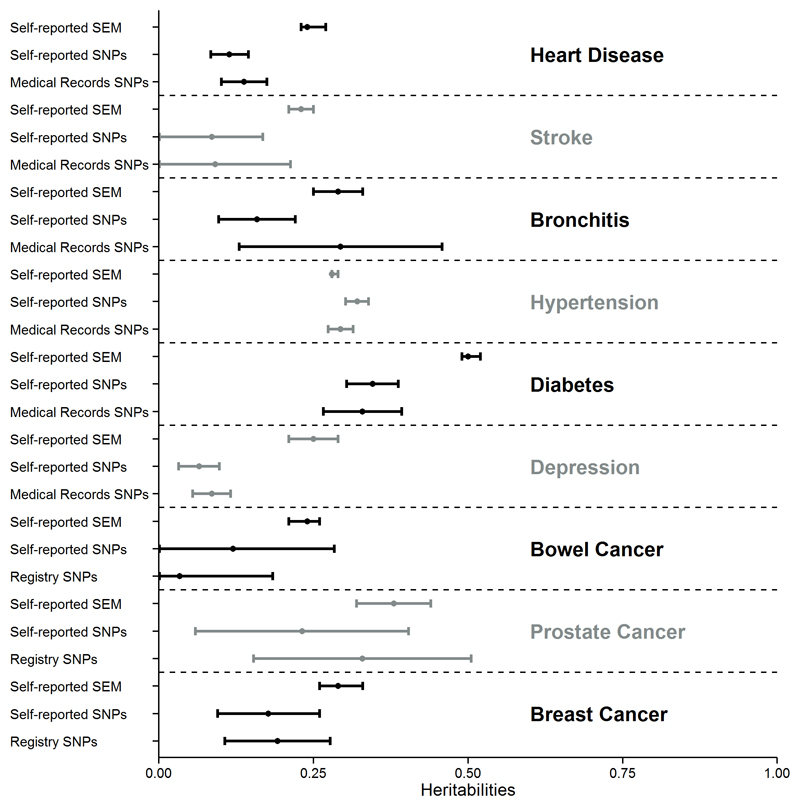
We obtained SNP heritability estimates using 525,242 SNPs in the genotyped subsample of 114,264 unrelated individuals for those diseases with prevalence higher than 0.50% (Methods). The SNPs explained an average of 44.2% of the Falconer’s method estimates, 44.0% of the SEM family–based heritability estimates using the AE model (Omitting family environmental factors - Supplementary Table 13) and 57.3% of the SEM family–based heritability estimates under the most parsimonious adequate model including family environmental factors, respectively, across diseases. The SNP heritability explained ~100% of the SEM heritability estimate for hypertension (Figure 1, Table 3), which suggests that, for this high-prevalence disease where we could model a large number of familial environmental factors, there might be little or no missing heritability. Conclusions from SNP heritability estimates were similar when SNPs were split into common and rare minor allele frequency (MAF) groups and the joint heritabilities of these two groups were estimated (Supplementary Table 14). However, as previously reported by Mancuso et al15 and Yang et al16 and) these estimates were generally slightly lower than estimates based on a single variance component of common and rare variants.
Figure 1. Heritability estimates using SEM family-based models (self-reported data) and SNPs (self-reported data and medical records).
Black and grey sets show the three heritability estimates for each disease using SEM family-based models (self-reported data) and SNPs (self-reported data and medical records).
Table 3. Heritability estimates of disease using common + rare SNPs and structural equation modelling (SEM) from self-reported data.
| Disease | h2C+R (CI95%) | h2SEM (CI95%) | %(h2 C+R /h2SEM) (SE) |
|---|---|---|---|
| Heart Disease | 0.11 (0.08-0.15) | 0.27 (0.24-0.27) | 40.74 (9.79) |
| Stroke | 0.09 (0.00-0.17) | 0.23 (0.21-0.25) | 39.13 (29.07) |
| Bronchitis | 0.16 (0.10-0.22) | 0.29 (0.25-0.33) | 54.43 (15.72) |
| Hypertension | 0.32 (0.30-0.34) | 0.28 (0.28-0.29) | 114.29 (3.29) |
| Diabetes | 0.35 (0.30-0.39) | 0.50 (0.49-0.52) | 70.00 (5.23) |
| Depression | 0.07 (0.03-0.10) | 0.25 (0.23-0.27) | 24.00 (13.79) |
| Bowel cancer | 0.12 (0.00-0.28) | 0.24 (0.21-0.26) | 50.0 (49.75) |
| Prostate cancer | 0.23 (0.06-0.40) | 0.38 (0.32-0.44) | 60.53 (30.42) |
| Breast cancer | 0.18 (0.10-0.26) | 0.29 (0.26-0.33) | 62.07 (19.05) |
h2C+R: Heritability estimates using SNPs in the liability scale; (CI95%): Confidence intervals; %(h2C+R /h2SEM): Percentage of SEM family-based estimate of heritability explained by SNPs. (SE): Standard error
SNP heritability estimates from self-reported and medical records (Supplementary Table 15) were not significantly different from each other, supporting the usefulness of the self-reported records. This was further confirmed by the similarity in the number of published GWAS hits with significant associations in the UK Biobank data using the self-reported definition of disease or the definition of disease from medical records (Supplementary Table 16).
Discusion
In the current study, we estimated the heritabilities of twelve diseases from family-based data using a model which does not take into account environmental factors shared by the family members (Falconer’s method) and a SEM method which enables joint estimation of these environmental factors and genetic factors. For most diseases, we obtained lower heritability values with the SEM method than with Falconer’s method associated with significant shared environmental effects. Therefore, the heritability estimates using SNPs were closer to SEM family-based heritability values than to those from Falconer’s method. Indeed for hypertension the heritability estimates using SNPs was similar to the SEM family-based heritability.
Recently, Yang et al16 have used information from simulated and observed data and analysis of high density imputed data to conclude that there is limited evidence for missing heritability for height and BMI once potential overestimation of heritability in family-based studies is taken into account. Zaitlen et al17 studied twenty three traits in the Icelandic population and suggested that most of the “missing heritability” is likely due to rare variants not included in the genotyping array but also reported that the excess correlation among close relatives was mostly accounted for by shared environment. Finally, Liu et al18 have also shown that models accounting for a diverse source of shared environmental effects should be tested to avoid bias in heritability estimation for a number of quantitative traits. In agreement with Zaitlen et al17, our very large study provides evidence that part of the missing heritability may be due to previous inflated heritability estimates and demonstrates this for important binary disease traits.
This study was based on a large cohort from the UK population, allowing us to estimate heritability with much narrower confidence intervals than in previous studies. In addition, models accounting for different environmental components shared by family members could be implemented due to the information available for different first-degree blood and adoptive relatives. The twelve diseases analyzed in this large cohort of individuals show significant but moderate values of heritability and an important impact of shared familial environmental effects and support the case for combining these factors with genetic marker information in order to improve the performance of disease-risk prediction methods19,20. Our results are very relevant when assessing the potential for the development of personalized medicine, providing realistic expectations of the value of genetic testing. In addition, demonstration of the importance of environmental risk factors that contribute to the aggregation of disease within families motivates research to identify and moderate these factors.
Online Methods
UK Biobank Data
The UK Biobank database) includes 502,682 participants who were aged between 49-69 years when recruited between 2006 and 2010 from across UK to take part of the project. The study was approved by the National Research Ethics Committee (REC reference: 11/NW/0382). The participants filled several questionnaires about their lifestyle, environmental risk factors and medical history and gave their informed consent4. The comprehension and acceptability of each question, the time taken to complete each of them, and their response distributions were examined in pilot studies, which aided the final selection and presentation of suitable questions. Self-reported medical history was confirmed by a trained nurse and where necessary by a medical doctor. Moreover, a pre-visit questionnaire was provided to participants before attending the assessment center, this questionnaire afforded participants the opportunity to record personal information such as family history before the visit to the assessment center to minimize problems of recalling. These details were entered directly into the assessment center computer and the questionnaire was not retained5. The UK Biobank contains information on about 445 types of diseases and 81 cancers in participants and the familial medical history of twelve broadly defined diseases among blood and adoptive fathers, mothers and siblings. Participants were considered as adopted when they answer “Yes” to the question: “Were you adopted as a child?”.
Family pairs (parent-offspring, sib-sib, parent-sibs and partners) were characterized for these twelve diseases, which include different subcategories in participants. The diseases analyzed were: heart disease (twenty-five subcategories), stroke (three subcategories), chronic bronchitis (three subcategories), hypertension (two subcategories), diabetes (four subcategories), Alzheimer's disease, Parkinson's disease, severe depression, lung cancer (two subcategories), bowel cancer (five subcategories), prostate and breast cancers (Supplementary Table 1). Those participant who answered “not to know” or “prefer not to answer” when they were asked about the disease status of relatives were removed from the analyses. Disease status of sibling was only considered when participants reported to have one sibling since they just had to report if at least one sibling had the corresponding disease and it was not possible to know how many siblings had suffered the disease when participants had more than one sibling. Disease status of 470,640 participants, 464,302 blood mothers, 459,716 blood fathers, 152,887 blood siblings, 4,962 adoptive mothers, 4,580 adoptive fathers and 1,819 adoptive siblings were used in the analyses. Those participants declared to have “white”, “British”, “Irish” or “Other white” ethnic background.
Medical Records
Data from medical records were used to test the accuracy of ten self-reported diseases. The type of medical record used to define a disease was different depending on the disease and was chosen because it was considered to be the best indicator of the disease available. Supplementary Table 2 shows the categories used to define each disease. There were available three kinds of medical records in the UK Biobank: data of hospitalization, medication/treatment and cancer register.
Data of hospitalization. Summary of the distinct main diagnoses codes a participant has had recorded across all their episodes in hospital. Heart disease, stroke, bronchitis, diabetes and Parkinson’s and Alzheimer’s diseases were defined with records from this register.
Medication/treatment. Medication self-reported by the participant used to treat the disease. Hypertension, diabetes and depression were defined with records form this registers.
Cancer Register. Data from the UK Cancer Register was used to define the cancer diagnoses.
Accuracy of self-reported data and family health status
Accuracy of self-reported health status was evaluated estimating the sensitivity, specificity, positive (PPV), and negative (NPV) predictive values among self-reported data and medical records from cancer register, hospitalization records and medication. The sensitivity was estimated as the percentage of individuals who self-reported having a disease among all those who appeared in the corresponding register as ill or taking the medication for the disease analyzed, the specificity was calculated as the percentage of those who self-reported being healthy for a particular disease among those who did not appear in the corresponding register or did not report taking medication for the corresponding disease. Positive predictive value (PPV) is the percentage of individuals who appeared in the corresponding register or were taking medication for a particular disease among those who self-reported having a disease, and the negative predictive value (NPV) is the proportion of those who did not appear in the registers or they did not report taking medication for the disease analyzed among those who did not report to have a particular disease. There were a total of 305,695 participants with hospitalization records that were used to estimate the accuracy of the self-reported phenotypes.
Prevalence
Prevalence of diseases in the UK Biobank were estimated as the number of people found to have a disease divided by the total number of individuals studied and their standard errors (SE) were estimated using the following formula:
where p is the prevalence and n is the total number of individuals studied.
Relative Risks
Relative risks of disease in the UK Biobank were estimated as follow21:
where a was the number of ill relatives of ill participants, b was the number of healthy relatives of ill participants, c was the number of ill relatives of healthy participants and d was the number of healthy relatives of healthy participants. The relative risk of parents (RRPO) and the relative risk of siblings (RRSIB) were estimated using this formula. The relative risks of partners, who are parents of participant, (RRPAR) was calculated in a similar way. The 95% confidence intervals (CI95%) were estimated as:
where RR is the corresponding relative risks and s is estimated as:
The minimum number of pairs in which both individuals are affected needed to estimate RR is one. In our dataset, the lowest number of pairs available to estimate RR was 33.
Heritability estimates
Diseases were treated as binary traits assumed to be determined by an underlying normal distribution of liability to disease. The correlation or regression among relatives (b) was used to estimate the heritability (h2 = 2b) of liability to disease. Method 4 described by Falconer1 was used to estimate b:
where pg is the prevalence of the disease in the relevant population within the UK Biobank, xc is the deviation of the threshold of liability that defines disease status from the mean of relatives of healthy participants, xr is the deviation of the threshold of liability that defines disease status from the mean of relatives of ill participants, and αg is the mean liability deviation of the ill participants from the mean liability of the relevant population within the UK Biobank. The sampling variance (Vb) of b was estimated according to appendix C of Falconer1 and confirmed by bootstrapping. The minimum number of pairs in which both individuals are affected needed to estimate b was one. In our data set the lowest number of pairs available to estimate b was three (in the adoptive pairs).
Across generation differences in disease prevalence were taken into account using an appropriate control population for comparison. Since prevalences among genders were different, four estimates according gender pairs were estimated using this method, which allows controlling for differences in gender and age prevalence when the variance in mean liability is different. The following sets of relatives were used: parent-offspring, sib-sib and parent-sib of participants (blood and adoptive) except for prostate and breast cancers where we only estimated same gender correlations. Moreover, b was estimated among the parents of the participants. For each relationship class, the correlations or regressions obtained from the four gender-parings were combined into a single weighted mean (bw), the weight being the reciprocal of the sampling variance of each regression coefficient. The sampling variance (Vbw) was calculated as the reciprocal of the sum of the weights and the standard error of the heritability was obtained as the square root of 4Vbw.
Genetic correlation
Genetic correlation (rG) between sexes was calculated for all the diseases except for prostate cancer and breast cancer which are expressed mostly in one sex. The following formula was used22:
where bFEMALE-MALE is the regression/correlation of mother-son or sister-brother bMALE-FEMALE, the regression/correlation of father-daughter or brother-sister, bFEMALE-FEMALE, the regression/correlation of mother-daughter or sister-sister and bMALE-MALE the regression/correlation of father-son or brother-brother.
Liability components
The liability to disease is the sum of genetic and different environmental effects. The distribution of the liability has a threshold value which differentiates between healthy and ill individuals. This threshold is based on the prevalence of the disease. As the prevalences are different in parents, siblings and participants, different thresholds must be assumed.
To estimate the liability parameters we can define the following structural equation:
where, A are genetic effects (assumed additive in the liability scale)23; C are environmental effects shared in common by all family members; S are environmental effects shared by siblings but not their parents which may include non-additive genetic effects; P are environmental effects shared among parents of participant (i.e. among partners) but not their children; and E are residual effects (including environmental effects specific to an individual and measurement error).
The correlations between each pair of blood and adoptive relatives for genetic and environmental components are set to fixed values according to their degree of genetic and environmental relationship. For example, blood parents-offspring pairs are correlated 0.5 for the genetic factors and 1 for common environmental effects. All the corresponding correlation values are shown in the Supplementary Figure 2. The relative importance of these components was evaluated using structural equation models (SEM) using OpenMx software version 1.4-353224.
Data of 210,787 blood and 4,184 adoptive families with one or two offspring (i.e. the participant and one sibling) were used to estimate liability components. A full model including all the effects (ACSPE) and all reduced models including genetic effects but removing one or more environmental effects were fitted. Each model was run 1,000 times and the run that converged with the maximum likelihood was chosen for model comparison. The relative fit of nested models was compared using hierarchic chi-square tests because the difference between the likelihood for a reduced model and that for the full model is approximately distributed, as a chi-square with df = df(full model) - df(reduced model). For each disease we started with the simplest model and included more parameters until we obtained the most parsimonious but adequate model that did not fit the data significantly worse than the full model.
Simulations
We simulated pedigrees with the same structure of families as in the real data comprising 210,787 blood and 4,184 adoptive families. To simulate the diseases, the prevalences of each disease in fathers, mothers, participant and siblings were used together with the parameters obtained using the full model (Supplementary Table 8). The full model was fitted using OpenMx following the same procedure as with real data. Analyses with 10 simulation replicates for each disease were performed to estimate liability parameters. The means and standard deviations of the 10 replicates for each of the liability components were estimated. Model comparison for each replicate was carried out in the same way as with real data.
Genotype Quality Control
We use data from the genotyped individuals in phase 1 of the UK Biobank genotyping program. In this phase, 49,979 individuals were genotyped using the Affymetrix UK BiLEVE Axiom array and 102,750 individuals using the Affymetrix UK Biobank Axiom array. Further details regarding genotyping procedure and genotype calling protocols are available at the UK Biobank website. We excluded multi-allelic markers, SNPs with an overall missing rate higher than 2% or with a strong platform specific missing bias (Fisher’s exact test, P < 10-100). We also excluded individuals with a missing rate higher than 5%, with a self-reported sex different from the genetic sex estimated from the X chromosome inbreeding or those with an excess of heterozygosity according to the UK Biobank internal QC procedures.
A reduced dataset of 151,532 individuals remained after filtering. In addition to this, common and rare variants (i.e. with a MAF > 0.0036) and those that did not exhibit departure from Hardy-Weinberg equilibrium (P < 10-50) in the unrelated (subset of 114,264 individuals with a relatedness below 0.0625) White-British cohort were kept. The genotype quality control and data filtering was performed using plink25.
SNP heritability estimates
SNP heritability estimates were estimated in a subset of 114,264 individuals for nine out of the twelve diseases with a prevalence in the population higher than 0.50% (heart disease, stroke, chronic bronchitis, hypertension, diabetes, severe depression and bowel, prostate and breast cancers) using self-reported data and medical records.
To estimate the heritability for each disease and data set, the genetic relationship matrices (GRMs) were computed fitting simultaneously 525,242 SNPs in the following mixed lineal model:
where y is the vector of phenotypes (diseases), β is the vector of fixed effects and covariates which included age of participant, the 20 first principal components and gender (except for prostate and breast cancer), u is the vector of SNP effects distributed as I is the identity matrix, and ϵ is a vector of residual effects distributed as W is a genotype matrix defined by the equation:
where sik is the number of copies of the reference allele for the SNP k of the individual i, and pk is the frequency of the reference allele for the SNP k. Under this model, the variance of y is:
Where A is the GRM, σ2g the genetic variance and σ2ε the residual variance. Variance components were estimated using restricted maximum likelihood (REML). These analyses were performed using DISSECT26.
In addition to this, a two variance component model splitting the SNPs into 319,037 common SNPs (MAF>0.05) and 206,205 rare SNPs (0.0036<MAF<0.05) was fitted for each disease.
where, ucommon and urare are the vectors of SNP effects for common and rare variants, respectively. Wcommon and Wrare are the genotype matrices defined for common and rare variants, respectively.
Under this model, the variance of y is:
where Acommon and the Arare are the GRMs computed using the common and rare variants, respectively. σ2gcommon and σ2grare are the genetic variances explained by the common and rare variants, respectively.
The heritability estimates were transformed to the liability scale using the following equation:
where h2L is the heritability in the liability scale is the heritability in the liability scale h2(0,1) is the heritability in the observed scale obtained from the REML analyses, P is the prevalence of the disease in the cohort and Z is the height of the standard normal probability density function at the threshold that truncates the proportion P23.
The percentage of SEM family-based estimates of heritability explained by SNPs was calculated as the ratio between h2SNPs and h2SEM multiplied by 100 and the standard error of the percentage was calculated according to Stuart et al27 as:
where h2C+RSNPS is the heritability explained by common and rare SNPs, h2SEM is the heritability using SEM family-based, σ2h2C+RSNPs is the standard deviation of h2C+RSNPS, σ2h2SEM is the standard deviation of h2SEM, C+RSNPs are related to the distribution of the estimates of h2C+RSNPS and SEM related to the distribution of the estimates of h2SEM. We cannot estimate 2COV(C+RSNPs,SEM) and assume this value is equal to 0.
Testing of GWAS hits for self-reported and clinical definitions of disease
GWAS hits for breast cancer, prostate cancer, bowel cancer, Type 2 diabetes, Hypertension, Stroke and Cardiovascular Artery Disease were downloaded from the GWAS catalogue. In total, we found that 278 of these SNPs were genotyped in our array, and tested them for association with our self-reported and clinical definitions of disease (breast cancer, prostate cancer, bowel cancer, Type 2 diabetes, Hypertension, Stroke and Heart Disease) using a chi-square test as implemented in the plink2 option (--assoc)25. Significant SNPs at a p-value of 0.05 and 0.00018 (i.e. 0.05/278) were counted for the two definitions of disease (self-reported and clinical). Only the subset of genotyped samples with clinical information was used to compare the power of the two alternative phenotype definitions.
Supplementary Material
Acknowledgements
This research has been conducted using the UK Biobank Resource, and funded by the Roslin Institute Strategic Programme Grant from the BBSRC (BB/J004235/1). CSH and AT also acknowledge funding from the Medical Research Council. We thank Ian White for his helpful comments. Heritability estimates using SNPs were performed using the ARCHER UK National Supercomputing Service.
Footnotes
URLs
UK Biobank, http://www.ukbiobank.ac.uk/; plink, https://www.cog-genomics.org/plink2; ARCHER UK National Supercomputing Service, http://www.archer.ac.uk; genotyping procedure and genotype calling protocols of the UK Biobank, http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=155580; UK Biobank internal QC procedures, http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=155580; DISSECT, https://www.dissect.ed.ac.uk; GWAS catalogue, https://www.genome.gov/gwastudies/
Author Contributions
AT and CSH conceived and designed the study. MM and AT performed the statistical analysis. OCX and KR carried out the SNP filtering and QC. MM, CSH and AT wrote the manuscript. RPW performed the simulations and contributed ideas and quantitative genetics expertise. All authors read and approved the manuscript.
Competing Interest Statement
The authors declare no competing financial interests.
References
- 1.Falconer DS. Inheritance of Liability to Certain Diseases Estimated from Incidence among Relatives. Annals of Human Genetics. 1965;29:51. [Google Scholar]
- 2.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tenesa A, Haley CS. The heritability of human disease: estimation, uses and abuses. Nat Rev Genet. 2013;14:139–49. doi: 10.1038/nrg3377. [DOI] [PubMed] [Google Scholar]
- 4.Allen N, et al. UK Biobank: Current status and what it means for epidemiology. Health Policy Technol. 2012;1:123–126. [Google Scholar]
- 5.Centre, U.B.C. UK Biobank: Protocol for a large-scale prospective epidemiological resource. Protocol No: UKBB-PROT-09-06 (Main Phase) 2007.
- 6.Kaprio J, et al. Concordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia. 1992;35:1060–7. doi: 10.1007/BF02221682. [DOI] [PubMed] [Google Scholar]
- 7.Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 8.Iliadou A, et al. Repeated blood pressure measurements in a sample of Swedish twins: heritabilities and associations with polymorphisms in the renin-angiotensin-aldosterone system. J Hypertens. 2002;20:1543–50. doi: 10.1097/00004872-200208000-00017. [DOI] [PubMed] [Google Scholar]
- 9.Gatz M, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–74. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 10.Zdravkovic S, Wienke A, Pedersen NL, de Faire U. Genetic influences on angina pectoris and its impact on coronary heart disease. Eur J Hum Genet. 2007;15:872–7. doi: 10.1038/sj.ejhg.5201846. [DOI] [PubMed] [Google Scholar]
- 11.Hallberg J, et al. Interaction between smoking and genetic factors in the development of chronic bronchitis. Am J Respir Crit Care Med. 2008;177:486–90. doi: 10.1164/rccm.200704-565OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korja M, et al. Genetic epidemiology of spontaneous subarachnoid hemorrhage: Nordic Twin Study. Stroke. 2010;41:2458–62. doi: 10.1161/STROKEAHA.110.586420. [DOI] [PubMed] [Google Scholar]
- 13.Polderman TJC, et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet. 2015;47:702–709. doi: 10.1038/ng.3285. [DOI] [PubMed] [Google Scholar]
- 14.Annest JL, Sing CF, Biron P, Mongeau JG. Familial aggregation of blood pressure and weight in adoptive families. II. Estimation of the relative contributions of genetic and common environmental factors to blood pressure correlations between family members. Am J Epidemiol. 1979;110:492–503. doi: 10.1093/oxfordjournals.aje.a112830. [DOI] [PubMed] [Google Scholar]
- 15.Mancuso N, et al. The contribution of rare variation to prostate cancer heritability. Nature Genetics. 2016;48:30–5. doi: 10.1038/ng.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015 doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaitlen N, et al. Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genetics. 2013;9:e1003520. doi: 10.1371/journal.pgen.1003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C, et al. Revisiting heritability accounting for shared environmental effects and maternal inheritance. Hum Genet. 2015;134:169–79. doi: 10.1007/s00439-014-1505-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aschard H, Vilhjalmsson BJ, Joshi AD, Price AL, Kraft P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. American Journal of Human Genetics. 2015;96:329–39. doi: 10.1016/j.ajhg.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tada H, et al. Risk prediction by genetic risk scores for coronary heart disease is independent of self-reported family history. European Heart Journal. 2016;37:561–7. doi: 10.1093/eurheartj/ehv462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Risch N. Linkage strategies for genetically complex traits. I. Multilocus models. American Journal of Human Genetics. 1990;46:222–8. [PMC free article] [PubMed] [Google Scholar]
- 22.Falconer DS, Mackay TF. Introduction to Quantitative Genetics. 1996 doi: 10.1093/genetics/167.4.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dempster ER, Lerner IM. Heritability of Threshold Characters. Genetics. 1950;35:212–36. doi: 10.1093/genetics/35.2.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boker S, et al. OpenMx: An Open Source Extended Structural Equation Modeling Framework. Psychometrika. 2011;76:306–317. doi: 10.1007/s11336-010-9200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canela-Xandri O, Law A, Gray A, Woolliams JA, Tenesa A. A new tool called DISSECT for analysing large genomic data sets using a Big Data approach. Nat Commun. 2015;6 doi: 10.1038/ncomms10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuart A, Ord JK, editors. Kendall's advanced theory of statistics. Hoder Arnold; London: 1994. 700. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.