Abstract
Microglia are fundamentally important immune cells within the central nervous system (CNS) that respond to environmental challenges to maintain normal physiological processes. Alterations in steady-state cellular function and over-activation of microglia can facilitate the initiation and progression of neuropathological conditions such as Alzheimer’s disease, Multiple Sclerosis, and Major Depressive Disorder. Alcohol consumption disrupts signaling pathways including both innate and adaptive immune responses that are necessary for CNS homeostasis. Coordinate expression of these genes is not ascertained from an admixture of CNS cell-types, underscoring the importance of examining isolated cellular populations to reveal systematic gene expression changes arising from mature microglia. Unbiased RNA-Seq profiling was used to identify gene expression changes in isolated prefrontal cortical microglia in response to recurring bouts of voluntary alcohol drinking behavior. The voluntary ethanol paradigm utilizes long-term consumption ethanol that results in escalated alcohol intake and altered cortical plasticity that is seen in humans. Gene coexpression analysis identified a coordinately regulated group of genes, unique to microglia, that collectively are associated with alcohol consumption. Genes within this group are involved in toll-like receptor signaling and transforming growth factor beta signaling. Network connectivity of this group identified Siglech as a putative hub gene and highlighted the potential importance of proteases in the microglial response to chronic ethanol. In conclusion, we identified a distinctive microglial gene expression signature for neuroimmune responses related to alcohol consumption that provides valuable insight into microglia-specific changes underlying the development of substance abuse, and possibly other CNS disorders.
Keywords: Microglia, transcriptome, ethanol, prefrontal cortex, transforming growth factor beta, toll-like receptors
1.1 Introduction
Microglia comprise approximately 5–15% of all cells present within the adult brain (Lawson et al., 1990), although abundance varies by brain region (De Biase et al., 2017), and serve as the principal cell type responsible for innate and adaptive immune processes. Mature microglia change morphological and expression profiles to participate in CNS homeostasis and respond to pathological insults.
Acute and chronic substance abuse cause persistent changes in gene expression, which can cause functional changes that contribute to maladaptive behavior (Nestler et al., 1993). Neurogenomic studies for substance abuse have primarily utilized whole tissue homogenates, representing a mixture of cell-types, including neurons, astrocytes, oligodendrocytes, and microglia. These cell types express many common genes; however, they each have a unique transcriptional landscape (Zhang et al., 2014). While studying the gene expression changes of whole tissue is important to understand communication within a cellular ensemble, this approach may fail to discern specific cellular responses occurring within an individual cell-type. Isolated microglial cells have been shown to undergo a unique set of gene expression changes associated with aging, neurodegeneration, and depression (Chiu et al., 2013; Gonzalez-Pena et al., 2016; Hickman et al., 2013). Exposure to in vitro alcohol leads to microglial activation and the release of specific proinflammatory cytokines, while long-term alcohol consumption results in microglial activation in rodent models and human postmortem brain (Fernandez-Lizarbe et al., 2009; He and Crews, 2008; Montesinos et al., 2016). These studies have led to the idea that alcohol exposure leads to an inflammatory state which further drives ethanol consumption. We hypothesize that microglial gene expression represents an important molecular phenotype correlated with alcohol consumption, and that characterizing the transcriptome of microglia versus total cell populations will provide a framework for the identification of specific alcohol-induced alterations in neuroimmune gene expression.
The prefrontal cortex (CTX) is an important brain region involved in alcohol use disorders (AUDs), serving as a key link in both positive and negative affect associated with substance abuse (Koob and Volkow, 2010). Alcohol exposure triggers a cascade of immune-related gene expression changes within adolescent and adult CTX (Osterndorff-Kahanek et al., 2013) suggesting that microglia may be critical mediators underlying the transition from abuse to dependence. We utilized RNA-sequencing to profile the transcriptome of isolated microglia in response to voluntary alcohol consumption and identified coordinated gene expression changes that were specific to the microglial cell population. These findings support the hypothesis that neuroimmune genes underlie the progression of alcohol use to AUDs.
1.2 Materials and Methods
1.2.1 Ethics Statement
All procedures were approved by the University of Texas at Austin Institutional Animal Care and Use Committee (animal protocol number AUP-2013-00061) and adhered to the NIH Guidelines. The University of Texas at Austin animal facility is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care.
1.2.2 Animals and voluntary alcohol consumption
Studies were conducted in adult (6–8 weeks old) C57BL/6J (B6) male mice (Jackson Laboratories, Bar Harbor, ME). Mice were individually housed and allowed to acclimate to upright bottles one week before the start of the experiment. The experimental rooms were maintained at an ambient temperature of 21±1°C, 40–60% humidity on a regular light/dark schedule, with ad libitum access to food and water.
An every-other-day 2-bottle choice (EOD-2BC) a paradigm (Osterndorff-Kahanek et al., 2013) was used with mice randomly assigned to control or alcohol-consumption treatment groups. Control and treatment groups each contained 12 mice per group (24 total). Control animals had daily access to water while treatment groups had every-other-day access to both water and a 15% (v/v) alcohol solution (access to water only on non-treatment days). An empty cage control was used to account for spillage. Alcohol bottle positions were alternated at each exposure to control for potential side preferences. Alcohol and water bottles were weighed on treatment days, and animals were weighed once per week to calculate consumption. The study concluded after 60 total days (30 drinking days) and blood alcohol concentrations were determined prior to sacrifice. Because mice consume most alcohol at the beginning of the dark phase (7 PM) and we sacrificed them between (9–10:30 AM), the blood alcohol levels were low to undetectable (average BAC 14 mg/dl, SD = 7.3 mg/dl, n=12).
1.2.3 Tissue harvest and microglial isolation
Mice were sacrificed immediately after the final alcohol drinking session and anaesthetized using isoflurane followed by transcardial perfusion with PBS. The CTX was dissected as previously described (Osterndorff-Kahanek et al., 2013) and put into cold Hank’s Balanced Salt Solution (HBSS). Microglial isolations (Nikodemova and Watters, 2012) were performed by first mincing individual CTX tissue on ice and suspending in cold HBSS. Approximately 1% of the minced tissue mixture per sample was taken as a total homogenate sample (includes all cell types). Homogenates were centrifuged at 1000 x g for 10 minutes at 4°C. The supernatant was removed and the cells were flash frozen in liquid nitrogen. The remaining minced tissue was used for microglial isolation with magnetic bead sorting technology. This method was chosen because it is shown to preserve cellular phenotype and RNA integrity as well as result in high purity compared to other methods (Ju et al., 2015; Nikodemova and Watters, 2012). Briefly, tissue solution was manually dissociated using the Neural Tissue Dissociation Kit (Papain) (Miltenyi Biotec, Germany). Dissociated tissue was passed through a 70 μM strainer (Miltenyi Biotec), centrifuged at 300 x g, then resuspended in 30% percoll (Sigma-Aldrich, St. Louis, MO). The percoll-cell suspension was centrifuged at 700 x g for 15 minutes at room temperature with the myelin fraction removed from the top of the suspended solution. Cells were incubated with Cd11b MicroBeads (Miltenyi Biotec) and eluted using MS columns to collect Cd11b+ cells. Previous studies have shown that microglial isolation using this protocol yields pure microglia and does not perturb gene expression (Ju et al., 2015; Nikodemova and Watters, 2012).
1.2.4 RNA Isolation and PCR validation of microglial isolation
RNA, from individual samples, was isolated using the RNeasy Micro Kit (Qiagen, Germany). RNA concentration and quality (RNA integrity number) was determined using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) using the Agilent RNA 6000 Pico Kit. All of the examined total homogenate and microglia samples met quality-control criterion. There were no significant different differences in RNA integrity number (RIN), evaluated based upon the 28S to 18S ratio, between total homogenate and microglia samples (P = 0.61). There were no significant differences in quality or quantity of microglial RNA between the control and ethanol groups. Total RNA was reverse transcribed into cDNA using the Applied Biosystems High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific Inc., Rockford, IL). Applied Biosystems Taqman® Gene Expression Assay primers were used for quantitative real-time PCR (q-rtPCR). Assay ID’s included Mm0434455_m1 (Itgam/Cd11b), Mm01253033_m1 (Gfap), Mm01248771_m1 (Rbfox3/Neun), Mm00443243_m1 (Tek), Mm00495930_m1 (Ugt8a), and 4333760T (18s). Q-rtPCR reactions were performed using SsoAdvanced™ Universal Probes Supermix (BioRad, Hercules, CA) in 10-μL reactions containing 1 ng of cDNA. All reactions were performed in technical triplicates for each biological replicate and included a negative no-template control. Q-rtPCR reactions were carried out using the CFX384 Real-Time System (BioRad). Samples were normalized to 18s and relative expression was determined using the CFX software (BioRad). Examining the five cellular makers with Q-rtPCR verified Cd11b positive cells were enriched compared to the Cd11b negative fraction and total homogenate (Supp. Fig. 1).
1.2.5 RNA Sequencing and Bioinformatics Analysis
Samples were Poly(A) selected using the Poly(A) Purist Kit (Thermo Fisher Scientific Inc.) and libraries were prepared using NEBNext® Ultra™ Directional RNA Library Preparation Kit (New England BioLabs, Ipswich, MA). Samples were sequenced on the NextSeq 500 (Illumina, San Diego, CA) using (75 bp; paired-end reads) to a minimum target depth of 20 million reads per sample. Bioinformatic analysis methods are shown in the supplementary information.
1.3 Results & Discussion
1.3.1 Transcriptome profiles of isolated microglia and total homogenate
Transcriptome profiles of the microglia (Cd11b+) and tissue homogenate (TH) from the CTX of individual samples were sequenced to a target depth of 20 million reads. A total of 17,377 genes were reliably detected in Cd11b+ samples and 19,876 genes detected in TH. Among the two preparations 17,172 genes were commonly expressed, with 205 unique transcripts in the Cd11b+ fraction and 2,704 unique transcripts in the TH. In agreement with single-cell sequencing of major CNS cell-types (Darmanis et al., 2015), the microglial population expressed not only a fewer number of genes, but the overall expression distribution of detected transcripts was lower in magnitude (Supp. Fig. 1). Although the isolated microglial population is being compared to a cumulative admixture of all major CNS cell-types present within CTX, this blunted profile further underscores the need for examining a solitary cellular population. Mean expression of genes present in isolated cells versus the mean expression of genes common to TH demonstrates the relative enrichment of genes more highly localized to microglia (Supp. Table 1). The cellular marker Cd11b+ (Itgam) used for separation of the microglial fraction showed nearly 50 times higher abundance relative to TH. Tmem119, an additional microglia-specific marker (Satoh et al., 2016), was elevated to approximately the same degree as Itgam; extending Q-rtPCR validations (Supp. Fig 2) and substantiating the enrichment protocol for assessing transcriptome-wide measurements derived from microglia. Consistent with the predominant function of microglia, gene ontology analysis of the top 100 microglial-enriched genes (Supp. Table 2) affirmed an over-representation for genes involved in mediating an ‘inflammatory response’ (GO:0006954) and ‘cytokine receptor binding’ (GO:0005126).
1.3.2 Differential expression following chronic alcohol consumption
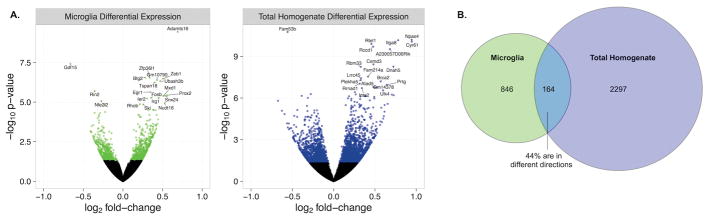
An every-other-day 2-bottle choice alcohol-drinking paradigm (EOD-2BC) was undertaken to evaluate gene expression changes related to chronic alcohol exposure (Fig. 1). EOD-2BC, compared to other voluntary alcohol drinking mouse models, leads to escalation of drinking over time and high alcohol consumption (Melendez, 2011). Previous work has shown that EOD-2BC leads to the greatest number of neuroimmune related changes in CTX (Osterndorff-Kahanek et al., 2013); however, a portion of these changes may not have been cell-type specific. RNA-Seq of an isolated microglial population using EOD-2BC was undertaken to determine the contribution of gene expression changes within this cell-type compared to tissue homogenate. Differential gene expression analysis for alcohol drinking exposure respectively identified 1,010 microglia and 2,461 total homogenate changes (Fig. 2). The reduced number of changes occurring in microglia is expected given the additional CNS cell-types present within the total homogenate.
Figure 1.
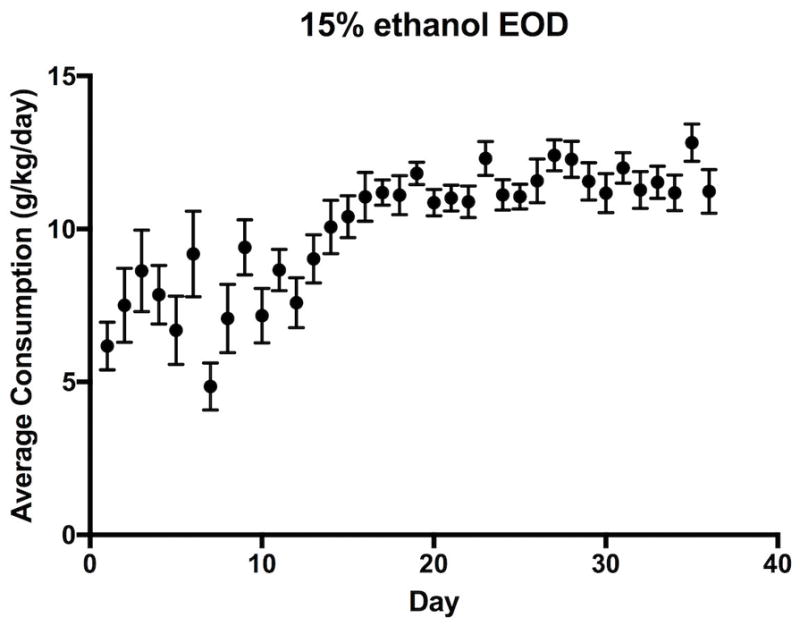
Ethanol consumption in voluntary every-other-day 2-bottle choice drinking paradigm. Values are shown as averages consumption (g/kg) over a 24-hour period with error bars showing SEM. Consumption is shown for days that alcohol was available only.
Figure 2.
Differential expression in response to chronic ethanol for microglia and total homogenate. A. Volcano plot showing log2 fold-change against log10 p-value. Differentially expressed genes (p < .05) are shown as green or blue dots. The top 20 differentially expressed genes are labeled. The total homogenate has larger fold-changes and lower p-values than the microglia. B. Venn Diagram showing the number of unique and overlapping differentially expressed genes (p < .05) in each of the preparations. 164 genes were differentially expressed with alcohol in both the microglia and total homogenate. Of these 164 genes, 44% (72 genes) were changed in different directions in the two preparations.
Contrasting the gene expression profiles for the two preparations shows isolated microglia have a similar range in fold-changes within the CTX due to alcohol (Fig. 2a). The observed changes are less than two-fold for either microglia or the total homogenate; however, labeling the top 20 statistically significant genes shows there are definitive biological differences between the experimental conditions due to EOD-2BC alcohol consumption. Small fold-change differences in alcohol-induced gene expression, and other biological responses, are not uncommon for the CNS (Lovinger and Crabbe, 2005). Chronic alcohol exposure in B6 mice evokes time-dependent changes in gene expression (Osterndorff-Kahanek et al., 2015), which may not achieve a chronic cumulative steady-state of induction suggested for some other drugs of abuse (Hope et al., 1994). The observed changes in gene expression may serve as a surrogate measure for cellular mechanisms activated as a result of EOD-2BC alcohol consumption, with select differences that are representative of microglial activation.
A subset of genes (164 genes in total) is regulated by EOD-2BC in both microglia and the total homogenate (Fig. 2b). Despite this mutual overlap, 44% of these changes (72 genes) occur in opposite directions. Additionally, although a small percentage of microglia are present within the total homogenate a total of 864 gene expression changes are uniquely discerned in the isolated cells. Alcohol-induced changes within microglia affected a broad spectrum of functional categories (Supp. Table 3), several of which were not enriched within the admixture of CNS cell-types. For example, alcohol-induced microglial changes were over-represented in genes attributed to ‘TGF-beta receptor signaling’ (GO:0007179, P = 9.08 E-05) and ‘chronic inflammatory response’ (GO:0002544, P = 1.57 E-03). Transforming growth factor beta (TGF-beta) is a cytokine expressed in brain, capable of controlling microglial activation and triggering neuroinflammation (Lodge and Sriram, 1996). Excessive and chronic alcohol abuse modulates similar inflammatory processes in the liver (Dooley and Dijke, 2012), involving TGF-beta and downstream signaling involving SMAD proteins (P = 5.21 E-04). The extent to which these systems are affected within the brain due to the alcohol abuse is largely unknown, but corresponding changes in CNS microglial may indicate their involvement may extend beyond liver dysfunction in an AUD. Importantly, these alterations in gene expression pathways witnessed in the isolated CNS microglial population are obscured within the total homogenate. The selective effects registered indicates the value of cellular separation to draw upon microglial-specific gene expression changes that take place in mouse CTX following alcohol drinking exposure.
1.3.3 Gene coexpression networks related to chronic alcohol exposure
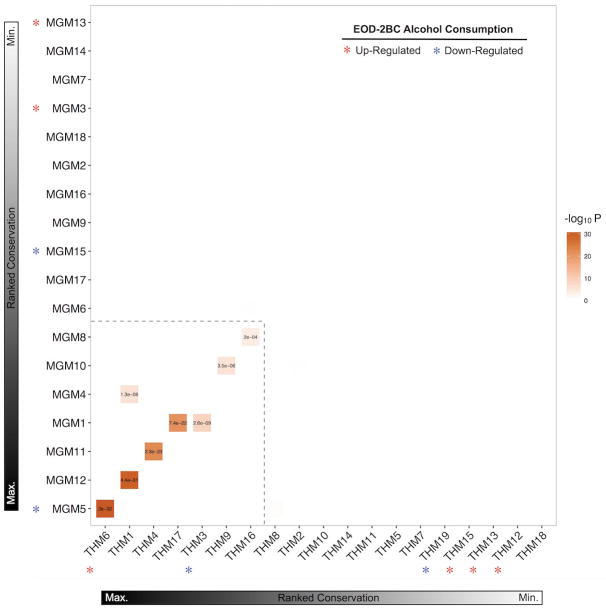
Coordinate expression of multiple genes enables biological function of intricate cellular systems. Weighted correlation networks, including all genes within their respective cellular populations, were evaluated to distinguish particular subsets of coordinately regulated gene expression clusters (i.e. modules) (Langfelder and Horvath, 2008). The assayed transcriptomes partitioned into 18 microglial modules (MGM) and 19 total homogenate modules (THM) (Supp. Figure 3). Each of the individual gene expression modules were representative of varied biological categories (Supp. Table 4), demonstrating the utility of this approach for delineating transcriptome substructure. Ranking the relative conservation of MGM and THM, based upon coinciding genes, illustrates the overall similarity and dissimilarity of the two experimental fractions (Fig. 3).
Figure 3.
Overlap between the microglia and total homogenate modules based on gene membership (Fishers-exact-test, correct for multiple comparisons using a false discovery rate less than .05). The modules contained in the hatched box show high overlap between the two fractions and likely represent shared genes and processes. The modules outside the hatched box are unique to that fraction. * denotes differentially expressed with ethanol (p < .05) and the color indicates the direction of the change.
Altogether, there are seven MGM and THM pairs sharing a significant number of common genes. Examining the top seven strongest MGM-THM pairwise relationship shows each of these shared overlapping gene sets denote a meaningful intracellular group, such as ‘chromatin modification’ (MGM5-THM6, P = 4.21E-03), ‘mitochondrial matrix’ (MGM1-THM3, P = 1.70 E-10), and ‘RNA splicing’ (THM16–MGM8, P = 4.60 E-12). Only one of the MGM-THM pairs, MGM5-THM6, showed evidence of chronic alcohol-responsive changes in gene expression; however, the direction of MGM5 change was opposite to THM16. Transcriptional regulation through epigenetic modification of chromatin is proposed as a major component in neuronal plasticity (Maze et al., 2015). DNA methylation alters chromatin structure, affecting persistent regulation of gene expression and CNS plasticity. Stimulation of neuronal firing leads to a redistribution of DNA methylation (Guo et al., 2011), suggesting pharmacological interactions affecting neuronal activity impacts epigenetic mechanisms of transcriptional regulation. Increased expression of genes involved in histone modification after chronic alcohol may be a surrogate indicator of epigenomic adaptations in neuronal circuits, or potentially other non-microglial cell types (Starkman et al., 2012).
EOD-2BC alcohol consumption altered the expression of seven THMs, four of which (THM19, THM15, THM13, THM12) were distinctly separate from MGMs. Alcohol-induced changes in these THM likely represent molecular processes set apart from microglial responses. THM19 is significantly decreased in EOD-2BC mice, representing an overall alcohol-driven reduction in kindred genes within CTX. THM19 is made-up of a significant number of oligodendrocytes-related genes (P = 3.31 E-03); which is the CNS cell responsible for myelinating neuronal axons. Human oligodendroglial genes are decreased in human alcoholic CTX (Lewohl et al., 2005), featuring cellular adaptations that are potentially conserved between human and mouse CTX subjected to repeated alcohol. THM19 is inversely correlated to THM15, 13, and THM12; wherein these three THM are intrinsically related with an average rank-based intermodule correlation equal to 0.88. THM15, THM13, and THM12 are collectively enriched for neuronal gene expression signatures (P = 1.72 E-05).
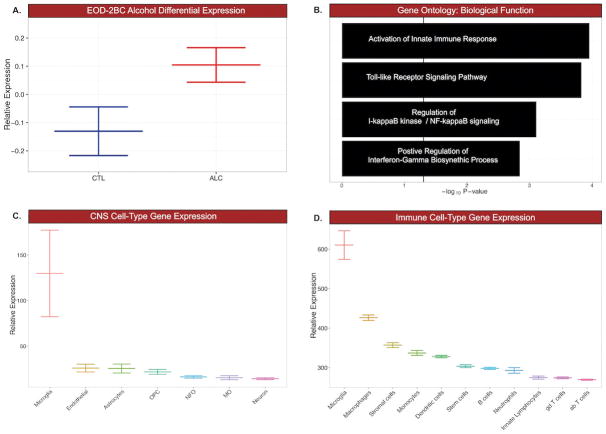
Gene expression for MGM5, MGM15, MGM3, and MGM13 are differentially regulated by EOD-2BC alcohol consumption. MGM15, MGM3, and MGM13 are by and large unrelated to THMs, indicating transcriptome measurements from an admixture of CNS cells may fail to notice genes affected by alcohol drinking behavior within these MGM groups. Cd11b is an integral membrane-associated protein that is a cellular marker of CNS microglia; however, Cd11b is also present on the surface of other monocyte-derived lineages, including macrophages. Macrophages are capable of invading the CNS, sparking neuroinflammation and neurodegeneration (Shemer and Jung, 2015). MGM15, MGM3, and MGM13 were further assessed for unintended non-microglial cellular gene expression arising from Cd11b+ enrichment. Expression of genes belonging to MGM13 and MGM15 were indistinguishable from other CNS or immune cell-types. The highest mean expression of MGM15 and MGM13 containing genes occurred in macrophage and stromal cellular populations (Supp. Figure 4). MGM3 encompassing 1,135 genes, showed increased expression following EOD-2BC alcohol exposure (Fig. 4A). MGM3 is significantly enriched for microglial-expressing genes involved in immune-related signaling systems (Fig. 4B, Supp. Table 4). Genes within MGM3 are positively correlated with the total amount of alcohol consumed (Pearson r=0.77, P = 0.01). Matching the same genes within the total homogenate obscured this correlation with alcohol consumption (Pearson r=−0.04, P = 0.88), stressing the importance of isolating a microglial cellular population in an animal model of alcohol consumption to unveil a genomic relationship with the neuroimmune system. Although additional cell-types are capable of relaying an immune response, MGM3 expression was significantly higher in microglia compared to other major CNS or immune cell-types (Fig. 4C & 4D). There are several known microglia genes within MGM3, including cell-type specific markers Tmem119, Cx3cr1, Aif1 (Iba1), suggesting MGM3 expression is mainly restricted to this particular CNS immune cell.
Figure 4.
Microglial module 3 (MGM3) was selected for further investigation based on its differential expression, biological function, and microglial specificity. A. MGM3 shows increased expression with chronic ethanol consumption (p < .05, Kruskal-Wallis Rank Sum Test). B. Genes contained in MGM3 are associated with several GO Biological Processes related to innate immune signaling. C. When compared to other CNS cell types, MGM3 shows highest relative expression of microglial genes (p < .05, ANOVA). D. When compared to other immune cell types, MGM3 shows highest relative expression of microglial genes (p < .05, ANOVA).
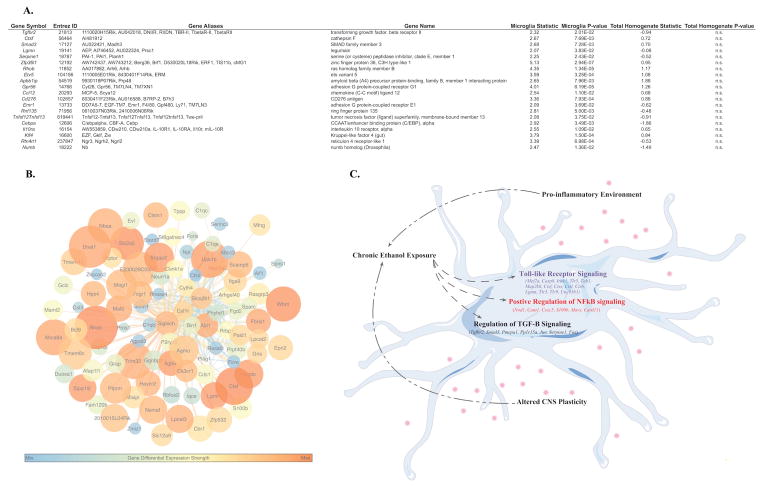
The top biological processes representative of MGM3 were ‘activation of innate immune response’ (GO:0002218) and ‘toll-like receptor signaling pathway’ (GO:0002224), which affect alcohol neurotoxicity and consumption (Blednov et al., 2011a) (Lippai et al., 2013). The MGM3 coexpression network (Fig. 5) was constructed to identify specific genes relevant to the interaction of microglia with EOD-2BC alcohol consumption. Central to MGM3 lies sialic acid binding Ig-like lectin H (Siglech). Siglech is a putative marker of isolated microglia (Chiu et al., 2013); which modulates type I Interferon secretion associated with microglial activation and phagocytosis of glioma cells (Blasius et al., 2006; Kopatz et al., 2013). Brain specific expression and function of Siglech has yet to be completely ascertained, however, because MGM3 shows ontologies related to TLR signaling, particularly endosomal TLRs which promote interferon secretion, it is possible that Siglech is regulating this response in brain. Although not all of the genes expressed within MGM3 are individually altered in relation to alcohol consumption, there is a dense interrelationship among MGM3 containing genes that may be important for the observed phenotype. Cathepsin proteases Ctsf and Ctss are examples of two MGM3 genes previously shown to causally affect multiple behavioral models of alcohol consumption (Blednov et al., 2011b). The presence of Ctsf and Ctss among the most connected genes within the MGM3 network may suggest there are additional microglial-immune genes involved in alcohol consumption that are unaddressed by examining the total homogenate. For example, legumain (Lgmn, also known as asparagine endopeptidase or AEP), 16.5-fold enriched within microglia, is differentially expressed and correlated with total alcohol consumption in the microglial but not in total homogenate. Acting as an asparagine endopeptidase, LGMN is important for the cleavage and activation of toll-like receptors (Sepulveda et al., 2009). LGMN works in concert with cathepsins on toll-like receptors, also expressed within MGM3, to bring about optimum signaling (Ewald and Barton, 2011). The joint presence of numerous cellular markers alongside genes with shared activity in immune function asserts MGM3 is a microglia-specific gene network, which may designate a convergence of an alcohol-responsive microglial signature.
Figure 5.
A. Table of differentially expressed genes in MGM3. Each listing contains the gene symbol., Entrez ID, Gene Aliases and name, the T-statistic and p-value for each gene in microglia and the total homogenate. All genes shown in the table were differentially expressed with ethanol in microglia but not in the total homogenate. B. Connectivity map containing the top 250 connections in MGM3. Warmer colored nodes have smaller p-values for differential expression with alcohol. Larger nodes have larger fold changes with alcohol. Larger connecting lines indicate higher topological overlap measure (TOM), or connectivity between 2 genes. Warmer colored lines indicate higher edge betweenness centrality, or importance of the connection in the network. Siglech is highly connected to many genes in MGM3. C. Hypothesis about effects of chronic ethanol on microglia. Chronic ethanol exposure leads to altered expression of genes involved in Toll-like Receptor Signaling, NF-κB signaling, and TGF-β signaling. Together, these alterations in immune signaling can result in a pro-inflammatory environment and altered CNS plasticity, which can drive further consumption.
In addition to highlighting ethanol-induced changes in TLR and immune signaling, the changes in MGM3 suggest that ethanol is perturbing microglial TGF-β signaling. Specifically, looking at the differentially expressed genes within MGM3 revealed enrichment for transforming growth factor beta receptor signaling pathway (GO:0007179) and R-SMAD binding (GO:0070412). Recent studies have highlighted the important role of TGF-β in microglial homeostasis and in the regulation of immune response. Butovsky et al. found that Tgfb1 was specific to microglia in the adult CNS and that treatment of cultured microglia with TGF-β1 resulted in increased expression of 60 genes, half of which were identified in MGM3 (Butovsky et al., 2013), including Lgmn, Siglech, and Tlr3. Furthermore, CNS-TGF-β1 −/ − mice had deficits in extracellular glutamate and synaptic plasticity, suggesting a key role for TGF-β1 in regulating these processes (Pribiag and Stellwagen, 2014). Investigation of microglial molecules that were suppressed in the CNS-TGF-β1 −/ − mice revealed 32 altered TGF-β signaling molecules, 22 of which (including Ctss, Ctsf) were identified in MGM3. These data further suggesting that MGM3 is involved in TGF-β signaling and that TGF-β could be regulating the observed immune changes.
The perturbation in TGF-β signaling in MGM3 has many functional implications for the effects of alcohol on microglia. TGF-β signals to SMADs, and SMAD3 knockout mice have impaired immune function, suggesting SMAD signaling is involved in regulating the immune response (Yang et al., 1999). Additionally, TGF-β and SMAD levels are elevated following brain injury. Thus, the perturbation in TGF-β signaling we observed in microglia could be responsible for the altered immune profile. TGF-β can also regulate excitotoxicity, which is often a consequence of glial activation and thought to be a mediator of ethanol-induced neurodegeneration.
Both microglia and TGF-β are also involved in regulation of important cellular processes such as proliferation, differentiation, maturation, and survival of CNS cells (Aarum et al., 2003; Gemma and Bachstetter, 2013; Villapol et al., 2013). SMAD3, which was changed on the transcript level in microglia after ethanol, is specifically known to regulate neurogenesis in the brain (Villapol et al., 2013). Thus, it is possible that alterations in microglial TGF-β expression mediate alcohol-induced changes like reduced neurogenesis and neurodegeneration. Furthermore, TGFβ release from microglia is important for survival of dopamine neurons (Polazzi et al., 2009; Roussa et al., 2009), which play an important role in the rewarding properties of ethanol. Therefore, altered microglia TGFβ signaling could be altering dopamine transmission and contributing to the dependent phenotype. In addition, there is an emerging role for microglia in modulating synaptic transmission (Salter and Beggs, 2014), a process which is also regulated by TGF-β signaling (Dobolyi et al., 2012). In particular, TGF-β2 appears to be an important regulator or synaptic function, and the mRNA encoding its receptor TGF-βR2 was differentially expressed in microglia following ethanol consumption. Therefore, altered microglial TGF-β signaling may contribute to alterations in synaptic function in response to chronic ethanol.
Although TGF-β has been implicated in ethanol-induced liver injury (Dooley and Dijke, 2012) and expression is increased in the blood of alcohol dependent subjects (Kim et al., 2009), no studies have investigated the role of TGF-β in ethanol-induced immune signaling in the brain. Thus, future studies would benefit from investigating TGF-β signaling in the brain following ethanol consumption. Futhermore, although there haven’t been any studies thus far investigating the interaction between TGFβ and Siglec-H, there is evidence for other Siglecs regulating TGFβ signaling in immune cells (Takamiya et al., 2013; Wu et al., 2016) and the addition of TGF-β1 to microglial cultures upregulates Siglech expression. Thus, the interaction between Siglec-H and TGFβ in microglia is also worthy of investigation.
1.4 Conclusions
The transcriptional landscape of microglia is a significant faction within the CNS immune system. Intermingled with other cell-types, microglia are instrumental in maintaining CNS homeostasis. The innate and adaptive immune systems are proposed as interrelated cellular mechanisms involved in chronic alcohol and excessive alcohol abuse (Vetreno and Crews, 2014). CNS cell composition varies by brain region, influencing enduring immune responses and neurobehavioral traits. Identifying the molecular response of individual cell-types provides a foundation for inferring the immunomodulatory effects that are instigated.
Neuroimmune signaling can affect neuronal transmission and actions of alcohol (Bajo et al., 2015; 2014). Although gene expression alterations are not entirely equivalent with protein levels due to cellular stochastic processes, increased coordinate gene expression within microglia following alcohol consumption is consistent with the activation and proliferation of microglia in alcoholic brain tissue (Dennis et al., 2014). The associated gene expression puts forth a broader portrait of an alcohol-responsive microglial gene network, which is notably distinct from other CNS- and immune-related cell-types. Microglial studies in alcohol dependence and other psychiatric conditions have mainly focused on a role in neurotoxicity (Alfonso-Loeches et al., 2010; Crews et al., 2006; Pascual et al., 2011), but our studies indicate that voluntary alcohol consumption, which is less toxic, has marked effects on the microglial transcriptome and suggest that microglia may be involved in alcohol consumption. Several genes within this microglial network (Ctss, Ctsf, Tlr3, Tlr9, Irak1, Casp8, and Ctsb) have been implicated in AUDs, further validating this module (Blednov et al., 2011b; Lippai et al., 2013; Qin and Crews, 2012). The cathepsins, Ctss and Ctsf, have been causally implicated in alcohol consumption, suggesting that other members of this module may regulate consumption. These results also suggest the involvement of many additional microglial genes that have not previously been studied with alcohol.
Of particular importance in this study, was the identification of MGM3, which contains co-expressing microglial genes that are altered by ethanol. Within this module, we highlighted TLR and immune signaling, possibly regulated by the hub gene Siglech. In addition, this module presented the novel hypothesis that chronic ethanol is regulating TGF-β signaling. The perturbations in TGF-β could be regulating the changes in immune signaling, as well as driving changes in synaptic plasticity. Further studies are necessary to elucidate to the role of TGF-β and Siglec-H in modulating the response to ethanol.
It is important to note that the genomic effects of alcohol consumption vary based on ethanol exposure model, duration, and time following exposure (McCarthy et al., 2017; Osterndorff-Kahanek et al., 2013; 2015; Whitman et al., 2013). Thus, the results from this study present a single snapshot of how alcohol impacts the microglial transcriptome. Although blood alcohol levels were low (~14 mg/dL) at the time tissue was collected, it is possible that some of the gene expression changes identified are a result of the acute effects of alcohol exposure or withdrawal. Future studies will be necessary to further define the time-course of the microglial response and to differentiate the acute and chronic effects of alcohol.
The transcriptome of isolated cells is informative for clarifying target genes and functional networks that may be important for ameliorating imbalanced systems (Lamb et al., 2006). Brain pathologies exemplified by disturbances in CNS immune function benefit from surveying the transcriptome of isolated microglia. Animal models incorporating such cellular preparations refine gene expression signatures procured from an admixture of cell-types present within homogenized tissue, permitting an in-depth characterization of particular cellular responses. Discriminating the microglia-specific perturbations initiated can call attention to a series of subtle molecular modifiers of immune system function relevant to human conditions. Our results emphasize a cluster of microglial genes aligned in relation to alcohol consumption. Further research into microglial gene networks, and the development of molecular tools to selectively manipulate microglia, will uncover CNS immune responses inherent to substance abuse and dependence.
Supplementary Material
Acknowledgments
The authors would like to thank Jillian Benavidez, Courtney Bridges, and the Genomic Sequencing and Analysis Facility (University of Texas at Austin) for their technical assistance.
Footnotes
Funding and Disclosure
The authors declare no conflicts of interest
This work was supported by the National Institutes of Alcohol Abuse and Alcoholism (NIAAA) through grants AA024654 (GMM), AA024836 (SPF), AA013520 (YAB & RAH), and AA020926 (RDM).
References
- Aarum J, Sandberg K, Haeberlein SLB, Persson MAA. Migration and differentiation of neural precursor cells can be directed by microglia. Proceedings of the National Academy of Sciences. 2003;100:15983–15988. doi: 10.1073/pnas.2237050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso-Loeches S, Pascual-Lucas M, Blanco AM, Sanchez-Vera I, Guerri C. Pivotal Role of TLR4 Receptors in Alcohol-Induced Neuroinflammation and Brain Damage. Journal of Neuroscience. 2010;30:8285–8295. doi: 10.1523/JNEUROSCI.0976-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajo M, Herman MA, Varodayan FP, Oleata CS, Madamba SG, Harris RA, Blednov YA, Roberto M. Role of the IL-1 receptor antagonist in ethanol-induced regulation of GABAergic transmission in the central amygdala. Brain Behavior and Immunity. 2015;45:189–197. doi: 10.1016/j.bbi.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajo M, Madamba SG, Roberto M, Blednov YA, Sagi VN, Roberts E, Rice KC, Harris RA, Siggins GR. Innate immune factors modulate ethanol interaction with GABAergic transmission in mouse central amygdala. Brain Behavior and Immunity. 2014;40:191–202. doi: 10.1016/j.bbi.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius AL, Cella M, Maldonado J, Takai T, Colonna M. Siglec-H is an IPC-specific receptor that modulates type I IFN secretion through DAP12. Blood. 2006;107:2474–2476. doi: 10.1182/blood-2005-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Benavidez JM, Geil C, Perra S, Morikawa H, Harris RA. Activation of inflammatory signaling by lipopolysaccharide produces a prolonged increase of voluntary alcohol intake in mice. Brain Behavior and Immunity. 2011a;25:S92–S105. doi: 10.1016/j.bbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Ponomarev I, Geil C, Bergeson S, Koob GF, Harris RA. Neuroimmune regulation of alcohol consumption: behavioral validation of genes obtained from genomic studies. Addiction Biology. 2011b;17:108–120. doi: 10.1111/j.1369-1600.2010.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu L, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, Weiner HL. Identification of a unique TGF-β–dependent molecular and functional signature in microglia. Nat Neurosci. 2013;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu IM, Morimoto ETA, Goodarzi H, Liao JT, O’Keeffe S, Phatnani HP, Muratet M, Carroll MC, Levy S, Tavazoie S, Myers RM, Maniatis T. A Neurodegeneration-Specific Gene-Expression Signature of Acutely Isolated Microglia from an Amyotrophic Lateral Sclerosis Mouse Model. Cell Reports. 2013;4:385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews F, Nixon K, Kim D, Joseph J, Shukitt-Hale B, Qin L, Zou J. BHT Blocks NF-?B activation and Ethanol-Induced Brain Damage. Alcoholism: Clinical and Experimental Research. 2006;30:1938–1949. doi: 10.1111/j.1530-0277.2006.00239.x. [DOI] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, Quake SR. A survey of human brain transcriptome diversity at the single cell level. Proceedings of the National Academy of Sciences. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biase LM, Schuebel KE, Fusfeld ZH, Jair K, Hawes IA, Cimbro R, Zhang HY, Liu QR, Shen H, Xi ZX, Goldman D, Bonci A. Local Cues Establish and Maintain Region-Specific Phenotypes of Basal Ganglia Microglia. Neuron. 2017;95:341–356e6. doi: 10.1016/j.neuron.2017.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis CV, Sheahan PJ, Graeber MB, Sheedy DL, Kril JJ, Sutherland GT. Microglial proliferation in the brain of chronic alcoholics with hepatic encephalopathy. Metab Brain Dis. 2014;29:1027–1039. doi: 10.1007/s11011-013-9469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobolyi A, Vincze C, Pál G, Lovas G. The neuroprotective functions of transforming growth factor beta proteins. Int J Mol Sci. 2012;13:8219–8258. doi: 10.3390/ijms13078219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley S, Dijke ten P. TGF-β in progression of liver disease. Cell Tissue Res. 2012;347:245–256. doi: 10.1007/s00441-011-1246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald SE, Barton GM. Nucleic acid sensing Toll-like receptors in autoimmunity. Curr Opin Immunol. 2011;23:3–9. doi: 10.1016/j.coi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Lizarbe S, Pascual M, Guerri C. Critical Role of TLR4 Response in the Activation of Microglia Induced by Ethanol. The Journal of Immunology. 2009;183:4733–4744. doi: 10.4049/jimmunol.0803590. [DOI] [PubMed] [Google Scholar]
- Gemma C, Bachstetter AD. The role of microglia in adult hippocampal neurogenesis. Front Cell Neurosci. 2013;7:229. doi: 10.3389/fncel.2013.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pena D, Nixon SE, O’Connor JC, Southey BR, Lawson MA, McCusker RH, Borras T, Machuca D, Hernandez AG, Dantzer R, Kelley KW, Rodriguez-Zas SL. Microglia Transcriptome Changes in a Model of Depressive Behavior after Immune Challenge. PLoS ONE. 2016;11:e0150858–28. doi: 10.1371/journal.pone.0150858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, Zhang K, Ming GL, Gao Y, Song H. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Crews FT. Increased MCP-1 and microglia in various regions of the human alcoholic brain. Experimental Neurology. 2008;210:349–358. doi: 10.1016/j.expneurol.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, El Khoury J. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope BT, Nye HE, Kelz MB, Self DW, Iadarola MJ, Nakabeppu Y, Duman RS, Nestler EJ. Induction of a long-lasting AP-1 complex composed of altered Fos-like proteins in brain by chronic cocaine and other chronic treatments. Neuron. 1994;13:1235–1244. doi: 10.1016/0896-6273(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Ju L, Zeng H, Chen Y, Wu Y, Wang B, Xu Q. Dual polarization of microglia isolated from mixed glial cell cultures. J Neurosci Res. 2015;93:1345–1352. doi: 10.1002/jnr.23563. [DOI] [PubMed] [Google Scholar]
- Kim Y-K, Lee BC, Ham BJ, Yang B-H, Roh S, Choi J, Kang T-C, Chai YG, Choi I-G. Increased transforming growth factor-beta1 in alcohol dependence. J Korean Med Sci. 2009;24:941–944. doi: 10.3346/jkms.2009.24.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35:217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopatz J, Beutner C, Welle K, Bodea LG, Reinhardt J, Claude J, Linnartz-Gerlach B, Neumann H. Siglec-h on activated microglia for recognition and engulfment of glioma cells. Glia. 2013;61:1122–1133. doi: 10.1002/glia.22501. [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson LJ, Perry VH, Dri P, Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. NEUROSCIENCE. 1990;39:151–170. doi: 10.1016/0306-4522(90)90229-w. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Wixey J, Harper CG, Dodd PR. Expression of MBP, PLP, MAG, CNP, and GFAP in the Human Alcoholic Brain. Alcoholism: Clinical and Experimental Research. 2005;29:1698–1705. doi: 10.1097/01.alc.0000179406.98868.59. [DOI] [PubMed] [Google Scholar]
- Lippai D, Bala S, Petrasek J, Csak T, Levin I, Kurt-Jones EA, Szabo G. Alcohol-induced IL-1 in the brain is mediated by NLRP3/ASC inflammasome activation that amplifies neuroinflammation. Journal of Leukocyte Biology. 2013;94:171–182. doi: 10.1189/jlb.1212659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge PA, Sriram S. Regulation of microglial activation by TGF-beta, IL-10, and CSF-1. Journal of Leukocyte Biology. 1996;60:502–508. doi: 10.1002/jlb.60.4.502. [DOI] [PubMed] [Google Scholar]
- Lovinger DM, Crabbe JC. Laboratory models of alcoholism: treatment target identification and insight into mechanisms. Nat Neurosci. 2005;8:1471–1480. doi: 10.1038/nn1581. [DOI] [PubMed] [Google Scholar]
- Maze I, Wenderski W, Noh KM, Bagot RC, Tzavaras N, Purushothaman I, Elsässer SJ, Guo Y, Ionete C, Hurd YL, Tamminga CA, Halene T, Farrelly L, Soshnev AA, Wen D, Rafii S, Birtwistle MR, Akbarian S, Buchholz BA, Blitzer RD, Nestler EJ, Yuan ZF, Garcia BA, Shen L, Molina H, Allis CD. Critical Role of Histone Turnover in Neuronal Transcription and Plasticity. Neuron. 2015;87:77–94. doi: 10.1016/j.neuron.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy GM, Warden AS, Bridges CR, Blednov YA, Harris RA. Chronic ethanol consumption: role of TLR3/TRIF-dependent signaling. Addiction Biology. 2017;91:289. doi: 10.1111/adb.12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez RI. Intermittent (every-other-day) drinking induces rapid escalation of ethanol intake and preference in adolescent and adult C57BL/6J mice. Alcoholism: Clinical and Experimental Research. 2011;35:652–658. doi: 10.1111/j.1530-0277.2010.01383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos J, Alfonso-Loeches S, Guerri C. Impact of the Innate Immune Response in the Actions of Ethanol on the Central Nervous System. Alcoholism: Clinical and Experimental Research. 2016;40:2260–2270. doi: 10.1111/acer.13208. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Hope BT, Widnell KL. Drug addiction: a model for the molecular basis of neural plasticity. Neuron. 1993;11:995–1006. doi: 10.1016/0896-6273(93)90213-b. [DOI] [PubMed] [Google Scholar]
- Nikodemova M, Watters JJ. Efficient isolation of live microglia with preserved phenotypes from adult mouse brain. Journal of Neuroinflammation. 2012;9:147. doi: 10.1186/1742-2094-9-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterndorff-Kahanek E, Ponomarev I, Blednov YA, Harris RA. Gene Expression in Brain and Liver Produced by Three Different Regimens of Alcohol Consumption in Mice: Comparison with Immune Activation. PLoS ONE. 2013;8:e59870–10. doi: 10.1371/journal.pone.0059870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterndorff-Kahanek EA, Becker HC, Lopez MF, Farris SP, Tiwari GR, Nunez YO, Harris RA, Mayfield RD. Chronic ethanol exposure produces time- and brain region-dependent changes in gene coexpression networks. PLoS ONE. 2015;10:e0121522. doi: 10.1371/journal.pone.0121522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual MA, Baliño P, Alfonso-Loeches S, Aragón CMG, Guerri C. Impact of TLR4 on behavioral and cognitive dysfunctions associated with alcohol-induced neuroinflammatory damage. Brain Behavior and Immunity. 2011;25:S80–S91. doi: 10.1016/j.bbi.2011.02.012. [DOI] [PubMed] [Google Scholar]
- Polazzi E, Altamira LEP, Eleuteri S, Barbaro R, Casadio C, Contestabile A, Monti B. Neuroprotection of microglial conditioned medium on 6-hydroxydopamine-induced neuronal death: role of transforming growth factor beta-2. J Neurochem. 2009;110:545–556. doi: 10.1111/j.1471-4159.2009.06117.x. [DOI] [PubMed] [Google Scholar]
- Pribiag H, Stellwagen D. Neuroimmune regulation of homeostatic synaptic plasticity. Neuropharmacology. 2014;78:13–22. doi: 10.1016/j.neuropharm.2013.06.008. [DOI] [PubMed] [Google Scholar]
- Qin L, Crews FT. Chronic ethanol increases systemic TLR3 agonist-induced neuroinflammation and neurodegeneration. Journal of Neuroinflammation. 2012;9:130. doi: 10.1186/1742-2094-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussa E, Bohlen und Halbach von O, Krieglstein K. TGF-beta in dopamine neuron development, maintenance and neuroprotection. Adv Exp Med Biol. 2009;651:81–90. doi: 10.1007/978-1-4419-0322-8_8. [DOI] [PubMed] [Google Scholar]
- Salter MW, Beggs S. Sublime Microglia: Expanding Roles for the Guardians of the CNS. Cell. 2014;158:15–24. doi: 10.1016/j.cell.2014.06.008. [DOI] [PubMed] [Google Scholar]
- Satoh JI, Kino Y, Asahina N, Takitani M, Miyoshi J, Ishida T, Saito Y. TMEM119 marks a subset of microglia in the human brain. Neuropathology. 2016;36:39–49. doi: 10.1111/neup.12235. [DOI] [PubMed] [Google Scholar]
- Sepulveda FE, Maschalidi S, Colisson R, Heslop L, Ghirelli C, Sakka E, Lennon-Duménil AM, Amigorena S, Cabanie L, Manoury B. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31:737–748. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Shemer A, Jung S. Differential roles of resident microglia and infiltrating monocytes in murine CNS autoimmunity. Semin Immunopathol. 2015;37:613–623. doi: 10.1007/s00281-015-0519-z. [DOI] [PubMed] [Google Scholar]
- Starkman BG, Sakharkar AJ, Pandey SC. Epigenetics-beyond the genome in alcoholism. Alcohol Res. 2012;34:293–305. [PMC free article] [PubMed] [Google Scholar]
- Takamiya R, Ohtsubo K, Takamatsu S, Taniguchi N, Angata T. The interaction between Siglec-15 and tumor-associated sialyl-Tn antigen enhances TGF-β secretion from monocytes/macrophages through the DAP12-Syk pathway. Glycobiology. 2013;23:178–187. doi: 10.1093/glycob/cws139. [DOI] [PubMed] [Google Scholar]
- Vetreno RP, Crews FT. Alcohol and the Nervous System. 1. Elsevier B.V; 2014. Current hypotheses on the mechanisms of alcoholism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villapol S, TT, JA . Trends in Cell Signaling Pathways in Neuronal Fate Decision. InTech; 2013. Role of TGF-β Signaling in Neurogenic Regions After Brain Injury. [DOI] [Google Scholar]
- Whitman BA, Knapp DJ, Werner DF, Crews FT, Breese GR. The Cytokine mRNA Increase Induced by Withdrawal from Chronic Ethanol in the Sterile Environment of Brain is Mediated by CRF and HMGB1 Release. Alcoholism: Clinical and Experimental Research. 2013;37:2086–2097. doi: 10.1111/acer.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Lan C, Ren D, Chen G-Y. Induction of Siglec-1 by Endotoxin Tolerance Suppresses the Innate Immune Response by Promoting TGF-β1 Production. J Biol Chem. 2016;291:12370–12382. doi: 10.1074/jbc.M116.721258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Letterio JJ, Lechleider RJ, Chen L, Hayman R, Gu H, Roberts AB, Deng C. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J. 1999;18:1280–1291. doi: 10.1093/emboj/18.5.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. Journal of Neuroscience. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.