Abstract
Structural variations (SVs) are the largest source of genetic variation, but remain poorly understood because of limited genomics technology. Single molecule long-read sequencing from Pacific Biosciences and Oxford Nanopore has the potential to dramatically advance the field, although their high error rates challenge existing methods. Addressing this need, we introduce open-source methods for long-read alignment (NGMLR, https://github.com/philres/ngmlr) and SV identification (Sniffles, https://github.com/fritzsedlazeck/Sniffles) that enable unprecedented SV sensitivity and precision, including within repeat-rich regions and of complex nested events that can have significant impact on human disorders. Examining several datasets, including healthy and cancerous human genomes, we discover thousands of novel variants using long-reads and categorize systematic errors in short-read approaches. NGMLR and Sniffles are further able to automatically filter false events and operate on low amounts of coverage to address the cost factor that has hindered the application of long-reads in clinical and research settings.
Introduction
Structural variations (SVs), including insertions, deletions, duplications, inversions and translocations at least 50bp in size, account for the largest number of divergent base-pairs across human genomes1. SVs contribute to polymorphic variation, pathogenic conditions, large-scale chromosome evolution2, and human diseases such as cancer3, autism4, or Alzheimer’s5. SVs also impact phenotypes across many other organisms6–10. One of the first reports of the prevalence of SVs came in 2004, when Sebat, et al. 11 used microarrays to discover large-scale copy number polymorphisms were common across healthy human genomes. Today, SV detection is most often performed using short paired-end reads. Copy number variations are observed as decreases (deletions) or increases (amplifications) in aligned read coverage12, and other types of SVs are identified by the arrangement of paired-end reads or split-read alignments13–16. Short-read approaches, however, have been reported as lacking sensitivity, with only 10%17 to 70%6,8 of SVs detected, very high (up to 89%) false positive rates 6,18–21 and misinterpreting complex or nested SVs6,22.
Long-read single molecule sequencing by Pacific Biosciences (PacBio) and Oxford Nanopore has the potential to substantially increase the reliability and resolution of detecting SVs. With average read lengths of 10kbp or higher, the reads can be more confidently aligned to repetitive sequences that often mediate the formation of SVs22. Long-reads are also more likely to span SV breakpoints with high-confidence alignments. Despite these advantages, long-reads introduce new challenges. Most significantly, they have a high sequencing error rate, currently 10% to 15% for PacBio, and 5% to 20% for Oxford Nanopore sequencing23, necessitating new methods. A few aligners are available, including LAST24, BlasR25, BWA-MEM26, GraphMap27, MECAT 28 and minimap229. Only one standalone method, PBHoney18, is available to detect all types of SV from long-read data, although others have been proposed for subset of SVs types e.g. SMRT-SV30.
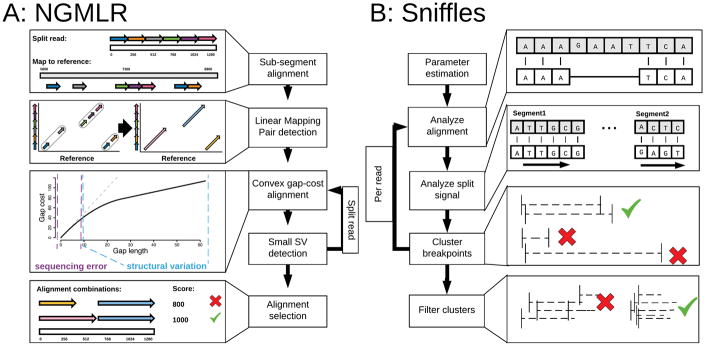
Addressing these challenges, we introduce two open-source analysis algorithms, NGMLR and Sniffles, for comprehensive long-read alignment and SV detection (Figure 1). NGMLR is a fast and accurate aligner for long-reads based on our previous short-read aligner NGM31 extended with a new convex gap-cost scoring model to align long-reads across SV breakpoints. Its partner algorithm Sniffles successively scans the alignments to identify all types of SVs. Sniffles employs a novel SV scoring scheme to exclude false SVs based on the size, position, type and coverage of the candidate SV to resolve the high indel error rates in long-read sequencing.
Figure 1.
Overview of the main steps implemented in NGMLR (left) and Sniffles (right). For details see Supplementary Notes 1 and 2 for NGMLR and Sniffles, respectively.
We apply our methods to simulated and genuine datasets of Arabidopsis, healthy human genomes, and a cancerous human genome to demonstrate the increased accuracy compared to alternate short- and long-read callers. A particularly innovative feature of Sniffles is its ability to detect nested SVs, such as inverted tandem duplications (INVDUP) or inversions flanked by indels (INVDEL). These are poorly studied classes of SVs, although both have been previously associated to genomic disorders32–34,35. However, as no alternative methods can routinely detect them, their full significance is currently unknown. Finally, we show that our methods reduce the sequencing and computational costs per sample, making it increasingly feasible to apply long-reads to large numbers of samples.
Results
Accurate mapping and detection of SVs using long-reads
Unlike most other aligners, NGMLR uses a convex gap scoring model36 to accurately align reads spanning genuine indel SVs in the presence of small indels (1–10bp) that commonly occur as sequencing errors (Figure 2). Larger or more complex SVs are captured through split-read alignments. To achieve both high performance and accuracy, NGMLR first partitions the long-reads into 256bp sub-segments and aligns them independently to the reference genome (Figure 1a). It then groups co-linear sub-segment alignments into long segments, which are then aligned using dynamic programming with our convex gap-cost scoring scheme. Finally, NGMLR selects the highest scoring non-overlapping combination of segments per read and outputs the results in standard SAM/BAM format. Overall NGMLR is among the fastest available methods while showing the highest accuracy. See Methods and Supplementary Note 1 for more details.
Figure 2.
Alignment improvements using NGMLR shown for a 228 bp deletion (left) and a 150 bp inversion (right) shown in IGV37. Upper track shows BWA-MEM alignments that indicate these events but is not able to localize the precise event and breakpoints. With the improved alignments of NGMLR, Sniffles can precisely pinpoint the location and type of the SV.
Sniffles detects all types of SVs (indels, duplications, inversions, translocations, and nested events) from long-read alignments. It can be used with any aligner, although it has the best performance with NGMLR as it produces the most accurate alignments. The principal steps consist of scanning the alignments of each read independently for potential SVs and then clustering the candidate SVs across all reads (Figure 1b). Sniffles uses both within-alignment and split-read information to detect SVs, as small indels can be spanned within a single alignment, but large or complex events lead to split-read alignments. The major advance of Sniffles is filtering false SV signals from the noisy reads. Like other variant detectors, minimum read support (default: 10 reads) is a critical feature, but it also considers the consistency of the breakpoint position and size. In addition, Sniffles can perform read-based phasing of SVs and report adjacent or nested events in the output VCF file. Overall Sniffles runs very fast and requires <3 hours for a deep coverage (50×) human genome analysis. See Methods and Supplementary Note 2 for more details.
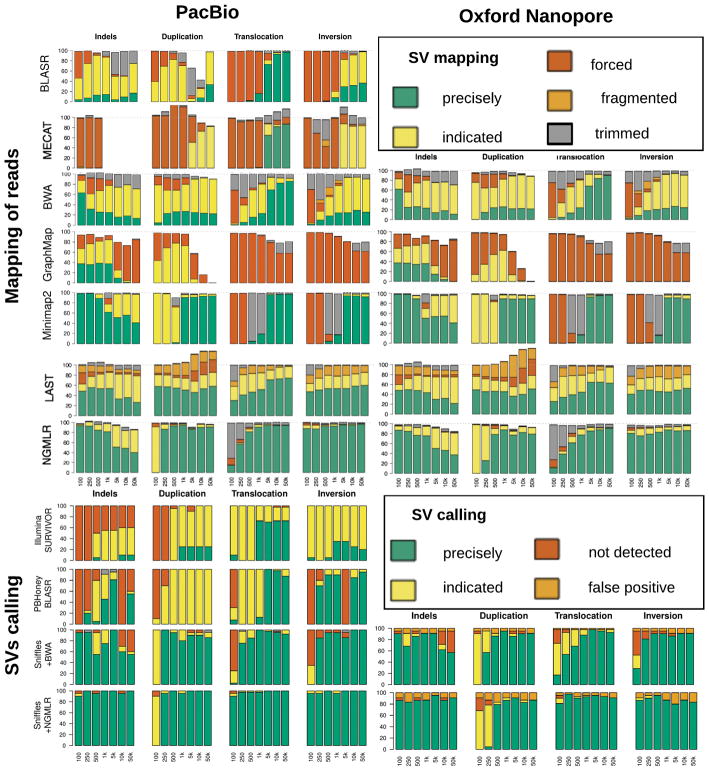
To establish the performance of NGMLR and Sniffles, we benchmarked them against widely used alternative approaches using simulated reads with SVs added of different sizes and types (Supplementary Notes 3, 4, & 5). Overall NGMLR and Sniffles showed the highest accuracy for alignments and SV calls (Figure 3). We also evaluated the performance using genuine sequencing reads mapped to modified reference genome with SVs embedded at known locations, and see similar superior results (Supplementary Note 5).
Figure 3.
Evaluation of NGMLR, Sniffles and related tools using simulated data with 840 SVs. X axis is showing the size of the simulated SVs. For read alignments (top), we simulated PacBio-like (left) and Oxford Nanopore-like reads (right), and distinguish between: precise (green), indicated (yellow), forced (red), unaligned reads (white), or trimmed but not aligned through the SV (grey). The SV analysis (bottom) used the same alignments as before, and distinguishes between.
With the accuracy established, we next used genuine sequencing of an Arabidopsis thaliana trio (Col-0, CVI and the Col-0 x CVI F1 progeny)38 and Ashkenazi human trio data set from Genome-in-a-Bottle (GiaB)39 to assess the recall and Mendelian consistency (Table 1 and Supplementary Notes 4 & 5). Overall, Sniffles and NGMLR had the highest recall rate, meaning the percentage of homozygous variants found in the parents that were found in the F1 (Arabidopsis trio: 99.75%, GiaB trio: 97.21%). The Mendelian discordance rate was also greatly improved: using NGMLR/Sniffles with PacBio reads resulted in 3.36% for Arabidopsis and 5.57% for GiaB, while state-of-the-art consensus calling with Illumina data had a 21.11% discordance rate for GiaB. Translocation calls were particularly erroneous for the short-read analysis and had an unreasonably high number of calls (1,550) in the son.
Table 1.
Summary of detected SVs across 15 different data sets. SVs were reported with a min. size of 50bp using SURVIVOR based on Delly, Lumpy and Manta for Illumina or Sniffles for PacBio (min 10 reads) or Oxford Nanopore (min 5 reads) due to the lower coverage. Supplementary Table 5 has the full details of all the data sets used.
| Data Set | Tech. | Cov. | Avg. read length(bp) | Total SVs | DEL | DUP | INS | INV | TRA |
|---|---|---|---|---|---|---|---|---|---|
| Arabidopsis Col-0 | PacBio | 127× | 6,482 | 355 | 67 | 63 | 106 | 68 | 51 |
| Arabidopsis CVI | PacBio | 123× | 6,073 | 9,652 | 3,822 | 904 | 1,823 | 478 | 2,625 |
| Arabidopsis Col-0 x CVI F1 | PacBio | 155× | 11,206 | 11,935 | 4,974 | 582 | 4,049 | 567 | 1,763 |
| Arabidopsis Col-0 X CVI F1 | Illumina | 40× | 250 | 10,950 | 4,324 | 643 | 0 | 671 | 5,312 |
| GiaB HG002 (son) | PacBio | 69× | 8,540 | 19,131 | 7,957 | 1,084 | 9,656 | 232 | 202 |
| GiaB HG002 (son) | Illumina | 80× | 148 | 10,822 | 5,018 | 863 | 0 | 823 | 4,118 |
| GiaB HG003 (father) | PacBio | 32× | 6,284 | 11,964 | 5,296 | 408 | 6,048 | 99 | 113 |
| GiaB HG003 (father) | Illumina | 80× | 148 | 11,395 | 5,553 | 869 | 0 | 818 | 4,155 |
| GiaB HG004 (mother) | PacBio | 30× | 7,285 | 10,463 | 4,590 | 276 | 5,436 | 93 | 68 |
| GiaB HG004 (mother) | Illumina | 80× | 148 | 8,901 | 5,000 | 868 | 0 | 829 | 2,204 |
| NA12878 (healthy female) | PacBio | 55× | 4,334 | 15,499 | 6,734 | 606 | 7,880 | 160 | 119 |
| NA12878 (healthy female) | Oxford Nanopore | 28× | 6,432 | 26,657 | 19,074 | 761 | 6,376 | 334 | 112 |
| NA12878 (healthy female) | Illumina | 50× | 101 | 7,275 | 3,744 | 553 | 0 | 731 | 2,247 |
| SKBR3 (Breast Cancer) | PacBio | 69× | 9,872 | 19,165 | 7,268 | 1,019 | 10,391 | 328 | 159 |
| SKBR3 (Breast Cancer) | Illumina | 25× | 101 | 5,046 | 2,776 | 483 | 0 | 627 | 1,160 |
Comparison of PacBio and Oxford Nanopore sequencing for human SV analysis
As a new sequencing technology, Oxford Nanopore has not yet been extensively tested for SV analysis, especially in human genomes. We investigated its capability in the well-studied NA12878 human genome using three publicly available datasets: 28× coverage of Oxford Nanopore data (release 3 and 4 from Jain et al40) analyzed with NGMLR/Sniffles, 55× coverage of PacBio data41 analyzed with NGMLR/Sniffles, and 50× coverage Illumina data42 analyzed by the consensus caller SURVIVOR used with Delly, Lumpy, and Manta (Table 1). We also compared these results to two previously published call sets, the GiaB indel call set based on PacBio sequencing41 and the Illumina-based deletion-only call set from the 1000 Genomes Project (1KGP)6.
Overall, Sniffles identified 15,499 SVs for the PacBio reads, and 26,657 SVs for the Oxford Nanopore reads, while SURIVOR reported 7,275 (Table 1, Supplementary Table 7). Comparing the 5 SVs sets resulted in a total of 40,601 SVs calls (Table 1, Supplementary Note 4). The majority (24,392) of the identified SVs are present in only one call set, while 16,209 SVs were identified in two or more call sets. Of the 15,499 PacBio calls, most (94.80%) were confirmed by Oxford Nanopore, Illumina or the existing call sets. Oxford Nanopore had substantially worse concordance, as Sniffles reports 11,433 calls unique to Oxford Nanopore, of which 10,977 (96.01%) were deletions and the majority (92.88%) were within homopolymers or other simple repeats. In contrast, the 773 calls only found by PacBio were mainly insertions (66.49%) and only 323 (41.79%) were overlapping with homopolymers or repeats. This systematic bias for deletions in the Oxford Nanopore data is most likely an error in the base-calling, as also reported by Jain, et al.40. The majority of these artifacts are small deletions, and by increasing the minimum SV size to 200bp, Sniffles reports only 38.57% of the SVs calls within homopolymers and low complexity regions. The Illumina-based SV calling had relatively low concordance to alternative approaches, and 49.71% of their calls were unique to the technology. Interestingly, the majority (54.10%) of the unique calls were translocations events, and most of these appear to be false positives (see below).
Detailed investigation of unique short-read vs. long-read events
Over all data sets, Sniffles detects far more indels than the short-read based callers (Table 1). Conversely, using the short-reads we detect, on average, 27 times more translocation events compared to using Sniffles within presumably healthy human data sets. We investigated these discrepancies using NA12878.
We first investigated the small insertion (50bp-300bp) and deletion (50bp–3kbp) calls from Sniffles using the orthogonal Illumina reads as evidence (Supplementary Note 4). We focused on these size ranges since they should be well captured by the paired-end Illumina data and used the compression-expansion statistic43 as an unbiased measure of the Illumina paired-end placements near predicted indels. This compares the genome-wide observed Illumina insert size (average 311bp) to the insert sizes spanning the indel breakpoints as aligned using BWA-MEM: real insertions in the sample should cause the pairs to map closer than expected, deletions further away. Using the Illumina data and a p-value threshold of 0.01 (two-sided, one sample t-test), we confirmed 3,415 and 3,879 deletions reported by Sniffles in the PacBio and Oxford Nanopore data, respectively (Supplementary Table 8). For insertions, we confirmed 2,685 and 1,703 for PacBio and Oxford Nanopore, respectively. For comparison, using SURVIVOR we could confirm 1,873 deletions, and only 10% of randomly selected regions show a significant alteration.
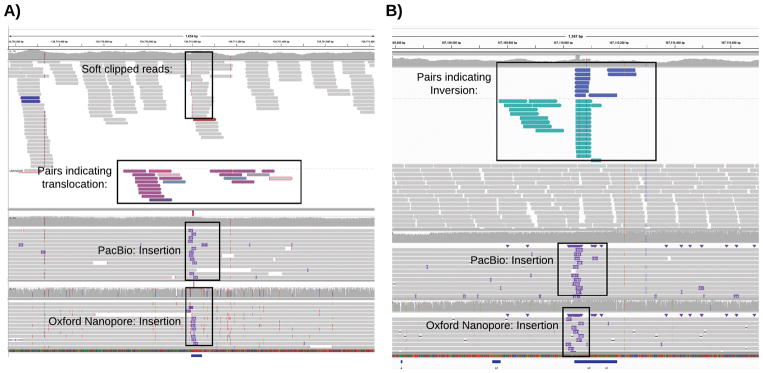
Next, we investigated the large number of translocations reported in the Illumina-based consensus calls (2,247) compared to Sniffles (PacBio: 119 and Oxford Nanopore: 43). (Supplementary Note 4 and Supplementary Table 9) We noted a large overlap (48.87%) of the Illumina-based translocation sites with insertion calls from Sniffles using both long-read technologies. Figure 4a shows a representative example, with an insertion called using both long-read data types overlapping the candidate short-read translocation. As the insertion falls within a low-complexity region, it causes the short-reads to be mis-mapped and mis-reported as a translocation, even when excluding low mapping quality reads (MQ < 20). Overall, we could rule out 1,869 (83.18%) of the Illumina-based translocation calls, with most overlapping an insertion (48.87%) or deletion (8.86%), or other SV types (1.20%). The remaining Illumina-based translocation calls are also questionable, with 404 (17.98%) in low complexity regions and 141 (6.28%) within a region with abnormally high coverage without any evidence in the long-read data. Inversions show a similar pattern, and 60% of the calls overlap with a different SV type identified by long-reads (Figure 4b) or align to low complexity sequences.
Figure 4.
Systematic error in short-read based SV calling. A) An example of a putative translocation identified in the short-read data (top alignments) that overlaps an insertion detected by both PacBio (middle) and Oxford Nanopore sequencing (bottom). B) An example of a putative inversion identified in the short-read data (top) that overlaps an insertion detected by both PacBio (middle) and Oxford Nanopore reads (bottom)
Overall, the majority of PacBio-based indels calls from Sniffles were validated by either the Oxford Nanopore or the Illumina paired-end reads. In contrast, the majority of calls unique to the Illumina-based methods were false, especially false translocations caused by mis-mapped reads across insertions.
Detection of Nested SVs
Next, we investigated the performance of Sniffles on complex, nested SV types such as inverted duplications (INVDUP) and inversions flanked by deletions (INVDEL). These variant types are poorly studied, but have been associated with a number of diseases, including INVDUPs with Pelizaeus-Merzbacher disease33 and other diseases32,44, and INVDELs with Haemophilia A genetic deficiency using long-range PCR35.
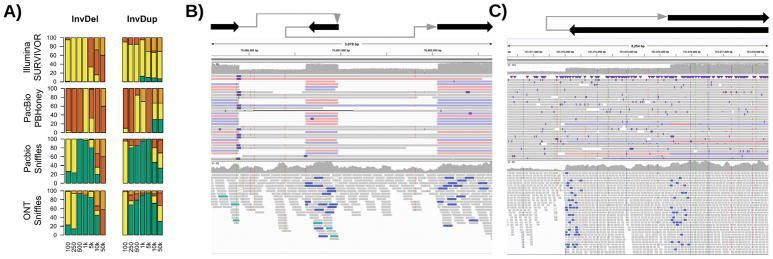
To start, we simulated 280 nested SVs of different sizes and types in the human genome along with simulated PacBio-like, Oxford Nanopore-like, and Illumina-like reads (Figure 5 and Supplementary Table 2). We evaluated each SV separately e.g. an inversion flanked by two deletions was evaluated as three SVs. Sniffles was able to detect the full range of types due to its dynamic splitting of events, and precisely called 67.88% of the nested SVs (Supplementary Note 2). This includes SVs that are larger than the read length, highlighting Sniffles’ ability to accurately infer complex events. With Oxford Nanopore-like reads, Sniffles’ ability is slightly reduced but was still able to precisely call 67.34% of SVs on average over INVDEL and INVDUP events of different length. None of the other methods could identify the full complexity of these events and only partially called the SVs (e.g. an inversion without the flanking deletions).
Figure 5.
Nested SVs in SKBR3 cancer cell line. A: Evaluation of Sniffles + NGMLR using simulated data to identify nested SVs. B: A 3kb region including two deletions flanking an inverted sequence clearly visible and detected by Sniffles using NGMLR (above) and not detected by the Illumina methods (below). C: The start of an inverted duplication. The breakpoints were reported by Sniffles as the start of an inverted duplication (above) and not correctly detected by short-read methods (below).
To highlight this capability in real data, we examined a PacBio-based data set for the SKBR3 breast cancer cell line45. Sniffles and NGMLR were used to investigate this data set revealing 15 gene fusions created by as many as 3 chained events, which were all validated by PCR. Figure 5 shows an INVDEL and INVDUP in SKBR3 in comparison to Illumina short-read data. The short-reads indicate an inversion but the poor resolution makes it impossible to detect and interpret the entire event. In contrast, Sniffles detects the events, and the read phasing allows for the complex regions to be fully resolved (Supplementary Note 2). Although these were the only two nested types in this sample, Sniffles is capable to detect and report any combination of SVs based on the IDs assigned in the reported VCF file.
How much coverage is required?
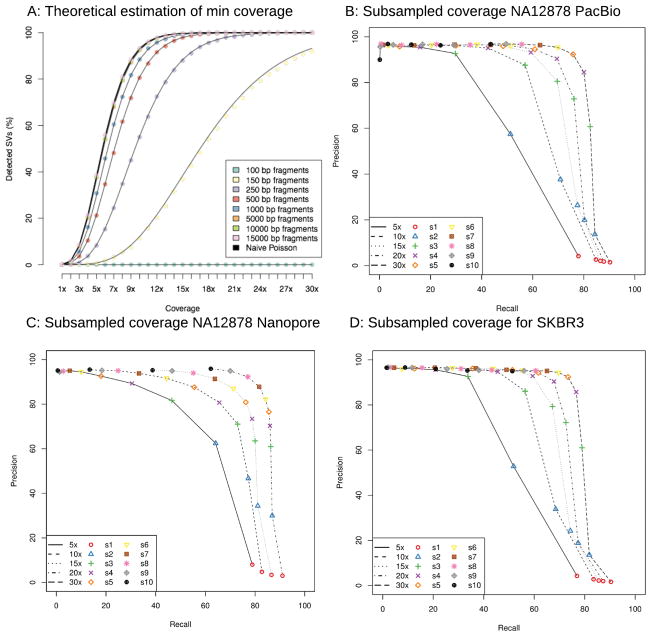
Finally, we assessed how much coverage is required to detect SVs using long-reads. This is an important consideration since these technologies are more expensive than short-read technologies to generate the same amount of coverage23. From a purely statistical analysis, about 10× coverage should be sufficient to infer all SV breakpoints using 10kbp reads whereas about 25× coverage is needed for 2×100bp short-reads (Figure 6a and Supplementary Note 4). However, this analysis represents an idealized case (e.g. lack of repeats or coverage biases) and underestimates the amount of coverage required.
Figure 6.
Analysis of SV detection accuracy with different amounts of coverage. A: Theoretical assessment of recall vs coverage for different read lengths requiring a 50bp overlap of each breakpoints for SV events. B: Subsampling experiment of the 55× PacBio NA12878 data; C: Subsampling experiment using 28× Oxford Nanopore NA12878 data; D: Subsampling experiment of the 70× PacBio SKBR3 breast cancer cell line dataset. For plots B–D, Sniffles and NGMLR were run on subsampled data (rate indicated by lines) and using different thresholds for Sniffles (s: 1–10 indicated in symbols and colors). In every data set we could show the success for Sniffles using NGMLR with only 10× to 30× coverage that recovers around 80% of the calls with a precision ~80% or higher.
To investigate this, we subsampled reads from the NA12878 PacBio and Oxford Nanopore datasets and the more complex SKBR3 PacBio sample to 5×, 10×, 15×, 20× and 30× coverage. We analyzed these subsets with NGMLR and Sniffles with different parameters (-s 1 to –s 10) to vary the minimum number of reads, and measured precision and recall with respect to the full coverage dataset (Figure 6b–d). As expected, using a minimum support of only one or two reads leads to many false positives.
Focusing on settings that have a precision rate of 80% or higher, we found 15× PacBio read coverage has a recall of 69.64% and 67.24% for NA12878 and SKBR3 for homozygous and heterozygous SVs of any type, respectively (Figure 6b,d). The difference in recall is largely due to the complexity of the SKBR3 cancer sample, which has some extreme copy (>20 fold) amplifications. Increasing the coverage to 30×, Sniffles has an 80.05% to 76.63% recall with a precision of ~85% for NA12878 and SKBR3, respectively.
For the Oxford Nanopore NA12878 data set, the highest recall rate (84.23%) had a precision of 82.24% for 20× coverage (Figure 6c). The higher apparent accuracy is largely because the original data set has only 28× coverage, so this constitutes a less dramatic down-sampling. Interestingly, we see a greater loss in precision than the PacBio data, due to the stringent minimum number of supporting reads (-s 10) used throughout the study. Overall, this shows NGMLR and Sniffles can detect the vast majority of heterozygous and homozygous SVs using only a fraction of the original coverage.
Discussion
NGMLR and Sniffles enable an unprecedented view into SVs using long-read sequencing. We demonstrated their capabilities over simulated and genuine data sets, where our methods outperformed existing tools in both sensitivity and specificity. In particular, we demonstrated that they can overcome the sensitivity issues reported for short-read callers, which miss 30%6,8 to 90%17 of the SVs. This allows us to detect many thousands of additional variants beyond what has been reported by large-scale short-read sequencing projects such as the 1000 Genomes Project. Indeed, prototype versions of our methods were used in a recent study to identify the causal, pathogenic SV in a patient who presented with multiple neoplasia and cardiac myxomata46. We also use the long-read data to identify systematic errors in short-read SV analysis, where the vast majority (>85%) of the translocations are false positives due to mis-mapped reads.
The identification of SVs from long-reads is challenging chiefly because of the high error rates involved. In addition to numerous small indels, we discovered that PacBio introduces larger false insertions at a low, but noticeable rate (Supplementary Note 2). We control for this artifact by requiring the size and composition of candidate SVs are consistent across the spanning reads. Within the Oxford Nanopore dataset, we have highlighted systematic artifacts in base-calling that form deletions in low complexity repeats. While we fully expect accuracy to improve through improved base-calling, it is currently necessary to exclude most small SV calls when using Nanopore sequencing. Beyond sequencing errors, we highlighted how alignment artifacts can lead to miscalling SVs. For example, some long-read mappers falsely align reads through a SV without indication of the underlying event. Although Sniffles recognizes the increase in mismatches, NGMLR alignments correct these issues more directly. Finally, we showed a deficiency in detecting nested variations such as INVDUP or INVDEL in all methods except Sniffles. Several diseases are already known to be associated with these SV types, and we expect their importance will grow as more samples are analyzed using our methods.
The last remaining barrier to routine analysis of SVs across large numbers of samples is cost. Long-read technologies are becoming less expensive every year, but remain more expensive than short-read sequencing23. We addressed this by investigating how much coverage is needed for accurate SV calling, and show high accuracy is possible with only 15× to 30× coverage for healthy or cancerous human genomes. These requirements will be reduced even more as the read lengths increase and error rates decrease. Altogether, these improvements, aided by our methods, will usher in a new era of high quality genome sequences for a broad range of research and clinical applications.
Online Methods
NGMLR: Fast, Accurate Mapping of Long Single Molecule Reads
NGMLR is designed to accurately map long single molecule sequencing reads from either Pacific Biosciences or Oxford Nanopore to a reference genome with the goal of enabling precise structural variation calls. We follow the terminology used by the SAM specification47 where a read mapping consists either of one linear alignment covering the full read length or multiple linear alignments covering non-overlapping segments of the read (i.e. split reads).
The main challenge when mapping high error long-reads is to evaluate whether a read should be mapped to the reference genome with one linear alignment, or must be split. For example, the correct mapping for a read that spans an inversion can only be found when splitting the read into three segments. Conversely, reads that do not span a structural variation should always be mapped with a single linear alignment. However, error rates are high, and are not always uniform with some regions having an error rate of over 30%. These segments can cause read mappers to falsely split a read. Furthermore, the high insertion and deletion sequencing error of long-read technologies cause current read aligners to falsely split up large SVs into several smaller ones and make it difficult to detect exact break points.
To address these challenges, NGMLR implements the following workflow (Figure 1a):
NGMLR identifies sub-segments of the read and of the reference genome that show high similarity and can be aligned with a single linear alignment. These segments can contain small insertions and deletions, but must not span a larger structural variation breakpoint. In reference to BLAST’s High-scoring Segment Pairs (HSPs), we call those segment linear mapping pairs (LMPs).
For each LMP, NGMLR extracts the read sequence and the reference sequence and uses the Smith-Waterman algorithm to compute a pairwise sequence alignment using a convex gap cost model that accounts for sequencing error and SVs at the same time.
NGMLR scans the sequence alignments for regions of low sequence identity to identify small SVs that were missed in step (1) and (3).
Finally, NGMLR selects the set of linear alignments with the highest joint score, computes a mapping quality for each alignment and reports them as the final read mapping in a SAM/BAM file.
Convex scoring model
When aligning high error long-reads it is crucial to choose an appropriate gap model as there are two sources of insertions and deletions (indels). Sequencing error predominantly causes very short randomly distributed indels (1–5bp) while longer indels (20bp+) are caused by genomic structural variations. A linear gap model appropriately models indels originating from sequencing error, but cannot account for longer indels from genomic variation as these large blocks occur as a single unit, not as the combination of multiple single base insertions or deletions. With affine gap models the gap-open penalty falsely causes short indels from sequencing error to cluster together for noisy long-reads, and has only little effect on longer indels, especially in repetitive regions of the genome. With the convex scoring model of NGMLR, extending an indel is penalized proportionally less the longer the indel is. Therefore, the convex scoring model encourages large alignment gaps, such as those occurring from a structural variation, to be grouped together into contiguous stretches (extending a large indel has relatively low cost), while small indels, which commonly occur as sequencing errors, remain separate (extending a 1 bp gap has almost the same cost as opening a new gap).
Using a convex gap model to compute optimal alignments increases computation time substantially as each cell in the alignment matrix not only depends on three other cells, but on the full row and column it is located in 36. This would make it infeasible to use convex gap costs for aligning large long-read datasets, so we adapted a heuristic implementation of the convex gap cost algorithm found in the swalign library (https://github.com/mbreese/swalign). Instead of scanning the full cell and row while filling the alignment matrix, we use two additional matrixes to store indel length estimations for each cell. Furthermore, we use the initial sub-segment alignments to identify the part of the alignment matrix that is most likely to contain the correct alignment and skip all other cells of the matrix during alignment computation. (Supplementary Note 1).
Sniffles: Robust Detection of Structural Variations from Long-read Alignments
Sniffles operates within and between the long-read alignments to infer SVs. It applies five major steps (Figure 1b).
Sniffles first estimates the parameters to adapt itself to the underlying data set, such as the distribution in alignment scores and distances between indels and mismatches on the read, as well as the ratios of the best and second best alignments scores.
Sniffles then scans the read alignments and segments to determine if they potentially represent SVs.
Putative SVs are clustered and scored based on the number of supporting reads, the type and length of the SV, consistency of the SV composition, and other features.
Sniffles optionally genotypes the variant calls to identify homozygous or heterozygous SVs.
Sniffles optionally provides a clustering of SVs based on the overlap with the same reads, especially to detect nested variants.
For details on each step see Supplementary Note 2. In the following, we focus on the methods that are unique to Sniffles, which are the detection and analysis of alignment artifacts to reduce falsely called variants and the clustering of variants.
Putative Variant Scoring
The high error rate of the long-reads induces many alignments that falsely appear as SVs. Sniffles addresses these by scoring each putative variant using several characteristics that we have determined to be the most relevant to detecting SVs. The two main user thresholds are the number of high quality reads supporting the variant (set using the –s parameter) as well as the standard deviation of the coordinates in the start and stop breakpoint across all supporting reads. The minimum variant size reported defaults to 50bp, but can also be adjusted using the –l parameter. To account for additional noise in the data and imprecision of the breakpoints we use a quantile filtering to ignore outliers given a coverage of more than 8 reads. The computed standard deviations for both breakpoints are compared to the standard deviation of a uniform distribution representing spurious SV breakpoints reported in the region. SVs are only reported if both breakpoints are below this threshold. If the standard deviation for both breakpoints is < 5bp, the coordinates are marked as PRECISE in the VCF file. See Supplementary Note 2.
Variant Scoring and Genotyping
At the start of the program the user may specify that the VCF output should be genotyped. In this case, Sniffles stores summary information (coordinates and orientation) about all high quality reads that do not include a SVs in a binary file. This includes those reads that support the reference sequence that pass the thresholds for MQ and alignment score ratio. After the detection of SVs, the VCF file is read in, and Sniffles constructs a self-balancing tree of the variants. With this information, Sniffles then computes the fraction of reads that support each variant versus those that support the reference. Variants below the minimum allele frequency (default: below 30%) are considered unreliable; variants with high allele frequency (default: above 80%) are considered homozygous; and variants with an intermediate allele frequency are considered heterozygous. Note Sniffles does not currently consider higher ploidy, however this will be the focus of future work. See Supplementary Note 2.
Clustering and Nested SVs
To enable the study of closely positioned or nested SVs, Sniffles optionally clusters SVs that are supported by the same set of reads. Note that Sniffles does not fully phase the haplotypes, as it does not consider SNPs or small indels, but rather identifies SVs that occur together. If this option is enabled, Sniffles stores the names of each read that supports a SVs in a hash table keyed by the read name, with the list of SVs associated with that read name as the value. The hash table is used to find reads that span more than one event, and later to cluster reads that span the one or more of the same variants. In this way Sniffles can cluster two or more events, even if the distance between the events is larger than the read length. Future work will include a full phasing of hapolotypes including SVs, SNPs and other small variants. See Supplementary Note 2.
Mapping and SV Evaluation
Simulation of SV and reads
As described above, SVs were randomly simulated on chromosome 21 and 22 of the human genome (GRCh37). Data sets were generated with 20 variants for each type of SV (tandem duplication, indel, inversion, translocation and nested) and sizes of these events (100, 250, 500, 1kb, 5kb, 10kb and 50kb). Illumina reads were simulated as 100bp paired end reads using the default parameters of dwgsim. For Pacbio and Oxford Nanopore we scanned the alignments of HG002 (GiaB) and NA12878, respectively, and measured their error profile using SURIVOR (option 2). The measured error profiles and read lengths were then used to simulate the reads for each simulated SV data set (Supplementary Note 3).
Modified reference analysis
To allow for a more realistic scenario, we also modified the human reference (GRCh37) and analyzed real reads to assess the introduced SVs. Here we could only simulate a subset of SV types to be insertions, deletions, inversions and translocations. We simulated 140 variants of each type on the human genome (GRCh37) using SURVIVOR (option 1) (Supplementary Note 5).
Evaluation of long-read mappings
All simulated reads were mapped to the human reference genome (GRCh37) using BWA-mem26, BLASR25, GraphMap27, MECAT28, LAST24, Minimap229, and NGMLR. Reads that overlap or map in close proximity to a simulated SV were extracted from the BAM files and used for evaluation. For the genuine datasets, we first mapped the reads to the unmodified reference genome (without SV) using BWA-MEM and extracted all reads that would span our simulated SV by at least 500 bp. Only these reads were then mapped to the modified reference genome using the four read mappers and used for evaluation.
Both simulated and genuine reads were then divided into six categories (Supplementary Figure 3.5):
Read mappings are considered “precise” if they fully identify the SV they cover. To fall into this category, read mappings have to cover all parts of the SV that are required for identification, e.g. a read mapping to an inversion has to cover the inverted part of the genome as well as the non-inverted sequences flanking the inversion. Furthermore, correct mappings have to be split at the simulated breakpoints (+/− 10bp) of the SV.
Read mappings are considered “indicated” if they show the presence of the correct SV but as the wrong type, e.g. a duplication that is represented as an insertion, or show the correct SV but do not show the exact borders (>10bp away).
Read mappings are considered “forced” if they indicated the wrong number of SVs (e.g. several small instead of a single long insertion) or contain a significant portion of mapping artifacts (eg. not simulated mismatches) over > 10% of the SV length. These include mappings such as a read that is aligned through a deletion or inversion (Figure 2, top).
Read mappings are considered “trimmed” if they have been soft-clipped or otherwise trimmed so that they cannot indicate the SV and do not contain randomly aligned base pairs (ie. noisy regions)
Read mappings that are split into more parts than required to cover the underlying SV are classified as “fragmented”.
Read mappings that are supposed to map across the SV but are not mapped are considered “unaligned”.
For all simulated SV types and sizes and all mappers, we count how many reads fall into the above categories, normalize by the number of simulated reads and visualize the result as barplots.
Evaluation of SV callers
After the SV calling each VCF file was evaluated using SURVIVOR 48 with appropriate parameter sets to compare the variants to the truth set. A SV is considered precise if its start and stop coordinate is within 10bp of the simulated start and stop coordinate and the caller predicted the correct type. A SV is considered indicated if the start and stop coordinate of the SV is within +-1kb of the simulated event regardless of the inferred type of SV. A simulated SV is considered not detected if there is no call that fulfill the two previous criteria. A SV is considered false-positive if the event was not simulated.
Data availability
The raw sequencing data used in this study are available from the respective publications listed in Supplementary Table 5. The alignments and structural variation calls produced in this study for NGMLR and Sniffles are available here: https://github.com/fritzsedlazeck/Sniffles.
Code availability
The source code, documentation and test data sets are available at: https://github.com/philres/ngmlr and https://github.com/fritzsedlazeck/Sniffles for the mapping and SVs calling method, respectively.
Software Versions and Parameter settings
BWA-MEM (version 0.7.12-r1039) 26 was used with “-M” parameter to map the short-reads and with “-X pacbio -M” to follow the recommended parameter settings for PacBio reads. The parameter –M is used to mark only one alignment as primary and the subsequent alignments as secondary. BlasR (version 1.3.1)25 was run using the parameters “-sam -bestn 1 -nproc 15” to obtain only the best alignment in SAM format using 15 threads. Furthermore, Blasr was run with the parameters suggested by PBHoney 18 “-nproc 15 -bestn 1 -sam -clipping subread -affineAlign -noSplitSubreads -nCandidates 20 -minPctIdentity 75 -sdpTupleSize 6”. SAMTools (version 0.1.19-44428cd) 47 was used to convert the SAM alignment files to BAM and to sort the aligned reads.
Delly (version v0.7.3) 15, Lumpy (version 0.2.13) 14 and Manta (version 1.0.3) 16 were used to call SVs over the high mapping quality aligned Illumina reads (MQ20+) followed by SURVIVOR (version 0.0.1) 48 to combine the calls and report the consensus variants. To allow for the uncertainty with short-read variant positioning, SVs were considered to be the same if their start and stop coordinates fell within 1kb of another and were of the same type. PBHoney (version PBSuite_15.8.24)18 with default parameters was used to infer SV based on the specified BlasR alignments. The output was converted into a VCF using SURVIVOR (option 10).
More general information can be obtained from the Life Sciences Reporting Summary.
Supplementary Material
Acknowledgments
We would like to thank W. Richard McCombie, Sarah Wheelan, Sara Goodwin, Heng Li and Bui Quang Minh for helpful discussions. This work was supported through National Science Foundation awards (DBI-1350041, IOS-1732253, and IOS-1445025) and National Institutes of Health award (R01-HG006677, UM1 HG008898). P.R. acknowledges support by the RNA-DK Biology (W1207-B09).
Footnotes
Competing interests
M.C.S. and F.J.S. have participated in PacBio sponsored meetings over the past few years and have received travel reimbursement and honoraria for presenting at these events. Since the initial submission, P.R. is an employee of Oxford Nanopore. PacBio and Oxford Nanopore had no role in decisions relating to the study/work to be published, data collection or analysis of data.
References
- 1.Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet. 2013;14:125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- 2.Lupski JR. Structural variation mutagenesis of the human genome: Impact on disease and evolution. Environ Mol Mutagen. 2015;56:419–436. doi: 10.1002/em.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macintyre G, Ylstra B, Brenton JD. Sequencing Structural Variants in Cancer for Precision Therapeutics. Trends Genet. 2016;32:530–542. doi: 10.1016/j.tig.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Hedges DJ, et al. Evidence of novel fine-scale structural variation at autism spectrum disorder candidate loci. Mol Autism. 2012;3:2. doi: 10.1186/2040-2392-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rovelet-Lecrux A, et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 6.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dennenmoser S, et al. Copy number increases of transposable elements and protein-coding genes in an invasive fish of hybrid origin. Mol Ecol. 2017 doi: 10.1111/mec.14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeffares DC, et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun. 2017;8:14061. doi: 10.1038/ncomms14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zichner T, et al. Impact of genomic structural variation in Drosophila melanogaster based on population-scale sequencing. Genome Res. 2013;23:568–579. doi: 10.1101/gr.142646.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imprialou M, et al. Genomic Rearrangements in Arabidopsis Considered as Quantitative Traits. Genetics. 2017;205:1425–1441. doi: 10.1534/genetics.116.192823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 12.Kadalayil L, et al. Exome sequence read depth methods for identifying copy number changes. Brief Bioinform. 2015;16:380–392. doi: 10.1093/bib/bbu027. [DOI] [PubMed] [Google Scholar]
- 13.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 17.Huddleston J, et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 2017;27:677–685. doi: 10.1101/gr.214007.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.English AC, Salerno WJ, Reid JG. PBHoney: identifying genomic variants via long-read discordance and interrupted mapping. BMC Bioinformatics. 2014;15:180. doi: 10.1186/1471-2105-15-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mills RE, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tattini L, D’Aurizio R, Magi A. Detection of Genomic Structural Variants from Next-Generation Sequencing Data. Front Bioeng Biotechnol. 2015;3:92. doi: 10.3389/fbioe.2015.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teo SM, Pawitan Y, Ku CS, Chia KS, Salim A. Statistical challenges associated with detecting copy number variations with next-generation sequencing. Bioinformatics. 2012;28:2711–2718. doi: 10.1093/bioinformatics/bts535. [DOI] [PubMed] [Google Scholar]
- 22.Lucas Lledo JI, Caceres M. On the power and the systematic biases of the detection of chromosomal inversions by paired-end genome sequencing. PLoS One. 2013;8:e61292. doi: 10.1371/journal.pone.0061292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kielbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv e-prints. 2013 arXiv:1303.3997 [q-bio.GN] [Google Scholar]
- 27.Sovic I, et al. Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat Commun. 2016;7:11307. doi: 10.1038/ncomms11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao CL, et al. MECAT: fast mapping, error correction, and de novo assembly for single-molecule sequencing reads. Nat Methods. 2017;14:1072–1074. doi: 10.1038/nmeth.4432. [DOI] [PubMed] [Google Scholar]
- 29.Li H. Minimap2: fast pairwise alignment for long nucleotide sequences. ArXiv e-prints. 2017 arXiv:1708.01492. [Google Scholar]
- 30.Chaisson MJ, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–611. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sedlazeck FJ, Rescheneder P, von Haeseler A. NextGenMap: fast and accurate read mapping in highly polymorphic genomes. Bioinformatics. 2013;29:2790–2791. doi: 10.1093/bioinformatics/btt468. [DOI] [PubMed] [Google Scholar]
- 32.Carvalho CM, et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet. 2011;43:1074–1081. doi: 10.1038/ng.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimojima K, et al. Pelizaeus-Merzbacher disease caused by a duplication-inverted triplication-duplication in chromosomal segments including the PLP1 region. Eur J Med Genet. 2012;55:400–403. doi: 10.1016/j.ejmg.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Carvalho CM, Lupski JR. Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet. 2016;17:224–238. doi: 10.1038/nrg.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muhle C, Zenker M, Chuzhanova N, Schneider H. Recurrent inversion with concomitant deletion and insertion events in the coagulation factor VIII gene suggests a new mechanism for X-chromosomal rearrangements causing hemophilia A. Hum Mutat. 2007;28:1045. doi: 10.1002/humu.9506. [DOI] [PubMed] [Google Scholar]
- 36.Gusfield D. Algorithms on strings, trees, and sequences : computer science and computational biology. Cambridge University Press; 1997. [Google Scholar]
- 37.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chin CS, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zook JM, et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data. 2016;3:160025. doi: 10.1038/sdata.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain M, et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 2018 doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zook JM, et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 2014;32:246–251. doi: 10.1038/nbt.2835. [DOI] [PubMed] [Google Scholar]
- 42.Eberle MA, et al. A reference data set of 5.4 million phased human variants validated by genetic inheritance from sequencing a three-generation 17-member pedigree. Genome Res. 2017;27:157–164. doi: 10.1101/gr.210500.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zimin AV, Smith DR, Sutton G, Yorke JA. Assembly reconciliation. Bioinformatics. 2008;24:42–45. doi: 10.1093/bioinformatics/btm542. [DOI] [PubMed] [Google Scholar]
- 44.Beri S, Bonaglia MC, Giorda R. Low-copy repeats at the human VIPR2 gene predispose to recurrent and nonrecurrent rearrangements. Eur J Hum Genet. 2013;21:757–761. doi: 10.1038/ejhg.2012.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nattestad M, et al. Complex rearrangements and oncogene amplifications revealed by long-read DNA and RNA sequencing of a breast cancer cell line. bioRxiv. 2017 doi: 10.1101/gr.231100.117. doi: https://doi.org/10.1101/174938. [DOI] [PMC free article] [PubMed]
- 46.Merker JD, et al. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genetics in medicine : official journal of the American College of Medical Genetics. 2017 doi: 10.1038/gim.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeffares DC, et al. Transient structural variations alter gene expression and quantitative traits in Schizosaccharomyces pombe. bioRxiv. 2016 doi: 10.1101/047266. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data used in this study are available from the respective publications listed in Supplementary Table 5. The alignments and structural variation calls produced in this study for NGMLR and Sniffles are available here: https://github.com/fritzsedlazeck/Sniffles.