Abstract
Nicotinic acetylcholine receptors (nAChR), the primary cell surface targets of nicotine, have implications in various neurological disorders. Here we investigate the proteome-wide effects of nicotine on human haploid cell lines (wildtype HAP1 and α7KO-HAP1) to address differences in nicotine-induced protein abundance profiles between these cell lines. We performed an SPS-MS3-based TMT10-plex experiment arranged in a 2-3-2-3 design with two replicates for the untreated samples and three for the treated samples for each cell line. We quantified 8,775 proteins across all 10 samples, of which several hundred differed significantly in abundance. Comparing α7KO-HAP1 and HAP1wt cell lines revealed significant protein abundance alterations, however we also measured differences resulting from nicotine treatment in both cell lines. Among proteins with increased abundance levels due to nicotine treatment included those previously identified: APP, APLP2, and ITM2B. The magnitude of these changes was greater in HAP1wt compared to the α7KO-HAP1 cell line, implying a potential role of the α7 nAChR in HAP1 cells. Moreover, the data revealed that membrane proteins and proteins commonly associated with neurons were predominant among those with altered abundance. This study, which is the first TMT-based proteome profiling of HAP1 cells, defines further the effects of nicotine on non-neuronal cellular proteomes.
Keywords: HAP-1, nAChR, Nicotine, TMT, SPS-MS3
1. INTRODUCTION
Tobacco use is among the leading causes of preventable illness and death in the United States. Tobacco products have long been associated with multiple cancers types and chronic diseases affecting virtually all organ systems [1]. For example, cigarette smoking is a prominent risk factor for heart attacks, strokes, chronic obstructive pulmonary disease (COPD), and cancer (most prominently, lung cancer, cancers of the larynx and mouth, and pancreatic cancer). Understanding the underlying mechanisms that dictate the course of disease is vital to developing counteracting intervention.
Cigarette smoke consists of a mixture of greater than 4,000 compounds, for which over 50 are suspected carcinogens [2]. Nicotine is a natural alkaloid present predominantly in tobacco-related products. As a major component of tobacco, nicotine is readily absorbed by the lungs, as well as distal organs via systemic circulation [3]. Aside from its presence in tobacco, nicotine is a main component in smoking cessation products and electronic cigarettes [4]. Growing evidence has linked nicotine to the disruption of cellular metabolic processes and its potential to be genotoxic and tumor-promoting [5]. Nicotine has been shown to induce cell proliferation and invasion in multiple lung and breast cancer lines [6], and to alter the phosphorylation states of proteins in pancreatic stellate cells [7]. Various cancers and chronic diseases have been linked to nicotine, yet the mechanisms thereof remain poorly understood [8].
Pharmacologically, nicotine binds specifically to nicotinic acetylcholine receptors (nAChR) with varying affinities. Downstream effectors involved in signal transduction result in the modulation of intracellular phosphorylation cascades. nAChR can bind distinct ligands and evidence suggests that each subtype has a unique set of interacting proteins, a wide array of ligands, and varying affinities to nicotine [9]. Nicotine binding to these cell surface receptors transduces extracellular signals to the intracellular space. This molecular mechanism is manifested by ion transport and/or initiation of phosphorylation cascades and other signaling pathways [10, 11]. nAChRs, particularly the α7 subtype, have been discovered in non-neuronal cells and are important regulators of cellular function [12]. For example, some nAChR subtype (including the α7 nAChR) are expressed in the HAP1 cell line.
The HAP1 cell line is near-haploid, having a single copy of each chromosome, aside from a 30-megabase fragment of chromosome 15 that was integrated into the long arm of chromosome 19 [13]. This immortalized human cell line was derived from the KBM-7 cell line which originated from a patient with chronic myeloid leukemia, a disease in which near haploid cells are often observed [14]. HAP1 cells are advantageous in biomedical research as typical cell lines are diploid and some even have multiple copies of a given allele. As such, genetic manipulation is simplified as only a single allele must be edited [15]. Here we compare the global proteome profile of the α7 nAChR null HAP1 cell line (α7KO) with wildtype HAP1 (HAP1wt) to study the effects of nicotine, using a multiplexed quantitative proteomics approach.
Multiplexing strategies in mass spectrometry-based quantitative analyses, such as tandem mass tags (TMT) and isobaric tags for relative and absolute quantitation (iTRAQ) have many advantages for whole proteome profiling [16]. Such strategies allow for samples to be analyzed simultaneously, thereby reducing instrument time and costs, while producing fewer missing values between samples and permitting multiple comparisons in a single experiment. Here we used a TMT-based strategy to quantitatively compare the global proteome abundance measurements of two cell lines under two conditions: mock-treated HAP1wt cells, HAP1wt cells treated with nicotine, mock-treated α7KO cells, and α7KO cells treated with nicotine. The TMT10-plex experiment is arranged in a 2-3-2-3 design with two replicates for the untreated samples and three for the treated samples. We present the first global proteome profiling analysis of HAP1 cell lines using TMT-based proteome profiling, with the aim of furthering our knowledge of the role of nicotine in cellular pathway dysregulation.
2. METHODS
Materials
Tandem mass tag (TMT) isobaric reagents were from Thermo Fisher Scientific (Waltham, MA). Nicotine was purchased from Sigma (St. Louis, MO). Water and organic solvents were from J.T. Baker (Center Valley, PA). Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) were from LifeTechnologies (Waltham, MA). Trypsin was purchased from Pierce Biotechnology (Rockford, IL) and Lys-C from Wako Chemicals (Richmond, VA). Unless otherwise noted, all other chemicals were from Pierce Biotechnology (Rockford, IL). The HAP1wt cell line (C859) and the CRISPR/Cas edited α7KO cell line (HZGHC003352c012) were from Horizon Discovery (Saint Louis, MO).
Cell growth and harvesting
Methods of cell growth and propagation followed techniques utilized previously [17]. In brief, cells were propagated in DMEM supplemented with 10% FBS. Upon achieving 80% confluency, the growth media was aspirated, and the cells were washed thrice with ice-cold phosphate-buffered saline (PBS). Designated cell culture dishes were supplemented with 1 μM nicotine and control cell culture dishes were mock-treated with an equal volume of sterile deionized water, in buffer adjusted to pH 7.4. We used the S(−)- enantiomer of nicotine (Sigma N3776), as it is present in this form in tobacco products and has been shown to be several times more potent than (+) or racemic nicotine [18]. Less than 0.2% of (+)-nicotine was present in our nicotine stock solution. Twenty-four hours after the addition of the drug, the cells were dislodged with a non-enzymatic reagent, harvested by trituration following the addition of 10 mL PBS, pelleted by centrifugation at 3,000 × g for 5 min at 4°C, and the supernatant was removed. One milliliter of 200 mM EPPS, 8M urea, pH 8.5 supplemented with 1X Roche Complete protease inhibitors was added per each 15 cm cell culture dish.
Cell lysis and protein digestion
Cells were homogenized by 10 passes through a 21-gauge (1.25 inches long) needle and incubated at 4°C with gentle agitation for 30 min. The homogenate was sedimented by centrifugation at 21,000 × g for 5 min and the supernatant was transferred to a new tube. Protein concentrations were determined using the bicinchoninic acid (BCA) assay (ThermoFisher Scientific). Proteins were subjected to disulfide bond reduction with 5 mM tris (2-carboxyethyl) phosphine (room temperature, 30 min) and alkylation with 10 mM iodoacetamide (room temperature, 30 min in the dark). Excess iodoacetamide was quenched with 10 mM dithiotreitol (room temperature, 15 min in the dark). Methanol-chloroform precipitation was performed prior to protease digestion. In brief, 4 parts of neat methanol were added to each sample and vortexed, 1-part chloroform was added to the sample and vortexed, and 3 parts water was added to the sample and vortexed. The sample was centrifuged at 14,000 RPM for 2 min at room temperature and subsequently washed twice with 100% methanol. Samples were resuspended in 200 mM EPPS, pH 8.5 and digested at room temperature for 13 h with Lys-C protease at a 100:1 protein-to-protease ratio. Trypsin was then added at a 100:1 protein-to-protease ratio and the reaction was incubated for 6 h at 37°C.
Tandem mass tag labeling
TMT reagents (0.8 mg) were dissolved in anhydrous acetonitrile (40 μL) of which 10 μL was added to the peptides (100 μg) with 30 μL of acetonitrile to achieve a final acetonitrile concentration of approximately 30% (v/v). Following incubation at room temperature for 1 h, the reaction was quenched with hydroxylamine to a final concentration of 0.3% (v/v). The TMT-labeled samples were pooled at a 1:1:1:1:1:1:1:1:1:1 ratio across the 10 samples. The pooled sample was vacuum centrifuged to near dryness and subjected to C18 solid-phase extraction (SPE) (Sep-Pak, Waters).
Off-line basic pH reversed-phase (BPRP) fractionation
We fractionated the pooled TMT-labeled peptide sample using BPRP HPLC [19]. We used an Agilent 1200 pump equipped with a degasser and a photodiode array (PDA) detector (set at 220 and 280 nm wavelength) from ThermoFisher Scientific (Waltham, MA). Peptides were subjected to a 50-min linear gradient from 5% to 35% acetonitrile in 10 mM ammonium bicarbonate pH 8 at a flow rate of 0.6 mL/min over an Agilent 300Extend C18 column (3.5 μm particles, 4.6 mm ID and 220 mm in length). The peptide mixture was fractionated into a total of 96 fractions, which were consolidated into 24, from which 12 non-adjacent samples were analyzed [20]. Samples were subsequently acidified with 1% formic acid and vacuum centrifuged to near dryness. Each consolidated fraction was desalted via StageTip, dried again via vacuum centrifugation, and reconstituted in 5% acetonitrile, 5% formic acid for LC-MS/MS processing.
LC-MS/MS analysis
All samples were analyzed on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled to a Proxeon EASY-nLC 1200 liquid chromatography (LC) pump (Thermo Fisher Scientific). Peptides were separated on a 100 μm inner diameter microcapillary column packed with 35 cm of Accucore C18 resin (2.6 μm, 150 Å, ThermoFisher). For each analysis, we loaded approximately 2 μg onto the column. Peptides were separated using a 150min gradient of 3 to 25% acetonitrile in 0.125% formic acid with a flow rate of 450 nL/min. Each analysis used an MS3-based TMT method [21], which has been shown to reduce ion interference compared to MS2 quantification [22]. Prior to starting our analysis, we perform two injections of trifluoroethanol (TFE) to elute any peptides that may be bound to the analytical column from prior injections to limit carry over. The scan sequence began with an MS1 spectrum (Orbitrap analysis, resolution 120,000, 350–1400 Th, automatic gain control (AGC) target 5E5, maximum injection time 100 ms). The top ten precursors were then selected for MS2/MS3 analysis. MS2 analysis consisted of: collision-induced dissociation (CID), quadrupole ion trap analysis, automatic gain control (AGC) 2E4, NCE (normalized collision energy) 35, q-value 0.25, maximum injection time 120 ms), and isolation window at 0.7. Following acquisition of each MS2 spectrum, we collected an MS3 spectrum in which multiple MS2 fragment ions are captured in the MS3 precursor population using isolation waveforms with multiple frequency notches. MS3 precursors were fragmented by HCD and analyzed using the Orbitrap (NCE 65, AGC 1.5E5, maximum injection time 150 ms, resolution was 50,000 at 400 Th). For MS3 analysis, we used charge state-dependent isolation windows: For charge state z=2, the isolation window was set at 1.3 Th, for z=3 at 1 Th, for z=4 at 0.8 Th, and for z=5 at 0.7 Th.
Data analysis
Mass spectra were processed using a Sequest-based pipeline [23]. Spectra were converted to mzXML using a modified version of ReAdW.exe. Database searching included all entries from the human UniProt database. This database was concatenated with one composed of all protein sequences in the reversed order. Searches were performed using a 50 ppm precursor ion tolerance for total protein level analysis. The product ion tolerance was set to 0.9 Da. TMT tags on lysine residues and peptide N termini (+229.163 Da) and carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.995 Da) was set as a variable modification.
Peptide-spectrum matches (PSMs) were adjusted to a 1% false discovery rate (FDR) [24]. PSM filtering was performed using a linear discriminant analysis (LDA), as described previously [23], while considering the following parameters: XCorr, ΔCn, missed cleavages, peptide length, charge state, and precursor mass accuracy. For TMT-based reporter ion quantitation, we extracted the summed signal-to-noise (S:N) ratio for each TMT channel and found the closest matching centroid to the expected mass of the TMT reporter ion. For protein-level comparisons, PSMs were identified, quantified, and collapsed to a 1% peptide false discovery rate (FDR) and then collapsed further to a final protein-level FDR of 1%, which resulted in a final peptide level FDR of <0.1%. Moreover, protein assembly was guided by principles of parsimony to produce the smallest set of proteins necessary to account for all observed peptides.
Proteins were quantified by summing reporter ion counts across all matching PSMs, as described previously [23]. PSMs with poor quality, MS3 spectra with more than eight TMT reporter ion channels missing, MS3 spectra with TMT reporter summed signal-to-noise of less than 100, or having no MS3 spectra were excluded from quantification [25]. Each reporter ion channel was summed across all quantified proteins and normalized assuming equal protein loading of all 10 samples. Student t-tests were used to determine statistical significance between each treatment and controls for each cell lines, as well as to compare protein abundance levels between cell lines. In all cases, a p-value <0.05 was considered statistically significant. A second threshold based on a log2 fold change of greater than 1.5-fold or less than -1.5-fold was chosen so as to focus the data analysis on a small set of proteins with the largest alterations in abundance. Protein quantification values were exported for further analysis in Microsoft Excel (dot plots and bar graphs), Mathematica (histograms and descriptive statistics), or SAS JMP (hierarchical clustering/heat maps). Each reporter ion channel was summed across all quantified proteins and normalized to assume equal protein loading of all 10 samples across the TMT10-plex.
Data access
RAW files will be made available upon request.
3. RESULTS and DISCUSSION
Over 8,700 proteins were quantified across all ten samples, some of which significantly differed in abundance between comparison groups
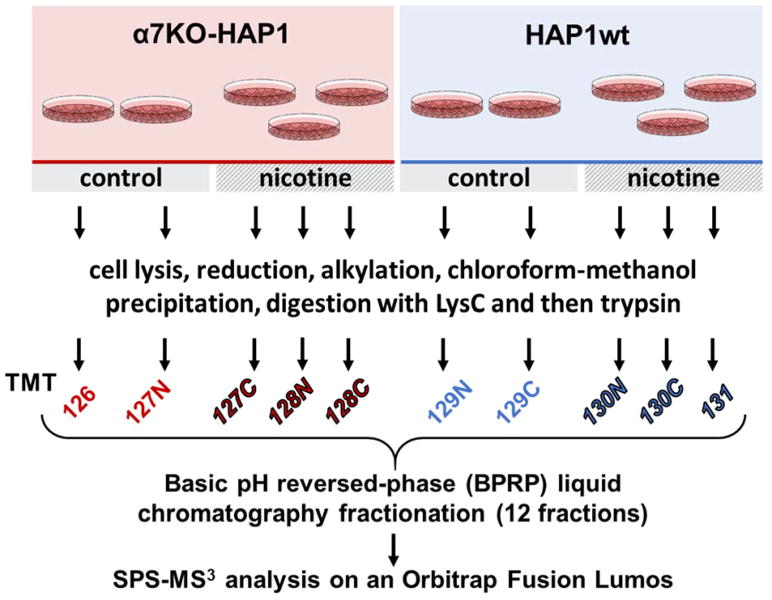
We used an SPS-MS3-based TMT10-plex experiment in a 2-3-2-3 arrangement: two biological replicates each of mock-treated HAP1wt and α7KO-HAP1 cells and three replicates each of nicotine-treated HAP1wt and α7KO-HAP1 cells (Figure 1). Our TMT10-plex analysis quantified a total of 126,810 peptides, of which 75,711 were unique. These peptides were assigned to a total of 8,775 non-redundant proteins at a 1% protein false discovery rate (FDR) (Table 1). We compared the overall protein abundance profiles among samples in this dataset. Using all quantified proteins, we performed unsupervised Ward’s hierarchical clustering to investigate sample grouping via the JMP software package. As illustrated by the dendrogram, clustering was the tightest within replicates and we observed the most deviation (earlier separation) between cell types (Figure 2A).
Figure 1. Experimental overview of the SPS-MS3 strategy.
Two cell types, α7KO-HAP1 and HAP1wt, were propagated and designated cultures were mock or nicotine-treated. Proteins were extracted and then digested with LysC and trypsin. The resulting peptides were labeled with TMT, pooled, and fractionated via basic pH reversed-phase (BPRP) high performance liquid chromatography (HPLC) prior to SPS-MS3 analysis on an Orbitrap Fusion Lumos mass spectrometer.
Table 1.
Dataset summary
| Category | |
|---|---|
| Quantified proteins across all samples | 8,775 |
| Unique quantified peptides | 75,711 |
| Total quantified peptides | 126,810 |
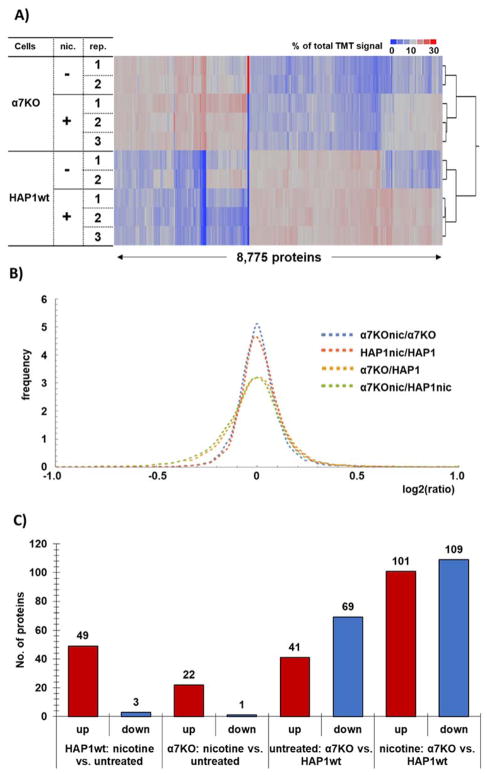
Figure 2. Protein abundance changes among cell lines and treatment.
A) The heat maps and associated dendrograms for each cell line and treatment. Across each row of the heat map, the relative protein expression levels are displayed, such that each row sums to 100. The scale corresponds to the percentage of total signal across all channels. B) Probability density function-smoothed histogram of protein alterations between α7KO+nicotine versus α7KO (untreated) (blue), HAP1wt+nicotine versus HAP1wt (untreated) (orange), α7KO (untreated) versus HAP1wt (untreated) (yellow), and α7KO+nicotine versus HAP1wt+nicotine (green). C) Bar chart representing significantly altered proteins in the different comparison groups within the TMT10-plex.
The layout of our TMT10-plex experiment lent itself to multiple comparisons, specifically, we focused on four: 1) α7KO versus HAP1wt (both untreated), 2) HAP1wt+nicotine versus HAP1wt (untreated), 3) α7KO+nicotine versus α7KO (untreated), and 4) α7KO+nicotine versus HAP1wt+nicotine. We defined significantly altered proteins as those with a two-tailed Student t-test p-value <0.05 and with a fold change exceeding 1.5 between two comparison groups. We plotted the fold changes as probability density function-smoothed distributions to visualize the differences among the four comparison groups. As expected, we noted broader distributions of protein abundance fold changes between the two cell lines than between nicotine-treated and untreated cells (Figure 2B). Nevertheless, we can compare similarities and differences in nicotine-induced proteins abundance alterations were measured in both cell lines. As such, we determined: 1) α7KO versus HAP1wt (both untreated) had 49 proteins of significantly up-regulated and 3 down-regulated proteins, 2) HAP1wt+nicotine versus HAP1wt (untreated) revealed 22 up-regulated and 1 down-regulated protein, 3) α7KO+nicotine versus α7KO (untreated) showed 41 up-regulated and 69 down-regulated proteins, and 4) α7KO+nicotine versus HAP1wt+nicotine were the least similar with 101 up-regulated and 109 down-regulated proteins (Figure 2C). Next, we compared alterations in protein abundance between comparison groups in relationship to gene ontology (GO) classifications.
Gene ontology analysis categorized proteins that differed in abundance between the two cell lines
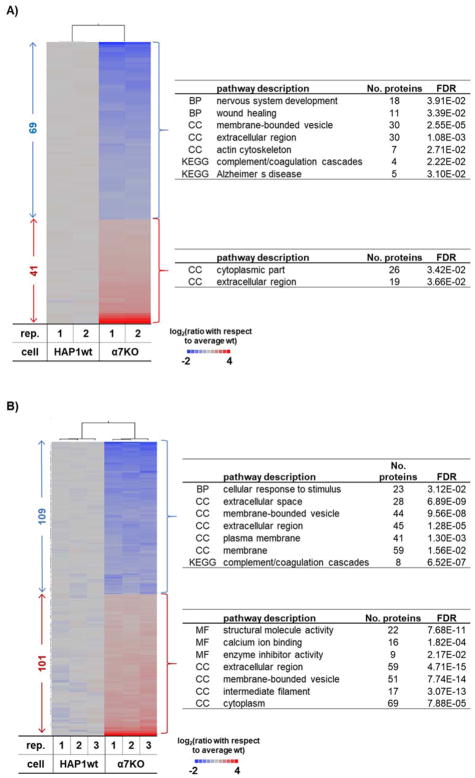
When comparing the α7KO and HAP1wt cell lines, we observed in total over 300 proteins with significant differences in abundance. Considering only statistically significant proteins from the binary comparison of the α7KO and HAP1wt cell lines, we performed gene ontology analysis using STRING [26] and EnrichR [27]. We compare untreated α7KO and HAP1wt cell lines (Figure 3A) and nicotine-treated α7KO and HAP1wt cell lines (Figure 3B). We isolated four clusters of proteins, which were then classified by GO categories. These clusters included proteins that were of: 1) lower abundance in untreated α7KO versus HAP1wt cells (n=69), 2) higher abundance in untreated α7KO versus HAP1wt cells (n=41), 3) lower abundance in nicotine-treated α7KO versus HAP1wt cells (n=109), and 4) higher abundance in nicotine-treated α7KO versus HAP1wt cells (n=101).
Figure 3. Cell-specific proteome differences between HAP1wt and α7KO HAP1 cell line.
Heat map of proteins with statistically significant differences between HAP1wt and α7KO cell lines in A) untreated and B) nicotine-treated cells. Heat maps include only proteins having a fold-change > |1.5| and a p-value of <0.05 as determined using a Student t-test. To the right of the heat map are the major gene ontology or KEGG pathway categories for which associated proteins are significantly (p-value<0.05) enriched.
Many proteins that were relatively lower in abundance in α7KO versus HAP1wt cells were localized to the extracellular regions, involved in nervous system development, and expressed in response to wound healing (Figure 3A top). Some of these proteins included: FERMT3 (fermitin family homolog 3), FN1 (fibronectin), BCHE (carboxylic ester hydrolase), BMP7 (bone morphogenetic protein 7), and IQGAP2 (Ras GTPase-activating-like protein). However, no significantly enriched categories were identified when comparing α7KO versus HAP1wt cells, other than 19% of those proteins being of extracellular origin (Figure 3A bottom). We next examined the protein expression profiles of HAP1wt and α7KO cell lines that were treated with nicotine. Proteins with lower abundance in nicotine-treated α7KO versus HAP1wt cells were categorized as both membrane and cytoplasmic proteins (Figure 3B top). Unlike the other categories examined, structural (n=22) and calcium binding (n=16) proteins were significantly enriched in this cluster of proteins. Example proteins include: CALB1 (calbindin), GCA (guanylyl cyclase), and CALML5 (calmodulin-like protein 5). However, proteins with higher abundance in nicotine-treated α7KO versus HAP1wt cells were categorized mainly as membrane origin, but many were also implicated in response to stimulus (Figure 3B bottom). Examples include: A2M (alpha-2-macroglobulin), ALPL (alkaline phosphatase), and MME (neprilysin). Although protein profiles differed extensively between the knockout and wildtype cell lines, the data from our TMT experiment can be used to determine proteins that are altered by nicotine in both cell lines and those unique to a specific cell line.
Nicotine treatment resulted in cell line-dependent differences in protein abundance
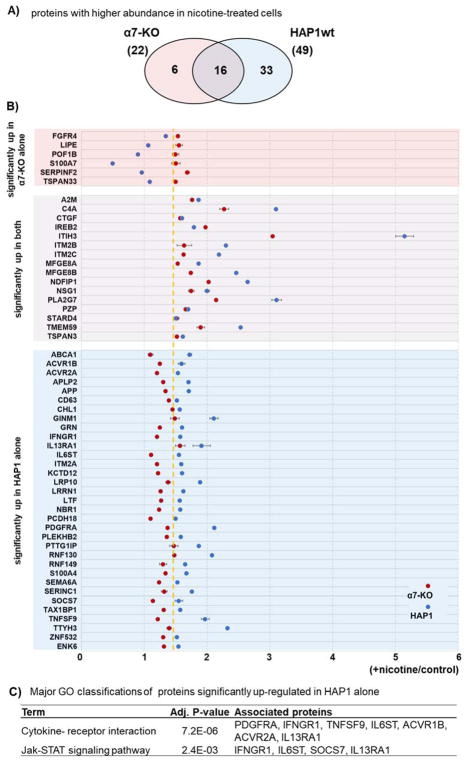
Our TMT-based experiment permitted us to investigate nicotine-induced protein abundance differences that were in common or unique to each cell line. In isobaric label-based quantification, as opposed to label-free analyses, all proteins have a measurement for every condition. This advantage eliminated much of the stochasticity associated with protein measurements across experiments as observed in label-free quantification and allowed us to determine the relative abundance for a given protein under numerous conditions. In the α7KO cell line, we determined 22 proteins as having significantly different abundance measurements (p-value <0.05, |fold change|>1.5), while in the HAP1wt cell line, 49 proteins were significant. Of these differentially expressed proteins, 16 overlapped between the cell lines, and 33 were exclusive to the HAP1wt cell line, while 6 proteins were exclusive to the α7KO line (Figure 4A). As such, we observed both common and different protein abundance profiles alterations between cell line due to nicotine treatment.
Figure 4. Nicotine-induced protein alterations within and between cell lines.
A) Venn diagram of proteins with statistically significant nicotine-induced up-regulation in α7KO and HAP1wt cell lines. B) Modified dot plot of proteins (n=6) that are of higher abundance in α7KO but not in HAP1wt cell lines (top), proteins (n=16) of higher abundance in both cell lines (middle), and proteins (n=33) of higher abundance in HAP1wt, but not in α7KO cell lines (bottom). Data points represent the average relative abundance measurement for nicotine-treated cells divided by that for untreated cells. Error bars represent standard deviations. Dashed yellow line represents the nicotine-to-control ratio of 1.5. C) The KEGG pathway categories that are enriched by the associated proteins listed on the right.
Using a modified dot plot, we illustrated those proteins (n=33) that were up-regulated in HAP1wt, but not in the α7KO cell line (Figure 4B). Similarly, we highlighted proteins up-regulated (n=6) in the α7KO, but not in the HAP1wt cell line (Figure 4C). Each data point represented the average of the relative abundance measurements for the nicotine-treated samples divided by that of the untreated samples for each cell line with error bars representing standard deviations. These lists of proteins were subjected to STRING gene ontology analysis to identify the key categories which were represented by these proteins. As expected, the majority, 26 of 33, proteins that were up-regulated only in HAP1wt cells were membrane proteins, and 7 were cytokine-cytokine interacting proteins (ACVR1B, ACVR2A, IFNGR1, IL13RA1, IL6ST, PDGFRA, TNFSF9). Within the smaller set of proteins (i.e., those up-regulated only in α7KO), 3 proteins (S100A7, FGFR4, SERPINF2) were positive regulators of ERK1 and ERK2 cascades. ERK signaling has a potential role in cell proliferation and migration, which are characteristic effects of nicotine treatment in several cell types [28, 29]. Future investigations may study further the α7KO cell line in relation to ERK, cell migration/proliferation, and the α7 nAChR.
Moreover, in the human body, nicotine is extensively metabolized by the liver, resulting in several metabolites which can alter protein abundance profiles. Like nicotine, these metabolites may alter protein abundance profiles and modulate signaling mechanisms. Using sample preparation and data analysis techniques analogous to those outlined herein, we can investigate the alterations in the downstream breakdown products of nicotine, such as cotinine, nicotine N′-oxide, and trans-3-hydroycotinine [30]. Accordingly, we can use the amalgamate of these data to better delineate the cellular effects of nicotine and its metabolites, first in cell and later expand this to an in vivo system.
Several proteins involved in neuronal function and signal transduction were among those up-regulated upon nicotine treatment
The role of nicotine has been comprehensively studied with respect to neuronal function in both in vivo and in vitro studies. In the current study, we measured nicotine-induced protein abundance differences in several neuronal function-related proteins. Protein abundance profiles from nAChR were not measured with our data-dependent acquisition (DDA) approach, as identification of these receptors are difficult in “shot-gun” proteomics experiments [31]. However, we expect that the development of integrated DDA and targeted strategies will eventually enable us to profile nAChR and other proteins consistently. Nonetheless, abundance profiles of several other proteins with known functions in neurons were altered by nicotine in the HAP1wt and α7KO cell lines.
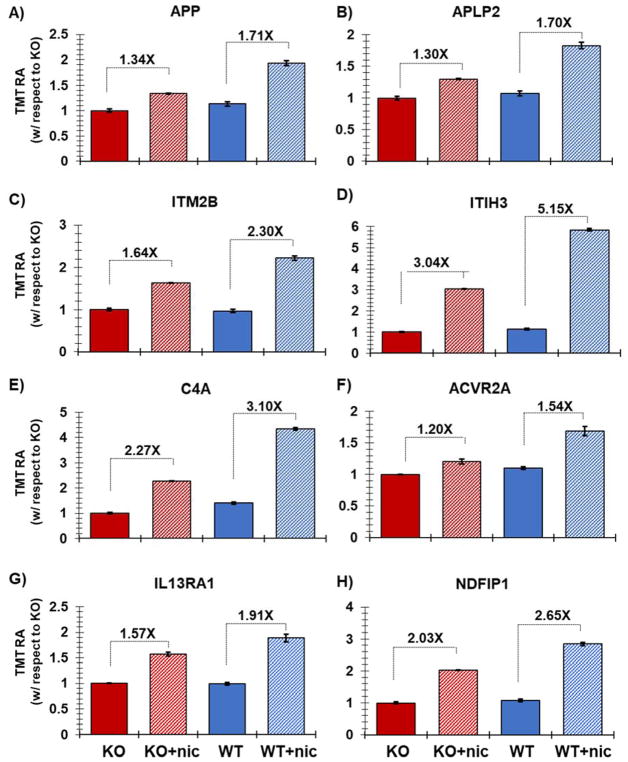
We highlighted eight proteins with altered expression due to nicotine treatment and plotted the relative TMT signal-to-noise levels for the two controls and three nicotine-treated samples for each cell line (Figure 5). Amyloid beta A4 protein (APP, Figure 5A) is well studied in the nervous system and is linked to the development of Alzheimer’s disease [32]. APP has been identified in an interactome study of the α7 nAChR from mouse brain tissue [11] and several studies have shown APP to bind and modulate the activity of this receptor [33]. APP has also been observed previously as up-regulated in nicotine-treated pancreatic cells in a cross-species analysis, which included pancreatic stellate cells (PaSC) from mouse, rat, and human species [34]. Moreover, an in vitro study using pancreatic ductal epithelial cells has indicated that increased APP expression in pancreatic cancer may influence cellular proliferation [35]. Like APP, Amyloid-like protein 2 (APLP2, Figure 5B) was also among those proteins demonstrating higher abundance across cell lines when treated with nicotine [36]. APLP2 may interact with proteins in cellular G-protein signaling pathways, which have been shown to associate with nAChR [37], and likewise are members of a murine nAChR interactome [11]. Integral membrane protein 2B (ITM2B, Figure 5C) plays a regulatory role in the processing of the beta-amyloid A4 precursor protein (APP), as an inhibitor of beta-amyloid peptide aggregation and fibril deposition [38]. ITM2B is up-regulated in nicotine-exposed cells. Related to neuronal functional pathways, inter-alpha-trypsin inhibitor heavy chain H3 (ITIH3, Figure 5D) stabilizes the extracellular matrix through its ability to bind hyaluronic acid. Polymorphisms of this gene may be associated with increased risk for schizophrenia and major depressive disorder [39]. Complement C4-A (C4A, Figure 5E) is another protein that is commonly observed to be up-regulated in nicotine-treated cell cultures. C4-A has been shown previously to predict survival of patients with metastatic renal cell carcinoma [40]. Members of the complement family have been also associated with nicotine exposure [41].
Figure 5. Representative proteins displaying altered expression due to nicotine treatment.
Plotted are the relative TMT signal-to-noise levels for the selected proteins across the two controls and three nicotine-treated samples for each cell line. The proteins highlighted include: A) Amyloid beta A4 protein (APP), B) Amyloid-like protein 2 (APLP2), C) ITM2B (integral membrane protein 2B), D) ITIH3 (inter-alpha-trypsin inhibitor heavy chain H3), E) complement C4-A (C4A), F) ACVR2A (Activin receptor type-2A), G) IL13RA1(interleukin 13 receptor, alpha 1), and H) NDFIP1 (NEDD4 Family Interacting Protein 1). Error bars represent standard deviations.
In addition, we highlighted several proteins with roles in signal transduction that were up-regulated upon nicotine treatment. Activin receptor type-2A (ACVR2A, Figure 5F) belongs to the TGF-beta superfamily of signaling proteins. This transmembrane has a cytoplasmic serine-threonine kinase domain [42]. As a kinase and being localized to the membrane, this protein may be important for early signal transduction from nAChR. Also involved in signal transduction, interleukin 13 receptor, alpha 1 (IL13RA1, Figure 5G) was up-regulated in nicotine-exposed cells. IL13RA1 binds tyrosine kinase TYK2, and thus may mediate the signaling processes that lead to the activation of JAK1, STAT3 and STAT6 [43], pathways which are known to be activated by α7 nAChR function. These pathways promote cellular proliferation. In fact, we observed that in both HAP1wt and α7 KO cells that cell proliferation was increased upon nicotine treatment, which agreed with previous studies [29, 44]. Similarly, NDFIP1 (NEDD4 Family Interacting Protein 1, Figure 5H) modulates EGFR signaling through multiple pathways. NDFIP1 may regulate the ratio of AKT1-to-MAPK8 signaling in response to EGF [45]. AKT signaling is associated with nAChR, however no direct association between nicotine and NDFIP1 has been described. Further experiments targeting these proteins, among others, and associated pathways are necessary to understand better these potential associations. Although many of the proteins with altered abundance profiles, particularly those in Figure 5, have been identified previously in other proteomic analyses, verification of proteins with altered abundance profiles with traditional or orthogonal biochemistry-based methods, such as Western blotting, ought to be performed before concerted effort is dedicated to its study. Moreover, the list of altered proteins is not comprehensive, but should support previous findings or lay the foundation for pursuing the study of certain proteins in cells perturbed by nicotine, nicotine metabolites, or other drugs.
4. CONCLUDING REMARKS
We investigated the effect of nicotine on HAP1wt and α7KO-HAP1 cell lines. We quantified over 8,700 proteins across all samples in a multiplexed TMT10-plex experiment. Of these, over 300 proteins differed significantly in abundance among comparison groups. Using gene ontology analysis, we determined that membrane proteins and those related to the nervous system were predominant among proteins with altered abundance. The abundance levels of several proteins, such as APP and C4A, were also altered by nicotine in previous studies. These data provided the protein profile for a single concentration of nicotine at a specific time point. The concentration used (1μM) is at least three times higher than blood nicotine concentrations in smokers (ranging from 0.06 to 0.31μM [46]), however this concentration and the time point chosen (24 h) have been used in previous studies [47] [48] [49]. Nonetheless, a far wider range of both parameters may be investigated which could result in proteomic alterations that would not be observed otherwise. Nicotine, for example, has been observed to have a more prominent effect on protein expression profiles at lower concentration (10 uM), than higher concentration (1 mM) in C. elegans [46]. In addition, shorter time of treatment is more informative for signaling changes, such as phosphorylation, than are longer times where the cells may have returned to equilibrium and only persistent protein profile alterations are observed. Further studies may be designed to expand the scope of our investigation by considering the dosage and temporal effects of nicotine, as well as changes in post-translational modifications.
The effects of nicotine are not limited to modulation of extracellular receptors, as nicotine can also have a direct intracellular role. Nicotine is a weak base that exists in two forms 1) the membrane-impermeable protonated form and 2) the membrane-permeable unprotonated form [47]. Approximately 30% of nicotine is unprotonated at physiological pH, allowing it to enter the cell. Studies have shown that nicotine can bind DNA and ultimately alter protein expression [48]. Nicotine has been shown also to accumulate in intracellular acidic vesicles [49]. More recently, mitochondrial nicotinic receptors have been identified on mitochondrial outer membrane and its activation by nicotine can facilitate cell survival by inhibiting apoptosis [50]. As such, some of the nicotine-induced protein profile alterations that are common between cell lines may reflect the intracellular effects of nicotine, which are not fully understood and merit further investigation.
Future studies may use techniques similar to those herein to assay other cell lines or specifically target proteins with parallel reaction monitoring (PRM) assays [51]. In addition to identifying individual candidate proteins for future studies, gene ontology analysis provided insight into the localization and functions of these proteins. Future studies may also use affinity-isolation and/or proximity labeling in the presence and absence of selected ligands to investigate the interactions of nicotinic receptors [52]. As most of the altered proteins were membrane-associated, additional studies using membrane-specific proteome analysis can examine further the differences in the membrane protein landscape of the cell upon nicotine treatment. Moreover, varying nicotine concentrations and treatment times may provide additional data concerning time- and dose-dependent nicotine-induced changes in cellular proteomes. Likewise, the multiplexing strategy itself can be expanded by incorporating the previously-published 3x3+1 arrangement to link multiple experiments [53]. In summary, our TMT-based quantitative proteomics strategy revealed nicotine-induced protein abundance differences in HAP1wt and α7KO-HAP1 cell lines. The methodologies that we used can be applied any cellular perturbation study to reveal differences in protein abundance profiles as was outlined herein.
Supplementary Material
Columns include: Uniprot protein identification number (proteinID), gene symbol (Gene Symbol), protein description/name (Description), number of peptides identified per protein (peptides), the normalized summed signal-to-noise for each of the 10 channels (126 to 131), and the log2 ratios (nicotine/null) for the summed signal-to-noise of the TMT relative abundance values for each cell type. We also include columns for the average of replicate conditions in this experiment, the log2 fold-change between comparison groups, and the associated p-values.
Columns include: Uniprot protein identification number (proteinID), gene symbol (Gene Symbol), description/name (Description), redundancy, peptide sequence (peptide sequence), number of quantified peptides (num_quant), and the summed signal-to-noise for each of the 10 channels (126 to 131).
SIGNIFICANCE.
Understanding protein function at the biomolecular level is essential to fully comprehend the intricacies of toxic stress on the cellular microenvironment. Tobacco product use has been linked to a wide array of cancers and chronic diseases. Here, we focus on a single toxin found in tobacco products, nicotine. Understanding the perturbations due to nicotine at the molecular level before marked changes appear, is paramount in diagnosing, modifying, or retarding associated diseases. Aside from its presence in tobacco, nicotine is a main component in smoking cessation products, and also increasingly popular electronic cigarettes. Growing evidence has linked nicotine to the disruption of cellular metabolic processes and its potential to be genotoxic and tumor-promoting. Nicotine has been shown to induce cell proliferation and invasion in multiple lung and breast cancer lines, and the phosphorylation states of proteins in pancreatic stellate cells. Here we show the effects of nicotine on a human haploid cell line (HAP1) and an associated nicotinic receptor-null cell line (α7KO-HAP1). These haploid cell lines offer a facile means of genetic manipulations and are valuable tools in nicotine-related research.
Acknowledgments
We would like to thank the members of the Gygi Lab at Harvard Medical School. This work was funded in part by an NIH/NIDDK grant K01 DK098285 (J.A.P.) and GM97645 (S.P.G.).
Footnotes
CONFLICT OF INTEREST
The authors have declared no conflict of interest
References
- 1.U.S. Dept. of Health and Human Services. The Health Consequences of Smoking-50 Years of Progress: A Report of the Surgeon General. Centers for Disease Control and Prevention; 2014. [Google Scholar]
- 2.National Toxicology Program, Report on carcinogens: carcinogen profiles. Vol. 12. U.S. Dept. of Health and Human Services, Public Health Service; 2011. p. 408. [PubMed] [Google Scholar]
- 3.Chowdhury P, Rayford PL. Eur J Gastroenterol Hepatol. 2000;12:869. doi: 10.1097/00042737-200012080-00006. [DOI] [PubMed] [Google Scholar]
- 4.McKee M, Capewell S. Lancet. 2015;386:1239. doi: 10.1016/S0140-6736(15)00146-4. [DOI] [PubMed] [Google Scholar]; McKee M, Chapman S, Daube M, Glantz S. Lancet. 2014;384:2107. doi: 10.1016/S0140-6736(14)62366-7. [DOI] [PubMed] [Google Scholar]
- 5.Grando SA. Nat Rev Cancer. 2014;14:419. doi: 10.1038/nrc3725. [DOI] [PubMed] [Google Scholar]
- 6.Dasgupta P, Rastogi S, Pillai S, Ordonez-Ercan D, Morris M, Haura E, Chellappan S. J Clin Invest. 2006;116:2208. doi: 10.1172/JCI28164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paulo JA, Gaun A, Gygi SP. Journal of proteome research. 2015;14:4246. doi: 10.1021/acs.jproteome.5b00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Momi N, Ponnusamy MP, Kaur S, Rachagani S, Kunigal SS, Chellappan S, Ouellette MM, Batra SK. Oncogene. 2013;32:1384. doi: 10.1038/onc.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chu KM, Cho CH, Shin VY. Curr Pharm Des. 2013;19:5. doi: 10.2174/13816128130103. [DOI] [PubMed] [Google Scholar]
- 9.Changeux JP. Nature reviews Neuroscience. 2010;11:389. doi: 10.1038/nrn2849. [DOI] [PubMed] [Google Scholar]
- 10.Al-Wadei MH, Al-Wadei HA, Schuller HM. Mol Cancer Res. 2012;10:239. doi: 10.1158/1541-7786.MCR-11-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]; Browne CJ, Sharma N, Waters KA, Machaalani R. International journal of developmental neuroscience: the official journal of the International Society for Developmental Neuroscience. 2010;28:1. doi: 10.1016/j.ijdevneu.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Paulo JA, Brucker WJ, Hawrot E. J Proteome Res. 2009;8:1849. doi: 10.1021/pr800731z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heeschen C, Weis M, Aicher A, Dimmeler S, Cooke JP. J Clin Invest. 2002;110:527. doi: 10.1172/JCI14676. [DOI] [PMC free article] [PubMed] [Google Scholar]; Macklin KD, Maus AD, Pereira EF, Albuquerque EX, Conti-Fine BM. J Pharmacol Exp Ther. 1998;287:435. [PubMed] [Google Scholar]; Sharma G, Vijayaraghavan S. Journal of neurobiology. 2002;53:524. doi: 10.1002/neu.10114. [DOI] [PubMed] [Google Scholar]
- 13.Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, Brummelkamp TR. Science (New York, NY) 2009;326:1231. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 14.Kotecki M, Reddy PS, Cochran BH. Exp Cell Res. 1999;252:273. doi: 10.1006/excr.1999.4656. [DOI] [PubMed] [Google Scholar]
- 15.Essletzbichler P, Konopka T, Santoro F, Chen D, Gapp BV, Kralovics R, Brummelkamp TR, Nijman SM, Burckstummer T. Genome Res. 2014;24:2059. doi: 10.1101/gr.177220.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Mol Cell Proteomics. 2004;3:1154. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]; Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Analytical chemistry. 2003;75:1895. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 17.Paulo JA, Urrutia R, Banks PA, Conwell DL, Steen H. J Proteomics. 2011;75:708. doi: 10.1016/j.jprot.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Paulo JA, Urrutia R, Banks PA, Conwell DL, Steen H. Journal of proteome research. 2011;10:4835. doi: 10.1021/pr2006318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yildiz D, Ercal N, Armstrong DW. Toxicology. 1998;130:155. doi: 10.1016/s0300-483x(98)00105-x. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T, Shen Y, Monroe ME, Lopez-Ferrer D, Reno T, Moore RJ, Klemke RL, Camp DG, 2nd, Smith RD. Proteomics. 2011;11:2019. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulo JA, O’Connell JD, Everley RA, O’Brien J, Gygi MA, Gygi SP. J Proteomics. 2016;148:85. doi: 10.1016/j.jprot.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ting L, Rad R, Gygi SP, Haas W. Nat Methods. 2011;8:937. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]; McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP. Analytical chemistry. 2014;86:7150. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulo JA, O’Connell JD, Gygi SP. Journal of the American Society for Mass Spectrometry. 2016;27:1620. doi: 10.1007/s13361-016-1434-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. Cell. 2010;143:1174. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elias JE, Gygi SP. Methods Mol Biol. 2010;604:55. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Elias JE, Gygi SP. Nat Methods. 2007;4:207. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 25.McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, Kuhn K, Pike I, Grothe RA, Blethrow JD, Gygi SP. Analytical chemistry. 2012;84:7469. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. Nucleic acids research. 2015;43:D447. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A. Nucleic acids research. 2016;44:W90. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manuela R, Mario M, Vincenzo R, Filippo R. Life Sci. 2015;135:49. doi: 10.1016/j.lfs.2015.04.027. [DOI] [PubMed] [Google Scholar]
- 29.Wang YY, Liu Y, Ni XY, Bai ZH, Chen QY, Zhang Y, Gao FG. Oncol Rep. 2014;31:1480. doi: 10.3892/or.2013.2962. [DOI] [PubMed] [Google Scholar]; Schaal C, Chellappan SP. Mol Cancer Res. 2014;12:14. doi: 10.1158/1541-7786.MCR-13-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benowitz NL, Hukkanen J, Jacob P., 3rd Handb Exp Pharmacol. 2009:29. doi: 10.1007/978-3-540-69248-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulcahy MJ, Blattman SB, Barrantes FJ, Lukas RJ, Hawrot E. PLoS One. 2015;10:e0134409. doi: 10.1371/journal.pone.0134409. [DOI] [PMC free article] [PubMed] [Google Scholar]; Paulo JA, Hawrot E. Anal Biochem. 2009;389:86. doi: 10.1016/j.ab.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Nature. 2002;416:535. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]; Hardy J, Selkoe DJ. Science (New York, NY) 2002;297:353. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 33.Dineley KT, Westerman M, Bui D, Bell K, Ashe KH, Sweatt JD. J Neurosci. 2001;21:4125. doi: 10.1523/JNEUROSCI.21-12-04125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang HY, Lee DH, D’Andrea MR, Peterson PA, Shank RP, Reitz AB. J Biol Chem. 2000;275:5626. doi: 10.1074/jbc.275.8.5626. [DOI] [PubMed] [Google Scholar]
- 34.Paulo JA, Urrutia R, Kadiyala V, Banks P, Conwell DL, Steen H. Proteomics. 2013;13:1499. doi: 10.1002/pmic.201200492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansel DE, Rahman A, Wehner S, Herzog V, Yeo CJ, Maitra A. Cancer research. 2003;63:7032. [PubMed] [Google Scholar]
- 36.Gutala R, Wang J, Hwang YY, Haq R, Li MD. Brain Res. 2006;1093:12. doi: 10.1016/j.brainres.2006.03.100. [DOI] [PubMed] [Google Scholar]
- 37.Inserra MC, Kompella SN, Vetter I, Brust A, Daly NL, Cuny H, Craik DJ, Alewood PF, Adams DJ, Lewis RJ. Biochem Pharmacol. 2013;86:791. doi: 10.1016/j.bcp.2013.07.016. [DOI] [PubMed] [Google Scholar]; Adams DJ, Callaghan B, Berecki G. Br J Pharmacol. 2012;166:486. doi: 10.1111/j.1476-5381.2011.01781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fotinopoulou A, Tsachaki M, Vlavaki M, Poulopoulos A, Rostagno A, Frangione B, Ghiso J, Efthimiopoulos S. J Biol Chem. 2005;280:30768. doi: 10.1074/jbc.C500231200. [DOI] [PubMed] [Google Scholar]
- 39.Diarra-Mehrpour M, Bourguignon J, Sarafan N, Bost F, Sesboue R, Muschio-Bonnet F, Martin JP. Biochim Biophys Acta. 1994;1219:551. doi: 10.1016/0167-4781(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 40.Zafar GI, Grimm EA, Wei W, Johnson MM, Ellerhorst JA. J Urol. 2009;181:1028. doi: 10.1016/j.juro.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paulo JA. JOP. 2014;15:465. doi: 10.6092/1590-8577/2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuchida K, Nakatani M, Matsuzaki T, Yamakawa N, Liu Z, Bao Y, Arai KY, Murakami T, Takehara Y, Kurisaki A, Sugino H. Mol Cell Endocrinol. 2004;225:1. doi: 10.1016/j.mce.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 43.Roy B, Bhattacharjee A, Xu B, Ford D, Maizel AL, Cathcart MK. J Leukoc Biol. 2002;72:580. [PubMed] [Google Scholar]
- 44.Yang X, Qi Y, Avercenc-Leger L, Vincourt JB, Hupont S, Huselstein C, Wang H, Chen L, Magdalou J. Biomed Mater Eng. 2017;28:S217. doi: 10.3233/BME-171644. [DOI] [PubMed] [Google Scholar]; Cucina A, Dinicola S, Coluccia P, Proietti S, D’Anselmi F, Pasqualato A, Bizzarri M. J Surg Res. 2012;178:233. doi: 10.1016/j.jss.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 45.Low LH, Chow YL, Li Y, Goh CP, Putz U, Silke J, Ouchi T, Howitt J, Tan SS. J Biol Chem. 2015;290:7141. doi: 10.1074/jbc.M114.613687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sobkowiak R, Zielezinski A, Karlowski WM, Lesicki A. Drug and chemical toxicology. 2017;40:470. doi: 10.1080/01480545.2016.1264411. [DOI] [PubMed] [Google Scholar]
- 47.Govind AP, Vallejo YF, Stolz JR, Yan JZ, Swanson GT, Green WN. Elife. 2017:6. doi: 10.7554/eLife.25651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu H, Wang F, Kranzler HR, Gelernter J, Zhang H. Sci Rep. 2017;7:41816. doi: 10.1038/srep41816. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ginzkey C, Kampfinger K, Friehs G, Kohler C, Hagen R, Richter E, Kleinsasser NH. Toxicol Lett. 2009;184:1. doi: 10.1016/j.toxlet.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Putney JW, Jr, Borzelleca JF. J Pharmacol Exp Ther. 1971;178:180. [PubMed] [Google Scholar]
- 50.Kalashnyk OM, Gergalova GL, Komisarenko SV, Skok MV. Life Sci. 2012;91:1033. doi: 10.1016/j.lfs.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 51.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Mol Cell Proteomics. 2012;11:1475. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu B, Li X, Chen H, Wang H, Zhu X, Hou H, Hu Q. Biochem Biophys Res Commun. 2017;486:971. doi: 10.1016/j.bbrc.2017.03.141. [DOI] [PubMed] [Google Scholar]
- 53.Paulo JA, McAllister FE, Everley RA, Beausoleil SA, Banks AS, Gygi SP. Proteomics. 2014 doi: 10.1002/pmic.201400154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Columns include: Uniprot protein identification number (proteinID), gene symbol (Gene Symbol), protein description/name (Description), number of peptides identified per protein (peptides), the normalized summed signal-to-noise for each of the 10 channels (126 to 131), and the log2 ratios (nicotine/null) for the summed signal-to-noise of the TMT relative abundance values for each cell type. We also include columns for the average of replicate conditions in this experiment, the log2 fold-change between comparison groups, and the associated p-values.
Columns include: Uniprot protein identification number (proteinID), gene symbol (Gene Symbol), description/name (Description), redundancy, peptide sequence (peptide sequence), number of quantified peptides (num_quant), and the summed signal-to-noise for each of the 10 channels (126 to 131).