Abstract
Aim:
Early life exposure to lead (Pb) has been shown to increase late life biomarkers involved in Alzheimer's disease (AD) pathology. Here, we tested the hypothesis that latent over expression of AD-related genes may be regulated through histone activation pathways.
Methods:
Chromatin immunoprecipitation sequencing was used to map the histone activation mark (H3K9Ac) to the mouse genome in developmentally Pb exposed mice on postnatal days 20, 270 and 700.
Results:
Exposure to Pb resulted in a global downregulation of H3K9Ac across the lifespan; except in genes associated with the Alzheimer pathway.
Discussion:
Early life exposure to Pb results in an epigenetic drift in H3K9Ac consistent with latent global gene repression. Alzheimer-related genes do not follow this trend.
Keywords: : Alzheimer's disease, epigenetics, histone acetylation, lead (Pb)
Environmental exposures have long been postulated to play a pivotal role in both promoting, as well as preventing disease. Numerous studies identify positive habits such as a healthy diet rich in polyphenols, regular exercise, caloric restriction and others as safeguarding against most cancers, metabolic syndrome and neurological disease [1,2]. Conversely, exposures from toxins and toxicants have been adversely associated with disease states. The critical window for exposure in adult diseases occurs during early development, including both perinatal and postnatal exposures. Evidence for an increased risk of disease states has been referred to as the developmental basis of disease, and has been tied to many adverse states including metabolic syndrome and neurological diseases [3–5]. Specifically in Alzheimer's disease (AD), where the vast majorities of cases are latent with no clear etiology, the environment is thought to play a major role [6]. One proposed mechanism by which the environment may alter the expression of genes and predispose to AD is via epigenetic pathways [6,7].
The developing brain is particularly susceptible to environmental exposures, as well as epigenetic modification [8]. Due to the properties of cells in the brain, such as low turnover of neurons, modifications that occur in the brain have the ability to stabilize and remain for a lifetime [9]. Epigenetic mechanisms refer to change in our genome that affect gene expression without altering the underlying DNA sequence [10]. Of these, the most commonly studied are DNA methylation and histone modifications. DNA methylation involves the addition of methyl groups to the cytosine residues of DNA. Recruitment of methyl groups to CpG islands of genes is usually involved in silencing of that particular region of the genome [11–13]. In the cell, DNA is organized around histone proteins, and histone tails can be modified by a number of post-translational modifications that will either repress or enhance gene expression [14]. Both DNA methylation and histone modifications work by recruiting other chromatin binding proteins to alter the structure of chromatin and recruit either activating or repressive elements [15,16]. Specifically, histone acetylation is involved in active gene expression, and is associated with loose binding of the histone proteins to the DNA, which in turn allows inclusion of transcriptional machinery [15,16].
Our lab has extensively shown that early life exposure to lead (Pb) can have significant impacts on behavior in mice, and that this exposure is also associated with an increase in biomarkers involved in Alzheimer's pathology [17,18]. Specifically, we have reported statistically significant increases in protein and mRNA levels of the APP and MAPT in those animals exposed to Pb [18,19]. These mice were also shown to have behavioral deficits at 18 and 24 months of age as measured by both the Morris water maze and Y-maze [17]. Recently, this same cohort was found to have alterations of proteins involved in modifying epigenetic marks such as DNA methyltransferases, as well as significant changes in epigenetic marks themselves [20].
Lifespan studies in mice also revealed changes in genome-wide gene expression and DNA methylation later in old age following developmental exposure to Pb. Methylation and gene expression profiles were overlaid and analyzed in animals at postnatal day (PND) 20 and 700 following Pb exposure. The results indicated a global drift of a repression profile of genes in old age, with a small subset of genes being overexpressed including AD-related genes [21,22]. Overall, reprogramming of gene expression influenced the genes related to immune and inflammatory functions, compromising the body's defense system to combat stressors in late age [9].
In this study, we have used a novel approach to examine H3K9Ac binding across the genome. We chose this acetylation mark because it has been widely shown to upregulate gene expression, and to be representative of methylation marks (H3K4me3 and H3K4me2) that also are indicative of active gene expression [23,24]. We performed chromatin immunoprecipitation sequencing analysis across the lifespan in animals previously exposed to Pb as infants and compared them with their age matched controls. Tissue samples from three time points, PND 20, 270 and 700, were profiled for H3K9Ac binding. The underlying hypothesis of our approach was to explore whether the small subset of latently overexpressed genes, due to early life exposure to Pb, were increased via histone activation pathways.
Materials & methods
Animal exposure
C57BL/6; wild type mice were bred in-house in the Animal Care Facility at the University of Rhode Island (URI) according to previously published protocols [20,25,26]. PND 1 was designated as 24 h after birth. Male pups from different dams were pooled and placed randomly to different dams (10 mice/dam). The mice were divided into two groups, the control group received regular tap water, and the second group (PbE, developmental exposure) was exposed to 0.2% Pb acetate from PND 1 to PND 20 through the drinking water of the dam. Brains were dissected at the following ages: PND 20, 270 and 700 were used in this study. These animals are from the subset of the same cohort that has been already characterized for Alzheimer's pathology and behavioral deficits [17]. Experiments were performed in accordance with the standard guidelines and protocol approved by the URI Institutional Animal Care and Use Committee with supervision of the university's veterinarian.
Tissue fractionation & shearing optimization
Chromatin isolation was performed using the reagents from the ChIP IT Express Kit (Cat # 53008, 53032 Active Motif, CA, USA) with modifications for working with tissue samples. All solutions were prepared according to the manufactures instructions. 100 mg of cerebellar tissue was pooled from mice (n = 5) and was homogenized in 4 ml of phosphate buffer saline (using a dounce homogenizer). The cerebellum was used due to its dense and abundant neuronal populations that would yield sufficient material for these experiments. Samples were fixed with 37% formaldehyde for 15 min at room temperature, following which fixation was halted using a glycine stop solution. Samples were lysed with ice cold lysis buffer, and the isolated nuclei were resuspended in shearing buffer.
Samples were sonicated using a Branson-sonifier 150 (CT, USA) while keeping the setting at level 2. Samples were sonicated four-times at 20 and 40 s intervals while on ice. A 50 μl aliquot of the supernatant was set aside for DNA cleanup and concentration measurement, and the rest aliquoted for future chromatin immunoprecipitation (ChIP) reactions. The DNA cleanup was performed per the manufactures instructions, Samples were run on a 2% agarose gel at 100 V for 45 min. The gel was visualized using the Amersham Typhoon Imager Scanner FLA 9000 (GE Life Sciences, NJ, USA) with the optimal fragment size between 200 and 500 bp.
Chromatin immunoprecipitation & validation
ChIP reactions were set up in 100 μl volumes as described in the ChIP IT Express Kit (Active Motif) with minor modifications, and 5 μg of ChIP grade H3K9Ac (Cat #39137 Active Motif) was incubated with 30 μg of sheared chromatin, and samples were incubated overnight on a rotating platform, at 4°C. The rest of the protocol was followed without any further modification. DNA purification was performed using the Agencourt® Ampure XP DNA Purification beads (Cat #A63880 Beckman Coulter, CA, USA). DNA was eluted using nuclease free water and quantified using the Nanodrop UV/VIS Spectrophotometer (Thermo Scientific, DE, USA). RT-PCR was performed using commercially available positive and negative control primers. A positive primer for the promoter region of GAPDH was utilized (Cat # 71018 Active Motif), the negative control primers used were for a gene desert on chromosome 6 (Cat # 71011, Active Motif) and gene desert on chromosome 17 (Cat # 71012, Active Motif). RT-PCR was performed in 20 μl reaction volumes, utilizing 10 ng of DNA, 10 μl SYBR green PCR master mix (Applied Biosystems, CA, USA) and 4 μl of the active motif primer. RT-PCR was performed using the ViiA7 Real-Time PCR System (Applied Biosystems) under the following conditions: 50°C for 2 min followed by 95°C for 10 min, then 40 cycles of 95°C for 15 s and 60°C for 1 min. Normalization of the data was performed using the ChIPed DNA of each sample against the inputs for each sample for both the positive and negative control primers using the 2-ΔΔCT method.
Library preparation, quality check & run conditions
The next generation sequencing library was prepared using the Ovation Ultralow System VS2 1–16 (Cat# 0344 NuGEN, CA, USA). The samples were prepared at the Brown University Sequencing Facility, according to the manufactures protocols. Briefly, 25 ng of ChIPed DNA was end repaired, and adapters were added to each sample and ligated to distinguish samples from one another on the flow cell. Following ligation, the samples were purified and amplified by 10 cycles of PCR and subsequently quantified using the Qubit dsDNA BR Assay Kit (Cat # Q32850 Life Technologies, CA, USA).
Fragment analysis was performed using the High Sensitivity NGS Fragment Analysis Kit (Cat# DNF-474 Advanced Analytical Technologies, IA, USA) using 2 μl of library input at a concentration of 4–6 ng/μl following the manufactures instructions. For each library 1:1000, 1:5000 and 1:10000 dilutions were made using the Kapa Library Quantification Kit (Cat #KK4873 Illumina, CA, USA), each dilution was run in triplicate on ViiA7 Real-Time PCR System (Applied Biosystems). Reaction volume of 20 μl were run under the following conditions: 95°C for 5 min, then 35 cycles of 95°C for 20 s, 60°C for 45 s. Samples were excluded from analysis if the standard deviation from the median was >0.1 Ct units. The total amount of library was determined for each library dilution using six know amplification standards with an amplification product size of 462 bp. The median concentration of the library was calculated using all three dilutions.
Samples were pooled and submitted for sequencing at 20 nM concentration to the Bauer Sequencing Facility at Harvard University. The samples were processed using the Illumina Hiseq 2500 system in rapid run mode producing 50 bp single-end reads (Illumina, CA, USA). One sample was run on two lanes of a flow cell according to standard protocol performed at the Bauer center.
Bioinformatics analysis
The raw data from chromatin immunoprecipitation sequencing was analyzed in-house at the URI by the Rhode Island IDeA Network for Excellence in Biomedical Research (INBRE) Bioinformatics and Rutgers Cores. Peak annotation was performed using the Homer program with gene ontology and genome ontology options [27]. Merged peak files were made in Homer and used for differential analysis with twofold difference and p-value < 0.01. Visualizations were made with EaSeq [28] or in R using the ChIP-seeker package [29].
Sequencing reads were aligned with mm10 mouse genome using Bowtie2 (v. 2.2.3) with default parameters and the ‘very-sensitive’ flag [30]. Alignments with a mapping quality (MAPQ) less than 10 were removed. Peaks were identified using MACS2 (v. 2.1.1.20160309) [31]. In one comparison, at each time point, the control was compared with treatment using ‘-g mm –bw 1000’ as settings. In another comparison, each sample was analyzed for peaks individually using ‘-g mm –bw 1000’. For two of the samples (AE3 and AE5), MACS2 warned about its fragment length model, so these samples were processed with ‘nomodel –extsize 400’.
Brain Pb concentration
Elemental profiling via inductively coupled plasma mass spectrometry (ICP–MS) was performed for Pb in cortical brain tissue. Samples were prepared by concentrated HNO3 and diluted in Milli-Q water to a final concentration of 2% HNO3. ICP–MS was performed using a ICAP-Q (Thermo-Fisher Scientific, MA, USA). The samples were introduced with a segmented flow technique (1.0 ml/min; 14 s for each sample) through a Meinhard nebulizer and a chilled spray chamber. No flow injection valve was used; the autosampler was simply programmed to stay in the sampling position for the specified time. The argon plasma conditions were: forward power 1350 W; reflected power <5 W; nebulizer gas flow rate 0.9, intermediate and outer gas flow rates 0.8 and 13.0 l/min, respectively. The instrument was calibrated using 2% HNO3 solution containing Pb at 100, 200 and 300 parts per billion.
Results
The Pb exposure model used in all these studies did not result in nutritional disturbances, or any overt changes in brain structure or anatomy in wild-type mice [17]. The estimated Pb concentration by ICP–MS at PND 20 in mice brain developmentally exposed to Pb was 1.26 μg/g of wet weight and in unexposed mice, an average of 0.13 μg/g of wet weight was measured in unexposed mice. On PND 700, the mean Pb concentration was 0.07 μg/g in unexposed mice, and 0.21 μg/g Pb in exposed ones.
Histone peak distribution
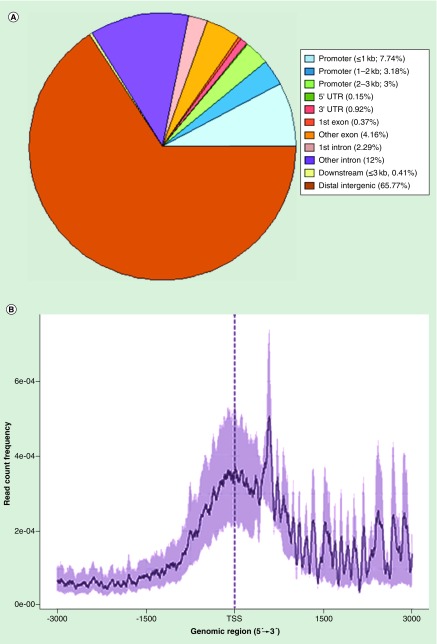
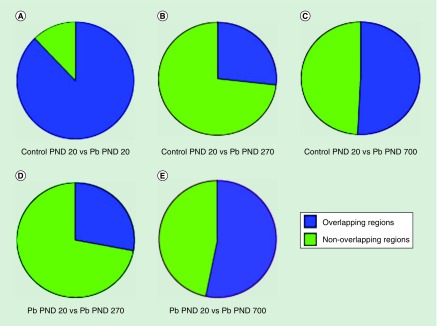
Our acetylation peaks tended to cluster within 3000 bp in either direction of the transcription start site of the promoter region of genes, with approximately 13% of our peaks found within the promoters of most genes (Figure 1). Pie graphs were generated for overlapping genomic regions with peaks between the samples (Figure 2). Regions with histone acetylation peaks were relatively unchanged between the control and developmentally exposed animals at PND 20 (Figure 2A). The vast majority of peaks among the day 20 time-points were overlapping. Surprisingly, Pb exposed PND 270 time points had the greatest difference in peak distribution when compared with the early PND 20 time points. The control day 20 (Figure 2B) and Pb day 20 (Figure 2D) against Pb day 270 had a major shift in acetylation peaks, with almost 75% of the regions presenting as unique and not overlapping between the samples. At last, Pb exposed PND 700 had approximately 50% nonoverlapping regions with both control PND 20 (Figure 2C) and Pb exposed PND 20 (Figure 2E). These results magnify the differences between the midlife (270) and late life (700) exposed animals across time points, against both day 20 samples (Pb exposed and control). They also highlight the minimal changes between PND 20 time points regardless of exposure.
Figure 1. . Distribution of histone peak regions within the genome.
(A) Displays the data in pie chart format, indicating the percentage of peaks obtained from the chromatin immunoprecipitation sequencing experiment in different genomic regions. (B) When examining 2000 base pairs in either direction of aggregated TSS, acetylation peak intensity tends to cluster around the TSS and promoter region.
TSS: Transcription start site.
Figure 2. . Overlapping H3K9Ac peak regions across the lifespan.
Figures display both overlapping and unique peaks among control and Pb exposed animals. (A) Control PND 20 versus Pb PND 20. (B) Control PND 20 versus Pb exposed PND 270. (C) Control PND 20 versus Pb exposed PND 700. (D) Pb exposed PND 20 versus Pb exposed PND 270. (E) Pb exposed PND 20 versus Pb exposed PND 700.
Pb: Lead; PND: Postnatal day.
Drift in histone acetylation peaks
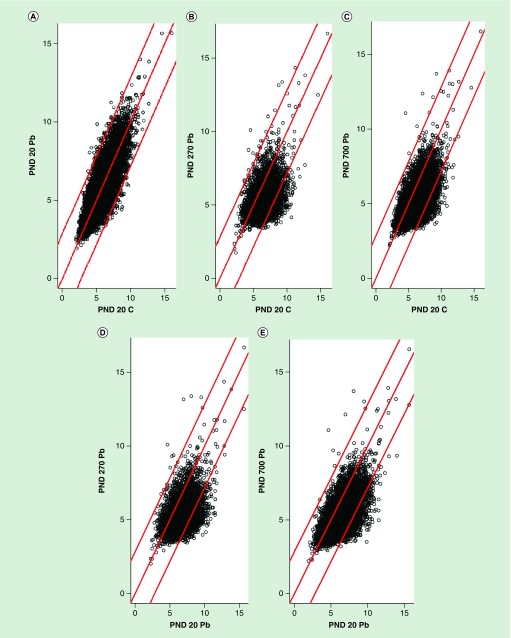
Differential scatter plots were created across the lifespan for control and Pb exposed animals (Figure 3). A relatively equal pattern was observed for PND 20 controls and PND 20 Pb exposed animals, indicating there is little or no difference in peak size between the two samples. However, when examining control PND 20 versus Pb exposed PND 270 (Figure 3B) and Pb exposed PND 700 (Figure 3C), displayed a downward shift indicating a greater peak size for the control PND 20 animals against both samples. Similar results were found when examining PND 20 Pb exposed animals against both PND 270 (Figure 3D) and PND 700 (Figure 3E) Pb exposed animals.
Figure 3. . Differential histone acetylation.
Scatterplots display raw number of acetylation sites between the samples. The red basal line indicates where the values are equal between samples. (A) PND 20 control against PND 20 Pb exposed animals. (B) PND 20 control against PND 270 Pb exposed animals. (C) PND 20 control against PND 700 Pb exposed animals. (D) PND 20 Pb exposed against PND 270 Pb exposed animals. (E) PND 20 Pb exposed against PND 700 Pb exposed animals.
Pb: Lead; PND: Postnatal day.
Histone acetylation peak intensity levels
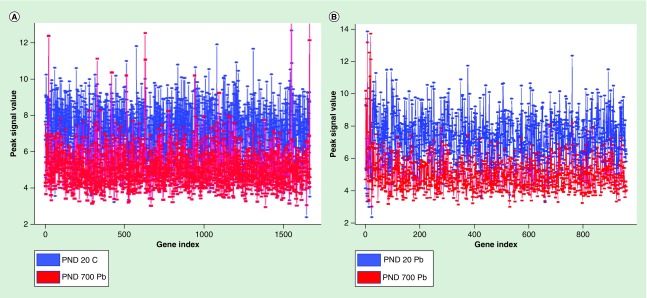
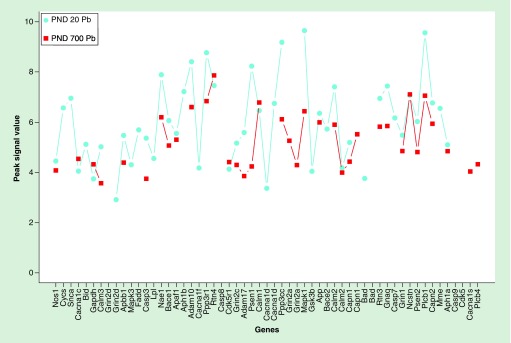
H3K9Ac peak signal levels were plotted for intensity for about 2000 genes (Figure 4). Peak intensity was much higher in control animals at PND 20 compared with developmentally exposed animals at PND 700 (Figure 4A), as well as Pb exposed animals at PND 20 compared with Pb exposed animals at PND 700 (Figure 4B). All but a few peaks reflected lower values for PND 700, which was consistent with global hypoacetylation of these brain samples. Furthermore, peak intensity values were plotted for a subset of genes from the ko0510 pathway, defined by the Kyoto Encyclopedia of Genes and Genomes database as genes involved in the pathogenesis of AD (Figure 5). In these results, we report PND 20 Pb exposed animals with higher peak intensity values compared with PND 700 Pb exposed animals. These specific genes were not globally hyperacetylated in late life exhibiting a different and minimally altered acetylation profile. All genes with >twofold difference between samples have been provided in the Supplementary Data.
Figure 4. . Line plot of common peak intensities with >twofold difference.
(A) Blue lines represent PND 20 control peak intensity values and red lines represent PND 700 Pb exposed peak intensity values. (B) Blue lines represent PND 20 Pb exposed peak intensity values and red lines represent PND 700 Pb exposed peak intensity.
Pb: Lead; PND: Postnatal day.
Figure 5. . Line plot of Ko5010 pathway genes with >twofold difference.
The turquoise points represent peak signal values of Pb exposed animals at PND 20, whereas the red points represent Pb exposed animals at PND 700.
Pb: Lead; PND: Postnatal day.
Discussion
ChIP sequencing is a powerful tool used to identify binding sites of transcription factors or histone marks to DNA regions. It is also the preferred method when working with larger genomes such as the human or mouse genome [32]. The design and sequence run of this experiment was specifically chosen based on the number of reads required to detect significant changes in histone marks in the genome. The Encyclopedia of DNA Elements (ENCODE) consortia has identified the appropriate number of reads for mapping histone marks at a threshold greater than 10 million reads per sample [33]. This experiment yielded between 17 and 19 million reads per sample for H3K9Ac, a mark well known for its role in activating gene expression.
The global finding in these studies is that H3K9Ac mark on the DNA is diminished over time following developmental exposure to Pb. The scatter plots covering the entire genome also show a time-dependent epigenetic drift in this mark (Figure 3) that is evident in early life and becomes more prominent at the end of life. This is consistent with the decreases in total protein levels of H3K9Ac, reported by us [20]. Furthermore, this global hypoacetylation and diminution of this mark on the genome is consistent with the global drift of gene repression and hypermethylation outcomes previously reported by us [21,22].
These genomic and epigenomic reference maps suggest that the overall impact of early life exposure to Pb is to repress genes in old age; however, select sets of genes appear to have altered expression not directly related to DNA methylation, or histone acetylation, and may involve other epigenetic pathways. Among these genes are those involved in AD pathogenesis for which we have extensively reported their latent induction in both mice and primates [19,34–36].
Here, we utilized the ko0510 to identify changes in our AD-related genes in animals that were developmentally exposed in late life, compared with PND 20. The Kyoto Encyclopedia of Genes and Genomes genomic database links genes and genetic information to higher order functional categories, such as molecular function and disease pathology [37,38]. Specifically, the ko0510 pathway identified 162 genes that have been associated with AD pathology. These genes are involved in amyloid precursor protein processing, Tau phosphorylation, as well as mitochondrial and calcium dysfunction and endoplasmic reticulum stress. We expected that histone activation might play a role in the latent expression of these genes previously reported by us [21,22]. However, the H3K9Ac histone activation map (Figure 5) did not correspond to the latent activation response of these genes to developmental Pb exposure. Since neither DNA methylation not histone activation seems to map to the latent expression of these AD-related genes, we believe that miRNA or other epigenetic mechanisms may be playing a major role in regulating the expression of App, tau and other genes in these wild-type mice [39].
We used the cerebellum, because it is packed with cells and thus would give a much better signal to noise ratio to validate these findings using Q-PCR. We understand that in humans, the cerebellum is relatively spared in terms of AD pathology, however, all the AD-related genes are expressed here and regulated in the same way. The objective here was not to relate pathology to target effects but rather to show that the Pb can act on its target in the same manner everywhere in the brain and that the epigenetic machinery can reprogram gene expression.
In conclusion, early life exposure to Pb results in a lifetime reprogramming of gene expression resulting in a global repression profile that may be mediated via both histone modification and DNA methylation pathways. Alzheimer related genes do not appear to undergo such reprogramming and their latent overexpression maybe mediated by alternate epigenetic pathways.
Summary points.
Most Alzheimer's disease (AD) cases have no clear genetic etiology; it is believed environmental exposure may substantially increase AD risk, specifically via epigenetic mechanisms.
We have previously shown in both mice and primates that developmental exposure to lead (Pb) increases biomarkers involved in AD pathology in late life.
In this manuscript, we test the hypothesis that latent over expression of AD-related genes may be regulated through histone activation pathways.
Chromatin immunoprecipitation sequencing was utilized to map the histone activation mark (H3K9Ac) to the mouse genome in developmentally Pb exposed mice on postnatal days 20, 270 and 700.
Our global findings suggest that early exposure to Pb is associated with a decrease in H3K9Ac over time, consistent with the latent global gene repression in late life.
Utilizing the ko0510 pathway, we examined 162 genes which have been associated with various aspects of AD pathology, including amyloid and tau pathology, neuroinflammation, as well as mitochondrial, calcium dysfunction and endoplasmic reticulum stress.
Alzheimer related genes do not appear to be upregulated by a reprogramming of H3K9Ac, and are likely to be mediated by different epigenetic pathways such as miRNA regulation.
These data suggest the global impact of early life exposure is to suppress genes in old age; this does not include genes involved in amyloid or tau pathology.
Supplementary Material
Acknowledgements
The authors would like to thank C Schorl of Brown University for his oversight and guidance with the DNA library preparation and B Renehan of the Ryan Institute at URI for advice and support given to A Eid.
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: https://www.futuremedicine.com/doi/suppl/10.2217/epi-2017-0143
Financial & competing interests disclosure
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, and by grant 5RO1ES015867-03. This research is based in part upon work conducted using the Rhode Island IDeA Network for Excellence in Biomedical Research Bioinformatics Core which is supported by the NIH under grant 2P20GM103430. JM Gaspar and RP Hart were supported by NIH grants 1R01AT009152 to A-N Kong of Rutgers University and 1R01ES026057 and R21DA035594 to RP Hart. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Ravussin E, Redman LM, Rochon J, et al. A 2-year randomized controlled trial of human caloric restriction: feasibility and effects on predictors of health span and longevity. J. Gerontol. A Biol. Sci. Med. Sci. 2015;70(9):1097–1104. doi: 10.1093/gerona/glv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Habauzit V, Morand C. Evidence for a protective effect of polyphenols-containing foods on cardiovascular health: an update for clinicians. Ther. Adv. Chronic Dis. 2012;3(2):87–106. doi: 10.1177/2040622311430006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Portha B, Fournier A, Kioon MD, Mezger V, Movassat J. Early environmental factors, alteration of epigenetic marks and metabolic disease susceptibility. Biochimie. 2014;97:1–15. doi: 10.1016/j.biochi.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Modgil S, Lahiri DK, Sharma VL, Anand A. Role of early life exposure and environment on neurodegeneration: implications on brain disorders. Transl. Neurodegener. 2014;3:9. doi: 10.1186/2047-9158-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanner CM, Goldman SM, Ross GW, Grate SJ. The disease intersection of susceptibility and exposure: chemical exposures and neurodegenerative disease risk. Alzheimers Dement. 2014;10(3 Suppl.):S213–S225. doi: 10.1016/j.jalz.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Lunnon K, Mill J. Epigenetic studies in Alzheimer's disease: current findings, caveats, and considerations for future studies. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2013;162b(8):789–799. doi: 10.1002/ajmg.b.32201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic mechanisms in Alzheimer's disease. Neurobiol. Aging. 2011;32(7):1161–1180. doi: 10.1016/j.neurobiolaging.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perera F, Herbstman J. Prenatal environmental exposures, epigenetics, and disease. Reprod. Toxicol. 2011;31(3):363–373. doi: 10.1016/j.reprotox.2010.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zawia NH. Unique aspects of the epigenetic code in the brain. Epigenomics. 2017;9(9):1157–1159. doi: 10.2217/epi-2017-0069. [DOI] [PubMed] [Google Scholar]; •• This commentary highlights epigenetic changes in the brain, specifically as to why the brain is more susceptible to these changes compared with other systems of the body, and its vulnerability to environmental exposures.
- 10.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23(7):781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 12.Ehrlich M, Gama-Sosa MA, Huang LH, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11(1):327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta. 2014;1839(8):627–643. doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espada J, Esteller M. Epigenetic control of nuclear architecture. Cell. Mol. Life Sci. 2007;64(4):449–457. doi: 10.1007/s00018-007-6358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014;15(11):703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 17.Bihaqi SW, Bahmani A, Subaiea GM, Zawia NH. Infantile exposure to lead and late-age cognitive decline: relevance to AD. Alzheimers Dement. 2014;10(2):187–195. doi: 10.1016/j.jalz.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bihaqi SW, Bahmani A, Adem A, Zawia NH. Infantile postnatal exposure to lead (Pb) enhances tau expression in the cerebral cortex of aged mice: relevance to AD. Neurotoxicology. 2014;44:114–120. doi: 10.1016/j.neuro.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basha MR, Wei W, Bakheet SA, et al. The fetal basis of amyloidogenesis: exposure to lead and latent overexpression of amyloid precursor protein and beta-amyloid in the aging brain. J. Neurosci. 2005;25(4):823–829. doi: 10.1523/JNEUROSCI.4335-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Original paper which identifies latent increases in the APP protein and mRNA in developmentally exposed animals.
- 20.Eid A, Bihaqi SW, Renehan WE, Zawia NH. Developmental lead exposure and lifespan alterations in epigenetic regulators and their correspondence to biomarkers of Alzheimer's disease. Alzheimer's Dementia. 2016;2:123–131. doi: 10.1016/j.dadm.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A research paper utilizing mice from the same cohort utilized in this study, providing evidence for a decrease in H3K9Ac at PND 700 in animals developmentally exposed to lead (Pb).
- 21.Alashwal H, Dosunmu R, Zawia NH. Integration of genome-wide expression and methylation data: relevance to aging and Alzheimer's disease. Neurotoxicology. 2012;33(6):1450–1453. doi: 10.1016/j.neuro.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dosunmu R, Alashwal H, Zawia NH. Genome-wide expression and methylation profiling in the aged rodent brain due to early-life Pb exposure and its relevance to aging. Mech. Ageing Dev. 2012;133(6):435–443. doi: 10.1016/j.mad.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A research study profiling gene expression changes and DNA methylation status in old age following developmental exposure to Pb.
- 23.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong X, Greven MC, Kundaje A, et al. Modeling gene expression using chromatin features in various cellular contexts. Genome Biol. 2012;13(9):R53. doi: 10.1186/gb-2012-13-9-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bihaqi SW, Bahmani A, Subaiea GM, Zawia NH. Infantile exposure to lead and late-age cognitive decline: relevance to AD. Alzheimers Dement. 2013;10(2):187–195. doi: 10.1016/j.jalz.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bihaqi SW, Bahmani A, Adem A, Zawia NH. Infantile postnatal exposure to lead (Pb) enhances tau expression in the cerebral cortex of aged mice: relevance to AD. Neurotoxicology. 2014;44:114–120. doi: 10.1016/j.neuro.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Original paper which identified latent increases in tau protein and mRNA in developmentally exposed animals.
- 27.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat. Struct. Mol. Biol. 2016;23(4):349–357. doi: 10.1038/nsmb.3180. [DOI] [PubMed] [Google Scholar]
- 29.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31(14):2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 30.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 33.Landt SG, Marinov GK, Kundaje A, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22(9):1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dosunmu R, Wu J, Adwan L, et al. Lifespan profiles of Alzheimer's disease-associated genes and products in monkeys and mice. J. Alzheimers Dis. 2009;18(1):211–230. doi: 10.3233/JAD-2009-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J, Basha MR, Brock B, et al. Alzheimer's disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): evidence for a developmental origin and environmental link for AD. J. Neurosci. 2008;28(1):3–9. doi: 10.1523/JNEUROSCI.4405-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bihaqi SW, Huang H, Wu J, Zawia NH. Infant exposure to lead (Pb) and epigenetic modifications in the aging primate brain: implications for Alzheimer's disease. J. Alzheimers Dis. 2011;27(4):819–833. doi: 10.3233/JAD-2011-111013. [DOI] [PubMed] [Google Scholar]
- 37.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Details the development of Kyoto Encyclopedia of Genes and Genomes and describes the tool which we utilized to examine genes identified as being involved or important in Alzheimer's.
- 38.Kanehisa M. The KEGG database. Novartis Found. Symp. 2002;247:91–101. [PubMed] [Google Scholar]
- 39.Masoud AM, Bihaqi SW, Machan JT, Zawia NH, Renehan WE. Early-life exposure to lead (Pb) alters the expression of microRNA that target proteins associated with Alzheimer's disease. J. Alzheimers Dis. 2016;51(4):1257–1264. doi: 10.3233/JAD-151018. [DOI] [PubMed] [Google Scholar]; • Supports the potential involvement of miRNA as participating in the mechanism of regulating the Alzheimer's disease related genes investigated within the manuscript.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.