Abstract
Astrocyte- and microglia-targeting peptides were identified and isolated using phage display technology. A series of procedures, including three cycles of both in vivo and in vitro biopanning, was performed separately in astrocytes and in M1 or M2 microglia, yielding 50–58 phage plaques in each cell type. Analyses of the sequences of this collection identified one candidate homing peptide targeting astrocytes (AS1[C-LNSSQPS-C]) and two candidate homing peptides targeting microglia (MG1[C-HHSSSAR-C] and MG2[C-NTGSPYE-C]). To determine peptide specificity for the target cell in vitro, each peptide was synthesized and introduced into the primary cultures of astrocytes or microglia. Those peptides could bind to the target cells and be selectively taken up by the corresponding cell, namely, astrocytes, M1 microglia, or M2 microglia. To confirm cell-specific gene delivery to M1 microglia, the complexes between peptide MG1 and siRNA-interferon regulatory factor 5 were prepared and intrathecally injected into a mouse model of neuropathic pain. The complexes successfully suppressed hyperalgesia with high efficiency in this neuropathic pain model. Here, we describe a novel gene therapy for the treatment neuropathic pain, which has a high potential to be of clinical relevance. This strategy will ensure the targeted delivery of therapeutic genes while minimizing side effects to non-target tissues or cells.
Keywords: homing peptides, gene delivery, microglia, interferon regulatory factor 5, IRF, astrocyte
Introduction
Homing peptides that selectively target many kinds of tissues have been identified and have received a great deal of attention for their potential application in cell-specific or tissue-specific delivery of drugs and genes by coupling and assembling into novel complexes.1, 2 These peptides are very short and typically consist of only 3 to 12 amino acids.1, 3 However, these peptides exhibit strong affinities and unique specificities against target ligands, such as the Arg-Gly-Asp (RGD) motif against αv-integrins.4, 5 By insertion of the homing peptides into viral vectors, tissue-specific targeting viral vectors can be produced for use in gene therapy and peptide-mediated drug delivery systems.6, 7, 8, 9 These tissue-specific viral vectors provide high transduction efficiency, and infected cells undergo a 100-fold increase in division efficiency of the objective genes.9 These homing peptides were identified by in vivo or in vitro phage display screening.10 M13 filamentous phages are used as the platform of the library, and they present short random peptides into a minor coat protein (pIII).10 The specific cell- or tissue-binding peptides are isolated by a binding-selection procedure, referred to as biopanning with phage.2
We had previously identified cell-specific homing peptides to target neurons in the dorsal root ganglion (DRG) in mice.11 Three kinds of peptides homing to DRG were lined up and recognized the different sizes of the neurons.11 Furthermore, those peptides were inserted into helper-dependent adenovirus vectors, known as gutless adenoviral vectors, and developed for clinical use. Building upon these findings, we established a novel technology of DRG-targeted tissue-specific gene therapy.9 Thus, homing peptides have a high potential for applicability toward, and being a powerful tool for, drug and gene delivery.
Here, phage display technology was applied to identify specific peptide motifs that recognized astrocytes and microglia. A combination of in vivo phage display in the spinal cord of mice and in vitro phage display in cultured cells identified peptides of interest that were then combined with small interfering RNA (siRNA) oligonucleotides for testing the capability of therapeutic gene delivery. Astrocytes and microglia in the spinal cord are potential targets for the treatment of many neurological diseases, such as motor neuron disease, spinal injury, spastic paraplegia, multiple sclerosis, sensory ataxia, and neuropathic pain.12, 13, 14, 15, 16, 17, 18
Studies have shown that interferon regulatory factor 5 (IRF5) is involved in the pathogenesis of neuropathic pain.19, 20 Specifically, IRF5 plays an important role in the pathogenesis of tactile allodynia induced by nerve injury, but not in that of allodynia involved with general sensations, such as thermal or movement allodynia.20 IRF5 is mainly expressed in M1 microglia21 and is upregulated by spinal nerve injury, which induces the expression of ATP receptors, such as the P2X4 receptor, to activate microglia and signal neuropathic pain in the spinal cord.19, 20 Therefore, we hypothesized that downregulation of IRF5 expression in microglia will lead to a reduction in neuropathic pain. With the ability to achieve targeted delivery of therapeutic genes to microglia in the spinal cord, homing peptides are considered powerful tools with potential for the discovery and design of diagnostic agents and novel therapeutics.
In this study, homing peptides to astrocytes and microglia were identified. Delivery of siRNA for the IRF5 gene by these homing peptides highlighted their potential application in the treatment of disease when combined with therapeutic siRNA oligonucleotides.
Results
Phage Display Screening of Homing Peptides Targeting Astrocytes and Microglia
After three rounds of in vivo phage display in mice and three rounds of in vitro phage display using KT-5 cells (astrocytes), 6-3 cells (M1; pro-inflammatory microglia), or Ra2 cells (M2; anti-inflammatory microglia), the DNA sequences were analyzed in phages with high affinities for astrocytes or microglia (Figure 1). The homing peptides targeting astrocytes were detected in six types of sequences (AS1–AS6) after three in vivo and three in vitro pannings. The AS1(C-LNSSQPS-C) peptide was the most frequent in those six types of homing peptides and was observed in 48 of the 58 phage plaques (frequency, 83%) (Table 1). In total microglia, 55 types of homing peptides were identified by the phage display screening. Thirty-one homing peptides (microglia-specific peptide [MG] 1–MG31) recognized M1-type microglia, and 30 peptides (MG1–MG3, MG8–MG10, and MG32–MG55) recognized M2-type microglia. In addition, 6 homing peptides (MG1–MG3 and MG8–MG10) were observed in both M1- and M2-type microglia (Table 1). The MG1(C-HHSSSAR-C) peptide was observed in 13 of the 51 phage plaques, most frequently in M1-type microglia, and was concurrently observed in 7 of the 50 phage plaques in M2-type microglia (Table 1). The next most frequent peptide sequence was MG2(C-NTGSPYE-C), which was present in 3 of the 51 plaques in M1-type microglia, was recognized at the highest frequency in M2-type microglia, and was observed in 8 of the 50 phage plaques (Table 1). The AS3(C-RGATPMS-C) peptide was present in 2 of the 58 phage plaques in astrocytes; it was observed in 1 of the 51 phage plaques in M1-type microglia and 1 of the 50 phage plaques in M2-type microglia. This peptide was also listed as MG10 in microglia homing peptides. Among all of them, AS1, MG1, and MG2 were strongly considered candidate homing peptides for targeting astrocytes, M1 microglia, and M2 microglia, respectively, and these three peptides were used for the additional analyses.
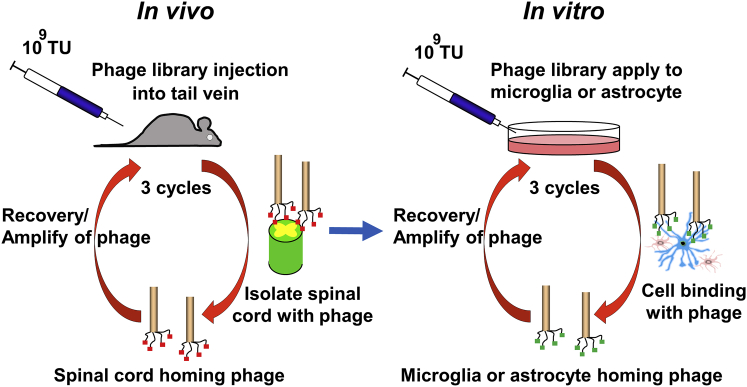
Figure 1.
Scheme for Screening Homing Peptides Targeting Astrocytes or Microglia
In vivo (left) and in vitro (right) screening of homing peptides with phage display methods. After three rounds of in vivo panning, three additional cycles of in vitro panning were performed. TU, titer unit.
Table 1.
Amino Acid Sequence of Astrocytes and Microglia (M1- and M2-Type)-Specific Homing Peptides and the Frequency of Phage Plaque
| Amino-Acid Sequence | Frequency/Total | ||
|---|---|---|---|
| Astrocyte Homing Peptides | |||
| AS1 | C-LNSSQPS-C | 48/58 | |
| AS2 | C-TMSAASH-C | 5/58 | |
| AS3 | C-RGATPMS-C | 2/58 | |
| AS4 | C-FPSHNMH-C | 1/58 | |
| AS5 | C-NHQPSEG-C | 1/58 | |
| AS6 | C-ASPFLCP-C | 1/58 | |
| Microglia Homing Peptides | |||
| M1 Type | M2 Type | ||
| MG1 | C-HHSSSAR-C | 13/51 | 7/50 |
| MG2 | C-NTGSPYE-C | 3/51 | 8/50 |
| MG3 | C-QTQNRSA-C | 3/51 | 4/50 |
| MG4 | C-ANATCPA-C | 2/51 | 0/50 |
| MG5 | C-GSKNSAV-C | 2/51 | 0/50 |
| MG6 | C-NGGTPNR-C | 2/51 | 0/50 |
| MG7 | C-LKLGEKW-C | 2/51 | 0/50 |
| MG8 | C-HHRAGVL-C | 1/51 | 2/50 |
| MG9 | C-HNETQKM-C | 1/51 | 1/50 |
| MG10 | C-RGATPMS-C | 1/51 | 1/50 |
| MG11 | C-DLNSDTQ-C | 1/51 | 0/50 |
| MG12 | C-DNPDRRK-C | 1/51 | 0/50 |
| MG13 | C-DDADQSR-C | 1/51 | 0/50 |
| MG14 | C-DFPSRGQ-C | 1/51 | 0/50 |
| MG15 | C-DGHDQSL-C | 1/51 | 0/50 |
| MG16 | C-EHSRPMS-C | 1/51 | 0/50 |
| MG17 | C-GPMSSKS-C | 1/51 | 0/50 |
| MG18 | C-LDLPNAM-C | 1/51 | 0/50 |
| MG19 | C-LNSRQPS-C | 1/51 | 0/50 |
| MG20 | C-MAPHSRV-C | 1/51 | 0/50 |
| MG21 | C-MCNSFTW-C | 1/51 | 0/50 |
| MG22 | C-NDSDTYT-C | 1/51 | 0/50 |
| MG23 | C-NTDAHRA-C | 1/51 | 0/50 |
| MG24 | C-QGERWMQ-C | 1/51 | 0/50 |
| MG25 | C-QQSMDPA-C | 1/51 | 0/50 |
| MG26 | C-SQLPWYS-C | 1/51 | 0/50 |
| MG27 | C-SDARSPK-C | 1/51 | 0/50 |
| MG28 | C-THGLTAS-C | 1/51 | 0/50 |
| MG29 | C-TQSSAMS-C | 1/51 | 0/50 |
| MG30 | C-TAKGLQA-C | 1/51 | 0/50 |
| MG31 | C-TCNQMAG-C | 1/51 | 0/50 |
| MG32 | C-MARYMSA-C | 0/51 | 2/50 |
| MG33 | C-PNSTHRN-C | 0/51 | 2/50 |
| MG34 | C-TAHDTNS-C | 0/51 | 2/50 |
| MG35 | C-DLLHRGA-C | 0/51 | 1/50 |
| MG36 | C-DFPKLRP-C | 0/51 | 1/50 |
| MG37 | C-EFSKFRS-C | 0/51 | 1/50 |
| MG38 | C-GKTSENS-C | 0/51 | 1/50 |
| MG39 | C-HDLNGSM-C | 0/51 | 1/50 |
| MG40 | C-IASVDSK-C | 0/51 | 1/50 |
| MG41 | C-IGNSNTL-C | 0/51 | 1/50 |
| MG42 | C-LGRTNGQ-C | 0/51 | 1/50 |
| MG43 | C-NRSQQPW-C | 0/51 | 1/50 |
| MG44 | C-PNMTNQW-C | 0/51 | 1/50 |
| MG45 | C-QFSKFRS-C | 0/51 | 1/50 |
| MG46 | C-QRATPGH-C | 0/51 | 1/50 |
| MG47 | C-RSANIYT-C | 0/51 | 1/50 |
| MG48 | C-SNSSLTH-C | 0/51 | 1/50 |
| MG49 | C-SPRPTQT-C | 0/51 | 1/50 |
| MG50 | C-SWQIGGN-C | 0/51 | 1/50 |
| MG51 | C-TTINQKV-C | 0/51 | 1/50 |
| MG52 | C-VPNLQGT-C | 0/51 | 1/50 |
| MG53 | C-WTDANRD-C | 0/51 | 1/50 |
| MG54 | C-YTDANRD-C | 0/51 | 1/50 |
| MG55 | C-YPHNTPN-C | 0/51 | 1/50 |
High Affinities and Specificities of AS1, MG1, and MG2 Peptides Targeting Cultured Cells
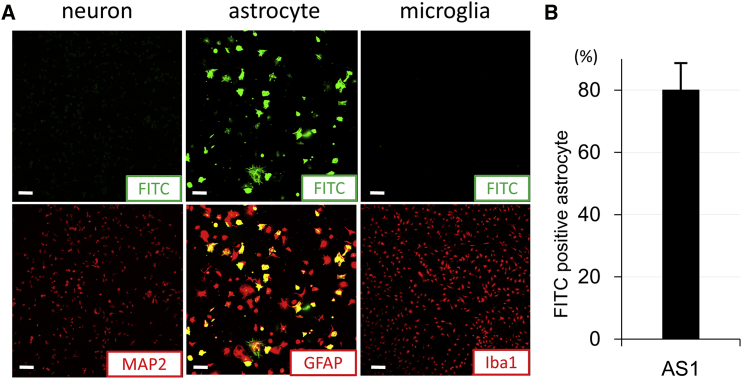
To determine the specificities of AS1, MG1, and MG2 against target cells, each synthesized peptide labeled with fluorescein isothiocyanate (FITC) was individually introduced into NSC-34 cells (neuron), primary cultured astrocytes, and microglia at a concentration of 1 μg/mL for 24 hr. AS1 peptides overlapped with glial fibrillary acidic protein (GFAP) immunostaining and were only observed in astrocytes, but not with MAP2 staining in neurons or Iba1 staining in microglia (Figure 2A). Among the GFAP-positive astrocytes, 80% were FITC positive, including the cells labeled in the partial area or only small spots (Figures 2A and 2B). 20% of GFAP-positive cells were not labeled with FITC. In addition, in those astrocytes labeled with AS1 peptides, various staining patterns were observed.
Figure 2.
Characterization of the Cell Type Targeted by AS1 Peptides
(A) The distribution of FITC-labeled, astrocyte-specific peptides (AS1; green) with MAP2 immunocytochemistry (red) in NSC-34 cells (neurons), with GFAP immunocytochemistry (red) in primary culture of astrocytes, and with Iba1 immunocytochemistry (red) in the primary culture of microglia after 24 hr of incubation with AS1 peptides (1 μg/mL). (B) The percentage of FITC-positive astrocytes binding to an astrocyte-specific peptide (AS1) in all GFAP-positive cells (red) (n = 6). Scale bars, 100 μm in (A). Error bar, mean + SD in (B).
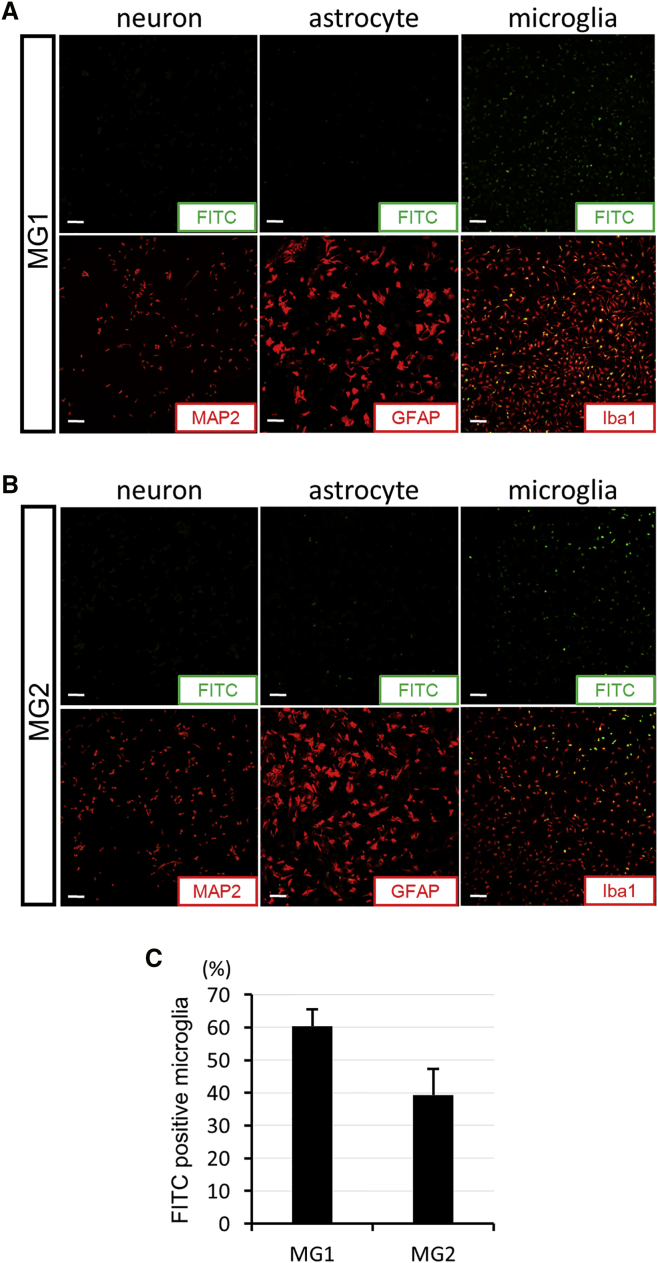
The affinities of MG1 and MG2 peptides are presented in Figure 3. MG1 peptides were mainly present in Iba1-positive microglia, and approximately 60% of microglia were labeled with MG1 peptides (Figures 3A and 3C). FITC-positive cells were not observed in NSC-34 cells or astrocytes upon MG1 administration. In the MG2 condition, the same pattern was observed. FITC fluorescence was observed only in the primary cultured microglia, not in the NSC-34 cells or astrocytes (Figure 3B). The population of MG2-labeled microglia was approximately 40%, which was lower than that in MG1 peptides (Figure 3C).
Figure 3.
Characterization of the Cell Type Targeed by MG1 and MG2 Peptides
(A and B) The distribution of FITC-labeled microglia-specific peptides (MG1 in A and MG2 in B; green) with MAP2 immunocytochemistry (red) in NSC-34 cells (neurons), with GFAP immunocytochemistry (red) in primary culture of astrocytes, and with Iba1 immunocytochemistry (red) in primary culture of microglia after 24 hr of incubation with MG1 or MG2 peptides (1 μg/mL). (C) The percentage of FITC-positive microglia binding with microglia-specific peptides (MG1 or MG2; n = 6 in each) in all Iba1-positive cells (red). Scale bars, 100 μm in (A) and (B). Error bars, mean + SD in (C).
Characterization of Cell Types Targeted by MG1 and MG2 Peptides
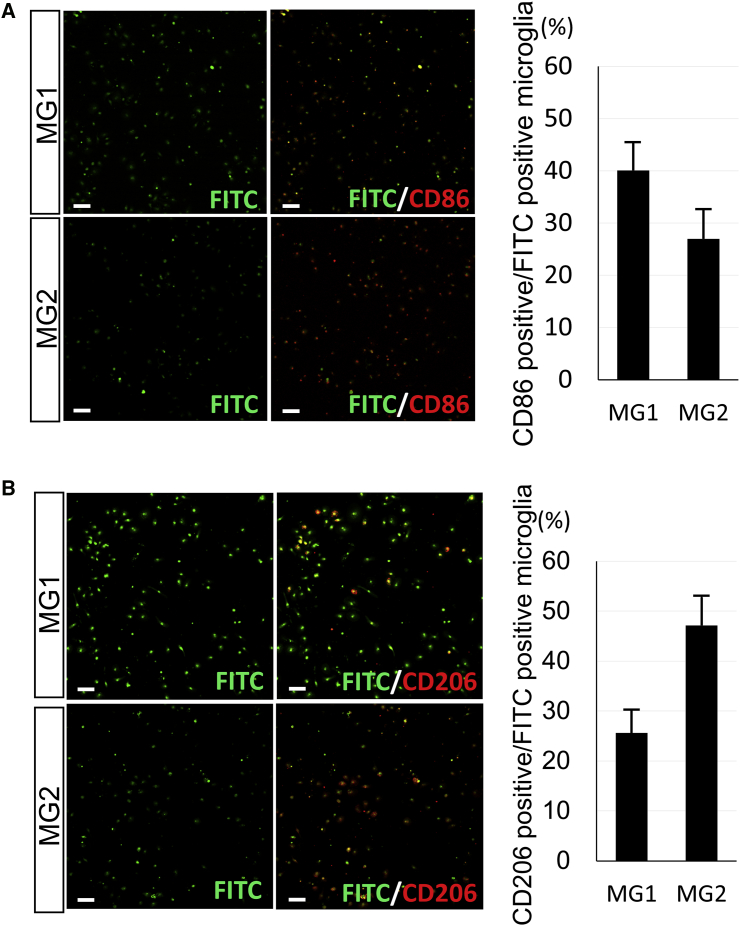
After introducing MG1 and MG2 peptides to microglia, immunofluorescent experiments were performed with anti-CD86 (M1) and anti-CD206 (M2) antibodies to identify the cell type labeled with each peptide. CD86 (M1)- and CD206 (M2)-positive cells were localized in total MG1 peptide-labeled microglia at rates of 40% and 26%, respectively (Figure 4). In addition, CD86 (M1) and CD206 (M2) were observed in 27% and 47% of MG2 peptide-positive cells, respectively (Figure 4). MG1 and MG2 peptides were both able to label M1- and M2-type cells; however, the MG1 peptide had greater binding affinity for CD86 (M1)-positive cells, and the MG2 peptide was predominantly observed in CD206 (M2)-positive cells (Figure 4). The difference in frequency between MG1 and MG2 peptides is potentially useful for cell-specific targeting in gene delivery or the development of imaging probes.
Figure 4.
Characterization of Microglia Targeted by MG1 and MG2 Peptides
Left: immunocytochemistry of CD86 (A) and CD206 (B) antibodies (red) in the primary microglia culture binding with FITC-labeled MG1 or MG2 (green) after 24 hr of incubation with the peptides (1 μg/mL). Right: the percentage of CD86-positive (A) or CD206-positive (B) cells (red) in the microglia labeled with MG1 or MG2 peptides (n = 6 in each). Scale bars, 100 μm. Error bars, mean + SD. MG, microglia-specific peptides.
In Vivo Targeting of the Spinal Cord by AS1, MG1, and MG2 Peptides
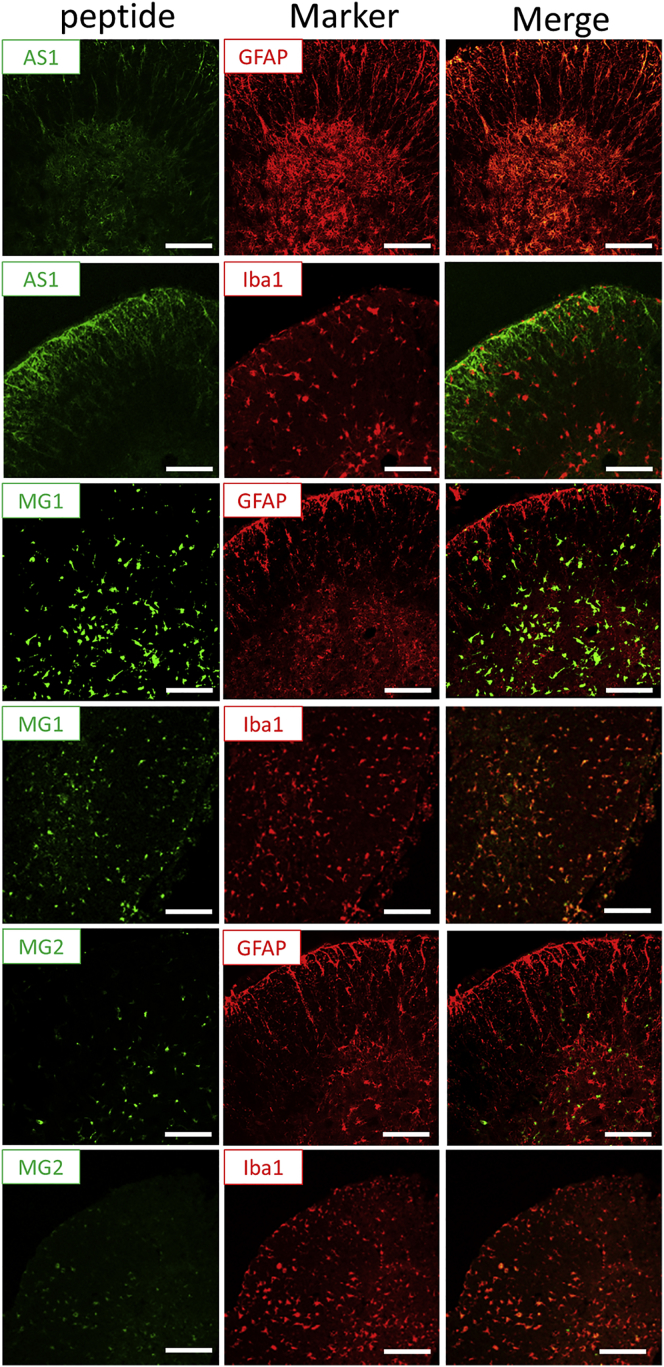
To confirm that these homing peptides can target the specific cells, in vivo experiments were performed in mice. A specified amount (3 μg) of AS1, MG1, and MG2 peptides labeled with FITC was intrathecally injected into mice. After 3 hr, the spinal cords were isolated from mice, and sections of the spinal cord were analyzed immunohistochemically. The AS1 peptides were represented by green fluorescence in the spinal cord and completely overlapped with the GFAP stain, an astrocyte marker, but not with the Iba1 stain, a microglia marker (Figure 5, upper two lines). The MG1 and MG2 peptides overlapped with Iba1, but not with GFAP, and there was a greater accumulation of MG1 peptides than of MG2 peptides in the spinal cord tissues (Figure 5, lower four lines). These findings suggest that AS1, MG1, and MG2 peptides could successfully target the spinal cord and its corresponding cells.
Figure 5.
In Vivo Targeting of the Spinal Cord by AS1, MG1, and MG2
The distribution of FITC-labeled, astrocyte-specific peptides (AS1; green) with GFAP or Iba1 stain (red) (upper two lines) or FITC-labeled microglia-specific peptides (MG1 or MG2; green) with GFAP or Iba1 stain (red) (lower four lines) in the spinal cord sections at 3 hr after injection of AS1, MG1, or MG2 peptides (3 μg in 10 μL of PBS). Scale bars, 100 μm.
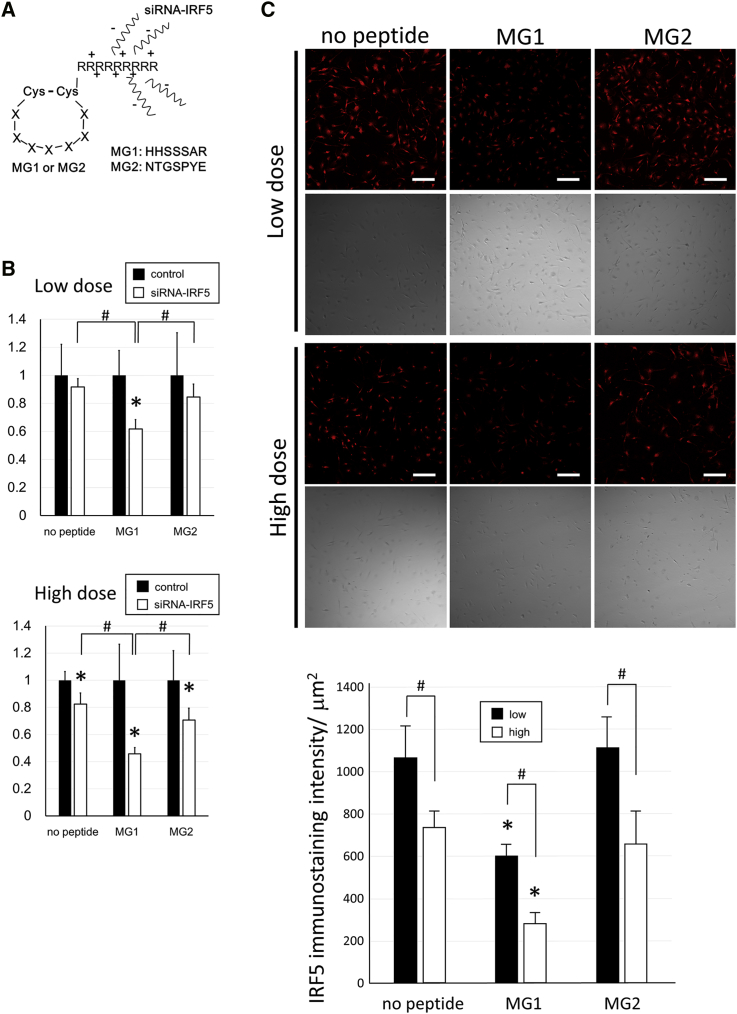
Knockdown Effect of Target Genes with MG1 or MG2 Peptides and siRNA Complex in Microglia
To clarify that the homing peptides have potential as vehicles for targeted gene delivery, gene delivery experiments were performed in microglia using the complex of MG1 or MG2 peptides and siRNA-IRF5. First, MG1-9R (MG1[C-HHSSSAR-C]+GGG+9 arginine residues [R]) or MG2-9R (MG2[C-NTGSPYE-C]+GGG+9R) peptides were synthesized and bound to siRNA oligonucleotides (Figure 6A). These complexes were bonded together by electrostatic interaction because of the negative electric charge of the oligonucleotides (Figure 6A). As a preliminary study, electrophoresis of the complexes was performed (Figure S1). Furthermore, to determine the appropriate ratio of peptides and siRNA, complexes (peptides and siRNA) at ratios of 0:1, 0.5:1, 1:1, 2:1, 3:1, 4:1, 5:1, and 5:0 were prepared and assessed by electrophoresis (Figure S1A). At ratios of more than 3:1, oligonucleotide bands were not observed even as smear bands in the agarose gel, because the negative electric charge of the oligonucleotides was neutralized by the binding with the peptides (Figure S1A). Therefore, a ratio of 3:1 was determined to be the minimal ratio for peptides to bind to siRNA without a residue. Next, the electrophoresis of the complexes of siRNA-IRF5 with MG1-3R (MG1[C-HHSSSAR-C]+GGG+3R), MG1-6R (MG1[C-HHSSSAR-C]+GGG+6R), or MG1-9R peptides was performed to confirm the binding acuities of arginine residues with oligonucleotides (Figure S1B). Proportionally, as the number of arginine residues increased from zero to three, six, or nine, the oligonucleotide bands became gradually weaker (Figure S1B). Most MG1-0R peptides labeled with FITC were observed as high signals near sample application wells in agarose gel, suggesting that MG1-0R could not bind with siRNA (Figure S1B). Therefore, nine arginine residues were used for binding between homing peptides and oligonucleotides. The siRNA-IRF5 with MG1-9R or MG2-9R complexes (siRNA, 66 ng; peptides, 200 ng) were administered to lipopolysaccharide (LPS)-stimulated microglia, and the levels of IRF5 mRNA expression were analyzed 3 hr after treatment. The mRNA expression of IRF5 in the MG1-9R+siRNA-IRF5 group significantly decreased to 60% of that in the MG1-9R+control group, whereas the siRNA-IRF5 (no peptide) and MG2-9R+siRNA-IRF5 groups failed to exhibit similar knockdown effects at low doses (Figure 6B, upper graph). In addition, the siRNA-only and MG2 groups exhibited knockdown effects only in the high-dose experiments (siRNA, 330 ng; peptides, 1 μg). However, the MG1 group showed >50% knockdown of IRF5 mRNA expression in high-dose experiments (Figure 6B, lower graph). The MG1-9R+siRNA-IRF5 group exhibited significant knockdown effects compared with siRNA-IRF5 (no peptide) and MG2-9R+siRNA-IRF5 groups in both low- and high-dose experiments (Figure 6B). The protein expression level of IRF5 was analyzed 24 hr after treatment through immunostaining of IRF5 and was demonstrated to be suppressed in the MG1 groups compared with the no-peptide and MG2 groups at both low and high doses (Figure 6C). In addition, the protein expression level of IRF5 in the MG1 group of the high-dose experiments decreased more than that in the three groups of the low-dose experiments (Figure 6C). The suppression of IRF5 protein expression exhibited the same pattern with mRNA knockdown effects, which were most effective in the MG1-9R+siRNA-IRF5 group, and a dose response was observed in the no-peptide, MG1, and MG2 groups (Figures 6B and 6C).
Figure 6.
Design of Complexes between Peptides and siRNAs and Knockdown Effects In Vitro
(A) Scheme of MG1 or MG2 peptide structures and their binding with siRNA-interferon regulatory factor 5 (IRF5) oligonucleotides. (B and C) Knockdown effects by a low dose (peptide, 200 ng; siRNA, 66 ng) or high dose (peptide, 1 μg; siRNA, 330 ng) of the complexes MG1 or MG2+siRNA-IRF5 in primary microglia. (B) Graphs show relative mRNA expression of IRF5 in the no-peptide, MG1, or MG2 group at 3 hr after administration of the complexes against each scrambled control or oligonucleotide group in low-dose (upper panel) or high-dose (lower panel) experiments (n = 5 in each group). IRF5 mRNA expression levels were standardized by β-actin mRNA expression and were shown as the relative ratio against each control oligonucelotide group. *p < 0.05 against the control oligonucleotide group. #p < 0.05. (C) IRF5 immunostainings (red) and phase images of primary microglia in the siRNA-IRF5 (no peptide), MG1+siRNA-IRF5, or MG2+siRNA-IRF5 group 24 hr after administration of a low or high dose of the complexes. The graph shows IRF5 immunostaining intensity (n = 5 in each group). *p < 0.05 against the other two groups in each dose experiment. #p < 0.05. Scale bars, 100 μm in (C). Error bars, mean + SD in (B) and (C). MG, microglia-specific peptides; Cys, cysteine; A, alanine; E, glutamate; G, glycine; H, histidine; N, asparagine; P, proline; R, arginine; S, serine; T, threonine; Y, tyrosine.
Gene Delivery of siRNA-IRF5 with MG1 Homing Peptides Targeting Microglia in Neuropathic Pain Mice
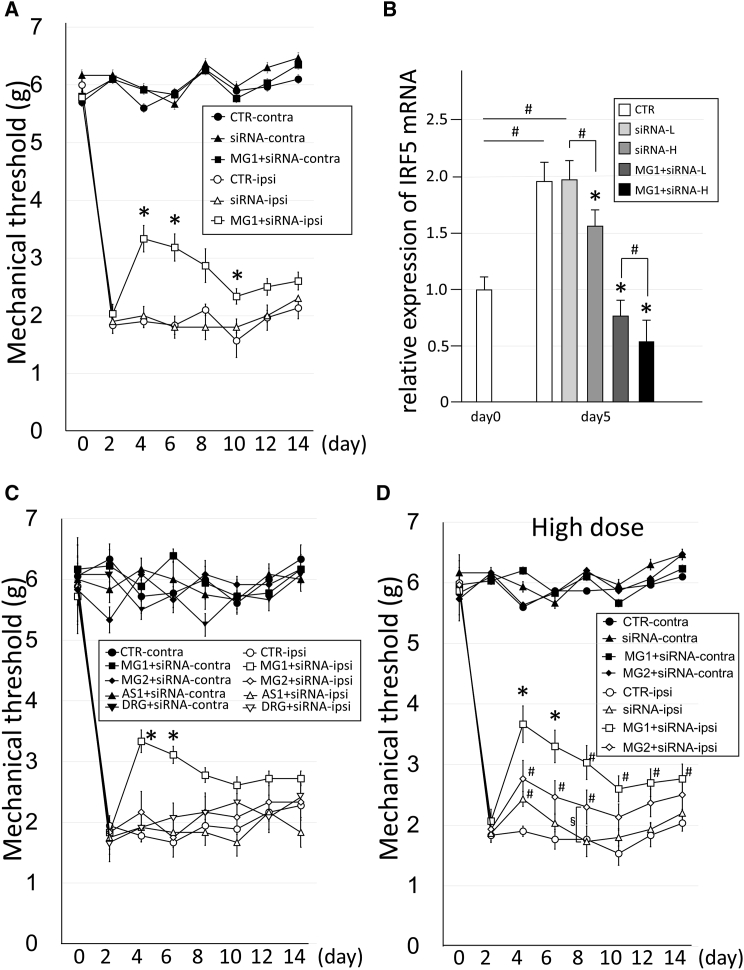
Gene therapy was performed for neuropathic pain by the knockdown of IRF5 using the MG1 peptide+siRNA-IRF5 complex (peptides, 3 μg; siRNA, 1 μg) and the comparative groups, which were MG2, AS1, and DRG complexes (peptides, 3 μg; siRNA, 1 μg) prepared with the same structure as the complex in Figure 6A. Because IRF5 is involved not in thermal or movement allodynia but rather in tactile allodynia, the effects of the complexes were evaluated by measuring the mechanical threshold with plantar tests. After spinal nerve transection, the reduction in the mechanical threshold against neuropathic pain was observed on the ipsilateral side of all mice on day 2 (Figure 7A). After confirmation of neuropathic pain, a therapeutic material was administered to mice through the intrathecal space on day 3. On days 4, 6, and 10, the mechanical threshold in the MG1+siRNA-IRF5 group significantly increased, whereas the threshold did not change from the pre-treatment baseline levels in the untreated control and in the siRNA-IRF5-only groups (Figure 7A).
Figure 7.
Gene Therapy for Neuropathic Pain Mice Using the Delivery of siRNA with Homing Peptides to the Spinal Cord
(A) The effects of the mechanical threshold in the neuropathic mice on the ipsilateral (ipsi) or contralateral sides by the injection of CTR (buffer only) (n = 5), siRNA-IRF5 only (1 μg/mouse, n = 5), and MG1+siRNA-IRF5 (peptide, 3 μg/mouse; siRNA, 1 μg/mouse; n = 10) on day 3 post-transection. Error bars, mean ± SD. *p < 0.05 versus CTR-ipsi and siRNA-ipsi. (B) Relative expression of IRF5 mRNA in the spinal cord of the neuropathic pain mice on day 5 post-transection in CTR (buffer only) (n = 5), siRNA-IRF5 only of the low-dose group (siRNA-L) (1 μg/mouse, n = 5) or the high-dose group (siRNA-H) (5 μg/mouse, n = 5), and MG1+siRNA-IRF5 of the low-dose group (MG1+siRNA-L) (peptide, 3 μg/mouse; siRNA, 1 μg/mouse; n = 10) or the high-dose group (MG1+siRNA-H) (peptide, 15 μg/mouse; siRNA, 5 μg/mouse; n = 5). IRF5 mRNA expression levels were standardized by β-actin mRNA expression and were shown as the relative ratio against the expression level at day 0 (n = 5) in the spinal cord. CTR, control mice; MG, microglia-specific peptides. Error bars, mean ± SD. *p < 0.05 versus CTR and another siRNA group in the same dose. #p < 0.05. (C) The effects of the mechanical threshold in the neuropathic pain mice on the ipsilateral or contralateral sides due to the injection of CTR (buffer only) (n = 6), MG1+siRNA-IRF5 (n = 6), MG2+siRNA-IRF5 (n = 6), AS1+siRNA-IRF5 (n = 6), and DRG+siRNA-IRF5 (n = 6) on day 3 post-transection. The concentration of administered peptide or siRNA is 3 or 1 μg in each mouse, respectively. Error bars, mean ± SD. *p < 0.05 versus all other ipsi groups. (D) The effects of the mechanical threshold in neuropathic pain mice on ipsilateral or contralateral sides by the high-dose injection of CTR (buffer only) (n = 5), siRNA-IRF5 only (n = 5), MG1+siRNA-IRF5 (n = 5), and MG2+siRNA-IRF5 (n = 5). The concentration of administered peptide or siRNA is 15 or 5 μg in each mouse. Error bars, mean ± SD. *p < 0.05 versus all other ipsi groups. #p < 0.05 versus the CTR-ipsi group. §p < 0.05.
In addition, non-target therapy was performed using MG2, AS1, and DRG peptides (targeting the DRG neuron) with siRNA-IRF5 (peptides, 3 μg; siRNA, 1 μg) and was compared with MG1 peptide treatment (Figure 7C). Only the MG1 peptide group exhibited a treatment effect for neuropathic pain; the other group of peptides did not exhibit even partial therapeutic effects (Figure 7C). Therefore, high-dose experiments (peptides, 15 μg; siRNA, 5 μg) were performed by using naked siRNA and MG2+siRNA complexes, which were compared with the MG1+siRNA group (Figure 7D). In naked siRNA on day 4 and in MG2+siRNA on days 4, 6, and 8, significant therapeutic effects were observed compared with the effects of the buffer control, which were partial and brief (Figure 7D). In addition, the effects in the MG2 peptide group were significantly higher than those in the naked siRNA group on day 8 alone (Figure 7D). This result suggested that the MG2 peptide had partial targeting potential to M1 microglia. The effect in the MG1 group was the highest among the four groups (buffer control, siRNA only, MG1, and MG2) (Figure 7D) and was slightly higher than that observed in the low-dose experiments, as shown in Figure 7A.
IRF5 mRNA expression in the spinal cord was upregulated in the buffer control and siRNA-IRF5 groups (low: siRNA, 1 μg; high: siRNA, 5 μg) 5 days after nerve transection (Figure 7B). In contrast, on day 5, the mRNA expression of IRF5 in both low-dose (peptides, 3 μg; siRNA, 1 μg) and high-dose (peptides, 15 μg; siRNA, 5 μg) MG1+siRNA-IRF5 groups was substantially suppressed to the levels seen before nerve transection (Figure 7B). The effect in the high-dose group was significantly greater than that in the low-dose group in MG1+siRNA-IRF5 administration. Simple siRNA-IRF5 injections slightly suppressed the mRNA expression of IRF5 only in the high-dose application of siRNA-IRF5, not in the low-dose application (Figure 7B). The effect of delivery and knockdown of the target gene was markedly higher in the MG1 peptides groups than in the no-peptide group (Figure 7B).
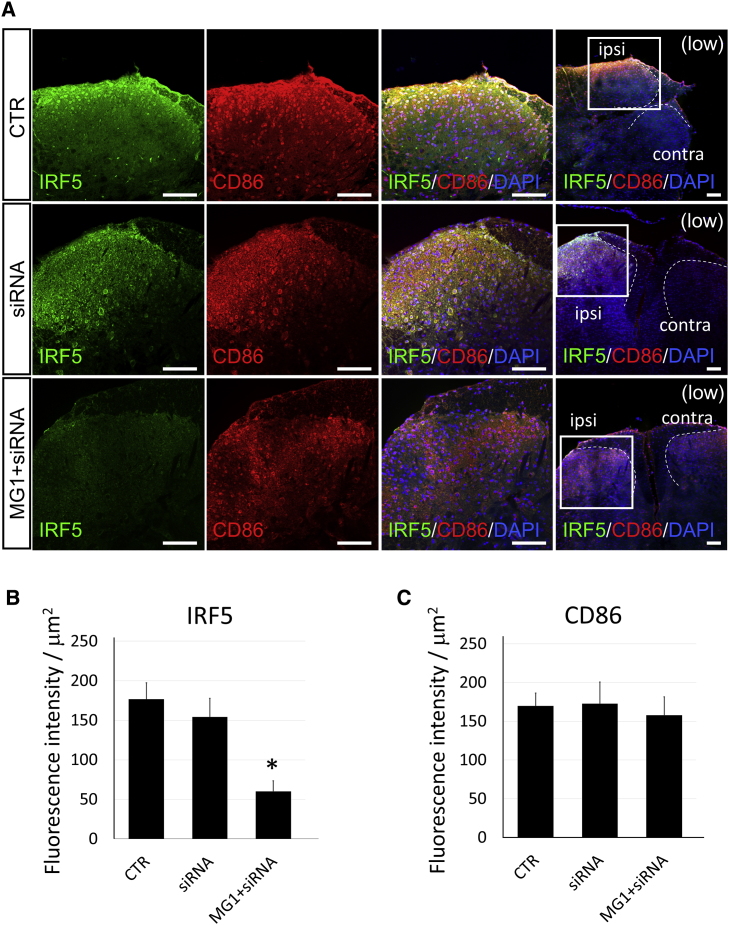
Immunohistochemistry experiments were performed using spinal cord sections from the neuropathic pain mouse model after gene therapy with MG1-siRNA-IRF5. In low-magnification pictures of the spinal cord (Figure 8A, extreme right), many CD86-positive cells accumulated on the ipsilateral side of the dorsal horn in the spinal cords, whereas few CD86-positive cells were observed on the contralateral side (Figure 8A). At a high magnification of the ipsilateral dorsal horn, numerous CD86-positive, pro-inflammatory microglia (Figure 8A, second column, red) were also observed in all groups (Figure 8A). The quantitation of immunofluorescent intensities revealed the same level of CD86 protein expression among control, siRNA, and MG1+siRNA groups (Figure 8C). However, IRF5 staining (Figure 8, first column, green) was markedly weaker in MG1+siRNA groups than in the control and siRNA-only groups, and quantitative analysis indicated the same pattern of IRF5 protein expression (Figures 8A and 8B). Both mRNA and protein levels of IRF5 were effectively suppressed by the MG1-siRNA-IRF5 complexes (Figures 7B and 8), which could result in the reduction of neuropathic pain in the mouse models (Figure 7A). Moreover, the reduction efficacy of the MG1 peptide was the highest among all groups (Figure 7). These findings highlight the selective targeting ability of MG1 peptides to deliver siRNA oligonucleotides to microglia in the spinal cord and may serve as a useful tool for developing novel gene therapy modalities.
Figure 8.
Histological Analysis of the Spinal Cord in Neuropathic Pain Mice after Gene Therapy
(A) Immunohistochemistry of IRF5 (green) and CD86 (red) in the spinal cord tissues from L5 5 days after treatment with the control buffer (CTR group), siRNA-IRF5 only (siRNA group) (1 μg/mouse), and MG1+siRNA-IRF5 (MG1+siRNA group) (peptide, 3 μg/mouse; siRNA, 1 μg/mouse). The left three panels (high, high magnification) show the enlargement of the ipsilateral dorsal horns in the spinal cord at the same section of the square portion of the panel farthest to the right (low, low magnification). Scale bars, 100 μm. Broken lines showed the margin of bilateral dorsal horns in spinal cords. Contra, contralateral side; ipsi, ipsilateral side. (B and C) Quantitative analysis of immunostainings in IRF5 (B) and CD86 (C). Bars showed the immunostaining intensity of IRF5 or CD86 in the CTR, siRNA, or MG1+siRNA group (n = 5 in each). Error bars, mean ± SD in (B) and (C). *p < 0.05 versus the CTR and siRNA groups.
Discussion
Effective gene delivery to selective target astrocytes and microglia requires the identification of homing peptides that can access the central nervous system. Therefore, in vitro and in vivo biopannings with phage libraries were used in this study to identify peptides that target astrocytes and microglia. We identified the AS1(C-LNSSQPS-C) peptide for astrocytes, which was the most frequent peptide in six types of peptide sequences detected in a total of 58 clones. Among the types of microglia homing peptides, 31 for M1 type, 30 for M2 type, and 6 for both M1 and M2 types were detected. The MG1(C-HHSSSAR-C) and MG2(C-NTGSPYE-C) peptides were identified as the two most frequent homing peptides. MG1 exhibited a higher binding capacity for M1-type microglia than did the MG2 peptide. In contrast, the MG2 peptide had a greater binding capacity for M2-type microglia. These results have potential applicability in cell-selective gene delivery. When the targeted cells consist of several subtypes, such as in microglia,14 cell-specific targeting is required for highly efficient gene therapy or imaging. In this context, MG1 with siRNA could feasibly be delivered selectively into CD86-positive cells in the spinal cord, as was the case in this study, which resulted in effective gene therapy for neuropathic pain.
In neuronal tissues, many types of cells function simultaneously to maintain normal physiological conditions and uphold the balance of homeostasis.22, 23, 24, 25, 26 A single molecule can induce variable cellular effects depending on the type of cell targeted. For example, brain-derived neurotrophic factor (BDNF) stimulates neurons to induce cell differentiation or growth.27, 28, 29 However, BDNF also induced neuropathic pain through astrocytes by activating the afferent signals from the peripheral nerves to the brain after injuries.30, 31 In addition to BDNF, many types of cytokines, such as interleukin (IL)-1, IL-6, and tumor necrosis factor alpha (TNF-α), have several functions and can stimulate many types of cells.32, 33, 34, 35, 36, 37 In the anti-TNF-α antibody therapy for rheumatoid arthritis or inflammatory bowel syndrome, the target tissue is the articular cavity or intestinal tract.38, 39 However, it occasionally induces a compromised host because the drug is systemically administered.38 Therefore, in the field of molecular therapy, it is crucial that the therapeutic molecule be delivered only to the target tissues and cells. In addition, this type of precise targeting can suppress the side effects of treatment or any unexpected phenomenon, which is ideal for developing therapeutic agents.
An important consideration when transferring drugs into cells is which of the many types of cell penetration peptides can be used for delivery into target cells.40, 41 Classically, the transactivator of transcription protein in HIV-1 was identified to be related to the translocation of proteins through the cell membrane.42, 43 An additional study demonstrated that polyarginine could transfer to the cell membrane because of the positive electric charge, which could bind to proteoglycan with a negative electric charge.44 In this study, nine arginine residues were connected to the MG1 homing peptide. This modification resulted in a powerful, high-performance tool for effective gene therapy. The MG1-9R peptide seems to have high potential as the delivery device for specific cells and cell penetration peptides. These findings could be used as a novel approach for the delivery of small oligonucleotides.
Many antisense oligonucleotides (ASOs) have been developed as the nucleic acid drugs and used in the trials targeting cancer and other incurable genetic diseases.45, 46, 47 In treatments using ASO, their stability is typically the most important problem. By their nature, ASO are immediately degraded by nuclease in vivo, because ASO is generally a single-stranded structure.45 Hence, to prevent the degradation of ASO, chemical modifications such as locked nucleic acids, 2′-O-methoxyethyl RNA, and constrained ethyl BNA have been devised for their stability.48, 49, 50, 51 In addition, the siRNAs have been developed as a device for novel gene therapy and are slightly more stable than ASO because of their double-stranded structure.48, 52, 53 The introduction of the drug delivery system is necessary for the siRNA gene therapy and requires coating with lipid nanoparticle or liposome, which were applied to protect siRNA from nuclease exposure.48, 53, 54 In addition, we used siRNA duplexes of 25-mer sense and 27-mer antisense, which are known to exhibit high stability and activity for knockdown compared with the duplex of 21-mer siRNA.55 The complexes of peptides and siRNA have been reported in several articles,52, 56 which demonstrated the effective gene delivery of siRNA conjugated with N-acetylgalactosamine (GalNAc) for hepatocytes56 or with rabies virus glycoprotein (RVG) for acetylcholine receptors.52 In this study, microglia homing peptides exhibited potential as a delivery device for siRNA and provided high knockdown efficacy of siRNA, although this MG1 peptide was shorter and smaller than the GalNAc and RVG peptides.52, 56
Here, we performed gene therapy for neuropathic pain using the complex formed between short peptides and small oligonucleotides. In clinical settings, repeat injections are occasionally required for sustained effects of treatment. However, our results provide a novel strategy for selective gene therapy targeted toward nervous system tissues, which might be safer than gene therapy with virus vectors57, 58, 59, 60, 61 or the chemical materials62, 63 used in lipofection. Moreover, intrathecal injection was used for direct delivery into the spinal cord. This procedure is routinely performed in clinical settings.64 In addition, local injection has merit with respect to antibody formation compared with intravenous systemic injection.60 Therefore, this strategy can be translated into clinical use for treating patients with neuropathic pain, although additional safety trials must be performed in vivo.
The IRF5 gene was selected as a therapeutic molecule in this study. Its expression increases in the microglia of the spinal cord following spinal nerve injury,21, 65 which was used to induce neuropathic pain in the disease model. Therefore, this molecule was well suited to confirm that microglia homing (MG1) peptides can selectively deliver the therapeutic gene to M1 microglia. The MG1 peptide was the most effective for neuropathic pain in all peptide groups. Surprisingly, only one shot of MG1+siRNA-IRF5 induced a remarkable knockdown effect with the same expression level of IRF5 in the pre-treatment condition. According to our in vitro study, MG1 could not bind to all of the microglia. Therefore, IRF5 mRNA might be strongly suppressed in CD86 cells targeted by MG1+siRNA-IRF5 until the level to offset the IRF5 expression in the untargeted cells. MG2 groups exhibited a certain level of therapeutic effect only with the high-dose administration study compared with that of the peptide-control and siRNA-control groups. These results are consistent with the in vitro study results, which indicated that the MG1 peptide was superior to the MG2 peptide for targeting M1 microglia, although the MG2 peptide was bound to a small population of M1 microglia. In contrast, the AS1 and DRG groups exhibited no therapeutic effects for neuropathic pain. These results suggest that the AS1 and DRG peptides could not target M1 microglia. In addition, it appears that IRF5 is expressed at low levels in neurons and astrocytes in the spinal cord. Overall, MG1 and siRNA-IRF5 complexes were highly effective for the successful suppression of hyperalgesia-induced spinal nerve injuries compared with that of other peptides or naked siRNA. This finding highlights the potential use of the MG1 peptide for developing targeting and delivery systems. IRF5 was identified as a key molecule involved in neuropathic pain. Many molecules have been reported to be associated with the pathogenesis of painful neuropathy or neuropathic pain.66, 67 Despite the multitude of drugs that have been developed to treat neuropathic pain, an effective medication has yet to be identified.67 In the present study, the selective suppression of IRF5 in microglia yielded effective results for reducing neuropathic pain, highlighting the pivotal role that IRF5 may play in potential treatment strategies. Our results emphasize the importance of recognizing IRF5 as a key factor in neuropathic pain, as previously demonstrated.20
This study highlights the inhibition of IRF5 with microglia homing peptides as a strategy for novel gene therapy for neuropathic pain in mice. The complexes between peptides and siRNA-IRF5 inhibited hyperalgesia induced by spinal nerve transection. The use of these complexes in the spinal cord may be considered a promising method for the treatment of neuropathic pain.
Materials and Methods
Animals
C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA). All animals were housed under a 12-hr light:dark cycle and were provided with water and mouse chow ad libitum. All animal experimental protocols were approved by the Institutional Animal Care and Usage Committee (IACUC) at Shiga University of Medical Science and were performed in accordance with the guidelines of the IACUC at Shiga University of Medical Science.
Preparation of Cultured Cells
NSC-34 cells were purchased from CELLutions Biosystems (Toronto, Canada) and were prepared as the representative neuronal cells. NSC-34 cell lines were maintained with 10% fetal calf serum in DMEM/F12 medium (Thermo Fisher Scientific, Waltham, MA, USA). KT-5 cells were obtained from the Japanese Collection of Research Bioresources (JCRB) cell bank at the National Institutes of Biomedical Innovation, Health and Nutrition. Ra2 and 6-3 cells were purchased from Cosmo Bio (Tokyo, Japan). These cells were maintained in culture according to the manufacturer’s protocol and were used to screen the homing peptides. Primary cultures of astrocytes and microglia were prepared from the brain tissue of C57BL/6 pups 1–2 days postpartum. Astrocytes were isolated by the passage of whole cells during the initial 5–7 days. After a 14-day incubation of mixed culture cells, microglia began to grow on the astrocyte cell sheet and were isolated in another culture dish. The isolated microglia were subsequently used for experiments.
In Vivo and In Vitro Phage Display and Screening of Homing Peptides Targeting Astrocytes and Microglia
The CX7C phage library was purchased from New England Biolabs (Ipswich, MA, USA) and was used for both in vivo and in vitro phage display to screen astrocyte or microglia homing peptides. The protocols described by Christianson et al.68 were used with minor modifications. The phage library was injected into C57BL/6 mice through the tail vein at a titer of 109 plaque-forming units (pfu)/μL in 100 μL Tris-buffered saline (50 mM Tris-HCl [pH 7.5]). Five minutes after the injections, whole spinal cords were isolated after the transcardiac removal of blood. After the weights of the harvested spinal cord tissues were recorded, each sample was homogenized in DMEM (Thermo Fisher Scientific) with protease inhibitor (Sigma-Aldrich, St. Louis, MO, USA). Aliquots were introduced into Escherichia coli ER2738 (New England Biolabs, Ipswich, MA, USA), and the number of phages in the spinal cord was titrated by plaque-forming units. After amplification to 109 pfu, the phages were injected into a new cohort of C57BL/6 mice. This process was performed three times to select the specific phages that bound to the spinal cord.
After three rounds of in vivo panning in the spinal cord, the phages were introduced separately to cultured astrocyte (KT-5) or microglia cell lines (6-3 cells for M1 [proinflammatory type] or Ra2 cells for M2 [anti-inflammatory type]) for 5 min, and three additional cycles of in vitro panning were performed for each cell line. In total, genomes were isolated from a collection of 50–58 phages with high affinities for each cell line and then analyzed for DNA sequences coding for pIII protein.
Analysis of Binding of Homing Peptides to Astrocytes and Microglia In Vitro and In Vivo
AS1(C-LNSSQPS-C), MG1(C-HHSSSAR-C), and MG2(C-NTGSPYE-C) peptides were synthesized and labeled with FITC by Invitrogen (Carlsbad, CA, USA). Isopropyl cyanide of FITC bound with the N terminus of each peptide by the intermediary of aminocaproic acid linker. These peptides labeled with FITC were refined to a purity of >95% by high-performance liquid chromatography, and their characteristics were confirmed by mass spectrometry. For in vitro experiments, the labeled peptides were applied to culture cells (NSC-34 as representative neuronal cells, primary culture of astrocytes and microglia) at a concentration of 1 μg/mL in 200 μL of culture medium in 24-well plates. After 24 hr of incubation with the peptides, cells were fixed with 4% paraformaldehyde (PFA) and were prepared for immunocytochemistry with each primary antibody: anti-MAP2 antibody (Merck Millipore, Darmstadt, Germany] for neurons, anti-GFAP antibody (Promega, Maddison, WI, USA) for astrocytes, anti-Iba1 antibody (Wako Pure Chemical Industries, Osaka, Japan) for microglia, anti-CD86 antibody (Abcam, Cambridge, UK) for M1 microglia, and anti-CD206 antibody (Abcam) for M2 microglia. Samples were subsequently incubated with species-matched secondary antibodies (Alexa Fluor 555 antibody; Molecular Probes, Eugene, OR, USA) to enable the visualization of stained cells with confocal laser microscopy (C1si; Nikon, Tokyo, Japan) using EZC1 3.90 (Nikon). FITC-positive cells were identified, counted in six independent wells, and calculated as the average of the percentage of positive staining cells in each peptide group. For microglia, CD86- and CD206-positive cells were counted, and their percentages were calculated in MG1- or MG2-positive microglia from six independent wells. In total, >1,000 cells were used for analysis. These in vitro experiments were performed in triplicate.
For in vivo experiments, 3 μg of AS1, MG1, and MG2 peptides labeled with FITC were separately injected into C57BL/6 mice through the intrathecal space at a concentration of 300 ng/μL in 10 μL of PBS. At 3 hr after the injection, the spinal cords were isolated from the mice after transcardiac perfusion fixation with 4% PFA. Sections of the spinal cord were cut and incubated with anti-GFAP primary antibodies (Promega) or anti-Iba1 primary antibodies (Wako Pure Chemical Industries) overnight at 4°C, washed, and then incubated with species-matched secondary antibodies tagged with Alexa Fluor 555 (Molecular Probes). All sections were visualized under a confocal laser microscope (C1si; Nikon) with EZC1 3.90 (Nikon). These in vivo experiments were performed with five mice in each peptide group.
Complex between Peptides and siRNA for Gene Delivery
MG1-3R (MG1[C-HHSSSAR-C]+GGG+3 arginine residue [R]), MG1-6R (MG1[C-HHSSSAR-C]+GGG+6 arginine residue [R]), MG1-9R, MG2-9R, AS1-9R (AS1[C-LNSSQPS-C]+GGG+9R), and DRG-9R (DRG[C-SPGARAF-C]+GGG+9R)11 peptides were synthesized by Invitrogen and the Peptide Institute (Osaka, Japan). These peptides were refined to a purity of >95% by high-performance liquid chromatography, and their characteristics were confirmed by mass spectrometry. The DRG peptide, which was previously reported to target the DRG neuron in mice, was prepared as a non-target control in this study. siRNA-IRF5 duplexes were synthesized by OriGene Technologies (Rockville, MD, USA), which consisted of 27-mer antisense and 25-mer sense of the following sequence: 5′-CGUAGACUGAUAUAGCAAUGUGGTG-3′. The fusion complexes between peptides and oligonucleotides were formulated by electrostatic interaction, and an appropriate molecular ratio of peptides to oligonucleotides was decided upon by the visualization of those oligonucleotides with ethidium bromide under UV excitation after electrophoresis in agarose gels (Figure S1). The final peptide-to-oligonucleotide molecular weight ratio of 3:1 was used for the in vitro and in vivo knockdown study. These electrophoreses were performed in triplicate.
Knockdown Experiments by Gene Delivery with Peptide-siRNA-IRF5 Complexes to Microglia
Primary cultures of microglia were prepared in 12-well plates 1 day before the experiments. The inflammation of the microglia was induced by LPS (1 μg/mL). One hour after LPS stimulation, MG1-9R+siRNA-IRF5, MG2-9R+siRNA-IRF5, or siRNA-IRF5 alone was applied to the cells at low (peptide, 200 ng or none; siRNA, 66 ng) or high (peptide, 1 μg or none; siRNA, 330 ng) doses. After a 3-hr period, the effects of the treatment were evaluated by measuring IRF5 mRNA expression with qRT-PCR (IRF5: forward primer 5′-CAGGTGAACAGCTGCCAGTA-3′, reverse primer 5′-GGCCTTGAAGATGGTGTTGT-3′; β-actin: forward primer 5′-ATGGATGACGATATCGCT-3′, reverse primer 5′-TCTGTCAGGTCCCGGCCA-3′) of each treatment group, in addition to the scrambled siRNA oligonucleotide, negative-control groups (MG1-9R+siRNA-control, MG2-9R+siRNA-control, or only siRNA-control) at the same dose as the treatment groups. After treatment with the complexes (peptides and oligonucleotides) for 24 hr, the cells were fixed with 4% PFA and incubated with IRF5 primary antibody for immunocytochemistry (Abcam). Samples were then incubated with species-matched secondary antibodies (Alexa Fluor 555 antibody; Molecular Probes) to enable the visualization of stained cells, and the immunostaining and phase images were observed through confocal laser microscopy (C1si; Nikon) with EZC1 3.90 (Nikon). Staining for IRF5 was quantified by ImageJ 1.51 (NIH, Bethesda, MD, USA) using at least 5 independent wells with low or high concentrations of peptide-siRNA complexes. These in vitro knockdown experiments were performed in triplicate.
Gene Therapy with Peptide-siRNA-IRF5 Complexes for Neuropathic Pain after Spinal Nerve Transection
Gene therapy experiments were performed using a mouse model of neuropathic pain, in which the spinal nerves of C57BL/6 mice were transected at L5 on the left side (ipsilateral).69 The peptide and oligonucleotide complexes (10 μL of a 0.1 M PBS solution including 3 μg of peptide [MG1-9R, MG2-9R, AS1-9R, or DRG-9R] and 1 μg of siRNA-IRF5) were injected once into mice through the intrathecal space 3 days after transection. For the experiments using the high dose, the peptide and oligonucleotide complexes of siRNA-IRF5 with MG1 or MG2 were used at the same volume (10 μL) of 0.1 M PBS with 15 μg of peptide and 5 μg of siRNA-IRF5. siRNA-IRF5- and buffer-only groups were used as controls for all gene therapy experiments. Plantar tests were performed before and after transection for 14 days to evaluate the mechanical threshold on the ipsilateral side (left) and contralateral side (right) by using a dynamic plantar aesthesiometer (Ugo Basil, Varese, Italy). Pressure applied to the bilateral hindpaw served as the mechanical stimuli. The level of pressure was measured at the point of paw withdrawal in response to the mechanical stimuli. The withdrawal threshold was determined by the mean of three trials.
Five days after transection, a subset of mice was used to analyze the efficacy of gene delivery by MG1 peptides. Total mRNA was purified from L1 to L5 spinal cord tissue, and transcript levels of the IRF5 gene were measured by quantitative RT-PCR.
For the histological analysis of gene delivery with MG1 peptides, spinal cords were sectioned after perfusion fixation with 4% PFA on day 5. The sections were incubated with anti-IRF5 primary antibodies (Abcam) and anti-CD86 primary antibodies (Abcam) overnight at 4°C, washed, and then incubated with species-matched secondary antibodies tagged with Alexa Fluor 488 and Alexa Fluor 555 (Molecular Probes). All sections were visualized under a confocal laser microscope (C1si; Nikon) with EZC1 3.90 (Nikon). The fluorescent intensity of IRF5 and CD86 immunostaining was quantified by ImageJ 1.51 in the sections from five independent mice in each control, siRNA, and MG1+siRNA group. The average was compared among those three groups.
Statistical Analyses
Data are expressed as mean ± SD. All in vitro experiments were performed in triplicate in at least three independent experiments. In vivo analyses were performed using a minimum of five mice per group, unless otherwise specified. Student’s unpaired two-tailed t tests were performed to determine the significant differences among the treatment groups. For multiple datasets, one-way ANOVA and Scheffe’s tests were used. p < 0.05 was considered statistically significant.
Author Contributions
T.T. conceived and performed the experiments, drafted the manuscript, and secured funding. N.O., Y.N., and M.K. performed the experiments. T.S. provided reagents and secured funding. J.O., H.M., and H.K. provided expertise and feedback.
Conflicts of Interest
T.S. is employed by Daiichi Sankyo, Co., Ltd. Other authors declare that the study was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank T. Yamamoto and F. Kimura for their technical support. This study was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (23790988 to T.T.), and by research funding from the Daiichi Sankyo TaNeDS Funding Program.
Footnotes
Supplemental Information includes one figure and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.02.007.
Supplemental Information
References
- 1.Kolonin M., Pasqualini R., Arap W. Molecular addresses in blood vessels as targets for therapy. Curr. Opin. Chem. Biol. 2001;5:308–313. doi: 10.1016/s1367-5931(00)00207-6. [DOI] [PubMed] [Google Scholar]
- 2.Giordano R.J., Cardó-Vila M., Lahdenranta J., Pasqualini R., Arap W. Biopanning and rapid analysis of selective interactive ligands. Nat. Med. 2001;7:1249–1253. doi: 10.1038/nm1101-1249. [DOI] [PubMed] [Google Scholar]
- 3.Arap W., Kolonin M.G., Trepel M., Lahdenranta J., Cardó-Vila M., Giordano R.J., Mintz P.J., Ardelt P.U., Yao V.J., Vidal C.I. Steps toward mapping the human vasculature by phage display. Nat. Med. 2002;8:121–127. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]
- 4.Pasqualini R., Koivunen E., Ruoslahti E. Alpha v integrins as receptors for tumor targeting by circulating ligands. Nat. Biotechnol. 1997;15:542–546. doi: 10.1038/nbt0697-542. [DOI] [PubMed] [Google Scholar]
- 5.Arap W., Pasqualini R., Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279:377–380. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 6.Müller O.J., Kaul F., Weitzman M.D., Pasqualini R., Arap W., Kleinschmidt J.A., Trepel M. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat. Biotechnol. 2003;21:1040–1046. doi: 10.1038/nbt856. [DOI] [PubMed] [Google Scholar]
- 7.White S.J., Nicklin S.A., Büning H., Brosnan M.J., Leike K., Papadakis E.D., Hallek M., Baker A.H. Targeted gene delivery to vascular tissue in vivo by tropism-modified adeno-associated virus vectors. Circulation. 2004;109:513–519. doi: 10.1161/01.CIR.0000109697.68832.5D. [DOI] [PubMed] [Google Scholar]
- 8.Work L.M., Büning H., Hunt E., Nicklin S.A., Denby L., Britton N., Leike K., Odenthal M., Drebber U., Hallek M., Baker A.H. Vascular bed-targeted in vivo gene delivery using tropism-modified adeno-associated viruses. Mol. Ther. 2006;13:683–693. doi: 10.1016/j.ymthe.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 9.Terashima T., Oka K., Kritz A.B., Kojima H., Baker A.H., Chan L. DRG-targeted helper-dependent adenoviruses mediate selective gene delivery for therapeutic rescue of sensory neuronopathies in mice. J. Clin. Invest. 2009;119:2100–2112. doi: 10.1172/JCI39038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koivunen E., Arap W., Rajotte D., Lahdenranta J., Pasqualini R. Identification of receptor ligands with phage display peptide libraries. J. Nucl. Med. 1999;40:883–888. [PubMed] [Google Scholar]
- 11.Oi J., Terashima T., Kojima H., Fujimiya M., Maeda K., Arai R., Chan L., Yasuda H., Kashiwagi A., Kimura H. Isolation of specific peptides that home to dorsal root ganglion neurons in mice. Neurosci. Lett. 2008;434:266–272. doi: 10.1016/j.neulet.2008.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell J.D., Borasio G.D. Amyotrophic lateral sclerosis. Lancet. 2007;369:2031–2041. doi: 10.1016/S0140-6736(07)60944-1. [DOI] [PubMed] [Google Scholar]
- 13.Jansen A.H.P., Reits E.A.J., Hol E.M. The ubiquitin proteasome system in glia and its role in neurodegenerative diseases. Front. Mol. Neurosci. 2014;7:73. doi: 10.3389/fnmol.2014.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graeber M.B. Changing face of microglia. Science. 2010;330:783–788. doi: 10.1126/science.1190929. [DOI] [PubMed] [Google Scholar]
- 15.González H., Elgueta D., Montoya A., Pacheco R. Neuroimmune regulation of microglial activity involved in neuroinflammation and neurodegenerative diseases. J. Neuroimmunol. 2014;274:1–13. doi: 10.1016/j.jneuroim.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 16.Malaspina A., Puentes F., Amor S. Disease origin and progression in amyotrophic lateral sclerosis: an immunology perspective. Int. Immunol. 2015;27:117–129. doi: 10.1093/intimm/dxu099. [DOI] [PubMed] [Google Scholar]
- 17.Hausmann O.N. Post-traumatic inflammation following spinal cord injury. Spinal Cord. 2003;41:369–378. doi: 10.1038/sj.sc.3101483. [DOI] [PubMed] [Google Scholar]
- 18.Hains B.C., Waxman S.G. Activated microglia contribute to the maintenance of chronic pain after spinal cord injury. J. Neurosci. 2006;26:4308–4317. doi: 10.1523/JNEUROSCI.0003-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuda M. Microglia in the spinal cord and neuropathic pain. J. Diabetes Investig. 2016;7:17–26. doi: 10.1111/jdi.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda T., Iwamoto S., Yoshinaga R., Tozaki-Saitoh H., Nishiyama A., Mak T.W., Tamura T., Tsuda M., Inoue K. Transcription factor IRF5 drives P2X4R+-reactive microglia gating neuropathic pain. Nat. Commun. 2014;5:3771. doi: 10.1038/ncomms4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krausgruber T., Blazek K., Smallie T., Alzabin S., Lockstone H., Sahgal N., Hussell T., Feldmann M., Udalova I.A. IRF5 promotes inflammatory macrophage polarization and TH1–TH17 responses. Nat. Immunol. 2011;12:231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 22.Suzumura A. Neuron-microglia interaction in neuroinflammation. Curr. Protein Pept. Sci. 2013;14:16–20. doi: 10.2174/1389203711314010004. [DOI] [PubMed] [Google Scholar]
- 23.Bélanger M., Allaman I., Magistretti P.J. Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab. 2011;14:724–738. doi: 10.1016/j.cmet.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 24.Streit W.J. Microglia as neuroprotective, immunocompetent cells of the CNS. Glia. 2002;40:133–139. doi: 10.1002/glia.10154. [DOI] [PubMed] [Google Scholar]
- 25.Wynn T.A., Chawla A., Pollard J.W. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tremblay M.-È., Stevens B., Sierra A., Wake H., Bessis A., Nimmerjahn A. The role of microglia in the healthy brain. J. Neurosci. 2011;31:16064–16069. doi: 10.1523/JNEUROSCI.4158-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soppet D., Escandon E., Maragos J., Middlemas D.S., Reid S.W., Blair J., Burton L.E., Stanton B.R., Kaplan D.R., Hunter T. The neurotrophic factors brain-derived neurotrophic factor and neurotrophin-3 are ligands for the trkB tyrosine kinase receptor. Cell. 1991;65:895–903. doi: 10.1016/0092-8674(91)90396-g. [DOI] [PubMed] [Google Scholar]
- 28.Binder D.K., Scharfman H.E. Brain-derived neurotrophic factor. Growth Factors. 2004;22:123–131. doi: 10.1080/08977190410001723308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McAllister A.K., Katz L.C., Lo D.C. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron. 1997;18:767–778. doi: 10.1016/s0896-6273(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 30.Pezet S., Malcangio M., McMahon S.B. BDNF: a neuromodulator in nociceptive pathways? Brain Res. Brain Res. Rev. 2002;40:240–249. doi: 10.1016/s0165-0173(02)00206-0. [DOI] [PubMed] [Google Scholar]
- 31.Trang T., Beggs S., Salter M.W. Brain-derived neurotrophic factor from microglia: a molecular substrate for neuropathic pain. Neuron Glia Biol. 2011;7:99–108. doi: 10.1017/S1740925X12000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alheim K., Andersson C., Tingsborg S., Ziolkowska M., Schultzberg M., Bartfai T. Interleukin 1 expression is inducible by nerve growth factor in PC12 pheochromocytoma cells. Proc. Natl. Acad. Sci. USA. 1991;88:9302–9306. doi: 10.1073/pnas.88.20.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giulian D., Young D.G., Woodward J., Brown D.C., Lachman L.B. Interleukin-1 is an astroglial growth factor in the developing brain. J. Neurosci. 1988;8:709–714. doi: 10.1523/JNEUROSCI.08-02-00709.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hama T., Kushima Y., Miyamoto M., Kubota M., Takei N., Hatanaka H. Interleukin-6 improves the survival of mesencephalic catecholaminergic and septal cholinergic neurons from postnatal, two-week-old rats in cultures. Neuroscience. 1991;40:445–452. doi: 10.1016/0306-4522(91)90132-8. [DOI] [PubMed] [Google Scholar]
- 35.Satoh T., Nakamura S., Taga T., Matsuda T., Hirano T., Kishimoto T., Kaziro Y. Induction of neuronal differentiation in PC12 cells by B-cell stimulatory factor 2/interleukin 6. Mol. Cell. Biol. 1988;8:3546–3549. doi: 10.1128/mcb.8.8.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavi E., Suzumura A., Murasko D.M., Murray E.M., Silberberg D.H., Weiss S.R. Tumor necrosis factor induces expression of MHC class I antigens on mouse astrocytes. J. Neuroimmunol. 1988;18:245–253. doi: 10.1016/0165-5728(88)90102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birdsall H.H. Induction of ICAM-1 on human neural cells and mechanisms of neutrophil-mediated injury. Am. J. Pathol. 1991;139:1341–1350. [PMC free article] [PubMed] [Google Scholar]
- 38.Bongartz T., Sutton A.J., Sweeting M.J., Buchan I., Matteson E.L., Montori V. Anti-TNF antibody therapy in rheumatoid arthritis and the risk of serious infections and malignancies: systematic review and meta-analysis of rare harmful effects in randomized controlled trials. JAMA. 2006;295:2275–2285. doi: 10.1001/jama.295.19.2275. [DOI] [PubMed] [Google Scholar]
- 39.Sandborn W.J. Therapy for Crohn disease. Curr. Opin. Gastroenterol. 2000;16:318–323. doi: 10.1097/00001574-200007000-00005. [DOI] [PubMed] [Google Scholar]
- 40.Schwarze S.R., Ho A., Vocero-Akbani A., Dowdy S.F. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 41.Derossi D., Joliot A.H., Chassaing G., Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J. Biol. Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 42.Frankel A.D., Pabo C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 43.Green M., Loewenstein P.M. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 44.Heitz F., Morris M.C., Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br. J. Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett C.F. Antisense oligonucleotide therapeutics. Expert Opin. Investig. Drugs. 1999;8:237–253. doi: 10.1517/13543784.8.3.237. [DOI] [PubMed] [Google Scholar]
- 46.Hong D., Kurzrock R., Kim Y., Woessner R., Younes A., Nemunaitis J., Fowler N., Zhou T., Schmidt J., Jo M. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med. 2015;7:314ra185. doi: 10.1126/scitranslmed.aac5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Deutekom J.C., Janson A.A., Ginjaar I.B., Frankhuizen W.S., Aartsma-Rus A., Bremmer-Bout M., den Dunnen J.T., Koop K., van der Kooi A.J., Goemans N.M. Local dystrophin restoration with antisense oligonucleotide PRO051. N. Engl. J. Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 48.Kanasty R., Dorkin J.R., Vegas A., Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 49.Elmén J., Thonberg H., Ljungberg K., Frieden M., Westergaard M., Xu Y., Wahren B., Liang Z., Ørum H., Koch T., Wahlestedt C. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teplova M., Minasov G., Tereshko V., Inamati G.B., Cook P.D., Manoharan M., Egli M. Crystal structure and improved antisense properties of 2′-O-(2-methoxyethyl)-RNA. Nat. Struct. Biol. 1999;6:535–539. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- 51.Blade H., Bradley D., Diorazio L., Evans T., Hayter B.R., Howell G.P. Modular synthesis of constrained ethyl (cEt) purine and pyrimidine nucleosides. J. Org. Chem. 2015;80:5337–5343. doi: 10.1021/acs.joc.5b00607. [DOI] [PubMed] [Google Scholar]
- 52.Kumar P., Wu H., McBride J.L., Jung K.-E., Kim M.H., Davidson B.L., Lee S.K., Shankar P., Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 53.Boado R.J. Blood-brain barrier transport of non-viral gene and RNAi therapeutics. Pharm. Res. 2007;24:1772–1787. doi: 10.1007/s11095-007-9321-5. [DOI] [PubMed] [Google Scholar]
- 54.Coelho T., Adams D., Silva A., Lozeron P., Hawkins P.N., Mant T., Perez J., Chiesa J., Warrington S., Tranter E. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 55.Kubo T., Zhelev Z., Ohba H., Bakalova R. Modified 27-nt dsRNAs with dramatically enhanced stability in serum and long-term RNAi activity. Oligonucleotides. 2007;17:445–464. doi: 10.1089/oli.2007.0096. [DOI] [PubMed] [Google Scholar]
- 56.Nair J.K., Willoughby J.L.S., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel’in A.V., Milstein S. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014;136:16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 57.Kootstra N.A., Verma I.M. Gene therapy with viral vectors. Annu. Rev. Pharmacol. Toxicol. 2003;43:413–439. doi: 10.1146/annurev.pharmtox.43.100901.140257. [DOI] [PubMed] [Google Scholar]
- 58.Bennett J. Immune response following intraocular delivery of recombinant viral vectors. Gene Ther. 2003;10:977–982. doi: 10.1038/sj.gt.3302030. [DOI] [PubMed] [Google Scholar]
- 59.Deyle D.R., Russell D.W. Adeno-associated virus vector integration. Curr. Opin. Mol. Ther. 2009;11:442–447. [PMC free article] [PubMed] [Google Scholar]
- 60.Pfeifer A., Verma I.M. Gene therapy: promises and problems. Annu. Rev. Genomics Hum. Genet. 2001;2:177–211. doi: 10.1146/annurev.genom.2.1.177. [DOI] [PubMed] [Google Scholar]
- 61.Howard D.B., Powers K., Wang Y., Harvey B.K. Tropism and toxicity of adeno-associated viral vector serotypes 1, 2, 5, 6, 7, 8, and 9 in rat neurons and glia in vitro. Virology. 2008;372:24–34. doi: 10.1016/j.virol.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Filion M.C., Phillips N.C. Toxicity and immunomodulatory activity of liposomal vectors formulated with cationic lipids toward immune effector cells. Biochim. Biophys. Acta. 1997;1329:345–356. doi: 10.1016/s0005-2736(97)00126-0. [DOI] [PubMed] [Google Scholar]
- 63.Stewart M.J., Plautz G.E., Del Buono L., Yang Z.Y., Xu L., Gao X., Huang L., Nabel E.G., Nabel G.J. Gene transfer in vivo with DNA-liposome complexes: safety and acute toxicity in mice. Hum. Gene Ther. 1992;3:267–275. doi: 10.1089/hum.1992.3.3-267. [DOI] [PubMed] [Google Scholar]
- 64.Roos K.L. Lumbar puncture. Semin. Neurol. 2003;23:105–114. doi: 10.1055/s-2003-40758. [DOI] [PubMed] [Google Scholar]
- 65.Weiss M., Byrne A.J., Blazek K., Saliba D.G., Pease J.E., Perocheau D., Feldmann M., Udalova I.A. IRF5 controls both acute and chronic inflammation. Proc. Natl. Acad. Sci. USA. 2015;112:11001–11006. doi: 10.1073/pnas.1506254112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nickel F.T., Seifert F., Lanz S., Maihöfner C. Mechanisms of neuropathic pain. Eur. Neuropsychopharmacol. 2012;22:81–91. doi: 10.1016/j.euroneuro.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 67.Baron R., Binder A., Wasner G. Neuropathic pain: diagnosis, pathophysiological mechanisms, and treatment. Lancet Neurol. 2010;9:807–819. doi: 10.1016/S1474-4422(10)70143-5. [DOI] [PubMed] [Google Scholar]
- 68.Christianson D.R., Ozawa M.G., Pasqualini R., Arap W. Techniques to decipher molecular diversity by phage display. Methods Mol. Biol. 2007;357:385–406. doi: 10.1385/1-59745-214-9:385. [DOI] [PubMed] [Google Scholar]
- 69.Ogawa N., Kawai H., Terashima T., Kojima H., Oka K., Chan L., Maegawa H. Gene therapy for neuropathic pain by silencing of TNF-α expression with lentiviral vectors targeting the dorsal root ganglion in mice. PLoS ONE. 2014;9:e92073. doi: 10.1371/journal.pone.0092073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.