Abstract
The APPswe (Swedish) mutation in the amyloid precursor protein (APP) gene causes dominantly inherited Alzheimer’s disease (AD) as a result of increased β-secretase cleavage of the amyloid-β (Aβ) precursor protein. This leads to abnormally high Aβ levels, not only in brain but also in peripheral tissues of mutation carriers. Here, we selectively disrupted the human mutant APPSW allele using CRISPR. By applying CRISPR/Cas9 from Streptococcus pyogenes, we generated allele-specific deletions of either APPSW or APPWT. As measured by ELISA, conditioned media of targeted patient-derived fibroblasts displayed an approximate 60% reduction in secreted Aβ. Next, coding sequences for the APPSW-specific guide RNA (gRNA) and Cas9 were packaged into separate adeno-associated viral (AAV) vectors. Site-specific indel formation was achieved both in primary neurons isolated from APPSW transgenic mouse embryos (Tg2576) and after co-injection of these vectors into hippocampus of adult mice. Taken together, we here present proof-of-concept data that CRISPR/Cas9 can selectively disrupt the APPSW allele both ex vivo and in vivo—and thereby decrease pathogenic Aβ. Hence, this system may have the potential to be developed as a tool for gene therapy against AD caused by APPswe and other point mutations associated with increased Aβ.
Keywords: Alzheimer's disease, Swedish mutation, adeno-associated virus, genome editing, CRISPR, amyloid precursor protein, amyloid-β
Introduction
Early-onset dominant familial forms of Alzheimer’s disease (AD) can be caused by point mutations or deletions in the genes for amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2). To date, more than 30 pathogenic APP mutations have been described, and several of these are located at or near the sites where β- and γ-secretase cleave APP to generate amyloid-β (Aβ), the peptide that accumulates as extracellular plaques in the AD brain1 (reviewed in Bertram and Tanzi2).
The KM670/671NL APP mutation, indigenous to Sweden (APPswe for the mutation and APPSW for the mutant allele), is located at the β-secretase site and results in increased enzymatic cleavage by β-secretase of APP and, thus, increased Aβ levels (Figures 1A and 1B).3 Individuals heterozygous for APPSW have been reported to display approximately three times higher Aβ levels in both brain and peripheral tissues, such as fibroblasts and plasma, as compared to non-mutation carriers.4, 5
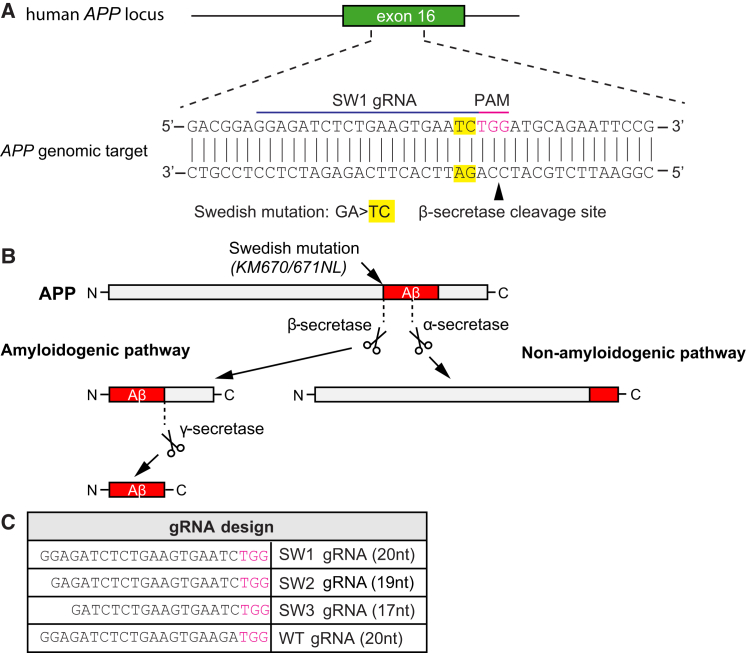
Figure 1.
CRISPR Targeting of Swedish KM670/671NL APP and Its Effect on Aβ Generation
(A) The Swedish KM670/671NL APP (APPswe) double-base change (yellow), causing familial Alzheimer’s disease (AD), is located near a potential Streptococcus pyogenes Cas9 PAM (NGG) site (purple). The mutation and gRNA target site is upstream of the β-secretase site. (B) In the amyloidogenic pathway, amyloid-β (Aβ) is produced via sequential cleavages of the amyloid precursor protein (APP) by β- and γ-secretases. No Aβ is generated upon α-secretase APP cleavage in the non-amyloidogenic pathway. The APPswe mutation is a better β-secretase substrate than the corresponding wild-type site, and individuals with this mutation develop AD as a consequence of elevated Aβ levels. (C) Three different gRNAs targeting the KM670/671NL site (SW1, SW2, and SW3) and one gRNA recognizing the wild-type sequence (WT) were evaluated. The PAM site is depicted in purple.
Gene editing by the CRISPR/Cas9 system is currently undergoing a rapid development and has also begun to be evaluated as a therapeutic strategy in various disease models6 (reviewed in Mali and Cheng7 and Hsu et al.8). Genomic DNA sequences with certain protospacer adjacent motif (PAM) sites (NGG in the case of Streptococcus pyogenes Cas9) can be targeted with short, approximately 20-nt-long, guide RNAs (gRNAs) that both base pair with these DNA sequences as well as mediate interaction with the Cas9 enzyme. This endonuclease then induces double-stranded DNA breaks 5′ of the PAM site that, depending on the conditions and cell type, will be repaired by cellular DNA repair pathways that include non-homologous end joining (NHEJ) and homology-directed repair (HDR).9 NHEJ is the predominant mechanism,10 which can lead to insertions or deletions (indels) at the targeted site. These indels can frequently lead to a frameshift in the coding sequence, thereby disrupting the gene expression (essentially by “knocking out” the gene).11 Thus, the combination of a targeted double-strand break and NHEJ-mediated repair constitutes a potential approach to disrupt dominantly inherited, disease-causing alleles. However, with dominant gene mutations, it will be important to ensure that disruption of the DNA sequence only occurs on the mutant and not on the wild-type allele.
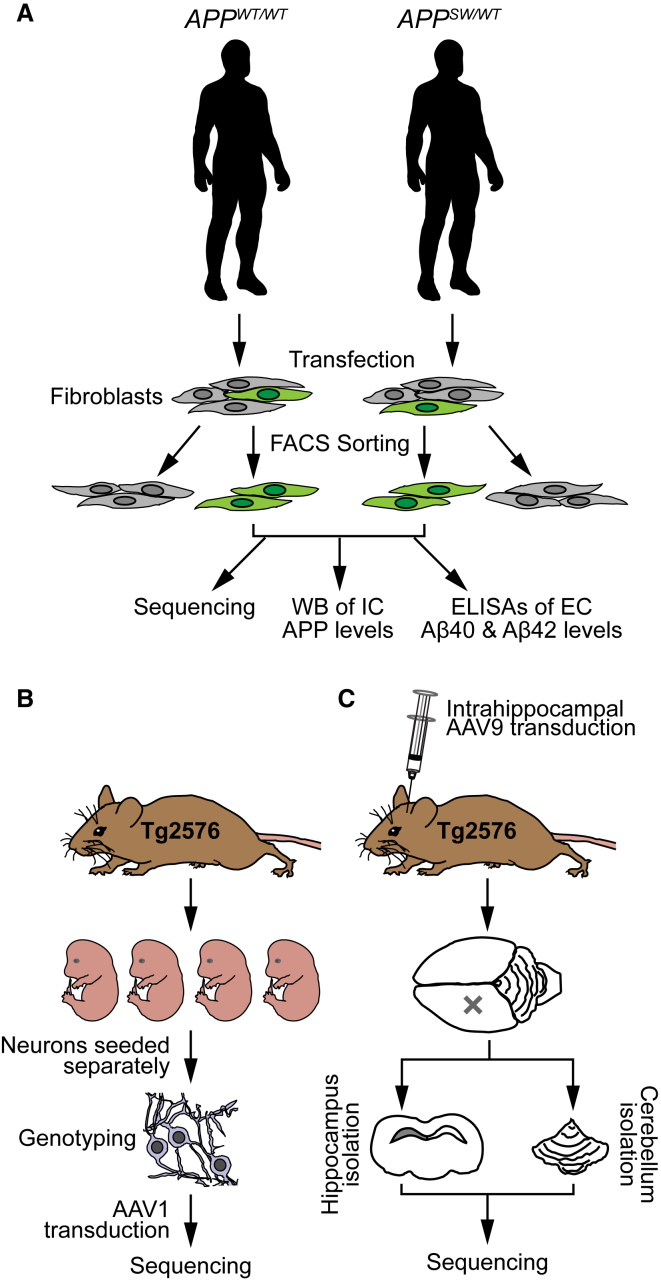
In this study, we developed a CRISPR/Cas9-based strategy to selectively target the mutant allele in the familial form of Alzheimer’s disease caused by the APPswe mutation. We hypothesized that disruption of the mutant allele would reduce the overproduction of Aβ in patient-derived cells. Moreover, we aimed to apply in vivo AAV delivery of CRISPR/Cas9 to disrupt the APPSW allele in transgenic APPswe mice. An overview of the study design is shown in Figure 2.
Figure 2.
Overview of the Study Design
(A) Fibroblasts were collected from human APPswe carriers and their non-affected relatives. The cells were transfected with S. pyogenes Cas9-2A-GFP and gRNAs. Successfully transfected cells were identified by GFP expression and sorted by FACS. Next, these cells were expanded in culture and analyzed by sequencing, western blot (WB) (for intracellular [IC] levels of APP), and ELISA (for extracellular [EC] secretion of Aβ40 and Aβ42). (B) Embryos from time-pregnant Tg2576 APPswe transgenic mice (embryonic day 14 [E14]–17) were used to generate primary cortical neuronal culture. The transgenic cultures were co-transduced for one day with AAV1-Cas9 and either AAV1-gRNA(SW1) at 3 days in vitro (DIV). At 21 DIV, the cells were collected for sequencing. (C) Adult Tg2576 transgenic mice were co-transduced unilaterally in hippocampus with AAV9-Cas9 and AAV9-gRNA(SW1). After 1 or 2 months, the mice were sacrificed and the injected hippocampi and non-injected cerebelli (as controls) were isolated for genomic DNA sequencing.
Results
CRISPR/Cas9-Mediated Knockout of APPSW or APPWT in Human Fibroblasts
To selectively disrupt APPSW or APPWT in the human patient-derived and control fibroblasts, we designed gRNAs against these alleles and transfected cell lines with an expression vector containing Cas9 and gRNA against APPSW or APPWT, respectively. We hypothesized that the APPswe mutation would be ideal for allele-specific recognition, as it consists of a double base change and the mutation is located immediately adjacent to an NGG PAM site in exon 16 of APP, placing it within the “seed” sequence (a 10- to 12-nt-long region proximal to the PAM site important for nuclease activity and specificity12) of a potential gRNA (Figure 1A). Because this mutation is close to the β-secretase site, we hypothesized that CRISPR-mediated disruption of APP upstream of this site would abrogate Aβ formation (Figure 1B).
We generated and tested three gRNAs with different lengths (SW1: 20 nucleotides, SW2: 19 nucleotides, and SW3: 17 nucleotides) against APPswe and one gRNA against APPwt, i.e., the same site in its non-mutated version (wild-type [WT]: 20 nucleotides; Figure 1C), to examine genome editing efficiency and specificity in cultured fibroblasts. We made these progressively more truncated gRNAs because our previous work had shown that decreasing the length of complementarity between the gRNA and its target DNA site can increase the specificity of recognition.13
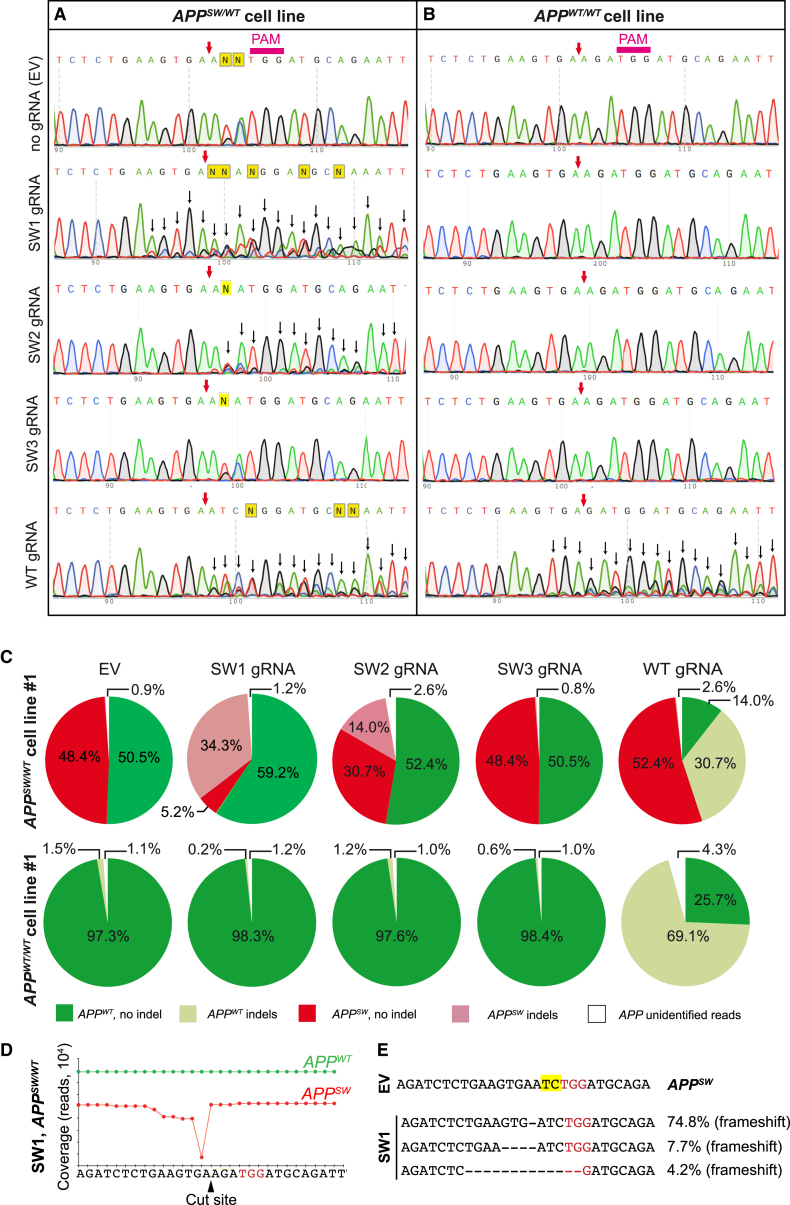
Fibroblasts from three mutation carriers and two non-mutated subjects from the same family were transfected with plasmids encoding the gRNA and SpCas9 co-translationally expressed with GFP. The GFP-positive cells was sorted by flow cytometry in order to enrich for CRISPR/Cas9-modified cells. Sanger sequencing of DNA from the transfected/sorted cells indicated site-specific gene disruption in GFP+ APPSW/WT cells treated with SW1, SW2, and WT gRNAs (Figure 3A). Additional peaks appearing around and downstream of the predicted cleavage site (Figure 3A, black arrows) indicated heterogeneity of the DNA samples (i.e., some reads showing indels were sequenced at the same time as reads without indels), characteristic of nuclease-induced mutations (Figure 3A). With SW3 gRNA, no indel formation could be detected (Figure 3A), suggesting that a 17-nt gRNA was inefficient in disrupting the APPSW allele. In contrast, there was no obvious gene disruption in APPWT/WT cells using the gRNA specific for APPswe (Figure 3B). However, we observed robust indel formation in APPWT/WT cells using the WT gRNA (Figure 3B).
Figure 3.
CRISPR/Cas9 Mediates Disruption of APPSW or APPWT Allele in Patient-Derived Fibroblasts
(A and B) Sanger sequencing of the APP gene from mutation carrier No. 1 and control line No. 1 transfected with Cas9-2A-GFP and gRNA targeting the APPSW (SW1, SW2, and SW3) or APPWT allele (WT), respectively. GFP-expressing cells were sorted 3 days after transfection. On the APPSW/WT cell line (A), SW1, SW2, and WT gRNA were disruptive and created indels (as indicated by background peaks [i.e., heterogeneity in reads] downstream of the cut site, black arrows). On the APPWT/WT cell line (B), WT gRNA led to indel formation, whereas SW1, SW2, or SW3 gRNA did not. PAM, protospacer-adjacent motif. The purple arrow shows the predicted CRISPR cut site. (C) Targeted deep sequencing of the APP allele from CRISPR-treated APPSW/WT fibroblasts (top pie charts) and control APPWT/WT fibroblasts (bottom pie charts) is shown. Colors indicate mutant (red), wild-type (green), mutant with indels (light red), and wild-type with indels (light green) reads. For some reads, we were not able to determine their origins as mutant or wild-type (due to sequencing errors or larger deletions), and these are marked as white areas on the pie charts. SW1 and SW2 gRNA resulted in indel formation in the APPSW, but not in the APPWT allele. WT gRNA resulted in indel formation only in the APPWT allele in APPSW/WT cells and APPWT/WT cells. SW3 did not lead to indel formation in any of the alleles. (D) Coverage of the APP allele in APPSW/WT cells, treated with Cas9 and SW1 gRNA, is shown. Deletions were detectable adjacent to the CRISPR cut site on the APPSW allele (red trace). No deletions were present on the APPWT allele. The PAM site is marked in red. (E) The most frequent reads in the APPSW/WT fibroblasts treated with Cas9 and SW1 gRNA are shown. PAM sites are marked with red, and the APPswe mutation is marked in yellow. The percentages refer to the fraction of reads that contained any indels.
Next, we characterized and quantified the extent of genome editing using targeted deep sequencing in CRISPR/Cas9-treated APPSW/WT and APPWT/WT fibroblasts (Figure 3C). Mutation-bearing cells transfected with an empty vector (EV) (Cas9-GFP without target gRNA) did not show any site-specific indels, and the number of mutant and WT reads in the samples were roughly equal (48.4% versus 50.5%, respectively; Figure 3C, top). However, APPSW/WT cells treated with Cas9 and SW1 or SW2 gRNAs showed a robust reduction in unperturbed APPSW reads, whereas the relative proportion of APPWT reads without indels did not decrease. Thus, CRISPR-induced indels were only detected in APPSW alleles and not in APPWT alleles when using gRNAs against the mutation. The SW3 gRNA did not show any detectable genome disruptions of either allele. Using the WT gRNA for APPSW/WT cells, we identified indels in APPWT alleles, but APPSW alleles were not affected by this gRNA (Figure 3C).
To further confirm the gRNA specificity, we performed targeted deep sequencing of the target site from CRISPR/Cas9-treated and control APPWT/WT cells. Importantly, we did not detect any indel formation in fibroblasts treated with SW1, SW2, and SW3 gRNAs (Figure 3C). However, indel formation was evident and very robust with the WT gRNA targeting the APPWT allele (Figure 3C, bottom). Taken together, these results confirm that the APPWT or APPSW alleles can be specifically targeted with the S. pyogenes CRISPR system.
Next, we characterized the type and location of the indels that occurred in SW1-treated APPSW/WT cells (Figures 3D and 3E). The most common indel event was a one-base-pair deletion at the predicted CRISPR cleavage site (74.8% of all APPSW reads), represented as a strong drop in coverage corresponding to this base pair position (Figure 3D). However, there was also a slight reduction in coverage of the positions preceding this nucleotide position, indicating that larger deletions had also occurred (Figure 3D). Approximately 7% of all mutant reads had a deletion of 4 nt, and about 4% of indels consisted of a deletion of 13 nt (Figure 3E). Some of these reads could not be assigned as either APPSW or APPWT as the mutation site itself was deleted. The presence of these larger deletions contributed to the number of “unidentified” reads in Figure 3C, the percentage of which was higher where CRISPR action was present (i.e., with SW1, SW2, and WT gRNAs in the case of APPSW/WT cells and WT gRNA in the case of APPWT/WT cells).
CRISPR/Cas9 Treatment Decreases Levels of Secreted Aβ
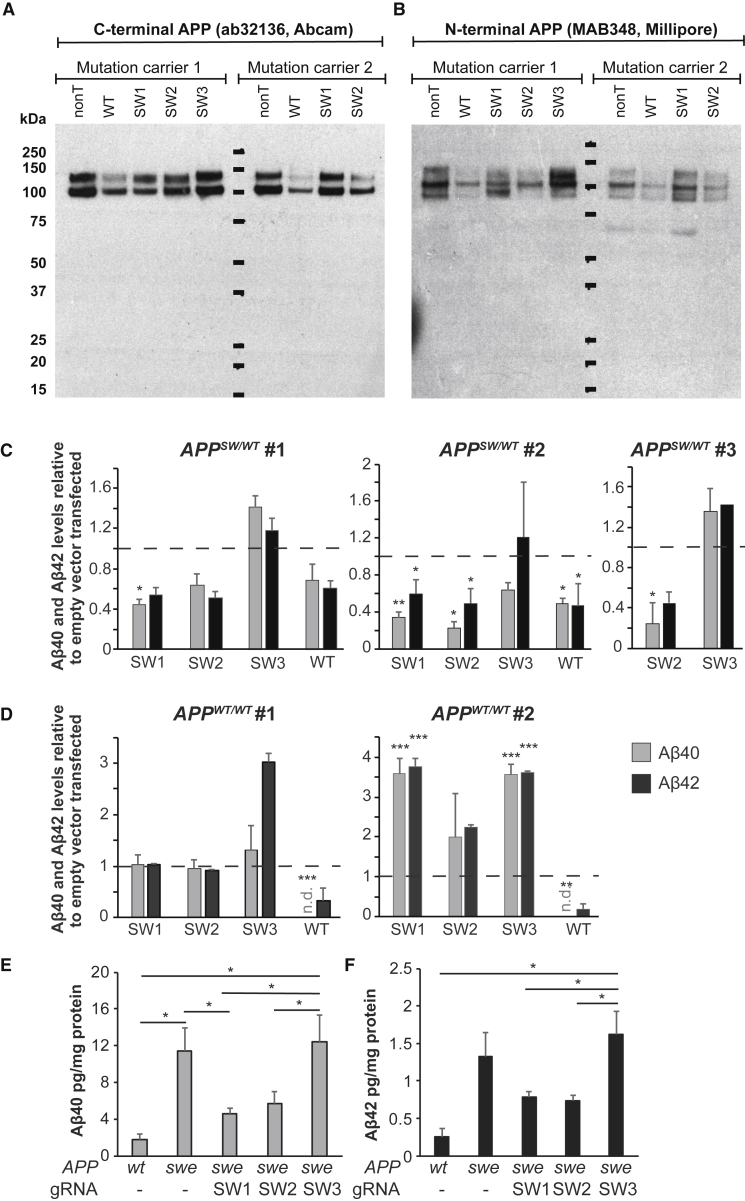
Next, we analyzed the effect of APP disruption at the protein level. Three different CRISPR/Cas9-treated cells lines were sorted for GFP expression and kept in culture for several weeks to generate enough cells for the measurements of intracellular APP and extracellular Aβ. Indel formation leading to a frameshift could potentially result in a protein that has a truncated C terminus. We did not see any statistically significant decrease of intracellular APP levels when probing for the C-terminal (Figures 4A and S1A) or N-terminal (Figures 4B and S1B) part of the protein, and we also did not see any truncated APP products by western blot using any of the gRNAs (Figures 4B and S1).
Figure 4.
CRISPR/Cas9 against APPSW and APPWT Reduces the Levels of Aβ
(A and B) Western blot of C terminus APP (C-APP) (A) and N terminus APP (N-APP) (B) of lysates from two mutation carriers. For quantification, see Figure S1. (C and D) Levels of secreted Aβ40 and Aβ42 in conditioned media of human CRISPR/Cas9-treated and control fibroblasts, as measured by ELISA. (C) shows APPSW/WT fibroblasts and (D) APPWT/WT fibroblasts. Data are normalized to total protein content and plotted as a relative value compared to empty vector transfected cells (value of empty vector [ = 1] is represented by dashed lines, mean ± SEM). The numbers (No. 1, No. 2, and No. 3) above the charts show cell lines from different patients or non-affected individuals. SW1, SW2, and SW3 are different gRNAs against the APPSW allele, whereas WT is a gRNA against the APPWT allele. n.d., not determined (below detection limit); *p < 0.05; **p < 0.01; ***p < 0.001. (E and F) Average of absolute levels (mean ±SEM) of Aβ40 (E) and Aβ42 (F) as measured by ELISA. swe, mutant lines; wt, wild-type lines; *p < 0.05 (one-way ANOVA with Tukey’s post hoc test).
Next, we analyzed whether CRISPR/Cas9 disruption of APPSW would lead to a decreased secretion of Aβ. Extracellular Aβ40 and Aβ42 peptide levels were assessed by ELISA in conditioned media from gRNA-treated cells, and the levels were normalized for total protein content determined by the bicinchoninic acid (BCA) assay (Figures 4C and 4D). In APPSW/WT cells expressing CRISPR/Cas9 (Figure 4C), we observed an approximately 60% reduction of Aβ40 levels (p < 0.05 and p < 0.01 for two separate cell lines, paired t test, respectively) when using the SW1 gRNA. The levels of Aβ42 were decreased by approximately 50% (significant for one of the cell lines; p < 0.05). The SW2 gRNA showed similar degrees of reduction (significant for 2 out of 3 lines for Aβ40; p < 0.01 and p < 0.05, respectively) and significant for 1 out of 3 lines for Aβ42 (p < 0.05). The SW3 gRNA treatment did not lead to any reduction in Aβ40 or Aβ42 levels in either of the mutation carrier fibroblasts tested (Figure 4C). Furthermore, the gRNA against the WT allele also reduced Aβ40 and Aβ42 levels by about 50% in APPSW/WT cells in one of the cell lines (p < 0.05; Figure 4C).
In contrast to the mutant cell lines, none of the gRNAs designed against APPswe led to any reduction of Aβ40 and Aβ42 in APPWT/WT cells (Figure 4D). However, the WT gRNA was highly effective in decreasing Aβ40 levels (non-detectable [n.d.] in both control cell lines; p < 0.001 and p < 0.01 for the two cell lines, respectively). Also, the Aβ42 levels seemed to be decreased, although the differences did not reach statistical significance (Figure 4D).
To compare the levels of secreted Aβ40 and Aβ42 between CRISPR/Cas9-treated and untreated patient fibroblasts and control fibroblasts, we averaged the ELISA data across all cell lines (from Figures 4C and 4D) and plotted Aβ40 and Aβ42 levels as normalized absolute protein concentrations (Figures 4E and 4F). In mutant cell lines, Aβ40 (Figure 4E) and Aβ42 (Figure 4F) were elevated roughly 6-fold compared to WT cells (p < 0.05 for Aβ40; not significant for Aβ42; one-way ANOVA; Tukey’s post hoc test). Conversely, the SW1 gRNA-treated mutation carriers showed significantly reduced levels of Aβ40 (p < 0.05; one-way ANOVA; Tukey’s post hoc test; Figure 4E), but not of Aβ42 (Figure 4F). The SW2 gRNA also seemed to result in a reduction of Aβ40, although the difference did not reach statistical significance. Finally, the SW3 gRNA did not change Aβ40 and Aβ42 levels as compared to the no-gRNA-treated mutation carriers (Figures 4E and 4F).
Overall, the ELISA analyses confirmed that both SW1 and SW2 were effective in decreasing Aβ levels in human cells.
CRISPR/Cas9 Treatment Leads to APP Indel Formation in Cultured Primary Neurons and Living Brains of APPswe Transgenic Mice
To extend the CRISPR/Cas9 technology against APPSW to the in vivo situation, we tested whether CRISPR/Cas9-mediated gene disruption could be performed in Tg2576 mice carrying multiple copies of the APPswe mutation. We aimed to directly inject the AAV-vector-packaged SW1 gRNA and Cas9 into the hippocampus of Tg2575 mice, which have multiple copies of human APPSW. We packaged Cas9/gRNA into exo-AAV1 (for in vitro experiments, as AAV1, but not AAV9 serotype, transduces neurons efficiently in vitro14) and exo-AAV9 vectors (for in vivo injections, as AAV9 serotype is highly efficient in transducing neurons after stereotactic injection in mice15).
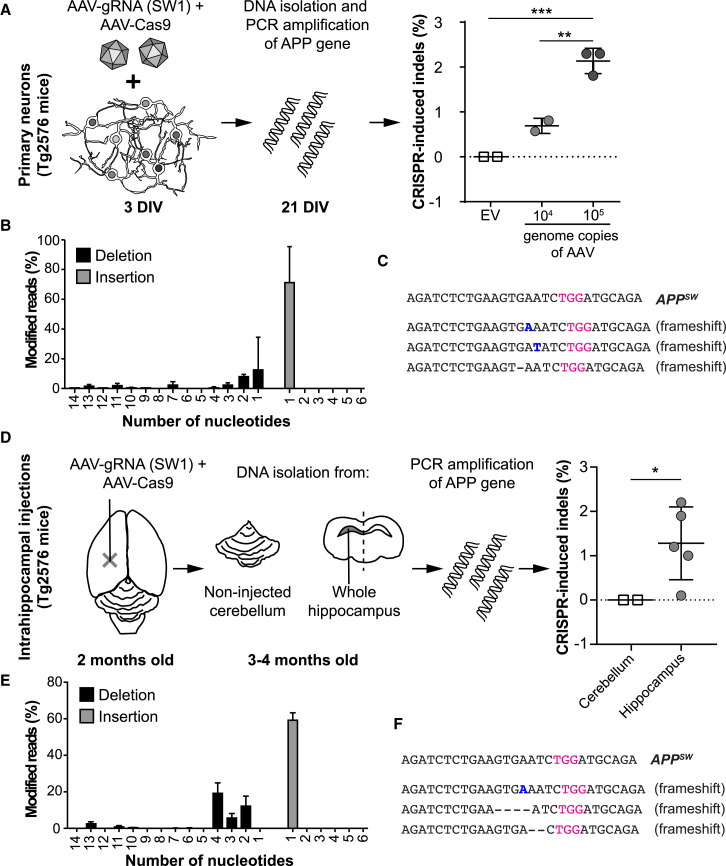
First, we evaluated the model by testing the CRISPR/Cas9 system on isolated primary cortical neurons from Tg2576 mice. Nineteen days after transduction, the neurons were analyzed by targeted high-throughput sequencing, which showed an average of 0.7% and 2.3% indel formation in APPSW alleles using 104 and 105 genome copies of AAV per cell, respectively. In contrast, neurons transduced with AAV-Cas9 and empty vector AAV-gRNA did not show any indel formation (Figure 5A). In contrast to the fibroblasts, where the predominant variants were one-base-pair deletions, one-base-pair insertions were the most highly enriched variants upon treatment of mouse cortical neurons (Figure 5B). The top three variants, representing over 90% of all reads containing indels, all led to frameshifts in the DNA sequence (Figure 5C). To determine whether this low percentage is a result of insufficient AAV delivery or CRISPR action, we also targeted an endogenous reference gene (mouse NC_000085.6 locus, using GGGTGGGACAGAACATCCCC as a gRNA). The AAV-mediated targeting of this site resulted in 13.0% ± 2.4% and 36.2% ± 5.4% indel formation when using 105 and 106 genome copies of AAV per cell, suggesting efficient AAV delivery and CRISPR action.
Figure 5.
CRISPR/Cas9 Can Specifically Target the APPSW Allele in Primary Neurons and in the Brain of Tg2576 Mice
(A) Primary cortical neurons from Tg2576 mice (embryonic day 14–17) were co-transduced with exo-AAV1-Cas9 and exo-AAV1-gRNA (SW1) or exo-AAV1-Cas9 with exo-AAV-gRNA (empty vector [EV]) at 3 days in vitro (DIV). At 21 DIV, total DNA was collected and analyzed for indel formation by targeted high-throughput sequencing. The SW1-gRNA-treated cells showed a 0.7% and 2.3% indel formation using 104 and 105 genomic copies of AAV per cell, respectively (mean ± SD). In contrast, EV-treated cells did not display indels (**p < 0.01; ***p < 0.001; one-way ANOVA with post hoc Tukey’s test). (B) Indel profile in vitro (mean ± SEM). (C) The most abundant CRISPR-induced changes in vitro on cortical neurons. Of all the indels presented after treatment with SW1, 70.9% were a single nucleotide insertion (inserted nucleotide is depicted in blue), whereas only about 12.6% were a single nucleotide deletion The target PAM site is marked in purple; inserted nucleotides are marked in blue. (D) Two-month-old Tg2576 mice were injected unilaterally into hippocampus with exo-AAV9-Cas9 and exo-AAV9-gRNA (SW1), 4–8 weeks post-injection, DNA was collected and analyzed from total hippocampus and from cerebellum as control. An average indel percent of 1.3% was calculated after high-throughput sequencing compared to no indels identified in cerebellum (*p < 0.05; Mann-Whitney test). Mean ± SD is plotted. (E) Indel profile in vivo (mean ± SD). (F) The most abundant CRISPR-induced changes in vivo. The most common indel was a single-nucleotide insertion (60.3%), the second most common indel a five-nucleotide deletion (21.5%), and the third most common a double deletion (10.1%).
After confirming ex vivo efficacy of the AAV-mediated delivery of SW1 gRNA, the AAV vectors encoding for CRISPR/Cas9 were injected into the hippocampus of adult Tg2576 mice (n = 5). After 6 weeks, the mice were sacrificed followed by DNA analyses. In addition to hippocampus (n = 5), non-injected cerebellum (n = 2) was also analyzed by high-throughput sequencing. All injected animals showed apparent indel formation in APPSW alleles in the hippocampus (an average of 1.3% of the transgenic APPSW alleles displayed indels), whereas the cerebellar tissue showed no indel formation (Figure 5D; significant; p < 0.05; Mann-Whitney test). The most abundant change was a one-base-pair insertion, similarly to the in-vitro-treated transgenic neurons (Figures 5E and 5F). We also observed larger deletions close to the target site (Figures 5E and 5F).
Taken together, these results confirm that, in primary cortical neurons and in the hippocampus of transgenic mice expressing the APPSW mutation, our CRISPR/Cas9 strategy targeting the mutant APP allele can disrupt this mutation leading to indel formations, resulting in frameshifts of the DNA sequence.
Discussion
There is an urgent need for novel therapies against AD. Although there have been great expectations for immunotherapy targeting Aβ, several recent trials have failed to show any clear efficacy.16, 17 However, other immunotherapy studies, specifically targeting aggregated toxic Aβ species, may prove to be more successful.18, 19 Yet other variations on Aβ immunotherapy protocols, as well as inhibitors of APP secretases, are also currently undergoing clinical trials.
Similar to these strategies, our approach also targets Aβ but instead at the genetic level. Gene therapy is currently in phase 1 trial for a number of neurological disorders, including Parkinson’s disease.20 Such efforts are propelled by novel tools enabling CNS delivery of agents that selectively target genetic factors underlying these conditions. For AD, the APPswe mutation, as well as other dominantly acting neurodegenerative disease genes, should be suitable targets.
Some of the familial forms of AD caused by mutations in the APP gene result in an increased generation and, hence, an accelerated deposition of the Aβ peptide in the affected brain. By targeting any of these mutations by gene therapy, Aβ levels could be reduced and pathology might thus be slowed down or even prevented. In addition to such familial disease forms, also conditions with an increased gene dosage could potentially be targeted by gene therapeutic strategies aimed at lowering gene expression. For example, both patients with APP duplications and subjects with Down syndrome develop Aβ brain pathology as a consequence of a 50% increase of APP expression.21, 22
Herein, we describe how the mutated allele in fibroblasts from AD patients with the APPswe mutation can be targeted with the CRISPR/Cas9 system. By designing gRNAs against this double-nucleotide mutation, we were able to achieve a strongly selective disruption of the mutated allele, leaving the wild-type allele intact. Importantly, we could also demonstrate a decreased generation of Aβ40 and Aβ42 in the conditioned media from patient cells treated with gRNA against the APPSW allele.
Among the gRNAs against the mutated site, SW1 and SW2 both showed a high efficacy of disrupting the APPSW allele and decreasing Aβ levels. These gRNAs were composed of 20 and 19 nt, respectively, whereas SW3 consisted of 17 nt. Both the 20- and 19-nt-long gRNAs were effective in disrupting the APP gene, predominantly by frameshift deletions with an associated decrease in the levels of both Aβ40 and Aβ42. In contrast, the 17-nt-long gRNA was completely ineffective, suggesting that the gRNA needs to exceed that length for this particular target site. Thus, our data suggest that 17-nt-long matching gRNAs are not universally functional.13
In this project, we also designed a gRNA against the corresponding non-mutated site. This 20-nt WT gRNA was found to be effective in terms of lowering Aβ in the non-mutated human cells by 50%–60%. As expected, also the heterozygous APPswe cells were affected by the WT gRNA treatment and displayed a moderate decrease of Aβ levels. Interestingly, the extent of Aβ reduction in non-mutated cells by WT gRNA slightly exceeds the 40% reduction in Aβ levels that has been reported for the protective APP A673T mutation, originally identified in the Icelandic population.23 However, in our cell-based model, the degree of Aβ reduction will to some extent depend on the stringency of cell sorting after gRNA/Cas9 transfection.
It has previously been described that PAM proximal bases confer higher specificity for the CRISPR system.24 Thus, the combined effect of its proximity to an NGG PAM site and its nature as a double-base-pair change makes the APPswe mutation very suitable for CRISPR targeting. Accordingly, we did not observe any indel formation in the APPSW allele when using a gRNA targeting the wild-type allele.
AAV-mediated gene editing by CRISPR has begun to be explored on animal models for various neurodegenerative diseases. Recently, it was demonstrated that targeting of SNPs in linkage disequilibrium with the pathogenic allele in a mouse model of Huntington’s disease (HD) could efficiently decrease both mRNA and protein levels of mutant huntingtin.25 In another recent study, Yang et al.26 demonstrated that CRISPR-mediated excision of the trinucleotide-repeat region of exon 1 in another HD mouse model could alleviate both brain pathology and motor disturbances in the mice.
So far, no study on the use of CRISPR/Cas9 on animal models for AD has been reported. However, it was recently shown that CRISPR-mediated correction of PSEN2 N141I could abolish the electrophysiological deficit and normalize the Aβ42/40 ratio in cultured basal forebrain cholingergic neuron derived from human induced pluripotent stem cells with this AD mutation.27
In this study, we investigated whether our CRISPR/Cas9 strategy could effectively disrupt the APPSW allele in vivo. For that reason, we utilized the Tg2576 mouse model, expressing the APPSW allele and displaying Aβ plaque pathology in the brain at 11 or 12 months of age.28 With AAV-mediated CRISPR/Cas9 delivery, we detected an approximate 2% indel formation in mutant alleles within the injected area. Although this number is relatively low, we have to take into account that we were not able to select for the CRISPR-expressing cells and that these mice harbor approximately 100 copies of the transgene.28, 29 Targeting a reference locus that is present only in two copies in the genome, as with the patient fibroblasts, we observed a much higher indel formation. This observation suggests that, in the case of APPswe, the limited gene disruption efficiency is a consequence of the high number of target alleles and not an insufficient AAV delivery of CRISPR. Thus, for this proof-of-concept part of the study, we did not attempt to assess functional restoration or reduced Aβ plaque pathology, because the behavioral changes in this mouse model are subtle and the CRISPR-disrupted alleles were relatively low in number. Hence, other genetic models, e.g., those based on knockin mutations of the human APP allele, will be needed to properly assess the in vivo efficiency of CRISPR-mediated gene disruption of APPswe or other mutations causing familial AD.
From our data, it is difficult to estimate the extent of cells that would have to be targeted to avoid disease development in the brain of a mutation carrier, but even a small degree of allelic disruption could be sufficient as the pathology is developing gradually over a long period of time. Thus, even if only a minor fraction of all APP alleles were affected in the AAV-injected brains, our approach could be therapeutically meaningful.
Taken together, our study provides the first experimental evidence that the CRISPR/Cas9 method can be used to specifically target an APP allele, causing an inherited form of AD, and normalize the increased expression of Aβ that is driving the pathogenesis. Thus, this method has the potential to be developed as a treatment strategy against AD caused by mutations in APP and other genes.
Materials and Methods
Please see Figure 2 for an overview of the study design.
Human Fibroblasts
Human APPswe fibroblasts (n = 3) and non-mutated control fibroblasts from subjects of the same family (n = 2) were isolated from skin biopsies and grown in 75-cm2 flasks (Corning, Corning, NY, USA) in DMEM (Gibco, Waltham, MA) with 10% of fetal bovine serum (FBS) (Gibco), 1× penicillin/streptomycin (Gibco), and 1× glutamine (Gibco). This part of the study was approved by the Ethical Committee at Massachusetts General Hospital, Boston and by the Regional Ethical Committee in Uppsala, Sweden (protocol number 2016/131). All experiments concerning human cells were carried out in accordance with the approved protocol.
Animals
Female (6 weeks old) B6;SJL-Tg(APPswe)2576Kha (Tg2576) mice that overexpress the human APP gene with the Swedish mutation were purchased from Taconic Biosciences (Hudson, NY). Mice were kept at a 12-hr light/dark cycle with access to food and water ad libitum. This part of the study was approved by the Institutional Animal Care and Use Committee at Massachusetts General Hospital, Boston (protocol number: 2004N000092). All experiments concerning animals were carried out in accordance with the approved protocol.
Generation of gRNAs and Cas9 Plasmids
The pSpCas9(BB)-2A-GFP (pX458) plasmid was used for the transfection experiments (gift from Dr. Feng Zhang; Addgene plasmid No. 48138).11 gRNA coding sequences were cloned into pX458 using BbsI (Thermo Scientific, Waltham, MA), as previously described.11 For AAV-mediated CRISPR delivery, we used the pAAV-pMecp2-SpCas9-spA and pAAV-U6sgRNA(SapI)_hSyn-GFP-KASH-bGH (pX551 and pX552, gifts from Dr. Feng Zhang; Addgene plasmid Nos. 60597 and 60958, respectively).30 gRNA coding sequences were cloned into pX552 using SapI (Thermo Scientific, Waltham, MA). Insertion of the gRNA cassette was verified by Sanger sequencing using the U6 sequencing primer: 5′-GACTATCATATGCTTACCGT-3′.
AAV Production
We isolated exosome-associated AAV (exo-AAV) vectors from cell culture media of virus producing HEK293T cells, as previously described.31, 32 Exo-AAV vectors have been shown to be superior compared to conventional AAV in transduction efficiency, both in cultured cells and living mouse brains.31, 33 Briefly, a conventional triple transfection protocol was used to introduce the transgene plasmids: the rep/cap plasmid (pXR1 for AAV134 and pAR9 for AAV9) and the adenovirus helper plasmid (pAdΔF635). Exo-AAV vectors were isolated from the medium three days after transfection using differential centrifugation, as previously described.31, 36 Vectors were stored at −80°C until use. The virus was titrated using qPCR with primers specific to the inverted terminal repeats of AAV after isolation of the AAV genomes using the Roche High Pure Viral Nucleic Acid Kit (Roche, Basel, Switzerland).37
Transfection and Sorting
To introduce CRISPR plasmids into human fibroblasts, we used electroporation (Nucleofector, Lonza, Basel, Switzerland). Approximately one million cells were used for transfection with pX458 containing different gRNAs against APPSW or APPWT. As a control, we used pX458 that did not contain any gRNA sequence (EV). Transfection efficiencies varied between 20% and 50%. Four days after transfection, GFP-positive and GFP-negative cells were sorted by fluorescence-activated cell sorting (FACS) (using BD FACS Aria II), enabling separation of Cas9-2A-GFP-expressing cells from non-expressing cells. We aimed at sorting out the approximately 5% highest GFP-expressing cells, and the same threshold was kept for every cell line. Each cell population was further cultured until confluency in the 12-well format was reached (approximately 2 months). Cells from one well were harvested for DNA extraction, and two additional wells were used to collect conditioned media for ELISA measurements.
Neuronal Culture
We set up timed pregnancies of the Tg2576 mice for embryo-derived, primary neuronal cultures. At embryonic day 14–17, the pregnant females were euthanized by CO2 and the embryos were collected. After dissection, the embryonic cortices were collected separately in Hank’s balanced salt solution with 1× penicillin/streptomycin and 10 mM HEPES buffer (HBSS) and placed on ice. The cortices were centrifuged at 200 × g for 5 min and dissociated in 500 μL plating medium (Neurobasal medium with 10 mM HEPES, 1× penicillin/streptomycin, 1× glutamine, and 10% FBS). Approximately 300,000 cells were plated in each well in plastic 12-well plates (Corning) coated with poly-L-ornithine (1:4 in H2O; Sigma-Aldrich) and laminin (1:1,000 in PBS). After 5 hr, the media was replaced with Neurobasal medium, similar to the plating medium but containing serum-free B-27 supplement instead of FBS. On day 3 after plating, the transgenic neurons were transduced with exo-AAV1-Mecp2-SpCas9-spA (referred to as AAV-Cas9) mixed with exo-AAV1- U6sgRNA(SapI)_hSyn-GFP-KASH-bGH (referred to as AAV-gRNA). We added 104 or 105 genomic copies per cell using a 2:1 AAV-Cas9 to AAV-gRNA ratio. Control cells were transduced with only AAV-Cas9 and AAV-gRNA construct without specific target gRNA (EV). The neurons were treated for 1 day with the AAV vectors, and DNA was isolated as described below at 21 days in vitro (DIV). All products were purchased from Thermo Fisher Scientific if not otherwise stated.
DNA Sequencing
DNA was extracted from human fibroblasts and mouse cortical neurons (Blood and Tissue Kit, QIAGEN, Valencia, CA) and resuspended in 10 mM TRIS-HCl (pH 8.5). We performed a PCR using a high-fidelity DNA polymerase (Phusion, New England Biolabs, Ipswich, MA) with intronic primers flanking exon 16 of the APP gene in human fibroblast cultures (forward: 5′-CAGGATGAACCAGAGTTAATAGGT-3′; reverse: 5′-CAGGATGAACCAGAGTTAATAGGT-3′). On genomic DNA samples from Tg2576 mouse neuronal cultures/brain samples, we performed PCR using primers in the coding sequence (as the tg2576 strain contains human cDNA without introns), flanking the mutation site (forward: 5′-CAGCCAACACAGAAAACGAA-3′; reverse: 5′-CACCTTTGTTTGAACCCACA-3′). The PCR products were run on a 1% agarose gel and then purified using a column-based precipitation method (QIAGEN). The purified PCR products (400–600 ng DNA) were submitted for Sanger sequencing and targeted high-throughput sequencing (Massachusetts General Hospital DNA Core, Cambridge, MA).
Next-Generation Sequencing and Bioinformatic Analyses
Following ligation of barcoding adapters and PCR amplification, DNA fragments were sequenced with MiSeq sequencer (Illumina, San Diego, CA, USA), generating 150-nt-long paired-end reads. Processing of DNA amplicon and sequencing was performed by the CCIB DNA Core Facility at Massachusetts General Hospital (Cambridge, MA). The quality of the reads was inspected with Fastqc application (version 0.11.3). The reads from heterozygous samples were segregated based on the presence of wild-type (“TCTGCATCCATC” and its reverse complement “GATGGATGCAGA”) and mutant (“TCTGCATCCAGA” and its reverse complement “TCTGGATGCAGA”; mutation site is underlined) sequences using custom Python script (version 3.4.2). An alignment of both segregated and non-segregated reads to the region of human APP gene (chr21: 27269744-27270179) was achieved with the bwa mem program (version 0.7.8; parameters bwa mem -M -t 6). The alignment was performed against the mutant sequence for all samples where gRNAs had been designed to target the mutant allele. This approach was chosen to prevent recognition of SNPs together with CRISPR-induced indels as a complex variant, which would have resulted in a less precise identification of particular indels. Conversion to the BAM format, sorting, and indexing were performed with the Samtools (version 1.3). The sequence variants representing indels present at an allelic frequency higher than 0.0001 were called by means of Vardict. The number of reads aligned to APP gene was calculated using BEDtools (version 2.23.0). Vcf. files generated with Vardict were parsed with VariantAnnotation package from Bioconductor to determine the type, length, frequency, and position of indels in wild-type and mutant reads. Homozygous samples were subjected to the same workflow without initial read segregation.
ELISA Measurements
The CRISPR/Cas9-modified human fibroblasts from patients and controls were maintained in DMEM with 10% FBS and 1× penicillin/streptomycin. For ELISA measurements, cells were replated and grown to confluency before they were further incubated for 48 hr in FBS-free cell culture medium. The conditioned medium was collected and subjected to ELISAs specific for Aβ40 (BNT77-BA27; No. 294-64701) and Aβ42 (BNT77-BC05; No. 292-64501; Wako Pure Chemicals Industries, Osaka, Japan). Both assays were performed according to the manufacturer’s instructions. A total of 90 μL serum-free conditioned media with 10 μL diluent buffer was analyzed in duplicates from each cell line. Two separate biological replicates were analyzed for all conditions. To normalize the Aβ levels to total protein levels, we performed Pierce BCA protein determination assay (Thermo Fisher Scientific) on the same samples.
Western Blotting
After having reached confluency in the 6-well format, fibroblasts were lysed using 50 μL RIPA buffer (Abcam, Cambridge, MA) supplemented with Complete protease inhibitor cocktail (Roche, Basel, Switzerland), before subjecting the samples to 15 min centrifugation at 12,000 × g (4°C). Total protein levels were measured on the supernatants using the Pierce BCA assay (Thermo Fisher Scientific). For each sample, 15 μg protein in lithium dodecyl sulfate loading buffer (Thermo Fisher Scientific) was loaded onto a 10-well 4%–12% Bis-Tris gel and run on ice for 1.5 hr at 150 V. Transfer to a nitrocellulose membrane was carried out on ice for 75 min at 30 V. Next, the membrane was incubated with one of the following primary antibodies (stripping in 0.4 M NaOH by 10 min with incubation at room temperature [RT] on a rocker was performed between each run): anti-C terminus (C-APP; ab32136; Abcam) and anti-N terminus (N-APP; clone 22C11; No. MAB348; Millipore, Billerica, MA) APP antibodies. Anti-GAPDH (No. NB100-56875; Novus Biologicals, Littleton, CO) and anti-vinculin (ab129002; Abcam) were both used as loading control antibodies. All antibodies were incubated in 5% milk in Tris-buffered saline with 0.1% Tween-20 (1:1,000) overnight at 4°C. Horseradish-peroxidase-linked secondary antibodies against rabbit (GE Healthcare, Chicago, IL) or mouse (Bio-Rad, Hercules, CA) immunoglobulin G (IgG) were used at a 1:20,000 dilution in 5% milk for 1 hr at RT, and ECL Prime (GE Healthcare) was used for development on Amersham hyperfilm ECL (GE Healthcare).
Intracerebral Injections in Mice
The Tg2576 mouse model, expressing Swedish KM670/671NL APP under the PrP promoter, was used for the in vivo experiments. Five mice were injected unilaterally into hippocampus (anteroposterior [AP] −2, mediolateral [ML] −1.5, and dorsoventral [DV] −2 related to bregma). A total of 2.3 × 109 genomic copies of AAV9-Mecp2-SpCas9-spA and AAV9-U6sgRNA(SW1 gRNA)-hSyn-GFP-KASH-bGH were injected at a 1:1 ratio in a total volume of 3 μL at a rate of 0.2 μL/min. After 1 or 2 months, the mice were sacrificed, followed by dissection of the injected hippocampus as well as of the cerebellum (as a non-transduced control region). Total DNA was extracted by QIAGEN Blood and Tissue kit with manual homogenization of the brain using disposable pestles (Fisherbrand; No. 12-141-363).
Author Contributions
B.G. designed the study, was responsible for the genetic analyses, and was one of the primary authors of the manuscript. C.L. performed the ex vivo and in vivo experiments on the transgenic mice and was one of the primary authors of the manuscript. M.P.Z. performed sequencing data analyses. S.T. performed most of the ELISAs. B.P.K. took part in the design and provided input to the study. C.C. and K.K. took part in the ELISA analyses. D.M. and A.V. performed work for the generation of AAV vectors. V.G. designed and interpreted some of the genetic analyses. L.L. provided the patient-based material and input to the study. C.A.M. was responsible for the generation of AAV vectors. J.K.J., B.T.H., and X.O.B. took part in the design and provided input to the study. M.I. planned the study, performed some of the cell based experiments, and was one of the primary authors of the manuscript. All authors have read and approved of the manuscript.
Conflicts of Interest
J.K.J. has financial interests in Beam Therapeutics, Editas Medicine, Monitor Biotechnologies, Pairwise Plants, Poseida Therapeutics, and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Acknowledgments
J.K.J. is the Desmond and Ann Heathwood Massachusetts General Hospital Research Scholar. B.G. is an Edward R. and Anne G. Lefler Center Postdoctoral Fellow. C.L. was supported by the Swedish Brain Foundation. M.P.Z. is a recipient of a scholarship from the Kosciuszko Foundation. B.P.K. acknowledges support from Banting (Natural Sciences and Engineering Research Council of Canada) and Charles A. King Trust Postdoctoral Fellowships. M.I. was supported financially by grants from The Marianne and Marcus Wallenberg Foundation. The authors thank the Massachusetts General Hospital, Center for Computational and Integrative Biology DNA Core for sequencing services. Funding was provided from U19 CA179563 by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director (XOB), and an NIH National Institute of General Medical Sciences Maximizing Investigators’ Research Award (MIRA) R35 118158 (J.K.J.). We also thank Bradley Chapman (Harvard Chan Bioinformatics Core) for reviewing our computational approach and scripts applied in the analysis. Analysis of next-generation sequencing data for this study was conducted on the Orchestra High Performance Compute Cluster at Harvard Medical School. The facility is supported by NIH and through NCRR grant 1S10RR028832-01. This environment is managed by the Research Computing group at Harvard Medical School (https://rc.hms.harvard.edu).
Footnotes
Supplemental Information includes one figure and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.03.007.
Supplemental Information
References
- 1.Bertram L., McQueen M.B., Mullin K., Blacker D., Tanzi R.E. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 2.Bertram L., Tanzi R.E. The genetics of Alzheimer’s disease. Prog. Mol. Biol. Transl. Sci. 2012;107:79–100. doi: 10.1016/B978-0-12-385883-2.00008-4. [DOI] [PubMed] [Google Scholar]
- 3.Mullan M., Crawford F., Axelman K., Houlden H., Lilius L., Winblad B., Lannfelt L. A pathogenic mutation for probable Alzheimer’s disease in the APP gene at the N-terminus of beta-amyloid. Nat. Genet. 1992;1:345–347. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- 4.Citron M., Vigo-Pelfrey C., Teplow D.B., Miller C., Schenk D., Johnston J., Winblad B., Venizelos N., Lannfelt L., Selkoe D.J. Excessive production of amyloid beta-protein by peripheral cells of symptomatic and presymptomatic patients carrying the Swedish familial Alzheimer disease mutation. Proc. Natl. Acad. Sci. USA. 1994;91:11993–11997. doi: 10.1073/pnas.91.25.11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnston J.A., Cowburn R.F., Norgren S., Wiehager B., Venizelos N., Winblad B., Vigo-Pelfrey C., Schenk D., Lannfelt L., O’Neill C. Increased beta-amyloid release and levels of amyloid precursor protein (APP) in fibroblast cell lines from family members with the Swedish Alzheimer’s disease APP670/671 mutation. FEBS Lett. 1994;354:274–278. doi: 10.1016/0014-5793(94)01137-0. [DOI] [PubMed] [Google Scholar]
- 6.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habin N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P., Cheng L. Concise review: human cell engineering: cellular reprogramming and genome editing. Stem Cells. 2012;30:75–81. doi: 10.1002/stem.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semenova E., Jore M.M., Datsenko K.A., Semenova A., Westra E.R., Wanner B., van der Oost J., Brouns S.J., Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. USA. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Royo N.C., Vandenberghe L.H., Ma J.Y., Hauspurg A., Yu L., Maronski M., Johnston J., Dichter M.A., Wilson J.M., Watson D.J. Specific AAV serotypes stably transduce primary hippocampal and cortical cultures with high efficiency and low toxicity. Brain Res. 2008;1190:15–22. doi: 10.1016/j.brainres.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alves S., Bode J., Bemelmans A.P., von Kalle C., Cartier N., Tews B. Ultramicroscopy as a novel tool to unravel the tropism of AAV gene therapy vectors in the brain. Sci. Rep. 2016;6:28272. doi: 10.1038/srep28272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salloway S., Sperling R., Fox N.C., Blennow K., Klunk W., Raskind M., Sabbagh M., Honig L.S., Porsteinsson A.P., Ferris S., Bapineuzumab 301 and 302 Clinical Trial Investigators Two phase 3 trials of bapineuzumab in mild-to-moderate Alzheimer’s disease. N. Engl. J. Med. 2014;370:322–333. doi: 10.1056/NEJMoa1304839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doody R.S., Thomas R.G., Farlow M., Iwatsubo T., Vellas B., Joffe S., Kieburtz K., Raman R., Sun X., Aisen P.S., Alzheimer’s Disease Cooperative Study Steering Committee. Solanezumab Study Group Phase 3 trials of solanezumab for mild-to-moderate Alzheimer’s disease. N. Engl. J. Med. 2014;370:311–321. doi: 10.1056/NEJMoa1312889. [DOI] [PubMed] [Google Scholar]
- 18.Lannfelt L., Möller C., Basun H., Osswald G., Sehlin D., Satlin A., Logovinsky V., Gellerfors P. Perspectives on future Alzheimer therapies: amyloid-β protofibrils - a new target for immunotherapy with BAN2401 in Alzheimer’s disease. Alzheimers Res. Ther. 2014;6:16. doi: 10.1186/alzrt246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sevigny J., Chiao P., Bussière T., Weinreb P.H., Williams L., Maier M., Dunstan R., Salloway S., Chen T., Ling Y. The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature. 2016;537:50–56. doi: 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- 20.Palfi S., Gurruchaga J.M., Ralph G.S., Lepetit H., Lavisse S., Buttery P.C., Watts C., Miskin J., Kelleher M., Deeley S. Long-term safety and tolerability of ProSavin, a lentiviral vector-based gene therapy for Parkinson’s disease: a dose escalation, open-label, phase 1/2 trial. Lancet. 2014;383:1138–1146. doi: 10.1016/S0140-6736(13)61939-X. [DOI] [PubMed] [Google Scholar]
- 21.Rovelet-Lecrux A., Hannequin D., Raux G., Le Meur N., Laquerrière A., Vital A., Dumanchin C., Feuillette S., Brice A., Vercelletto M. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat. Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 22.Rumble B., Retallack R., Hilbich C., Simms G., Multhaup G., Martins R., Hockey A., Montgomery P., Beyreuther K., Masters C.L. Amyloid A4 protein and its precursor in Down’s syndrome and Alzheimer’s disease. N. Engl. J. Med. 1989;320:1446–1452. doi: 10.1056/NEJM198906013202203. [DOI] [PubMed] [Google Scholar]
- 23.Jonsson T., Atwal J.K., Steinberg S., Snaedal J., Jonsson P.V., Bjornsson S., Stefansson H., Sulem P., Gudbjartsson D., Maloney J. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 24.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monteys A.M., Ebanks S.A., Keiser M.S., Davidson B.L. CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Mol. Ther. 2017;25:12–23. doi: 10.1016/j.ymthe.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang S., Chang R., Yang H., Zhao T., Hong Y., Kong H.E., Sun X., Qin Z., Jin P., Li S., Li X.J. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Invest. 2017;127:2719–2724. doi: 10.1172/JCI92087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortiz-Virumbrales M., Moreno C.L., Kurglikov I., Marazuela P., Sproul A., Jacob S., Zimmer M., Paull D., Zhang B., Schadt E.E. CRISPR/Cas-9Correctable mutation-related molecular and physiological phenotypes in iPSC-derived Alzheimer’s PSEN2 N141I neurons. Acta Neuropathol. Commun. 2017;5:77. doi: 10.1186/s40478-017-0475-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsiao K., Chapman P., Nilsen S., Eckman C., Harigaya Y., Younkin S., Yang F., Cole G. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 29.Hsiao K.K., Borchelt D.R., Olson K., Johannsdottir R., Kitt C., Yunis W., Xu S., Eckman C., Younkin S., Price D. Age-related CNS disorder and early death in transgenic FVB/N mice overexpressing Alzheimer amyloid precursor proteins. Neuron. 1995;15:1203–1218. doi: 10.1016/0896-6273(95)90107-8. [DOI] [PubMed] [Google Scholar]
- 30.Swiech L., Heidenreich M., Banerjee A., Habib N., Li Y., Trombetta J., Sur M., Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.György B., Fitzpatrick Z., Crommentuijn M.H., Mu D., Maguire C.A. Naturally enveloped AAV vectors for shielding neutralizing antibodies and robust gene delivery in vivo. Biomaterials. 2014;35:7598–7609. doi: 10.1016/j.biomaterials.2014.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maguire C.A., Balaj L., Sivaraman S., Crommentuijn M.H., Ericsson M., Mincheva-Nilsson L., Baranov V., Gianni D., Tannous B.A., Sena-Esteves N. Microvesicle-associated AAV vector as a novel gene delivery system. Mol. Ther. 2012;20:960–971. doi: 10.1038/mt.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hudry E., Martin C., Gandhi S., György B., Scheffer D.I., Mu D., Merkel S.F., Mingozzi F., Fitzpatrick Z., Dimant H. Exosome-associated AAV vector as a robust and convenient neuroscience tool. Gene Ther. 2016;23:819. doi: 10.1038/gt.2016.65. [DOI] [PubMed] [Google Scholar]
- 34.Rabinowitz J.E., Rolling F., Li C., Conrath H., Xiao W., Xiao X., Samulski R.J. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 2002;76:791–801. doi: 10.1128/JVI.76.2.791-801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao X., Li J., Samulski R.J. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.György B., Sage C., Indzhykulian A.A., Scheffer D.I., Brisson A.R., Tan S., Wu X., Volak A., Mu D., Tamvakologos P.I. Rescue of hearing by gene delivery to inner-ear hair cells using exosome-associated AAV. Mol. Ther. 2017;25:379–391. doi: 10.1016/j.ymthe.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aurnhammer C., Haase M., Muether N., Hausl M., Rauschhuber C., Huber I., Nitschko H., Busch U., Sing A., Ehrhardt A., Baiker A. Universal real-time PCR for the detection and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum. Gene Ther. Methods. 2012;23:18–28. doi: 10.1089/hgtb.2011.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.