Abstract
Transcription by RNA polymerase can induce the formation of hypernegatively supercoiled DNA in vitro and in vivo. This phenomenon has been nicely explained by a “twin-supercoiled-domain” model of transcription where a positively supercoiled domain is generated ahead the RNA polymerase and a negatively supercoiled domain behind it. In E. coli topA strains, DNA gyrase selectively converts the positively supercoiled domain into negative supercoils to produce hypernegatively supercoiled DNA. In this article, in order to examine whether promoter strength affects transcription-coupled DNA supercoiling (TCDS), we developed a two-plasmid system in which a linear, non-supercoiled plasmid was used to express lac repressor constitutively while a circular plasmid was used to gauge TCDS in E. coli cells. Using this two-plasmid system, we found that TCDS in topA strains is dependent on promoter strength. We also demonstrated that transcription-coupled hypernegative supercoiling of plasmid DNA did not need the expression of a membrane-insertion protein for strong promoters; however, it might require co-transcriptional synthesis of a polypeptide. Furthermore, we found that for weak promoters the expression of a membrane-insertion tet gene was not sufficient for the production of hypernegatively supercoiled DNA. Our results can be explained by the “twin-supercoiled-domain” model of transcription where the friction force applied to E. coli RNA polymerase plays a critical role in the generation of hypernegatively supercoiled DNA.
Keywords: Transcription-coupled DNA supercoiling, promoter strength, E. coli topA strains, twin-supercoiled-domain model of transcription, hypernegatively supercoiled DNA
1. Introduction
DNA supercoiling plays fundamental roles in a number of essential DNA metabolic pathways, such as DNA replication, recombination, and transcription (Bates and Maxwell 2005; Cozzarelli and Wang 1990; James C.Wang 2009). In E. coli, DNA is typically negatively supercoiled. DNA supercoiling status inside E. coli cells is primarily set by counter actions of two DNA topoisomerases, DNA gyrase, and topoisomerase I (Champoux 2001; Snoep et al 2002; Wang 1996; Zechiedrich et al 2000). Inactivating DNA gyrase or toposipomerase I results in the production of positively (Lockshon and Morris 1983) or hypernegatively (Pruss 1985) supercoiled DNA, respectively.
Since the 1980s, it has been demonstrated that transcription by RNA polymerase could introduce supercoils to plasmid DNA templates in vitro and in vivo (Leng et al 2004; Leng and McMacken 2002; Lockshon and Morris 1983; Pruss 1985; Tsao et al 1989; Wu et al 1988). Liu and Wang proposed a “twin-supercoiled-domain” model of transcription to explain how transcription by RNA polymerase is able to supercoil the plasmid DNA templates (Liu and Wang 1987). This elegant model hypothesizes that a transcribing RNA polymerase becomes increasingly more difficult to rotate around the axis of the DNA double helix as the size of the growing RNA transcript increases. At a critical point, energetically, it is more feasible for the DNA molecule to rotate around its own helix axis to produce a positively supercoiled domain in front of the RNA polymerase and a negative supercoiled domain behind it. These two transient supercoiled domains may be relaxed by DNA topoisomerases or cancel each other by diffusion (Leng and McMacken 2002; Mielke et al 2004; Nelson 1999; Tsao, Wu, and Liu 1989; Wu, Shyy, Wang, and Liu 1988).
So far, there is substantial experimental evidence to support the “twin-supercoiled-domain” model of transcription (Albert et al 1996; Cook et al 1992; Dunaway and Ostrander 1993; Leng and McMacken 2002; Lodge et al 1989; Lynch and Wang 1993a; Ma et al 1994; Stupina and Wang 2004; Tsao, Wu, and Liu 1989; Wu, Shyy, Wang, and Liu 1988). For instance, in E. coli topoisomerase I-deficient (topA) strains, transcription by RNA polymerases is capable of driving the plasmid DNA templates to hypernegatively supercoiled status (Cook, Ma, Pon, and Hearst 1992; Lodge, Kazic, and Berg 1989; Pruss 1985; Wang and Lynch 1993). It was shown that transcription-coupled hypernegative supercoiling of plasmid DNA required co-transcriptional synthesis of a membrane-associated protein or polypeptide for plasmid pBR322 and derivatives (Cook, Ma, Pon, and Hearst 1992; Lodge, Kazic, and Berg 1989; Lynch and Wang 1993b; Ma, Cook, Pon, and Hearst 1994). A possible explanation for this requirement is that co-transcriptional synthesis of a membrane-associated protein or polypeptide substantially increased the friction force against the transcribing RNA polymerase. In this scenario, a significant amount of “twin-supercoiled-domains” are generated. After the positively supercoiled domain is converted into negative supercoils by DNA gyrase, the transcribed DNA templates become hypernegatively supercoiled.
Using a similar approach, we recently demonstrated that transcription by T7 RNA polymerase strikingly stimulated DNA supercoiling; transcription-coupled DNA supercoiling (TCDS) was dependent on the length of RNA transcripts in E. coli topA strains VS111(DE3) and DM800(DE3) (Samul and Leng 2007). Additionally, we found that hypernegative supercoiling of plasmid DNA by T7 RNA polymerase did not require anchoring of DNA to the bacterial cytoplasmic membrane (Samul and Leng 2007). We attributed these results to the fact that a much stronger T7 promoter and a much faster T7 RNA polymerase (comparing with E. coli RNA polymerase (Seidel and Dekker 2007)) were used in our transcription-supercoiling (T-S) assays. In this case, the “twin-supercoiled-domains” were efficiently generated and, as a result, TCDS did not need transcriptional machinery to couple to translation and membrane-insertion. These results also suggested that promoter strength is important to TCDS in E. coli cells. In order to further study how promoter strength affects the efficiency of TCDS in E. coli topA strains, herein we developed a new two-plasmid system: the first plasmid is a linear plasmid derived from coliphage N15 (Ravin and Ravin 1999) and was used to express lac repressor constitutively. In addition, the linear plasmids cannot be supercoiled (Deneke et al 2000) and therefore will not interfere with the supercoiling assays; the second plasmid is a circular plasmid that was used to examine TCDS by E. coli RNA polymerase. Using this unique two-plasmid system, we found that transcription-coupled hypernegative supercoiling of plasmid DNA templates was dependent on promoter strength and did not require the expression of a membrane-insertion protein for strong promoters, which is consistent with our results for T7 RNA polymerase (Samul and Leng 2007).
2. MATERIAL AND METHODS
2.1. Materials
Ethidium bromide, kanamycin, lysozyme, and chloroquine were purchased from Sigma-Aldrich Corp. (St. Louis, MO). Ampicillin and bovine serum albumin (BSA) were obtained from Fisher Scientific (Fairlawn, NJ). Tetracycline was purchased from EMI Science (Gibbstown, NJ). Isopropyl-β-D-thiogalactopyranoside (IPTG) was obtained from Anatrace, Inc (Maumee, Ohio). All restriction enzymes, T4 DNA ligase, and T4 polynucleotide kinase were bought from New England Biolabs, Inc. (Beverly, MA). Pfu DNA polymerase was purchased from Stratagene, Inc. (La Jolla, CA). All synthetic oligonucleotides used as primers were obtained from MWG-Biotech, Inc. (Huntsville, AL). QIAprep Spin Miniprep Kit, QIAquick Gel Extraction Kit, RNeasy Mini Kit, and QIAquick Nucleotide Removal Kit were bought from QIAGEN, Inc. (Valencia, CA). ThermoScript RT-PCR System plus Platinum® Taq DNA polymerase was purchased from Invitrogen, Inc. (Carlsbad, CA). Power SYBR Green PCR Master Mix was obtained from Applied Biosystems, Inc. (Carlsbad, CA). GFP-Ab2 Mouse Monoclonal Antibody is a product of Thermo Fisher Scientific, Inc. (Fremont, CA). Horseradish peroxidase (HRP)-conjugated anti-mouse antibody was obtained from EMD Biosciences, Inc. (Madison, WI). Supersignal West Pico Chemiluminescent Substrate was bought from Thermo Scientific, Inc. (Rockford, IL).
2.2. Bacterial strains and plasmids
E. coli strain VS111 [F−LAM- rph-I ΔtopA] as described in Stupina and Wang (Stupina and Wang 2005) was obtained from the Coli Genetic Stock Collection/E. coli Genetic Resource Center (CGSC) at Yale University. E. coli strain DM800 [F−Δ(topAcysB)204 arcA13 gyrB225] was kindly provided by Dr. Marc Drolet at Universite de Montreal. All linear plasmids were derived from coliphage N15-based, linear plasmid pG591 (Ravin and Ravin 1999), which was kindly provided by Dr. Nikolai V. Ravin at Centre “Bioengineering” RAS, Russia. Plasmid pZXD4 was constructed by inserting a 33 bp synthetic DNA fragment containing a multiple cloning site into the unique Bgl II site of pG591. Plasmid pZXD51 (Fig. 1A) was constructed in two steps. First, promoter Placi controlling the expression of LacI in pET-30a(+) was mutated to the strong promoter Placiq using PCR-based, site-directed mutagenesis. Then the laci gene including promoter Placiq was amplified by PCR and inserted between NheI and AflII sites of pZXD4 to generate plasmid pZXD51. E. coli strains carrying pZXD51 express LacI constitutively.
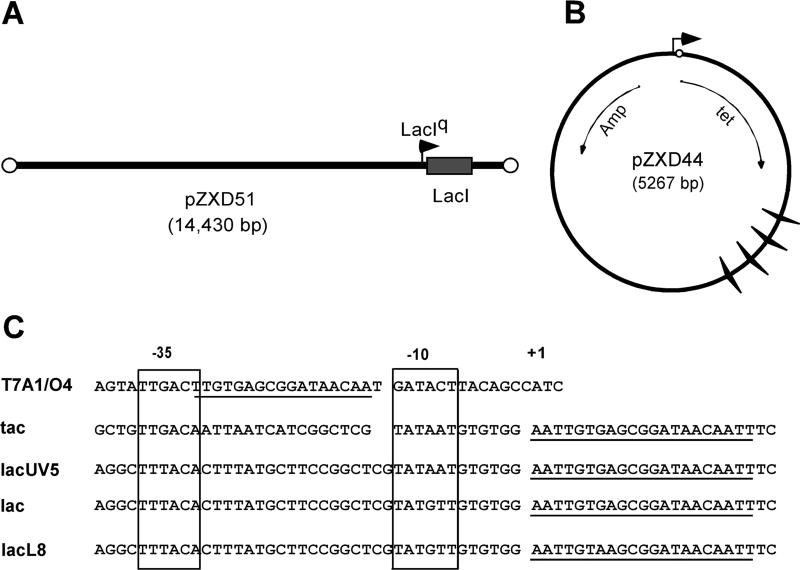
Figure 1. A two-plasmid system to study TCDS in vivo.
This system contains two plasmids, a linear plasmid (A), i.e., pZXD51 and a circular, supercoiling-reporter plasmid (B), such as pZXD44. The linear plasmid is derived from linear coliphage N15-based plasmid pG591 and carries a laci gene under the control of the strong PlacIq promoter. E. coli cells containing pZXD51 over-express lac repressor (LacI) constitutively, which binds to the lac O1 operator (the open circle) on the supercoiling-reporter plasmids. The supercoiling-reporter plasmids were derived from plasmid pBR322 and constructed as detailed under Experimental Procedures. They harbor an IPTG-inducible promoter with different strengths and a transcription unit between the promoter and a set of 4 Rho-independent E. coli rrnB T1 terminators (winged triangles). (C)The DNA sequence of five different E. coli promoters PT7A1/O4, Ptac, PlacUV5, Plac, and PlacL8. The underlines represent the lac O1 operators.
All circular plasmids constructed in this work were derived from plasmid pBR322. Plasmid pBR322se1 was constructed by converting the −35 region of promoter Panti-tet into an XhoI site using PCR-based, site-directed mutagenesis. In this case, promoter Panti-tet was removed. Plasmid pBR322se2 was created after a Shine-Dalgarno sequence (5’-AAGGAGG-3’) was inserted to the upstream region of the open reading frame of the tet gene. Plasmid pBR322se3 was made by introducing KpnI and SacI sites into the plasmid surrounding the weak promoter Pbla. In this scenario, promoter Pbla may easily be replaced by other promoters. Plasmid pZXD7 was created by removing a Dcm sequence of pBR322se3 associated with the unique MscI recognition site using PCR-based, site-directed mutagenesis. Plasmid pZXD8 was generated after a BglII site was inserted to the downstream region of the tet gene of plasmid pZXD7. A 37 bp synthetic deoxyoligonucleotide containing a multiple cloning site was then inserted into the unique KpnI site of pZXD8 to yield plasmid pZXD9. Plasmid pZXD11 was constructed after a 36 bp synthetic DNA fragment containing a T7 promoter was inserted into the EcoRI and XhoI sites to replace promoter Ptet. Plasmid pZXD12 was made after a 64 bp synthetic DNA fragment containing an inactive promoter Pleu500 was inserted into the HindIII and KpnI sites of pZXD11. Plasmid pZXD14 was created when an 813 bp DNA fragment of pLUC1 carrying four tandem copies of rrnB T1 transcription terminators (Leng and McMacken 2002) was inserted into the unique MscI site of pZXD12. Plasmids pZXD44, 50, 49, 47, and 48 were constructed by replacing the T7 promoter with E. coli promoters PT7A1/O4, Ptac, PlacUV5, Plac, and PlacL8, respectively, between the EcoRI and XhoI sites of pZXD14.
Plasmids pZXD57, 58, 56, 55, and 54, each carrying a GFPuv gene under the control of E. coli promoters PT7A1/O4, Ptac, PlacUV5, Plac and PlacL8, respectively, were constructed in a few steps. First, an AgeI site was introduced between the Shine-Dalgarno sequence and the start codon of the tet gene of plasmid pXZD48 to generate plasmid pXZD52 using PCR-based, site-directed mutagenesis. Second, the unique XhoI site in the GFPuv gene of plasmid pGFPuv (Stratagene, Inc., La Jolla, CA) was silently removed without changing the open reading frame of GFPuv gene using PCR-based, site-directed mutagenesis to yield plasmid pZXD53. Third, a 737 bp PCR product containing the GFPuv gene was cloned into the AgeI and BsmI sites of plasmid pZXD52 to produce plasmid pZXD54. In this case, the tet gene was replaced by the GFPuv gene under the control of E. coli promoter PlacL8. Plasmids pZXD57, 58, 56, and 55 were constructed by replacing the E. coli promoter PlacL8 with promoters PT7A1/O4, Ptac, PlacUV5, and Plac between the EcoRI and XhoI sites of pZXD54, respectively.
Plasmid pZXD59 was created when a 735 bp DNA fragment containing the GFPuv gene in reverse orientation amplified from plasmid pZXD53 was inserted into the AgeI and BsmI sites of plasmid pZXD57. In this scenario, E. coli cells carrying pZXD59 are not able to express GFPuv protein after IPTG induction. Plasmids pZXD60 and 61 were produced when lacZ gene in the forward and reverse orientations was amplified from plasmid pYC2/CTlacZ and inserted into the AgeI and BsmI sites of pZXD57. In this case, E. coli cells carrying pZXD60 are able to express β-galactosidase after IPTG induction. However, E. coli cells carrying pZXD61 are not able to express β-galactosidase after IPTG induction. Plasmid pZXD62 was constructed after a 2.3 kb PCR fragment of lacZ gene was inserted between the HindIII and KpnI sites of pZXD57. Plasmid pZXD63 was produced after a 1.8 kb PCR fragment of lacZ gene was inserted into the HindIII and KpnI sites of pZXD44. Plasmids pZXD60A and pZXD63A were constructed where the start codon (ATG) of the lacZ and tet genes were, respectively, mutated to the stop codon TAG using PCR-based, site-directed mutagenesis. In this scenario, E. coli cells carrying plasmids pZXD60A and 63A are not able to express β-galactosidase and tetracycline resistance protein, respectively. Please notice that plasmids pZXD60, 61, 62, 63, 60A, and 63A have the same size, i.e., 7,055 bp.
2.3. In vivo T-S assays
E. coli cells carrying different plasmids were grown overnight in LB containing 50 µg/ml of ampicillin and kanamycin. The overnight culture was then diluted (1:100) in fresh LB containing 50 µg/ml of ampicillin and kanamycin, and grown until optical density of the cells at 600 nm reached approximately 0.5. IPTG (final concentration, 1 mM) was added to the cell culture to initiate transcription by different promoters, i.e., PT7A1/O4, Ptac, PlacUV5, Plac, and PlacL8. Plasmid DNA was purified using QIAGEN Miniprep Kit. The topological state of each DNA preparation was analyzed by electrophoresis in a 1% agarose gel in 1×TAE buffer (40 mM Tris-acetate, 1 mM EDTA, and pH 7.8) containing 2.5 µg/ml of chloroquine. After electrophoresis, agarose gels were stained with ethidium bromide, destained, and photographed under UV light. The net intensity of DNA topoisomers was determined using KODAK 1D Image Analysis Software. The percentage of hypernegative DNA supercoils was calculated by dividing the intensity of the hypernegatively supercoiled DNA band by the total intensity of all DNA topoisomers.
2.4. Western blotting experiments
Western blotting experiments were used to verify the expression of GFPuv protein in E. coli topA strain VS111 after 1 mM of IPTG induction. Total protein purified from E. coli cells was analyzed by electrophoresis in a 15% SDS-PAGE and electrophoretically transferred to a 0.45-nm nitrocellulose membrane. The membrane blot was then blocked with a solution containing 5% nonfat skim milk in TBST (50 mM Tris-HCl, pH 8.0, 138 mM NaCl, 2.7 mM KCl, and 0.05% Tween-20) for 45 min at room temperature and incubated with the primary antibody, GFP Ab-2 mouse monoclonal antibody, diluted 1:1000 in TBST solution overnight at 4°C. After the overnight incubation, the membrane blot was washed three times with TBST, blocked with a solution containing 5% nonfat skim milk in TBST for 15 min at room temperature, and then incubated for 1 hour with an HRP-conjugated anti-mouse IgG secondary antibody (diluted 1:5000) at room temperature. The immunoreactive GFPuv protein was detected with Supersignal West Pico Chemiluminescent Substrate.
2.5. RNA isolation, cDNA synthesis, and polymerase chain reaction (PCR)
Total RNA was isolated from E. coli cells using QIAGEN RNeasy Kit as described by the manufacturer. To determine the integrity of the total RNA samples, 16S and 23S rRNA were resolved by electrophoresis in a 1.2% agarose gel in 1×MOPS buffer containing formaldehyde (20 mM MOPS, 8 mM sodium acetate, 1 mM EDTA, 1% formaldehyde, and pH 7.0). After electrophoresis, agarose gels were stained with ethidium bromide, destained, and photographed under UV light. cDNA were synthesized from total RNA samples using ThermoScript RT-PCR System. 2.76 µg of RNA was mixed with a sequence-specific primer (final concentration, 0.5 µM) or random hexamer primers (50 ng/µl) and four deoxynucleotide triphosphates (dNTPs; final concentration, 1 mM). The mixtures were incubated at 65°C for 5 min and transferred to ice for another 5 min to remove secondary structures of RNA. The denatured RNA samples were then mixed with 1×cDNA synthesis buffer with a total volume of 20 µl containing 5 mM DTT, 40 units of RNaseOut, and 15 units of ThermoScript Reverse Transcriptase, and incubated at 60°C for 1 hour to synthesize cDNA. The cDNA synthesis mixtures were transferred to an 85°C water bath for 5 min to terminate the reactions. After the synthesis step, 2 units of RNase H were added to the reaction mixtures and incubated at 37°C for 20 min to remove the RNA templates.
PCR reactions were carried out using cDNA samples synthesized as described above. A 50 µl PCR reaction contains 1×PCR Buffer without Mg2+, 1.58 mM MgCl2, 0.2 mM dNTPs, 0.2 µM of each primer (Table 1), 0.5 µl cDNA, and 2 units of Platinum Taq DNA polymerase. The reactions started at 94°C for 2 min, proceeded for 16 cycles of 94°C for 30 sec, 55°C for 30 sec and 72°C for 1 min, and terminated at 72°C for 10 min. Subsequently, the PCR products were analyzed by electrophoresis in a 12% polyacrylamide gel in 1×TAE buffer. After electrophoresis, polyacrylamide gels were stained with ethidium bromide or SYBR gold, destained, and photographed under UV light.
Table 1.
DNA oligonucelotides used for primers of the RT-PCR experiments
| Oligoa | Sequence (5’-3’) | Location in the gene | Gene | PCR products |
|---|---|---|---|---|
| FL594F | ATTATGGCCCACACCAGTGGCGC | 2917–2939 | LacZ | 173bp, distalb |
| FL594R | TGACGGGCTCCAGGAGTCGTC | 3069–3089 | LacZ | 173bp, distalb |
| FL654F | CACCGATCGCCCTTCCCAACAGTTG | 212–236 | LacZ | 172bp, proximalc |
| FL654R | GTAGATGGGCGCATCGTAACCG | 362–383 | LacZ | 172bp, proximalc |
| FL580F | GCGAGGCTGGATGGCCTTCC | 990–1009 | tet | 176bp, distalb |
| FL580R | CCGTGACGATCAGCGGTCCAG | 1145–1165 | tet | 176bp, distalb |
| FL657F | GGCCTCTTGCGGGATATCGTCCATTCC | 184–210 | tet | 168bp, proximalc |
| FL657R | GATAGTGGCTCCAAGTAGCGAAGCG | 327–351 | tet | 168bp, proximalc |
| FL590F | TCCAATTGGCGATGGCCCTGT | 660–680 | GFPuv | 101bp |
| FL590R | GGACCATGTGGTCACGCTTTTCGT | 737–760 | GFPuv | 101bp |
| FL586F | AGTTATCCCCCTCCATCAGG | 154–135 | 16S rRNA | 99bp |
| FL586R | TGCAAGTCGAACGGTAACAG | 56–75 | 16S rRNA | 99bp |
FLXXXF and FLXXXR represent the forward and reverse primers of the PCR reactions, respectively.
Distal indicates the PCR products that locate in the distal region of the gene.
Proximal indicates the PCR products locating in the proximal region of the gene.
2.6. Real-time PCR Assays
Real-time PCR assays were carried out using MiniOpticon Real-time PCR system (Bio-rad, Hercules, CA). A 20 µl reaction contains 0.5 µl cDNA, 0.5 µM of each primer (Table 1), and 10 µl of Power SYBR Green PCR Master Mix (2×). The reaction started at 95°C for 10 min and continued for 40 cycles at 95°C for 15 sec and 60°C for 1 min. The Cq values (quantification cycle values) were calculated from the exponential phase of each PCR amplification reaction as recommended by the manufacturer.
3. Results
3.1. Establish an IPTG-inducible, two-plasmid system to study TCDS in E. coli topA strains
In this study we established a two-plasmid system to examine effects of different factors on TCDS in E. coli topA strains VS111 and DM800. The first plasmid pZXD51 (Fig. 1A) is a linear plasmid derived from coliphage N15-based, low-copy-number plasmid pG591 (Ravin and Ravin 1999) where a laci gene was cloned under the control of the strong Placiq promoter. In this case, E. coli strains carrying pZXD51 produce ~3,000 molecules of LacI per cell constitutively (Lutz and Bujard 1997). The second plasmid is a circular plasmid that serves as a supercoiling-reporter (Fig. 1B). Here we constructed a series of plasmid DNA templates that contain different strengths of E.coli promoters (Lanzer and Bujard 1988), i.e., PT7A1/O4, Ptac, PlacUV5, Plac, and PlacL8 (Fig. 1C), to examine effects of promoter strength on TCDS by E. coli RNA polymerase. Since each promoter region contains a lac O1 operator (Fig. 1C), transcription initiated from these promoters is IPTG-inducible in E. coli cells overexpressing LacI, e.g., E. coli cells carrying the linear plasmid pZXD51. We also added a set of Rho-independent, rrnB T1 transcription terminators to each plasmid. The presence of multiple rrnB T1 terminators enabled us to restrict transcription to selected regions of supercoiling-reporter plasmids and to modulate the length of RNA transcripts produced. Since supercoiling-reporter plasmids have a different DNA replication origin (pMB1 origin), they can co-exist with the linear plasmid pZXD51 in E. coli cells. Indeed, these two types of plasmids were able to simultaneously transform E. coli topA strains VS111 and DM800. Additionally, as mentioned above, an advantage of using a linear plasmid to express LacI is that linear plasmids cannot be supercoiled (Deneke, Ziegelin, Lurz, and Lanka 2000) and, as a result, will not interfere with the supercoiling assays. In this case, this new system will be ideal for our in vivo supercoiling studies. Plasmids constructed in this study are summarized in Fig. 1 and Table 2.
Table 2.
Plasmids constructed in this study
| Plasmid | E. coli Promoter | Gene under control of the cloned promoter |
|---|---|---|
| pZXD44 | T7A1/O4 | teta |
| pZXD50 | tac | teta |
| pZXD49 | lacUV5 | teta |
| pZXD47 | lac | teta |
| pZXD48 | lacL8 | teta |
| pZXD57 | T7A1/O4 | GFPuvb |
| pZXD58 | tac | GFPuvb |
| pZXD56 | lacUV5 | GFPuvb |
| pZXD55 | lac | GFPuvb |
| pZXD54 | lacL8 | GFPuvb |
| pZXD59 | T7A1/O4 | Reverse GFPuvb |
| pZXD60 | T7A1/O4 | lacZc |
| pZXD60A | T7A1/O4 | N/Ad |
| pZXD61 | T7A1/O4 | Reverse lacZc |
| pZXD62 | T7A1/O4 | GFPuvb |
| pZXD63 | T7A1/O4 | teta |
| pZXD63A | T7A1/O4 | N/Ad |
The tetracycline resistance gene of pBR322 (tet) encodes a 41 kD transmembrane protein TetA.
The green fluorescence protein UV (GFPuv) gene encodes a cytosolic protein GFPuv.
The lacZ gene encodes a cytosolic protein β-D-galactosidase.
N/A represents not applicable. Plasmid pZXD60A and 63A have an amber mutation in the start codon of lacZ and tet, respectively.
3.2. Transcription-coupled hypernegative supercoiling of plasmid DNA is dependent on promoter strength
Having established the two-plasmid system, we proceeded to examine whether TCDS is dependent on promoter strength after IPTG induction. As mentioned above, we simultaneously introduced the linear plasmid pZXD51 and a supercoiling-reporter plasmid into E. coli topA strains VS111 and DM800. Since previous studies showed that hypernegative supercoiling of plasmid pBR322 and its derivatives is dependent on the expression of the co-transcription and translation of membrane-associated tet gene (Cook, Ma, Pon, and Hearst 1992; Lodge, Kazic, and Berg 1989; Lynch and Wang 1993a; Ma, Cook, Pon, and Hearst 1994; Pruss 1985; Pruss and Drlica 1986; Stupina and Wang 2005), we decided to use a set of five supercoiling-reporter plasmids that carry a tet gene under the control of an IPTG-inducible promoter with different strengths, i.e., promoters PT7A1/O4, Ptac, PlacUV5, Plac, and PlacL8. As expected, our RT-PCR experiments (Fig. 2) demonstrated that the transcription level of E. coli strains harboring different plasmids after IPTG induction is correlated with promoter strength in vivo (Lanzer and Bujard 1988). Interestingly, E. coli cells carrying plasmids pZXD44 and 50 were able to grow on agar plates containing 10 µg/ml of tetracycline even in the absence of IPTG (Fig. S1A). This resistance is most likely due to the leaky expression of the tetracycline resistance protein from the strong PT7A1/O4 and Ptac promoters, since pZXD44 and 50 contain PT7A1/O4 and Ptac, respectively. However, only E. coli cells carrying plasmids pZXD50 and 49 were able to grow on agar plates containing 10 µg/ml of tetracycline and 1 mM of IPTG (Fig. S1B). As demonstrated previously, overexpression of tetracycline resistance protein results in cell death (Eckert and Beck 1989), which is likely the reason for E. coli cells harboring plasmid pZXD44 carrying the strong PT7A1/O4 promoter being unable to grow on agar plates in the presence of 1 mM of IPTG. Indeed, our results showed that IPTG was able to inhibit cell growth for E. coli strains VS111 and MG1655 harboring plasmid pZXD44 (Fig. S1C and D). Because promoters Plac and PlacL8 are too weak, E. coli cells carrying plasmids pZXD47 and 48 could not produce enough tetracycline resistance protein to overcome the antimicrobial activities of tetracycline.
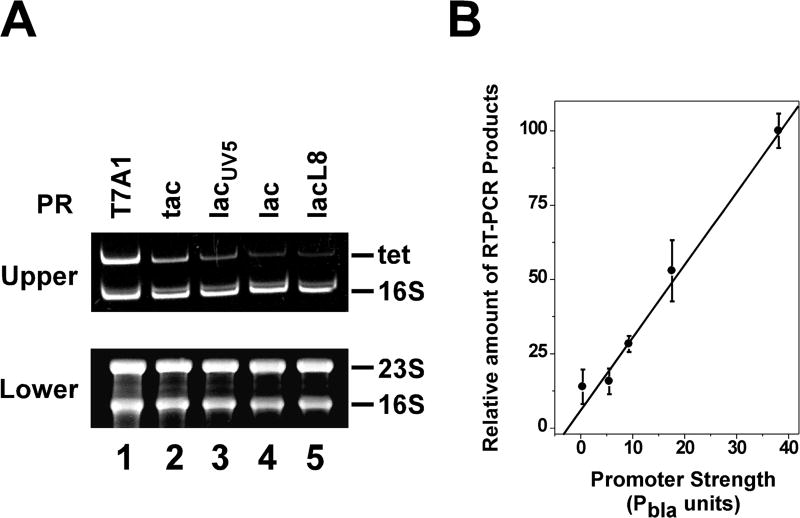
Figure 2. RT-PCR analysis of cDNA products of mRNA transcribed from different supercoiling-reporter plasmids pZXD44, 50, 49, 47, and 48 in E. coli topA strain VS111 harboring the linear plasmid pZXD51 after 10 min of IPTG induction (1 mM).
(A) RT-PCR experiments were performed as described under Experimental Procedures. The lower panel is a 1.2% agarose gel containing 1% formaldehyde to show the integrity of the RNA samples used for the RT-PCR experiments. The upper panel is a 12% polyacrylamide gel in 1×TAE buffer to show the PCR products of 16S rRNA and tet gene cDNA synthesized from the RNA samples isolated from E. coli strain VS111 carrying supercoiling-reporter plasmids pZXD44, 50, 49, 47, and 48 after 10 min of IPTG induction (lanes 1–5 respectively). Labels: PR, promoter; T7A1, the T7A1/O4 promoter; tac, the tac promoter; lacUV5, the lacUV5 promoter; lac, the lac promoter; lacL8, the lacL8 promoter. (B) Real-time RT-PCR analyses of the tet gene mRNA for E. coli strain VS111 carrying different supercoiling-reporter plasmids pZXD44, 50, 49, 47, and 48 after 10 min of IPTG induction (mean±SD, three independent experiments). The relative level of RT-PCR products is proportional to the promoter strength. Promoter strength in Pbla units was obtained from ref.(Lanzer and Bujard 1988)
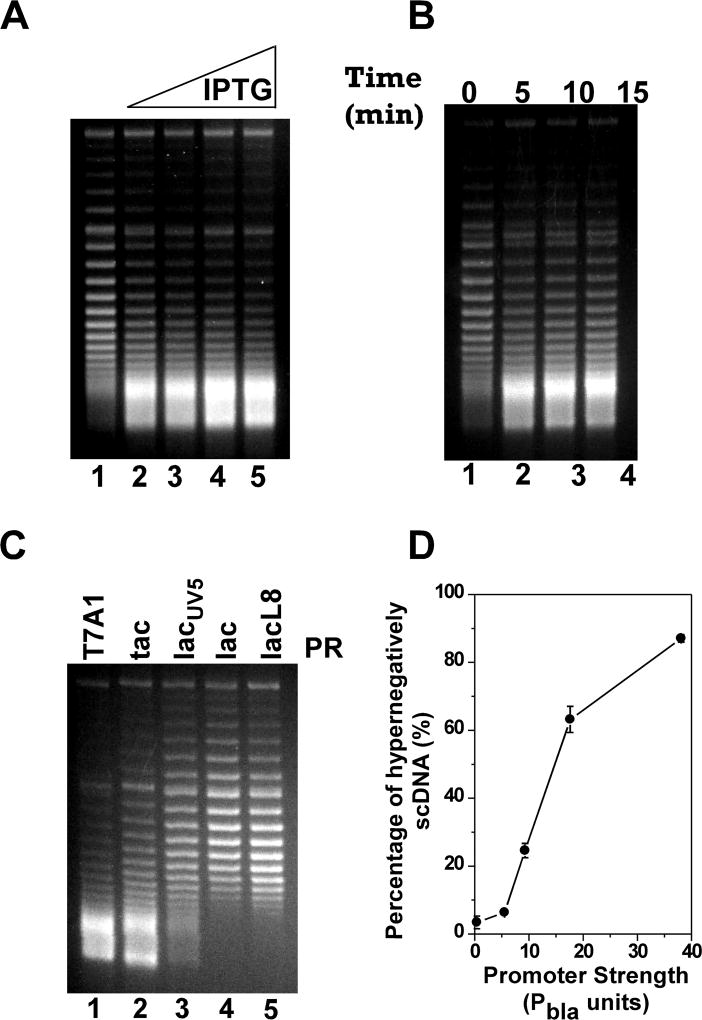
We next determined the topological status of the set of five supercoiling-reporter plasmids pZXD44, 50, 49, 47, and 48 in E. coli topA strain VS111 harboring the linear plasmid pZXD51. Fig. 3 shows the results. Before IPTG induction, plasmid pZXD44, which carries a strong PT7A1/O4 promoter, had a superhelical density, σ, of approximately −0.06 to −0.07 (Fig. 3A and 3B, lane 1). In the presence of the DNA intercalator chloroquine (2.5 µg/ml), this plasmid migrated during agarose gel electrophoresis as if it contained a few negative supercoils. After IPTG induction, as expected, some topoisomers quickly became hypernegatively supercoiled (estimated σ<−0.09; hypernegatively supercoiled DNA is the fastest moving band in the gels where DNA topoisomers are no longer resolvable under our experimental conditions): the amount of hypernegatively supercoiled DNA was dependent on the IPTG concentration added to the cell culture (Fig. 3A and S2) and the induction time (Fig. 3B and S2). These results clearly demonstrated that the induction of expression of the membrane-insertion tet gene under the control of the strong PT7A1/O4 promoter was able to drive the formation of hypernegatively supercoiled DNA, which is consistent with previous published results (Cook, Ma, Pon, and Hearst 1992; Lynch and Wang 1993a; Pruss 1985). Similar results were obtained for plasmids pZXD50 and 49 which carry Ptac and PlacUV5, respectively (Fig. 3C and S2). However, to our surprise, IPTG was not able to induce the production of hypernegatively supercoiled DNA for plasmids pZXD47 and 48, which harbor the weak promoters, Plac and PlacL8, respectively (lanes 4 and 5 of Fig. 3C). These results demonstrated that the expression of a membrane-insertion tet gene is not sufficient for the production of hypernegatively supercoiled DNA. Intriguingly, our results showed that transcription-coupled hypernegative supercoiling of plasmid DNA is dependent on promoter strength: the stronger the promoter, the more hypernegatively-supercoiled DNA produced (Fig. 3C and 3D). Similar results were also obtained using E. coli topA strain DM800 as the host strain (Fig. S2D).
Figure 3. TCDS in E. coli strain VS111 is dependent on promoter strength.
The in vivo T-S assays were performed as described under Experimental Procedures. DNA topoisomers were resolved by electrophoresis in a 1% agarose gel containing 2.5 µg/ml chloroquine and stained with ethidium bromide. The fastest moving band in the gels where DNA topoisomers are no longer resolvable under our experimental conditions represents the hypernegatively supercoiled DNA. (A) Dependence of TCDS on IPTG concentration for plasmid pZXD44 carrying a tet gene under the control of the strong PT7A1/O4 promoter. Lane 1 contained the DNA sample isolated from E. coli cells prior to IPTG induction. Lanes 2–5 contained the DNA samples isolated from E. coli cells after 10 min of induction with 25, 50, 100, and 1000 µM IPTG, respectively. (B) Time course of the hypernegative supercoiling of plasmid pZXD44 in E. coli strain VS111 after 1 mM of IPTG induction. Lanes 1–4 contained, respectively, DNA samples isolated from VS111 after 0, 5, 10, and 15 min of IPTG induction. (C) Dependence of TCDS on promoter strength. Lanes 1–5 contained, respectively, DNA samples isolated from E. coli topA strain VS111 containing plasmids pZXD44, 50, 49, 47, and 48 after 5 min of IPTG (1 mM) induction. These plasmids carry a tet gene under the control of IPTG-inducible promoters with different strengths, i.e., promoters PT7A1/O4, Ptac, PlacUV5, Plac, and PlacL8. Labels: PR, promoter; T7A1, the T7A1/O4 promoter; tac, the tac promoter; lacUV5, the lacUV5 promoter; lac, the lac promoter; lacL8, the lacL8 promoter. (D) The percentage of hypernegatively supercoiled DNA is proportional to promoter strength (the values are the average of at least three independent determinations and the standard deviations are shown). These results were calculated as described under Experimental Procedures using the TCDS data shown in (C). Promoter strength in Pbla units was obtained from ref. (Lanzer and Bujard 1988)
3.3. Transcription-coupled hypernegative supercoiling of plasmid DNA in E. coli topA strains did not require the expression of a membrane-insertion protein for strong promoters
As mentioned above, we recently found that hypernegative supercoiling of plasmid DNA by T7 RNA polymerase did not require anchoring of DNA to bacterial cytoplasmic membrane (Samul and Leng 2007). Thus, we decided to examine whether TCDS by E. coli RNA polymerase in E. coli topA strains VS111 and DM800 requires anchoring of DNA to bacterial cytoplasmic membrane through co-transcriptional synthesis of polypeptides encoding membrane proteins in this new two-plasmid system. Our results showed that, for strong promoters, TCDS did not require anchoring of DNA to bacterial cytoplasmic membrane through the expression of a membrane-insertion protein.
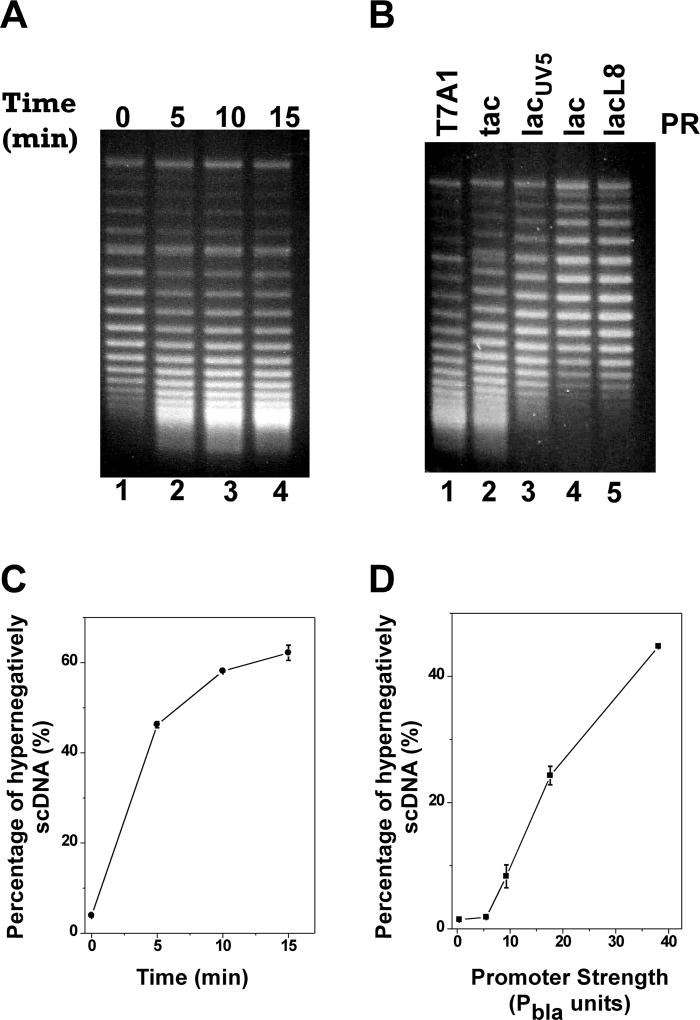
We first constructed a set of five supercoiling-reporter plasmids, pZXD57, 58, 56, 55, and 54 that carry a cytosolic green fluorescence protein UV (GFPuv) gene under the control of IPTG-inducible promoters with different strengths, i.e., promoters PT7A1/O4, Ptac, PlacUV5, Plac, and PlacL8 (Fig. S3 and Table 2). These plasmids were introduced to E. coli topA strains VS111 and DM800 that also harbor the linear plasmid pZXD51. As expected, after IPTG induction, GFPuv gene products in E. coli strains harboring different plasmids at transcription and expression levels were correlated with promoter strength (Fig. S4). Interestingly and also as expected, IPTG was able to induce the generation of hypernegatively supercoiled DNA for plasmid pZXD57 that carries a strong PT7A1/O4 promoter (Fig. 4). These results suggest that the expression of a cytosolic GFPuv protein was sufficient to induce the production of hypernegatively supercoiled DNA although the amount of generating hypernegatively supercoiled DNA was lower than that for plasmid pZXD44 (compare Fig. 3A and 4A; also compare Fig. S2E and 4C). Consistent with above described results, transcription-coupled hypernegative supercoiling of plasmid DNA for this set of five supercoiling-reporter plasmids, i.e., pZXD57, 58, 56, 55, and 54, was also dependent on promoter strength (Fig. 4B and 4D). Similar results were obtained for plasmids by using E. coli topA strain DM800 as the host strain (data not shown).
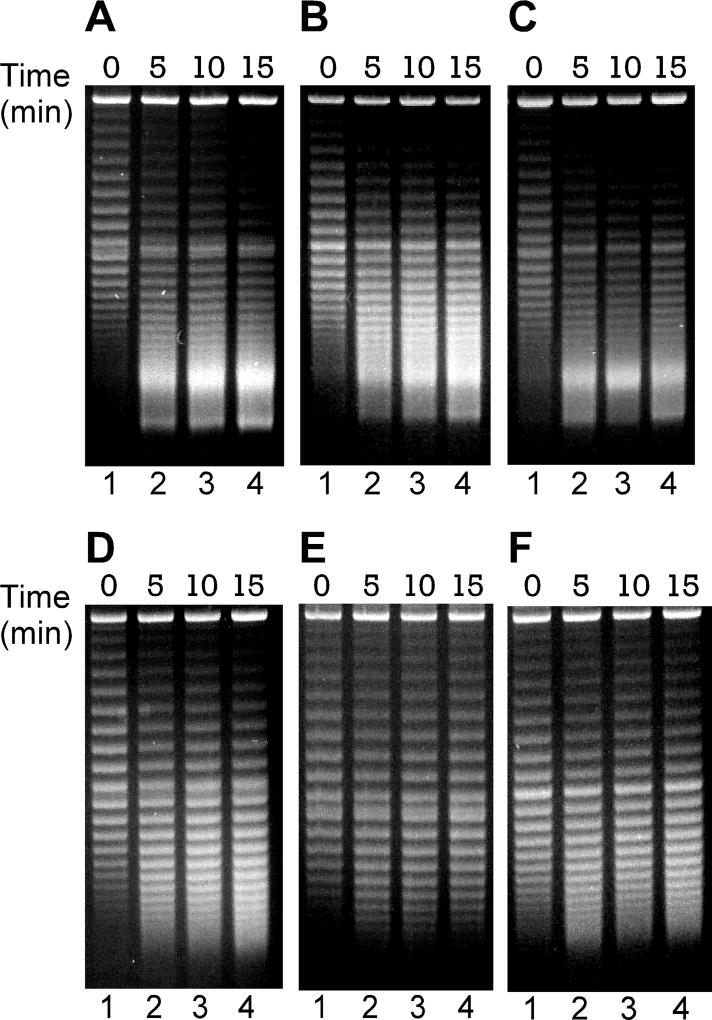
Figure 4. For strong promoters, TCDS in E. coli topA strain VS111 did not require the expression of a membrane insertion protein.
The in vivo T-S assays for plasmids carrying a GFPUV gene under the control of IPTG-inducible promoters with different strengths were performed as described under Experimental Procedures. DNA topoisomers were resolved by electrophoresis in a 1% agarose gel containing 2.5 µg/ml chloroquine and stained with ethidium bromide. (A) and (C) The time course of the hypernegative supercoiling of plasmid pZXD57 carrying a GFPUV gene under the control of the strong PT7A1/O4 promoter in E. coli strain VS111 after 1 mM of IPTG induction. Lanes 1–4 contained, respectively, DNA samples isolated from VS111 after 0, 5, 10, and 15 min of IPTG induction. (B) and (D) Dependence of TCDS on promoter strength. Lanes 1–5 contained, respectively, DNA samples isolated from E. coli topA strain VS111 containing plasmids pZXD57, 58, 56, 55, and 54, that carry a cytosolic GFPuv gene under the control of IPTG-inducible promoters with different strengths, i.e., promoters PT7A1/O4, Ptac, PlacUV5, Plac, and PlacL8 after 5 min of IPTG (1 mM) induction. Labels: PR, promoter; T7A1, the T7A1/O4 promoter; tac, the tac promoter; lacUV5, the lacUV5 promoter; lac, the lac promoter; lacL8, the lacL8 promoter. (C) The percentage of hypernegatively supercoiled DNA for pZXD57 is a function of IPTG induction time. These results were calculated as described under Experimental Procedures using the TCDS data shown in (A). (D) The percentage of hypernegatively supercoiled DNA is proportional to promoter strength (the values are the average of at least three independent determinations and the standard deviations are shown). These results were calculated as described under Experimental Procedures using the TCDS data shown in (B). The promoter strength in Pbla units was obtained from ref. (Lanzer and Bujard 1988).
In order to further study transcription-coupled hypernegative supercoiling of plasmid DNA in our two-plasmid system, we next constructed six supercoiling-reporter plasmids of identical size which also carry a strong, IPTG-inducible PT7A1/O4 promoter controlling the expression of different genes (Fig. S5 and Table 2). Plasmid pZXD60 carries a lacZ gene under the control of PT7A1/O4. E. coli cells carrying pZXD60 are able to overexpress β-galactosidase after PTG induction. Plasmid pZXD60A is identical with pZXD60 except the start codon (ATG) of lacZ was mutated to the stop codon TAG (amber mutation). In this case, IPTG is not able to induce the expression of β-galactosidase for E. coli cells harboring pZXD60A although the transcripts from both plasmids should be almost identical. Plasmid pZXD61 contains a lacZ gene in the reverse orientation, which cannot direct the expression of β-galactosidase. Plasmid pZXD62 carries a GFPuv gene under the control of PT7A1/O4, which is able to direct the overexpression of GFPuv protein after IPTG induction. Plasmids pZXD63 and 63A are identical except for the start codon of tet gene. pZXD63 carries a tet gene under the control of PT7A1/O4 and is able to direct the overexpression of membrane-insertion, tetracycline resistance protein. pZXD63A, however, has an amber mutation in the start codon of the tet gene (ATG to TAG). In this scenario, E. coli cells containing pZXD63A are not able to express tetracycline resistance protein. These six plasmids were transformed into E. coli topA strain VS111 carrying the linear plasmid pZXD51 and their superhelical states were examined after IPTG induction. Fig. 5 shows the results of these in vivo T-S assays. As expected, IPTG was able to induce the production of hypernegatively supercoiled DNA for plasmids pZXD60, 62, and 63 which express β-galactosidase, GFPuv protein, and tetracycline resistance protein, respectively (Fig. 5A, 5B, and 5C; Fig. S5A). Interestingly, our results showed that IPTG was not able to induce the generation of hypernegatively supercoiled DNA for plasmids pZXD60A, 61, and 63A although transcription added a few negative supercoils to these plasmids (Fig. 5D, 5E, and 5F). Since plasmids pZXD60A, 61, and 63A are not able to direct the expression of a polypeptide after IPTG induction, although each plasmid carries a strong PT7A1/O4 promoter, these results suggest that the co-transcriptional synthesis of a protein is able to facilitate the generation of hypernegatively supercoiled DNA. Nevertheless, a close inspection of these gel images revealed that transcription added 5 to 6 negative supercoils to plasmids pZXD60A and 63A, and only 2 supercoils to pZXD61. We examined the DNA sequences of these plasmid DNA templates and found a downstream open reading frame for both pZDX60A and pZDX63A but not for pZDX61. Although both open reading frames do not contain a SD sequence, it is possible that they are still able to direct the synthesis of a polypeptide with low efficiency. These downstream open reading frames may be the reason for the topological difference among these DNA templates. Alternatively, the different amounts of friction torque caused by various mRNA secondary structures may also result in the topological difference. Similar results were also obtained when E. coli topA strain DM800 was used as the host strain (Fig. S6).
Figure 5. TCDS in E. coli topA strain VS111 for different plasmid DNA templates with identical size carrying a strong, IPTG-inducible PT7A1/O4 promoter.
The in vivo T-S assays were performed as described under Experimental Procedures. Transcription was induced with 1 mM of IPTG. Lane 1 contained the DNA sample before IPTG induction. Lanes 2–4 contained the plasmid DNA samples after 5, 10, and 15 min of IPTG induction, respectively. DNA topoisomers were resolved by electrophoresis in a 1% agarose gel containing 2.5 µg/ml chloroquine and stained with ethidium bromide. Plasmids pZXD60 (A), 62 (B), and 63 (C) carry a lacZ, GFPuv, and tet gene under the control of the strong PT7A1/O4 promoter, respectively. (D) Plasmid pZXD60A is identical with pZXD60 except the start codon (ATG) of lacZ was mutated to the stop codon TAG (amber mutation). (E) Plasmid pZXD61 contains a lacZ gene in the reverse orientation, which cannot direct the expression of β-galactosidase. (F) Plasmid pZXD63A is similar to pZXD63 except the start codon of the tet gene was mutated to the stop codon TAG (amber mutation).
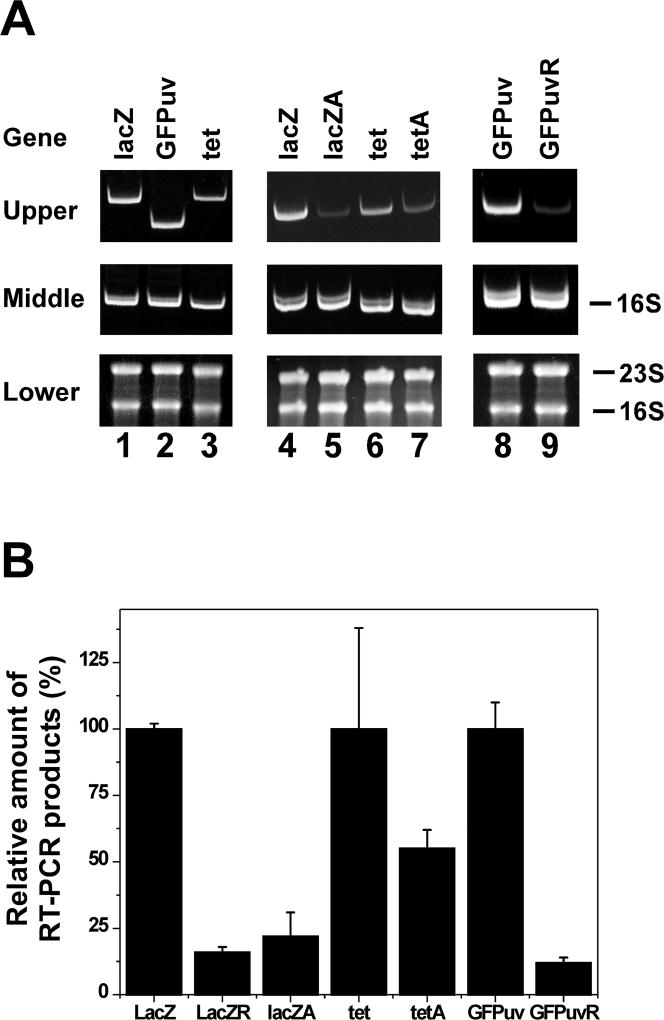
It has been demonstrated that the stability of E. coli mRNA was strongly affected by their association with ribosomes and ribosome-free mRNA was rapidly degraded in vivo (Deana and Belasco 2005; Nilsson et al 1987; Pedersen et al 2011). Therefore, we decided to examine whether transcription-coupled hypernegative supercoiling of DNA in the new two-plasmid system is correlated with the stability of mRNA produced by these plasmids after IPTG induction. Fig. 6 shows the results of our RT-PCR experiments. E. coli cells harboring plasmids pZXD60, 62 and 63, which are able to direct the overexpression of a polypeptide, i.e., β-galactosidase, GFPuv protein, and tetracycline resistance protein, respectively, produced almost the same amount of mRNA after 10 min of IPTG induction (compare lanes 1 to 3 of Fig. 6A), suggesting that mRNA of the lacZ, GFPuv, and tet genes had similar stability. The introduction of an amber mutation to the start codon of lacZ and tet genes or reversing the orientation of the open reading frame of lacZ and GFPuv gene greatly reduced the stability of the RNA transcripts (Fig. 6 and S7). Coincidently, transcription was not able to drive these plasmids into hypernegatively supercoiled status (Fig. 5D, E, and F). These results suggest that transcription-coupled hypernegative supercoiling of DNA is related to the stability of mRNA produced by these plasmids after IPTG induction.
Figure 6. RT-PCR analyses of cDNA products of mRNA transcribed from different supercoiling-reporter plasmids pZXD60, 60A, 61, 62, 63 and 63A in E. coli topA strain VS111 harboring the linear plasmid pZXD51 after 10 min of IPTG induction.
(A) RT-PCR experiments were performed as described under Experimental Procedures. The lower panel is a 1.2% agarose gel containing 1% formaldehyde to show the integrity of the RNA samples used for the RT-PCR experiments. The middle panel is a 12% polyacrylamide gel in 1×TAE buffer to show the PCR products of the cDNA synthesized from 16S rRNA samples isolated from E. coli strain VS111 carrying different supercoiling-reporter plasmids pZXD60, 60A, 62, 63, 63A, 57, and 59 after 10 min of IPTG induction. The upper panel is also a 12% polyacrylamide gel in 1×TAE buffer to show the PCR products of the cDNA samples synthesized from the mRNA samples isolated from E. coli strain VS111 carrying different supercoiling-reporter plasmids pZXD60 (lanes 1 and 4), 60A (lane 5), 62 (lane 2), 63 (lanes 3 and 6), 63A (lane 7), 57 (lane 8), and 59 (lane 9) after 10 min of IPTG induction. (B) Real-time RT-PCR analyses of the mRNA samples for E. coli strain VS111 carrying different supercoiling-reporter plasmids pZXD60, 60A, 61, 62, 63, 63A, and 59 after 10 min of IPTG induction (mean±SD, three independent experiments). Labels: lacZ, the lacZ gene; lacZA, the lacZ gene with an amber mutation in the start codon; lacZR, the lacZ gene in the reverse orientation; GFPuv, the GFPuv gene; GFPuvR, the GFPuv gene in the reverse orientation; tet, the tet gene; tetA, the tet gene with an amber mutation in the start codon.
4. Discussion
In this article, we have presented strong evidence demonstrating that transcription-coupled hypernegative supercoiling of plasmid DNA in E. coli topA strains is dependent on promoter strength, a functional property that has not been revealed previously. Not only did we show that transcription-coupled hypernegative supercoiling of plasmids carrying a tet gene encoding a membrane-insertion protein required a strong promoter (Fig. 3 and S2), but also we demonstrated that transcription from strong promoters, such as PT7A1/O4 and Ptac, was able to induce plasmid DNA templates into hypernegatively supercoiled status for those harboring a GFPuv or a lacZ gene encoding a cytosolic protein in E. coli topA strains VS111 and DM800 (Fig. 4, 5, and S6). Transcription from weak promoters, such as Plac and PlacL8, however, was not able to induce topological changes to plasmids carrying either a membrane-associated tet gene (Fig. 3 and S2) or a cytosolic-associated gene, such as GFPuv or lacZ (Fig. 4). Since promoter strength is correlated with transcription initiation (Brunner and Bujard 1987; Lutz et al 2001; McClure 1985; Saecker et al 2011), these results suggest that transcription initiation plays a critical role in TCDS in E. coli cells. However, our results also showed that transcription initiation alone was not capable of inducing plasmid DNA templates into hypernegatively supercoiled status. For instance, transcription from the strong PT7A1/O4 promoter was not able to induce plasmids pZXD60A, 61, and 63A into hypernegatively supercoiled status although transcription added a few supercoils to these plasmids after IPTG induction (Fig. 5D, E, and F). Nevertheless, our results are consistent with the “twin-supercoiled-domain” model of transcription (Liu and Wang 1987) (please see below for more discussion).
In this study, we also showed that, for strong promoters, transcription-coupled hypernegative supercoiling of plasmid DNA in E. coli topA strains did not need the expression of a membrane-insertion protein although it required co-transcriptional synthesis of a polypeptide (Fig. 4, 5, and S6). These results are consistent with our previously published results for T7 RNA polymerase where the strong T7 RNA polymerase efficiently drove plasmid DNA templates to hypernegatively supercoiled status even when the transcriptional machinery did not couple to translation and membrane insertion (Samul and Leng 2007). We noticed that these results appear inconsistent with the previously published results showing that plasmid hypernegative supercoiling by E. coli RNA polymerase required the anchoring or insertion of the coupled transcription-translation complex into the cytoplasmic membrane (Cook, Ma, Pon, and Hearst 1992; Lodge, Kazic, and Berg 1989; Lynch and Wang 1993a; Ma, Cook, Pon, and Hearst 1994; Pruss 1985; Pruss and Drlica 1986; Stupina and Wang 2005). However, a careful analysis showed that both situations can be explained by the “twin-supercoiled-domain” model of transcription where the friction force (Liu and Wang 1987) applied to E. coli RNA polymerase is different for promoters with different strengths. Under our experimental conditions, weak promoters with very low rates of transcription initiation might not lead to the formation of an active transcriptional ensemble including a transcribing RNA polymerase, a newly-transcribed RNA, the associated ribosomes, and a newly generated polypeptide for most plasmids, which are not capable of generating sufficient friction force on E. coli RNA polymerase to produce the “twin-supercoiled-domains” on plasmids. In this case, TCDS is negligible. For example, recent studies showed that a synthetic weak promoter Plar, a derivative of Plac, only produced 4 RNAs on average in one hour in E. coli cells after maximum induction (Golding et al 2005; Kandhavelu et al 2011). For most times, there was no transcription initiation from Plar promoter (Kandhavelu, Mannerstrom, Gupta, Hakkinen, Lloyd-Price, Yli-Harja, and Ribeiro 2011). Since DNA gyrase is also limited in E. coli cells (Taniguchi et al 2010), the chance for weak promoters to drive the plasmid DNA templates to hypernegatively supercoiled status is low. Promoters with moderate strength, such as the Ptet promoter (Stuber and Bujard 1981), may be able to generate one or two transcriptional ensembles per plasmid. Because the transcription elongation rate of E. coli RNA polymerase is relatively low (Golding and Cox 2004; Uptain et al 1997), it is possible that transcription alone may not be able to generate enough friction torque to fully prevent E. coli RNA polymerase from rotating against the DNA double helix and therefore cannot induce the formation of significant amounts of localized supercoiled domains. In this case, in order to generate “twin-supercoiled-domains,” the transcriptional ensemble has to anchor to the bacterial cytoplasmic membrane through co-transcriptional synthesis of polypeptide encoding membrane proteins to maximize friction resistance. This interpretation explains why TCDS for plasmid pBR322 and its derivatives carrying the Ptet promoter depends on the expression of a membrane-insertion tetracycline resistance protein in E. coli topA strains (Cook, Ma, Pon, and Hearst 1992; Lodge, Kazic, and Berg 1989; Lynch and Wang 1993a; Ma, Cook, Pon, and Hearst 1994; Pruss 1985; Pruss and Drlica 1986; Stupina and Wang 2005). For strong promoters, such as PT7A1/O4 and Ptac, E. coli RNA polymerase is able to rapidly initiate transcription from them (Brunner and Bujard 1987; Lanzer and Bujard 1988). Therefore, each plasmid may have multiple RNA polymerases (more than two RNA polymerases) simultaneously transcribing along the DNA template. It is possible that the friction force against multiple transcribing RNA polymerases is significantly increased and sufficient to cause the formation of the “twin-supercoiled-domains.” In this scenario, transcription-coupled hypernegative supercoiling of plasmid DNA did not need the expression of a membrane-insertion protein. Regardless, our results showed that co-transcriptional synthesis of a polypeptide is still required for the formation of hypernegative supercoiling of plasmid DNA in E. coli topA strains (Fig. 5). There are two possibilities for this requirement. The first possibility is that co-transcriptional synthesis of a polypeptide significantly increases the size of a transcriptional ensemble (including a transcribing RNA polymerase, the newly synthesized RNA transcript, the associated ribosomes, and the newly synthesized polypepides) and therefore increases the friction torque against the transcription ensemble, which prevents or retards the transcribing RNA polymerase from rotating around the DNA double helix and helps generate the “twin-supercoiled-domains.” If this explanation is correct, anchoring of plasmid DNA to bacterial cytoplasmic membrane should increase the efficiency of TCDS. Indeed, our results showed that the efficiency of TCDS is higher for plasmids expressing tetracycline resistance protein than that for plasmids expressing GFPuv protein although the sizes of both proteins are similar (compare Fig. 3D and 4D). The second possibility is that ribosomes protect mRNA from degradation by ribonucleases. Because the length of RNA transcripts, which should be proportional to the friction force applied to E. coli RNA polymerase, plays a critical role in the production of the “twin-supercoiled-domains” (Leng, Amado, and McMacken 2004; Liu and Wang 1987; Samul and Leng 2007), co-transcriptional synthesis of a polypeptide should greatly stabilize the mRNA (Deana and Belasco 2005; Pedersen, Nissen, Mitarai, Lo, Sneppen, and Pedersen 2011) and therefore increase the efficiency of TCDS in E. coli cells. Our results demonstrating that transcription-coupled hypernegative supercoiling of DNA is correlated with the stability of mRNA produced by these plasmids after IPTG induction strongly support this explanation (Fig. 6). Additionally, the facts that transcription alone added a few supercoils to plasmids pZXD60A, 61, and 63A also support this interpretation (Fig. 5D, E, and F). Nevertheless, these two possibilities are not mutually exclusive and may contribute together to TCDS in vivo.
RNA polymerases are powerful motor proteins (Bai et al 2006; Seidel and Dekker 2007; Wang et al 1998) which are able to rapidly move along chromosomes and remodel chromosome structures through TCDS (Albert, Spirito, Figueroa-Bossi, Bossi, and Rahmouni 1996; Cook, Ma, Pon, and Hearst 1992; Dunaway and Ostrander 1993; Leng and McMacken 2002; Lodge, Kazic, and Berg 1989; Lynch and Wang 1993a; Ma, Cook, Pon, and Hearst 1994; Stupina and Wang 2004; Tsao, Wu, and Liu 1989; Wu, Shyy, Wang, and Liu 1988). The chromosomal remodeling by RNA polymerase in E. coli cells is directly link to the activation of transcription and DNA replication. For example, in the ilvYC operon of E. coli, the ilvY promoter is divergently coupled to the ilvC promoter (Rhee et al 1999). Results from Hatfield Laboratory clearly demonstrated that the transcriptional activities of the ilvY and ilvC promoters are dependent on the localized superhelical density around the promoter region and can be activated by each other (Opel and Hatfield 2001; Rhee, Opel, Ito, Hung, Arfin, and Hatfield 1999). Another well-characterized example is the activation of the S. typhimurium leu-500 promoter by divergently-coupled transcription. Results from Wu’s and Lilley’s Laboratories showed that transcription-driven localized supercoiling rather than the global superhelical density is responsible for activation of the leu-500 promoter (Chen et al 1992; Tan et al 1994). An additional example demonstrating the biological functions of TCDS stems from the studies of bacteriophage λ DNA replication initiation. Previous studied showed that bacteriophage λ DNA replication initiation is dependent on transcription in a nearby promoter in vivo (Hase et al 1989). Our recent results showed that TCDS is responsible for the activation of λ DNA replication (Leng et al 2011). Specifically, the O-some assembled from the DNA replication initiator O protein binding to the DNA replication origin functions as a DNA topological barrier blocks, confines, and captures TCDS. In this scenario, λ DNA replication origin is unwound and DNA replication is initiated. All these examples demonstrated that TCDS plays a critical role in certain biological events.
Supplementary Material
Acknowledgments
We would like to thank Dr. Sankar Adhya for helpful discussion and suggestions. We also thank Steven Eichelbaum for technical support and for critical reading of the manuscript before submission. We are grateful to Drs. Marc Drolet and Nikolai V. Ravin for providing us with E. coli strain DM800 and linear plasmid pG591, respectively. This work was supported by grant 1SC1HD063059-01A1 from the National Institutes of Health (to F.L.).
Abbreviations
- TCDS
transcription-coupled DNA supercoiling
- T-S
transcription-supercoiling
- IPTG
Isopropyl-β-D-thiogalactopyranoside
- LacI
lac repressor
Footnotes
Supplementary Data associated with this article can be found at Gene Web page. The supplementary data comprises seven Figures.
References
- Albert AC, Spirito F, Figueroa-Bossi N, Bossi L, Rahmouni AR. Hyper-negative template DNA supercoiling during transcription of the tetracycline-resistance gene in topA mutants is largely constrained in vivo. Nucleic Acids Res. 1996;24:3093–3099. doi: 10.1093/nar/24.15.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Santangelo TJ, Wang MD. Single-molecule analysis of RNA polymerase transcription. Annu. Rev. Biophys. Biomol. Struct. 2006;35:343–360. doi: 10.1146/annurev.biophys.35.010406.150153. [DOI] [PubMed] [Google Scholar]
- Bates AD, Maxwell A. DNA Topology. 2. Oxford University Press; Oxford, UK: 2005. [Google Scholar]
- Brunner M, Bujard H. Promoter recognition and promoter strength in the Escherichia coli system. EMBO J. 1987;6:3139–3144. doi: 10.1002/j.1460-2075.1987.tb02624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Chen D, Bowater R, Dorman CJ, Lilley DM. Activity of a plasmid-borne leu-500 promoter depends on the transcription and translation of an adjacent gene. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8784–8788. doi: 10.1073/pnas.89.18.8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DN, Ma D, Pon NG, Hearst JE. Dynamics of DNA supercoiling by transcription in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1992;89:10603–10607. doi: 10.1073/pnas.89.22.10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzarelli NR, Wang JC. DNA Topology and Its Biological Effects. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1990. [Google Scholar]
- Deana A, Belasco JG. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 2005;19:2526–2533. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- Deneke J, Ziegelin G, Lurz R, Lanka E. The protelomerase of temperate Escherichia coli phage N15 has cleaving-joining activity. Proc. Natl. Acad. Sci. U. S. A. 2000;97:7721–7726. doi: 10.1073/pnas.97.14.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau E, Margolin P. Suppression of promoter mutations by the pleiotropic supx mutations. Mol. Gen. Genet. 1972;117:91–112. doi: 10.1007/BF00267607. [DOI] [PubMed] [Google Scholar]
- Dunaway M, Ostrander EA. Local domains of supercoiling activate a eukaryotic promoter in vivo. Nature. 1993;361:746–748. doi: 10.1038/361746a0. [DOI] [PubMed] [Google Scholar]
- Eckert B, Beck CF. Overproduction of transposon Tn10-encoded tetracycline resistance protein results in cell death and loss of membrane potential. J. Bacteriol. 1989;171:3557–3559. doi: 10.1128/jb.171.6.3557-3559.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding I, Cox EC. RNA dynamics in live Escherichia coli cells. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11310–11315. doi: 10.1073/pnas.0404443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Hase T, Nakai M, Masamune Y. Transcription of a region downstream from lambda ori is required for replication of plasmids derived from coliphage lambda. Mol. Gen. Genet. 1989;216:120–125. doi: 10.1007/BF00332239. [DOI] [PubMed] [Google Scholar]
- Wang James C. Untangling the Double Helix: DNA Entanglement and the Action of the DNA Topoisomerases. Cold Spring Harbor Laboratory Press; 2009. [Google Scholar]
- Kandhavelu M, Mannerstrom H, Gupta A, Hakkinen A, Lloyd-Price J, Yli-Harja O, Ribeiro AS. In vivo kinetics of transcription initiation of the lar promoter in Escherichia coli. Evidence for a sequential mechanism with two rate-limiting steps. BMC. Syst. Biol. 2011;5:149. doi: 10.1186/1752-0509-5-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzer M, Bujard H. Promoters largely determine the efficiency of repressor action. Proc. Natl. Acad. Sci. U. S. A. 1988;85:8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng F, Amado L, McMacken R. Coupling DNA supercoiling to transcription in defined protein systems. J. Biol. Chem. 2004;279:47564–47571. doi: 10.1074/jbc.M403798200. [DOI] [PubMed] [Google Scholar]
- Leng F, Chen B, Dunlap DD. Dividing a supercoiled DNA molecule into two independent topological domains. Proc. Natl. Acad. Sci. U. S. A. 2011;108:19973–19978. doi: 10.1073/pnas.1109854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng F, McMacken R. Potent stimulation of transcription-coupled DNA supercoiling by sequence-specific DNA-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9139–9144. doi: 10.1073/pnas.142002099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U. S. A. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshon D, Morris DR. Positively supercoiled plasmid DNA is produced by treatment of Escherichia coli with DNA gyrase inhibitors. Nucleic Acids Res. 1983;11:2999–3017. doi: 10.1093/nar/11.10.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge JK, Kazic T, Berg DE. Formation of supercoiling domains in plasmid pBR322. J. Bacteriol. 1989;171:2181–2187. doi: 10.1128/jb.171.4.2181-2187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Lozinski T, Ellinger T, Bujard H. Dissecting the functional program of Escherichia coli promoters: the combined mode of action of Lac repressor and AraC activator. Nucleic Acids Res. 2001;29:3873–3881. doi: 10.1093/nar/29.18.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch AS, Wang JC. Anchoring of DNA to the bacterial cytoplasmic membrane through cotranscriptional synthesis of polypeptides encoding membrane proteins or proteins for export: a mechanism of plasmid hypernegative supercoiling in mutants deficient in DNA topoisomerase I. J. Bacteriol. 1993a;175:1645–1655. doi: 10.1128/jb.175.6.1645-1655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch AS, Wang JC. Anchoring of DNA to the bacterial cytoplasmic membrane through cotranscriptional synthesis of polypeptides encoding membrane proteins or proteins for export: a mechanism of plasmid hypernegative supercoiling in mutants deficient in DNA topoisomerase I. J. Bacteriol. 1993b;175:1645–1655. doi: 10.1128/jb.175.6.1645-1655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Cook DN, Pon NG, Hearst JE. Efficient anchoring of RNA polymerase in Escherichia coli during coupled transcription-translation of genes encoding integral inner membrane polypeptides. J. Biol. Chem. 1994;269:15362–15370. [PubMed] [Google Scholar]
- McClure WR. Mechanism and control of transcription initiation in prokaryotes. Annu. Rev. Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Mielke SP, Fink WH, Krishnan VV, Gronbech-Jensen N, Benham CJ. Transcription-driven twin supercoiling of a DNA loop: a Brownian dynamics study. J. Chem. Phys. 2004;121:8104–8112. doi: 10.1063/1.1799613. [DOI] [PubMed] [Google Scholar]
- Mukai FH, Margolin P. ANALYSIS OF UNLINKED SUPPRESSORS OF AN O degrees MUTATION IN SALMONELLA. Proc. Natl. Acad. Sci. U. S. A. 1963;50:140–148. doi: 10.1073/pnas.50.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P. Transport of torsional stress in DNA. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14342–14347. doi: 10.1073/pnas.96.25.14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson G, Belasco JG, Cohen SN, von GA. Effect of premature termination of translation on mRNA stability depends on the site of ribosome release. Proc. Natl. Acad. Sci. U. S. A. 1987;84:4890–4894. doi: 10.1073/pnas.84.14.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opel ML, Hatfield GW. DNA supercoiling-dependent transcriptional coupling between the divergently transcribed promoters of the ilvYC operon of Escherichia coli is proportional to promoter strengths and transcript lengths. Mol. Microbiol. 2001;39:191–198. doi: 10.1046/j.1365-2958.2001.02249.x. [DOI] [PubMed] [Google Scholar]
- Pedersen M, Nissen S, Mitarai N, Lo SS, Sneppen K, Pedersen S. The functional half-life of an mRNA depends on the ribosome spacing in an early coding region. J. Mol. Biol. 2011;407:35–44. doi: 10.1016/j.jmb.2011.01.025. [DOI] [PubMed] [Google Scholar]
- Pruss GJ. DNA topoisomerase I mutants. Increased heterogeneity in linking number and other replicon-dependent changes in DNA supercoiling. J. Mol. Biol. 1985;185:51–63. doi: 10.1016/0022-2836(85)90182-2. [DOI] [PubMed] [Google Scholar]
- Pruss GJ, Drlica K. Topoisomerase I mutants: the gene on pBR322 that encodes resistance to tetracycline affects plasmid DNA supercoiling. Proc. Natl. Acad. Sci. U. S. A. 1986;83:8952–8956. doi: 10.1073/pnas.83.23.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin NV, Ravin VK. Use of a linear multicopy vector based on the mini-replicon of temperate coliphage N15 for cloning DNA with abnormal secondary structures. Nucleic Acids Res. 1999;27:e13. doi: 10.1093/nar/27.17.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee KY, Opel M, Ito E, Hung S, Arfin SM, Hatfield GW. Transcriptional coupling between the divergent promoters of a prototypic LysR-type regulatory system, the ilvYC operon of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14294–14299. doi: 10.1073/pnas.96.25.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saecker RM, Record MT, Jr, deHaseth PL. Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011;412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samul R, Leng F. Transcription-coupled hypernegative supercoiling of plasmid DNA by T7 RNA polymerase in Escherichia coli topoisomerase I-deficient strains. J. Mol. Biol. 2007;374:925–935. doi: 10.1016/j.jmb.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel R, Dekker C. Single-molecule studies of nucleic acid motors. Curr. Opin. Struct. Biol. 2007;17:80–86. doi: 10.1016/j.sbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Snoep JL, van der Weijden CC, Andersen HW, Westerhoff HV, Jensen PR. DNA supercoiling in Escherichia coli is under tight and subtle homeostatic control, involving gene-expression and metabolic regulation of both topoisomerase I and DNA gyrase. Eur. J. Biochem. 2002;269:1662–1669. doi: 10.1046/j.1432-1327.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- Stuber D, Bujard H. Organization of transcriptional signals in plasmids pBR322 and pACYC184. Proc. Natl. Acad. Sci. U. S. A. 1981;78:167–171. doi: 10.1073/pnas.78.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupina VA, Wang JC. DNA axial rotation and the merge of oppositely supercoiled DNA domains in Escherichia coli: effects of DNA bends. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8608–8613. doi: 10.1073/pnas.0402849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupina VA, Wang JC. Viability of Escherichia coli topA mutants lacking DNA topoisomerase I. J. Biol. Chem. 2005;280:355–360. doi: 10.1074/jbc.M411924200. [DOI] [PubMed] [Google Scholar]
- Tan J, Shu L, Wu HY. Activation of the leu-500 promoter by adjacent transcription. J. Bacteriol. 1994;176:1077–1086. doi: 10.1128/jb.176.4.1077-1086.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao YP, Wu HY, Liu LF. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989;56:111–118. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- Wang JC. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- Wang JC, Lynch AS. Transcription and DNA supercoiling. Curr. Opin. Genet. Dev. 1993;3:764–768. doi: 10.1016/s0959-437x(05)80096-6. [DOI] [PubMed] [Google Scholar]
- Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DM, Cozzarelli NR. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.