Abstract Abstract
Three new Fusarium species, F. convolutans, F. fredkrugeri, and F. transvaalense (Ascomycota, Hypocreales, Nectriaceae) are described from soils collected in a catena landscape on a research supersite in the Kruger National Park, South Africa. The new taxa, isolated from the rhizosphere of three African herbaceous plants, Kyphocarpa angustifolia, Melhania acuminata, and Sida cordifolia, are described and illustrated by means of morphological and multilocus molecular analyses based on sequences from five DNA loci (CAL, EF-1 α, RPB1, RPB2 and TUB). According to phylogenetic inference based on Maximum-likelihood and Bayesian approaches, the newly discovered species are distributed in the Fusarium buharicum, F. fujikuroi, and F. sambucinum species complexes.
Keywords: Natural parks, phylogeny, fungi, multigene, morphology, diversity
Introduction
Fungi are common colonisers of the plant rhizobiome and endosphere, where they play a key role in modulating the interactions between plant roots and soil (Zachow et al. 2009; Visioli et al. 2014). The direct and indirect interaction between fungal growth in the rhizosphere and its effect on plant growth and health is well documented (Havlicek and Mitchell 2014; Hargreaves et al. 2015; Lareen et al. 2016). Such effects include either a positive feedback by producing plant growth promoting factors, solubilising and stimulating nutrient uptake by plant roots or by inhibiting the growth of concomitant pathogenic organisms (Schippers et al. 1987; Mommer et al. 2016). Conversely, deleterious effects have also been observed, either related to the presence of pathogenic fungal species or caused by fungal-induced modifications of plant root functions, impeding root growth or negatively altering nutrient availability (Schippers et al. 1987; Mommer et al. 2016). Likewise, plants can select and harbour a particular fungal community on its roots via root exudates (Lareen et al. 2016; Sasse et al. 2018), while abiotic influences including water availability, climate and season, soil type, grazers and other animals, orchestrate the development of a unique fungal diversity (Philippot et al. 2013; Havlicek and Mitchell 2014; Hargreaves et al. 2015; Lareen et al. 2016).
The genus Fusarium Link (Hypocreales, Nectriaceae) includes a vast number of species, commonly recovered from a variety of substrates including soil, air, water and decaying plant materials; being also able to colonise living tissues of plants and animals, including humans; acting as endophytes, secondary invaders or becoming devastating plant pathogens (Nelson et al. 1994). In addition to their ability to colonise a multiplicity of habitats, Fusarium is a cosmopolitan genus, present in almost any ecosystem in the world, including human-made settings such as air and dust in the indoor environment or even in hospitals (Perlroth et al. 2007; Aydogdu and Asan 2008; Pinheiro et al. 2011).
Being common inhabitants of plant root ecosystems, fusaria and, particularly Fusarium graminearum Schwabe, F. proliferatum (Matsush.) Nirenberg ex Gerlach & Nirenberg, F. verticillioides (Sacc.) Nirenberg (Syn. F. moniliforme J. Sheld.), F. oxysporum Schltdl., as well as species recently segregated from Fusarium, including Neocosmospora phaseoli (Burkh.) L. Lombard & Crous (Syn. Fusarium phaseoli Burkh.) and N. virguliforme (O’Donnell & T. Aoki) L. Lombard & Crous (Syn. F. virguliforme O’Donnell & T. Aoki), have been regularly studied for their interactions with the rhizobiome, motivated mainly by the importance of these organisms as soil-borne plant pathogens and the need to develop effective control mechanisms (Larkin et al. 1993; Hassan Dar et al. 1997; Pal et al. 2001; Fravel et al. 2003; Idris et al. 2006; Díaz Arias et al. 2013). Similarly, abundant data is available regarding the ecology and distribution of plant-associated fusaria, particularly related to pathogenic species or commonly isolated endophytes (Leslie and Summerell 2006). Little attention has however been given to the occurrence of non-pathogenic fungal species, including Fusarium spp. in root microbial communities (Zakaria and Ning 2013; Jumpponen et al. 2017; LeBlanc et al. 2017), while comprehensive DNA sequence-based surveys have been directed mostly to the study of highly relevant and abundant rhizosphere fungal genera such as Trichoderma Pers., Verticillium Nees or mycorrhizal fungi (Zachow et al. 2009; Bent et al. 2011; Ruano-Rosa et al. 2016; Saravanakumar et al. 2016).
The Kruger National Park (KNP) in South Africa is one of the largest natural reserves in Africa, encompassing a number of non-manipulated landscapes, with almost no human alteration (Carruthers 2017). Recently, four research “supersites” have been identified and established in KNP, each of these supersites representing unique geological, ecological and climatic features of the park (Smit et al. 2013). A multidisciplinary study was conducted in KNP aimed to determine functioning and interaction between abiotic and biotic components, as well as soil properties, hydrology and other processes that determine the structure, biodiversity and heterogeneity of a catena or hill slope ecosystem on one of these “supersites”, located deep inside the KNP (data not published). In order to assess the microbial soil population and community dynamics, mainly focused on bacteria, several rhizosphere samples were obtained from diverse African plants on one of these exceptional protected savannah landscapes. From these collections, interesting fusaria were isolated from the root ecosystem of three native African herbaceous plants i.e. Kyphocarpa angustifolia (Moq.) Lopr. (Amaranthaceae), Melhania acuminata Mast. (Malvaceae) and Sida cordifolia Linn. (Malvaceae). According to their unique morphological traits and clear phylogenetic delimitations, these isolates are described here as three new Fusarium species.
Methods
Study site and sampling
During March 2015, rhizosphere soil from three herbaceous plants was collected in the Southern Granites “supersite” catena (Stevenson-Hamilton supersite) in the KNP, between 25°06'28.6S, 31°34'41.9E and 25°06'25.7S, 31°34'33.7E (Fig. 1). A catena consists of different soil types observed from a crest to a valley bottom with a wetland or drainage exhibiting different water retention capabilities due to the slope or aspect (topography) and the depth of underlying geological rocks (Brown et al. 2004, Van Zijl and Le Roux 2014). The main characteristics of the Stevenson-Hamilton supersite are described in detail by Smit et al. (2013). Briefly, in this site, a single catena landscape covers approximately 1 km from top to bottom and consists of a hill slope, a sodic site (or grazing lawn), a riparian and floodplain area and a dry drainage line. Three species of plants were selected for sampling occurring at the two extremes of the catena. Two of these species (Kyphocarpa angustifolia and Sida cordifolia) occurred at both top and bottom sites while Melhania acuminata only occurred at the top site. The soil (100 mm depth) at the top of the slope is Clovelly with a high percentage of sand (90%) and a low cation exchange capacity (CEC) (mean sodium concentration of 1062 mg/kg) and pH (mean 5.85). The soil at the bottom of the slope is of the Sterkspruit type, with higher clay content thus higher CEC (mean sodium concentration of 3802 mg/kg) and higher pH (mean 6.4). Rhizosphere soil of 10 plants of the same species occurring at each top or bottom site was sampled using a core soil sampler. A total of 50 samples consisting of ca. 200 g of soil from the roots of each plant were taken, deposited in zip-lock plastic bags and kept on ice in a cool bag at approximately 5 °C until analysed in the laboratory.
Figure 1.
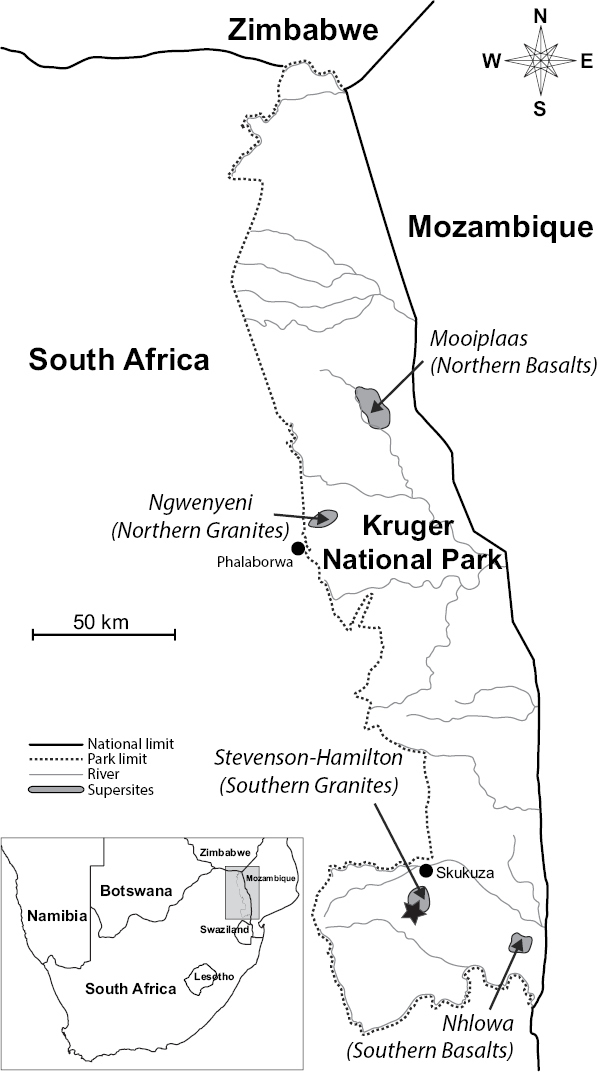
Map of the Kruger National Park (KNP) in South Africa. The arrows indicate the location of the four research “supersites” (adapted from Smit et al. 2013). Sampling site is indicated with a black star. The inset shows the location of the KNP within South Africa, indicated by a grey box.
Isolation of Fusarium strains
Soil samples were mixed thoroughly and sieved to remove large elements. Fine soil particles were uniformly spread and distributed over the surface of pentachloronitrobenzene agar (PCNB; also known as the Nash-Snyder medium, recipe in Leslie and Summerell 2006) supplemented with streptomycin (0.3 g/l) and neomycin sulphate (0.12 g/l) and malt-extract agar (MEA; recipes on Crous et al. 2009) on 9 mm Petri dishes and incubated at 24 °C for 10 d under a natural day/night photoperiod. Each soil sample was processed in duplicate. Fungal growth was evaluated daily and growing colonies were transferred to fresh Potato Dextrose Agar (PDA; recipe in Crous et al. 2009). Colonies were evaluated for their macro- and microscopic characteristics and a total of 19 fungal cultures showing features typical of Fusarium were subjected to single spore isolation as described previously (Sandoval-Denis et al. 2018). Single spore isolates were finally transferred and maintained in Oatmeal Agar plates and slants (OA; recipe in Crous et al. 2009). Fungal strains isolated in this study were deposited in the collection of the Westerdijk Fungal Biodiversity Institute (CBS; Utrecht, the Netherlands), the working collection of Pedro W. Crous (CPC), held at CBS (Table 1); and voucher specimens were deposited in The South African National Collection of Fungi (NCF) (Mycology Unit, Biosystematics Division, Plant Protection Institute, Agricultural Research Council, Pretoria, South Africa).
Table 1.
Origin, strain and GenBank/ENA accession number of strains and DNA sequences included in this study.
| Species name | Strain†‡ | Country | Host | Sequence accession number§ | ||||
|---|---|---|---|---|---|---|---|---|
| CAL | EF-1α | RPB1 | RPB2 | TUB | ||||
| Fusarium agapanthi | NRRL 54463T | Australia | Agapanthus sp. | KU900611 | KU900630 | KU900620 | KU900625 | KU900635 |
| Fusarium ananatum | CBS 118516T | South Africa | Ananas comosus fruit | LT996175 | LT996091 | LT996188 | LT996137 | LT996112 |
| Fusarium andiyazi | CBS 119857T = NRRL 31727 | South Africa | Sorghum bicolor soil debris | LT996176 | LT996092 | LT996189 | LT996138 | LT996113 |
| Fusarium anthophilum | CBS 737.97 = NRRL 13602 | Germany | Hippeastrum sp. | LT996177 | LT996093 | LT996190 | LT996139 | LT996114 |
| Fusarium armeniacum | NRRL 6227 | USA | Fescue hay | JX171446 | JX171560 | |||
| Fusarium asiaticum | CBS 110257 = NRRL 13818 | Japan | Barley | JX171459 | JX171573 | |||
| Fusarium bactridioides | NRRL 20476 | USA | Cronartium conigenum | AF158343 | AF160290 | Not public | Not public | U34434 |
| Fusarium begoniae | CBS 403.97T = NRRL 25300 | Germany | Begonia elatior hybrid | AF158346 | AF160293 | LT996191 | LT996140 | U61543 |
| Fusarium buharicum | CBS 178.35 = NRRL 25488 | USSR | Gossypium rotting stem base | KX302912 | KX302920 | KX302928 | ||
| CBS 796.70 = NRRL 13371 | Iran | Hibiscus cannabinus stalk | JX171449 | JX171563 | ||||
| Fusarium bulbicola | CBS 220.76T = NRRL 13618 | Germany | Nerine bowdenii | KF466327 | KF466415 | KF466394 | KF466404 | KF466437 |
| Fusarium brachygibbosum | NRRL 13829 | Japan | River sediments | JX171460 | JX171574 | |||
| Fusarium circinatum | CBS 405.97T = NRRL 25331 | USA | Pinus radiata | KM231393 | KM231943 | JX171510 | HM068354 | KM232080 |
| Fusarium coicis | NRRL 66233T | Australia | Coix gasteenii | LT996178 | KP083251 | KP083269 | KP083274 | LT996115 |
| Fusarium concentricum | CBS 450.97T = NRRL 25181 | Costa Rica | Musa sapientum fruit | AF158335 | AF160282 | LT996192 | JF741086 | U61548 |
| Fusarium continuum | F201128 | China | Zanthoxylum bungeanum stem | KM236720 | KM520389 | KM236780 | ||
| Fusarium convolutans | CBS 144207T = CPC 33733 | South Africa | Kyphocarpa angustifolia rhizophere | LT996094 | LT996193 | LT996141 | ||
| CBS 144208 = CPC 33732 | South Africa | Kyphocarpa angustifolia rhizophere | LT996095 | LT996194 | LT996142 | |||
| Fusarium culmorum | CBS 417.86 = NRRL 25475 | Denmark | Moldy barley kernel | JX171515 | JX171628 | |||
| Fusarium denticulatum | CBS 735.97 = NRRL 25302 | USA | Ipomoea batatas | AF158322 | AF160269 | LT996195 | LT996143 | U61550 |
| Fusarium dlaminii | CBS 119860T = NRRL 13164 | South Africa | Soil debris in cornfield | AF158330 | AF160277 | KU171681681 | KU171701 | U34430 |
| Fusarium fracticaudum | CBS 137234PT | Colombia | Pinus maximonoii stem | LT996179 | KJ541059 | LT996196 | LT996144 | KJ541051 |
| Fusarium fractiflexum | NRRL 28852T | Japan | Cymbidium sp. | AF158341 | AF160288 | Not public | LT575064 | AF160315 |
| Fusarium fredkrugeri | NRRL 26152 | Niger | Unknown | AF160306 | AF160321 | |||
| CBS 144209T = CPC 33747 | South Africa | Melhania acuminata rhizophere | LT996181 | LT996097 | LT996199 | LT996147 | LT996117 | |
| CBS 144210 = NRRL 26061 | Madagascar | Striga hermonthica | AF158356 | AF160303 | LT996197 | LT996145 | AF160319 | |
| CBS 144495 = CPC 33746 | South Africa | Melhania acuminata rhizophere | LT996180 | LT996096 | LT996198 | LT996146 | LT996116 | |
| Fusarium fujikuroi | NRRL 13566 | China | Oryza sativa | AF158332 | AF160279 | JX171456 | JX171570 | U34415 |
| Fusarium globosum | CBS 428.97T = NRRL 26131 | South Africa | Zea mays | KF466329 | KF466417 | KF466396 | KF466406 | KF466439 |
| Fusarium goolgardi | NRRL 66250T = RBG 5411 | Australia | Xanthorrhoea glauca | KP083270 | KP083280 | |||
| Fusarium graminearum | CBS 123657 = NRRL 31084 | USA | Corn | JX171531 | JX171644 | |||
| Fusarium konzum | CBS 119849T | USA | Sorghastrum nuttans | LT996182 | LT996098 | LT996200 | LT996148 | LT996118 |
| Fusarium kyushuense | NRRL 25349 | Japan | Triticum aestivum | GQ915492 | ||||
| Fusarium lactis | CBS 411.97NT = NRRL 25200 | USA | Ficus carica | AF158325 | AF160272 | LT996201 | LT996149 | U61551 |
| Fusarium langsethiae | NRRL 54940 | Norway | Oats | JX171550 | JX171662 | |||
| Fusarium lateritium | NRRL 13622 | USA | Ulmus sp. | AY707173 | JX171457 | JX171571 | ||
| Fusarium longipes | NRRL 13368 | Australia | Soil | JX171448 | JX171562 | |||
| Fusarium mangiferae | NRRL 25226 | Israel | Mangifera indica | AF158334 | AF160281 | JX171509 | HM068353 | U61561 |
| Fusarium mexicanum | NRRL 47473 | Mexico | Mangifera indica inflorescence | GU737389 | GU737416 | Not public | Not public | GU737308 |
| Fusarium napiforme | CBS 748.97T = NRRL 13604 | Namibia | Pennisetum typhoides | AF158319 | AF160266 | HM347136 | EF470117 | U34428 |
| Fusarium nygamai | CBS 749.97T = NRRL 13448 | Australia | Sorghum bicolor necrotic root | AF158326 | AF160273 | LT996202 | EF470114 | U34426 |
| Fusarium oxysporum | CBS 716.74 = NRRL 20433 | Germany | Vicia faba vascular bundle | AF158366 | AF008479 | JX171469 | JX171583 | U34435 |
| CBS 744.97 = NRRL 22902 | USA | Pseudotsuga menziesii | AF158365 | AF160312 | LT996203 | LT575065 | U34424 | |
| Fusarium palustre | NRRL 54056T | USA | Spartina alterniflora | KT597718 | KT597731 | |||
| Fusarium parvisorum | CBS 137236T | Colombia | Pinus patula roots | LT996183 | KJ541060 | LT996150 | KJ541055 | |
| Fusarium phyllophilum | CBS 216.76T = NRRL 13617 | Italy | Dracaena deremensis leaf | KF466333 | KF466421 | KF466399 | KF466410 | KF466443 |
| Fusarium poae | NRRL 13714 | Unknown | Unknown | JX171458 | JX171572 | |||
| Fusarium proliferatum | CBS 217.76 = NRRL 22944 | Germany | Cattleya pseudobulb, hybrid | AF158333 | AF160280 | JX171504 | HM068352 | U34416 |
| Fusarium pseudocircinatum | CBS 449.97T = NRRL 22946 | Ghana | Solanum sp. | AF158324 | AF160271 | LT996204 | LT996151 | U34427 |
| Fusarium pseudograminearum | CBS 109956T = NRRL 28062 | Australia | Hordeum vulgare crowns | JX171524 | JX171637 | |||
| Fusarium pseudonygamai | CBS 417.97T = NRRL 13592 | Nigeria | Pennisetum typhoides | AF158316 | AF160263 | LT996205 | LT996152 | U34421 |
| Fusarium ramigenum | CBS 418.98T = NRRL 25208 | USA | Ficus carica | KF466335 | KF466423 | KF466401 | KF466412 | KF466445 |
| Fusarium sacchari | CBS 223.76 = NRRL 13999 | India | Saccharum officinarum | AF158331 | AF160278 | JX171466 | JX171580 | U34414 |
| Fusarium sambucinum | NRRL 22187 = NRRL 20727 | England | Solanum sp. | JX171493 | JX171606 | |||
| Fusarium sarcochroum | CBS 745.79 = NRRL 20472 | Switzerland | Viscum album | JX171472 | JX171586 | |||
| Fusarium sibiricum | NRRL 53430T | Russia | Avena sativa | HQ154472 | ||||
| Fusarium sororula | CBS 137242T | Colombia | Pinus patula stems | LT996184 | KJ541067 | LT996206 | LT996153 | KJ541057 |
| Fusarium sp. | NRRL 66179 | USA | Hibiscus moscheutos | KX302913 | KX302921 | KX302929 | ||
| NRRL 66180 | USA | Hibiscus moscheutos | KX302914 | KX302922 | KX302930 | |||
| NRRL 66181 | USA | Hibiscus moscheutos | KX302915 | KX302923 | KX302931 | |||
| NRRL 66182 | USA | Hibiscus moscheutos | KX302916 | KX302924 | KX302932 | |||
| NRRL 66183 | USA | Hibiscus moscheutos | KX302917 | KX302925 | KX302933 | |||
| NRRL 66184 | USA | Hibiscus moscheutos | KX302918 | KX302926 | KX302934 | |||
| CBS 201.63 = NRRL 36351 | Portugal | Arachis hypogaea stored nut | GQ915484 | |||||
| Fusarium sporotrichioides | NRRL 3299 | USA | Corn | JX171444 | HQ154454 | |||
| Fusarium sterilihyphosum | NRRL 25623 | South Africa | Mango | AF158353 | AF160300 | Not public | Not public | AF160316 |
| Fusarium stilboides | NRRL 20429 | Nyasaland | Coffee bark | JX171468 | JX171582 | |||
| Fusarium subglutinans | CBS 747.97 = NRRL 22016 | USA | Corn | AF158342 | AF160289 | JX171486 | JX171599 | U34417 |
| Fusarium sublunatum | CBS 190.34 = NRRL 20897 | Unknown | Unknown | KX302919 | KX302927 | KX302935 | ||
| CBS 189.34T = NRRL 13384 | Costa Rica | Soil of banana plantation | JX171451 | JX171565 | ||||
| Fusarium succisae | CBS 219.76 = NRRL 13613 | Germany | Succisa pratensis flower | AF158344 | AF160291 | LT996207 | LT996154 | U34419 |
| Fusarium sudanense | CBS 454.97T = NRRL 25451 | Sudan | Striga hermonthica | LT996185 | KU711697 | LT996208 | LT996155 | KU603909 |
| Fusarium temperatum | NRRL 25622 = NRRL 26616 | South Africa | Zea mays | AF158354 | AF160301 | Not public | Not public | AF160317 |
| Fusarium terricola | CBS 483.94T | Australia | Soil | KU603951 | KU711698 | LT996209 | LT996156 | KU603908 |
| Fusarium thapsinum | CBS 733.97 = NRRL 22045 | South Africa | Sorghum bicolor | LT996186 | AF160270 | JX171487 | JX171600 | U34418 |
| Fusarium tjaetaba | NRRL 66243T | Australia | Sorghum interjectum | LT996187 | KP083263 | KP083267 | KP083275 | LT996119 |
| Fusarium torreyae | NRRL 54149 | USA | Torreya sp. | HM068337 | JX171548 | HM068359 | ||
| Fusarium transvaalense | CBS 144211T = CPC 30923 | South Africa | Sida cordifolia rhizosphere | LT996099 | LT996210 | LT996157 | LT996120 | |
| CBS 144212 = CPC 30929 | South Africa | Melhania acuminata rhizophere | LT996100 | LT996211 | LT996158 | LT996121 | ||
| CBS 144213 = CPC 33751 | South Africa | Melhania acuminata rhizophere | LT996159 | LT996122 | ||||
| CBS 144214 = CPC 30946 | South Africa | Sida cordifolia rhizosphere | LT996101 | LT996212 | LT996160 | LT996123 | ||
| CBS 144215 = CPC 33723 | South Africa | Sida cordifolia rhizosphere | LT996102 | LT996161 | LT996124 | |||
| CBS 144216 = CPC 30918 | South Africa | Sida cordifolia rhizosphere | LT996103 | LT996213 | LT996162 | LT996125 | ||
| CBS 144217 = CPC 30919 | South Africa | Sida cordifolia rhizosphere | LT996104 | LT996214 | LT996163 | LT996126 | ||
| CBS 144218 = CPC 30922 | South Africa | Sida cordifolia rhizosphere | LT996105 | LT996215 | LT996164 | LT996127 | ||
| CBS 144219 = CPC 30926 | South Africa | Sida cordifolia rhizosphere | LT996106 | LT996216 | LT996165 | LT996128 | ||
| CBS 144220 = CPC 30927 | South Africa | Sida cordifolia rhizosphere | LT996107 | LT996217 | LT996166 | LT996129 | ||
| CBS 144221 = CPC 33740 | South Africa | Kyphocarpa angustifolia rhizophere | LT996167 | LT996130 | ||||
| CBS 144222 = CPC 30939 | South Africa | Kyphocarpa angustifolia rhizophere | LT996108 | LT996218 | LT996168 | LT996131 | ||
| CBS 144223 = CPC 30941 | South Africa | Kyphocarpa angustifolia rhizophere | LT996109 | LT996169 | LT996132 | |||
| CBS 144224 = CPC 30928 | South Africa | Melhania acuminata rhizophere | LT996110 | LT996219 | LT996170 | LT996133 | ||
| CBS 144496 = CPC 33750 | South Africa | Melhania acuminata rhizophere | LT996171 | LT996134 | ||||
| NRRL 31008 | Australia | Soil | JX171529 | JX171642 | ||||
| Fusarium tupiense | NRRL 53984 | Brazil | Mangifera indica | GU737377 | GU737404 | Not public | Not public | GU737296 |
| Fusarium udum | CBS 178.32 = NRRL 22949 | Germany | Lactarius pubescens | AF158328 | AF160275 | LT996220 | LT996172 | U34433 |
| Fusarium venenatum | CBS 458.93T | Austria | Winter wheat halm base | KM232382 | ||||
| Fusarium verticillioides | CBS 734.97 = NRRL 22172 | Germany | Zea mays | AF158315 | AF160262 | LT996221 | EF470122 | U34413 |
| Fusarium xanthoxyli | F201114 | China | Zanthoxylum bungeanum | KM236706 | KM520380 | KM236766 | ||
| Fusarium xylarioides | CBS 258.52 = NRRL 25486 | Ivory Coast | Coffea sp. trunk | AY707136 | JX171517 | HM068355 | AY707118 | |
†CBS: Westerdijk Fungal Biodiversity Institute. CPC: Collection of Pedro W. Crous, held at CBS. F: College of Forestry, Northwest A&F University, Taicheng Road, Yangling, Shaanxi China. NRRL: Agricultural Research Service, Peoria, IL, USA.
‡ IT: ex-isotype culture. PT: ex-paratype culture. T: ex-type culture. NT: ex-neotyype culture.
§CAL: Calmodulin. EF-1α: Translation elongation factor 1-alpha. RPB1: RNA polymerase largest subunit. RPB2: RNA polymerase second largest subunit. TUB: Tubulin. New sequences are shown in bold. Sequences marked as “Not public” were obtained from Kerry O’Donnell’s alignment datasets.
Morphological characterisation
Fusarium isolates were characterised morphologically according to procedures described elsewhere (Aoki et al. 2013; Leslie and Summerell 2006, Sandoval-Denis et al. 2018). Colonial growth rates and production of diffusible pigments were evaluated on PDA, colony features were also recorded on corn-meal agar (CMA; recipe in Crous et al. 2009) and OA. Colour notations followed those of Rayner (1970). For the study of micro-morphological features, cultures were grown for 7–10 d at 24 °C, using a 12 h light/dark cycle with near UV and white fluorescent light. Aerial and sporodochial conidiophores and conidia and formation of chlamydospores were evaluated on Synthetic Nutrient-poor Agar (SNA; Nirenberg 1976) and on Carnation Leaf Agar (CLA; Fisher et al. 1982). Measurements and photomicrographs were recorded from a minimum of 30 elements for each structure, using sterile water as mounting medium and a Nikon Eclipse 80i microscope with Differential Interference Contrast (DIC) optics and a Nikon AZ100 dissecting microscope, both equipped with a Nikon DS-Ri2 high definition colour digital camera and the Nikon software NIS-elements D software v. 4.30.
DNA isolation, amplification and sequencing
Isolates were grown for 7 d on MEA at 24 °C using the photoperiod described above. Fresh mycelium was scraped from the colony surface and subjected to total DNA extraction using the Wizard® Genomic DNA purification Kit (Promega Corporation, Madison, WI, USA), according to the manufacturer’s instructions. Fragments of five DNA loci were amplified using primers and PCR conditions described by O’Donnell et al. (2009) for calmodulin (CAL), O’Donnell et al. (2010) for the RNA polymerase largest subunit (RPB1) and second largest subunit (RPB2), O’Donnell et al. (1998) for the translation elongation factor 1-alpha (EF-1α) and Woudenberg et al. (2009) for beta-tubulin (TUB). Sequencing was made in both strand directions using the same primer pairs as for PCR amplification on an Applied Biosystems, Hitachi 3730xl DNA analyser (Applied Biosystems Inc., Foster City, California, USA). Consensus sequences were assembled using Seqman Pro v. 10.0.1 (DNASTAR, Madison, WI, USA). All DNA sequences generated in this study were lodged in GenBank and the European Nucleotide Archive (ENA) (Table 1).
Molecular identification and phylogenetic analyses
A first analysis was based on pairwise alignments and blastn searches on the Fusarium MLST (http://www.westerdijkinstitute.nl/fusarium/) and NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) databases, respectively, using EF-1α and RPB2 sequences in order to resolve the position of the KNP isolates amongst the different species complexes recognised in Fusarium (O’Donnell et al. 2013). Sequences from individual loci were aligned using MAFFT (Katoh and Standley 2013), on the web server of the European Bioinformatics Institute (EMBL–EBI; http://www.ebi.ac.uk/Tools/msa/mafft/) (Li et al. 2015).
Phylogenetic analyses were based on Maximum-likelihood (ML) and Bayesian (B) analyses, both algorithms run on the CIPRES Science Gateway portal (Miller et al. 2012). Evolutionary models were calculated using MrModelTest v. 2.3 using the Akaike information criterion (Nylander 2004; Posada and Crandall 1998). For ML, RAxML-HPC2 v. 8.2.10 on XSEDE was used (Stamatakis 2014), clade stability was tested with a bootstrap analysis (BS) using the rapid bootstrapping algorithm with default parameters. The B analyses were run using MrBayes v. 3.2.6 on XSEDE (Ronquist and Huelsenbeck 2003) using four incrementally heated MCMC chains for 5M generations, with the stop-rule option on and sampling every 1000 trees. After convergence of the runs (average standard deviation of split frequencies below 0.01) the first 25% of samples were discarded as the burn-in fraction and 50% consensus trees and posterior probabilities (PP) were calculated from the remaining trees.
Phylogenies were first made individually for each locus dataset and visually compared for topological incongruence amongst statistically supported nodes (ML-BS ≥ 70% and B-PP ≥ 0.95) (Mason-Gamer and Kellogg 1996, Wiens 1998), before being concatenated for multi-locus analyses using different locus combinations according to strains and DNA sequences currently available in public databases, in addition to previously published phylogenies (O’Donnell et al. 2000, 2013; Herron et al. 2015; Lupien et al. 2017; Moussa et al. 2017, Sandoval-Denis et al. 2018). A further 232 sequences representing 72 taxa were retrieved from GenBank and included in the phylogenetic analyses, while an additional 58 DNA sequences were obtained from 24 fungal strains requested from the CBS and NRRL (Agricultural Research Service, Peoria, IL, USA) culture collections (Table 1). All alignments and trees generated in this study were uploaded to TreeBASE (https://treebase.org).
Results
Phylogenetic analyses
Pairwise DNA alignments and BLAST searches using EF-1α and RPB2 sequences showed that the 19 isolates from KNP belonged to three different species complexes of the genus Fusarium i.e. the F. buharicum Jacz. ex Babajan & Teterevn.-Babajan species complex (FBSC; two isolates), the F. fujikuroi Nirenberg species complex (FFSC; two isolates) and the F. sambucinum Fuckel species complex (FSAMSC; 15 isolates). According to these results, sequences of related taxa and lineages were retrieved from GenBank and incorporated into individual phylogenetic analyses for each species complex.
Multi-locus analyses were carried out in order to further delimit the KNP Fusarium isolates amongst the known diversity in their respective species complexes. With the exception of the FFSC, the topologies observed from ML and B analyses of single and multi-locus datasets were highly congruent, with only minor differences affecting unsupported nodes on the trees (all trees available in TreeBASE). The characteristics of the different alignments and tree statistics for all the species complexes are shown in Table 2.
Table 2.
Characteristics of the different datasets and statistics of phylogenetic analyses used in this study.
| Analysis† | Locus‡ | Number of Sites§ | Evolutionary model| | Number of trees sampled in B | Maximum-likelihood statistics | ||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Conserved | Phylogenetically informative | B unique patterns | Best tree optimised likelihood | Tree length | ||||
| Fusarium buharicum SC | EF-1α | 495 | 300 | 119 | 198 | GTR+G | 414 | -11313.23702 | 0.598675 |
| RPB1 | 930 | 682 | 203 | 211 | SYM+G | ||||
| RPB2 | 1663 | 1251 | 330 | 310 | GTR+I+G | ||||
| Fusarium fujikuroi SC | CAL | 545 | 423 | 67 | 167 | SYM+G | 282 | -20603.30043 | 0.567054 |
| EF-1α | 677 | 428 | 127 | 295 | GTR+I+G | ||||
| RPB1 | 1534 | 1219 | 185 | 137 | SYM+I+G | ||||
| RPB2 | 1551 | 1211 | 227 | 315 | GTR+I+G | ||||
| TUB | 488 | 351 | 66 | 336 | SYM+G | ||||
| Fusarium sambucinum SC | RPB1 | 854 | 594 | 201 | 213 | SYM+I+G | 241 | -9871.793718 | 0.740271 |
| RPB2 | 1580 | 1128 | 346 | 396 | GTR+G | ||||
† SC: Species complex.
‡CAL: Calmodulin. EF-1α: Translation elongation factor 1-alpha. RPB1: RNA polymerase largest subunit. RPB2: RNA polymerase second largest subunit. TUB: Tubulin.
§ B: Bayesian inference.
| G: Gamma distributed rate variation among sites. GTR: Generalised time-reversible. I: Proportion of invariable sites. SYM: Symmetrical model.
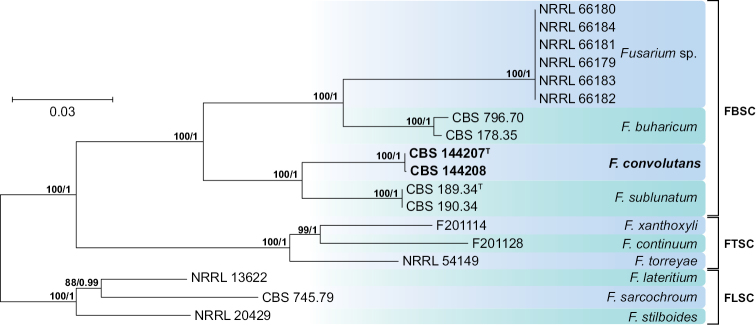
The analysis of the FBSC included sequences of EF-1α, RPB1 and RPB2 loci from 18 isolates representing 10 taxa, including members of the Fusarium torreyae T. Aoki, J.A. Sm., L.L. Mount, Geiser & O’Donnell species complex (FTYSC) and Fusarium lateritium Nees species complex (FLSC) as outgroup (Fig. 2). The four ingroup taxa resolved with high statistical support. Two KNP isolates from K. angustifolia obtained from the bottom site of the catena (CBS 144207 and 144208) clustered in a sister relationship with the clade representing Fusarium sublunatum Reinking, but were genetically clearly delimited.
Figure 2.
Maximum-likelihood (ML) phylogram obtained from combined EF-1α, RPB1 and RPB2 sequences of 18 strains belonging to the Fusarium buharicum (FBSC), Fusarium tricinctum (FTSC) and Fusarium lateritium (FLSC) species complexes. Numbers on the nodes are ML bootstrap values above 70% and Bayesian posterior probability values above 0.95. Branch lengths are proportional to distance. Ex-type strains are indicated with T. Strains corresponding to new species described here are shown in bold.
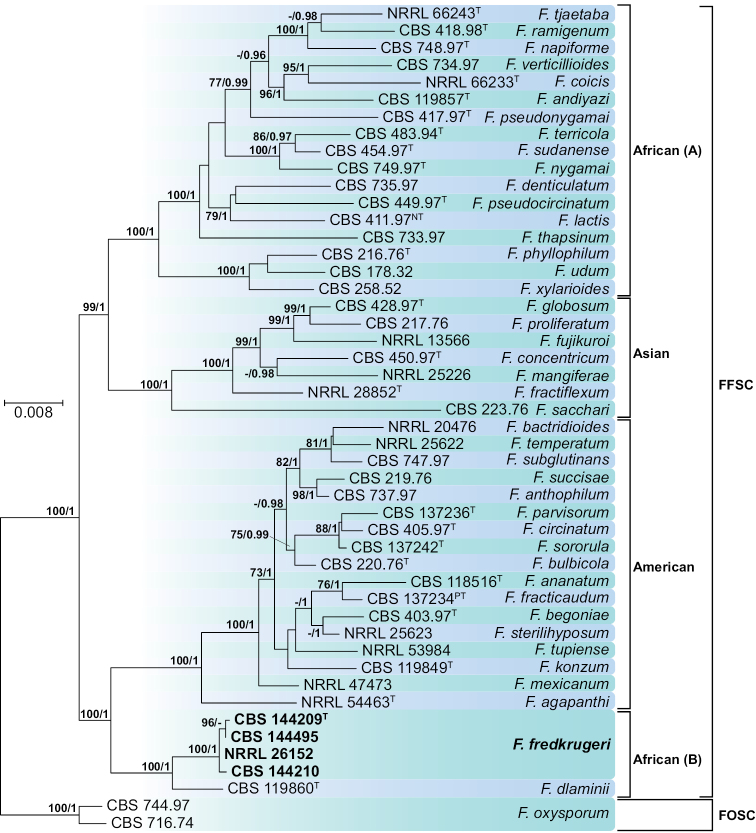
The phylogeny of the FFSC included sequences of CAL, EF-1α, RPB1, RPB2 and TUB loci from 48 strains and 44 taxa, including two outgroups (F. oxysporum CBS 716.74 and 744.97) (Fig. 3). The phylogeny showed a clear delimitation between the biogeographic clades recognised in this species complex (African, American and Asian clades sensu O’Donnell et al. 1998). Both American and Asian clades where shown as monophyletic with high ML-BS and B-PP support; in contrast, the African clade was resolved as polyphyletic, comprising two distinct and highly supported lineages. A terminal, speciose clade (African A) encompassing 17 taxa and a basal clade (African B), close to the American clade which included the ex-type of Fusarium dlaminii Marasas, P.E. Nelson & Toussoun (CBS 119860) and a sister terminal clade (ML-BS=100, B-PP=1) comprising two KNP isolates from M. acuminata (CBS 144209 and 144495) and two unidentified African Fusarium isolates (CBS 144210 and NRRL 26152). From the loci used here, only TUB resolved both African clades as sister groups; however, its monophyly was not supported by clade stability measurements (data not shown). Conversely, individual CAL, EF-1α and RPB2 phylogenies resolved African B as basal to the ingroup, while RPB1 allocated this clade as basal to the American clade. Nonetheless, all the individual phylogenies, in addition to the combined dataset, clearly demonstrated genealogical uniqueness of the terminal clade encompassing KNP isolates.
Figure 3.
Maximum-likelihood (ML) phylogram obtained from combined CAL, EF-1α, RPB1, RPB2 and TUB sequences of 48 strains belonging to the Fusarium fujikuroi (FFSC) and Fusarium oxysporum (FOSC) species complexes. Numbers on the nodes are ML bootstrap values above 70% and Bayesian posterior probability values above 0.95. Branch lengths are proportional to distance. Ex-type, ex-neotype and ex-paratype strains are indicated with T, NT and PT, respectively. Strains corresponding to new species described here are shown in bold.
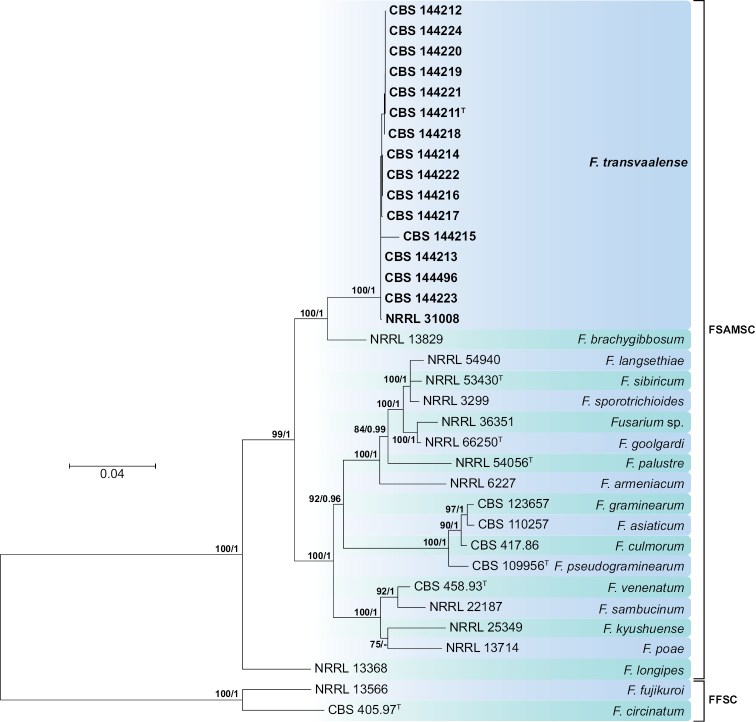
The FSAMSC was studied using combined RPB1 and RPB2 sequences. The phylogeny included 35 isolates from 20 taxa, including the two outgroups Fusarium circinatum Nirenberg & O’Donnell (CBS 405.97) and Fusarium fujikuroi Nirenberg (NRRL 13566) (Fig. 4). Fifteen KPN Fusarium isolates from the three sampled plant species (three isolates from K. angustifolia, four isolates from M. acuminata and eight isolates from S. cordifolia), all obtained from the top site of the catena, clustered with an unidentified Fusarium isolate (NRRL 31008) in a distinct clade (ML-BS=100, B-PP=1), close to Fusarium brachygibbosum Padwick (strain NRRL 13829).
Figure 4.
Maximum-likelihood (ML) phylogram obtained from combined RPB1 and RPB2 sequences of 35 strains belonging to the Fusarium sambucinum (FSAMSC) and Fusarium fujikuroi (FFSC) species complexes. Numbers on the nodes are ML bootstrap values above 70% and Bayesian posterior probability values above 0.95. Branch lengths are proportional to distance. Ex-type strains are indicated with T. Strains corresponding to new species described here are shown in bold.
The clades including KNP isolates and corresponding to previously undisclosed lineages of Fusarium are described in the taxonomy section as the three novel species, F. convolutans, F. fredkrugeri and F. transvaalense.
Taxonomy
Fusarium convolutans
Sandoval-Denis, Crous & W.J. Swart sp. nov.
MB825102
Figure 5.
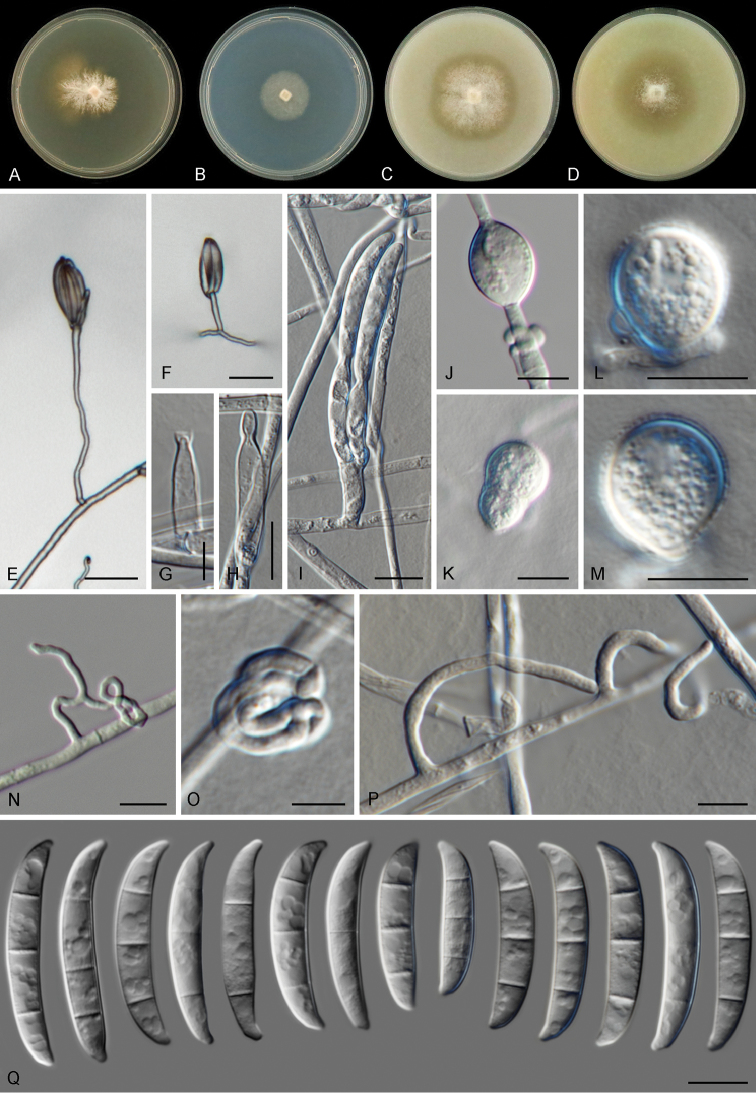
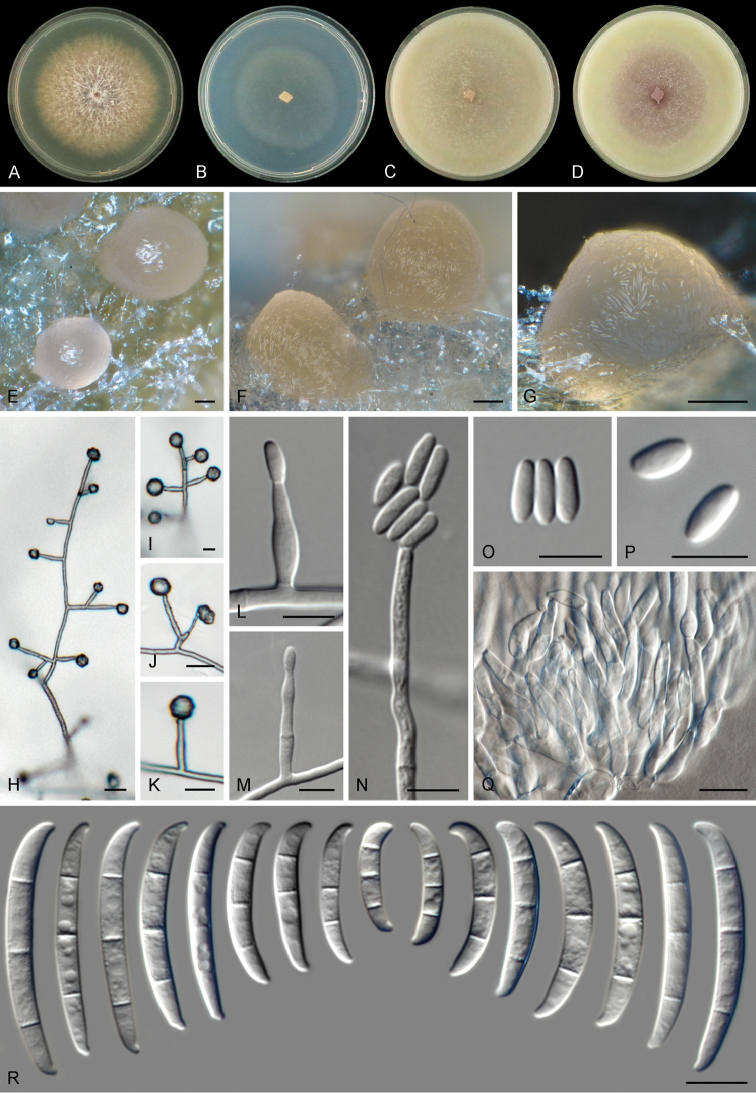
Fusarium convolutans sp. nov. A–D Colonies on PDA, SNA, OA and CMA, respectively, after 7 d at 24 °C in the dark E–I Conidiophores, phialides and conidia J–M Chlamydospores N–P Sterile hyphal projections Q Conidia. Scale bars: 20 μm (E, F); 5 μm (G–I); 10 μm (J–Q).
Diagnosis.
Different from F. circinatum, F. pseudocircinatum O’Donnell & Nirenberg and F. sterilihyphosum Britz, Marasas & M.J. Wingf. by the absence of aerial conidia (microconidia) and the presence of chlamydospores. Different from F. buharicum Jacz. ex Babajan & Teterevn.-Babajan and F. sublunatum by its shorter, less septate and less curved conidia and by the presence of sterile hyphal coils.
Type.
South Africa, Kruger National Park, Skukuza, Granite Supersite, 25°06'33.9"S, 31°34'40.9E, from rhizosphere soil of Kyphocarpa angustifolia, 23 Mar 2015, W.J. Swart, holotype CBS H-23495, dried culture on OA, ex-holotype strain CBS 144207 = CPC 33733.
Description.
Colonies on PDA growing in the dark with an average radial growth rate of 2.1–4.8 mm/d, 4.4–5.8 mm/d and 4.6–6.3 mm/d at 24, 27 and 30 °C, respectively; reaching 11–28 mm diam. in 7 d at 24 °C and a maximum of 23–37 mm diam. in 7 d at 30 °C. Minimum temperature for growth 12 °C, maximum 36 °C, optimal 27–33 °C. Colony surface white to cream coloured, flat and highly irregular in shape, velvety to felty, with scant and short aerial mycelium; colony margins highly irregular to rhizoid, with abundant white to grey submerged mycelium. Reverse white, straw to yellow diffusible pigment produced between 21–33 °C, scarcely produced and turning luteous to orange at 36 °C. Colonies on CMA and OA incubated in the dark reaching 40–48 mm diam. in 7 d at 24 °C. Colony surface white to cream coloured, flat or slightly elevated at the centre, velvety to dusty; aerial mycelium abundant, short and dense, concentrated on the colony centre; margins membranous and regular, buff to honey coloured, without aerial mycelium. Reverse ochreous without diffusible pigments. Sporulation scant from conidiophores formed on the aerial mycelium, sporodochia not formed. Conidiophores on the aerial mycelium straight or flexuous, smooth- and thin-walled, simple, mostly reduced to conidiogenous cells borne laterally on hyphae or up to 50 μm tall, bearing terminal single or paired monophialides; phialides subulate to subcylindrical, smooth- and thin-walled, 15.5–22 μm long, (3.5–)4–5 μm at the widest point, with inconspicuous periclinal thickening and a short- flared collarette; conidia clustering in discrete false heads at the tip of monophialides, lunate to falcate, curved or somewhat straight, tapering gently toward the basal part, robust; apical cell often equal in length or slightly shorter than the adjacent cell, blunt to conical; basal cell papillate to distinctly notched, (1–2–)3-septate, hyaline, thin- and smooth-walled. One-septate conidia: 24 × 4.5 μm; two-septate conidia: 24.5 × 6 μm; three-septate conidia: (25.5–)29–36.5(–38.5) × (4–)5–6.5(–7.5) μm. Chlamydospores abundantly formed, globose to subglobose, smooth- and thick-walled, (9.5–)11–13.5(–14) μm diam.; terminal or intercalary in the hyphae or conidia, often borne laterally at the tip of elongated, cylindrical, stalk-like projections, solitary or in small clusters. Sterile, coiled, sometimes branched hyphal projections abundantly formed laterally from the substrate and aerial mycelium.
Distribution.
South Africa.
Etymology.
From Latin, “convolutans”, participle of convolutare, coiling, in reference to the abundant sterile, coiled lateral hyphal projections.
Additional isolate examined.
South Africa, Kruger National Park, Skukuza, Granite Supersite, 25°06'33.9"S, 31°34'40.9E, from rhizosphere soil of Kyphocarpa angustifolia, 23 Mar 2015, W.J. Swart, CBS 144208 = CPC 33732.
Notes.
The main morphological feature of F. convolutans, namely the production of sterile, coiled hyphal projections, grossly resembles other Fusarium species producing similar structures i.e. F. circinatum, F. pseudocircinatum and F. sterilihyphosum. The three latter species, however, are genetically unrelated to F. convolutans, being allocated in the FFSC; and are also easily differentiable by the characteristics of the aerial conidia (typical Fusarium microconidia are absent in the new species) and the lack of chlamydospores (present in the new species) (Leslie and Summerell 2006). Fusarium convolutans can be easily differentiated morphologically from their phylogenetically closely related species, F. buharicum and F. sublunatum. It has relative simple conidiophores and shorter, less septate and markedly less curved conidia (up to 38.5 μm long and 1–3-septate vs. up to 87 and 81 μm long, 0–8-septate in F. buharicum and F. sublunatum, respectively) (Gerlach and Nirenberg 1982). Fusarium buharicum and F. sublunatum also lack sterile hyphal coils.
Fusarium fredkrugeri
Sandoval-Denis, Crous & W.J. Swart sp. nov.
MB825103
Figure 6.
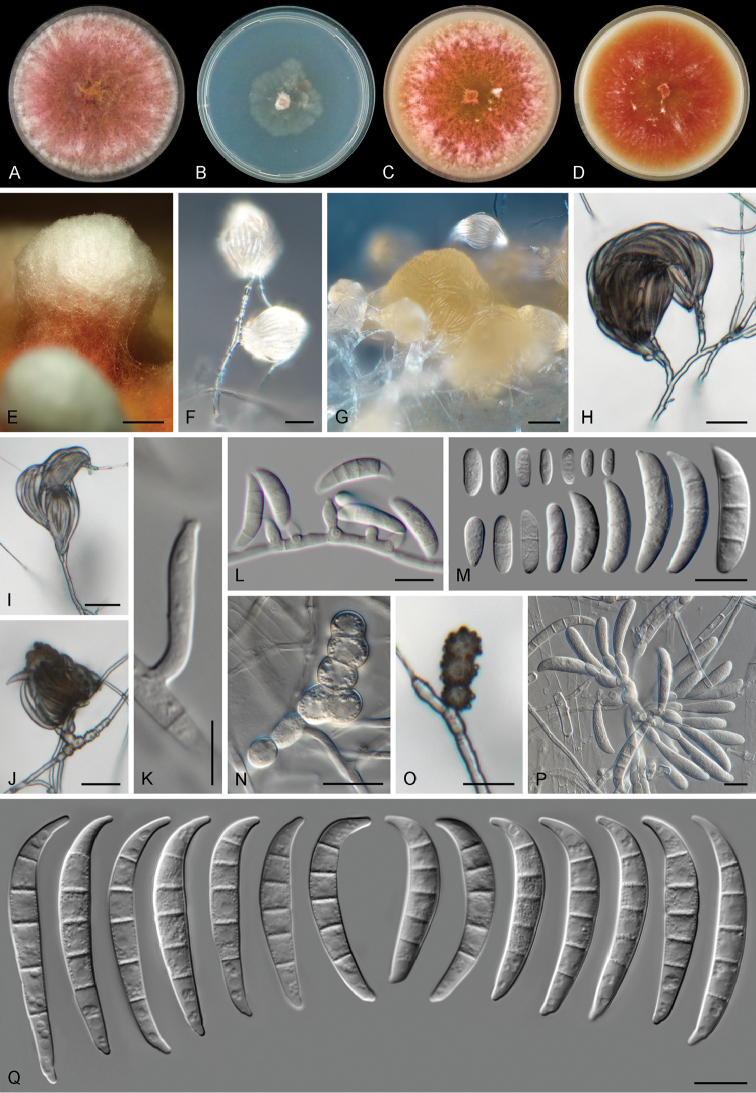
Fusarium fredkrugeri sp. nov. A–D Colonies on PDA, SNA, OA and CMA, respectively, after 7 d at 24 °C in the dark E–G Sporodochia formed on the surface of carnation leaves H–N Aerial conidiophores, phialides and conidia O, P Aerial conidia Q Sporodochial conidiophores and phialides R Sporodochial conidia. Scale bars: 100 μm (E–G); 10 μm (H–R).
Diagnosis.
Differs from Fusarium dlaminii Marasas, P.E. Nelson & Toussoun by producing only one type of aerial conidia, shorter sporodochial conidia and the absence of chlamydospores.
Type.
South Africa, Kruger National Park, Skukuza, Granite Supersite, 25°06'48.6"S, 31°34'36.5"E, from rhizosphere soil of Melhania acuminata, 23 Mar 2015, W.J. Swart, holotype CBS H-23496, dried culture on OA, culture ex-holotype CBS 144209 = CPC 33747.
Description.
Colonies on PDA growing in the dark with an average radial growth rate of 4.7–5.8 mm/d and reaching 22–35 mm diam. in 7 d at 24 °C, filling an entire 9 cm Petri dish in 7 d at 27 and 30 °C. Minimum temperature for growth 12 °C, maximum 36 °C, optimal 27–30 °C. Colony surface at first white to cream coloured, later turning bay to chestnut with pale luteous to luteous periphery; flat, felty to cottony with abundant erect- aerial mycelium forming white patches; colony margins regular and filiform with abundant submerged mycelium. Reverse pale luteous, a blood sepia to chestnut coloured diffusible pigment is scarcely produced at 24 °C, pigment production is markedly enhanced at 27–30 °C, becoming greyish-sepia at 33 °C. Colonies on CMA and OA incubated at 24 °C in the dark reaching 65–67 mm diam. or occupying an entire 9 cm Petri dish in 7 d, respectively. Colony surface pale bay coloured, flat, felty to velvety, aerial mycelium scant, forming white to cream patches; margins regular. Reverse pale bay to pale vinaceous. Sporulation abundant from conidiophores formed on the substrate and aerial mycelium and from sporodochia. Conidiophores on the aerial mycelium straight or flexuous, erect or prostrate, septate, smooth- and thin-walled, often appearing rough by accumulation of extracellular material, commonly simple or reduced to conidiogenous cells borne laterally on hyphae or up to 200 μm tall and irregularly branched at various levels, branches bearing lateral and terminal monophialides borne mostly single or in pairs; phialides subulate, ampulliform, lageniform to subcylindrical, smooth- and thin-walled, (8.5–)9.5–17.5(–24.5) μm long, 2–3(–3.5) μm at the widest point, without periclinal thickening, collarets inconspicuous; conidia formed on aerial conidiophores, hyaline, obovoid, ellipsoidal to slightly reniform or allantoid, smooth- and thin-walled, 0-septate, (4.5–)5–8.5(–12.5) × (1.5–)2–3.5(–6) μm, clustering in discrete false heads at the tip of monophialides. Sporodochia pale orange to pink coloured, often somewhat translucent, formed abundantly on the surface of carnation leaves and on the agar surface. Conidiophores in sporodochia 26–46 μm tall, densely aggregated, irregularly and verticillately branched up to three times, with terminal branches bearing 2–3 monophialides; sporodochial phialides doliiform to subcylindrical, (9–)11.5–15.5(–18.5) × (2.5–)3–4(–4.5) μm, smooth- and thin-walled, with periclinal thickening and an inconspicuous apical collarette. Sporodochial conidia falcate, tapering toward the basal part, robust, moderately curved and slender; apical cell more or less equally sized than the adjacent cell, blunt to slightly papillate; basal cell papillate to distinctly notched, (1–)3–4-septate, hyaline, thin- and smooth-walled. One-septate conidia: 13–17(–18) × (2.5–)3–4 μm; two-septate conidia: 15 × 4.5 μm; three-septate conidia: (16–)28.5–39(–45) × (3–)4–5(–5.5) μm; four-septate conidia: 39.5–40(–41) × 4.5–5 μm; overall (13–)27.5–39.5(–45) × (3–)3.5–5.5 μm. Chlamydospores absent.
Distribution.
Madagascar, Niger and South Africa.
Etymology.
In honour and memory of Dr. Frederick J. Kruger, pioneer of forest hydrology, fynbos ecology and invasive species and fundamental for the collections included in this study.
Additional isolates examined.
Madagascar, from Striga hermonthica, unknown date, A.A. Abbasher, CBS 144210 = NRRL 26061 = BBA 70127. South Africa, Kruger National Park, Skukuza, Granite Supersite,25°06'48.6"S, 31°34'36.5"E, from rhizosphere soil of Melhania acuminata, 23 Mar 2015, W.J. Swart, CBS 144495 = CPC 33746.
Notes.
This species is genetically closely related to F. dlaminii, both species having similar colonial morphology, optimal growth conditions and biogeography. Moreover, both species exhibit relatively short aerial phialides producing conidia in heads, somewhat resembling those produced by F. oxysporum rather than most members of the FFSC (Leslie and Summerell 2006; Marasas et al. 1985). However, besides exhibiting much faster growth rates, F. fredkrugeri presents clearly distinctive morphological features such as the production of only one type of aerial conidia (vs. two types in F. dlaminii: allantoid to fusiform and 0-septate; and napiform 0–1-septate); orange to pink sporodochia, produced on carnation leaves but also abundantly on the agar surface (vs. orange sporodochia, produced only on the surface of carnation leaves in F. dlaminii) (Leslie and Summerell 2006). Additionally, F. fredkrugeri produces shorter and less septate sporodochial conidia ((1–)3–4-septate and up to 45 μm long in the latter species vs. mostly 5-septate and up to 54 μm long in F. dlaminii) while chlamydospores are not produced. The latter feature, coupled with the somewhat more complex conidiophores also clearly differentiates F. fredkrugeri from F. oxysporum.
Fusarium transvaalense
Sandoval-Denis, Crous & W.J. Swart sp. nov.
MB825104
Figure 7.
Fusarium transvaalense sp. nov. A–D Colonies on PDA, SNA, OA and CMA, respectively, after 7 d at 24 °C in the dark E Pustule-like growth on OA F, G Sporodochia formed on the surface of carnation leaves H–L Aerial conidiophores phialides and conidia M Aerial conidia N, O Chlamydospores P Sporodochial conidiophores and phialides Q Sporodochial conidia. Scale bars: 2 mm (E); 20 μm (F–J); 5 μm (K); 10 μm (L–Q).
Diagnosis.
Different from most species in FSAMSC by its slender sporodochial conidia with tapered and somewhat rounded apex; its smooth- to tuberculate, often pigmented chlamydospores and the formation of large mycelial tufts on OA.
Type.
South Africa, Kruger National Park, Skukuza, Granite Supersite, 25°06'45.5"S, 31°34'35.0"E, from rhizosphere soil of Sida cordifolia, 23 Mar 2015, W.J. Swart, holotype CBS H-23497, dried culture on SNA, culture ex-holotype CBS 144211 = CPC 30923.
Description.
Colonies on PDA growing in the dark with an average radial growth rate of 8.5–9.3 mm/d, reaching 34–37 mm diam. in 7 d at 24 °C, filling an entire 9 cm Petri dish in 7 d at 27–33 °C. Minimum temperature for growth 12 °C, maximum 36 °C, optimal 27–30 °C. Colony surface at first white, turning coral to dark vinaceous with white periphery and abundant yellow hyphae at the centre; flat, velvety to woolly, with abundant aerial mycelium and erect hyphal strings reaching several mm tall; colony margins regular and filiform. Reverse with yellow, coral or dark vinaceous patches, coral diffusible pigments strongly produced between 15–30 °C, turning scarlet to orange at 33–36 °C. Colonies on CMA and OA incubated at 24 °C in the dark occupying an entire 9 cm Petri dish in 7 d. Colony surface coral, rust to chestnut coloured in irregular patches, flat, felty to woolly, aerial mycelium scarce on CMA, mostly as radially dispersed white patches, on OA aerial mycelium abundant, especially on the periphery of the colony, forming dense, pustule-like, white mycelial tufts, formed by abundant intermingled hyphae and chlamydospores, 1–1.5 cm tall, with flesh to coral coloured stipes; margins on CMA and OA regular. Reverse pale luteous with red to coral periphery. Sporulation abundant from conidiophores formed on the aerial mycelium, at the agar level and from sporodochia. Conidiophores on the aerial mycelium straight or flexuous, septate, smooth- and thin-walled, up to 150 μm tall, sometimes emerging from irregular, swollen, pigmented and rough-walled cells on the hyphae; simple or sparingly and irregularly branched, branches bearing terminal, rarely lateral monophialides or reduced to conidiogenous cells borne laterally on hyphae; phialides on the aerial conidiophores short ampulliform, subulate to subcylindrical, smooth- and thin-walled, (7–)9–14(–15) μm long, (3–)4–5 μm at the widest point, without periclinal thickening and with a minute, inconspicuous collarette; conidia formed on aerial conidiophores of two types: a) hyaline, obovoid, ellipsoidal to clavate, smooth- and thin-walled, 0–1-septate, 2–14 × 2–4 μm; b) lunate to short falcate with a pointed apex and a somewhat flattened base, smooth- and thin-walled, 3–5-septate. Three-septate conidia: (16–)18–27(–29) × 5–6 μm; four-septate conidia: 21–24(–25) × 5–6 μm; five-septate conidia: (25–)27–33 × 5–6 μm. Sporodochia cream to orange coloured, formed abundantly on the surface of carnation leaves and rarely on the agar surface, at first very small and sparse later becoming aggregated. Conidiophores in sporodochia 22–31 μm tall, irregularly branched, bearing clusters of 3–6 monophialides; sporodochial phialides doliiform to ampulliform, (5–)9–14(–18) × (3–)4–5 μm, smooth- and thin-walled, with periclinal thickening and a short apical collarette. Sporodochial conidia falcate, wedge-shaped, tapering towards both ends, markedly curved and robust; apical cell longer than the adjacent cell, pointed; basal cell distinctly notched, sometimes somewhat extended (1–)3–5(–6)-septate, hyaline, smooth- and thick-walled. One-septate conidia: 19 × 4 μm; three-septate conidia: 20–27(–28) × 5–7 μm; four-septate conidia: (29–)30–32 × 5–7 μm; five-septate conidia: (26–)29–41(–53) × 4–5(–6) μm; six-septate conidia: 36 × 7 μm; overall (19–)25.9–40(–53) × (3.5–)4–6(–7) μm. Chlamydospores abundant, hyaline or pigmented, smooth- to rough-walled or tuberculate, 7–8 μm diam., terminal or intercalary, solitary, in chains or in clusters.
Distribution.
Australia and South Africa
Etymology.
After Transvaal, the name of a former colony and Republic located between the Limpopo and Vaal rivers, currently a province of South Africa and where this species was found. From Latin trans meaning “on the other side of” and Vaal a South African river.
Additional isolates examined.
South Africa, Kruger National Park, Skukuza, Granite Supersite, 25°06'48.6"S, 31°34'36.5"E, from rhizosphere soil of Melhania acuminata, 23 Mar 2015, W.J. Swart, CBS 144224 = CPC 30928, CBS 144212 = CPC 30929); 25°06'45.6"S, 31°34'37.7"E, CBS 144496 = CPC 33750, CBS 144213 = CPC 33751; 25°06'48.8"S, 031°34'36.6"E, from rhizosphere soil of Sida cordifolia, 23 Mar 2015, W.J. Swart, CBS 144214 = CPC 30946; 25°06'45.7"S, 31°34'35.1"E, CBS 144215 = CPC 33723; 25°06'45.5"S, 31°34'35.0"E, CBS 144216 = CPC 30918, CBS 144217 = CPC 30919, CBS 144218 = CPC 30922, , CBS 144219 = CPC 30926, CBS 144220 = CPC 30927); 25°06'51.4"S, 31°34'37.5"E, from rhizosphere soil of Kyphocarpa angustifolia, 23 Mar 2015, W.J. Swart, CBS 144221 = CPC 33740; 25°06'51.8"S, 31°34'38.1"E, CBS 144222 = CPC 30939, CBS 144223 = CPC 30941.
Notes.
Fusarium transvaalense exhibits a sporodochial conidial morphology typical of members of FSAMSC with marked dorsiventral curvature and tapered ends. Several species in FSAMSC form comparable conidia in culture i.e. F. crookwellense L.W. Burgess, P.E. Nelson & Toussoun, F. sambucinum, F. sporotrichioides Sherb., F. venenatum Nirenberg and F. culmorum (Wm.G. Sm.) Sacc. However, with the exception of F. sporotrichioides, the conidia of most species above-mentioned, differ by being more robust and often more pointed apically. Fusarium transvaalense differs from F. sporotrichioides by the absence of pyriform aerial conidia.
Two strains NRRL 13829 and NRRL 31008, previously identified as F. brachygibbosum Padwick showed different degrees of genetic similitude with the new species. While NRRL 31008 clustered within F. transvaalense, NRRL 13829 formed a clearly delimited sister linage. Morphologically, F. transvaalense exhibits significant differences allowing its separation from F. brachygibbosum. Both species produce sporodochial conidia with similar septation and sizes; however, F. brachygibbosum commonly exhibits a bulge in the middle portion of the conidia (Padwick 1945), a feature not present in F. transvaalense. In addition, the latter species produces comparatively larger sporodochial conidia, when elements with the same degree of septation are compared; its chlamydospores are smaller, smooth-walled to markedly tuberculate and pigmented (7–8 μm vs. 10.7–15.3 μm, smooth-walled and hyaline in F. brachygibbosum) and has a distinctive colonial growth on OA, forming large, pustule-like hyphal tufts, a feature not reported for F. brachygibbosum (Padwick 1945).
Discussion
In this study, three new Fusarium spp. were introduced, isolated from rhizosphere soils of three native African shrubs in a protected savannah ecosystem deep inside the Kruger National Park, South Africa.
Some remarkable differences were noted regarding the distribution of the novel fungal species and their respective hosts on this particular site. For instance, F. transvaalense, which exhibited the greatest relative abundance, was found in high quantities from the rhizospheres of the three hosts sampled, showing a considerable genetic diversity. Interestingly, this species was only on the top of the catena, even when two of its hosts, K. angustifolia and S. cordifolia, were found and sampled either at the top and bottom sites. Similarly, F. fredkrugeri was recovered only from soils under M. acuminata, a host species which occurred only at the top location. In contrast, F. convolutans was found in the rhizosphere of K. angustifolia, occurring only at the bottom of the catena, while none of the three fungal species was found associated with S. cordifolia at the bottom of the site. Nevertheless, not being an objective of this work, it was not possible to categorically assign these new species to specific hosts or locations. Likely, these fungi could be in low abundance and thus not detectable using the current methods. However, plant species composition varies considerably through a catena ecosystem, in relation to the different soil characteristics, pH gradient and water availability, which also greatly influence microbial and animal biodiversity (Lareen et al. 2016; Mohammadi et al. 2017). However, the full patterns of variation between locations on this particular catena still need to be systematically assessed and compared. As evidenced here, certain differences do exist between the soils at the upper and bottom locations of the Stevenson-Hamilton supersite, which might explain the fungal diversity variation observed here. The cation exchange capacity (CEC; capacity of a soil to hold exchangeable cations) varies considerably between sampling sites, basically depending on the proportion of sand versus clay content of each soil type (Ketterings et al. 2007; Van Zijl and Le Roux 2014). It is known that CEC greatly impacts the soil’s ability to retain essential nutrients and prevents soil acidification (Ketterings et al. 2007). Nutrient content also increased from the top to the bottom of the slope which is consistent with the increase in CEC. Nutrient poor soils are also a driver of biological diversity and most likely influenced fungal diversity in these particular locations (Havlicek and Mitchell 2014, Mapelli et al. 2017).
The three Fusarium species, described here, were not associated with any visible symptomatology on their hosts. However, they cannot be ruled out as pathogens since they were not assessed for pathogenicity against the sampled plants nor any other putative host species at the same locations. Likewise, it is unknown if these fungi exert any beneficial or deleterious effect on their ecosystems. These are important unsolved questions that need further evaluation. However, as shown by phylogenetic analyses, each of the three new species was in close genetic proximity with well-known plant pathogenic Fusarium spp. on their respective species complexes, which could suggest a potential pathogenic role. Fusarium convolutans clustered within the FBSC, together with three known plant pathogenic Fusarium spp. i.e. F. buharicum, a pathogen of Hibiscus cannabinus L. and Gossypium L.; F. sublunatum, known to affect banana and Theobroma cacao L. in Central America (Gerlach and Nirenberg 1982, Leslie and Summerell 2006) and a newly discovered although unnamed phylogenetic species causing wilt, crown and root rot of Hibiscus moscheutos L. (Lupien et al. 2017). Fusarium transvaalense belonged to the FSAMSC, a genetically diverse group common in temperate and subtropical zones (Leslie and Summerell 2006). Fusarium sambucinum, the conserved type species of the genus (Gams et al. 1997) being an aggressive plant pathogen and one of the most important agents of potato dry rot (Peters et al. 2008); while the latter species and several others in the complex have been reported causing disease on diverse crops, including many cereals and fruits (Leslie and Summerell 2006).
Fusarium fredkrugeri is here recognised and formally proposed as a new species. Although the clade representing this taxon had already been identified as a distinct unnamed phylogenetic species by O’Donnell et al. (2000), it had not been given a formal description pending the collection of additional isolates. Two other African isolates previously determined to belong to this clade i.e. CBS 144210 from Striga hermonthica (Del.) Benth. in Madagascar and NRRL 26152 from an unknown substrate in Niger, were incorporated into the analyses, although the latter strain is not viable anymore (NRRL, pers. comm.), thus not available for morphological assessment. Strain CBS 144210, however, is known as a pathogen of the ‘purple witchweed’, a parasite plant common to sub-Saharan Africa and known to devastate Sorghum bicolor (L.) Moench and Oryza sativa L. plantations (O’Donnell et al. 2000; Yoshida et al. 2010). As previously demonstrated by O’Donnell et al. (2000), our phylogenetic results showed that the clade comprising F. fredkrugeri and its sister species F. dlaminii does not cluster within the main African core of species in the FFSC. Thus, despite the African origin of our isolates, the predicted biogeographic patterns did not match the observed phylogeny. It has been hypothesised that this should not be the result of genetic markers tracing different phylogenies, but the consequence of losing the phylogenetic signal due to saturated sites and introns (O’Donnell et al. 2000). However, the inclusion in our analysis of additional, highly informative and slowly evolving loci such as RPB1 and RPB2 yielded similar results, which points out the need to re-evaluate the phylogeographic arrangement of this important species complex including the vast new data generated during the last 20 years that challenges the established assumptions (Kvas et al. 2009; Walsh et al. 2010; O’Donnell et al. 2013; Laurence et al. 2015). Nevertheless, although rather unlikely, alternative factors such as anthropogenic dispersion of F. fredkrugeri, its host or additional invasive alternative hosts, cannot be rejected as an explanation for the discordance between biogeography and phylogenetic results. However, these scenarios are difficult to imagine given the characteristics of the sampled site, not being an agroecosystem but a protected, isolated zone, with minimal human intervention (Smit et al. 2013).
This study is a new example of how easily new Fusarium spp. can be found when mycological studies are directed to neglected natural ecosystems of minimal anthropogenic disturbance (Phan et al. 2004; Leslie and Summerell 2011; Summerell et al. 2011; Burgess 2014, Laurence et al. 2015). Although irrelevant for some researchers, finding and properly describing new species, regardless of whether they have little or no pathogenic or mycotoxigenic potential, is of utmost importance to improve our understanding on the diversity, biogeographic and phylogeographic patterns of such a complex and heterogeneous genus as Fusarium. In addition, this study remarks on the significance and need to further stimulate the exploration of conserved, non-manipulated natural environments (supersites) and their potential impact on biodiversity research on the fungal kingdom.
Supplementary Material
Aknowledgments
Todd J. Ward and James Swezey (Agricultural Research Service, Peoria, IL, USA) are thanked for providing strains. We kindly thank Kerry O’Donnell (Mycotoxin Prevention and Applied Microbiology Research Unit, Agricultural Research Service, US Department of Agriculture, Peoria, IL, USA) for providing DNA sequence datasets. Mercia Coetzee (Central University of Technology, Bloemfontein, South Africa) is thanked for her technical support in the field. Alejandra Giraldo (Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands) is thanked for her assistance with fungal isolation. Eddie Riddell and Navashni Govender (SANParks) are acknowledged for their research support in the Kruger National Park. We also thank Konstanze Bensch (Mycobank curator) and Uwe Braun (Geobotanik und Botanischer Garten, Martin-Luther-Universität Halle-Wittenberg, Halle, Germany) for their help regarding Latin names.
Citation
Sandoval-Denis M, Swart WJ, Crous PW (2018) New Fusarium species from the Kruger National Park, South Africa. MycoKeys 34: 63–92. https://doi.org/10.3897/mycokeys.34.25974
References
- Aoki T, Smith JA, Mount LL, Geiser DM, O’Donnell K. (2013) Fusarium torreyae sp. nov., a pathogen causing canker disease of Florida torreya (Torreya taxifolia), a critically endangered conifer restricted to northern Florida and southwestern Georgia. Mycologia 105: 312–319. https://doi.org/10.3852/12-262 [DOI] [PubMed] [Google Scholar]
- Aydogdu H, Asan A. (2008) Airborne fungi in child day care centers in Edirne City, Turkey. Environmental Monitoring and Assessment 147: 423–444. https://doi.org/10.1007/s10661-007-0130-4 [DOI] [PubMed] [Google Scholar]
- Bent E, Kiekel P, Brenton R, Taylor DL. (2011) Root-associated ectomycorrhizal fungi shared by various boreal forest seedlings naturally regenerating after a fire in interior Alaska and correlation of different fungi with host growth responses. Applied and Environmental Microbiology 77: 3351–3359. https://doi.org/10.1128/AEM.02575-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DJ, Clayton MK, McSweeney K. (2004) Potential terrain controls on soil color, texture contrast and grain-size deposition for the original catena landscape in Uganda. Geoderma 122: 51–72. http://doi.org/10.1016/j.geoderma.2003.12.004 [Google Scholar]
- Burgess LW. (2014) 2011 McAlpine Memorial Lecture – A love affair with Fusarium. Australasian Plant Pathology 43: 359–368. https://doi.org/10.1007/s13313-013-0261-8 [Google Scholar]
- Carruthers J. (2017) National Park Science: A Century of Research in South Africa (Ecology, Biodiversity and Conservation). Cambridge University Press, 554 pp. https://doi.org/10.1017/9781108123471
- Crous PW, Verkley GJM, Groenewald JZ, Samson RA. (2009) Fungal Biodiversity. CBS Laboratory Manual Series (CBS-KNAW Fungal Biodiversity Centre, Utrecht) 1: 1−270.
- Díaz Arias MM, Leandro LF, Munkvold GP. (2013) Aggressiveness of Fusarium species and impact of root infection on growth and yield of soybeans. Phytopathology 103: 822−832. https://doi.org/10.1094/PHYTO-08-12-0207-R [DOI] [PubMed]
- Fisher NL, Burguess LW, Toussoun TA, Nelson PE. (1982) Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology 72: 151–153. https://doi.org/10.1094/Phyto-72-151 [Google Scholar]
- Fravel D, Olivain C, Alabouvette C. (2003) Fusarium oxysporum and its biocontrol. New Phytologist 157: 493–502. https://doi.org/10.1046/j.1469-8137.2003.00700.x [DOI] [PubMed] [Google Scholar]
- Gams W, Nirenberg HI, Seifert KA, Brayford D, Thrane U. (1997) (1275) Proposal to conserve the name Fusarium sambucinum (Hyphomycetes). Taxon 46: 111–113. https://doi.org/10.2307/1224298 [Google Scholar]
- Gerlach W, Nirenberg HI. (1982) The genus Fusarium – a pictorial atlas. Mitteilungen der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 209: 1–406. [Google Scholar]
- Hargreaves SK, Williams RJ, Hofmockel KS. (2015) Environmental filtering of microbial communities in agricultural soil shifts with crop growth. PLoS One 30: e0134345. https://doi.org/10.1371/journal.pone.0134345 [DOI] [PMC free article] [PubMed]
- Hassan Dar GH, Zargar MY, Beigh GM. (1997) Biocontrol of Fusarium root rot in the common bean (Phaseolus vulgaris L.) by using symbiotic Glomus mosseae and Rhizobium leguminosarum. Microbial Ecology 34: 74–80. https://doi.org/10.1007/s002489900036 [DOI] [PubMed] [Google Scholar]
- Havlicek E, Mitchell EAD. (2014) Soils supporting biodiversity. In: Dighton J, Krumins JA. (Eds) Interactions in Soil: Promoting Plant Growth, Biodiversity, Community and Ecosystems. Springer, Dordrecht, 27–28. https://doi.org/10.1007/978-94-017-8890-8_2
- Herron DA, Wingfield MJ, Wingfield BD, Rodas CA, Marincowitz S, Steenkamp ET. (2015) Novel taxa in the Fusarium fujikuroi species complex from Pinus spp. Studies in Mycology 80: 131–150. https://doi.org/10.1016/j.simyco.2014.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris HA, Labuschagne N, Korsten L. (2007) Screening rhizobacteria for biological control of Fusarium root and crown rot of sorghum in Ethiopia. Biological Control 40: 97−106. https://doi.org/10.1016/j.biocontrol.2006.07.017
- Jumpponen A, Herrera J, Porras-Alfaro Rudgers J. (2017) Biogeography of root-associated fungal endophytes. In: Tedersoo L (Ed.) Biogeography of Mycorrhizal Symbiosis. Ecological Studies 230 (Springer), 195−222. https://doi.org/10.1007/978-3-319-56363-3
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. https://doi.org/10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketterings Q, Reid S, Rao R. (2007) Cation Exchange Capacity (CEC), Agronomy Fact Sheet Series (22). Cornel University Cooperative Extension.
- Kvas M, Marasas WFO, Wingfield BD, Wingfield MJ, Steenkamp ET. (2009) Diversity and evolution of Fusarium species in the Gibberella fujikuroi complex. Fungal Diversity 34: 1–21. [Google Scholar]
- Lareen A, Burton F, Schäfer P. (2016) Plant root-microbe communication in shaping root microbiomes. Plant Molecular Biology 90: 575–587. https://doi.org/10.1007/s11103-015-0417-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin RP, Hopkins DL, Martin FN. (1993) Effect of successive watermelon plantings on Fusarium oxysporum and other microorganisms in soils suppressive and conducive to Fusarium wilt of watermelon. Phytopathology 83: 1097–1105. https://doi.org/10.1094/Phyto-83-1097. [Google Scholar]
- Laurence MH, Walsh JL, Shuttleworth LA, Robinson DM, Johansen RM, Petrovic T, Vu TTH, Burgess LW, Summerell BA, Liew ECY. (2015) Six novel species of Fusarium from natural ecosystems in Australia. Fungal Diversity 77: 349–366. https://doi.org/10.1007/s13225-015-0337-6 [Google Scholar]
- LeBlanc N, Essarioui A, Kinkel L, Kistler HC. (2017) Phylogeny, plant species, and plant diversity influence carbon use phenotypes among Fusarium populations in the rhizosphere microbiome. Phytobiomes 1: 150–157. https://doi.org/10.1094/PBIOMES-06-17-0028-R [Google Scholar]
- Leslie JF, Summerell BA. (2006) The Fusarium laboratory manual. Blackwell Publishing, Ames. 1–388. https://doi.org/10.1002/9780470278376
- Leslie JF, Summerell BA. (2011) In search of new Fusarium species. Plant Breeding and Seed Science 63: 94–101. https://doi.org/10.2478/v10129-011-0020-3 [Google Scholar]
- Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R. (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Research 43: W580–584. https://doi.org/10.1093/nar/gkv279 [DOI] [PMC free article] [PubMed]
- Lupien SL, Dugan FM, Ward KM, O’Donnell K. (2017) Wilt, crown, and root rot of common rose mallow (Hibiscus moscheutos) caused by a novel Fusarium sp. Plant Disease 101: 354–358. https://doi.org/10.1094/PDIS-05-16-0717-RE [DOI] [PubMed] [Google Scholar]
- Mapelli F, Marasco R, Fusi M, Scaglia B, Tsiamis G, Rolli E, Fodelianakis S, Bourtzis K, Ventura S, Tambone F, Adani F, Borin S, Daffonchio D. (2017) The stage of soil development modulates rhizosphere effect along a High Arctic desert chronosequence. The ISME Journal: 1–11. https://doi.org/10.1038/s41396-017-0026-4 [DOI] [PMC free article] [PubMed]
- Marasas WFO, Nelson PE, Toussoun TA. (1985) Fusarium dlamini, a new species from Southern Africa. Mycologia 77: 971–975. https://doi.org/10.2307/3793311 [Google Scholar]
- Mason-Gamer R, Kellogg E. (1996) Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Systematic Biology 45: 524–545. https://doi.org/10.1093/sysbio/45.4.524 [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. In: Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the extreme to the campus and beyond, Association for Computing Machinery, Chicago, USA, 1–8. https://doi.org/10.1145/2335755.2335836
- Mohammadi MF, Jalali SW, Kooch Y, Theodose TA. (2017) Tree species composition, biodiversity and regeneration in response to catena shape and position in a mountain forest. Scandinavian Journal of Forest Research 32: 80–90. https://doi.org/10.1080/02827581.2016.1193624 [Google Scholar]
- Mommer L, Kirkegaard J, van Ruijven J. (2016) Root-root interactions: towards a rhizosphere framework. Trends in Plant Science 21: 209–217. https://doi.org/10.1016/j.tplants.2016.01.009 [DOI] [PubMed] [Google Scholar]
- Moussa TAA, Al-Zahrani HS, Kadasa NMS, Ahmed SA, de Hoog GS, Al-Hatmi AMS. (2017) Two new species of the Fusarium fujikuroi species complex isolated from the natural environment. Antonie Van Leeuwenhoek 110: 819–832. https://doi.org/10.1007/s10482-017-0855-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PE, Dignani MC, Anaissie EJ. (1994) Taxonomy, biology, and clinical aspects of Fusarium species. Clinical Microbiology Reviews 7: 479–504. https://doi.org/10.1128/CMR.7.4.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg HI. (1976) Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 169: 1–117. https://doi.org/10.1002/jpln.19771400220 [Google Scholar]
- Nylander JAA. (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC. (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 95: 2044–2049. https://doi.org/10.1073/pnas.95.5.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Nirenberg HI, Aoki T, Cigelnik E. (2000) A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience 41: 61–78. https://doi.org/10.1007/BF02464387 [Google Scholar]
- O’Donnell K, Rooney AP, Proctor RH, Brown DW, McCormick SP, Ward TJ, Frandsen RJ, Lysøe E, Rehner SA, Aoki T, Robert VA, Crous PW, Groenewald JZ, Kang S, Geiser DM. (2013) Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genetics and Biology 52: 20–31. https://doi.org/10.1016/j.fgb.2012.12.004 [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Sutton DA, Rinaldi MG, Gueidan C, Crous PW, Geiser DM. (2009) Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum – F. equiseti and F. chlamydosporum species complexes within the United States. Journal of Clinical Microbiology 47: 3851–3861. https://doi.org/10.1128/JCM.01616-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Sutton DA, Rinaldi MG, Sarver BA, Balajee SA, Schroers HJ, Summerbell RC, Robert VA, Crous PW, Zhang N, Aoki T, Jung K, Park J, Lee YH, Kang S, Park B, Geiser DM. (2010) Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. Journal of Clinical Microbiology 48: 3708–3718. https://doi.org/10.1128/JCM.00989-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padwick GW. (1945) Notes on Indian fungi III. Mycological Papers 12: 1–15. [Google Scholar]
- Pal KK, Tilak KVBR, Saxcna AK, Dey R, Singh CS. (2001) Suppression of maize root diseases caused by Macrophomina phaseolina, Fusarium moniliforme and Fusarium graminearum by plant growth promoting rhizobacteria. Microbiological Research 156: 209–223. https://doi.org/10.1078/0944-5013-00103 [DOI] [PubMed] [Google Scholar]
- Perlroth J, Choi B, Spellberg B. (2007) Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Medical Mycology 4: 321–346. https://doi.org/10.1080/13693780701218689 [DOI] [PubMed] [Google Scholar]
- Peters JC, Lees AK, Cullen DW, Sullivan L, Strouda GP, Cunnington AC. (2008) Characterization of Fusarium spp. responsible for causing dry rot of potato in Great Britain. Plant Pathology 57: 262–271. https://doi.org/10.1111/j.1365-3059.2007.01777.x [Google Scholar]
- Phan HT, Burgess LW, Summerell BA, Bullock S, Liew ECY, Smith-White JL, Clarkson JR. (2004) Gibberella gaditjirrii (Fusarium gaditjirrii) sp. nov., a new species from tropical grasses in Australia. Studies in Mycology 50: 261–272. [Google Scholar]
- Philippot L, Raaijmakers JM, Lemanceau P, Van Der Putten WH. (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nature Reviews Microbiology 11: 789–799. https://doi.org/10.1038/nrmicro3109 [DOI] [PubMed] [Google Scholar]
- Pinheiro AC, Macedob MF, Jurado V, Saiz-Jimenez C, Viegas C, Brandão J, Rosado L. (2011) Mould and yeast identification in archival settings: preliminary results on the use of traditional methods and molecular biology options in Portuguese archives. International Biodeterioration & Biodegradation 65: 619–627. https://doi.org/10.1016/j.ibiod.2011.02.008 [Google Scholar]
- Posada D, Crandall KA. (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. https://doi.org/10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rayner RW. (1970) A Mycological Colour Chart. CMI and British Mycological Society, Kew, Surrey, 34 pp. [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. https://doi.org/10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ruano-Rosa D, Prieto P, Rincón AM, Gómez-Rodríguez MV, Valderrama R, Barroso JB, Mercado-Blanco J. (2016) Fate of Trichoderma harzianum in the olive rhizosphere: time course of the root colonization process and interaction with the fungal pathogen Verticillium dahliae. BioControl 61: 269–282. https://doi.org/10.1007/s10526-015-9706-z [Google Scholar]
- Sandoval-Denis M, Guarnaccia V, Polizzi G, Crous PW. (2018) Symptomatic Citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Persoonia 40: 1–25. https://doi.org/10.3767/persoonia.2018.40.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanakumar K, Fan L, Fu K, Yu C, Wang M, Xia H, Sun J, Li Y, Chen J. (2016) Cellulase from Trichoderma harzianum interacts with roots and triggers induced systemic resistance to foliar disease in maize. Scientific Reports 6: 35543. https://doi.org/10.1038/srep35543 [DOI] [PMC free article] [PubMed]
- Sasse J, Martinoia E, Northen T. (2018) Feed your friends: do plant exudates shape the root microbiome? Trends in Plant Science. 23: 25–41. https://doi.org/10.1016/j.tplants.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Schippers B, Bakker AW, Bakker PAHM. (1987) Interactions of deleterious and beneficial rhizosphere microorganisms and the effect of cropping practices. Annual Review of Phytopathology 25: 339–358. https://doi.org/10.1146/annurev.py.25.090187.002011 [Google Scholar]
- Smit IPJ, Riddell ES, Cullum C, Petersen R. (2013) Kruger National Park research supersites: establishing long-term research sites for cross-disciplinary, multiscaled learning. Koedoe – African Protected Area Conservation and Science 55: Art. 1107 https://doi.org/10.4102/koedoe.v55i1.1107
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerell B, Leslie J, Liew E, Laurence M, Bullock S, Petrovic T, Bentley AR, Howard CG, Peterson SA, Walsh JL, Burgess LW. (2011) Fusarium species associated with plants in Australia. Fungal Diversity 46: 1–27. https://doi.org/10.1007/s13225-010-0075-8 [Google Scholar]
- Van Zijl G, Le Roux P. (2014) Creating a conceptual hydrological soil response map for the Stevenson Hamilton Research Supersite, Kruger National Park, South Africa. Water SA 40: 331–336. http://doi.org/10.4314/wsa.v40i2.15 [Google Scholar]
- Visioli G, D’Egidio S, Sanangelantoni AM. (2014) The bacterial rhizobiome of hyperaccumulators: future perspectives based on omics analysis and advanced microscopy. Frontiers in Plant Science 5: 752. https://doi.org/10.3389/fpls.2014.00752 [DOI] [PMC free article] [PubMed]
- Walsh J, Laurence M, Liew E, Sangalang A, Burgess L, Summerell B, Petrovic T. (2010) Fusarium: two endophytic novel species from tropical grasses of northern Australia. Fungal Diversity 44: 149–159. https://doi.org/10.1007/s13225-010-0035-3 [Google Scholar]
- Wiens JJ. (1998) Testing phylogenetic methods with tree congruence: phylogenetic analysis of polymorphic morphological characters in phrynosomatid lizards. Systematic Biology 47: 427–444. https://doi.org/10.1080/106351598260806 [Google Scholar]
- Woudenberg JHC, Aveskamp MM, De Gruyter J, Spiers AG, Crous PW. (2009) Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia 22: 56–62. https://doi.org/10.3767/003158509X427808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Maruyama S, Nozaki H, Shirasu K. (2010) Horizontal gene transfer by the parasitic plant Striga hermonthica Science 328: 1128. https://doi.org/10.1126/science.1187145 [DOI] [PubMed]
- Zachow C, Berg C, Müller H, Meincke R, Komon-Zelazowska M, Druzhinina IZ, Kubicek CP, Berg G. (2009) Fungal diversity in the rhizosphere of endemic plant species of Tenerife (Canary Islands): relationship to vegetation zones and environmental factors. The ISME Journal 3: 79–92. https://doi.org/10.1038/ismej.2008.87 [DOI] [PubMed] [Google Scholar]
- Zakaria L, Ning CH. (2013) Endophytic Fusarium spp. from roots of lawn grass (Axonopus compressus). Tropical Life Sciences Research 24: 85–90. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.