Abstract
We previously demonstrated that long non-coding RNA cytoskeleton regulator RNA (CYTOR), also known as Linc00152, was significantly overexpressed in colon cancer and conferred resistance to oxaliplatin-induced apoptosis. At the same time, elevated CYTOR expression was also reported in gastric cancer and exerted influences on epithelial-mesenchymal transition (EMT) markers. However, the precise mechanism by which CYTOR promotes the EMT phenotype and cancer metastasis remains poorly understood. Here, we showed that loss of epithelial characteristics and simultaneous gain of mesenchymal features correlated with CYTOR expression. Knockdown of CYTOR attenuated colon cancer cell migration and invasion. Conversely, ectopic expression of CYTOR induced an EMT program and enhanced metastatic properties of colon cancer cells. Mechanistically, the binding of CYTOR to cytoplasmic β-catenin impeded casein kinase 1 (CK1)-induced β-catenin phosphorylation that enabled it to accumulate and translocate to the nucleus. Reciprocally, β-catenin/TCF complex enhanced the transcription activity of CYTOR in nucleus, thus forming a positive feed-forward circuit. Moreover, elevated CYTOR, alone or combined with overexpression of nuclear β-catenin, was predictive of poor prognosis. Our findings suggest that CYTOR promotes colon cancer EMT and metastasis by interacting with β-catenin, and the positive feed-forward circuit of CYTOR-β-catenin might be a useful therapeutic target in antimetastatic strategy.
Keywords: CYTOR, colon cancer, β-catenin, feed-forward loop, metastasis
In this study, we found that LncRNA-CYTOR promoted colon cancer epithelial-mesenchymal transition (EMT) and metastasis by interacting with β-catenin. Moreover, elevated CYTOR, alone or combined with overexpression of nuclear β-catenin, was predictive of poor prognosis, and targeting the CYTOR/β-catenin feed-forward loop might be a useful therapeutic strategy.
Introduction
Human colon cancer is one of the most common and aggressive malignancies worldwide, accounting for one-tenth of all cancer-related deaths.1 Colon cancer cells exert distant invasive potential and metastatic ability. These characteristics have been considered to be responsible for at least 90% of colon cancer-associated mortality.2 One of the crucial molecular steps in the process of colon cancer distant metastasis includes epithelial-mesenchymal transition (EMT), which enables the formation of migratory mesenchymal cells with invasive phenotype. Loss of cellular adhesion proteins such as E-cadherin, tight junction proteins, as well as concomitant, acquire expression of mesenchymal markers such as vimentin, fibronectin, and N-cadherin, are generally accepted as hallmarks of the EMT process.3 As a complex process, EMT is also associated with a poor prognosis.4 Therefore, revealing the underlying mechanism of the EMT process involved in colon cancer metastasis is necessary for developing therapeutic strategies.
In fact, several significant signaling pathways are known to be involved in the acceleration of EMT program and tumor metastasis, such as the nuclear factor-kappa B (NF-κB) pathway,5 transforming growth factor β (TGF-β) pathway,6 Notch pathway,7 and Wnt/β-catenin pathway.8 Owing to the constitutively activated status and profound impact on EMT during cancer progression, the canonical Wnt/β-catenin signaling has become one of the most important molecular therapy targets for colon cancer.9 The canonical Wnt/β-catenin signaling has been considered as one of the most vital pathways accelerating the process of EMT and enhancing metastatic properties of colon cancer.10 Without active stimuli, β-catenin would not accumulate in the cytoplasm, since APC/Axin/GSK-3 complex would normally degrade it via the ubiquitin-proteasome pathway. During above process, casein kinase 1 (CK1) phosphorylates β-catenin at Ser45. This phosphorylation event primes β-catenin for subsequent phosphorylation by GSK-3β. GSK-3β destabilizes β-catenin by phosphorylating it at Ser33, Ser37, and Thr41. Following activation of the Wnt signaling, β-catenin accumulates and enters the nucleus.11 In the nucleus, β-catenin binds to T cell factor (TCF) and then trans-activate target genes through TCF-binding elements (TBEs).12, 13 Mutations in the key regulatory factors of the Wnt/β-catenin signaling have been widely reported in a variety of tumors, especially in metastatic colon cancer. Notably, nuclear accumulation of β-catenin has been found in up to 80% of colon cancer patients.10, 14 Thus, further investigations of novel regulators of the Wnt/β-catenin signaling and underlying mechanisms may potentiate the development of antimetastatic strategies for colon cancer.
Long non-coding RNAs (LncRNAs) are newly identified regulatory RNA members with length greater than 200 nucleotides and characterized by lack of protein-coding capacity. They modulate biological process through diverse mechanisms, including not only the regulation of transcription and post-transcription, but also the process of genomic imprinting and chromatin modification.15, 16, 17 Recently, the regulatory role of LncRNAs in tumorigenesis and progression has been increasingly recognized. A number of LncRNAs have been found to be linked to invasion-metastasis cascade of multiple malignancies.18, 19 Nevertheless, the role of LncRNAs in the regulation of colon cancer metastasis remains unclear up to now. We previously demonstrated that LncRNA cytoskeleton regulator RNA (CYTOR), also known as Linc00152, was significantly overexpressed in colon cancer and conferred resistance to oxaliplatin-induced apoptosis.20 Interestingly, elevated CYTOR expression was also reported in gastric cancer and exerted influences on EMT markers, but the precise mechanism by which CYTOR promotes the EMT phenotype and cancer metastasis remains poorly understood.21, 22
In this study, we demonstrated that CYTOR promoted the EMT program and metastasis of colon cancer. Mechanistically, CYTOR blocked CK1-mediated cytoplasmic β-catenin phosphorylation, facilitating β-catenin nuclear translocation. Reciprocally, β-catenin enhanced the transcription of CYTOR in nucleus, thus forming a feed-forward loop. Furthermore, the clinical value of combining CYTOR and nuclear β-catenin to predict the prognosis for colon cancer patients was also evaluated. Collectively, our findings identified CYTOR as a novel regulator of the canonical Wnt/β-catenin signaling and a promising therapeutic target for antimetastatic strategies for colon cancer.
Results
CYTOR Levels Positively Correlate with EMT Phenotype in Colon Cancer Cells
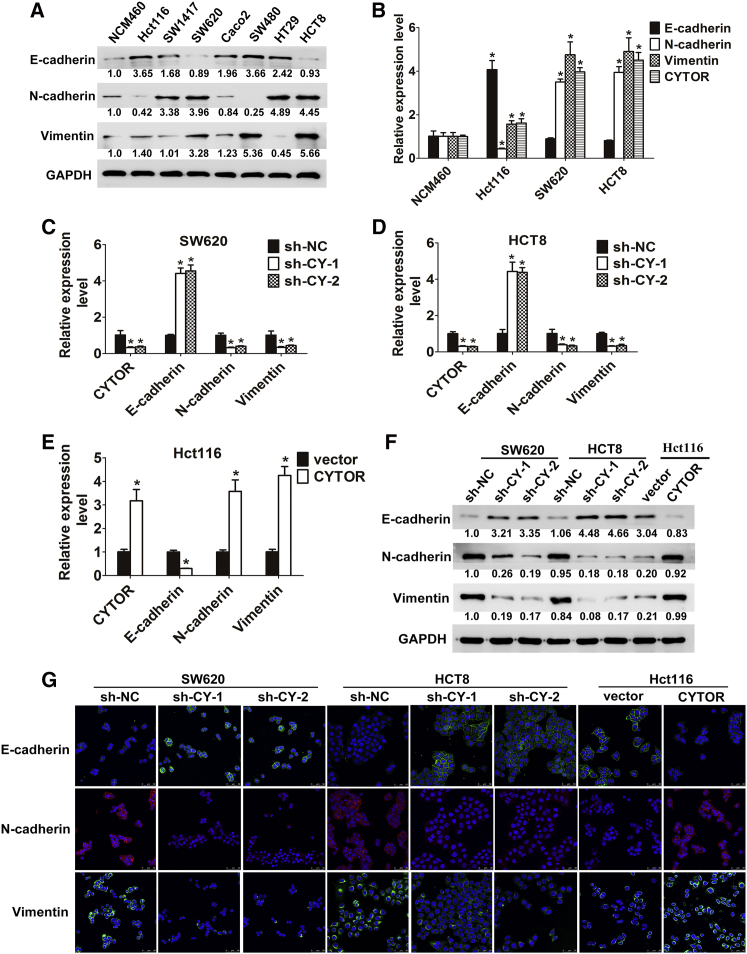
To explore the possible effects of CYTOR on the colon cancer EMT process, we first determined the EMT phenotype of human colon mucosal epithelial cell line NCM460 and seven colon cancer cell lines through evaluating the expression of epithelial marker E-cadherin and mesenchymal markers N-cadherin and vimentin. We found that the Hct116 cell line shows robust expression of E-cadherin and low levels of N-cadherin and vimentin, which is representative of its epithelial character. On the other hand, SW620 and HCT8 primarily expressed N-cadherin and vimentin, with a concurrent low E-cadherin level, which is consistent with a mesenchymal phenotype (Figure 1A). The two distinguishing features of the above three cell lines are essential for the further investigation of metastasis. Furthermore, a negative correlation was observed between CYTOR and E-cadherin expression, while the endogenous levels of CYTOR and mesenchymal markers N-cadherin and vimentin were comparable (Figures 1B and S1A). Thus, CYTOR levels positively correlate with the EMT phenotype in colon cancer cells.
Figure 1.
Loss of Epithelial Features and Simultaneous Gain of Mesenchymal Characteristics Correlate with CYTOR Expression in Colon Cancer Cell Lines
(A) Western blot analysis of EMT markers in different cell lines. (B) CYTOR and EMT marker levels were evaluated by qRT-PCR in three colon cancer cells; colon mucosal epithelial cell line NCM460 was used as control. (C and D) The shRNA-mediated CYTOR repression was confirmed by qRT-PCR after lentivirus infection in both SW620 (C) and HCT8 (D) cells, and the mRNA levels of EMT markers E-cadherin, N-cadherin, and Vimentin were also assessed by qRT-PCR. (E) Overexpression of CYTOR was confirmed by qRT-PCR after lentivirus infection in Hct116 cells; related EMT markers mRNA levels were also assessed. (F and G) CYTOR knockdown or overexpression affected the protein levels of EMT markers in both western blot analysis (F) and confocal immunofluorescent assay (G). Error bars, ±SD. *p < 0.05.
CYTOR Facilitates EMT Program in Colon Cancer Cells
To explore functional links between CYTOR and EMT, loss- and gain-of-function studies were performed. As shown in Figures 1C–1E, knockdown of CYTOR resulted in a significant upregulation of E-cadherin, accompanied by a prominent downregulation of N-cadherin and Vimentin in HCT8 and SW620 cells at mRNA levels. Reciprocally, CYTOR overexpression triggered the reduced expression of E-cadherin, together with the increased expression of N-cadherin and vimentin in Hct116 cells. Altered CYTOR expression also led to marked changes in the EMT markers at protein level as determined by western blot analysis (Figure 1F). Additionally, above findings were further confirmed by confocal immunofluorescent microscopy assay (Figure 1G). It is noteworthy that knocking down CYTOR effectively also enhances E-cadherin levels and decreases N-cadherin and Vimentin levels in another colon cancer cell line, Hct116, which originally exhibits little CYTOR expression (Figure S1B). Collectively, CYTOR facilitates EMT program in colon cancer cells.
CYTOR Promotes Colon Cancer Cell Invasion and Metastasis In Vitro and In Vivo
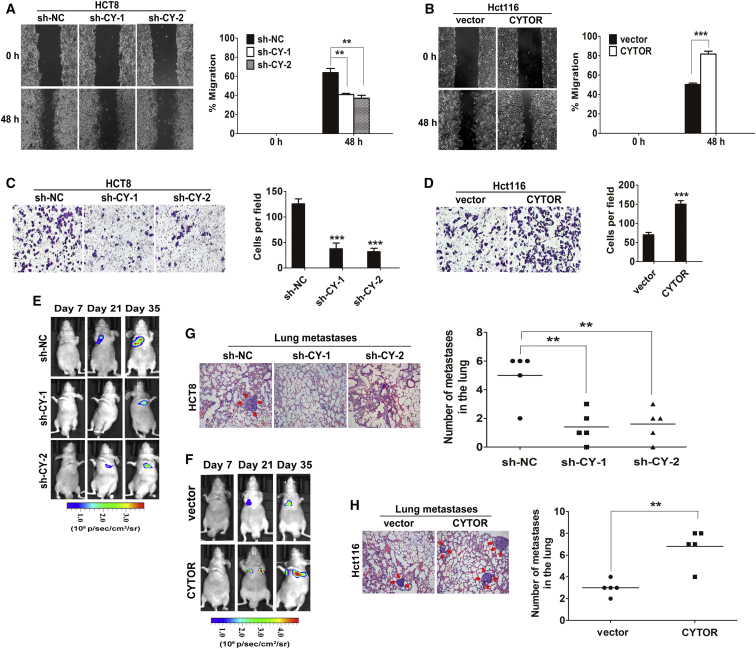
As EMT plays a critical promoted role in the initial stage of colon cancer metastasis,23 we further explored the physiological relevance of CYTOR to the colon cancer cell motility. The wound-healing assay demonstrated that CYTOR knockdown significantly impeded the migratory ability of HCT8 and SW620 cells compared with sh-NC control (Figure 2A and S1C). In addition, ectopic expression of CYTOR in Hct116 cells induced more extensive wound closure areas (Figure 2B). Similarly, the invasive ability of colon cancer cells was dramatically suppressed in response to CYTOR depletion, while markedly enhanced by ectopic expression of CYTOR (Figures 2C and 2D). Thus, CYTOR significantly increased the migration and invasion potential of colon cancer cells in vitro. To further confirm the above findings in vivo, a lung metastasis model was introduced by inoculating colon cancer cells with modified CYTOR expression directly into the tail veins of nude mice. Luciferase signals were monitored weekly to observe location and growth of tumor xenografts in the lung. As a result, at 35 days, luciferase signals are lower in CYTOR knockdown group compared with those in control group, CYTOR depletion significantly reduced the lung metastases burden of HCT8 cells (Figures 2E and 2G). Conversely, luciferase signals in the CYTOR-overexpressing group increase over time, a significant difference was noted in the size and number of lung metastatic tumor nodules between the Hct116-CYTOR group and control group (Figures 2F and 2H). Moreover, immunochemistry analysis revealed that EMT markers N-cadherin and Vimentin were reduced, and E-cadherin was upregulated in the lung metastatic tumor tissues with CYTOR knockdown. An inverse result was observed in the CYTOR-overexpressing group (Figure S1D). All these findings suggest a crucial role of CYTOR in promoting colon cancer cells invasion and metastasis and inducing EMT process.
Figure 2.
CYTOR Promotes Colon Cancer Cell Metastasis In Vitro and In Vivo
(A and B) Stable HCT8 (A) and Hct116 (B) cells were subjected to wound healing assay. The wound space was photographed at 0 and 48 hr. (C) The invasive ability of HCT8 cells was suppressed in response to knockdown of CYTOR. (D) Ectopic expression of CYTOR significantly enhanced Hct116 cells invasion into Matrigel-coated transwell membranes. (E and F) Representative mice injected with modified CYTOR expressing HCT8 (E) or Hct116 cells (F) (n = 5 per group). The luciferase signal intensity from days 7 to 35 is on equivalent scales in lung metastasis model. (G and H) Representative results of H&E staining of the metastatic nodules in the lung. The stained metastatic nodules are indicated by arrows, and the number of metastatic nodules for each mouse were counted under the microscope. Error bars, ±SD. **p < 0.01, ***p < 0.001.
CYTOR Activates the Wnt/β-Catenin Signaling Pathway in Colon Cancer Cells
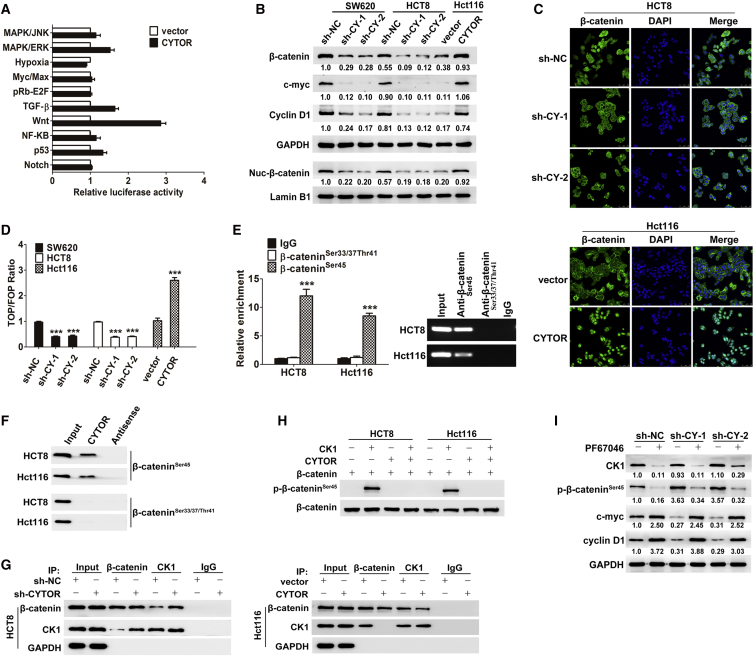
To unveil the molecular mechanism behind the process, a pathway reporter array including 10 selected cancer-related signaling pathways was used to test which pathways were associated with CYTOR in Hct116 cells. As a result, Wnt/β-catenin signaling was significantly activated by CYTOR (Figure 3A). Nowadays, the canonical Wnt/β-catenin signaling is known as one of the most vital pathways to accelerate the process of EMT and metastasis in colon cancer.10, 24 Interestingly, the expression level of β-catenin, the central molecule of Wnt/β-catenin signaling, is relatively higher with concurrent elevated CYTOR in all the colon cancer cell lines used in our study (Figure S1E). According to the above results, we further examined whether CYTOR activates the Wnt/β-catenin pathway in colon cancer cells. Western blot analyses showed that knockdown of CYTOR caused a decreased expression of total β-catenin, nuclear β-catenin, and several Wnt/β-catenin pathway transcriptional targets, such as c-myc and cyclin D1, whereas overexpression of CYTOR significantly increased their expression (Figure 3B). Furthermore, the role of CYTOR in promoting the redistribution of cytoplasmic β-catenin to nuclear localization was also confirmed through immunofluorescence assays. Depletion of CYTOR resulted in a redistribution of nuclear β-catenin to the cytoplasmic localization, whereas overexpression of CYOTR leaded to substantial nuclear accumulation of β-catenin (Figure 3C). As expected, TOPflash and FOPflash luciferase reporter analyses revealed that the transactivation of TCF reporter was inhibited by the depletion of CYTOR and enhanced via overexpression of CYTOR (Figure 3D). All these results demonstrate that upregulation of CYTOR is able to accelerate β-catenin nuclear translocation then enhance the transcriptional activity of the β-catenin/TCF complex in nucleus and consequently activate the Wnt/β-catenin signaling pathway.
Figure 3.
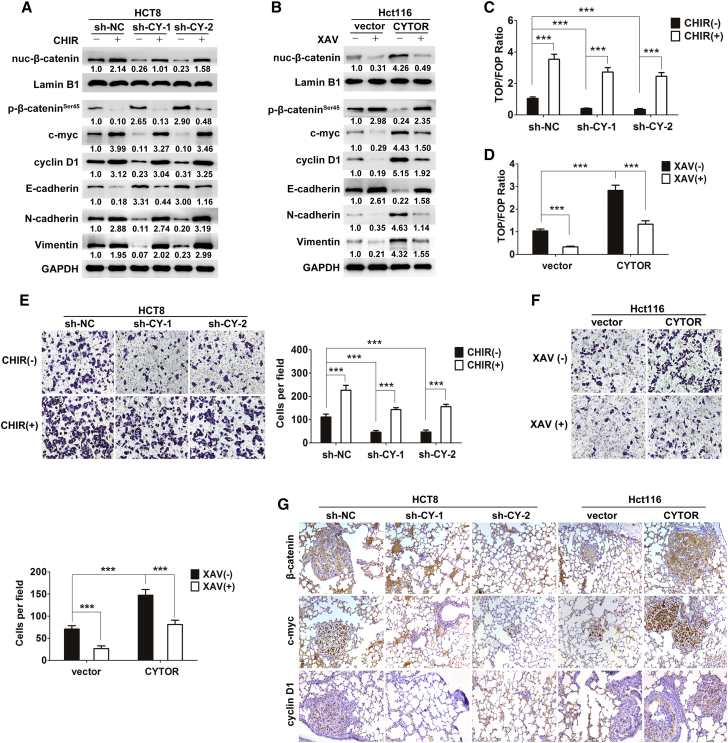
CYTOR Activates the Wnt/β-Catenin Signaling by Blocking CK1-Mediated β-Catenin Phosphorylation
(A) Signaling pathway reporter array was used for seeking the relative pathway associated with CYTOR on Hct116 cells. (B) Knockdown of CYTOR caused a decreased expression of total β-catenin, nuclear β-catenin, c-myc, and cyclin D1, whereas overexpression of CYTOR significantly increased their expression. (C) The role of CYTOR in promoting the redistribution of cytoplasmic β-catenin to nuclear localization was confirmed through immunofluorescence assays. Depletion of CYTOR resulted in a redistribution of nuclear β-catenin to the cytoplasmic localization, whereas overexpression of CYOTR caused substantial nuclear accumulation of β-catenin. (D) TOPflash and FOPflash luciferase reporter analyses revealed that the transactivation of TCF reporter was inhibited by the depletion of CYTOR and enhanced via overexpression of CYTOR. (E and F) RIP assay (E) and RNA pull-down assay (F) revealed that non-phospho-β-cateninSer45, rather than other active non-phospho sites, was required for β-catenin interaction with CYTOR. (G) Coimmunoprecipitation of CK1 and β-catenin in lysates of colon cancer cells with modified CYTOR expression. (H) The role of CYTOR in protecting β-catenin from phosphorylation by CK1 was confirmed via an in vitro phosphorylation assay. (I) HCT8 cells were treated with 10 nmol/L PF670462 for 24 hr, inhibiting CK1 activity by PF670462 abolished the effects of CYTOR-shRNA on β-cateninSer45 phosphorylation and Wnt/β-catenin signaling activity. Error bars, ±SD. ***p < 0.001.
CYTOR Activates the Wnt/β-Catenin Signaling by Blocking CK1-Mediated β-Catenin Phosphorylation
In the absence of Wnt signals, β-catenin is known to be phosphorylated and degraded in the cytoplasm via the ubiquitin-proteasome pathway.25 Given that CYTOR mainly locates in the cytoplasm,26 we hypothesized that CYTOR might enhance the Wnt/β-catenin signaling activity by attenuating the phosphorylation status of β-catenin. As expected, increased phosphorylation level of β-catenin at Ser45 was observed in colon cancer cells transfected with CYTOR small hairpin RNA (shRNA), whereas overexpression of CYTOR significantly decreased phospho-β-cateninSer45 expression (Figure S1F). Cytoplasmic β-catenin has been reported to interact with CK1, which could induce β-cateninSer45 phosphorylation. This phosphorylation event primes β-catenin for subsequent phosphorylation at other sites by GSK-3β.11, 27 Interestingly, CK1 levels showed no obvious changes in all colon cancer cells with modified CYTOR expression used in our study (Figure S1F). Cytoplasmic LncRNAs are reported to be involved in molecular pathways through their interactions with proteins and modulation of protein function.28 RNA immunoprecipitation (RIP) assay was performed, and the results revealed that non-phospho-β-cateninSer45, rather than other active non-phospho sites, was required for β-catenin interaction with CYTOR (Figure 3E), which was confirmed by RNA pull-down assay (Figure 3F). All above clues intrigued us to speculate that CYTOR might block CK1-β-catenin interaction by competitively binding non-phospho-β-cateninSer45 to attenuate the phosphorylation status of β-catenin and facilitate β-catenin nuclear translocation. As shown in Figure 3G, CYTOR knockdown promoted CK1-β-catenin interaction in HCT8 cells, and ectopic expression of CYTOR attenuated this interaction in Hct116 cells. The role of CYTOR in protecting β-catenin from phosphorylation by CK1 was also confirmed via an in vitro phosphorylation assay (Figure 3H). Notably, inhibiting CK1 activity by PF670462 abolished the effects of CYTOR-shRNA on β-cateninSer45 phosphorylation and Wnt/β-catenin signaling activity (Figure 3I). Taken together, these data suggest that CYTOR-β-catenin interaction prevents the phosphorylation of β-catenin by CK1 and facilitates β-catenin nuclear translocation to transactivate the downstream target genes.
The β-Catenin/TCF4 Complex Promotes CYTOR Transcription in Colon Cancer Cells
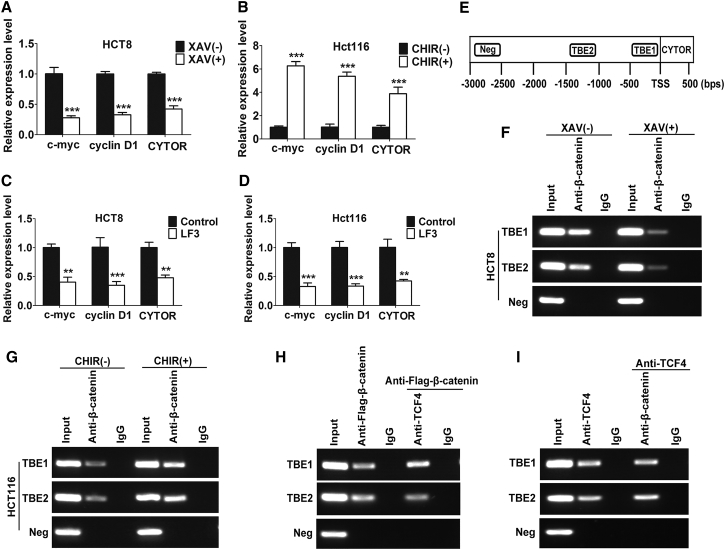
Following the Wnt/β-catenin signaling activation, β-catenin enters the nucleus, and associates with TCF. The β-catenin/TCF complex then directly activates the target genes via TCF binding elements (TBEs) present in their cis-regulatory regions.29 Importantly, bioinformatics analysis predicted three TBEs (−1,097 to −1,092, -TCAAAG-; −552 to −546 bp, -GATCACA-; −464 to −459 bp, -ATTCAA-) in the CYTOR promoter that specifically bind to the β-catenin/TCF complex, indicating that β-catenin/TCF complex might modulate CYTOR transcription. As shown in Figure 4A, CYTOR expression was repressed when Wnt/β-catenin signaling was blocked by using a specific signaling inhibitor XAV939.30 Inversely, CHIR99021, an activator of Wnt/β-catenin signaling,31 markedly increased the levels of CYTOR (Figure 4B). Moreover, treatment with LF3, a small molecule antagonist selectively disrupting the β-catenin-TCF4 interaction,32 significantly decreased CYTOR transcription in both HCT8 and Hct116 cells (Figures 4C and 4D). Furthermore, the enrichment of β-catenin on two TBEs in the CYTOR promoter was confirmed by chromatin immunoprecipitation (ChIP) assay. Consistently, the enrichment was significantly downregulated in HCT8 cells when Wnt/β-catenin signaling was blocked, an inverse result was observed in Hct116 cells after activating the Wnt/β-catenin signaling (Figures 4E–4G). Finally, the co-occupancy of β-catenin and TCF4 on the CYTOR promoter was identified via ChIP-re-ChIP assay (Figures 4H and 4I). These findings indicate that the β-catenin/TCF complex binds to the CYTOR promoter and promotes CYTOR transcription, thus forming a positive feedback loop in colon cancer cells.
Figure 4.
β-Catenin/TCF4 Complex Promotes CYTOR Transcription in Colon Cancer Cells
(A) CYTOR expression was repressed in HCT8 cells treated with 15 μmol/L XAV939 for 24 hr. (B) CYTOR expression was markedly increased in Hct116 cells treated with 8 μmol/L CHIR99021 for 24 hr. (C and D) Treatment with 1.6 μmol/L LF3 for 24 hr significantly decreased CYTOR transcription in both Hct8 (C) and HCT116 (D) cells. (E) The enrichment of β-catenin on two TBEs in the CYTOR promoter was confirmed. (F and G) The enrichment was significantly downregulated in HCT8 cells when Wnt signaling was blocked (F); an inverse result was observed in Hct116 cells after activating the Wnt signaling (G). (H and I) The co-occupancy of β-catenin and TCF4 on CYTOR promoter was identified via ChIP-re-ChIP assay. Error bars, ±SD. **p < 0.01, ***p < 0.001.
Wnt/β-Catenin Signaling Is Responsible for CYTOR-Mediated EMT and Metastasis
Although we have established the promotive role of CYTOR in EMT and metastasis, whether this effect is specifically attributed to the Wnt/β-catenin signaling needs to be further determined. As shown in Figure 5A, activation of Wnt/β-catenin signaling recapitulated the levels of mesenchymal markers N-cadherin and Vimentin and decreased the expression of epithelial marker E-cadherin in CYTOR-knockdown cells. Conversely, the blockage of Wnt/β-catenin signaling abolished the acceleration of EMT caused by CYTOR overexpression (Figure 5B). Furthermore, the inhibition of TCF/lymphoid enhancer factor (LEF) activity caused by CYTOR knockdown was significantly rescued by CHIR99021; on the other hand, ectopically expressed CYTOR enhanced the TCF/LEF activity, which was suppressed by XAV939 (Figures 5C and 5D). The transwell invasion assay showed that depletion of CYTOR decreased the invasive ability of colon cancer cells, and this effect was rescued by activating Wnt/β-catenin signaling. Reciprocally, overexpression of CYTOR increased the cells’ invasion, and inhibition of Wnt/β-catenin signaling abrogated this effect (Figures 5E and 5F). Consistent with in vitro results, β-catenin, c-myc, and cyclin D1 were reduced with decreased lung metastasis burden and upregulated with more lung colonization caused by modified CYTOR expression according to the immunohistochemistry (Figure 5G). Taken together, these data suggest that Wnt/β-catenin signaling is responsible for CYTOR-induced EMT and metastasis.
Figure 5.
Wnt/β-Catenin Signaling Is Responsible for CYTOR-Mediated EMT and Metastasis
(A) Stable HCT8 cells were treated with 8 μmol/L CHIR99021 for 24 hr; indicated protein levels were assayed by western blotting. (B) Stable Hct116 cells were treated with 15 μmol/L XAV939 for 24 hr; indicated protein levels were assayed by western blotting. (C and D) The inhibition of TCF/LEF activity caused by CYTOR knockdown was significantly rescued by CHIR99021 in HCT8 cells (C); on the other hand, ectopically expressed CYTOR enhanced the TCF/LEF activity, which was suppressed by XAV939 in Hct116 cells (D). (E and F) The transwell assay showed different cell invasive capacities in stable HCT8 (E) and Hct116 (F) cells treated with CHIR99021 (8 μmol/L) or XAV939 (15 μmol/L) for 24 hr. (G) β-catenin, c-myc, and cyclin D1 were reduced with decreased lung metastasis burden and upregulated with more lung colonization caused by modified CYTOR expression according to the immunohistochemistry. Error bars, ±SD. ***p < 0.001.
The Prognostic Value of Combining CYTOR and β-Catenin for Colon Cancer Patients
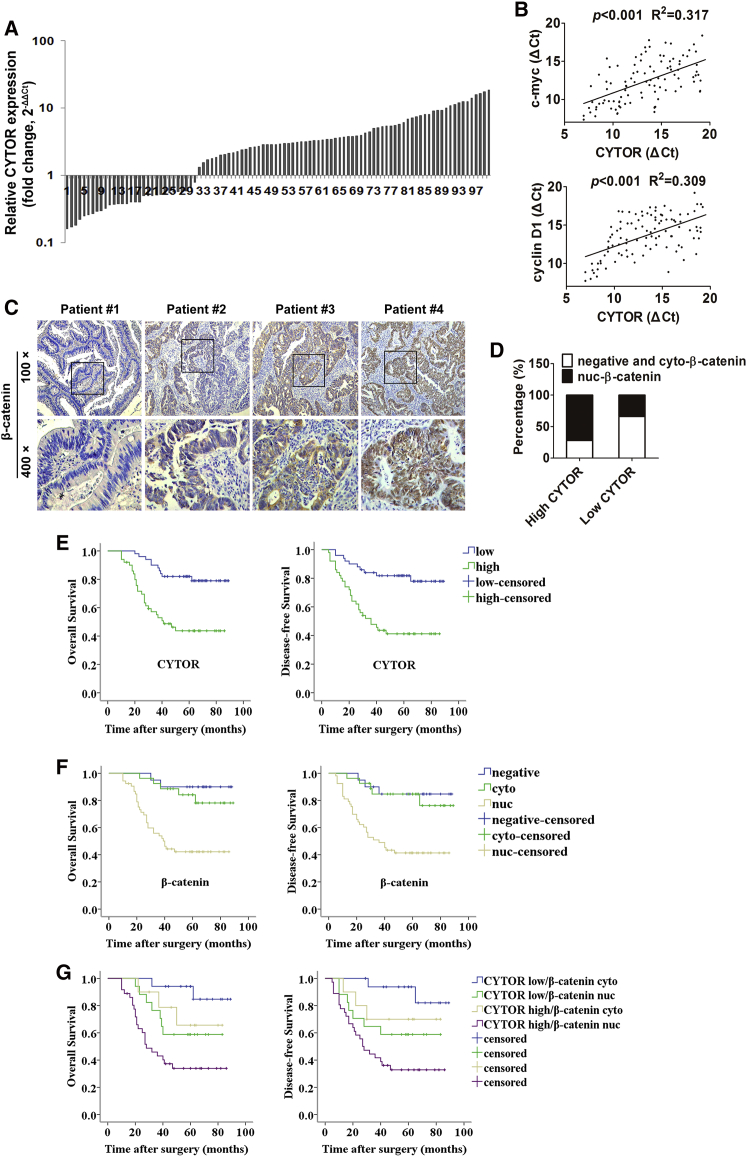
To evaluate the clinical significance of association between CYTOR and β-catenin, 100 pairs of colon cancer tissues were used for further study. As shown in Figure 6A, elevated expression of CYTOR was observed in 69.0% (69/100) of colon cancer tissues by using qRT-PCR compared with adjacent normal mucosa. In addition, the levels of CYTOR were positively correlated with the levels of Wnt/β-catenin target genes c-myc and cyclin D1 (Figure 6B). Furthermore, patients were divided into high and low CYTOR group according to the median value to investigate the correlation between of CYTOR expression and clinicopathologic parameters. CYTOR expression was significantly associated with pT stage (p = 0.033), pN stage (p < 0.001), pM stage (p = 0.006), and TNM stage (p < 0.001) (Table S1). Univariate and multivariate analysis indicated that increased CYTOR expression was an independent prognostic indicator for both overall survival (OS) and disease-free survival (DFS) in colon cancer patients (Table S2). Immunochemistry staining was also performed to measure the β-catenin expression in colon cancer tissues (Figure 6C). We found that CYTOR transcript level was significantly positively correlated with nuclear accumulation of β-catenin in colon cancer tissues (Figure 6D; Table S3). Although either elevated CYTOR or nuclear β-catenin predicts a poor prognosis (Figures 6E and 6F), cases with both high CYTOR level and nuclear β-catenin expression exhibited an even worse prognosis (Figure 6G). Moreover, the expression of E-cadherin was significantly lower in CYTORhigh/β-cateninnuc tumor tissues than in CYTORlow/β-catenincyto tumor tissues according to immunochemistry. The staining of N-cadherin and Vimentin were significantly higher in CYTORhigh/β-cateninnuc tumor tissues than in CYTORlow/β-catenincyto tumor tissues (Figure S1G). Above findings suggested an induction of EMT program in CYTORhigh/β-cateninnuc patients. Collectively, these results unravel that combination of CYTOR and nuclear β-catenin that displays a better prognostic value and may be useful predictor for colon cancer patients.
Figure 6.
The Prognostic Value of Combining CYTOR and β-Catenin for Colon Cancer Patients
(A) Elevated expression of CYTOR was observed by using qRT-PCR in colon cancer tissues when compared with adjacent normal mucosa. (B) The levels of CYTOR were positively correlated with the levels of Wnt/β-catenin target genes c-myc and cyclin D1. (C) Immunochemistry staining was performed to measure the β-catenin expression in colon cancer tissues. (D) Relative percentages of nuclear β-catenin in high (72.0%) and low (34.0%) CYTOR groups. (E) Kaplan-Meier analysis of OS (log-rank test, p < 0.001) or DFS (log-rank test, p < 0.001) with cytoplasmic or nuclear β-catenin in colon cancer patients. (F) Kaplan-Meier analysis of OS (log-rank test, β-cateninnegative versus β-cateninnuc, p = 0.001; β-catenincyto versus β-cateninnuc, p = 0.001) or DFS (log-rank test, β-cateninnegative versus β-cateninnuc, p = 0.002; β-catenincyto versus β-cateninnuc, p = 0.001) with negative or cytoplasmic or nuclear β-catenin in colon cancer patients. (G) Kaplan-Meier analysis of OS (log-rank test, CYTORlowβ-catenincyto versus CYTORlowβ-cateninnuc, p = 0.041; CYTORhighβ-catenincyto versus CYTORhighβ-cateninnuc, p = 0.046) or DFS (log-rank test, CYTORlowβ-catenincyto versus CYTORlowβ-cateninnuc, p = 0.033; CYTORhighβ-catenincyto versus CYTORhighβ-cateninnuc, p = 0.066) with high or low CYTOR and cytoplasmic or nuclear β-catenin in colon cancer patients.
Discussion
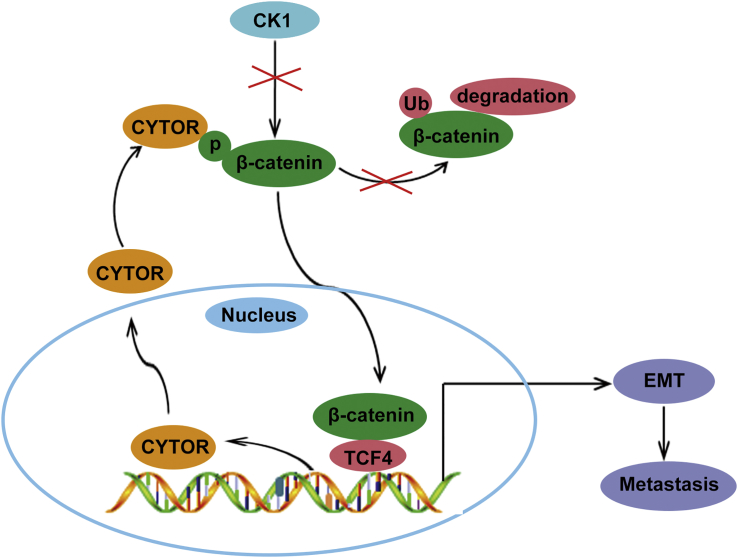
In this study, we provided several new insights into the function of LncRNA CYTOR in EMT and colon cancer metastasis. Our results clearly showed a positive feedback mechanism involving CYTOR and β-catenin, thus promoting the understanding of deregulated Wnt/β-catenin signaling in colon cancer (Figure 7). More importantly, the combination of CYTOR and nuclear β-catenin conferred prognostic significance and potential therapeutic target of novel antimetastatic treatment for colon cancer.
Figure 7.
A Schematic Model of CYTOR-β-Catenin Signaling Circuit in Colon Cancer Cells
As one of the most important malignant tumor hallmarks, tumor metastasis is a complex and multistep process influenced by genetic and epigenetic changes.33, 34 EMT initiates the early step of tumor cell metastatic dissemination via endowing cells with increased motility and invasiveness.35 Notably, aberrant activation of EMT has been demonstrated as the dominant program of human colon cancer.36 Recently, the involvement of LncRNAs in modulation of EMT and metastatic cascade has been widely reported.6, 18 However, the understanding of underlying regulatory mechanisms remains largely unknown.21 We previously reported that LncRNA CYTOR was significantly overexpressed in colon cancer and conferred resistance to oxaliplatin-induced apoptosis.20 In this study, we revealed that CYTOR contributed to the EMT program and metastasis of colon cancer both in vitro and in vivo. These findings are consistent with previous studies showing that modified CYTOR expression exerted influences on EMT markers of gastric cancer in vitro. Multiple oncogenic events, mediated, for example, by Wnt, Notch, and TGF-β signaling, are implicated in the induction of EMT.6, 8, 37 Different from mutations in other cancer types, mutations in the key regulatory factors of Wnt/β-catenin signaling are frequent events in colon cancer.38 Constitutive activation of Wnt/β-catenin signaling, which was reflected by the translocation of β-catenin into the nucleus, may cause the reduction of E-cadherin and subsequent induction of EMT.39 Until now, LncRNAs participating in the Wnt/β-catenin signaling-induced EMT are still seldom reported. Our findings indicated that CYTOR facilitates the Wnt/β-catenin signaling through enhancing the nuclear translocation of β-catenin and transactivation of TCF reporter activity. Notably, the promotive effect of CYTOR on EMT and metastasis was significantly blocked by XAV939, and the inhibition of EMT and metastasis caused by CYTOR depletion was markedly rescued by CHIR99021. Thus, CYTOR-induced EMT and colon cancer metastasis is indeed mediated by the Wnt/β-catenin signaling.
Latest evidences showed that LncRNAs could exert impacts through affecting their binding proteins’ stability. For instance, lncARSR was found to interact with YAP and protect YAP from phosphorylation by LATS1.40 LncRNA FAL1 was shown to associate with the epigenetic repressor BMI1 and enhance its stability.41 In this study, we found that CYTOR bound specifically to the Ser45 of β-catenin, thus protecting β-catenin from interaction and phosphorylation by CK1. Recently, the involvement of regulatory feed-forward loops in tumor progression have been reported.40, 42 Considering that β-catenin/TCF complex fulfills oncogenic functions in the nucleus by binding to the promoter of target genes and TBEs were predicted in the CYTOR promoter, we speculated that Wnt/β-catenin signaling might enhance CYTOR transcription via interaction between β-catenin/TCF complex and CYTOR. Consistently, our speculation was confirmed by the ChIP and ChIP-re-ChIP assays.
Lots of LncRNAs involved in the Wnt pathway are aberrantly expressed in colon cancer, for example CCAT2, CASC11, TINCR, CCAL, cir-ITCH, CTD903, and H19.43 It is possible that these LncRNAs interrupt or counteract the function of CYTOR and the feed-forward loop. On the other hand, despite the positive feed-forward loop of CYTOR-β-catenin that exerts accelerative effects on metastasis of colon cancer, the master regulator of this loop have not been identified. Whether other LncRNAs compensate their roles if CYTOR is genetically inhibited in colon cancer cells remains unknown. Thus, further investigations are warranted to advance our understanding of their potential reciprocal modulation, especially in cancer recrudescence and drug resistance study.
Nowadays, optimal prognostic biomarkers for colon cancer have not been established. Here, we found that CYTOR was clinically significant in colon cancer patients. In addition, β-catenin nuclear accumulation has also been suggested to be linked to poor patient outcome. Our findings uncovered that high CYTOR expression could serve as an independent prognostic factor for OS and DFS in colon cancer. Finally, high CYTOR expression combined with nuclear β-catenin accumulation predicted a worse outcome than either marker alone, indicating a potential combinational marker for colon cancer prognosis.
Collectively, we identify the positive feed-forward loop between CYTOR and Wnt/β-catenin signaling that involves in the regulation of EMT and metastasis of colon cancer. Based on the clinical evidence, targeting the CYTOR/β-catenin axis might be a useful therapeutic strategy, as such, may hold a potential value for clinical transformation. Taken together, the discovery of CYTOR and its interaction with Wnt/β-catenin signaling may provide a promising option for facilitating the investigation of colon cancer metastasis.
Materials and Methods
Specimens and Cell Culture
Between January 2001 and December 2003, a total of 100 fresh colon cancer tissues and paired adjacent normal mucosa were collected from colon cancer patients, who underwent radical resection. The diagnosis of all specimens was histopathologically confirmed by two pathologists. None of the patients received any preoperative treatment. This study was approved by the Ethics Committee of Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, and signed informed consent was obtained from each patient enrolled in the study. Human colon mucosal epithelial cell line NCM460 and colon cancer cell lines Hct116, SW1417, SW620, Caco2, SW480, HT29, and HCT8 were obtained from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (IBCB, Shanghai, China), and kept in culture for no more than 3 months in DMEM (Gibco BRL, Grand Island, NY), 10% fetal bovine serum (Invitrogen, Camarillo, CA). Cells were maintained in a humidified atmosphere at 37°C with 5% CO2. All cell lines were authenticated via DNA fingerprinting.
qRT-PCR and Plasmid Transfection
Total RNA from all tissues and cells was extracted using Trizol regent (Ambition Technology, Melbourne, Australia). For LncRNA and mRNA, real-time PCR was carried out as previously described.44 The relative gene expression was calculated by using 2−ΔΔCt method in which higher 2−ΔΔCt value reflects higher expression. The primer sequences can be found in Table S4. The shRNAs specifically targeting CYTOR were obtained from GenPharma (Shanghai, China). For construction of lentiviral vector expressing exogenous CYTOR, CYTOR cDNA was PCR amplified and inserted into the lentiviral vector (Obio Technology, Shanghai). All constructs were confirmed via DNA sequencing. Transfections were performed using the Lipofectamine 2000 kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Animal Studies
6-week-old male athymic BALB/c nude mice were fed under standard pathogen-free conditions in the Experimental Animal Department of Fudan University. For the in vivo metastasis model, representative mice injected with modified CYTOR expressing HCT8 or Hct116 cells (n = 5 per group). The luciferase signal intensity from days 7 to 35 is on equivalent scales in the lung metastasis model. All the mice were killed after 6 weeks, and the lungs were subjected to immunohistochemical analysis and H&E staining. Metastatic progression was monitored and quantified using an IVIS-100 system (Caliper Life Sciences, Boston, MA, USA) as described previously.10 All animal experiments were performed in accordance with the Shanghai Medical Experimental Animal Care Commission Animal Care Guidelines.
Immunohistochemistry
Immunohistochemistry analysis was performed using a GT Vision III Kit (Genetech, Shanghai, China) following the manufacturer’s instructions. Relative gene expressions were analyzed using mouse lung tissues and human colon cancer tissues. The final stainings were scored as described previously.20 Cytoplasmic β-catenin specimens were defined by no visible nuclear staining. Nuclear β-catenin specimens were defined by at least weak positive nuclear staining in ≥25% of tumor cells.
Western Blot Analysis
Equivalent amounts of protein were separated by SDS-PAGE at 80 V for 2.5 hr and transfected to polyvinylidene fluoride (PVDF) membranes for 2 hr. The membranes were washed using 1% Tris-buffered saline Tween (TBST) for 30 min after incubation with specific antibodies targeting E-cadherin, N-cadherin, Vimentin, β-catenin, non-phospho-β-cateninSer45, non-phospho-β-cateninSer31/37/Thr41, Lamin B1, c-myc, cyclin D1, CK1, and GAPDH (Cell Signaling Technology and Abcam) at 4°C overnight, then incubated with secondary antibodies (1:5,000, BioTNT, China) for 2 hr. Finally, they were washed by 1% TBST and detected by a chemiluminescence system (Amersham Biosciences, Piscataway, NJ).
RIP and RNA Pull-Down
For RIP assay, Magna RIP RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) was used following the manufacturer’s instructions. Antibodies specific to β-cateninSer45 and β-cateninSer33/37/Thr41 (Cell Signaling Technology, USA) were diluted as 1:50 and 1:100, respectively. The RNAs precipitated by RIP were detected by reverse transcription PCR.
For RNA pull-down assay, biotin-labeled RNAs were transcribed in vitro using the Biotin RNA Labeling Mix (Roche, Indianapolis, USA) and T7 RNA polymerase (Roche), then treated with RNase-free DNase I (Roche), and purified with an RNeasy Mini Kit (QIAGEN, Valencia, CA). Purified biotinylated transcripts were incubated with whole-cell lysates from colon cancer cells for 1 hr. Streptavidin agarose beads (Invitrogen, USA) were washed briefly and boiled in SDS buffer. The retrieved protein was detected by western blot detection.
Confocal Immunofluorescent Assay
The following antibodies were used: goat anti-rabbit immunoglobulin G (IgG) (Alexa Fluor 488, Invitrogen), goat anti-rabbit IgG (Alexa Fluor 647, Invitrogen), E-cadherin (Cell Signaling Technology, USA), N-cadherin (Cell Signaling Technology, USA), Vimentin (Cell Signaling Technology, USA). Cells were fixed in 1% formaldehyde for 20 min and permeabilized with 0.2% Triton X-100 for 5 min at room temperature, respectively. After being washed three times in PBS and blocked for 1 hr with 2% BSA, cells were incubated with primary antibodies for 2 hr at room temperature and washed three times in PBS and incubated with secondary antibodies for 1 hr at room temperature. Photographs were taken using a confocal microscope (Olympus).
Luciferase Assay
The Cignal Finder Reporter Array (QIAGEN) was used to identify the signaling pathways. CYTOR cDNA was PCR amplified and inserted into the firefly luciferase plasmid.
TOPflash or FOPflash (Millipore) with pRL-TK plasmid (reference) were cotransfected into colon cancer cells (1 × 105) in 24-well plates, then, the activities of both Renilla and firefly luciferase reporters were determined after 48 hr using a dual-luciferase reporter gene assay system (Promega, Madison, WI, USA). The relative ratio of firefly luciferase activity to Renilla luciferase activity was determined as the TOPflash reporter activity.
ChIP
EZ ChIP Chromatin Immunoprecipitation Kit (Millipore, Bedford, MA) was used following the manufacturer’s protocol. ChIP-re-ChIP was performed as described previously.45 Cross-linked chromatin was sonicated into fragments, and then the fragments were immunoprecipitated using TCF4 or β-catenin antibodies. The immunoprecipitated complexes were eluted with re-ChIP buffer. The primer sequences for ChIP-PCR can be found in Table S5.
Statistical Analysis
All statistical analyses were performed with SPSS 19.0 Software package (SPSS, Chicago, IL). Between-group differences were analyzed using Student’s t test and one-way ANOVA. Correlations between gene expression levels were determined by Spearman correlation. The relative gene expression levels and clinicopathologic parameters were compared by χ2 test. Colon cancer patient survival curves were plotted by the Kaplan-Meier method with log-rank test. Cox regression was utilized to estimate the effect of clinical variables on patient survival via analyzing the univariate and multivariate hazards models. All statistical analyses were defined as significant for p values < 0.05.
Author Contributions
B.Y., C.L., and M.Z. designed the experiments; M.L., C.S., and R.C. conducted the experiments; H.S. and S.Q. provided clinical samples and patient data.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the Doctoral Innovation Fund Projects from School of Medicine, Shanghai Jiao Tong University (BXJ201630).
Footnotes
Supplemental Information includes one figure and five tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.02.024.
Contributor Information
Shenglong Qiu, Email: qsl65@126.com.
Ming Zhong, Email: drzhongming@hotmail.com.
Supplemental Information
References
- 1.Siegel R., Desantis C., Jemal A. Colorectal cancer statistics, 2014. CA Cancer J. Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 3.Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spaderna S., Schmalhofer O., Hlubek F., Berx G., Eger A., Merkel S., Jung A., Kirchner T., Brabletz T. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131:830–840. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 5.Liu B., Sun L., Liu Q., Gong C., Yao Y., Lv X., Lin L., Yao H., Su F., Li D. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Yuan J.H., Yang F., Wang F., Ma J.Z., Guo Y.J., Tao Q.F., Liu F., Pan W., Wang T.T., Zhou C.C. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J., Kuang Y., Wang Y., Xu Q., Ren Q. Notch-4 silencing inhibits prostate cancer growth and EMT via the NF-kappaB pathway. Apoptosis. 2017;22:877–884. doi: 10.1007/s10495-017-1368-0. [DOI] [PubMed] [Google Scholar]
- 8.Yang S., Liu Y., Li M.Y., Ng C.S.H., Yang S.L., Wang S., Zou C., Dong Y., Du J., Long X. FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol. Cancer. 2017;16:124. doi: 10.1186/s12943-017-0700-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weng W., Feng J., Qin H., Ma Y. Molecular therapy of colorectal cancer: progress and future directions. Int. J. Cancer. 2015;136:493–502. doi: 10.1002/ijc.28722. [DOI] [PubMed] [Google Scholar]
- 10.Liu C.C., Cai D.L., Sun F., Wu Z.H., Yue B., Zhao S.L., Wu X.S., Zhang M., Zhu X.W., Peng Z.H., Yan D.W. FERMT1 mediates epithelial-mesenchymal transition to promote colon cancer metastasis via modulation of β-catenin transcriptional activity. Oncogene. 2017;36:1779–1792. doi: 10.1038/onc.2016.339. [DOI] [PubMed] [Google Scholar]
- 11.Polakis P. Casein kinase 1: a Wnt’er of disconnect. Curr. Biol. 2002;12:R499–R501. doi: 10.1016/s0960-9822(02)00969-7. [DOI] [PubMed] [Google Scholar]
- 12.Grumolato L., Liu G., Haremaki T., Mungamuri S.K., Mong P., Akiri G., Lopez-Bergami P., Arita A., Anouar Y., Mlodzik M. β-Catenin-independent activation of TCF1/LEF1 in human hematopoietic tumor cells through interaction with ATF2 transcription factors. PLoS Genet. 2013;9:e1003603. doi: 10.1371/journal.pgen.1003603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu F.I., Sun Y.H., Wei C.Y., Thisse C., Thisse B. Tissue-specific derepression of TCF/LEF controls the activity of the Wnt/β-catenin pathway. Nat. Commun. 2014;5:5368. doi: 10.1038/ncomms6368. [DOI] [PubMed] [Google Scholar]
- 14.White B.D., Chien A.J., Dawson D.W. Dysregulation of Wnt/β-catenin signaling in gastrointestinal cancers. Gastroenterology. 2012;142:219–232. doi: 10.1053/j.gastro.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan Y., Shen B., Tan M., Mu X., Qin Y., Zhang F., Liu Y. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 19.Li Z., Hou P., Fan D., Dong M., Ma M., Li H., Yao R., Li Y., Wang G., Geng P. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24:59–71. doi: 10.1038/cdd.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yue B., Cai D., Liu C., Fang C., Yan D. Linc00152 functions as a competing endogenous RNA to confer oxaliplatin resistance and holds prognostic values in colon cancer. Mol. Ther. 2016;24:2064–2077. doi: 10.1038/mt.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J., Liu Y., Zhang W., Zhou Z., Wu J., Cui P., Zhang Y., Huang G. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112–3123. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai Q., Wang Z., Wang S., Weng M., Zhou D., Li C., Wang J., Chen E., Quan Z. Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial-mesenchymal transition by regulating HIF-1α via miR-138. Open Biol. 2017;7:160247. doi: 10.1098/rsob.160247. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Weyemi U., Redon C.E., Choudhuri R., Aziz T., Maeda D., Boufraqech M., Parekh P.R., Sethi T.K., Kasoji M., Abrams N. The histone variant H2A.X is a regulator of the epithelial-mesenchymal transition. Nat. Commun. 2016;7:10711. doi: 10.1038/ncomms10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Q., Abraham A.D., Li L., Babalmorad A., Bagby S., Arcaroli J.J., Hansen R.J., Valeriote F.A., Gustafson D.L., Schaack J. Topoisomerase IIα mediates TCF-dependent epithelial-mesenchymal transition in colon cancer. Oncogene. 2016;35:4990–4999. doi: 10.1038/onc.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slyper M., Shahar A., Bar-Ziv A., Granit R.Z., Hamburger T., Maly B., Peretz T., Ben-Porath I. Control of breast cancer growth and initiation by the stem cell-associated transcription factor TCF3. Cancer Res. 2012;72:5613–5624. doi: 10.1158/0008-5472.CAN-12-0119. [DOI] [PubMed] [Google Scholar]
- 26.Zhou J., Zhi X., Wang L., Wang W., Li Z., Tang J., Wang J., Zhang Q., Xu Z. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J. Exp. Clin. Cancer Res. 2015;34:135. doi: 10.1186/s13046-015-0250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C., Li Y., Semenov M., Han C., Baeg G.H., Tan Y., Zhang Z., Lin X., He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 28.Schmitt A.M., Chang H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murgan S., Kari W., Rothbächer U., Iché-Torres M., Mélénec P., Hobert O., Bertrand V. Atypical transcriptional activation by TCF via a Zic transcription factor in C. elegans neuronal precursors. Dev. Cell. 2015;33:737–745. doi: 10.1016/j.devcel.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang S.M., Mishina Y.M., Liu S., Cheung A., Stegmeier F., Michaud G.A., Charlat O., Wiellette E., Zhang Y., Wiessner S. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 31.Zhao J., Li S., Trilok S., Tanaka M., Jokubaitis-Jameson V., Wang B., Niwa H., Nakayama N. Small molecule-directed specification of sclerotome-like chondroprogenitors and induction of a somitic chondrogenesis program from embryonic stem cells. Development. 2014;141:3848–3858. doi: 10.1242/dev.105981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang L., Zhu Q., Neuenschwander M., Specker E., Wulf-Goldenberg A., Weis W.I., von Kries J.P., Birchmeier W. A small-molecule antagonist of the β-catenin/TCF4 interaction blocks the self-renewal of cancer stem cells and suppresses tumorigenesis. Cancer Res. 2016;76:891–901. doi: 10.1158/0008-5472.CAN-15-1519. [DOI] [PubMed] [Google Scholar]
- 33.Gupta G.P., Massagué J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Tiwari N., Tiwari V.K., Waldmeier L., Balwierz P.J., Arnold P., Pachkov M., Meyer-Schaller N., Schübeler D., van Nimwegen E., Christofori G. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23:768–783. doi: 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 35.Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 36.Loboda A., Nebozhyn M.V., Watters J.W., Buser C.A., Shaw P.M., Huang P.S., Van’t Veer L., Tollenaar R.A., Jackson D.B., Agrawal D. EMT is the dominant program in human colon cancer. BMC Med. Genomics. 2011;4:9. doi: 10.1186/1755-8794-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sieiro D., Rios A.C., Hirst C.E., Marcelle C. Cytoplasmic NOTCH and membrane-derived β-catenin link cell fate choice to epithelial-mesenchymal transition during myogenesis. eLife. 2016;5:e14847. doi: 10.7554/eLife.14847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blaj C., Bringmann A., Schmidt E.M., Urbischek M., Lamprecht S., Frohlich T., Arnold G.J., Krebs S., Blum H., Hermeking H. ADNP is a therapeutically inducible repressor of WNT signaling in colorectal cancer. Clin. Cancer Res. 2017;23:2769–2780. doi: 10.1158/1078-0432.CCR-16-1604. [DOI] [PubMed] [Google Scholar]
- 39.Goto H., Kimmey S.C., Row R.H., Matus D.Q., Martin B.L. FGF and canonical Wnt signaling cooperate to induce paraxial mesoderm from tailbud neuromesodermal progenitors through regulation of a two-step epithelial to mesenchymal transition. Development. 2017;144:1412–1424. doi: 10.1242/dev.143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu L., Wu Z., Li Y., Xu Z., Liu B., Liu F., Bao Y., Wu D., Liu J., Wang A. A feed-forward loop between lncARSR and YAP activity promotes expansion of renal tumour-initiating cells. Nat. Commun. 2016;7:12692. doi: 10.1038/ncomms12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu X., Feng Y., Zhang D., Zhao S.D., Hu Z., Greshock J., Zhang Y., Yang L., Zhong X., Wang L.P. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rokavec M., Öner M.G., Li H., Jackstadt R., Jiang L., Lodygin D., Kaller M., Horst D., Ziegler P.K., Schwitalla S. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Invest. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo J., Qu J., Wu D.K., Lu Z.L., Sun Y.S., Qu Q. Long non-coding RNAs: a rising biotarget in colorectal cancer. Oncotarget. 2017;8:22187–22202. doi: 10.18632/oncotarget.14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yue B., Qiu S., Zhao S., Liu C., Zhang D., Yu F., Peng Z., Yan D. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J. Gastroenterol. Hepatol. 2016;31:595–603. doi: 10.1111/jgh.13206. [DOI] [PubMed] [Google Scholar]
- 45.Pillai S., Kovacs M., Chellappan S. Regulation of vascular endothelial growth factor receptors by Rb and E2F1: role of acetylation. Cancer Res. 2010;70:4931–4940. doi: 10.1158/0008-5472.CAN-10-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.