To the Editor
The 17q21 asthma-susceptibility locus, identified through genome-wide association studies, contains the zona pellucida binding protein 2 (ZPBP2), gasdermin B (GSDMB), and ORMDL sphingolipid biosynthesis regulator 3 (ORMDL3) genes.1 The single nucleotide polymorphisms (SNPs) associated with asthma in this region are correlated with increased expression of both GSDMB and ORMDL3.2 However, genetic risk factors alone cannot account for the overall risk for developing asthma nor explain the observed increases in asthma prevalence over the past three decades. Indeed, epidemiologic studies support a substantial role for environmental factors in the development of asthma, such as early life exposure to tobacco smoke and respiratory viral infections, both of which have also been shown to modify the genetic risk attributable to the 17q21 locus.3,4 One possible mechanism by which environmental exposures may alter genetic risk is through epigenetic modifications, such as DNA methylation. One recent, small study demonstrated an association between asthma and differential methylation of ORMDL3 from peripheral blood leukocytes in children; however, these findings have not been replicated.5 Furthermore, gene expression of GSDMB and ORMDL3 has been associated with CpG methylation in a larger population.6 To understand better the relative contribution of CpG methylation and asthma-associated SNPs in the cis-regulation of 17q21 candidate gene expression, we performed an integrative genomic analysis in two large, racially diverse cohorts collected as part of the Asthma BioRepository for Integrative Genomic Exploration (Asthma BRIDGE) study.7 Genotype, gene-expression and methylation were available for 293 and 264 non-overlapping subjects with data from whole blood (WB) and CD4+ T cells, respectively. Detailed methods and characteristics of the study population (Table E1) as well as an overview of the main findings (Figure E1) can be found in this article’s Online Repository at www.jacionline.org (Table E1).
Our analysis of cis-regulatory variants was limited to within 50 kb of ZPBP2, GSDMB and ORMDL3 as those further away have not been associated with asthma-risk. The 17q21 region under study showed a remarkable degree of architectural similarity between the WB and CD4+ cohorts, both with respect to correlation of minor allele frequency (Pearson correlation coefficient of 0.941, 95% CI 0.928-0.952, p-value < 2.2E-16) and pairwise linkage disequilibrium (LD, r^2; Pearson correlation coefficient of 0.928, 95% CI 0.927-0.928, p-value < 2.2E-16), despite being compromised of different proportions of European Americans, African Americans and Hispanics (Table E1). We first confirmed the association between SNP genotype and target gene expression through expression quantitative trait loci (eQTL) mapping. Of the 1638 SNP-gene expression probes pairs tested, 117 SNPs were significantly associated with ORMDL3 and 127 with GSDMB in both WB and CD4+ T-cells at a false discovery rate (FDR) of less than 0.05. Figure 1A shows the overlap of eQTL SNPs across tissues and genes, and demonstrates that a plurality of SNPs (n=73) was associated with expression of both genes in both cohorts. Most significant eQTLs identified were in strong LD (data not shown) and included the most commonly replicated asthma-susceptibility SNP (rs7216389; Figure 1B) and the putative functional variants affecting CTCF (rs12936231) and transcription factor (rs8076131 and rs4065275) binding (Table E2).2,8
Figure 1.
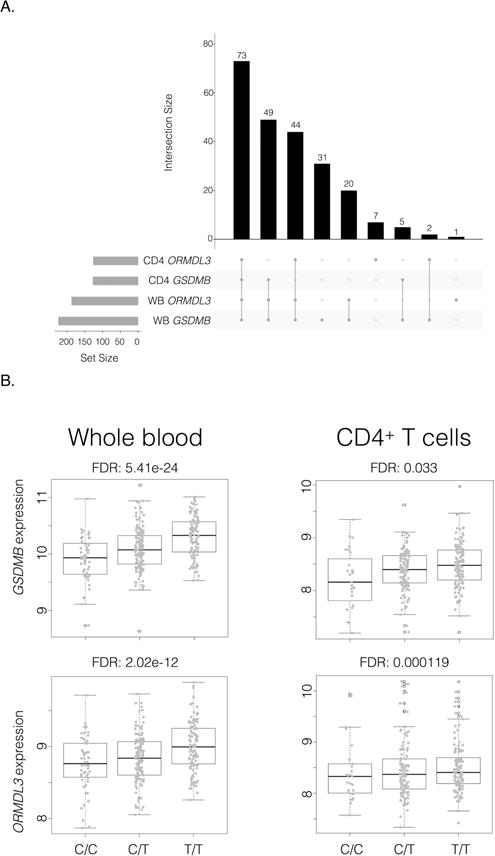
Summary of 17q21 eQTL analyses.
A. Main bar graph (black vertical bars) denotes counts of SNPs significantly associated (FDR < 0.05) with ORMDL3 or GSDMB expression in CD4+ T cells or whole blood (WB). The vertical gray line and dot graph directly underneath denotes the count of significant SNP common to combinations of tested groups, with combinations indicated by connected circles. The gray bar graph on the bottom left shows the number of eQTLs for the specified groups. Plot generated with upset function from “UpSetR” R package.
B. Box plots for gene expression for GSDMB (top) and ORMDL3 (bottom) according to rs7216389 genotype in whole blood (left; C/C [n=52], C/T [n=131], T/T [n=105]) and CD4+ T cells (right; C/C [n=27], C/T [n=115], T/T [n=122]).
We next evaluated the effect of methylation on gene expression for CpG sites within 50 kb of the target genes, a total of 265 CpG-gene expression probe pairs. Five CpG sites (cg12655416, cg22144450, cg18711369, cg10909506, cg26162295) were significantly associated with ORMDL3 expression in both the WB and CD4+ T cell cohorts. These five sites and a sixth (cg24910161) were also associated with GSDMB expression in both groups, with increased methylation at these sites associated with decreased expression of ORMDL3 and GSDMB (Figure 2). Furthermore, we found evidence of strong correlation between these six CpG sites, two of which are upstream of gasdermin A (GSDMA) and four within the ORMDL3 gene body.
Figure 2.
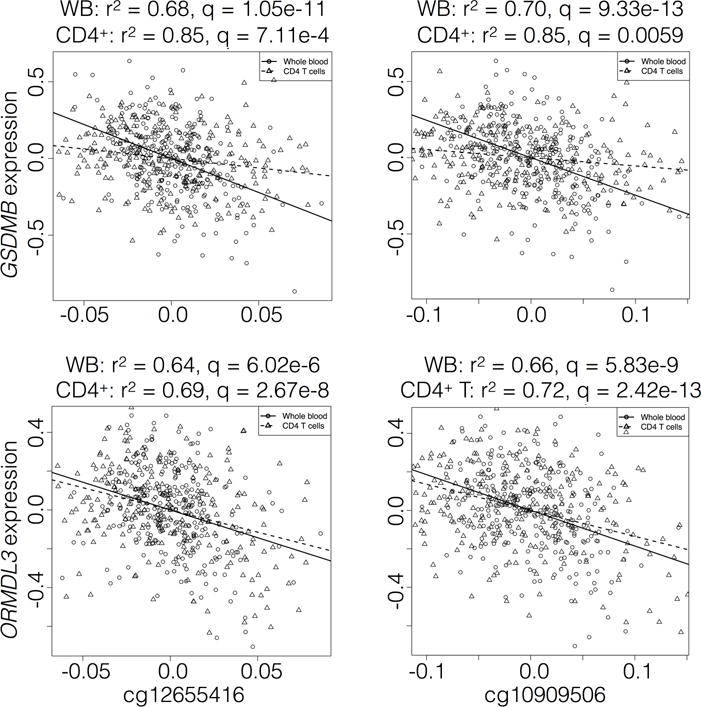
Correlation of CpG methylation with GSDMB and ORMDL3 gene expression. Scatter plots of residuals, after adjusting for covariates, for gene expression of GSDMB (top panel) and ORMDL3 (bottom panel) and methylation of selected CpG sites in whole blood (black open circles) and CD4+ T cell (black open triangles) cohorts. Linear regression fits for whole blood (solid lines) and CD4+ T cell cohorts (dashed lines) are provided.
Given that both genotype and methylation were associated with gene expression at this locus, we next evaluated the relationship between 17q21 SNPs and CpG sites. Testing of 9,207 CpG-SNP pairs within 50 kb of each other across the region identified 1,505 significant associations, representing various combinations between 19 CpG sites and 310 SNPs, present in both the WB and CD4+ cohorts. Notably, these 310 methylation QTL (mQTL) SNPs included all 171 replicated eQTL SNPs in our cohorts. The six expression-associated CpG sites were associated with 196 mQTL SNPs, most of which were eQTL SNPs as well (164/196 [84%]). Thus, there was a strong association between SNPs and CpG sites associated with gene expression for the 17q21 locus. Importantly, these associations were robust to several sensitivity analyses, including tests for confounding by residual population stratification and asthma affection status (by repeated analysis restricting either to subjects from only the major racial subgroup in each cohort, or those with asthma, respectively). In addition, to determine if our observed associations extended to neighboring genes, we expanded the region under study to include IKZF3, GSDMA and PSMD3 but no additional associations were detected.
In light of the considerable associations among SNPs, CpGs, and gene expression, we then performed causal inference testing (CIT)9 to evaluate whether the observed SNP eQTL effects could be attributed to the associated methylation marks, or whether genetic and epigenetic marks impact target gene expression independently. We limited this analysis to the subset of SNPs and CpG sites significantly associated with the gene expression probes for GSDMB and ORMDL3 in both cohorts. Of 1343 gene expression probe-SNP-CpG combinations tested, 862 (64%) supported methylation as a potential mediator of the eQTL effect (overall CIT FDR < 0.05 in both cohorts). These associations included all 127 SNPs and six CpG sites associated with GSDMB, as well as all 117 SNPs and five CpG sites associated with ORMDL3. Thus, regardless of which SNP was considered by the CIT, its effects on target gene expression could be mediated by at least one associated CpG site. This is exemplified for the three putative functional variants (rs12936231, rs8076131 and rs4065275) and the most replicated asthma-susceptibility variant (rs7216389) in Table E3 in the Online Repository. In addition, Figure E2 in the Online Repository demonstrates how the correlation between ORMDL3 gene expression and cg12655416 methylation differs by rs12936231 genotype. Furthermore, the CIT suggests that CpG methylation influences gene expression independent of genotype as shown in Table E3.
Given the strong correlations of methylation at the tested CpG sites and the extensive LD at 17q21, the CIT could not localize the putative regulatory signals to one specific SNP-CpG combination. Nonetheless, our results demonstrated a higher percentage of eQTL SNPs showing evidence of mediation through CpG sites within the ORMDL3 gene body (cg10909506, cg12655416, cg18711369, and cg22144450) compared with those upstream of GSDMA (cg24910161 and cg26162295). Indeed, the four CpG sites found within ORMDL3 were found to mediate the observed eQTL associations.
In conclusion, our population-based integrative genomics analysis of the 17q21 asthma-susceptibility locus illustrates the complex relationship between sequence variation, CpG methylation and target gene expression. We show, in two well-powered, independent cohorts that (i) local CpG methylation mediates a proportion, but not all, of the functional effects of cis-acting asthma-susceptibility regulatory variants on GSDMB and ORMDL3 gene expression; and that in addition to these effects, (ii) residual differences in CpG methylation not explained by SNP genotype, also influence target gene expression. In vitro studies that dissociate and isolate the genetic and epigenetic determinants of GSDMB and ORMDL3 expression could confirm these observations, could determine if these findings are applicable to other cells and tissues associated with asthma, and would motivate epigenetic manipulation of this locus as a potential therapeutic strategy.
Supplementary Material
Capsule Summary.
An integrative analysis of genotype, methylation and gene expression of the 17q21 locus reveals complex interactions with CpG methylation functioning as an intermediary of the genotypic effect on ORMDL3 and GSDMB transcription.
Acknowledgments
Funding
This work was supported by extramural grants from the National Institutes of Health (NIH) through the National Heart, Lung, and Blood Institute [RC2 HL101543 (America Recovery and Reinvestment Act “Grand Opportunity”), and R01 HL086601] and National Institute of Allergy and Infectious Diseases [T32 AI007306 to P.H.K.]. Additional support provided by the NIH [K01 HL127265 to D.C.C.-C, P30 ES007048, R01 ES023262, R01 HL061768, R01 ES021801, and P01 ES011627 to F.D.G. and the Intramural Research Program ZO1 ES49019 to S.J.L].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A Large-Scale, Consortium-Based Genomewide Association Study of Asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verlaan DJ, Berlivet S, Hunninghake GM, Madore A-M, Larivière M, Moussette S, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–93. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flory JH, Sleiman PM, Christie JD, Annaiah K, Bradfield J, Kim CE, et al. 17q12-21 variants interact with smoke exposure as a risk factor for pediatric asthma but are equally associated with early-onset versus late-onset asthma in North Americans of European ancestry. J Allergy Clin Immunol. 2009;124:605–7. doi: 10.1016/j.jaci.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 4.Çalışkan M, Bochkov YA, Kreiner-Møller E, Bønnelykke K, Stein MM, Du G, et al. Rhinovirus Wheezing Illness and Genetic Risk of Childhood-Onset Asthma. N Engl J Med. 2013;368:1398–407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acevedo N, Reinius LE, Greco D, Gref A, Orsmark-Pietras C, Persson H, et al. Risk of childhood asthma is associated with CpG-site polymorphisms, regional DNA methylation and mRNA levels at the GSDMB/ORMDL3 locus. Hum Mol Genet. 2015;24:875–90. doi: 10.1093/hmg/ddu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2017;49:131–8. doi: 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 7.Croteau-Chonka DC, Qiu W, Martinez FD, Strunk RC, Lemanske RF, Liu AH, et al. Gene Expression Profiling in Blood Provides Reproducible Molecular Insights into Asthma Control. Am J Respir Crit Care Med. 2017;195:179–88. doi: 10.1164/rccm.201601-0107OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schedel M, Michel S, Gaertner VD, Toncheva AA, Depner M, Binia A, et al. Polymorphisms related to ORMDL3 are associated with asthma susceptibility, alterations in transcriptional regulation of ORMDL3, and changes in TH2 cytokine levels. J Allergy Clin Immunol. 2015;136:893–903.e14. doi: 10.1016/j.jaci.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 9.Millstein J, Chen GK, Breton CV. cit: hypothesis testing software for mediation analysis in genomic applications. Bioinformatics. 2016;32:2364–5. doi: 10.1093/bioinformatics/btw135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


