Abstract
Mammalian physiology exhibits 24-hour cyclicity due to circadian rhythms of gene expression controlled by transcription factors (TF) that comprise molecular clocks. Core clock TFs bind to the genome at enhancer sequences to regulate circadian gene expression, but not all binding sites are equally functional. Here we demonstrate that circadian gene expression in mouse liver is controlled by rhythmic chromatin interactions between enhancers and promoters. Rev-erbα, a core repressive TF of the clock, opposes functional loop formation between Rev-erbα-regulated enhancers and circadian target gene promoters by recruitment of the NCoR-HDAC3 corepressor complex, histone deacetylation, and eviction of the elongation factor BRD4 and the looping factor MED1. Thus, a repressive arm of the molecular clock operates by rhythmically modulating chromatin loops to control circadian gene transcription.
Circadian rhythms of mammalian physiology are orchestrated by core clock TFs functioning as activators or repressors in interlocking feedback loops that drive daily oscillations of gene expression (1, 2). These TFs generate circadian rhythms of histone modification at circadian enhancers (3–6). Enhancer activities are mediated by looping to promoters within insulated topologically associating domains (TADs) (7–9). Previous studies with cultured cells have described circadian regulation of higher-order chromatin organization (10), long-range interchromosomal interactions (11), and short-range chromatin loops (12) at specific loci, but this has not been examined at a genome-wide level in native tissues under normal physiology.
We performed in situ Hi-C (13, 14) on C57BL/6J mouse livers harvested 12 hours apart, at zeitgeber time 22 (ZT22, 5 AM) and ZT10 (5 PM) to examine whether chromatin interactions change in a circadian manner. Hi-C identified megabase-size TADs whose boundaries were highly similar to those identified in mouse embryonic stem cells (Fig. S1A). This was expected since TADs are largely conserved among different tissues (7), and indeed the overall TAD organization was also highly similar between ZT22 and ZT10 (Fig. S1B). Within each TAD we also observed sub-megabase structures (sub-TADs) of different lengths (Fig. S1C) that were flanked by CTCF and cohesin (RAD21) and internally occupied by Mediator (MED1) (Fig. S1D), as described previously (14–17). Globally, genomic occupancy of CTCF and cohesin at ZT10 versus ZT22 was very similar at sub-TAD boundaries (Fig. S1E–F).
Since TFs bind at enhancers to regulate genes confined within sub-TAD boundaries (16), we next searched for interactions occurring within sub-TADs (“intra-TAD”). Of 6510 intra-TAD interactions, only 349 were ZT22-specific while 527 were ZT10-specific (Wilcoxon signed rank test, p < 0.001). For example, the circadian Npas2 gene exhibited increased intra-TAD interactions at ZT22, including extensive looping between the transcriptional start site and four non-coding regions (E1-4) (Fig. 1A). These non-coding regions mapped to regions of divergent transcriptional activity characteristic of enhancer RNA (eRNA) that were similarly regulated (Fig. S2A). Intra-TAD interaction was also observed within the gene body as previously reported at actively transcribed genes (18, 19), and this was also enhanced at ZT22 (Fig. 1A).
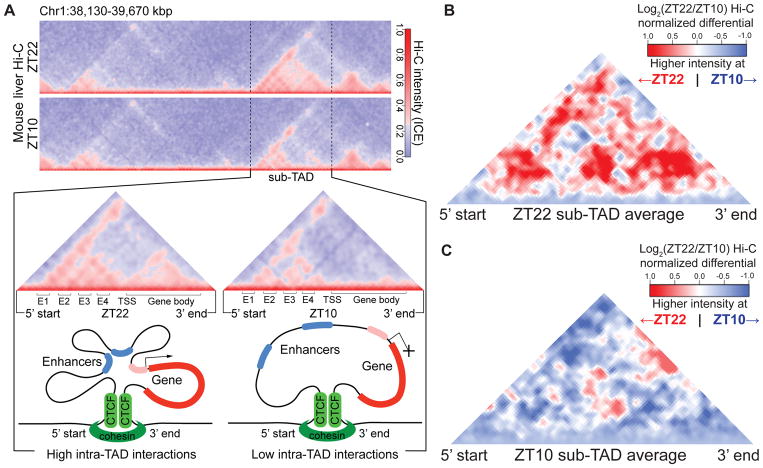
Figure 1. Circadian sub-TADs undergo rhythmic intra-TAD compaction within stable boundaries.
(A) Heat maps of ZT22 and ZT10 Hi-C demonstrating circadian intra-TAD interactions within sub-TAD boundaries (dotted lines), as represented by Hi-C intensity normalized by Iterative Correction and Eigenvector decomposition (ICE). Transcriptional start site of the Npas2 gene (TSS) forms rhythmic intra-TAD loops with upstream enhancers (E1-4) as well as with the gene body, as illustrated by schematics below. (B) ZT22 sub-TAD and (C) ZT10 sub-TAD averaged differential changes in intra-TAD interactions visualized as log2 ratio within size-normalized sub-TAD 5’ and 3’ boundaries (red= higher interaction ratio at ZT22, blue=higher interaction ratio at ZT10).
Genome-wide, we identified hundreds of sub-TAD containing circadian genes whose expression has been previously demonstrated to peak between ZT21-24 (“ZT22 sub-TADs”) or ZT9-12 (“ZT10 sub-TADs”) (6). The overall structure of these sub-TADs (Fig. S2B), and the binding of CTCF and RAD21 at their boundaries (Fig. S2C–D) changed very little between ZT22 and ZT10 (Fig. S2B–D). However, ZT22 and ZT10 sub-TADs exhibited greater intra-TAD interactions corresponding to their transcriptional activities (Fig. 1B–C), whereas non-circadian sub-TADs did not (Fig. S3A). Similar conclusions were obtained when the circadian windows were adjusted by one hour (Fig. S3B–C). Interactions within gene bodies were also circadian (Fig. S3D–H).
The circadian Cry1 locus is located within a ZT22 sub-TAD, and Hi-C revealed an interaction between the gene promoter and an intronic enhancer that was increased at ZT22 (Fig. S4A–B), as best visualized by differential analysis of the data (Fig. 2A). The nuclear receptor Rev-erbα, a repressive component of the mammalian clock whose expression peaks at ZT10 to confer circadian expression of genes in the opposite phase (6, 20, 21), binds at this intronic enhancer (Fig. 2A). The enhancer-promoter (E-P) loop identified in ZT22 Hi-C was confirmed by chromatin conformation capture (3C) experiments (Fig. 2B). Tandem 3C and Rev-erbα chromatin immunoprecipitation (ChIP) at 6 time points throughout the day revealed that this E-P loop was indeed circadian and in phase with Cry1 mRNA expression, with a peak that was anti-phase to Rev-erbα binding (Fig. 2C). At ZT10, both looping from the Rev-erbα site to the Cry1 promoter (Fig. 2D) and Cry1 gene expression (Fig. 2E) were enhanced by genetic depletion of Rev-erbα, which attenuated the rhythmicity of these parameters over the course of 24 hours (Fig. S4C). Reciprocally, ectopic expression of Rev-erbα in liver was sufficient to reduce looping as well as mRNA expression at ZT22 (Fig. 2F–G), consistent with an active role of Rev-erbα in opposing loop formation. We next performed Hi-C on livers from mice genetically lacking Rev-erbα and harvested at ZT10, which confirmed the enhanced E-P looping at the Cry1 locus (Fig. S4D). Moreover, throughout the genome intra-TAD interactions that were normally favored at ZT22 were increased in the genetic absence of Rev-erbα at ZT10 (Fig. 2H).
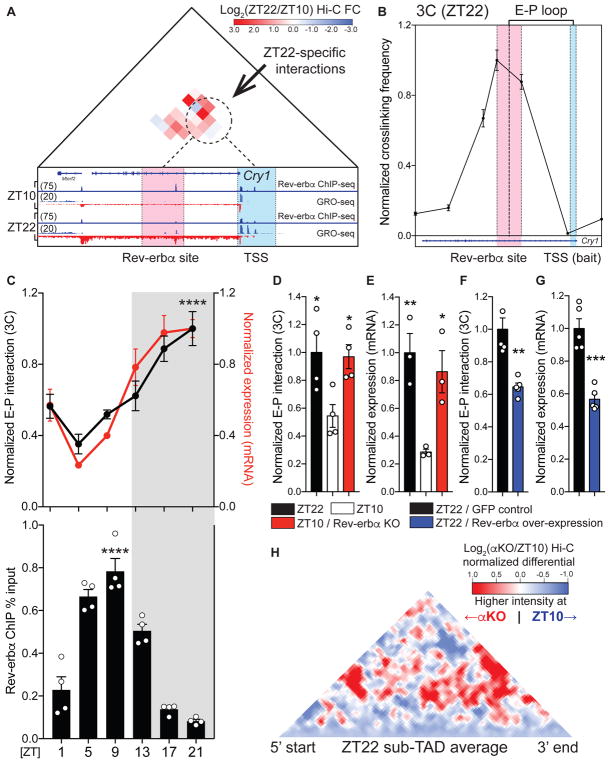
Figure 2. Rev-erbα causally opposes enhancer-promoter loop formation.
(A) Differential Hi-C analysis at the Cry 1 locus revealing ZT22-specific interactions, represented as log2 ratio (ZT22 Hi-C/ZT10 Hi-C). ZT22-specific interactions (dotted circle) occur between a region around the intronic Rev-erbα site (red) and the Cry1 TSS (blue). Global Run-On seq (GRO-seq) demonstrates circadian nascent transcription as well as the presence of bidirectional eRNA at Rev-erbα site at ZT22. (B) 3C validation of enhancer-promoter loop (E-P loop) identified at ZT22 between Rev-erbα site (red) and TSS (blue) (n=5, mean ±SEM). (C) Circadian plot demonstrating Cry1 E-P loop, mRNA expression, and Rev-erbα ChIP ±SEM (n=4–5, p values shown for 3C and ChIP peaks compared to troughs, one-way ANOVA followed by multiple comparisons correction with the Tukey method) (D) E-P loop and (E) mRNA expression of Cry1 at ZT22 (black), ZT10 (white), and ZT10 Rev-erbα KO (red), represented as mean ±SEM (n=4, one-way ANOVA followed by Dunnett’s multiple comparisons test). (F) E-P loop and (G) mRNA expression of Cry1 at ZT22 with control GFP expression (black) versus Rev-erbα over-expression (blue) expressed as mean ±SEM (n=5, two-tailed Student’s t-test). (H) Same analysis as in Fig. 1B, but comparing ZT10 Rev-erbα KO (αKO) to ZT10 WT at ZT22 sub-TADs (red= higher interaction ratio at ZT10 αKO, blue=higher interaction ratio at ZT10 WT). * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001
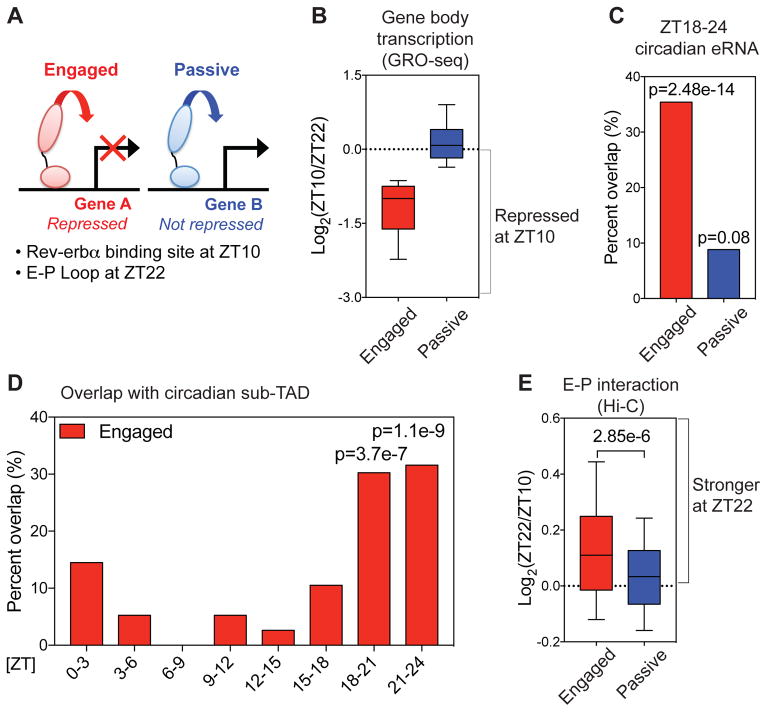
These findings suggested that the ability of Rev-erbα to oppose loop formation is a critical feature at binding sites from which Rev-erbα actively represses transcription. To explore the relationship between regulation of looping and functional repression, we identified Rev-erbα binding sites that directly loop to promoters at ZT22 in the physiological absence of Rev-erbα. E-P loops at Rev-erbα binding sites were defined as “engaged” when transcription of the gene body was repressed at ZT10, or “passive” if not repressed (Fig. 3A–B). Engaged sites were highly correlated with circadian eRNAs whose activity peaked around ZT18-24 antiphase to Rev-erbα binding (Fig. 3C), demonstrating that engaged sites are direct links between circadian eRNAs and genes repressed by Rev-erbα. De novo motif analysis at engaged sites revealed enrichment of DNA motifs known to be bound by Rev-erbα directly (RORE and RevDR2) (22) or indirectly (HNF6) (21) (Fig. S5A), while these motifs were not enriched at passive sites (Fig. S5B). While the number of sub-TADs containing circadian genes from each phase was similar (Fig. S5C), only engaged Rev-erbα sites were enriched in ZT18-24 circadian sub-TADs (Fig. 3D and S5D). Most importantly, E-P interactions at engaged Rev-erbα binding sites were strengthened at ZT22 relative to ZT10 (Fig. 3E). Together, these findings demonstrate that the ability of Rev-erbα to oppose E-P loops within sub-TADs is a likely determinant of active repression by Rev-erbα.
Figure 3. Rev-erbα attenuates enhancer-promoter looping at functional binding sites.
(A) Rev-erbα sites at E-P loops were classified as “engaged” when looped to genes whose transcription was repressed at ZT10 relative to ZT22, and otherwise classified as “passive”. (B) Gene body transcription change between ZT22 and ZT10 at engaged vs. passive sites. Engaged target genes were defined based on nascent gene body transcription fold change ≥1.5 between ZT22 and ZT10. (C) Engaged Rev-erbα sites were highly correlated with ZT18-24 circadian eRNAs (6) (within ± 2kbp, one-tailed hypergeometric tests). (D) Engaged Rev-erbα sites were confined within sub-TADs that contain circadian genes peaking at ZT18-24 (one-tailed hypergeometric tests). (E) E-P loops between engaged Rev-erbα binding sites and target gene promoters were stronger at ZT22 than ZT10 (Mann-Whitney test). For boxplots, whiskers drawn at 10th and 90th percentiles
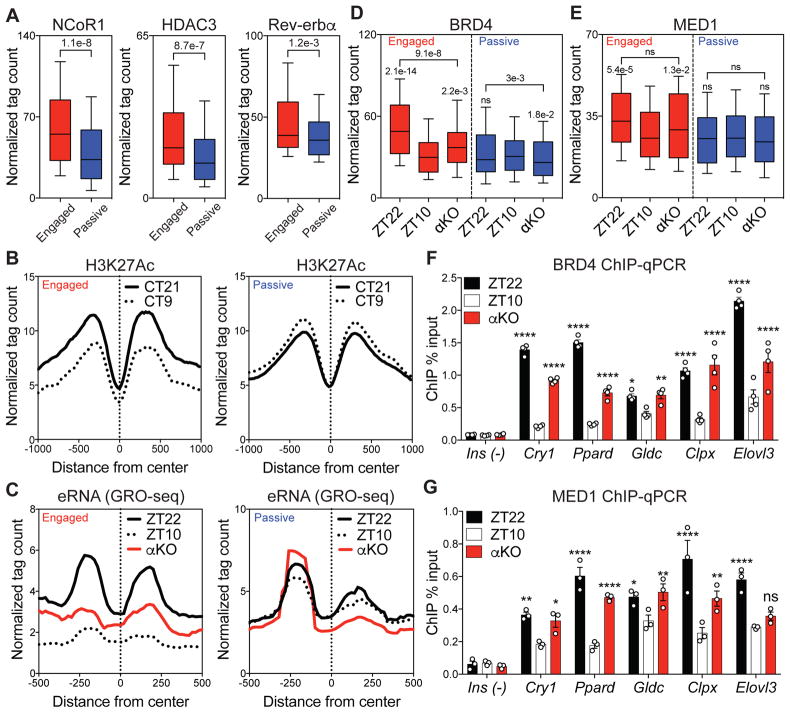
We next addressed the mechanism by which Rev-erbα binding controls circadian E-P interactions. The repressive action of Rev-erbα is often mediated by recruitment of a corepressor complex containing Nuclear Receptor Corepressor (NCoR) and Histone Deacetylase 3 (HDAC3) (23), leading to circadian histone deacetylation associated with repressed enhancers (3). Indeed, recruitment of NCoR and HDAC3 was greater at engaged Rev-erbα binding sites, with a modest average increase in Rev-erbα binding relative to passive sites (Fig. 4A). Consistent with this, the previously demonstrated circadian acetylation of histone H3 on lysine 27 (H3K27Ac) in mouse liver (5) is exaggerated at engaged Rev-erbα binding sites, while nearly absent at passive sites (Fig. 4B). eRNA transcription also demonstrated an enhanced circadian rhythm at engaged sites, which was abrogated in livers lacking Rev-erbα (Fig. 4C), demonstrating that Rev-erbα was required for the epigenomic rhythms that occurred selectively at engaged sites.
Figure 4. Functional Rev-erbα binding evicts BRD4 and MED1 from sites of looping.
(A) Higher recruitment of NCoR1 and HDAC3 at engaged Rev-erbα sites associated with a slight average increase in Rev-erbα binding (Mann-Whitney tests). (B) Circadian deacetylation of histone 3 lysine 27 (H3K27Ac) at circadian time 21 (CT21, black) and CT9 (dotted) at engaged vs. passive sites. (C) Circadian eRNA transcription between ZT22 (black) and ZT10 (dotted), with increased transcription at ZT10 in αKO (red) at engaged sites. (D) Circadian eviction of BRD4 and (E) MED1 between ZT22 and ZT10, with enhanced binding at ZT10 in αKO at engaged sites (Dunn’s multiple comparisons tests after one-way ANOVA/Friedman test). (F) ChIP-qPCR validation of BRD4 and (G) MED1 eviction at ZT10 and enhanced binding at ZT10 in αKO at engaged sites (Ins as a negative control, n=3–4, two-way ANOVA followed by Dunnett’s multiple comparisons test). For boxplots, whiskers drawn at 10th and 90th percentiles, ns p≥0.05, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001
The transcriptional regulator BRD4 acts as a reader of acetylated histone (24–26) , and forms a functional transcriptional complex with the well-established looping factor MED1 (27–30). In agreement with the changes in H3K27Ac at engaged sites, BRD4 binding was higher at ZT22 than at ZT10, and this difference was attenuated in the genetic absence of Rev-erbα (Fig. 4D). Similarly, MED1 was also evicted at engaged sites but not in livers lacking Rev-erbα (Fig. 4E). The opposition of Rev-erbα to the circadian binding of BRD4 and MED1 was confirmed by ChIP-qPCR in both the genetic absence (Fig. 4F–G) and ectopic expression of Rev-erbα (Fig. S6A–C). Focusing on Rev-erbα binding sites where BRD4 is evicted at ZT10 (Fig. S6D) revealed the concurrent eviction of MED1 (Fig. S6E) and also independently predicted functional sites with circadian eRNA transcription (Fig. S6F). However, the binding of CTCF and RAD21 was low and not circadian at Rev-erbα binding sites (Fig. S6G–J).
Our findings demonstrate genome-wide organizational plasticity at the level of sub-TADs that occurs in a circadian manner as a component of normal mammalian physiology. The mechanisms by which Rev-erbα functionally opposes E-P loop formation, leading to circadian repression of gene transcription within sub-TADs, are likely applicable to other transcriptional repressors whose function in controlling chromatin architecture is currently not as well defined as for transcriptional activators.
Supplementary Material
Acknowledgments
We acknowledge Bin Fang, Dongyin Guan, Romeo Papazyan, Raymond Soccio, Yann Aubert and the Lazar laboratory for helpful discussion. We also thank Jennifer Phillips-Cremins (University of Pennsylvania) and Peng Huang and Chris Edwards (Childrens’ Hospital of Philadelphia) for technical advice. ChIP-seq and Hi-C data have been deposited in the Gene Expression Omnibus (GEO) (GSE104129). We thank the Functional Genomics Core of the Penn Diabetes Research Center (P30 DK19525) and the Penn Epigenetics Sequencing Core for next-generation sequencing, and the Viral Vector Core of the Penn Diabetes Research Center (P30 DK19525) for recombinant virus preparation. This work was supported by NIH R01 DK45586 (MAL), the JPB Foundation (MAL), NIH R01 DK106027 (KJW), NIH T32GM007170 (YHK), NIH T32 GM008216 (YHK), and NIH F30 DK112507 (YHK).
References
- 1.Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science. 2016;354:994–998. doi: 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2016;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng D, et al. A Circadian rhythm orchestrated by histone deacetylase 3 controls Hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–54. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vollmers C, et al. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 2012;16:833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang B, et al. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014;159:1140–1152. doi: 10.1016/j.cell.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Zhao H, et al. PARP1- and CTCF-Mediated Interactions between Active and Repressed Chromatin at the Lamina Promote Oscillating Transcription. Mol Cell. 2015:984–997. doi: 10.1016/j.molcel.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 11.Aguilar-Arnal L, et al. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat Struct Mol Biol. 2013;20:1206–13. doi: 10.1038/nsmb.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, et al. Long-Range Chromosome Interactions Mediated by Cohesin Shape Circadian Gene Expression. PLoS Genet. 2016;12:1–26. doi: 10.1371/journal.pgen.1005992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieberman-aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao SSP, Huntley MH, Durand NC, Stamenova EK. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips-Cremins JE, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–95. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowen JM, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siersbæk R, et al. Dynamic Rewiring of Promoter-Anchored Chromatin Loops during Adipocyte Differentiation. Mol Cell. 2017;66:420–435. doi: 10.1016/j.molcel.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Tan-Wong SM, et al. Gene loops enhance transcriptional directionality. Science. 2012;338:671–675. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grosso AR, De Almeida SF, Braga J, De Almeida F, Carmo-fonseca M. Dynamic transitions in RNA polymerase II density profiles during transcription termination Dynamic transitions in RNA polymerase II density profiles during transcription termination. Genome Res. 2012;22:1447–1456. doi: 10.1101/gr.138057.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Preitner N, et al. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science. 2015;348:1488–92. doi: 10.1126/science.aab3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harding HP, Lazar MA. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol Cell Biol. 1995;15:4791–4802. doi: 10.1128/mcb.15.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin L, Lazar MA. The Orphan Nuclear Receptor Rev-erbα Recruits the N-CoR/Histone Deacetylase 3 Corepressor to Regulate the Circadian Bmal1 Gene: Molecular Endocrinology: Vol 19, No 6. Mol Endocrinol. 2005;19:1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 24.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Mol Cell. 2015;58:1028–1039. doi: 10.1016/j.molcel.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon KJ, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 27.Jiang YW, et al. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 30.Bhagwat AS, et al. BET Bromodomain Inhibition Releases the Mediator Complex from Select cis-Regulatory Elements. Cell Rep. 2016;15:519–530. doi: 10.1016/j.celrep.2016.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.