Abstract
The Tat machinery catalyzes the transport of folded proteins across the cytoplasmic membrane in bacteria and the thylakoid membrane in plants. Transport occurs only in the presence of an electric field (Δψ) and/or a pH (ΔpH) gradient, and thus, Tat transport is considered to be dependent on the proton motive force (pmf). This presents a fundamental and major challenge, namely, that the Tat system catalyzes the movement of large folded protein cargos across a membrane without collapse of ion gradients. Current models argue that the active translocon assembles de novo for each cargo transported, thus providing an effective gating mechanism to minimize ion leakage. A limited structural understanding of the intermediates occurring during transport and the role of the pmf in stabilizing and/or driving this process have hindered the development of more detailed models. A fundamental question that remains unanswered is whether the pmf is actually ‘consumed’, providing an energetic driving force for transport, or alternatively, whether its presence is instead necessary to provide the appropriate environment for the translocon components to become active. Including addressing this issue in greater detail, we explore a series of additional questions that challenge current models, and, hopefully, motivate future work.
Keywords: twin-arginine translocation, secretion, protein export, proton motive force, signal peptide, precursor protein
New findings about the Tat protein transport system as well as basic structural and energetic features that remain unresolved are discussed.
ABBREVIATIONS
- Tat
twin arginine translocation
- Δψ
electric field gradient
- ΔpH
pH gradient
- pmf
proton motive force
INTRODUCTION
In prokaryotes and plants, proteins are transported out of the cytoplasm or into the thylakoid lumen via two general translocation systems: the Sec and Tat machineries. The Sec system transports unfolded proteins, whereas the Tat (twin arginine translocation) pathway transports folded proteins (Berks 1996; Clark and Theg 1997; Santini et al.1998; Sargent et al.1998; Palmer and Berks 2012; Cline 2015). The distribution of protein cargos between the two pathways varies considerably. For example, >90% of proteins are exported by the Tat pathway in Haloarchaea (Rose et al.2002; Dilks, Gimenez and Pohlschroder 2005), whereas a functional Tat system is not required for growth of Escherichia coli and other γ-proteobacteria under most conditions (Lee, Tullman-Ercek and Georgiou 2006). The diversity in Tat usage is also seen within a phylum, for example, ∼20% of Streptomyces coelicolor secreted proteins use the Tat pathway but Mycobacteria leprae have <10 Tat substrates (Dilks et al.2003). Initial analyses of Tat substrates revealed that many are co-factor containing redox proteins and/or oligomeric complexes. Thus, the Tat pathway enables co-factors, such as molybdopterins or metal centers, to be integrated into proteins during folding in the cytoplasm prior to transport (Palmer and Berks 2012). For oligomeric Tat substrates, only one protein in the complex requires a signal peptide, but the oligomeric structure must be achieved in the cytoplasm and is retained during Tat transport (‘hitchhiker’ transport) (Rodrigue et al.1999). Considering that not all Tat substrates have co-factors or form heterooligomers (Berks, Palmer and Sargent 2005), and moreover, that a folded structure is not essential for transport (Richter et al.2007), it remains uncertain what selective pressures favor the Tat pathway over the Sec system.
Mechanistically, the Tat machinery is unique in that it transports folded proteins across energetic membranes without collapsing ion gradients (Mould and Robinson 1991; Cline, Ettinger and Theg 1992; Clark and Theg 1997; Santini et al.1998; Sargent et al.1998; Alder and Theg 2003). It is the only known protein transport system for which a proton motive force (pmf) is essential for all substrates (Cline, Ettinger and Theg 1992; Bageshwar and Musser 2007; Braun, Davis and Theg 2007). This minireview addresses recent advances made towards understanding the Tat transport mechanism, critically re-examines some basic assumptions and discusses some currently unresolved issues. Recent detailed reviews on Tat transport are available, and the reader is referred to these for more comprehensive summaries (Palmer and Berks 2012; Berks 2015; Cline 2015). Here, we have taken a forward-looking approach, concentrating on unknowns and challenges to motivate future work.
Tat COMPONENTS
A minimal Tat system contains the two membrane proteins: TatA and TatC. This two protein system is common in certain gram-positive bacteria with low G + C content, such as Bacillus, and in some archaea (Jongbloed et al.2004; Jongbloed, van der Ploeg and van Dijl 2006). However, the best-studied Tat systems are found in E. coli and in plant thylakoids, both of which contain TatB (Hcf106 in plants), a TatA-like membrane protein, in addition to TatA and TatC (Tha4 and cpTatC, respectively, in plants). Escherichia coli also contain a fourth protein, TatE, which is homologous to TatA and TatB (Sargent et al.1998, 1999; Eimer et al.2015). The presence of multiple TatA-like proteins is considered to be a consequence of gene duplication followed by sequence divergence, resulting in specialized functions (Yen et al.2002). Here, we focus on the E. coli TatABCE system and the corresponding chloroplast Tat (cpTat) system.
THE RECEPTOR COMPLEX
TatB and TatC form a 1:1 heterodimer (Cline and Mori 2001; McDevitt et al.2006) that forms the basis of a receptor complex, which recognizes and binds Tat signal peptides (Fig. 1). Whether TatA is a normal constituent of this receptor complex has been controversial, though the weight of evidence leans toward some TatA in this complex (Mangels et al.2005; Aldridge et al.2014; Blummel et al.2015; Alcock et al.2016). Since TatABC forms a minimal Tat transport system (Sargent et al.1998; Yahr and Wickner 2001), these three Tat proteins have been used for most bacterial transport studies. However, a recent report finding that TatA, TatB and TatE occupy very similar positions on TatC (Eimer et al.2018) raises the possibility that TatE is also part of the receptor complex, or that TatA takes the place of TatE when the latter is absent. It further suggests that inclusion of TatE in bacterial transport studies is important for deciphering the precise role of the various Tat proteins. Due to the uncertainty in receptor complex composition, Tat(AE)BC will be used to designate this complex.
Figure 1.
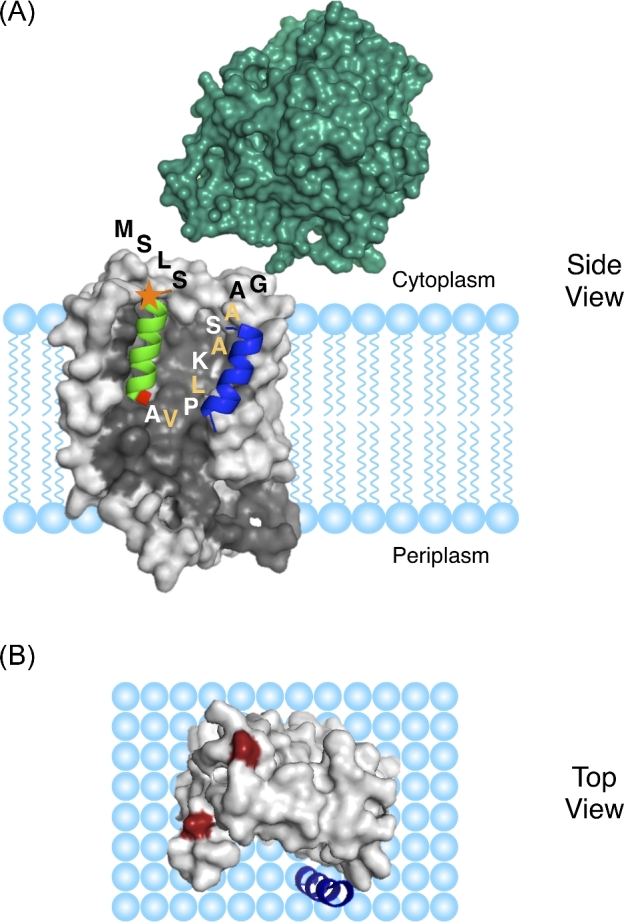
The receptor–substrate interaction. (A) Model of the interaction of the native substrate pre-SufI with TatBC. A signal peptide hairpin penetrates about halfway across the membrane in a deep groove on the side of TatC (gray; SWISS-MODEL ID: P69423) with both RR-motif (orange star) and mature domain (teal; PDB ID: 2UXV) on the cytoplasmic side of the membrane. Modeling suggests that the first part of the hairpin is helical (green), and the second half is disordered and interacts mostly with the TatB membrane domain (blue; residues L7-G21, Zhang et al.2014). Adapted from Hamsanathan et al. (2017). (B) Top view of TatBC. TatC residues E15 and E103 (red) interact with the RR-motif on Tat signal peptides (Rollauer et al.2012).
TatC has six transmembrane (TM) helices and forms the core of the Tat(AE)BC receptor complex. X-ray structures for Aquifex aeolicus TatC reveal a glove-like shape with a deep groove (the ‘palm’ of the ‘glove’) on a membrane-spanning face that seems ideally suited to accommodate signal peptides (Rollauer et al.2012; Ramasamy et al.2013). TM5 of TatC interacts with the N-terminal membrane domain of TatB, which positions it just outside of the TatC groove (Kneuper et al.2012; Rollauer et al.2012; Zoufaly et al.2012; Aldridge et al.2014; Alcock et al.2016).
OLIGOMERIZATION OF THE RECEPTOR COMPLEX
The Tat(AE)BC receptor complex forms higher order oligomers. However, the oligomeric state remains under debate, with dimer, trimer, tetramer, heptamer and octamer models having been proposed (Mangels et al.2005; Tarry et al.2009; Celedon and Cline 2012; Aldridge et al.2014; Blummel et al.2015; Alcock et al.2016), though even a monomer may be sufficient for polypeptide translocation (Hamsanathan et al.2017). The oligomeric organization of the receptor complex is expected to provide clues about the location of bound signal peptides, which would provide insight into, or provide constraints on, the translocation mechanism. While the visibility of TatB and TatC oligomers in live E. coli (Alcock et al.2013) suggests that the receptor oligomers are on the larger size, the number of Tat(AE)BC units in these clusters has not been established. Moreover, it has not been demonstrated that these clusters are where polypeptide translocation across the membrane occurs.
THE SIGNAL PEPTIDE BINDING SITE
Tat substrates interact with the Tat(AE)BC receptor complex via their signal peptides (Fig. 1), which are removed by signal peptidase after transport to release the folded mature domain (Luke et al.2009). The two arginine residues (RR-motif) (Berks 1996; Sargent et al.1998) within the highly conserved N-terminal RRXFLK consensus sequence likely interact with E15 and E105 (E. coli residues) of TatC (Rollauer et al.2012). Other than the RRXFLK consensus sequence, the primary structures of Tat signal peptides are highly variable, but nonetheless share a few common features. Instead of binding in a defined pocket in a distinct orientation, signal sequences are more likely accommodated in the receptor binding pocket in slightly different configurations. A hydrophobic helical domain (h-domain; typically 12–18 residues), following the RRXFLK motif, is generally thought to bind in the groove on the side of TatC (Ramasamy et al.2013; Hamsanathan et al.2017). The disordered C-terminal domain of signal peptides interacts with the membrane domain of TatB (Alami et al.2003; Gerard and Cline 2006; Maurer et al.2010; Hamsanathan et al.2017). Consistent with these structural interactions, the receptor complex plays an active role as an insertase, catalyzing the insertion of a signal peptide hairpin into the membrane (Fröbel et al.2012). While this initial report concluded that the signal peptide hairpin extends all the way across the membrane such that the signal peptide can be cleaved by signal peptidase (Fröbel et al.2012), a more recent study found that the signal peptide hairpin penetrates about halfway across the membrane (Hamsanathan et al.2017).
The finding that a signal peptide hairpin penetrates partway, rather than all the way, across the membrane solves multiple confounding issues. First, while membrane spanning peptides are typically helical, consistent with the predicted helical propensities of Tat h-domains, Tat signal peptides are generally too short to span the membrane twice, as is required for a fully penetrating hairpin, if part is helical. In contrast, a partially penetrating hairpin structure readily explains how the RR-motif and fully folded mature domain remain on the cis-side of the membrane in the receptor–substrate complex (Hamsanathan et al.2017). Second, a fully penetrating signal peptide hairpin is expected to be substantially more stable thermodynamically than a partially penetrating hairpin. Thus, reversibility of the receptor-substrate binding interaction (KD ∼ 10–20 nM) (Bageshwar et al.2009; Whitaker, Bageshwar and Musser 2012) is consistent with a partially penetrating hairpin. And third, it has been unclear whether precursor proteins continuously maintain specific interactions with the Tat translocon during translocation of the mature domain across the membrane, which would presumably enhance efficiency, or whether substrate–translocon interactions are transient and fluid during this stage of the turnover cycle. Flexibility at the tip of a signal peptide hairpin would allow the N-terminal portion of the hairpin (including the RR motif) to remain continuously bound to the receptor complex, while the C-terminal portion of the hairpin and the mature domain migrate across the membrane. While such continuously maintained signal peptide interactions can occur for a fully inserted signal peptide hairpin, migration of the signal peptide cleavage site across the membrane to yield a fully penetrating signal peptide hairpin requires partial unfolding of the mature domain, which does not occur, at least for pre-SufI (Hamsanathan et al.2017). These interactions and structural dynamics form the basis of the recently proposed Hairpin-Hinge model (Hamsanathan et al.2017) (Fig. 2). Notably, the membrane insertion of a signal peptide hairpin and the hairpin-hinge translocation mechanism are consistent with all of the receptor complex oligomerization models postulated thus far.
Figure 2.
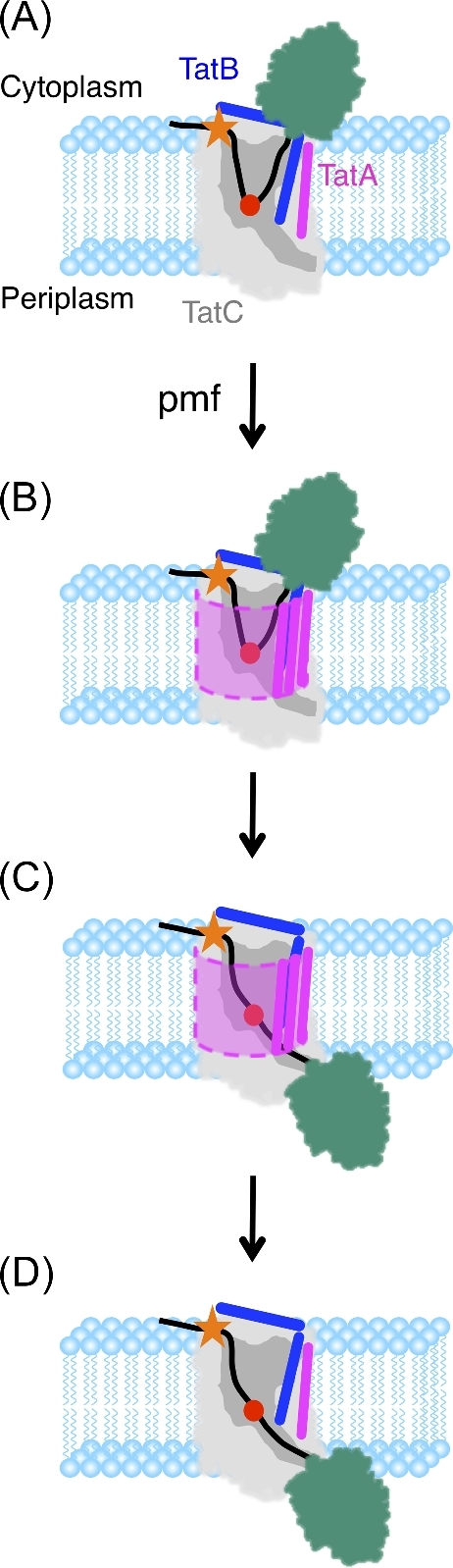
The Hairpin-Hinge model of Tat mature domain translocation. The Hairpin-Hinge model (Hamsanathan et al.2017) explains how the precursor protein remains continuously bound to the receptor complex while the C-terminal portion of the signal peptide hairpin and the mature domain migrate across the membrane. The membrane insertion of a signal peptide hairpin and the hairpin-hinge translocation mechanism are consistent with all of the receptor complex oligomerization models postulated thus far. For simplicity, a monomeric receptor complex is shown here. (A) The receptor complex inserts the signal peptide (black) of the precursor protein (mature domain in teal) into the membrane in a hairpin configuration that extends about halfway across the bilayer. Hairpin insertion is pmf-independent. The RR-motif (orange star) interacts with E15 and E103 (see Fig. 1) on the surface of TatC (gray) and the C-terminal end of the signal peptide after the hinge (red dot) interacts with TatB (dark blue) (see Fig. 1). (B) In the presence of a pmf, TatA (magenta) is recruited to the receptor–substrate complex, resulting in formation of a translocation conduit (dashed outline). (C) Unhinging of the signal peptide hairpin allows the mature domain to translocate through the pore across the membrane. (D) The translocation conduit disassembles after transport. The mature domain is released to the periplasm upon signal peptide cleavage.
OLIGOMERIZATION OF TatA WITH THE RECEPTOR–SUBSTRATE COMPLEX
Translocation of the folded mature domain of a Tat precursor protein requires a conduit across the membrane. TatA is an important, and likely primary, contributor to generating and maintaining this conduit. Early work on detergent-solubilized complexes revealed that TatA forms ring-like structures, and suggested that oligomerization between a receptor–substrate complex and a pre-existing TatA oligomer could therefore lead to transfer of the mature domain through a TatA pore (Gohlke et al.2005). However, this picture no longer appears valid. Instead, measurements within membranes indicate that TatA is highly dispersed, but oligomerizes in the presence of receptor–substrate complexes and the pmf (Dabney-Smith, Mori and Cline 2006; Alcock et al.2013; Rose et al.2013). While there are numerous competing models (Fig. 3), the current general consensus is that the Tat(AE)BC receptor complex and additional recruited TatA together generate the active translocon (termed herein as the TatAn(E)BC complex), which promotes mature domain transport. The oligomeric state of dispersed TatA is unclear. Though some reports have suggested tetrameric TatA protomers (Leake et al.2008; Dabney-Smith and Cline 2009), comparison with in vivo fluorescence intensities observed for tagged TatB and TatC proteins suggests that dispersed TatA has a lower oligomerization state (Alcock et al.2013).
Figure 3.
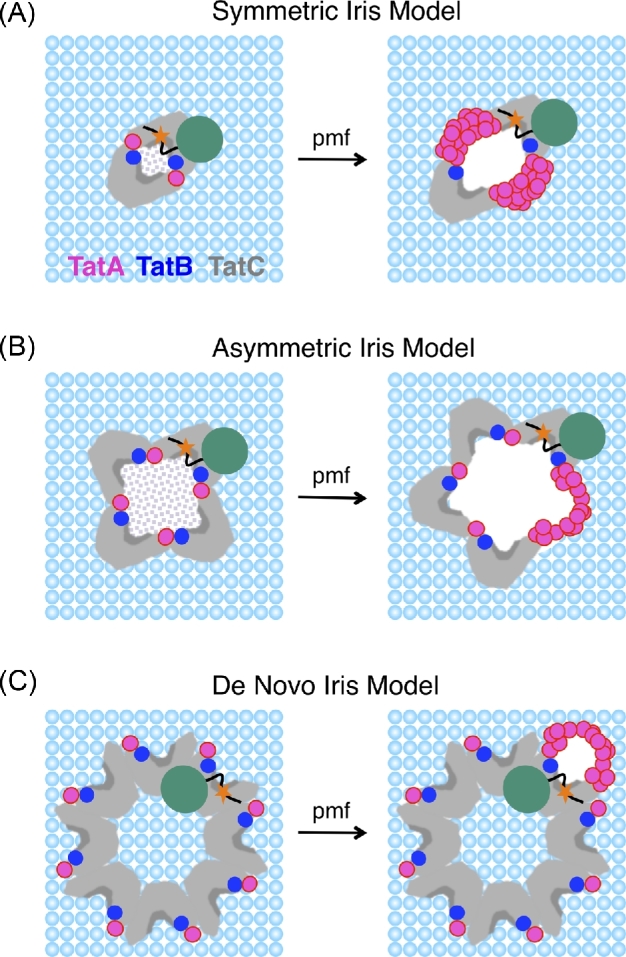
Models of the role of TatA in the Tat translocation mechanism. The number and arrangement of the TatA (magenta), TatB (dark blue) and TatC (gray) proteins in the receptor complex is unknown, though it is generally agreed that the signal peptide (black) of the precursor protein (orange star, RR-motif; teal, mature domain) binds in the groove on the side of TatC (see also Fig. 1). Three general types of current models are depicted. In all cases, the receptor complex oligomerization state shown is one of multiple reasonable possibilities, and TatA is recruited to the receptor–substrate complex in the presence of a pmf. Substantially different conformational rearrangements of the receptor complex and/or different TatA recruitment sites are feasible. The primary features distinguishing the different models is whether TatA is recruited to form one or multiple oligomeric structures and whether the signal peptide binding site is located within or outside of the receptor complex oligomer. (A) Symmetric iris model: this model assumes that the signal peptide binding pockets (dark gray) on TatC face inside the oligomer (face to face), and TatA is recruited symmetrically to the interfaces between protomers. A dimeric receptor arrangement is depicted here, as was originally postulated (Aldridge et al.2014), although a trimeric or tetrameric stoichiometry is also reasonable (Blummel et al.2015; Alcock et al.2016). (B) Asymmetric iris model: similar to the symmetric iris model, although in this case, the translocation conduit grows asymmetrically. While a tetrameric receptor arrangement is depicted here, either a tetrameric or trimeric stoichiometry was initially postulated (Alcock et al.2016), although a dimeric stochiometry is also reasonable. The central hole in the iris models (A and B) is likely blocked in some manner (indicated by the dotted shading) to prevent leakage before recruitment of additional TatA, most likely by the cytoplasmic domains of TatA and/or TatB in the receptor complex, or via lipids. (C) De novo iris model: for this model, the signal peptide binding sites of the receptor complex face outside, and recruited TatA forms a translocation conduit around the precursor bound to the receptor complex. An octomeric arrangement is shown here, based on biochemical evidence (Celedon and Cline 2012), though, in principle, any receptor complex oligomerization state is feasible.
TatA consists of an N-terminal TM domain, followed by an amphipathic helix, and then a highly charged, intrinsically disordered C-terminal tail (Hu et al.2010; Walther et al.2010). Though the monomeric unit is small (<100 residues) and relatively simply organized, the changes in TatA oligomerization state during Tat transport and the significantly different interactions and functional differences in these intermediates provide substantial structural challenges. First, TatA oligomerization requires the receptor complex, substrate and the pmf, and yet most interactions within the resulting oligomers are typically proposed to occur between TatA molecules. It is not clear what changes within TatA could be induced to trigger this self-assembly. And second, a translocation conduit is presumably an aqueous channel through the membrane, which, considering thermodynamics, is expected to be lined by a hydrophilic surface. The hydrophobic membrane domain of TatA seems incompatible with such a channel. These issues have made it challenging to postulate detailed structural models of the Tat translocation cycle.
MODELS OF THE TRANSLOCATION MECHANISM
Unlike most protein translocation pathways, the Tat proteins do not seem to generate a stable pre-existing translocation pore through which the precursor can migrate, but rather, a translocation conduit seems to assemble on demand. The dominant picture that has emerged is that the active translocon assembles from a receptor–substrate complex and TatA protomers in the presence of a pmf, and that this TatAn(E)BC complex disassembles after the mature domain is translocated across the membrane (Leake et al.2008; Alcock et al.2013). While both TatB and TatA have multiple interaction sites on TatC (Blummel et al.2015; Alcock et al.2016; Habersetzer et al.2017), the composition and structural organization of the Tat(AE)BC receptor complex and the active translocon are unknown, and this remains a fundamental barrier to understanding the translocation mechanism. Importantly, it has not been established whether the signal peptide binding pocket is on the inside or outside of the receptor complex oligomer. If inside, the receptor oligomer likely dilates (e.g. by recruitment of TatA) to accommodate mature domain passage through the middle of the complex, or, alternatively, a major structural rearrangement is necessary to expose the signal peptide binding pocket toward the lipids so that a channel generated by TatA recruitment can be accessed. In contrast, if the signal peptide binding pocket is on the outside of the Tat(AE)BC oligomer, a major conformational rearrangement of the receptor complex is not necessary. Rather than discussing proposed models in detail, we instead present the three general classes of current models. While there are numerous potential variations, the fundamental differences between the models that we seek to illustrate here lie in two features: (i) whether TatA forms one or multiple homooligomeric structures when it is recruited by and appended on the Tat(AE)BC oligomer; and (ii) whether the signal peptide binding site is located within or outside of the receptor complex oligomer.
Symmetric Iris Models
In symmetric iris models, the signal peptide binding site is on the inside of a Tat(AE)BC oligomer. Recruitment of additional TatA to multiple identical sites within the Tat(AE)BC oligomer would enable the central opening in the middle of the complex to swell into a pore with a diameter sufficient for accommodating the mature domain during transport across the membrane (Fig. 3A). A dimeric oligomerization model was postulated (Aldridge et al.2014), although trimeric (Blummel et al.2015; Alcock et al.2016) or tetrameric (Blummel et al.2015; Alcock et al.2016) stoichiometries, in principle, are also reasonable. The pore diameter expansion generated by recruitment of TatA could provide an effective gating mechanism for preventing ion leakage.
Asymmetric Iris Models
Asymmetric iris models are similar to the symmetric iris models, except that additional TatA would be recruited to a single interface between Tat(AE)BC protomers. Thus, the Tat proteins would be asymmetrically distributed around the expanded pore that develops (Fig. 3B). Trimeric and tetrameric Tat(AE)BC stoichiometries have been proposed (Alcock et al.2016), although a dimer or larger receptor oligomers are formally possible.
De Novo Iris Models
In de novo iris models, the signal peptide binding site is on the outside of the Tat(AE)BC complex, and recruitment of additional TatA around this binding site creates the necessary translocation pore. Thus, recruitment of TatA to the receptor–substrate complex would create an entirely new pore where there was none before (Fig. 3C). The Tat(AE)BC oligmerization state is not critical here—monomers up to octomers and larger would all be reasonable (Bageshwar et al.2009; Hamsanathan et al.2017).
Two features of de novo iris models make them particularly attractive. First, the only available receptor–substrate structure suggests a heptameric complex of TatBC with the natural substrate SufI (and presumably the signal peptide binding site) on the outside of this complex (Tarry et al.2009). And second, an external signal peptide binding site enables easy access for precursor bound to the membrane surface (Bageshwar et al.2009; Hamsanathan et al.2017). However, numerous crosslinking studies (Punginelli et al.2007; Ma and Cline 2010; Ma and Cline 2013; Aldridge et al.2014; Blummel et al.2015) and evolutionary covariance data (Alcock et al.2016) point toward a receptor complex with the signal peptide binding site on the interior of the oligomer, and thus, at this juncture, such models are favored.
THE NATURE OF THE TRANSLOCATION CONDUIT
The nature of the translocation conduit remains an intriguing question. Most current models assume that the translocation conduit across the membrane is generated by some combination of the various Tat proteins arranged to generate a proteinaceous wall of an aqueous pore. There are multiple difficulties with this assumption. First, there are no unambiguous structural data including all three proteins as well as a substrate undergoing transport. And second, the lack of hydrophilic residues within membrane domains suggests extreme thermodynamic instability of an aqueous pore. One possibility is that these issues simply point toward the conclusion that the translocation conduit is transient and highly unstable, and therefore it disassembles after each translocation event. However, a potential consequence of a high-energy intermediate is a low efficiency, or low flux, translocation pathway. Two alternate conceptual models seek to address these issues.
The ‘charge zipper’ model postulates that the amphipathic helix and the C-terminal densely charged region of TatA fully insert into the membrane as a salt-bridge-stabilized hairpin. Such TatA hairpins are predicted to contribute to a self-assembled pore with a hydrophilic interior surface, as expected for an aqueous translocation conduit (Walther et al.2013). Though charge-inversion substitutions do not support the charge zipper model (Aldridge et al.2012; Alcock et al.2017), these data are, in fact, indirect tests, as it must be assumed that the tested charge pairs do indeed form a salt bridge and that this is required to form the postulated TatA inserted hairpin, and, in addition, the involved residues do not play any alternate role, which would be abrogated by charge inversion. Since current data do not explicitly establish whether a TatA hairpin does or does not form, or whether TatA inserts into the membrane at any point in the translocation cycle, the substantial topological rearrangement of TatA in the ‘charge zipper’ model cannot be ruled out at this time. Nonetheless, multiple studies argue against TatA hairpin insertion based on the accessibility of specific residues to chemical reagents (Aldridge et al.2012; Koch et al.2012). However, it is not clear that such methods are capable of detecting the transient intermediate proposed in the charged zipper model (which would likely exist briefly, to minimize leakage). More importantly, the purpose of the proposed TatA hairpin insertion in the charged zipper model is to generate a pore, which can significantly alter the accessibility of inserted residues to aqueous chemical reagents.
The ‘membrane weakening’ model hypothesizes that the recruitment of TatA reduces the ability of the lipids to seal around the translocation system, thus enabling the mature domain to penetrate across the membrane (Bruser and Sanders 2003; Hou and Bruser 2011). The fundamental concept introduced by this model is that there is an enhanced local membrane permeability introduced by an increase in the local TatA concentration. What this implies, for example, is that the TatA molecules do not need to have strongly adherent interfaces, as found in stable membrane-spanning pore structures, but rather that their interactions can be many, weak and transient, but sufficient for clustering. Exactly what physical mechanism could disrupt the membrane seal has not been clarified, though a recent report suggests that the amphipathic helix of TatA and the Tat(AE)BC oligomer work together to modulate the insertion depth of the TatA TM domain, and hence, the strength of the membrane weakening (Hou et al.2018). Alternatively, a process similar to that used by antimicrobial peptides (Yang et al.2001; Sengupta et al.2008; Ebenhan et al.2014) is reasonable, particularly for those TatA molecules not directly contacting Tat(AE)BC, suggesting that a significant or substantial fraction of the edges of the translocation conduit could be lipid. Lipid bilayers with a sufficient percentage of positive curvature lipids can form spontaneous pores (Karatekin et al.2003; Rodriguez, Cribier and Pincet 2006; Hamai, Cremer and Musser 2007), and TatA may be designed to promote and/or stabilize such structures. Note that the membrane weakening hypothesis and pore models are not completely distinct pictures. For example, the lateral escape of lipids from within an assembling pore before closure (Hamsanathan et al.2017) would fundamentally be a consequence of membrane weakening.
WHY IS THE PMF REQUIRED FOR PROTEIN TRANSLOCATION?
After transport, precursor proteins are converted into mature proteins on the trans side of the membrane due to signal peptide cleavage. Consequently, the precursor protein concentration is higher on the cis side of the membrane (the precursor concentration tends toward 0 on the trans side). Therefore, precursor proteins migrate from high concentration to low concentration. As only precursor proteins, not mature proteins, are substrates for the Tat translocation machinery (mature proteins cannot bind to the Tat translocon and migrate backwards across the membrane), the transport process becomes irreversible upon signal peptide cleavage. Thus, the fundamental overall process catalyzed by the Tat machinery, moving a folded mature protein domain (or protein complex, in the case of ‘hitchhiker’ transport) across a membrane, is energetically downhill (precursor protein moving from high concentration to low concentration). Though uncleavable precursor proteins are transport competent (Maurer et al.2010; Hamsanathan et al.2017), it has not been established whether such proteins can be transported against a concentration gradient (in vitro transport efficiencies are typically <50%). Why, then, is a pmf required for Tat-dependent protein translocation? The common implicit assumption is that there is at least one step in the translocation process that is thermodynamically unfavorable, and thus, it requires an input (consumption) of energy (thermodynamic control). An alternate possibility is that the pmf is required for a translocation step, but that energy consumption does not actually occur (kinetic control). Considering that there are multiple pmf-dependent steps (Bageshwar and Musser 2007), both of these potential roles for the pmf may, in fact, hold true. The following sections address these issues in more detail and pose a series of fundamental open questions and conundrums about the Tat transport process, primarily focusing on the role of the pmf, which reveal that our understanding of the physical mechanisms involved is still quite poor.
DOES Tat TRANSPORT THROUGH THE PORE REQUIRE THE CONSUMPTION OF ENERGY?
For Tat transport to consume energy stored in the pmf, ions must flow down their electrochemical gradient. If these ions are protons, they must move across the membrane in the direction opposite to that in which the mature domain moves. How could this occur? Considering the vast size difference between the protein cargos and protons, and their different transport directions, one reasonable explanation is that protons and cargo travel through distinct, independently controlled channels. These translocation events would not need to occur simultaneously. Alternately, protons and cargo could migrate through the same channel, but in opposite directions. In this case, it is unclear if the proton and cargo transport processes could be independently controllable and, more importantly, how direct coupling between proton and cargo movement could be distinguished from proton leakage through an open channel. For either situation, there do not seem to be a sufficient number of strictly conserved residues in the Tat proteins with pKa’s suitable for forming a proton wire across the membrane (Hicks et al.2003; Walther et al.2010; Rollauer et al.2012; Ramasamy et al.2013; Alcock et al.2017).
Both components of the pmf, the electric field (Δψ) and pH (ΔpH) gradients, store energy (Silverstein 1993), which can potentially be coupled to Tat protein transport. The most detailed studies of the energetics of the Tat transport process have been carried out using the cpTat system (Cline, Ettinger and Theg 1992; Finazzi et al.2003; Braun, Davis and Theg 2007). For the OE17 cargo, which requires a ΔpH ≥ 1, ∼8 × 104 protons cross the thylakoid membrane per cargo translocated (Alder and Theg 2003), establishing that the translocation of a large number of protons occurs concomitant with the translocation of cargo, and providing an upper limit for the energy input required for transport. However, it has not been determined whether proton movement is necessary for the protein translocation event (energetic driving force) or whether such proton movement is a consequence of an open channel during cargo translocation (leakage through the translocation channel). While a leak analysis performed in the presence of various transport incompetent mutant precursor proteins indicated a steady-state proton leak rate of ∼10% of that observed in the presence of wild-type precursor protein (Alder and Theg 2003), the leak rate when a precursor protein is within the translocation channel undergoing active translocation has not been established. Thus, while the presence of a pmf is clearly required for Tat transport, it has not been established whether transport requires the consumption of the energy stored within the pmf.
One might argue that surely the movement of a large folded protein through a hole in a membrane bilayer is a complex energy intensive process. In fact, the opposite argument can be readily made. Diffusion on the nanoscale is rapid—most proteins move around the cell over many micrometers in seconds (diffusion constants of ∼1–10 μm2/s; (Milo, Phillips and Orme 2016)). Therefore, once an open channel exists, why should migration through a 5-nm long channel (approximate thickness of the membrane bilayer) be slow or require energy? Taking an example from a system where the translocation times of folded proteins are known, single molecule measurements have revealed that the time required for energy-independent passage though the nuclear pore complex is <10 ms for numerous cargos (Tu and Musser 2011). The nuclear pore is about an order of magnitude longer than a membrane bilayer thickness (∼50 nm vs ∼5 nm), and its interior is highly crowded (Fu et al.2017; Konishi et al.2017; Sakiyama, Panatala and Lim 2017). The single molecule nuclear transport data indicate, therefore, that the thermal fluctuations driving diffusional motion are sufficient for translocation through a pore in the bilayer on the millisecond timescale. Note that the free energy available within a protein's concentration gradient does not determine whether a single molecule transports in either direction though a pore. Rather, the concentration gradient determines the frequency with which translocation can occur in both directions. That is, translocations from the higher concentration to the lower concentration occur more frequently (leading to a higher net rate) simply because more molecules impinge on the pore from the higher concentration side (Kopito and Elbaum 2007)—the molecular movement through the pore itself is determined by diffusional constraints. In short, there is no apparent theoretical biophysical requirement that the transport of a Tat cargo through an open pore must be coupled to energy input. Notably and critically important, however, the opening (creation) and/or closing (destruction) of a pore (gating) can certainly require, and for many systems certainly does require, the input of energy.
WHY DOES TatA OLIGOMERIZATION WITH Tat(AE)BC REQUIRE THE PMF?
The recruitment of TatA molecules to the Tat(AE)BC–substrate complex requires a pmf (Dabney-Smith, Mori and Cline 2006; Alcock et al.2013; Rose et al.2013). As far as we know, it has not been proposed that the pmf is consumed to drive this oligomerization process (e.g. by proton transfer across the membrane), nor are we aware of a potential mechanism. What, then, might be the function of the pmf? It is well accepted that a membrane protein may not retain its native conformation when it is removed from its native lipid environment (e.g. upon extraction with detergent or reconstituted into non-native membranes). For proteins embedded in energetic membranes, the pmf can also be considered to be part of their native environment. It stands to reason, then, that the native, fully functional conformation of a protein in an energetic membrane may only be achieved in the presence of a pmf. The conformational rearrangements of voltage-gated ion channels provide ample examples of significant pmf-driven conformational changes (Tombola, Pathak and Isacoff 2006; Bezanilla 2008; Catterall, Wisedchaisri and Zheng 2017). There are numerous potential mechanisms whereby conformations or interactions could be altered by the pmf: (i) charged amino acid side chains and polypeptide segments can move in response to changes in the Δψ or local pH; (ii) α-helices can reorient in response to the Δψ due to their inherent dipole moments (which are approximately equivalent to ± 0.5 charges separated by the length of the helix), a process that can also be modulated by (de)protonation; (iii) ions within a proteinaceous channel can redistribute in response to a pmf changes and thereby induce a conformational change; and (iv) the Δψ can directly influence pKa values, resulting in (de)protonation events that promote conformational rearrangements (Sengupta et al.2005; Bezanilla 2008). The finding that various charge mutants of TatA inhibit disassembly of TatABC oligomers when the pmf is collapsed (Alcock et al.2017) supports the picture that changes in the protonation state of charged residues directly influences the thermodynamics of TatA oligomerization. However, an alternate possibility that the mutated residues are involved in pmf-independent interactions, such as those required for the structural stability of an intermediate.
DO THE CHLOROPLAST AND BACTERIAL Tat SYSTEMS USE THE PMF IN THE SAME WAY?
Parsimony suggests that the Tat systems from different species require the pmf for the same reason(s). However, there is some evidence suggesting that the chloroplast and bacterial Tat systems may be influenced by the pmf in different ways. Both the Δψ and ΔpH increase cpTat transport efficiency (Alder and Theg 2003; Braun, Davis and Theg 2007). In contrast, a ΔpH alone does not promote bacterial Tat transport (Bageshwar and Musser 2007), arguing against a role for the ΔpH in promoting transport. However, since the ΔpH is significantly lower in bacteria (≤1 vs ≥3 in plants) (Quintanilha and Mehlhorn 1978; Krulwich, Sachs and Padan 2011), it may be that the thermodynamic threshold required for transport (Alder and Theg 2003) was not achieved in these experiments. More importantly, bacterial Tat transport efficiency can be increased by a collapse of the ΔpH, presumably due to a measurable increase in the Δψ (Bageshwar and Musser 2007). While it could be argued that this finding supports the concept of interchangeability of the two components of the pmf, as is expected for a pmf-driven machine (Wiedenmann, Dimroth and von Ballmoos 2008; Soga et al.2012), the fact that transport became more efficient suggests that the additional thermodynamic driving force generated by the increase in Δψ was significantly higher than the thermodynamic loss from the collapse of the ΔpH. It is unclear how this latter point could be true under the chemiosmotic hypothesis, which assumes that the thermodynamic loss from one gradient will be compensated in the other (Mitchell 2011). Alternatively, if bacterial Tat transport were driven solely by the Δψ, an increase in Δψ resulting from the collapse of the ΔpH would result in a straightforward increase in transport efficiency. While this picture argues that the bacterial Tat system requires only the Δψ and not the ΔpH gradient, we will continue to generalize and consider Tat transport as pmf dependent for simplicity.
HOW MANY TRANSPORT SUBSTEPS ARE INFLUENCED BY THE PMF?
The Tat transport process can be broken down into the following seven steps: (1) binding of a precursor protein to the Tat(AE)BC receptor complex (possibly precluded by a lipid-bound precursor intermediate (Musser and Theg 2000; Bageshwar et al.2009)); (2) recruitment of TatA to the receptor–substrate complex; (3) gating the channel open; (4) translocation of the mature domain through the channel; (5) gating the channel closed; (6) disassembly of the TatAn(E)BC complex; and 7) cleavage of the signal peptide. The gating steps (3 and 5) may or may not be distinct intermediates, particularly if rapid assembly and disassembly of TatA is the primary mechanism to prevent ion leakage across the membrane. However, when disassembly of TatA from the TatAn(E)BC complex is inhibited by mutation of one or more charged residues in TatA, the cells survive (Alcock et al.2017), suggesting the absence of a permanent leak, and therefore, the presence of some gating mechanism.
How many of these seven steps are influenced by the pmf? For the bacterial system, at least two distinct steps require a Δψ—a short-duration, relatively high magnitude Δψ is required early in the transport process and a long-lived, low magnitude Δψ is needed for a later step (Bageshwar and Musser 2007). Surprisingly, efficient transport was observed when the precursor protein was added after collapse of the short-duration, high magnitude Δψ (Bageshwar and Musser 2007; Whitaker, Bageshwar and Musser 2012), indicating that the precursor protein does not need to be present during the intermediate state that makes use of this Δψ. The most straightforward conclusion is that something that occurs before step 1, the precursor binding step, is the step influenced by the high magnitude Δψ. For example, a conformational change or assembly step could be induced by the Δψ before the precursor binding interaction. Energy recovered from this Δψ could be stored (e.g. via high energy conformational structures) and then used in one or more later steps. Such an energy storage mechanism undoubtedly precludes utilizing the flow of a large number of ions to drive translocation events. The fundamentally important consequence of these findings and conclusions is that steps 2–7 (as outlined earlier) require no further energetic input other than a low magnitude Δψ (<∼50 mV, below the detection limit) (Bageshwar and Musser 2007). Since it is well established that step 2 requires a pmf (Dabney-Smith, Mori and Cline 2006; Alcock et al.2013; Rose et al.2013), and the second Δψ-requiring step seems most likely to occur before step 1, steps 3–7 could in fact be pmf-independent. This possibility does not necessarily conflict with the finding that tens of thousands of protons cross the membrane during translocation in the chloroplast system (Alder and Theg 2003) since, as discussed earlier, it was not determined whether this results from leakage or whether these protons are necessary to drive translocation. Not ruled out, then, is that a third pmf-dependent step may exist within steps 3–7.
ARE THERE ‘RESTING’ AND ‘ACTIVE’ CONFORMATIONS?
Utilization of a high magnitude Δψ by the Tat transport system without the precursor protein being present (Bageshwar and Musser 2007) is an intriguing finding. How can this be explained? One possibility is that the pmf induces a conformational change within one or more Tat proteins (or a Tat oligomer), hence converting a ‘resting’ configuration into an ‘active’ configuration (Hamsanathan et al.2017). For example, the Tat(AE)BC receptor complex may be in a resting (inactive) configuration in the absence of a pmf. A high magnitude Δψ could induce the Tat(AE)BC receptor complex to convert into an active configuration. In such a model, both resting and active conformations may bind precursor proteins (possibly with different affinities), but only the active form would be competent to recruit additional TatA. This model is consistent with the earlier argument that the pmf is an essential environmental requirement for at least some proteins in energetic membranes to achieve their native structure. Intriguingly, the effect of a high magnitude Δψ on transport is observed many minutes after its collapse (Bageshwar and Musser 2007), suggesting a slow conversion from an active to an inactive configuration. In short, it may only be feasible to understand the true nature of the Tat translocation system under conditions of continuous pmf, which more accurately resembles the in vivo conditions (Takizawa et al.2007; Krulwich, Sachs and Padan 2011).
DO TatABC OLIGOMERS DISASSEMBLE AND REASSEMBLE FOR EACH CARGO TRANSPORTED?
In contrast to the rapid (tens of milliseconds) conformational cycling of voltage-gated ion channels, which respond to frequent large fluctuations in the Δψ (Hoop and Peng 2000), the Tat machinery is expected to experience a steady-state pmf (Takizawa et al.2007; Krulwich, Sachs and Padan 2011), and thus, conformational changes during turnover are not expected to be driven by fluctuations in the pmf. This conclusion suggests that the disassembly of TatAn(E)BC oligomers that occurs upon collapsing the pmf is a useful experimental tool, but it is unlikely to reflect conditions normally experienced during the translocation process. It is therefore important to question how accurately this disassembly process reflects what occurs during turnover. According to most current models, the TatAn(E)BC oligomer disassembles after mature domain translocation even in the presence of a pmf, but this has not been demonstrated experimentally. More precisely, and of fundamental importance, it is not known whether full (or even partial) disassembly is essential before the next precursor molecule binds to the receptor complex. Thus, it is possible that multiple precursor proteins can bind to and be transported successively by the same TatAn(E)BC oligomer with little or no disassembly/reassemble between cycles. Based on quantitative mass spectrometry of 10 Tat substrates, an E. coli cell must transport over 6000 molecules via the Tat system (Soufi et al.2015). Considering a doubling time of ∼20 min (Sezonov, Joseleau-Petit and D’Ari 2007) and ∼5 Tat translocons/cell (Alcock et al.2013), these values predict that each translocon must transport a cargo protein in <∼1 s. It is uncertain whether this time is sufficient for a complete cycle of TatA and Tat(AE)BC assembly and disassembly. Note that while the number of translocons/cell is likely an underestimate, as entire E. coli cells were not visible in 2D images (Alcock et al.2013), this is potentially compensated by an underestimate of the number of Tat cargos transported per cell division, as there are ∼28 Tat substrates in E. coli (Berks, Palmer and Sargent 2003) (http://www.lifesci.dundee.ac.uk/groups/tracy_palmer/docs/signals.htm). More importantly, since numerous Tat substrates are required for anaerobic respiration (Berks, Palmer and Sargent 2005; Lee, Tullman-Ercek and Georgiou 2006), the total number of Tat cargos is most likely higher under anoxic conditions (Jack et al.2001) compared with the aerobic conditions of the cells analyzed by mass spectrometry (Soufi et al.2015), suggesting an even higher capacity of the Tat system, which has similar expression levels (within a factor of two) under aerobic and anaerobic conditions (Jack et al.2001).
OUTLOOK
The Tat transport system solves a seemingly complex problem, namely, transporting a large folded protein through a membrane without the collapse of ion gradients. Current models propose a series of complex conformational changes and rearrangements that are linked in a poorly defined manner to the required pmf. In this minireview, we have presented a series of questions that challenge numerous basic assumptions with the goal of motivating future efforts to obtain more refined answers. As a guiding principle, we note that complex models and explanations can always be generated; however, nature tends toward simplicity in mechanism and rules. So, as a final question, we ask, is Tat transport as complicated as it seems, or are we missing some basic simplifying principles?
Acknowledgements
We thank Burkely Pettijohn and Umesh Bageshwar for critical comments on the manuscript.
FUNDING
This research was supported by the National Institutes of Health [R01GM116995 to SMM]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest. None declared.
REFERENCES
- Alami M, Luke I, Deitermann S et al. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol Cell 2003;12:937–46. [DOI] [PubMed] [Google Scholar]
- Alcock F, Baker MAB, Green NP et al. Live cell imaging shows reversible assembly of the TatA component of the twin-arginine protein transport system. Proc Natl Acad Sci USA 2013;110:E3650–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock F, Damen MP, Levring J et al. In vivo experiments do not support the charge zipper model for Tat translocase assembly. Elife 2017;6:e30127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock F, Stansfeld PJ, Basit H et al. Assembling the Tat protein translocase. Elife 2016;5:e20718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder NN, Theg SM. Energetics of protein transport across biological membranes. a study of the thylakoid DeltapH-dependent/cpTat pathway Cell 2003;112:231–42. [DOI] [PubMed] [Google Scholar]
- Aldridge C, Ma X, Gerard F et al. Substrate-gated docking of pore subunit Tha4 in the TatC cavity initiates Tat translocase assembly. J Cell Biol 2014;205:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge C, Storm A, Cline K et al. The chloroplast twin arginine transport (Tat) component, Tha4, undergoes conformational changes leading to Tat protein transport. J Biol Chem 2012;287:34752–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bageshwar UK, Musser SM. Two electrical potential-dependent steps are required for transport by the Escherichia coli Tat machinery. J Cell Biol 2007;179:87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bageshwar UK, Whitaker N, Liang FC et al. Interconvertibility of lipid- and translocon-bound forms of the bacterial Tat precursor pre-SufI. Mol Microbiol 2009;74:209–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berks BC. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol 1996;22:393–404. [DOI] [PubMed] [Google Scholar]
- Berks BC. The twin-arginine protein translocation pathway. Annu Rev Biochem 2015;84:843–64. [DOI] [PubMed] [Google Scholar]
- Berks BC, Palmer T, Sargent F. The Tat protein translocation pahway and its role in microbial physiology. Adv Microbiol Physiol 2003;47:187–254. [DOI] [PubMed] [Google Scholar]
- Berks BC, Palmer T, Sargent F. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr Opin Microbiol 2005;8:174–81. [DOI] [PubMed] [Google Scholar]
- Bezanilla F. How membrane proteins sense voltage. Nat Rev Mol Cell Bio 2008;9:323–32. [DOI] [PubMed] [Google Scholar]
- Blummel AS, Haag LA, Eimer E et al. Initial assembly steps of a translocase for folded proteins. Nat Commun 2015;6:7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun NA, Davis AW, Theg SM. The chloroplast Tat pathway utilizes the transmembrane electric potential as an energy source. Biophys J 2007;93:1993–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruser T, Sanders C. An alternative model of the twin arginine translocation system. Microbiol Res 2003;158:7–17. [DOI] [PubMed] [Google Scholar]
- Catterall WA, Wisedchaisri G, Zheng N. The chemical basis for electrical signaling. Nat Chem Biol 2017;13:455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celedon JM, Cline K. Stoichiometry for binding and transport by the twin arginine translocation system. J Cell Biol 2012;197:523–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SA, Theg SM. A folded protein can be transported across the chloroplast envelope and thylakoid membranes. Mol Biol Cell 1997;8:923–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K. Mechanistic aspects of folded protein transport by the twin arginine translocase (Tat). J Biol Chem 2015;290:16530–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Ettinger WF, Theg SM. Protein-specific energy requirements for protein transport across or into thylakoid membranes. Two lumenal proteins are transported in the absence of ATP. J Biol Chem 1992;267:2688–96. [PubMed] [Google Scholar]
- Cline K, Mori H. Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J Cell Biol 2001;154:719–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabney-Smith C, Cline K. Clustering of C-terminal stromal domains of Tha4 homo-oligomers during translocation by the Tat protein transport system. Mol Biol Cell 2009;20:2060–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabney-Smith C, Mori H, Cline K. Oligomers of Tha4 organize at the thylakoid Tat translocase during protein transport. J Biol Chem 2006;281:5476–83. [DOI] [PubMed] [Google Scholar]
- Dilks K, Gimenez MI, Pohlschroder M. Genetic and biochemical analysis of the twin-arginine translocation pathway in halophilic archaea. J Bacteriol 2005;187:8104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilks K, Rose RW, Hartmann E et al. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J Bacteriol 2003;185:1478–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebenhan T, Gheysens O, Kruger HG et al. Antimicrobial peptides: their role as infection-selective tracers for molecular imaging. Biomed Res Int 2014;2014:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimer E, Frobel J, Blummel AS et al. TatE as a regular constituent of bacterial twin-arginine protein translocases. J Biol Chem 2015;290:29281–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimer E, Kao WC, Frobel J et al. Unanticipated functional diversity among the TatA-type components of the Tat protein translocase. Sci Rep 2018;8:1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finazzi G, Chasen C, Wollman FA et al. Thylakoid targeting of Tat passenger proteins shows no DeltapH dependence in vivo. EMBO J 2003;22:807–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröbel J, Rose P, Lausberg F et al. Transmembrane insertion of twin-arginine signal peptides is driven by TatC and regulated by TatB. Nat Commun 2012;3:1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G, Tu LC, Zilman A et al. Investigating molecular crowding within nuclear pores using polarization-PALM. Elife 2017;6:e28716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard F, Cline K. Efficient twin arginine translocation (Tat) pathway transport of a precursor protein covalently anchored to its initial cpTatC binding site. J Biol Chem 2006;281:6130–5. [DOI] [PubMed] [Google Scholar]
- Gohlke U, Pullan L, McDevitt CA et al. The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc Natl Acad Sci USA 2005;102:10482–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habersetzer J, Moore K, Cherry J et al. Substrate-triggered position switching of TatA and TatB during Tat transport in Escherichia coli. Open Biol 2017;7:170091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamai C, Cremer PS, Musser SM. Single giant vesicle rupture events reveal multiple mechanisms of glass-supported bilayer formation. Biophys J 2007;92:1988–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamsanathan S, Anthonymuthu TS, Bageshwar UK et al. A hinged signal peptide hairpin enables Tat-dependent protein translocation. Biophys J 2017;113:2650–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks MG, de Leeuw E, Porcelli I et al. The Escherichia coli twin-arginine translocase: conserved residues of TatA and TatB family components involved in protein transport. FEBS Lett 2003;539:61–7. [DOI] [PubMed] [Google Scholar]
- Hoop B, Peng CK. Fluctuations and fractal noise in biological membranes. J Membr Biol 2000;177:177–85. [DOI] [PubMed] [Google Scholar]
- Hou B, Bruser T. The Tat-dependent protein translocation pathway. Biomol Concepts 2011;2:507–23. [DOI] [PubMed] [Google Scholar]
- Hou B, Heidrich ES, Mehner-Breitfeld D et al. The TatA component of the twin-arginine translocation system locally weakens the cytoplasmic membrane of Escherichia coli upon protein substrate binding. J Biol Chem 2018, DOI: 10.1074/jbc.RA118.002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhao E, Li H et al. Solution NMR structure of the TatA component of the twin-arginine protein transport system from gram-positive bacterium Bacillus subtilis. J Am Chem Soc 2010;132:15942–4. [DOI] [PubMed] [Google Scholar]
- Jack RL, Sargent F, Berks BC et al. Constitutive expression of Escherichia coli tat genes indicates an important role for the twin-arginine translocase during aerobic and anaerobic growth. J Bacteriol 2001;183:1801–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongbloed JD, Grieger U, Antelmann H et al. Two minimal Tat translocases in Bacillus. Mol Microbiol 2004;54:1319–25. [DOI] [PubMed] [Google Scholar]
- Jongbloed JD, van der Ploeg R, van Dijl JM. Bifunctional TatA subunits in minimal Tat protein translocases. Trends Microbiol 2006;14:2–4. [DOI] [PubMed] [Google Scholar]
- Karatekin E, Sandre O, Guitouni H et al. Cascades of transient pores in giant vesicles: line tension and transport. Biophys J 2003;84:1734–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneuper H, Maldonado B, Jager F et al. Molecular dissection of TatC defines critical regions essential for protein transport and a TatB-TatC contact site. Mol Microbiol 2012;85:945–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S, Fritsch MJ, Buchanan G et al. Escherichia coli TatA and TatB proteins have N-out, C-in topology in intact cells. J Biol Chem 2012;287:14420–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi HA, Asai S, Watanabe TM et al. In vivo analysis of protein crowding within the nuclear pore complex in interphase and mitosis. Sci Rep 2017;7:5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RB, Elbaum M. Reversibility in nucleocytoplasmic transport. Proc Natl Acad Sci USA 2007;104:12743–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 2011;9:330–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leake MC, Greene NP, Godun RM et al. Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc Natl Acad Sci USA 2008;105:15376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PA, Tullman-Ercek D, Georgiou G. The bacterial twin-arginine translocation pathway. Annu Rev Microbiol 2006;60:373–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke I, Handford JI, Palmer T et al. Proteolytic processing of Escherichia coli twin-arginine signal peptides by LepB. Arch Microbiol 2009;191:919–25. [DOI] [PubMed] [Google Scholar]
- Ma X, Cline K. Multiple precursor proteins bind individual Tat receptor complexes and are collectively transported. EMBO J 2010;29:1477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Cline K. Mapping the signal peptide binding and oligomer contact sites of the core subunit of the pea twin arginine protein translocase. Plant Cell 2013;25:999–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt CA, Buchanan G, Sargent F et al. Subunit composition and in vivo substrate-binding characteristics of Escherichia coli Tat protein complexes expessed at native levels. FEBS J 2006;273:5656–68. [DOI] [PubMed] [Google Scholar]
- Mangels D, Mathers J, Bolhuis A et al. The core TatABC complex of the twin-arginine translocase in Escherichia coli: TatC drives assembly whereas TatA is essential for stability. J Mol Biol 2005;345:415–23. [DOI] [PubMed] [Google Scholar]
- Maurer C, Panahandeh S, Jungkamp A-C et al. TatB functions as an oligomeric binding site for folded Tat precursor proteins. MBoC 2010;21:4151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Phillips R, Orme N. Cell Biology by the Numbers. New York, NY; Abingdon: Garland Science, Taylor & Francis Group, 2016. [Google Scholar]
- Mitchell P. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. 1966. Biochim Biophys Acta 2011;1807:1507–38. [DOI] [PubMed] [Google Scholar]
- Mould RM, Robinson C. A proton gradient is required for the transport of two lumenal oxygen-evolving proteins across the thylakoid membrane. J Biol Chem 1991;266:12189–93. [PubMed] [Google Scholar]
- Musser SM, Theg SM. Characterization of the early steps of OE17 precursor transport by the thylakoid ΔpH/Tat machinery. Eur J Biochem 2000;267:2588–98. [DOI] [PubMed] [Google Scholar]
- Palmer T, Berks BC. The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol 2012;10:483–96. [DOI] [PubMed] [Google Scholar]
- Punginelli C, Maldonado B, Grahl S et al. Cysteine scanning mutagenesis and topological mapping of the Escherichia coli twin-arginine translocase TatC Component. J Bacteriol 2007;189:5482–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilha AT, Mehlhorn RJ. pH gradients across thylakoid membranes measured with a spin-labeled amine. FEBS Lett 1978;91:104–8. [Google Scholar]
- Ramasamy S, Abrol R, Siuloway CJM et al. The glove-like structure of the conserved membrane protein TatC provides insight into signal sequence recognition in twin-arginine translocation. Structure 2013;21:777–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Lindenstrauss U, Lucke C et al. Functional Tat transport of unstructured, small, hydrophilic proteins. J Biol Chem 2007;282:33257–64. [DOI] [PubMed] [Google Scholar]
- Rodrigue A, Chanal A, Beck K et al. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial tat pathway. J Biol Chem 1999;274:13223–8. [DOI] [PubMed] [Google Scholar]
- Rodriguez N, Cribier S, Pincet F. Transition from long- to short-lived transient pores in giant vesicles in an aqueous medium. Phys Rev E 2006;74:061902. [DOI] [PubMed] [Google Scholar]
- Rollauer SE, Tarry MJ, Graham JE et al. Structure of the TatC core of the twin-arginine protein transport system. Nature 2012;492:210–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose P, Frobel J, Graumann PL et al. Substrate-dependent assembly of the Tat translocase as observed in live Escherichia coli cells. PLoS One 2013;8:e69488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose RW, Bruser T, Kissinger JC et al. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol Microbiol 2002;45:943–50. [DOI] [PubMed] [Google Scholar]
- Sakiyama Y, Panatala R, Lim RYH. Structural dynamics of the nuclear pore complex. Semin Cell Dev Biol 2017;68:27–33. [DOI] [PubMed] [Google Scholar]
- Santini CL, Ize B, Chanal A et al. A novel sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J 1998;17:101–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent F, Bogsch EG, Stanley NR et al. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J 1998;17:3640–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent F, Stanley NR, Berks BC et al. Sec-independent protein translocation in Escherichia coli. A distinct and pivotal role for the TatB protein. J Biol Chem 1999;274:36073–82. [DOI] [PubMed] [Google Scholar]
- Sengupta D, Behera RN, Smith JC et al. The alpha helix dipole: screened out? Structure 2005;13:849–55. [DOI] [PubMed] [Google Scholar]
- Sengupta D, Leontiadou H, Mark AE et al. Toroidal pores formed by antimicrobial peptides show significant disorder. Biochim Biophys Acta 2008;1778:2308–17. [DOI] [PubMed] [Google Scholar]
- Sezonov G, Joseleau-Petit D, D’Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol 2007;189:8746–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein TP. Transmembrane measurements across bioenergetic membranes. Biochim Biophys Acta 1993;1183:1–3. [Google Scholar]
- Soga N, Kinosita K Jr, Yoshida M et al. Kinetic equivalence of transmembrane pH and electrical potential differences in ATP synthesis. J Biol Chem 2012;287:9633–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi B, Krug K, Harst A et al. Characterization of the E. coli proteome and its modifications during growth and ethanol stress. Front Microbiol 2015;6:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa K, Cruz JA, Kanazawa A et al. The thylakoid proton motive force in vivo. Quantitative, non-invasive probes, energetics, and regulatory consequences of light-induced pmf. Biochim Biophys Acta 2007;1767:1233–44. [DOI] [PubMed] [Google Scholar]
- Tarry MJ, Schafer E, Chen S et al. Structural analysis of substrate binding by the TatBC component of the twin-arginine protein transport system. Proc Natl Acad Sci USA 2009;106:13284–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola F, Pathak MM, Isacoff EY. How does voltage open an ion channel? Annu Rev Cell Dev Biol 2006;22:23–52. [DOI] [PubMed] [Google Scholar]
- Tu LC, Musser SM. Single molecule studies of nucleocytoplasmic transport. Biochim Biophys Acta 2011;1813:1607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TH, Gottselig C, Grage SL et al. Folding and self-assembly of the TatA translocation pore based on a charge zipper mechanism. Cell 2013;152:316–26. [DOI] [PubMed] [Google Scholar]
- Walther TH, Grage SL, Roth N et al. Membrane alignment of the pore-forming component TatA(d) of the twin-arginine translocase from Bacillus subtilis resolved by solid-state NMR spectroscopy. J Am Chem Soc 2010;132:15945–56. [DOI] [PubMed] [Google Scholar]
- Whitaker N, Bageshwar UK, Musser SM. Kinetics of precursor interactions with the bacterial Tat translocase detected by real-time FRET. J Biol Chem 2012;287:11252–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenmann A, Dimroth P, von Ballmoos C. Deltapsi and DeltapH are equivalent driving forces for proton transport through isolated F(0) complexes of ATP synthases. Biochim Biophys Acta 2008;1777:1301–10. [DOI] [PubMed] [Google Scholar]
- Yahr TL, Wickner WT. Functional reconstitution of bacterial Tat translocation in vitro. EMBO J 2001;20:2472–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Harroun TA, Weiss TM et al. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys J 2001;81:1475–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen MR, Tseng YH, Nguyen EH et al. Sequence and phylogenetic analyses of the twin-arginine targeting (Tat) protein export system. Arch Microbiol 2002;177:441–50. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang L, Hu Y et al. Solution structure of the TatB component of the twin-arginine translocation system. Biochim Biophys Acta 2014;1838:1881–8. [DOI] [PubMed] [Google Scholar]
- Zoufaly S, Frobel J, Rose P et al. Mapping precursor-binding site on TatC subunit of twin arginine-specific protein translocase by site-specific photo cross-linking. J Biol Chem 2012;287:13430–41. [DOI] [PMC free article] [PubMed] [Google Scholar]


