Abstract
Ultraconserved (UCEs) are popular markers for phylogenomic studies. They are relatively simple to collect from distantly-related organisms, and contain sufficient information to infer relationships at almost all taxonomic levels. Most studies of UCEs use partitioning to account for variation in rates and patterns of molecular evolution among sites, for example by estimating an independent model of molecular evolution for each UCE. However, rates and patterns of molecular evolution vary substantially within as well as between UCEs, suggesting that there may be opportunities to improve how UCEs are partitioned for phylogenetic inference. We propose and evaluate new partitioning methods for phylogenomic studies of UCEs: Sliding-Window Site Characteristics (SWSC), and UCE Site Position (UCESP). The first method uses site characteristics such as entropy, multinomial likelihood, and GC content to generate partitions that account for heterogeneity in rates and patterns of molecular evolution within each UCE. The second method groups together nucleotides that are found in similar physical locations within the UCEs. We examined the new methods with seven published data sets from a variety of taxa. We demonstrate the UCESP method generates partitions that are worse than other strategies used to partition UCE data sets (e.g., one partition per UCE). The SWSC method, particularly when based on site entropies, generates partitions that account for within-UCE heterogeneity and leads to large increases in the model fit. All of the methods, code, and data used in this study, are available from https://github.com/Tagliacollo/PartitionUCE. Simplified code for implementing the best method, the SWSC-EN, is available from https://github.com/Tagliacollo/PFinderUCE-SWSC-EN.
Keywords: phylogenomics, UCEs, partitioning, partitioning methods, sliding-window site characteristics, PartitionFinder
Introduction
Ultraconserved Elements (UCEs) are becoming one of the most popular DNA markers used for phylogenomic studies (Crawford et al. 2012; Faircloth et al. 2013, 2015; Jarvis et al. 2014; Meiklejohn et al. 2016). UCEs are highly conserved regions of DNA found throughout the genomes of many distant-related species. The functions of UCEs are not completely understood; however, their frequent proximity to transcriptional regulators or developmental genes has led to suggestions that they are directly involved in transcriptional regulation (Woolfe et al. 2005; Pennacchio et al. 2006). The conserved core regions of UCEs contain little variation, but the adjacent flanking regions contain more variation, and numerous studies have demonstrated that this variation is useful for inferring phylogenetic relationships between individuals, species, and higher clades (Crawford et al. 2012; Faircloth et al. 2012, 2015; Smith et al. 2014; Harrington et al. 2016; Moyle et al. 2016).
The accuracy of phylogenetic inferences often depends on choosing an appropriate model of molecular evolution, and many studies have demonstrated that accounting for variation in rates and patterns of evolution among sites is of primary importance (Shapiro et al. 2006; Li et al. 2008; Ho and Lanfear 2010; Lanfear et al. 2012, 2014). There are multiple methods that account for variation of molecular evolution among sites (Le et al. 2008; Zhang and Townsend 2009; Goremykin et al. 2010; Cummins and McInerney 2011; Soubrier et al. 2012). The simplest and most widely-used in phylogenomics are the partitioning methods, which rely on defining groups of sites that have evolved under similar conditions. Specifically, partitioning methods assume a priori that each group of sites has evolved under the same Markov model of DNA sequence evolution, but that different groups may have evolved under different models. Partitioning has been shown to improve estimates of topologies, branch lengths, and divergence dates (Lanfear et al. 2014; Kainer and Lanfear 2015; Hoff et al. 2016).
Most phylogenomic studies of UCE markers use partitioning to account for variation in rates and patterns of evolution across sites. Typically, researchers choose one of two strategies: either they assign all UCEs in the alignment to a single partition (e.g., Faircloth et al. 2015), or they assign each UCE to a separate partition (e.g., Faircloth et al. 2013). The former strategy makes the assumption that every site in the alignment has evolved under a common Markov process, which is inadvisable and has been shown to increase the risk of inferring unreliable phylogenetic trees (Kainer and Lanfear 2015). The latter strategy accounts for variation in rates and patterns of evolution between UCEs, but makes the assumption that all of the sites within each UCE have evolved under the same Markov process. Given that the rates of molecular evolution within each UCE are known to vary predictably, with near-zero variation in the conserved core of the UCE increasing to large amounts of variation at the edges, it seems sensible to ask whether it is possible to improve upon the assumption that all sites within each UCE have evolved under a single Markov model.
Our focus in this paper is on asking whether it is possible to estimate partitioning schemes for UCE data sets which improve upon the assumption that all sites in each UCE have evolved under a single Markov model. One way to approach this would be to use a recently-proposed k-means clustering approach which iteratively divides alignments into groups comprised of nucleotide sites sharing similar substitution rates (Frandsen et al. 2015). However, it has recently come to light that this approach systematically generates a partition comprised of all the invariant sites in the data set, which can subsequently mislead phylogenetic inference methods (Baca et al. 2017). We sought instead to develop approaches that avoid this problem, leverage the known molecular evolutionary patterns of UCEs, and allow us to assess whether it is better to group all sites in a UCE into a single partition, or to split each UCE into more than one group of sites.
This study proposes and evaluates two partitioning methods for phylogenomic studies of UCEs: UCE Site Position (UCESP) and Sliding-Window Site Characteristics (SWSC). The UCESP method groups nucleotide sites across UCEs using their physical location within the UCE, for example by grouping all of the central sites of each UCE into a single partition. The SWSC method uses proxies of rates and patterns of molecular evolution [e.g., entropy (EN), multinomial likelihoods (MUs), GC content] and a sliding-window approach to determine whether a central region of a UCE (which may loosely correspond to what is often called the ‘core’) evolves in a different way to the two flanking regions. This approach naturally splits each UCE into three data blocks corresponding to the underlying structure of these phylogenomic markers (i.e., conserved cores and more variable flanks).
Each of these methods conducts partitioning independently of estimating phylogenetic trees. One strength of these methods is their specificity to UCEs, whose use is increasing in phylogenomics. A key aim of phylogenetic model selection is to find models that capture the key biological features of the underlying data with a modest number of parameters (Kolaczkowski and Thornton 2004; Steel 2005). Here, we seek to achieve this by leveraging the known biological properties of UCEs themselves. In this respect, our approach is similar to biologically inspired models such as codon-based partitioning, the power of which in largely derived from building on the known biological properties of protein-coding DNA sequences.
New Approaches
Partitioning methods are based on the assumption that sites within a partition have evolved under similar conditions. We first sought to ascertain whether this is the case for UCEs by measuring three properties of each site in each UCE: the EN of a site, which can serve as a rough proxy for the rate of evolution of that site in the absence of a known phylogenetic tree; the GC content (GC) of a site; and the MU of a site, which describes the likelihood of observing a particular site pattern given the observed base frequencies of a particular UCE. Visualizations of the patterns of EN, GC, and MU with each of the 1000s of UCEs across seven diverse data sets (table 1, fig. 1) show that all three properties contain considerable and predictable variation within UCEs. EN and MU are low in the central region of the UCE and higher in the flanks, and the GC is high in the central region and lower in the flanks. Figure 1 shows that this variation can be very large—for example, in many cases GC content is <20% at the end of a UCE but >50% in the center.
Table 1.
Data Set Names and References, Clade Names, and Summary Information of the UCE Alignments Used to Evaluate the Two New Partitioning Methods.
| UCE data set | Clade (scientific name) | Clade (common name) | Size (bp) | UCEs | OTUs | GC% | References |
|---|---|---|---|---|---|---|---|
| Branstetter | Aculeata | Stinging wasps | 183747 | 807 | 187 | 46 | Branstetter et al. (2017) |
| Crawford | Sauria | Diapsids | 465241 | 1143 | 10 | 38 | Crawford et al. (2012) |
| Harrington | Pleuronectiformes | Flatfishes | 235232 | 999 | 55 | 44 | Harrington et al. (2016) |
| McCormack | Neoaves | Birds | 539526 | 1537 | 33 | 37 | McCormack et al. (2013) |
| Meiklejohn | Phasianidae | Gallopheasants | 599627 | 1479 | 18 | 43 | Meiklejohn et al. (2016) |
| Moyle | Oscines | Songbirds | 375172 | 515 | 106 | 40 | Moyle et al. (2016) |
| Prebus | Temnothorax | Acorn ants | 1561581 | 2098 | 50 | 44 | Prebus (2017) |
Fig. 1.
Rates and patterns of evolution vary within UCEs. These plots highlight the predictable and sometimes dramatic variation in rates and patterns of molecular evolution within UCEs. Each line in each plot represents a single UCE. Each row represents a single data set (see table 1), and each column represents a metric measured for each site in a UCE. EN denotes the EN of a site, GC denotes the GC content of a site, and MU denotes the multinomial likelihood of a site. A value of 0 on the X-axis represents the central site of each UCE. Values among neighboring sites in each UCE are smoothed using the geom_smooth() function of ggplot2.
Commonly-used models of molecular evolution (e.g., GTR models) can account for variation in rates of molecular evolution among sites within a partition using approximations such as the gamma distribution, proportion of invariant sites, and free-rates models. However, all commonly-used models of molecular evolution assume that the base frequencies (and thus GC) of all sites in a single partition are drawn from a single distribution. Large and predictable variation of GC within UCEs appears to be the rule rather than the exception (fig. 1). Together with the predictable variation in EN (fig. 1) this suggests that dividing UCEs up into more than one partition might improve models of molecular evolution by accounting for variation in GC and rates of molecular evolution among sites in ways that are not possible with standard un-partitioned Markov models.
We propose two new methods to automatically divide individual UCEs into multiple partitions based on their characteristic patterns of molecular evolution: SWSC, and UCESP. These two methods represent pragmatic approaches to potentially improve on treating each UCE as a single partition. The SWSC uses a sliding-window approach to divide each UCE into three data blocks. The choice of three data blocks is motivated by the observed patterns of variation in figure 1: all three measured site characteristics show that predictable variation focused on the center of each UCE. The SWSC is graphically summarized in figure 2. We describe its algorithm in detail in the Materials and Methods. The UCESP method takes a different approach, and groups nucleotides sharing similar locations in the UCEs into data blocks. In contrast to the SWSC, which divides the sites of each UCE into three data blocks, the UCESP places almost every site of a UCE into a different data block that contains similarly-positioned sites from almost every other UCE. This approach is motivated by the relatively predictable variation in the three measured site characteristics within UCEs and across diverse data sets (table 1, fig. 1). The UCESP is graphically summarized in figure 3. We describe its algorithm in detail in the Materials and Methods.
Fig. 2.
SWSC method for partitioning UCEs. Schematic diagrams illustrating major steps of the SWSC algorithm. This diagram includes three hypothetical alignments: UCE-1 (green), UCE-2 (yellow), and UCE-3 (blue). The alignments include the same patterns of molecular evolution (i.e., EN, GC, and MU rates) as seen in the UCE markers. Note the alignments are comprised of conserved cores and variable flanking regions. The SWSC algorithm includes three steps. First, it proposes all combinations of three-window models in the alignments. It delimitates windows by locating all conceivable pairs of nucleotide sites in the alignments. Second, it estimates site-wise molecular evolution and nucleotide proportions/counts across the alignments. We used three alternatives properties of UCEs: (1) EN (SWSC-EN), (2) GC content (SWSC-GC), and (3) MUs (SWSC-MU). See further details in the New Approaches section. Third, it calculates the SSE statistics across windows and sums them up to obtain the sum of SSEs for every three data block model. The SWSC algorithm selects the best models by minimizing the squared residuals among three windows. The diagrams illustrate the best three-window models as indicated by the SWSC-GC, SWSC-EN, and SWSC-MU, respectively. The vertical bars delimit the three data blocks. The windows (i.e., size of the data blocks) are indicated by the pair of numbers in parenthesis.
Fig. 3.
UCESP for partitioning UCE alignments. Diagram illustrating the second new method of proposing partitions using the locations of nucleotide sites among UCEs. This method relies on the assumption the UCE regions (e.g., cores and flanking regions) have evolved under similar evolutionary processes. See further details in the New Approaches section. This diagram includes three hypothetical alignments: UCE-1 (green), UCE-2 (yellow), and UCE-3 (blue). The position of nucleotide sites in the alignments are indicated by the numbers of each site. The UCESP algorithm includes two steps. First, define the central site of the UCE as site 0 (zero) and sites to the left of this site are labeled with negative numbers, and sites to the right of this site are labeled with positive numbers. Second, it is to create one data block for each label, by combining all the sites with the same label into a single data block. Note that there are nine partitions, all sharing the same locations of sites among the UCEs.
Both the SWSC and UCESP methods group together putatively similar sites into data blocks, but neither asks whether any of the resulting data blocks are similar to one another and so both risk over-partitioning the data. For this reason we use PartitionFinder 2 (Lanfear et al. 2016) to estimate optimal partitioning schemes by grouping together similar data blocks from the output of the SWSC and UCESP. Given the optimal partitioning scheme for each of the evaluated new method, we then define the best method for each data set as the one that results in the partitioning scheme with the best (i.e., lowest) AICc score.
Results
Visual Inspection of the Properties of UCEs
We measured three properties of UCE sites: the EN of a site, the GC content (GC) of a site, and the MU of a site. We observed that EN and MU are lower at the center of each UCE and higher at the edges, and that this pattern is reversed for GC (figure 1). Figure 1 also shows that the absolute values of EN, GC, and MU can vary substantially between data sets, as expected given that each data set represents a particular taxonomic clade sampled at a particular phylogenetic depth. For example, the mean EN varies roughly 6-fold from 0.03 in the gallopheasant data set (Meiklejohn et al. 2016) to 0.19 in the flatfish data set (Harrington et al. 2016), and the mean GC content varies from 37% in the bird data set to 44% in the stinging wasp data set (table 1).
The SWSC Algorithm Divides UCEs into Data Blocks That Reflect Site Properties
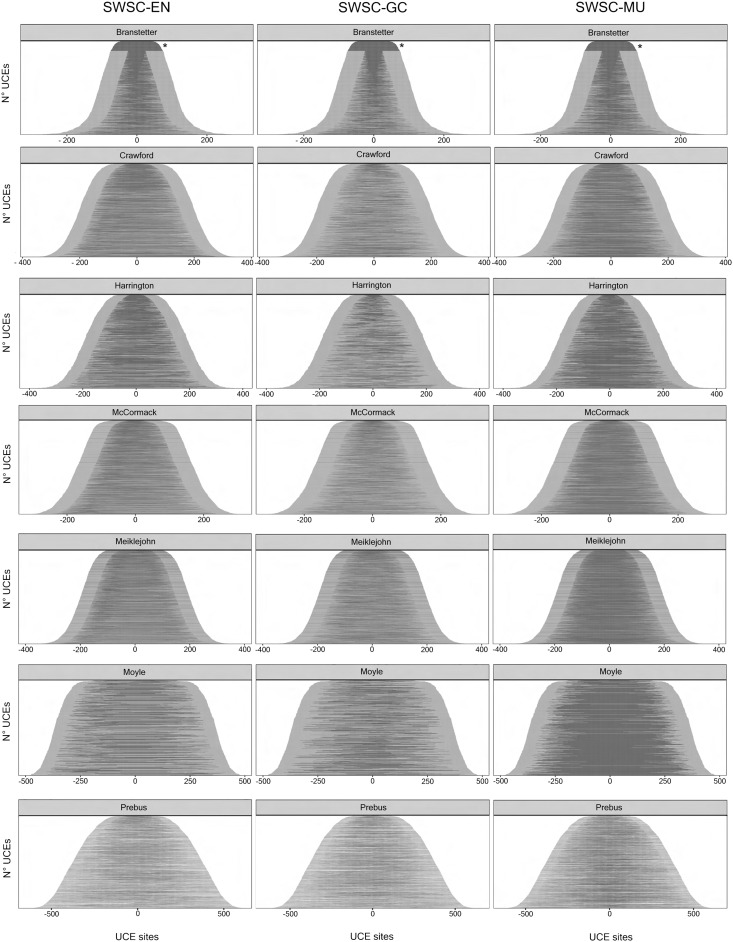
The SWSC partitioning method uses a site property (i.e., EN, GC, or MU) to split each UCE into three data blocks (fig. 4). As expected given our observations in figure 1, the EN and MU are lower in the central data blocks and higher in the edge data blocks, and GC shows the opposite pattern (table 2). In a small proportion of cases (∼16% or 910 UCEs across all seven data sets) a UCE could not be split into three data blocks because there was no solution that satisfied the criteria that each data block had to contain at least 50 sites, all four nucleotides, and at least one variable site (see Materials and Methods). Most of the UCEs set to single partitions were smaller than 150 bp and were found in the stinging wasp data set (table 1, fig. 4).
Fig. 4.
Best three data blocks for each UCE. Graphical representations of the best three data block for each UCE of each data set (see table 1) as indicated by the SWSC method. Each line in each plot represents a single UCE split in three data blocks. Black indicates the position of the central window, which may loosely correspond to what is often called the core region of UCEs, and gray indicates the position of the edge windows, which may loosely correspond to what is often called the flanking regions of UCEs. Each row of panels represents a single data set (see table 1), and each column of panels represents a metric measured for each site in a UCE. EN denotes the entropy of a site, GC denotes the GC content of a site, and MU denotes the multinomial likelihood of a site. UCEs are organized by increasing length. Asterisks (*) indicate UCEs smaller than 150 bp, which were not split into three data blocks but kept as a single data block (see details in the Materials and Methods: Sliding-Window Site Characteristics).
Table 2.
Means of EN and GC of Each Window and Data Set Produced by the SWSC Method.
| Branstetter | Crawford | Harrington | McCormack | Meiklejohn | Moyle | Prebus | Overall | |
|---|---|---|---|---|---|---|---|---|
| SWSC-EN | ||||||||
| Left | 0.828 | 0.212 | 0.278 | 0.097 | 0.055 | 0.140 | 0.182 | 0.256 |
| Center | 0.236 | 0.072 | 0.090 | 0.031 | 0.015 | 0.054 | 0.122 | 0.088 |
| Right | 0.831 | 0.145 | 0.267 | 0.059 | 0.036 | 0.090 | 0.181 | 0.230 |
| SWSC-GC | ||||||||
| Left | 32.91 | 31.39 | 40.12 | 28.42 | 36.10 | 33.61 | 36.58 | 34.16 |
| Center | 43.10 | 41.02 | 46.68 | 34.09 | 43.38 | 44.38 | 48.04 | 42.96 |
| Right | 32.24 | 30.99 | 40.47 | 28.75 | 36.61 | 33.5 | 36.94 | 34.21 |
The SWSC-EN Method Outperforms Other Approaches
The SWSC approach using site EN to derive data blocks (SWSC-EN) outperformed all other methods on all seven empirical data sets (table 3, fig. 5). We compared seven approaches to partitioning UCEs, comprising the new methods proposed here and three partitioning methods widely used in phylogenomic studies: (1) SWSC-EN; (2) SWSC-GC; (3) SWSC-MU; (4) UCESP; (5) Single (i.e., all UCEs treated as one partition); (6) UCE (i.e., each UCE treated as a partition); (7) and PF-UCE (i.e., a partitioning scheme estimated by defining each UCE as an initial data block and optimizing the partitioning scheme in PartitionFinder). We used PartitionFinder 2 (Lanfear et al. 2016) to optimize the partitioning schemes for methods (1)–-(4) and method (7), see Materials and Methods. In all cases, the SWSC-EN method received the lowest AICc score by a considerable margin. The closest AICc score to the SWSC-EN method for a single data set was more than 2,000 points higher, for the SWSC-MU approach on the Branstetter data set (table 3). The three standard approaches (Single, UCE, and PF-UCE) performed a great deal worse than all of the SWSC methods on all data sets. For example, the smallest difference between any of the three standard approaches and the SWSC-EN method was >60,000 AICc units for the Meiklejohn data set (table 3). The UCESP method performed poorly on all data sets, achieving the second-worst AICc scores in more than half of the seven data sets (table 3, fig. 5); that is worse only than treating the entire alignment as a single partition.
Table 3.
AICc Scores, Number of Subsets, and Parameters of Partitioning Schemes Inferred From Seven Partitioning Strategies Across Seven UCE Data Sets.
| Data set | Partitioning strategy | AICc | ΔAICc | Subsets | Parameters |
|---|---|---|---|---|---|
| Branstetter | Single | 17672426 | −336594 | 1 | 380 |
| UCESP | 17591017 | −255186 | 158 | 1950 | |
| UCE | 17581458 | −245627 | 807 | 8440 | |
| PF−UCE | 17575239 | −239408 | 475 | 5120 | |
| SWSC-GC | 17415498 | −79666 | 1018 | 10550 | |
| SWSC-MU | 17338096 | −2264 | 948 | 9850 | |
| SWSC-EN | 17335832 | 0 | 948 | 9850 | |
| Crawford | Single | 2481257 | −100299 | 1 | 26 |
| UCESP | 2458689 | −77731 | 85 | 866 | |
| UCE | 2467719 | −86761 | 1145 | 11466 | |
| PF-UCE | 2456063 | −75104 | 222 | 2236 | |
| SWSC-GC | 2410728 | −29770 | 456 | 4576 | |
| SWSC-MU | 2397171 | −16213 | 461 | 4626 | |
| SWSC-EN | 2380958 | 0 | 452 | 4536 | |
| Harrington | Single | 3833182 | −145228 | 1 | 116 |
| UCESP | 3800797 | −112843 | 114 | 1246 | |
| UCE | 3796629 | −108675 | 596 | 6066 | |
| PF-UCE | 3793471 | −105517 | 267 | 2776 | |
| SWSC-GC | 3736560 | −48606 | 513 | 5236 | |
| SWSC-MU | 3695067 | −7112 | 493 | 5036 | |
| SWSC-EN | 3687954 | 0 | 493 | 5036 | |
| McCormack | Single | 3192533 | −137223 | 1 | 72 |
| UCESP | 3166286 | −110976 | 101 | 1072 | |
| UCE | 3162976 | −107666 | 1541 | 15472 | |
| PF-UCE | 3147458 | −92148 | 318 | 3242 | |
| SWSC-GC | 3088673 | −33363 | 615 | 6212 | |
| SWSC-MU | 3094082 | −38772 | 638 | 6442 | |
| SWSC-EN | 3055310 | 0 | 586 | 5922 | |
| Meiklejohn | Single | 2339168 | −100584 | 1 | 42 |
| UCESP | 2314874 | −76290 | 97 | 1002 | |
| UCE | 2316404 | −77820 | 1479 | 14822 | |
| PF-UCE | 2299674 | −61091 | 249 | 2522 | |
| SWSC-GC | 2255398 | −16814 | 463 | 4662 | |
| SWSC-MU | 2254129 | −15545 | 471 | 4742 | |
| SWSC-EN | 2238583 | 0 | 458 | 4612 | |
| Moyle | Single | 6005672 | −230918 | 1 | 218 |
| UCESP | 5936152 | −161398 | 200 | 2208 | |
| UCE | 5937377 | −162623 | 515 | 5358 | |
| PF-UCE | 5935114 | −160361 | 271 | 2918 | |
| SWSC-GC | 5839663 | −64910 | 520 | 5408 | |
| SWSC-MU | 5820918 | −46165 | 496 | 5168 | |
| SWSC-EN | 5774753 | 0 | 496 | 5168 | |
| Prebus | Single | 20339794 | −625451 | 1 | 106 |
| UCESP | 20234314 | −519970 | 178 | 1876 | |
| UCE | 20173076 | −458732 | 2098 | 21076 | |
| PF-UCE | 20161619 | −447276 | 806 | 8156 | |
| SWSC-GC | 19835806 | −121462 | 1141 | 11506 | |
| SWSC-MU | 19766846 | −52503 | 1138 | 11476 | |
| SWSC-EN | 19714343 | 0 | 1125 | 11346 |
Note.—(1) Single: treating all sites as belonging to a single subset; (2) UCE: one subset for each UCE alignment; (3) PF-UCE: each UCE was defined as a data block; (4) SWSC-GC: data blocks defined by the SWSC-GC algorithm; (5) SWSC-EN: data blocks defined by the SWSC-EN algorithm; (6) SWSC-MU: data blocks defined by the SWSC-MU algorithm; and (7) UCESP: data blocks defined by the UCESP algorithm. In (3)–(7), the final partitioning scheme was optimized in PartitionFinder 2. Bold: best partitioning scheme.
Fig. 5.
The SWSC partitioning method leads to substantial improvements in model-fit. AICc scores of partitioning schemes derived from the SWSC outperform UCESP and other methods applied in phylogenomics. The plots show the AICc scores (Y-axis) of seven partitioning schemes (X-axis). The seven partitioning strategies were: (1) Single: treating all sites as belonging to a single subset; (2) UCE: one subset for each UCE alignment; (3) PF-UCE: each UCE was defined as a data block; (4) SWSC-GC: data blocks defined by the SWSC-GC algorithm; (5) SWSC-EN: data blocks defined by the SWSC-EN algorithm; (6) SWSC-MU: data blocks defined by the SWSC-MU algorithm; and (7) UCESP: data blocks defined by the UCESP algorithm. In (3)–(7), the final partitioning scheme was optimized in PartitionFinder 2. The AICc scores indicate that the SWSC, particularly the SWSC-EN, produces partitioning schemes that outperform all other partitioning strategies investigated here. Surprisingly, the partitioning schemes derived from the UCESP obtained AICc scores that were worse than partitioning schemes widely used in phylogenomics (i.e., UCE and UCE-PF).
Comparison of Observed and Shuffled UCE Alignments
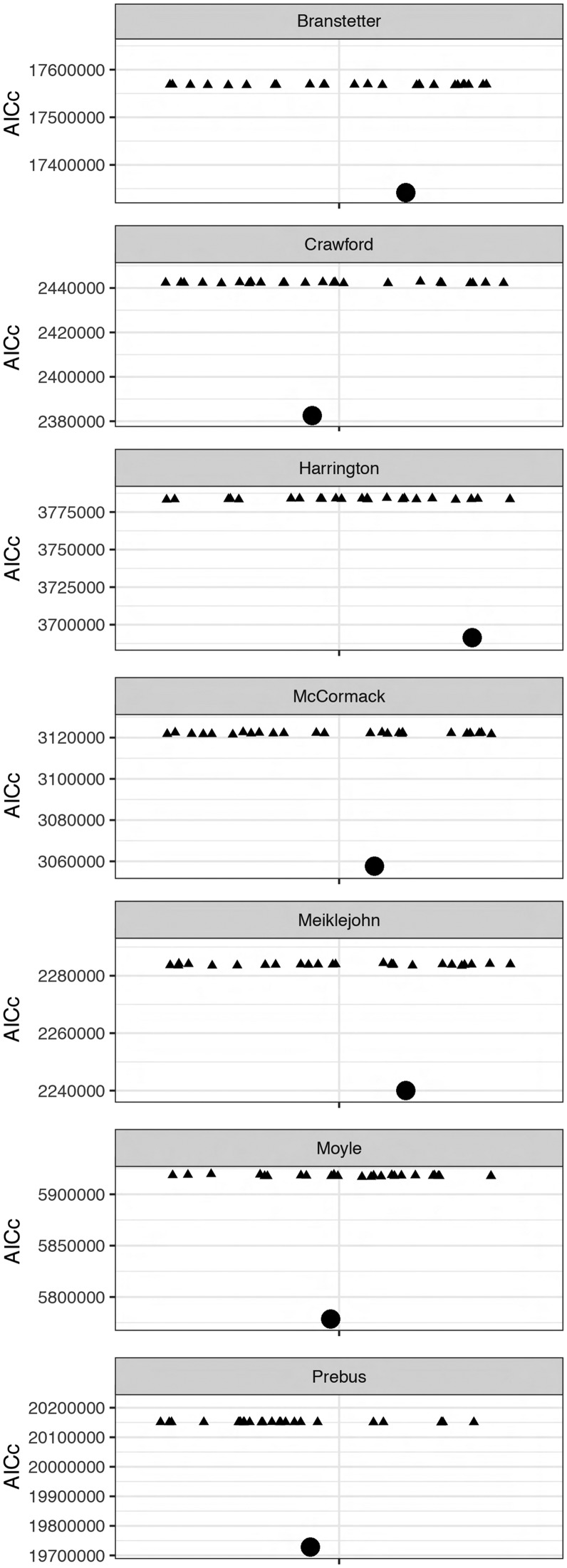
A potential concern with the SWSC and UCESP approaches is that they may achieve their improvement in AICc scores simply by defining a large number of starting subsets for PartitionFinder. This would define a larger search space of potential partitioning schemes for a data set, potentially accounting for the improved AICc scores. To assess this possibility, we created 25 permutations of all 7 data sets, in which we shuffled the sites within each UCE. We then re-ran all of the SWSC-EN analyses on the permuted alignments. This compares the observed SWSC-EN solution to one in which the assignment of sites to data blocks within each UCE is randomized, while other characteristics of the partitioning scheme such as the number and size of the starting subsets are held constant. Across all seven empirical data sets (table 1), and for all 25 permutations of the UCE alignments, the shuffled UCE alignments produced consistently much worse (i.e., higher) AICc scores than the un-shuffled alignments (fig. 6). This suggests that the improvement in AICc score seen in the SWSC-EN method is not due solely to having an increased number of starting subsets.
Fig. 6.
SWSC-EN split UCEs into meaningful partitions. Comparisons between the AICc scores of empirical data sets (circle) and 25 permuted data sets (triangles) of partitioning schemes derived from the SWSC-EN and optimized in PartitionFinder 2. For each of the randomizations, the sites within each UCE were shuffled and the new alignments were used with the original data block definitions from the un-shuffled data as input to PartitionFinder 2. Each panel includes a data set and 25 AICc scores of partitioning schemes optimized in PartitionFinder 2. The AICc scores suggest that the SWSC-EN method improves model-fit and parameter estimates not because it searches a larger space of possible partitioning schemes, but primarily because splitting each UCE into three data blocks is uncovering biological differences in rates and/or patterns of molecular evolution within each UCE.
Phylogenetic Inference
We estimated ML tree topologies for each of the seven data sets (table 1) under each of the seven partitioning schemes used in this study (i.e., SWSC-EN, SWSC-GC, SWSC-MU, UCESP, Single, UCE, PF-UCE). The resulting trees are presented in the Supplementary Material online. For each data set, we calculated the tree lengths of every tree, and the Path Difference (PD) between the SWSC-EN topology and all of the other topologies inferred under the other six partitioning schemes (table 4).
Table 4.
Measurements of TL (the Sum of All Branch Lengths in Substitutions Per Site) and PD of Each Topology Compared to the SWSC-EN Topology for Trees Estimated with the Seven Different Partitioning Schemes Evaluated in this Study.
| Partition strategy | TL | PD |
|---|---|---|
| Branstetter | ||
| Single | 1.081 | 89.5 |
| UCESP | 1.038 | 514.2 |
| UCE | 0.982 | 91.7 |
| PF-UCE | 1.052 | 80.7 |
| SWSC-GC | 1.006 | 112.2 |
| SWSC-MU | 1.005 | 170.5 |
| SWSC-ENa | 1.000 | 0.0 |
| Crawford | ||
| Single | 1.289 | 0.0 |
| UCESP | 1.252 | 0.0 |
| UCE | 1.213 | 0.0 |
| PF-UCE | 1.250 | 0.0 |
| SWSC-GC | 0.983 | 0.0 |
| SWSC-MU | 0.807 | 0.0 |
| SWSC-ENa | 1.000 | 0.0 |
| Harrington | ||
| Single | 1.141 | 0.0 |
| UCESP | 1.174 | 0.0 |
| UCE | 0.837 | 0.0 |
| PF-UCE | 0.958 | 0.0 |
| SWSC-GC | 0.903 | 0.0 |
| SWSC-MU | 1.028 | 0.0 |
| SWSC-ENa | 1.000 | 0.0 |
| McCormack | ||
| Single | 1.540 | 24.6 |
| UCESP | 1.437 | 28.4 |
| UCE | 1.388 | 25.5 |
| PF-UCE | 1.467 | 0.0 |
| SWSC-GC | 1.088 | 12.7 |
| SWSC-MU | 0.961 | 24.8 |
| SWSC-ENa | 1.000 | 0.0 |
| Meiklejohn | ||
| Single | 1.580 | 0.0 |
| UCESP | 1.506 | 0.0 |
| UCE | 1.523 | 0.0 |
| PF-UCE | 1.543 | 0.0 |
| SWSC-GC | 1.234 | 0.0 |
| SWSC-MU | 1.389 | 0.0 |
| SWSC-ENa | 1.000 | 0.0 |
| Moyle | ||
| Single | 0.973 | 117.5 |
| UCESP | 0.971 | 85.4 |
| UCE | 0.969 | 20.2 |
| PF-UCE | 0.970 | 112.8 |
| SWSC-GC | 0.957 | 99.8 |
| SWSC-MU | 0.941 | 27.1 |
| SWSC-ENa | 1.000 | 0.0 |
| Prebus | ||
| Single | 1.021 | 0.0 |
| UCESP | 1.010 | 23.7 |
| UCE | 1.010 | 0.0 |
| PF-UCE | 1.021 | 11.9 |
| SWSC-GC | 0.980 | 11.9 |
| SWSC-MU | 0.754 | 16.2 |
| SWSC-ENa | 1.000 | 0.0 |
Note.—To aid comparison of tree lengths, the tree lengths of all trees for a given data set are scaled such that the tree length of the tree estimated under the SWSC-EN partitioning scheme is equal to 1.0. Larger PD indicate larger differences between tree topologies, but they cannot be directly compared between data sets.
The tree topology estimated under the partitioning scheme obtained by the SWSC-EN was used as reference. (1) Single: treating all sites as belonging to a single subset; (2) UCE: one subset for each UCE alignment; (3) PF-UCE: each UCE was defined as a data block; (4) SWSC-GC: data blocks defined by the SWSC-GC algorithm; (5) SWSC-EN: data blocks defined by the SWSC-EN algorithm; (6) SWSC-MU: data blocks defined by the SWSC-MU algorithm; and (7) UCESP: data blocks defined by the UCESP algorithm. In (3)–(7), the final partitioning scheme was optimized in PartitionFinder 2.
Overall, tree lengths were similar among all seven partitioning schemes, and the SWSC-EN tree length showed no clear pattern of difference to the tree lengths from other partitioning schemes (table 4, fig. 7). The PDs show that the choice of partitioning scheme influenced the inferred phylogenetic tree in four out of seven data sets (table 4). Visual examination of the differences (see Supplementary Material online) reveals that most of the observed topological differences are associated with nodes with low bootstrap support (< 70, see Supplementary Material online), suggesting that the choice of partitioning scheme rarely influenced the resulting tree topology in ways that would affect biological inference. However, in at least one case, the SWSC-EN partitioning scheme led to a strongly-supported and biologically important change in the tree topology when compared to trees estimated with traditional approaches to partitioning. In this case, the phylogenetic position of the Sclerogibbidae, a family of wasps analyzed in the study of Branstetter et al. (2017), was sensitive to the partitioning scheme used. The original study Branstetter et al. (2017) and two of our analyses using standard partitioning schemes (i.e., a single partition, or one partition for each UCE) place the Sclerogibbidae together with a clade comprising the Embolemidae + Dryinidae, with the three families forming the sister group to the remaining nonchrysidoid lineages in the phylogeny [i.e., ((Sclerogibbidae, (Embolemidae, Dryinidae)), remainder)]. Bootstrap support for the former grouping varies considerably depending on the details of the analysis both in the original study (59–90% support) and in our analyses (52% support using a single partition, and 100% support using a partition for each UCE). In contrast, the SWSC-EN partitioning scheme places the Sclerogibbidae as sister to the remaining nonchrysidoid lineages, with the other two families as sister to this grouping [i.e., ((Embolemidae, Dryinidae), (Sclerogibbidae, remainder))], with 100% bootstrap support for both groupings. Notably, the same grouping was recovered with 100% bootstrap support in the original study when they explicitly accounted for variation among gene trees when estimating the species tree (Branstetter et al. 2017). It would be premature to suggest that any of these analyses are definitive, but it may be prudent to lean towards preferring the relationships supported by the model that best fits the data, which in the case of our analyses is the SWSC-EN partitioning scheme. These results show that on some occasions, the new methods we propose here may contribute to clarifying phylogenetic relationships.
Fig. 7.
Tree lengths (TL) of phylogenetic trees estimated under seven partitioning schemes and across seven data sets. To aid comparison of tree lengths, the tree lengths of all trees for a given data set are scaled such that the tree length of the tree estimated under the SWSC-EN partitioning scheme is equal to 1.0. There is no clear trend between TL values and partitioning schemes across the data sets. (1) Single: treating all sites as belonging to a single subset; (2) UCE: one subset for each UCE alignment; (3) PF-UCE: each UCE was defined as a data block; (4) SWSC-GC: data blocks defined by the SWSC-GC algorithm; (5) SWSC-EN: data blocks defined by the SWSC-EN algorithm; (6) SWSC-MU: data blocks defined by the SWSC-MU algorithm; and (7) UCESP: data blocks defined by the UCESP algorithm. In (3)–(7), the final partitioning scheme was optimized in PartitionFinder 2.
Discussion
Model selection is an important part of phylogenetic inference (Sullivan and Joyce 2005; Shapiro et al. 2006; Lanfear et al. 2014). In this study, we focused on selecting partitioned models of molecular evolution for data set comprised of UCEs. We first showed that patterns of molecular evolution within UCEs vary in predictable ways (fig. 1). Importantly, this variation is likely to violate at least one key assumption of most commonly-used models of molecular evolution: that base composition within a UCE can be adequately modeled as a single vector of base frequencies. This suggests that model violation might limit the accuracy of inferences made from UCE data sets, as long as each UCE is assumed to have evolved under a single Markov model.
We proposed four new approaches to partitioning UCEs, and showed that an approach based on site entropies to divide each UCE into three data blocks dramatically improved the fit of the models to the data when compared to three commonly-used approaches to partitioning in UCE studies (fig. 5). Phylogenetic trees estimated using partitioning schemes from this new method (which we call the SWSC-EN method) showed some differences in tree length and tree topology when compared to trees estimated using standard approaches (table 4, fig. 7). These differences tended to be relatively minor, but since it is hard to predict a priori the effect of model choice on tree inference, and since the models estimated with the SWSC-EN method have much improved AICc scores, it seems prudent to employ these new models where appropriate. Indeed, in a single case we observed a strongly-supported change in a topology when comparing the tree estimated using the SWSC-EN method to trees estimated using more traditional methods such as treating all data as a single data block, or assigning each UCE to its own data block. The SWSC-EN method we describe here can be implemented for any UCE data set using the scripts we provide on Github at https://github.com/Tagliacollo/PFinderUCE-SWSC-EN (doi: 10.5281/zenodo.1028743).
Phylogenetics is no longer limited by our ability to sequence sufficient loci from a sample (Delsuc et al. 2005; Posada 2016). Rather, the current challenges have shifted to finding appropriate ways to model the vast quantities of data that we have (Kumar et al. 2012). We hope that the new methods we propose here will help improve the accuracy of phylogenetic inferences made from UCEs. However, we note that our approach has many limitations. For example, our observations (fig. 1) suggest that rates of evolution and GC content vary in quite predictable ways within each UCE. Dividing each UCE up into three data blocks provides a crude but pragmatic way to model this variation within the current phylogenetic workflow, but it might be more appropriate to model this variation directly. For example, the patterns of rates of evolution and GC content within UCE could be modeled with four additional parameters added to a standard model from the GTR family: a minimum rate and a maximum GC content that applied to the center of the UCE, and a maximum rate and a minimum GC content that applied to the two ends of the UCE. Such an approach would likely provide a better description of the observed patterns of variation within each UCE, and would require fewer parameters than dividing each UCE up into three separate data blocks.
Conclusion
Partitioning remains a popular method for accounting for variation in rates of molecular evolution; however, it relies on the assumptions that predefined groups of sites have evolved under the same process. We present a new method that improves phylogenomic estimates of species relationships by improving partitioning schemes of UCE alignments. This method can be implemented using scripts available from GitHub at https://github.com/Tagliacollo/PFinderUCE-SWSC-EN (doi: 10.5281/zenodo.1028743). All of the methods we describe here can be implemented using scripts available at https://github.com/Tagliacollo/PartitionUCE (doi: 10.5281/zenodo.1027526).
Materials and Methods
Empirical Data Sets
We evaluated the performance of the two new partitioning methods described here using seven empirical UCE data sets available from the Dryad Digital Repository. The data sets range from 515 to 2,098 UCEs, from 10 to 187 taxa, and from 183,747 to 1,561,581 nucleotide sites (table 1). Non-UCE markers were excluded from one data set (Meiklejohn et al. 2016). The modified version is available from GitHub along with all of the other data sets we used for this study at https://github.com/Tagliacollo/PartitionUCE (doi: 10.5281/zenodo.1027526).
Visualizing Patterns of Molecular Evolution in UCEs
We measured three properties of molecular evolution of each site in each UCE of the seven empirical UCE data sets used in this study (table 1): the EN of a site, the GC content of a site; and the MU of a site. We calculate the entropy (H) of each site using the following formula:
| (1) |
where j = 1, 2, 3, and 4 corresponds to nucleotide A, C, G, and T, and pj corresponds to the proportion of a nucleotide j at site i. The Hi varies between 0 and 2, where 0 indicates invariant sites and 2 indicates sites with an equal frequency of all four nucleotides.
We calculate the GC content as the ratio of the sum of the counts of G and C to the sum of the counts of all four nucleotides, ignoring gaps and ambiguous nucleotides. Finally, we calculate the multinomial likelihood (M) using the following formula:
| (2) |
where
| (3) |
where N corresponds to the number of species, j = 1, 2, 3, and 4 corresponds to nucleotides A, C, G, and T, respectively, pj represents the proportion of nucleotide j at site i, and bj represents the counts of nucleotides in the alignment of the UCE.
New Methods for Automated Partitioning of UCEs
In what follows, we use the word ‘data block’ to refer to a user- or algorithmically defined set of sites that are assumed to have evolved in similar ways. We refer to a ‘subset’ as a group of one or more data blocks, and we refer to a ‘partitioning scheme’ as a group of subsets in which each site in the alignment is included only once.
The new partitioning methods, the SWSC and UCESP, were developed specifically for phylogenomic studies of UCE markers. The SWSC uses a sliding-window approach to divide each UCE into three data blocks, which can then be combined across UCEs using standard algorithms in PartitionFinder 2 (Lanfear et al. 2016). The UCESP takes a different approach, and groups nucleotide sites sharing similar locations in the alignments into partitions. We implemented both methods using Python scripts that are available on GitHub at https://github.com/Tagliacollo/PartitionUCE (doi: 10.5281/zenodo.1027526).
Both methods require the following input: a concatenated nexus alignment comprised of UCE markers and including nexus-formatted character sets (charsets) that define the location of each UCE in the alignment. The methods export PartitionFinder configuration files including data blocks defined by the individual methods. We avoid defining data blocks with fewer than 50 nucleotide sites that do not contain all four nucleotides, or have no variable sites. These conditions avoid the creation of very small data blocks, which can provide unreliable parameter estimates and thus mislead algorithms such as PartitionFinder 2 (Lanfear et al. 2016). The occurrence of all four nucleotide bases and at least one variable site in each data block was necessary to ensure that RAxML (Stamatakis et al. 2008) could analyze that data block. Example input files can be found in the ‘raw_data’ folder available on GitHub at https://github.com/Tagliacollo/PartitionUCE (doi: 10.5281/zenodo.1027526).
Sliding-Window Site Characteristics (SWSC)
The SWSC is graphically summarized in figure 2, and includes the following four steps which we describe in more detail below:
Calculate a metric (i.e., EN, GC content, or MU) for each UCE site.
Define all valid ways of dividing the UCE into three contiguous groups of sites.
Calculate the total sum of squared errors (SSE) for each of the splits defined in (2).
Select the split from (3) with the smallest SSE.
Step 1 involves simply calculating a metric for each site of the UCE, as described in the New Approaches section: the EN of a site, which can serve as a rough proxy for the rate of evolution of that site; the GC content (GC); and the MU of a site, which describes the likelihood of observing a particular site pattern given the observed base frequencies of a particular UCE.
Step 2 of the SWSC algorithm is to propose all valid ways of dividing a UCE into three contiguous groups of sites, which we hereafter refer to as a split. We do this by first listing all possible splits for a single UCE. The number of such splits is large, and is defined by , where N is the number of sites in the UCE, and the 2 refers to the positions of the two cuts required to split a single UCE into three contiguous parts. Given the list of all possible splits, we then reject splits that do not meet the following conditions: (1) all three sections defined by the split must contain at least 50 bases; (2) all three sections of the split must have at least one variable site; (3) all three sections of the split must contain at least one of each of the four nucleotide bases (see above). This results in a set of all valid splits for a single UCE.
Step 3 of the SWSC algorithm is to calculate the SSE for each of the splits defined in step 2. To do this, we first calculate for each valid split the SSE within each of the three contiguous groups sites, by summing the absolute differences of the metric at each site and the mean of the metric for that group of sites (e.g., the sum of the absolute differences between the GC content of each site and the mean GC content for the window). We then sum the SSEs for the three groups of sites to obtain a total SSE for the split.
Step 4 of the SWSC algorithm is to define the best split as the one with the smallest total SSE. In cases where we have more than one split with the same minimum SSE, we choose the split that has the lowest variance in the lengths of the three contiguous groups of sites. This maximizes the sizes of each window and avoids small data blocks which could provide unreliable parameter estimates.
The outcome of the SWSC algorithm for a single UCE is a single split that defines three groups of contiguous sites, each of which is then used as a data block for input into PartitionFinder 2 (see below).
UCE Site Position (UCESP)
The UCESP is graphically summarized in figure 3, and includes the following three steps, which we describe in more detail below:
Find the central nucleotide site for each UCE.
Create data blocks based on their locations relative to the central site in each UCE.
Merge small data blocks.
Step 1 of the UCESP is to define the central site of the UCE as site 0. Sites to the left of this site are labeled with negative numbers, and sites to the right of this site are labeled with positive numbers. Step 2 of the UCESP is to create one data block for each label, by combining all the sites with the same label into a single data block. For a data set with 1,000 UCEs, this will create many data blocks with 1,000 sites (one from each UCE). However, the longest UCEs in the data set will lead to the creation of much smaller data blocks, because they will have sites labeled with large numbers that are rarely found in other UCEs. Small data blocks cannot be practically analyzed; therefore, step 3 of the UCESP is to progressively merge small data blocks with their nearest neighbors until each data block meets the same criteria previously described in the step 2 of the SWSC method. This algorithm results in a single set of data blocks which can be used for downstream analyses in PartitionFinder 2.
Partitioning Schemes: PartitionFinder
We used PartitionFinder 2 (Lanfear et al. 2016) to estimate the optimal partitioning schemes and models of molecular evolution for seven different approaches to partitioning each of the seven data sets analyzed here (table 1): four of the partitioning approaches are described here (i.e., SWSC-EN, SWSC-GC, SWSC-MU, UCESP), and we compare these to three partitioning approaches that are widely used in phylogenomic studies of UCEs (treating the entire alignment as one partition; assigning one partition to each UCE; and assigning one partition to each UCE and optimizing this scheme in PartitionFinder). Given an optimal partitioning scheme for each method, we then define the best method for each data set as the method that results in the partitioning scheme with the best (i.e., lowest) AICc score. We used PartitionFinder 2 with the following settings: start with a tree estimated through maximum parsimony, linked branch lengths, and GTR + G model of nucleotide evolution, and the relaxed clustering algorithm with default settings. For each data set, we calculated the AICc scores of the following partitioning schemes: (1) Single: treating all sites as belonging to a single subset; (2) UCE: one subset for each UCE alignment; (3) PF-UCE: each UCE was defined as a data block; (4) SWSC-GC: data blocks defined by the SWSC-GC algorithm; (5) SWSC-EN: data blocks defined by the SWSC-EN algorithm; (6) SWSC-MU: data blocks defined by the SWSC-MU algorithm; and (7) UCESP: data blocks defined by the UCESP algorithm. In (3)–(7), the final partitioning scheme was optimized in PartitionFinder 2.
Checking Reliability of SWSC-EN
To evaluate whether the performance of the SWSC-EN method results from simply from splitting the UCEs into smaller subsets, we compared the observed AICc score for each of the seven empirical data sets to 25 AICc scores calculated from the same data sets in which the sites of each UCE were shuffled. Shuffling the sites of each UCE serves to hold the number and size of initial data blocks input to PartitionFinder 2 constant, but within each UCE it randomizes the sites that are assigned to each of the three data blocks. For each of the 25 randomizations for each of the seven empirical data sets (175 analyses in total) we (1) shuffled the sites within each UCE; (2) used the shuffled alignments with the original data block definitions from the un-shuffled data as input to PartitionFinder 2; (3) optimized the partitioning as described above. The scripts and permuted data sets are available from GitHub at https://github.com/Tagliacollo/PartitionUCE (doi: 10.5281/zenodo.1027526).
Phylogenomic Inferences: Branch Order and Branch Lengths
We used IQ-Tree (Nguyen et al. 2015) to infer phylogenetic trees for each of the seven empirical data sets (table 1), under each of the seven partitioning schemes. The aim was to evaluate whether more appropriate partitioning schemes for UCE data sets leads to changes in branch order and/or branch lengths. The IQ-Tree runs used default parameters, including linked branch lengths among partitions (option –spp) and 100 nonparametric bootstrap replicates to investigate node support. We compared the branching orders of topologies through visual inspection of the trees, and by calculating the PD between pairs of topologies. The PD is a measure of the difference between two topologies, which is similar in principle to the more commonly-used Robinson–Foulds distance, but has more attractive statistical properties such as not giving maximal differences to pairs of topologies that differ by the placement of a single taxon (Steel and Penny 1993). We compared branch lengths between topologies by plotting the pairs of topologies facing each other, and by calculating and comparing the total tree lengths (i.e., the sum of all branch lengths) of topologies estimated from the same data using different models. The latter approach may reveal any systematic differences of the methods to over- or under-estimate branch lengths relative to each other.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We particularly thank Andy Magee for sharing the multinomial likelihood equation. V.A.T. was supported by Brazilian National Postdoctoral Program (PNPD/CAPES) and Australian Endeavour Program. RL was supported by an Australian Research Council Future Fellowship.
References
- Baca SM, Toussaint EF, Miller KB, Short AE.. 2017. Molecular phylogeny of the aquatic beetle family Noteridae (Coleoptera: Adephaga) with an emphasis on data partitioning strategies. Mol Phylogenet Evol. 107:282–292. [DOI] [PubMed] [Google Scholar]
- Branstetter MG, Danforth BN, Pitts JP, Faircloth BC, Ward PS, Buffington ML, Gates MW, Kula RR, Brady SG.. 2017. Phylogenomic insights into the evolution of stinging wasps and the origins of ants and bees. Curr Biol. 277:1019–1025. [DOI] [PubMed] [Google Scholar]
- Crawford NG, Faircloth BC, McCormack JE, Brumfield RT, Winker K, Glenn TC.. 2012. More than 1000 ultraconserved elements provide evidence that turtles are the sister group of archosaurs. Biol Lett. 85:783–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins CA, McInerney JO.. 2011. A method for inferring the rate of evolution of homologous characters that can potentially improve phylogenetic inference, resolve deep divergence and correct systematic biases. Syst Biol. 606:833–844. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Herve P.. 2005. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 65:361.. [DOI] [PubMed] [Google Scholar]
- Faircloth BC, Branstetter MG, White ND, Brady SG.. 2015. Target enrichment of ultraconserved elements from arthropods provides a genomic perspective on relationships among Hymenoptera. Mol Ecol Resour. 153:489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faircloth BC, McCormack JE, Crawford NG, Harvey MG, Brumfield RT, Glenn TC.. 2012. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst Biol. 615:717–726. [DOI] [PubMed] [Google Scholar]
- Faircloth BC, Sorenson L, Santini F, Alfaro ME.. 2013. A phylogenomic perspective on the radiation of ray-finned fishes based upon targeted sequencing of ultraconserved elements (UCEs). PLoS ONE 86:e65923.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen PB, Calcott B, Mayer C, Lanfear R.. 2015. Automatic selection of partitioning schemes for phylogenetic analyses using iterative k-means clustering of site rates. BMC Evol Biol. 151:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goremykin VV, Nikiforova SV, Bininda-Emonds OR.. 2010. Automated removal of noisy data in phylogenomic analyses. J Mol Evol. 71(5–6):319–331. [DOI] [PubMed] [Google Scholar]
- Harrington RC, Faircloth BC, Eytan RI, Smith WL, Near TJ, Alfaro ME, Friedman M.. 2016. Phylogenomic analysis of carangimorph fishes reveals flatfish asymmetry arose in a blink of the evolutionary eye. BMC Evol Biol. 161:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SYW, Lanfear R.. 2010. Improved characterisation of among-lineage rate variation in cetacean mitogenomes using codon-partitioned relaxed clocks. Mitochondrial DNA 21(3–4):138–146. [DOI] [PubMed] [Google Scholar]
- Hoff M, Orf S, Riehm B, Darriba D, Stamatakis A.. 2016. Does the choice of nucleotide substitution models matter topologically? BMC Bioinformatics 171:1471–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SYW, Faircloth BC, Nabholz B, Howard JT et al. , . 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 3466215:1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainer D, Lanfear R.. 2015. The effects of partitioning on phylogenetic inference. Mol Biol Evol. 326:1611–1627. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B, Thornton JW.. 2004. Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature 4317011:980–984. [DOI] [PubMed] [Google Scholar]
- Kumar S, Filipski AJ, Battistuzzi FU, Kosakovsky Pond SL, Tamura K.. 2012. Statistics and truth in phylogenomics. Mol Biol Evol. 292:457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S.. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 296:1695–1701. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A.. 2014. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol Biol. 141:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B.. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34:772–773. [DOI] [PubMed] [Google Scholar]
- Le SQ, Lartillot N, Gascuel O.. 2008. Phylogenetic mixture models for proteins. Philos Trans R Soc Lond B Biol Sci. 3631512:3965–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Lu G, Orti G.. 2008. Optimal data partitioning and a test case for ray-finned fishes (Actinopterygii) based on ten nuclear loci. Syst Biol. 574:519–539. [DOI] [PubMed] [Google Scholar]
- McCormack JE, Harvey MG, Faircloth BC, Crawford NG, Glenn TC, Brumfield RT.. 2013. A phylogeny of birds based on over 1,500 loci collected by target enrichment and high-throughput sequencing. PLoS ONE 81:e54848.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn KA, Faircloth BC, Glenn TC, Kimball RT, Braun EL.. 2016. Analysis of a rapid evolutionary radiation using ultraconserved elements: evidence for a bias in some multispecies coalescent methods. Syst Biol. 654:612–627. [DOI] [PubMed] [Google Scholar]
- Moyle RG, Oliveros CH, Andersen MJ, Hosner PA, Benz BW, Manthey JD, Travers SL, Brown RM, Faircloth BC.. 2016. Tectonic collision and uplift of Wallacea triggered the global songbird radiation. Nat Commun. 7:12709.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 321:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD et al. , . 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 4447118:499–502. [DOI] [PubMed] [Google Scholar]
- Posada D. 2016. Phylogenomics for systematic biology. Syst Biol. 653:353–356. [DOI] [PubMed] [Google Scholar]
- Prebus M. 2017. Insights into the evolution, biogeography and natural history of the acorn ants, genus Temnothorax Mayr (hymenoptera: Formicidae). BMC Evol Biol. 171:250.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B, Rambaut A, Drummond AJ.. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol. 231:7–9. [DOI] [PubMed] [Google Scholar]
- Smith BT, Harvey MG, Faircloth BC, Glenn TC, Brumfield RT.. 2014. Target capture and massively parallel sequencing of ultraconserved elements (UCEs) for comparative studies at shallow evolutionary time scales. Syst Biol. 631:83–95. [DOI] [PubMed] [Google Scholar]
- Soubrier J, Steel M, Lee MS, Der Sarkissian C, Guindon S, Ho SY, Cooper A.. 2012. The influence of rate heterogeneity among sites on the time dependence of molecular rates. Mol Biol Evol. 2911:3345–3358. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J.. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 575:758–771. [DOI] [PubMed] [Google Scholar]
- Steel MA. 2005. Should phylogenetic models be trying to ′fit an elephant′? Trends Genet. 216:307–309. [DOI] [PubMed] [Google Scholar]
- Steel MA, Penny D.. 1993. Distributions of tree comparison metrics—some new results. Syst Biol. 422:126–141. [Google Scholar]
- Sullivan J, Joyce P.. 2005. Model selection in phylogenetics. Annu Rev Ecol Evol Syst. 361:445–466. [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K et al. , . 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 31:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1996. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol Evol. 119:367–372. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Townsend JP.. 2009. Maximum-likelihood model averaging to profile clustering of site types across discrete linear sequences. PLoS Comput Biol. 56:e1000421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.