Abstract
Microgravity-induced alterations in the autonomic nervous system (ANS) contribute to derangements in both the mechanical and electrophysiological function of the cardiovascular system, leading to severe symptoms in humans following space travel. Because the ANS forms embryonically from neural crest (NC) progenitors, we hypothesized that microgravity can impair NC-derived cardiac structures. Accordingly, we conducted in vitro simulated microgravity experiments employing NC genetic lineage tracing in mice with cKitCreERT2/+, Isl1nLacZ, and Wnt1-Cre reporter alleles. Inducible fate mapping in adult mouse hearts and pluripotent stem cells (iPSCs) demonstrated reduced cKitCreERT2/+-mediated labeling of both NC-derived cardiomyocytes and autonomic neurons (P < 0.0005 vs. controls). Whole transcriptome analysis, suggested that this effect was associated with repressed cardiac NC- and upregulated mesoderm-related gene expression profiles, coupled with abnormal bone morphogenetic protein (BMP)/transforming growth factor beta (TGF-β) and Wnt/β-catenin signaling. To separate the manifestations of simulated microgravity on NC versus mesodermal-cardiac derivatives, we conducted Isl1nLacZ lineage analyses, which indicated an approximately 3-fold expansion (P < 0.05) in mesoderm-derived Isl-1+ pacemaker sinoatrial nodal cells; and an approximately 3-fold reduction (P < 0.05) in cardiac NC-derived ANS cells, including sympathetic nerves and Isl-1+ cardiac ganglia. Finally, NC-specific fate mapping with a Wnt1-Cre reporter iPSC model of murine NC development confirmed that simulated microgravity directly impacted the in vitro development of cardiac NC progenitors and their contribution to the sympathetic and parasympathetic innervation of the iPSC-derived myocardium. Altogether, these findings reveal an important role for gravity in the development of NCs and their postnatal derivatives, and have important therapeutic implications for human space exploration, providing insights into cellular and molecular mechanisms of microgravity-induced cardiomyopathies/channelopathies.
Keywords: : neural crest cells, microgravity, cardiomyopathy, space travel, cardiac autonomic nervous system, pacemaker cells
Introduction
Space exploration is physically challenging and studies on humans returning from spaceflight, as well as earth-based simulated microgravity and space radiation experiments, have identified numerous hazards associated with low earth orbit and deep space travel [1]. These exposures may pose a risk to cardiovascular health, including, but not limited to, hemodynamic changes and body fluid shifts [2]; loss of myocardial mass [3]; reduced vascular resistance [1,2]; abnormal heart rate variability [4–6]; and orthostatic intolerance [5,7,8]. Importantly, increased mortality rates due to cardiovascular disease have also been reported [1].
Although the underlying mechanisms remain unclear and controversial, clinical and experimental data suggest that the detrimental effects of microgravity on cardiovascular health are related at least in part to alterations in the autonomic nervous system (ANS) and conduction systems [4–7,9–12]. The cardiac ANS develops from a migratory population of cells, the neural crest (NC) [13], which we [14–17] and others [13,18–20] have shown to play an important role in the plasticity and pathophysiology of the developing and adult heart. The cardiac conduction system, which is largely operated by pacemaker cells in the sinoatrial node (SAN), arises instead from cardiac mesoderm progenitors [21]. Importantly, despite their distinct embryonic origins, both NC and SAN lineages are produced under a cardiac regulatory network involving the stage-specific modulation of the bone morphogenetic protein (BMP) and Wnt signaling pathways [13,22,23]. Recent studies indicate that exposure of mouse embryonic stem cells (mESCs) to simulated microgravity [24] or spaceflight [25] significantly alters their capacity to differentiate into cardiogenic and neurogenic cells, through mechanisms that involve altered Wnt and BMP signaling pathways. However, whether the impact of microgravity on cardiovascular health involves abnormalities in the molecular and cellular mechanisms of the ANS and SAN lineages, remains unknown.
Accordingly, we employed simulated microgravity experiments, combined with cKitCreERT2/+, Isl1nLacZ, and Wnt1-Cre lineage mapping approaches in mice and induced pluripotent stem cell (iPSC) models of cardiogenesis, to study the impact of microgravity on the ANS and SAN lineages.
Materials and Methods
Animals
All animal studies were performed in an AAALAC-approved animal facility at the University of Miami, Miller School of Medicine, and procedures were performed using IACUC-approved protocols according to NIH standards. The cKitCreERT2/+, IRG, Isl1nLacZ, Isl1MerCreMer, Wnt1-Cre, and tdTomato mice have been described previously [14,15].
iPSC modeling experiments
The iPSCKit-Cre and iPSCWnt1-Cre mouse iPSC lines have been described elsewhere [14,15]. Mouse iPSCs were propagated without feeders on 0.1% gelatin-coated plates (Millipore), with NDiff227 (Clontech), supplemented with 1,000 U/mL LIF (Millipore), 1 μM PD0325901 (Tocris), and 3 μM CHIR99021 (Tocris). For cardiac NC differentiation, iPSCs were resuspended in differentiation medium [IMDM (Thermo), l-glutamine, 20% FBS (Thermo), 0.1 mM nonessential aminoacids, and 0.1 mM 2-mercaptoethanol] at 25,000 cells/mL and aggregated into embryoid bodies (EBs) in the presence of the small molecule BMP antagonist dorsomorphin (2 μM; Tocris), or 100 μg/mL ascorbic acid (Sigma), using the hanging drop method, as described previously [14,15]. To simulate microgravity, day 4 EBs were randomly transferred into 2-mL high-aspect ratio vessels (HARVs), with or without Sephadex microcarrier beads (Sigma), or in 0.1% gelatin-coated six-well plates (Corning), as control. A total of 30 EBs were cultured per HARV or per well. HARVs were attached into a rotary cell suspension culture system (RCCS; Synthecon), and speed was adjusted every other day at 12–20 rpm, as previously described [24]. To induce cKitCreERT2/+ recombination, cultures were supplemented every other day with 1 μM (Z)-4-hydroxytamoxifen (Abcam), from day 6 until the end of the experiment. To explore the effect of transitioning into normal gravity, RCCS-grown EBs were collected on day 10 and plated on gelatin-coated six-well plates, until day 21. Medium was exchanged every other day. Quantification of beating and Cre-reporter gene expression were performed as described previously [14,15]. Live-tissue epifluorescence imaging was performed in an Olympus IX81 fluorescent microscope, as described before [14,15].
Adult lineage tracing and heart tissue culturing
For tamoxifen-inducible lineage tracing of adult c-Kit+ cardiac cells, female cKitCreERT2/+;IRG mice (8–12 weeks old) were treated for 3 consecutive days with intraperitoneal injections of tamoxifen, as previously described [15]. For cKitCreERT2/+;IRG tissue culture experiments, heart explants were collected from the interventricular septal wall of adult cKitCreERT2/+;IRG mice of both sexes (8–12 weeks old) as previously described [14]. Tissues were minced into ∼2–3 mm3 fragments and digested in a solution of DMEM/F12 (Thermo), 20% FBS (Thermo), 1% penicillin/streptomycin (Thermo), and 200 U/mL Collagenase-Type II solution (Worthington) at 37°C. Digested tissue explants were then collected and washed twice with DMEM (Thermo) to remove residual enzyme. Single tissue fragments were then handpicked under sterile conditions with a stereomicroscope and a micropipette, and randomly transferred into six-well plates [static culture (SC)] or 2 mL-HARVs (RCCS), coated with 2.5 × 105 gamma-irradiated mouse embryonic fibroblasts (MEFs; Millipore). According to manufacturer's instructions (Synthecon), all tissue samples cultured in HARVs were supplemented with MEF-coated Sephadex microcarrier beads (Sigma). Samples were fed every other day with DMEM/F12, 15% FBS (HyClone), 1% penicillin/streptomycin (Thermo), 1% β-mercaptoethanol (Thermo), 1,000 U/mL recombinant mouse LIF (Millipore), 100 ng/mL recombinant mouse SCF (PeproTech) 1 ng/mL recombinant mouse bFGF (PeproTech), 0.1 mM nonessential aminoacids (Thermo), and 1 μM (Z)-4-hydroxytamoxifen. Quantification of Cre reporter gene expression in heart explants was performed as described before [14].
For Isl1nLacZ tissue culture experiments, Isl1nLacZ hearts of both sexes (8–12 weeks old) were harvested and rinsed in ice cold Hanks' balanced salt solution (HBSS; Thermo Scientific), before dissecting both ventricles and atria under a stereomicroscope (Discovery V8; Zeiss). The remaining tissue, containing the intact inflow and outflow tracts, the outflow tract base, and sinus node, was cleared from any epicardial fat, esophageal and pharyngeal tissue fragments, and placed in SC with DMEM/F12 (Thermo), 20% FBS (Thermo), 1% penicillin/streptomycin (Thermo), 0.1 mM nonessential aminoacids (Thermo), and 1% β-mercaptoethanol (Thermo), at 37°C/5% CO2. After 1 h, tissues were collected in fresh medium and randomized into SC or RCCS cultures. Medium was changed every 2 days. MEF-coated microcarrier beads were not used in this set of experiments. After a total period of 2 weeks, samples were collected, fixed for 10 min in 4% paraformaldehyde, and processed for X-gal staining as previously described [15]. X-gal-stained samples were subsequently imaged in a Zeiss, Discovery V8 stereomicroscope equipped with a Nikon D7200 digital camera. For X-gal quantification, jpeg images were used to select the X-gal+ areas in Adobe Photoshop Elements (version 12.1) using the semiautomated color range selection tool, and converted into pixel values.
Immunostaining
Confocal immunofluorescence was performed in adult heart tissue and iPSC-derived EBs as previously described [14,15]. The following antibodies were used: anti-enhanced green fluorescent protein (EGFP) (chicken monoclonal, 1:500; Aves); Nkx2.5 (goat polyclonal, 1:50; Santa Cruz biotechnologies); vesicular acetylcholine transporter (1:200; Novus Biologicals); Tyrosine Hydroxylase (rabbit polyclonal, 1:500; Novus Biologicals), Tuj1 (Rabbit polyclonal, 1:500; Covance). Subsequently, the antibodies were visualized by incubating the cells for 45 min at RT with Alexa 488, Alexa 546, or Alexa 647 secondary antibodies (Molecular probes).
TaqMan gene expression analysis
TaqMan gene expression analysis was performed as described previously [14,15]. Briefly, RNA was isolated from EBs using the RNeasy Plus Mini Kit (Qiagen), according to the manufacturer's instructions. RNA quality was assessed using NanoDrop (Thermo). cDNA was generated using the High-Capacity cDNA Reverse Transcription Kit (Thermo), and gene expression was analyzed using the TaqMan Universal PCR Master Mix (Thermo) in a Bio-Rad Q5 Real-Time PCR. For the analysis, all values were normalized to GAPDH. The following probes were used: Mm99999915_g1, GAPDH; Mm00432087_m1, BMP4; Mm01300555_g1, WNT1; Mm00440359_m1, MYH7; Mm01297833_s1, NOGGIN; Mm00550265_m1, LEF1; Mm00435491_m1, PAX3; Mm01309813_s1, NKX2.5; Mm00445212_m1, CKIT; Mm00486938_m1, CDH5; Mm00443610_m1, AXIN2; Mm00443121_m1, SEMA3C; Mm01224783_m1, SEMA3D; Mm01222421_m1, KDR; Mm00486299_s1, SOX1; Mm00517585_m1, ISL1; Mm00447557_m1, TH; and Mm01221880_m1, CHAT.
Microarray analysis
RNA was collected from ascorbic acid-treated iPSCKit-Cre-derived EBs, on days 10 (n = 3 SC and n = 3 RCCS) and 21 (n = 3 SC and n = 3 RCCS), using the RNeasy Plus Mini Kit (Qiagen), according to the manufacturer's instructions. RNA quality was assessed in an Agilent Bioanalyzer, at the University of Miami, Hussman Institute of Human Genomics, Center for Genome Technology (HIHG-CGT), Gene Expression Core Facility. One sample from the day 10 RCCS group failed the quality test and was therefore excluded from the array. A total of n = 11 samples were analyzed on the Affymetrix GeneChip Mouse Gene 2.0 ST array, at the HIHG-CGT. Analysis of array data was conducted in R package [26]. The CEL files of 11 samples were read into R and sample quality was evaluated in R before normalization. The normalized data were tested by F-test to remove genes whose expression values are not different among groups. Hierarchical clustering and PCA were applied on filtered gene set to evaluate the relationship among samples. Limma package was used to fit the expression values of four groups, and then the genes of significantly differential expression and adjusted P < 0.05 between each two groups were collected. The enrichment analyses on DE genes were conducted in GeneGo (MetaCore Bioinformatics software, Thomson Reuters), and top 10 enriched pathways and gene ontology processes were reported.
Statistics
All statistical analyses, except microarrays, were performed using GraphPad Prism version 5.00 for Windows. Values were analyzed using Student's t-test, Mann–Whitney, or ANOVAs (one- or two-way) followed by Tukey's post hoc tests. All data met the assumptions of the tests. A P < 0.05 was considered statistically significant. All values are reported as mean ± SEM.
Results
Effects of simulated microgravity on adult and iPSC-derived cKit+ cardiac cells
Contrary to the harmful effects of spaceflight in human cardiovascular pathophysiology [1,6], exposure to microgravity exerts beneficial effects on the cardiac mesoderm differentiation of pluripotent stem cell-derived progenitors [24,25].
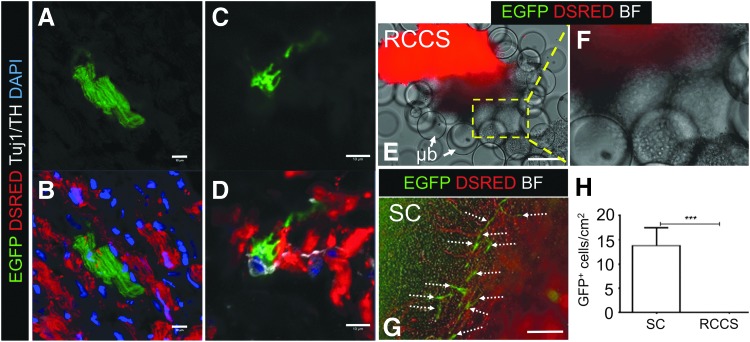
To address this paradox, we first tested the effect of simulated microgravity on the fate of adult cardiac precursor cells, by combining a RCCS with a cKitCreERT2/+; IRG-inducible genetic fate-mapping approach in adult mice, under which the expression of the cell surface marker cKit results in Cre-mediated irreversible replacement of Discosoma sea anemones-derived red fluorescent protein (DSRED) with the EGFP fluorescent reporter gene, upon induction with tamoxifen [14,15]. In agreement with earlier reports [14,15], cKitCreERT2/+;IRG fate mapping marked a rare pool of cKit+ cells in the adult heart which contributed to postnatal turnover of cardiomyocytes and autonomic neurons (Fig. 1A–D). To study the responses of adult cKit+ cells to microgravity, myocardial tissue was extracted from the interventricular septum of cKitCreERT2/+;IRG adult mice and randomized to be cultured on MEF feeder layers for up to 2 months, either under SC or RCCS conditions, in the presence of 4-hydroxytamoxifen, as described previously [14,23]. Both SC and RCCS cultures promoted the outgrowth of proliferative cardiac cells from within the myocardial explants (Fig. 1E–G). However, although the SC group became enriched in migratory EGFP+ cKit+ cells over time (Fig. 1G, H), there was complete absence of EGFP expression, both within the explanted tissues and in explant-derived cells, in the RCCS group (P < 0.005) (Fig. 1E, F, H).
FIG. 1.
Rotary Cell Culture System (RCCS) impairs cKit+ cells in the adult heart. (A–D) An enhanced green fluorescent protein (EGFP)+ cardiomyocyte (A–B) and Tuj1+/TH+ neuron (C, D) following 2-week cKitCreERT2/+;IRG lineage fate mapping (n = 3), indicates contribution of adult cKit+ cells to postnatal cardiomyocyte and ANS renewal. (E, F) Live tissue imaging of EGFP and Discosoma sea anemone-derived red fluorescent protein (DSRED) in cardiac explants from a cKitCreERT2/+;IRG mouse heart, cultured for 24 days in RCCS in the presence of 4-hydroxytamoxifen, illustrates loss of the cKit-CreERT2 lineage, as indicated by the lack of EGFP expression in the explant as well as in explant-derived cells (boxed region) migrating onto the MEF-coated microbeads (μb). (F) Is a higher magnification of the boxed region in (E). (G) In contrast to RCCS, culture under static (SC) conditions promotes the outgrowth of EGFP+ cardiac explant-derived cells (arrows). (H) Quantification of EGFP+ cells under SC and RCCS (n = 5 mice/group). Values are mean ± SEM. ***P < 0.0005. Scale bars 10 μm (A–D), and 150 μm (E, F). ANS, autonomic nervous system; BF, brightfield; MEF, mouse embryonic fibroblast; RCCS, rotary cell suspension culture system; TH, tyrosine hydroxylase.
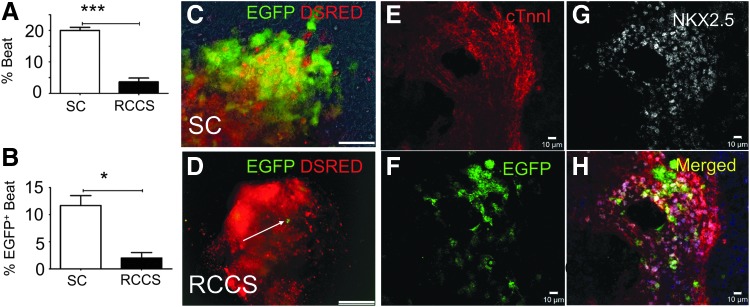
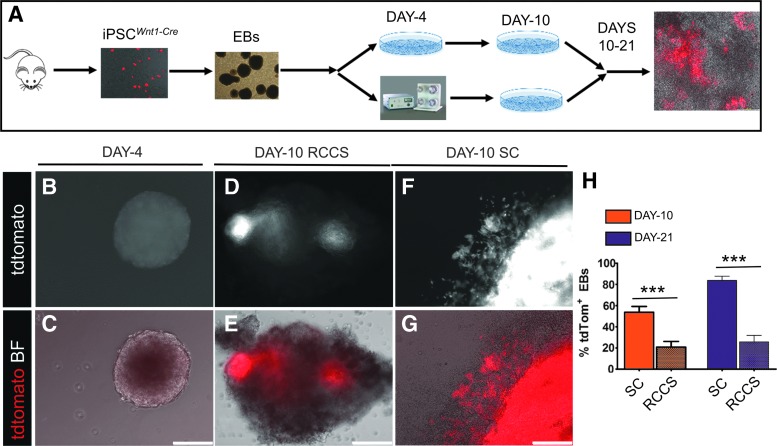
To better understand this observation, we developed an iPSC approach to model cardiac cKit+ cell development and differentiation, by generating iPSCs from cKitCreERT2/+;IRG reporter mice (iPSCsKit-Cre) [14,15]. To direct differentiation toward the cardiac lineages, iPSCsKit-Cre were first aggregated into EBs under SC, in the presence of ascorbate or the BMP antagonist Dorsomorphin (DM) as previously described [14,15]. To genetically fate-map the impact of simulated microgravity on iPSCsKit-Cre-derived cKit+ cells, EBs were randomized onto gelatin-coated plates (SC) or RCCS on EB day 4and subjected to treatment with 4-hydroxytamoxifen until their full cardiac differentiation on EB day 10. Analysis of EGFP expression and spontaneous contractions on EB day 10, indicated an approximately 5-fold decrease in the rate of spontaneously beating EBs (P < 0.005), and an approximately 6-fold decrease in the number of EBs containing EGFP+/Nkx2.5+ derivatives (P < 0.05) in RCCS compared with SC (Fig. 2A–H, and Supplementary Movies S1–S2; Supplementary Data are available online at www.liebertpub.com/scd).
FIG. 2.
RCCS impairs embryonic development of cKit+ cells in an iPSCkit-Cre model of cardiogenesis. (A, B) Quantification of spontaneously beating EBs (A) and spontaneously beating EBs expressing enhanced green fluorescent protein (EGFP) (B) under SC or RCCS (n = 3/group). (C, D) Live tissue imaging of EGFP and Discosoma sea anemone-derived red fluorescent protein (DSRED) in SC (C) and RCCS-grown (D) iPSCkit-Cre-derived EBs, following NC differentiation and induction of Cre-mediated recombination with 4-hydroxytamoxifen. (E–H) Confocal immunofluorescence showing colocalization of EGFP, NKX2.5, and cardiac troponin I in iPSCkit-Cre-derived cKit+ NCs, grown under SC. Two-tailed t-test, *P < 0.05, ***P < 0.0005. Values are mean ± SEM. Scale bars 150 μm (C, D) and 10 μm (E–H). EBs, embryoid bodies; iPSC, induced pluripotent stem cells; NC, neural crest.
Altogether, these findings suggest that, contrary to its beneficial effects on the cardiac mesoderm differentiation of pluripotent stem cells [24,25], simulated microgravity exerts detrimental effects in both adult and iPSC-derived cardiac cKit+ cells.
Abnormal Wnt and BMP signaling is associated with cKit+ cell defects
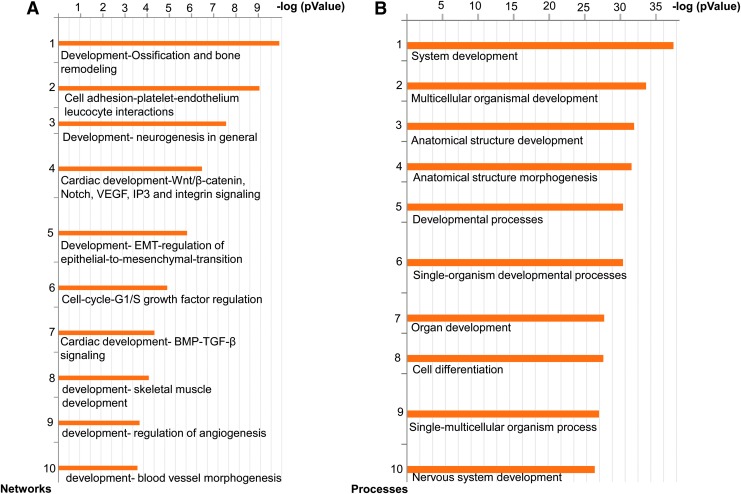
To investigate the potential mechanisms underlying the negative effects of simulated microgravity on cKit+ cells, we performed gene expression microarray analysis of day 10 iPSCKit-Cre-derived EBs (Fig. 2A). Overall, a total of 391 genes were found to be differentially expressed between SC and RCCS-grown day 10 EBs (Supplementary Fig. S2 and Supplementary Table S1). More specifically, compared with SC, EBs subjected to RCCS exhibited significant enrichment in cardiogenic mesoderm-related genes, such as Mef2C, Msx1, Hand2, Kdr, pecam1, and Tbx3 (Supplementary Table S2); whereas, expression of neuroectodermal lineage-related genes, such as Sox1, Pax3, Sema3C, Sema3D, Sema3E, and Zic3 (Supplementary Table S3), were significantly downregulated. Furthermore, enrichment analysis by sorting for the 10 most significantly affected gene networks and gene ontology processes, revealed a significant impact of RCCS on cardiovascular, neurogenic, and epithelial-to-mesenchymal transition gene programs compared with SC; and these effects were accompanied by significant differences in the activities of Wnt/β-catenin and BMP/TGF-β signaling pathways (Fig. 3).
FIG. 3.
RCCS affects cardiogenic and neurogenic programs by altering the Wnt and bone morphogenetic protein (BMP) signaling pathways. (A) The 10 process networks most significantly affected (statistically significant), based on microarray enrichment analysis of day 10 iPSCkit-Cre-derived EBs, grown under SC or RCCS. (B) The 10 GO processes most significantly affected, based on microarray enrichment analysis of day 10 iPSCkit-Cre-derived EBs, grown under SC or RCCS. (n = 2 biological replicates for day 10 RCCS, and n = 3 biological replicates for day 10 SC). GO, gene ontology.
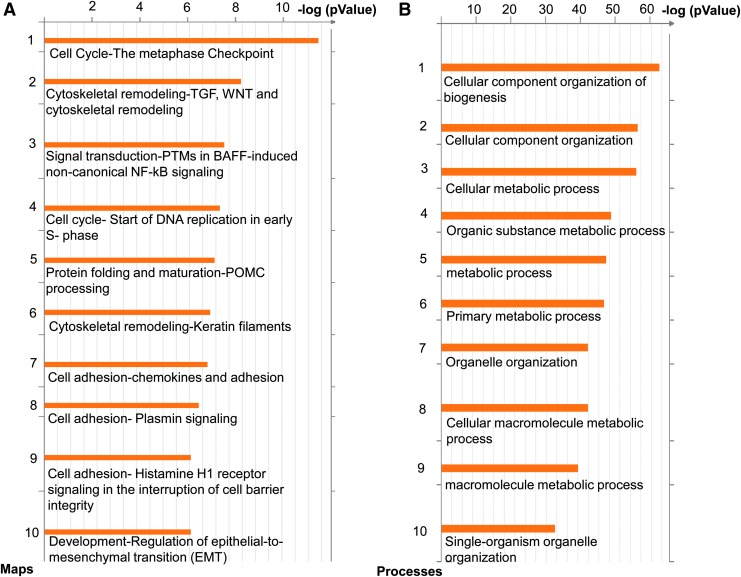
To explore whether the effects of simulated microgravity on cardiac cKit+ cells are reversible, day 10 RCCS iPSCKit-Cre-derived EBs were transferred to SC and subcultured until EB day 21. Microarray analysis indicated a total of 7,337 genes to be differentially expressed between SC and RCCS (Supplementary Fig. S1 and Supplementary Table S4). Enrichment analysis showed that, compared with SC, the abnormalities in the TGF-β/BMP, Wnt/β-catenin, and epithelial-to-mesenchymal transition pathways, persisted in RCCS EBs even after their transfer to SC (Fig. 4A). Moreover, we noted significant differences in gene ontology processes related to biogenesis and organization of subcellular organelles, as well as cell metabolism (Fig. 4B). Notably, similar metabolic abnormalities have been described before on stem cells following actual spaceflight experiments [25].
FIG. 4.
Abnormal BMP/TGF-β and Wnt/β-catenin following transition from RCCS to SC. (A) The 10 signaling pathways most significantly affected based on microarray enrichment analysis of day 21 iPSCkit-Cre-derived EBs, grown continuously under SC or following transfer from RCCS to SC on day 10. (B) The 10 GO processes most significantly affected, based on microarray enrichment analysis of day 21 iPSCkit-Cre-derived EBs, grown continuously under SC or following transfer from RCCS to SC on day 10 (n = 3 biological replicates per group).
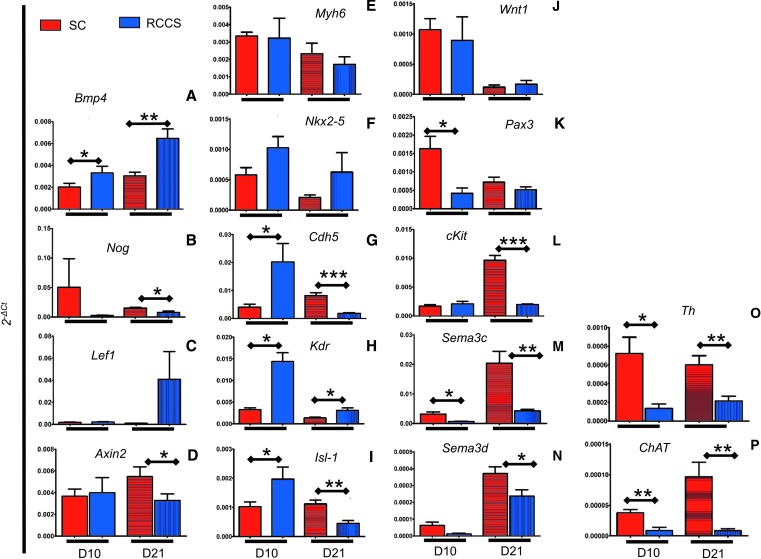
To verify the microarray results, we performed quantitative polymerase chain reaction (qPCR) analysis. Expression of Bmp4 and its antagonist Noggin corroborated that, compared with SC, the RCCS group was associated with abnormal BMP signaling (Fig. 5A, B). Similarly, differences in Lef1 and Axin2 transcription between the two groups corroborated the RCCS-induced abnormalities in Wnt/β-catenin signaling indicated by the gene expression microarray (Fig. 5C, D).
FIG. 5.
Impact of RCCS on NC- and cardiogenic mesoderm-related genes. qPCR in NC and cardiogenic mesoderm-related genes found to be differentially expressed between SC and RCCS, at EB days 10 and 21. (A, B) RCCS-mediated activation of bone morphogenetic protein (BMP) signaling indicated by upregulated Bmp4 and downregulated Noggin (Nog) expression. (C, D) RCCS-mediated activation of Wnt signaling indicated by upregulated Lef1 and downregulated Axin2 expression. (E, F) Comparison of cardiomyogenic genes Myh6 and Nkx2-5 between SC and RCCS, before and after transition to SC. (G, H) Comparison of vasculogenic genes Cdh5 and Kdr between SC and RCCS, before and after transition to SC. (I) Expression of Isl-1 is upregulated in RCCS at day 10, and downregulated after transition to SC. (J–N) Comparison of NC-related genes Wnt1, Pax3, cKit, Sema3c, and Sema3d between SC and RCCS, before and after transition to SC. (O, P) Expression of Th and ChAT on day 10 and 21 iPSCKit-Cre EBs. (n = 2 biological replicates for day 10 RCCS, and n = 3 for all other groups). Two-tailed t-test, *P < 0.05, **P < 0.005, ***P < 0.0005. Values are mean ± SEM. qPCR, quantitative polymerase chain reaction.
Analysis of the cardiomyogenic genes, Myh6 and Nkx2.5, did not show significant differences between groups, (Fig. 5E, F). However, compared with SC, the day 10 RCCS group exhibited significantly higher expression of the vasculogenic genes, Kdr and Cdh5, which remained upregulated following transition to SC (Fig. 5G, H). Interestingly, compared with the SC group, the transcription factor Isl-1, which is commonly expressed in mesoderm- and NC- derived cardiac cells, was significantly upregulated in the RCCS group at day 10, but was significantly downregulated by day 21, following transition to SC (Fig. 5I).
Expression of the NC-related genes, Pax3 and Wnt1, were downregulated at day 10 in the RCCS group, but were expressed at similar levels between groups at day 21 (Fig. 5J, K). In contrast, the NC-related genes, cKit, Sema3C, and Sema3d, were also downregulated at day 10 in the RCCS group, but remained repressed throughout the period of the study (Fig. 5L–N). Consequently, analysis of the adrenergic and cholinergic nerve markers, Tyrosine Hydroxylase (Th) (Fig. 5O) and Choline Acetyltransferase (ChAT) (Fig. 5P), indicated that sympathetic and parasympathetic neurogenesis, respectively, from NCs were significantly downregulated in the RCCS group compared with SC, both at days 10 and 21 of the study.
Collectively, these findings support that the detrimental effects of RCCS on cardiac cKit+ cells are associated with a repressed NC gene expression profile and impaired autonomic neurogenesis, which are not fully reversible upon transition to SC, likely due to underlying abnormalities in Wnt/β-catenin and BMP/TGF-β signaling pathways.
Effects of simulated microgravity on NC versus mesoderm-derived Isl-1+ cardiac cells
The cKitCreERT2/+ fate mapping studies suggest that the effects of simulated microgravity are manifested differently between mesoderm and NC-derived cardiac tissues. To test this hypothesis, we investigated the response of postnatal Isl-1+ cardiac cells, which comprise a diverse pool of second heart field (SHF) mesodermal derivatives and NC cells, expressing the ISL LIM homeobox 1 transcription factor Isl-1 [17,27–29].
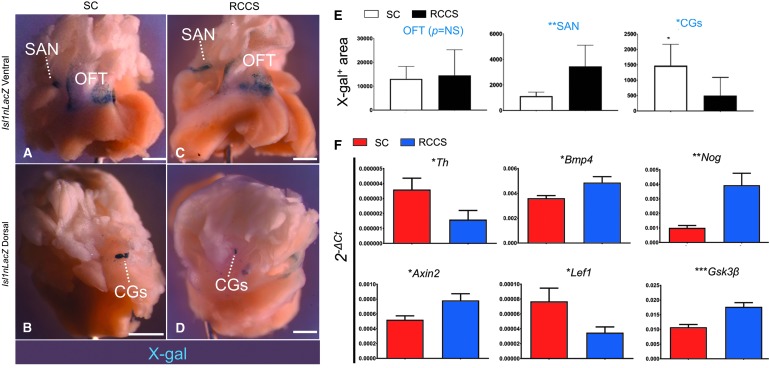
First, we employed a previously described Isl1nLacZ/+ indicator mouse line [27], to verify the expression of Isl-1 in postnatal SHF and NC cardiac cells. In agreement with previous reports [17,21,27], X-gal analysis indicated that postnatal Isl1nLacZ expression identifies a group of posterior SHF-derived pacemaker cells in the SAN; a mixed pool of NC- and SHF-derived cells in the proximal outflow tract (OFT); and finally, a group of NC cells in the cardiac ganglia (CGs), located in the dorsal aspect of the heart (Supplementary Fig. S2A). Furthermore, analysis with a tamoxifen-inducible Isl1MerCreMer/+ reporter allele [17,27,28] indicated that postnatal Isl-1+ cells retain plasticity and contribute to postnatal turnover of NC-derived sympathetic neurons (Supplementary Fig. S2B), and rarely cardiomyocytes (Supplementary Fig. S2C).
Next, to test the effects of simulated microgravity on postnatal Isl-1+ cells, the cardiac bases of adult Isl1nLacZ mice, which included the SAN, OFT, and CGs, were dissociated from the atria and ventricles and randomized to the SC or RCCS groups. Two weeks later, X-gal analysis indicated that Isl1nLacZ expression in the OFT was similar between the two groups (Fig. 6A, C, E). However, compared with SC, the RCCS group displayed an approximately 3-fold expansion (P < 0.05) in the pool of X-gal+ pacemaker cells in the SAN (Fig. 6A, C, E), and an approximately 3-fold reduction (P < 0.05) in the pool of X-gal+ cells in the CGs (Fig. 6B, D, E). Furthermore, qPCR analysis indicated that these manifestations were accompanied by a significant reduction in the expression of the sympathetic nerve marker Th (Fig. 6F); and more importantly, by abnormalities in BMP signaling as indicated by differences in Bmp4 and Noggin expression (Fig. 6F) and Wnt/β-catenin signaling, as indicated by differences in Axin2, Lef1, and Gsk3β expression (Fig. 6F).
FIG. 6.
RCCS expands the pool of Isl1nLacZ+ pacemaker cells and reduces the pool of Isl1nLacZ+ ANS cells. (A, B) X-gal analysis in Isl1nLacZ adult heart tissues following 2 weeks in SC, indicates that Isl-1 is abundantly expressed in sinoatrial node pacemaker cells (A, SAN) and the proximal outflow tract (A, OFT). Dorsally, Isl-1 is abundantly expressed in cardiac ganglia (B, CGs). (C, D) In contrast, tissues cultured for 2 weeks in RCCS exhibit an expansion of X-gal staining in the SAN (C) and diminished X-gal staining in the CGs (D). No differences are noted in the proximal outflow tract (C). (E) Quantification of X-gal+area in the OFT, SAN, and CGs, between groups. (F) qPCR analysis of Isl1nLacZ adult heart tissues indicates that compared with SC, the RCCS group exhibits reduced sympathetic neurogenesis, as indicated by Th expression. Furthermore, these differences are accompanied by abnormal bone morphogenetic protein (BMP) signaling, as indicated by the expression of Bmp4 and Noggin (Nog); as well as abnormal canonical Wnt signaling, as indicated by the expression of Axin2, Lef1, and Gsk3β. n = 5/group. *P < 0.05; **P < 0.005; ***P < 0.0005, two-tailed T-test. Values are mean ± SEM. Scale bars 5 mm.
Taken together, these findings suggest that RCCS-induced abnormalities in Wnt and BMP signaling are manifested as a significant expansion of posterior SHF-derived Isl-1+ conduction system cells in the SAN; and repression of NC-derived ANS cells, including sympathetic nerves and Isl-1+ CGs.
Effects of simulated microgravity on iPSCs-derived cardiac NC cells and ANS
Although the experiments with the cKit and Isl-1 alleles support the hypothesis that simulated microgravity induces NC-based abnormalities of the ANS, neither cKit or Isl-1 are NC-specific markers. Accordingly, we sought to specifically test the effects of simulated microgravity on cardiac NCs, by generating a NC-specific Cre-reporter iPSC line (iPSCWnt1-Cre) from Wnt1-Cre;tdTomato neonatal mouse cardiac fibroblasts [15].
As with the iPSCsKit-Cre, iPSCWnt1-Cre were differentiated into EBs before being transferred into SC or RCCS on day 4, for up to 21 days (Fig. 7A). By day 9, a migratory population of tdTomato+ cells emerged from within the iPSCWnt1-Cre EBs, indicating differentiation into NC cells (Fig. 7B, C). Quantification of tdTomato+ EBs on day 10 indicated that NC differentiation was significantly impaired in the RCCS group compared with SC (20.8% ± 5.3% vs. 53.7% ± 5.5% tdTomato+ EBs in RCCS and SC groups, respectively, P < 0.0001) (Fig. 7D–H)].
FIG. 7.
RCCS specifically compromises NC lineage development. (A) Schematic of the iPSCsWnt1-Cre model of microgravity-induced cardiomyopathy. (B, C) Live epifluorescence imaging demonstrating absence of tdTomato expression in day 4 iPSCWnt1-Cre EBs, immediately before transfer to RCCS, indicating that NC differentiation has not commenced. (D–G) tdTomato expression in day 10 iPSCWnt1-Cre EBs, indicative of NC differentiation. Expression of tdTomato is less abundant in RCCS- (D, E) than SC-grown (F, G) EBs. (H) Quantification of tdTomato epifluorescence under RCCS and SC on day 10; and day 21, after transition on day 10 EBs from SC to RCCS (n = 17/group at day 10; n = 7 for RCCS on day 21; n = 12 for SC at day 21). ***P < 0.0001, two-tailed T-test. Values are mean ± SEM. Scale bars 150 μm.
To test whether the RCCS-induced defects in NC development can be rescued following transition to SC, iPSCsWnt1-Cre-derived EBs were transitioned from RCCS to SC on day 10, and subcultured until EB day 21. However, tdTomato quantification indicated that the degree of NC differentiation was still significantly lower in the RCCS group compared with SC (25.7% ± 6.1% vs. 83.8% ± 3.8% tdTomato+ EBs at day 21, between RCCS and SC groups, respectively; P < 0.0001) (Fig. 7H).
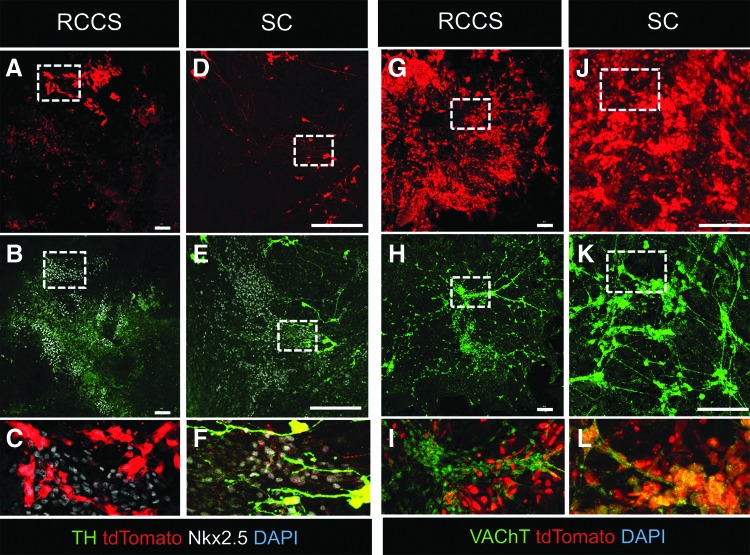
Live-cell imaging and confocal immunofluorescence did not show any differences in the capacity of iPSCsWnt1-Cre-derived tdTomato+ NCs to differentiate into NKX2.5+ spontaneously beating cardiomyocytes (Fig. 8A–C and Supplementary Movies S3–S4). However, although tdTomato+ NCs in the SC group contributed extensively to the cardiac autonomic innervation of spontaneously beating EBs with tdTomato+ sympathetic and parasympathetic nerves (Fig. 8D–F, J–L, and Supplementary Movie S5), differentiation of iPSCsWnt1-Cre-derived NCs into cardiac ANS derivatives was diminished in the RCCS group (Fig. 8A–C, G–I, and Supplementary Movie S6).
FIG. 8.
RCCS compromises NC differentiation into cardiac ANS. (A–F) Confocal immunofluorescence of TH, Nkx2.5, and tdTomato on day 19 iPSCWnt1-Cre EBs, transitioned from RCCS on day 10 (A–C) or continuously grown in SC (D–F). Exposure to RCCS supports differentiation of EBs into tdTomato+/Nkx2.5+ myocardium, which however lacks sympathetic innervation, as indicated by the complete lack of TH+ neurons (A–C). In contrast, SC-differentiated Nkx2.5+ myocardium is innervated with tdTomato+/TH+ sympathetic neurons (D–F). (C, F) Are blownup images of the areas depicted in insets in (A, B, D, E), respectively. (G, H) Confocal immunofluorescence of vesicular acetylcholine transporter (VAChT) and tdTomato on day 19 iPSCWnt1-Cre EBs, transitioned from RCCS on day 10 (G–I) or continuously grown in SC (J–L). Exposure to RCCS results in EB differentiation into VAChT+/tdTomato-negative parasympathetic neurons (G–I). In contrast, parasympathetic neurons in SC-differentiated EBs are derived from NCs, as indicated by colocalization of tdTomato+/VAChT+ (J–L). (I, L) Are higher magnification images of the areas depicted in insets in (G, H, J, K), respectively. Scale bars 100 μm.
Collectively, these findings indicate that RCCS directly impacts the development of the NC lineage from iPSCs, and consequently, their contribution to sympathetic and parasympathetic innervation of iPSC-derived myocardium; and further suggest that these defects may not be fully reversible upon transition to SC.
Discussion
We employed a series of earth-based in vitro experiments to separate the effects of microgravity on NC versus mesodermal cardiac precursors. The major new finding of this study is that simulated microgravity exerts adverse effects on the autonomic and pacemaking pathways of the heart, by directly repressing NC while enhancing mesoderm-derived cardiac lineages (Supplementary Fig. S3). More specifically, we found that simulated microgravity produced perturbations in the expression of cKitCreERT2/+, Isl1nLacZ, and Wnt1-Cre reporter alleles in adult as well as iPSC-derived cardiac NCs and ANS derivatives, whereas promoting the expansion of Isl1nLacZ reporter gene expression in SHF-derived pacemaker cells of the sinus node. Furthermore, gene expression microarrays and qPCR analyses suggested that these effects were associated with abnormalities in BMP/TGF-β and Wnt/β-catenin signaling; and may not be fully reversible upon transition to normal gravity.
In agreement with our findings, microgravity-induced abnormalities associated with the cardiac ANS and SAN have been previously reported in humans subjected to spaceflight [2,3,5,6,10,11,30–33]. Particularly, transition to microgravity has been reported to induce cardiovascular deconditioning, an acute syndrome manifested as abnormal cardiac preload, stroke volume, and output; loss of myocardial mass; reduced vascular resistance; abnormal baroreflex responses; heart rate variability; and orthostatic intolerance [4–7,11]. Furthermore, a retrospective analysis recently reported significantly higher mortality rates of the Apollo lunar astronauts due to cardiovascular disease [1]. Importantly—although the etiologies for the disease remain undetermined—in support to our study, a cohort of the lunar astronauts reportedly experienced unstable autonomic nervous function and cardiac rhythm problems, including bigeminy as well as premature auricular and ventricular contractions, both during and after space travel [12,34]. Thus, the detrimental effects of microgravity in postnatal cardiac NCs and their ANS derivatives, in conjunction to the abnormal expansion of Isl-1+ conduction system cells in the SAN described in this study, provide a possible explanation for the development of spaceflight-induced cardiomyopathies in humans.
Our findings on simulated microgravity-induced abnormalities in adult and iPSC-derived NC cells are also in agreement with previous developmental studies, reporting neural tube and other neurological defects in urodeles [35,36] and rodents [37,38] in response to actual spaceflight. In addition, consistent with our findings that these effects were associated with abnormal BMP/TGF-β and Wnt/β-catenin signaling, Lei et al. reported enhanced mesendoderm and repressed neuroectodermal differentiation of mESCs in RCCS, due to increased expression of Wnt3 [24]. Similarly, Blaber et al. recently investigated the effects of microgravity during spaceflight on mESCs, and reported abnormalities in several signaling pathways, including Wnt/β-catenin and TGF-β/BMP [25]. Remarkably, compared with our findings and previous simulated microgravity experiments by others [24], the effects of spaceflight on mESCs were associated with retention of pluripotency and inhibition of differentiation into both mesendodermal and neuroectodermal lineages, suggesting that earth-based simulated microgravity experiments may not recapitulate the full spectrum of spaceflight-induced abnormalities on stem/progenitor cells.
Finally, several limitations should be taken into account when interpreting our findings. First, the data described in this study are derived from earth-based simulated microgravity experiments. Second, the impact of simulated microgravity in cardiovascular development and plasticity were assessed ex vivo, in explanted heart tissues and iPSC-based in vitro models of cardiogenesis. Third, our experiments were conducted in murine tissues and cell lines. Therefore, further studies are warranted to elucidate whether the NC-based molecular and cellular mechanisms described herein, are associated with microgravity-induced cardiovascular disorders in humans following space travel.
In summary, we employed a series of earth-based experiments to simulate and study the impact of microgravity on the cellular and molecular mechanisms underlying the embryonic development and postnatal plasticity of the mammalian heart. Our findings support that microgravity impacts cardiac NC cells and their ANS derivatives, as well as the cardiac conduction system, by altering the BMP/TGF-β and Wnt/β-catenin signaling pathways. Taken together, our study provides novel mechanistic insights on the effects of microgravity in cardiovascular development and health; and therefore may lead to novel therapeutic strategies for the prevention and treatment of microgravity-induced cardiomyopathies.
Supplementary Material
Acknowledgments
This research is supported by the United States National Institutes of Health grants R01 HL094849, P20 HL101443, R01 HL084275, R01 HL107110, and R01 HL11073 (J.M.H.); Award No. GA2014131 from the Center for the Advancement of Science in Space (CASIS) to J.M.H; and by the University of Miami Miller School of Medicine, Interdisciplinary Stem Cell Institute. The authors thank Dr. Sylvia M. Evans from the Skaggs School of Pharmacy and Pharmaceutical Sciences, UCSD, La Jolla, California, USA and Dr. Chenleng Cai from the Icahn School of Medicine at Mount Sinai, New York, USA for providing the Isl1-nLacZ mice.
Author Disclosure Statement
Dr. Hare discloses a relationship with Vestion Inc. that includes equity, board membership, and consulting. Dr. Joshua Hare is the Chief Scientific Officer, a compensated consultant, and advisory board member for Longeveron and holds equity in Longeveron. Dr. Hare is also the coinventor of intellectual property licensed to Longeveron. Dr. Hatzistergos and Mrs. Valasaki disclose a relationship with Vestion Inc. that includes equity. Vestion did not contribute funding to this study. The other authors report no conflicts.
References
- 1.Delp MD, Charvat JM, Limoli CL, Globus RK. and Ghosh P. (2016). Apollo lunar astronauts show higher cardiovascular disease mortality: possible deep space radiation effects on the vascular endothelium. Sci Rep 6:29901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norsk P, Asmar A, Damgaard M. and Christensen NJ. (2015). Fluid shifts, vasodilatation and ambulatory blood pressure reduction during long duration spaceflight. J Physiol 593:573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perhonen MA, Franco F, Lane LD, Buckey JC, Blomqvist CG, Zerwekh JE, Peshock RM, Weatherall PT. and Levine BD. (2001). Cardiac atrophy after bed rest and spaceflight. J Appl Physiol (1985) 91:645–653 [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Li Y, Verheyden B, Chen S, Chen Z, Gai Y, Liu J, Gao J, Xie Q, et al. (2015). Is autonomic modulation different between European and Chinese astronauts? PLoS One 10:e0120920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vandeput S, Widjaja D, Aubert AE. and Van Huffel S. (2013). Adaptation of autonomic heart rate regulation in astronauts after spaceflight. Med Sci Monit 19:9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otsuka K, Cornelissen G, Kubo Y, Hayashi M, Yamamoto N, Shibata K, Aiba T, Furukawa S, Ohshima H. and Mukai C. (2015). Intrinsic cardiovascular autonomic regulatory system of astronauts exposed long-term to microgravity in space: observational study. Npj Microgravity 1:15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandsager KT, Robertson D. and Diedrich A. (2015). The function of the autonomic nervous system during spaceflight. Clin Auton Res 25:141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung AS, Harrison R, Lee KH, Genut J, Nyhan D, Brooks-Asplund EM, Shoukas AA, Hare JM. and Berkowitz DE. (2005). Simulated microgravity produces attenuated baroreflex-mediated pressor, chronotropic, and inotropic responses in mice. Am J Physiol Heart Circ Physiol 289:H600–H607 [DOI] [PubMed] [Google Scholar]
- 9.Anzai T, Frey MA. and Nogami A. (2014). Cardiac arrhythmias during long-duration spaceflights. J Arrhythm 30:139–149 [Google Scholar]
- 10.Baevsky RM, Baranov VM, Funtova , II, Diedrich A, Pashenko AV, Chernikova AG, Drescher J, Jordan J. and Tank J. (2007). Autonomic cardiovascular and respiratory control during prolonged spaceflights aboard the International Space Station. J Appl Physiol (1985) 103:156–161 [DOI] [PubMed] [Google Scholar]
- 11.Arzeno NM, Stenger MB, Bloomberg JJ. and Platts SH. (2013). Spaceflight-induced cardiovascular changes and recovery during NASA's Functional Task Test. Acta Astronautica 92:10–14 [Google Scholar]
- 12.Rowe WJ. (1998). The Apollo 15 space syndrome. Circulation 97:119–120 [DOI] [PubMed] [Google Scholar]
- 13.Hasan W. (2013). Autonomic cardiac innervation: development and adult plasticity. Organogenesis 9:176–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatzistergos KE, Saur D, Seidler B, Balkan W, Breton M, Valasaki K, Takeuchi LM, Landin AM, Khan A. and Hare JM. (2016). Stimulatory effects of MSCs on cKit+ cardiac stem cells are mediated by SDF1/CXCR4 and SCF/cKit signaling pathways. Circ Res 119:921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatzistergos KE, Takeuchi LM, Saur D, Seidler B, Dymecki SM, Mai JJ, White IA, Balkan W, Kanashiro-Takeuchi RM, Schally AV. and Hare JM. (2015). cKit+ cardiac progenitors of neural crest origin. Proc Natl Acad Sci U S A 112:13051–13056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White IA, Gordon J, Balkan W. and Hare JM. (2015). Sympathetic reinnervation is required for mammalian cardiac regeneration. Circ Res 117:990–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatzistergos KE. and Hare JM. (2016). Abstract 322: Postnatal Islet-1 cardioblasts are of neural crest and not second heart-field lineage. Circ Res 119:A322–A322 [Google Scholar]
- 18.Kimura K, Ieda M. and Fukuda K. (2012). Development, maturation, and transdifferentiation of cardiac sympathetic nerves. Circ Res 110:325–336 [DOI] [PubMed] [Google Scholar]
- 19.Tamura Y, Matsumura K, Sano M, Tabata H, Kimura K, Ieda M, Arai T, Ohno Y, Kanazawa H, et al. (2011). Neural crest-derived stem cells migrate and differentiate into cardiomyocytes after myocardial infarction. Arterioscler Thromb Vasc Biol 31:582–589 [DOI] [PubMed] [Google Scholar]
- 20.Tomita Y, Matsumura K, Wakamatsu Y, Matsuzaki Y, Shibuya I, Kawaguchi H, Ieda M, Kanakubo S, Shimazaki T, et al. (2005). Cardiac neural crest cells contribute to the dormant multipotent stem cell in the mammalian heart. J Cell Biol 170:1135–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang X, Wang G, Lin L, Lowe J, Zhang Q, Bu L, Chen Y, Chen J, Sun Y. and Evans SM. (2013). HCN4 dynamically marks the first heart field and conduction system precursors. Circ Res 113:399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brade T, Pane LS, Moretti A, Chien KR. and Laugwitz KL. (2013). Embryonic heart progenitors and cardiogenesis. Cold Spring Harb Perspect Med 3:a013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohan R, Boukens B. and Christoffels V. (2017). Lineages of the Cardiac Conduction System. J Cardiovasc Dev Dis 4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei X, Deng Z, Zhang H, Zhao H, Zhou J, Liu S, Chen Q, Ning L, Cao Y, et al. (2014). Rotary suspension culture enhances mesendoderm differentiation of embryonic stem cells through modulation of Wnt/beta-catenin pathway. Stem Cell Rev 10:526–538 [DOI] [PubMed] [Google Scholar]
- 25.Blaber EA, Finkelstein H, Dvorochkin N, Sato KY, Yousuf R, Burns BP, Globus RK. and Almeida EA. (2015). Microgravity reduces the differentiation and regenerative potential of embryonic stem cells. Stem Cells Dev 24:2605–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Development Core Team. (2008). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 27.Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, Chen J. and Evans SM. (2007). Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol 304:286–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, et al. (2005). Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature 433:647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engleka KA, Manderfield LJ, Brust RD, Li L, Cohen A, Dymecki SM. and Epstein JA. (2012). Islet1 derivatives in the heart are of both neural crest and second heart field origin. Circ Res 110:922–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lees PJ. (2005). Cardiology in space. Hellenic J Cardiol 46:320–323 [PubMed] [Google Scholar]
- 31.White RJ. and Averner M. (2001). Humans in space. Nature 409:1115–1118 [DOI] [PubMed] [Google Scholar]
- 32.Leguay G. and Seigneuric A. (1981). Cardiac arrhythmias in space. Role of vagotonia. Acta Astronaut 8:795–801 [DOI] [PubMed] [Google Scholar]
- 33.Charles JB, Frey MA, Fritsch-Yelle JM. and Fortner. GW. (1996). Chapter 3. cardiovascular and cardiorespiratory function. In: Space Biology and Medicine—Volume III Books 1 & 2—Humans in Spaceflight. American Institute of Aeronautics and Astronautics, Inc; Reston, Virginia, pp 63–88 [Google Scholar]
- 34.Johnston RS, Dietlein LF. and Berry CA. Biomedical Results of Apollo. (1975). Scientific and Technical Information Office, National Aeronautics and Space Administration: for sale by the Supt. of Docs., U.S. Govt. Print. Off., Washington [Google Scholar]
- 35.Gualandris-Parisot L, Husson D, Foulquier F, Kan P, Davet J, Aimar C, Dournon C. and Duprat AM. (2001). Pleurodeles waltl, amphibian, Urodele, is a suitable biological model for embryological and physiological space experiments on a vertebrate. Adv Space Res 28:569–578 [DOI] [PubMed] [Google Scholar]
- 36.Gualandris-Parisot L, Husson D, Bautz A, Durand D, Kan P, Aimar C, Membre H, Duprat AM. and Dournon C. (2002). Effects of space environment on embryonic growth up to hatching of salamander eggs fertilized and developed during orbital flights. Biol Sci Space 16:3–11 [DOI] [PubMed] [Google Scholar]
- 37.Jamon M. (2014). The development of vestibular system and related functions in mammals: impact of gravity. Front Integr Neurosci 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ronca AE, Fritzsch B, Bruce LL. and Alberts JR. (2008). Orbital spaceflight during pregnancy shapes function of mammalian vestibular system. Behav Neurosci 122:224–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.