Abstract
In studying how stem cells make and maintain tissues, nearly every chapter of a cell biology textbook takes on special interest. The field even allows us to venture where no chapters have yet been written. In studying this basic problem, we are continually bombarded by nature's surprises and challenges. Stem cell biology has captured my interest for nearly my entire scientific career. Below, I focus on my laboratory's contributions to this fascinating field, to which so many friends and colleagues have made seminal discoveries equally deserving of this award.
Keywords: stem cells, skin, hair follicle, cancer, wound healing, epidermis
In this Perspective, Elaine Fuchs describes her laboratory's contributions to the field of skin stem cells, giving a historical overview of their first isolation and characterization, the discovery of extrinsic and intrinsic factors regulating skin stem cell states, and their role in skin disease.
Main Text
As a student in physical chemistry, my initial view of biology was that it was a science with too many variables to design a well-controlled experiment. However, the urge to venture into biomedical research was too great, and as a graduate student, I dove into biology head first, never haven taken a biology course in college. At Princeton University, I worked on how bacterial spores become activated to a vegetative state. In retrospect, the parallel to understanding how quiescent stem cells transition to an active tissue-generating mode seems striking. And yet it was not until I heard a lecture by Howard Green, who was then at MIT, that my excitement about stem cells was launched. Green spoke about taking a piece of human skin and identifying cells that he could passage clonally for hundreds of generations without losing their diploidy or their ability to make tissue (Rheinwald and Green, 1975). Wow! He did not call them stem cells, but in fact, these human keratinocytes fit the definition of stem cells as we know it today. I was passionate to learn more about this system and was excited to be accepted to his laboratory.
At the time, Green had begun to exploit for clinical purposes the amazing ability of these long-lived keratinocytes to create sheets of epidermis for the treatment of burn patients (Green, 1991). The highlight of these studies demonstrated that cultured stem cell therapy was successful in saving two children whose body surface was >90% burned. Twenty-five years later, Graziella Pellegrini and Michele De Luca showed that keratinocyte stem cells could be genetically engineered to successfully perform whole-body epidermal replacement for a child suffering from junctional epidermolysis bullosa (Hirsch et al., 2017). Collectively, these findings underscore the clinical potential for epidermal stem cells.
For my own research, I have always been interested in how stem cells work. What gives them their long-term potential for self-renewal and tissue regeneration during homeostasis and wound repair, and what goes awry when stem cells acquire oncogenic mutations that will lead to malignancy? In Howard's lab, I began by characterizing the main structural proteins, keratins, that these stem cells produced (Fuchs and Green, 1978). I found that the stem cells expressed K5 and K14, and that these were markers of progenitors of other stratified squamous epithelia. As the stem cells differentiated to make tissue, they displayed distinguishing features: epidermal cells expressed K1 and K10, whereas differentiating esophageal cells expressed other keratins (Fuchs and Green, 1980).
When I began at the University of Chicago in 1980, it was at the cusp of DNA recombinant technology. I made a cDNA library to epidermal stem cells and cloned and characterized first the keratin mRNAs and then their genes (Fuchs et al., 1981, Hanukoglu and Fuchs, 1982, Hanukoglu and Fuchs, 1983, Marchuk et al., 1984). Illuminating how these protein pairs form heterodimers and assemble into 10 nm cytoskeletal filaments, we then systematically worked our way toward the autosomal dominant blistering disorders, which we showed were caused by mutations in the keratins we had cloned: Epidermolysis bullosa simplex, a disorder of keratins 5 and 14, was rooted in epidermal stem cell fragility (Coulombe et al., 1991, Vassar et al., 1991); Epidermolytic hyperkeratosis, a disorder of keratins 1 and 10, was rooted in a skin barrier breach due to loss of integrity of the differentiating skin layers (Cheng et al., 1992). The keratin networks provided the stem cells and progeny with the mechanical integrity to withstand the physical traumas to which we subject our skin daily.
In the 1990s, we continued to focus on the cytoskeletal, intercellular, and integrin-mediated adhesion of epidermal stem cells, gaining inroads into understanding how stem cells make tissues. Upon my transition to Rockefeller University, we began focusing on the other epithelial stem cells within the skin that give rise to the hair follicles (HFs) and sebaceous and sweat glands. HFs seemed particularly interesting, as in the mouse, they undergo synchronized, cyclical bouts of quiescence, active tissue regeneration, and destruction. As such, they offered a unique opportunity in the stem cell field to explore how stem cells, which fuel these bouts, transition from a quiescent to a tissue-regenerating mode. At the time, Cotsarelis et al. (1990) had posited that the HF stem cells reside in the bulge, an anatomical structure positioned strategically at the base of the non-cycling portion of the resting hair follicle. We engineered a mouse that allowed us to label all the skin epithelial progenitors with a fluorescent histone, and then switch off the gene and trace the dilution of the histone with each cell division. This enabled us to label the layer of less frequently dividing K5/K14+ progenitors within the bulge, purify and characterize them, and document their stemness by picking colonies cultured from single bulge cells and showing that when engrafted, they gave rise to epidermis, sebaceous glands, and HFs (Blanpain et al., 2004, Tumbar et al., 2004).
Over the past 12 years, we have probed deeper into the biology of these cells and their progeny. Among our major findings is that hair follicle stem cells (HFSCs) spend much of their life in quiescence due to high levels of BMP6 and FGF18 signaling within the inner bulge layer of terminally differentiated cells, which derive from the HFSCs at the end of each hair cycle (Blanpain et al., 2004, Hsu et al., 2011). NFATc1 and FOXC1 are two HFSC transcription factors that are downstream of BMP signaling and which function in HFSC quiescence (Horsley et al., 2008, Lay et al., 2016). With age, BMP signaling and its downstream effectors increase, as HFs spend more time in quiescence (Keyes et al., 2013), and when NFATc1 or FOXC1 are removed, HFs undergo many more cycles than normal (Lay et al., 2016). Under continuous cycling, the HFSC pool is depleted, raising the question as to whether in their native state, HFSCs may need to conserve their proliferative potential to have a sufficient output during their lifetime.
HFSCs must overcome the threshold of BMP in order to transition to a tissue-regenerating mode. During the resting phase, they undergo a buildup of WNT signals and BMP inhibitory signals at the base of the bulge, a region now molecularly distinguished as the hair germ. At the transition to the growth phase, nuclear β-catenin and loss of pSMAD1 are seen in the hair germ, concomitant with proliferation (Genander et al., 2014, Greco et al., 2009). Soon thereafter, SHH is expressed by early stem cell progeny, and for a brief period, these cells become a key signaling component of the niche (Hsu et al., 2014). They signal to the mesenchymal stimulus, the dermal papilla to elevate BMP inhibitory and FGF7/10 proactivating signals to fuel hair growth. They also signal to the bulge to briefly cause self-renewal and fuel production of the outer root sheath of K14/K5 progenitors. As the new follicle grows, the bulge is distanced from the dermal papilla and SHH signaling center and it returns to quiescence (Figure 1). At the end of the hair cycle, some of these outer root sheath cells will survive the destructive phase to form the new bulge and hair germ for the next hair cycle. The old bulge serves to anchor the hair until it is eventually shed, when it merges with the newer bulges. In this way, the stem cells ensure a healthy hair coat, and their ability to generate stimulatory signals in their early progeny and inhibitory signals in their late stage progeny serve as autoregulators of the hair cycle (Hsu et al., 2011, Hsu et al., 2014).
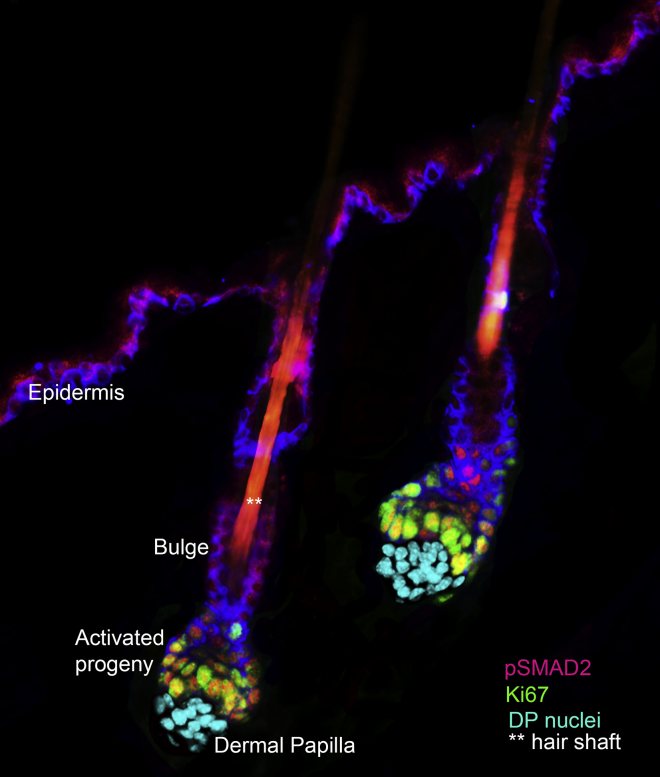
Figure 1.
Early Stages of Stem Cell Activation in Two Hair Follicles
The stem cells that fuel the hair cycle are located within the outermost layer of the bulge niche. At the base of this niche (hair germ) are “primed” stem cells. These stem cells become activated at the start of each new hair cycle to form TGF-β-signaling progenitors that progress to produce the hair shaft and its channel. Several days later, the bulge stem cells begin to proliferate to self-renew and fuel the outer root sheath production that pushes the signaling center with the dermal papilla away from the bulge, returning the bulge to quiescence. Note that other cells within the skin have been removed for the purposes of illustration here, and that the image has been pseudocolored to highlight the mesenchymal stimulus (dermal papilla): Cyan marks nuclei of dermal papilla; red, pSmad2 denotes proliferative progeny that emerged from the base of the bulge stem cell niche at the start of the hair cycle. Green denotes Ki67, a marker of cycling cells. The image was prepared by Naoki Oshimori, when he was a postdoctoral fellow in my laboratory.
More recently, we have focused on how niche signals work together with the stem cell transcription factors to coordinate the balance between stem cell quiescence and activation. We conditionally ablated each of the HFSC transcription factors and showed that not only NFATc1 and FOXC1 but also SOX9, LHX2, and TCF3/4 are required to maintain the HFSCs in their quiescent niche (Lien and Fuchs, 2014, Nguyen et al., 2009, Nowak et al., 2008, Rhee et al., 2006). Upon in vivo chromatin immunoprecipitation and high-throughput sequencing (ChIP-seq), we learned that all of these transcription factors bind to highly clustered binding sites that reside within larger open chromatin regions, coined by the Young lab at MIT as being “super-enhancers” (Adam et al., 2015). While only ∼5% of all the genes expressed by HFSCs are regulated by super-enhancers, they include most of the genes that we know to regulate stemness, including the stem cell transcription factor genes themselves, and also the BMP genes and their receptors, and the WNT receptors and their inhibitory factors, as well as a number of human skin disease genes. Moreover, these regulatory elements also contain binding sites for WNT regulation (TCF3/4) and for BMP regulation (pSMAD1). Our recent mutagenesis studies reveal that when either TCF3/4 or pSMAD1 sites are mutated, enhancer activity is diminished (Adam et al., 2018). During natural HFSC activation, these enhancers are silenced, as BMP signaling is shut off, and WNT signaling promotes the loss of TCF3/4. Interestingly, concomitant with the silencing of these HFSC super-enhancers is the activation of a new set of super-enhancers needed to make the new hair follicle and grow hair (Lien et al., 2014, Yang et al., 2017). The activation of these new super-enhancers appears to be positively regulated by WNT signaling and involves new WNT effectors, LEF1 and TCF1 (Adam et al., 2015). As multipotent progenitors diverge to form the eight lineages of the hair follicle, additional super-enhancers open, regulated by LEF1 and by other signaling effectors, e.g., those of BMP and NOTCH signaling (Adam et al., 2018, Yang et al., 2017). Together, these findings begin to illuminate how stem cells are able to integrate their own transcription factors with the various signaling pathways they are exposed to.
Elaborating on this information, we have focused on how stem cells respond to unforeseen changes in their microenvironment, such as that which occurs when there are placed in tissue culture or face upon a skin injury. Intriguingly, the super-enhancer landscape again changes, enabling these normally quiescent stem cells to survive in a growth-promoting microenvironment. The wound and culture states are very similar, resulting in a downregulation of the repertoire of quiescent stem cell transcription factors and the upregulation of wound-induced stress factors (Adam et al., 2015, Ge et al., 2017). Interestingly, the stem cells enter a transient state of “lineage infidelity,” where they adopt a molecular profile that resembles part HF and part epidermal behavior. Eventually once tissue is repaired, the lineage infidelity resolves, although for HFSCs repairing epidermal wounds, the result is a fate change, as thereafter, the HFSCs appear to look and behave like epidermal stem cells (Ge et al., 2017).
During malignant transformation, the lineage infidelity state becomes permanent, as high levels of oncogenic RAS-MAPK lead to the phosphorylation and super-activation of SOX2 and phosphorylation of pETS2, and new regulatory elements that appear to be controlled by these transcription factors become permanently activated (Ge et al., 2017, Schober and Fuchs, 2011, Yang et al., 2015). Two of my former postdoctoral fellows, Cedric Blanpain (University of Brussels) and William Lowry (UCLA) showed that epidermal and HF stem cells are a source of squamous cell carcinomas (SCCs) in the skin. SCCs are the second most common cancer world-wide, and when SCCs are considered as a broader class of cancers, including those of head and neck, lung, esophagus, breast, and cervix, they are one of the most deadly cancers, for which treatments are often inadequate to halt disease progression and metastasis.
In recent years, we have been exploring the role of stem cells in malignant progression of SCCs. In 2011, we isolated and characterized two populations of tumor-initiating, so-called “cancer stem cells,” which differed in their proliferation rates. Given our prior work showing that loss of transforming growth factor β (TGF-β) signaling in skin stem cells renders them prone to SCC formation (Guasch et al., 2007), we devised a method where we could mark and trace the TGF-β-responding stem cells at the tumor-stroma interface to probe its other role as a tumor-promoter. By selectively marking and tracing these stem cells, we learned that the TGF-β signal emanates from the perivasculature, and wherever a blood vessel approaches the tumor-stroma interface, the stem cells respond by reducing their cycling activity but becoming invasive and resistant to chemotherapy (Oshimori et al., 2015). Although their reduced proliferative activity may play some role in protection (Brown et al., 2017), the ability of TGF-β to lead to a cascade culminating in upregulation of the glutathione pathway, allows these stem cells to counter reactive oxygen species, as encountered by radiotherapy, as well as chemotherapies, for example cisplatin (Oshimori et al., 2015). These new findings provide insights and begin to suggest possible combinatorial drugs for targeting this deadly cancer.
In addition, we have performed large-scale genetic screens for oncogenes, tumor suppressors, and microRNAs in skin cancers, as well as screens for genes that control the balance between epidermal stem cell growth and proliferation (Asare et al., 2017, Beronja et al., 2013, Ge et al., 2017, Schramek et al., 2014). These screens exploit a powerful genetic method developed by my lab, which enables us to expose a living mouse embryo to lentivirus just after gastrulation, when skin exists as a single layer of unspecified progenitors that will give rise to the epidermis, HFs, mammary, sweat and sebaceous glands, and head and neck epithelium (Beronja et al., 2010). Once integrated, the lentiviral DNA is stably propagated, resulting in selective transduction of the entire surface epithelium, including their stem cells. Thus, in a very short time, we can perform complicated genetics that would take years to achieve with conventional genetics. With the advent of tools for CRISPR/CAS and tetracycline-inducible methods, we can now knock down (shRNA), knock out (sgRNA), or inducibly express (tetracycline enhancer-driven cDNAs) genes in the skin stem cells. We have exploited these methods to test the functionality and develop new drivers for skin stem cells in their homeostatic, wound-induced, inflammatory, and cancerous states (Adam et al., 2015, Ge et al., 2017, Naik et al., 2017). These methods will allow us to expedite our understanding of stem cell biology for decades to come.
Although much of our studies have focused on how stem cells integrate signaling pathways at the transcriptional level, we are cognizant of other levels of regulation. In the past year, we have used in vivo ribosomal profiling to study how stem cells respond when they receive an oncogenic stress, e.g., elevated SOX2, frequently amplified in human SCCs. We discovered that stem cells phosphorylate and dampen activity of the canonical initiation pathways and allow a less efficient EIF2A initiator to take over. Intriguingly, this switch is required for stem cells to survive oncogenic stress and develop tumors. Moreover, it selectively acts on a group of oncogenic mRNAs with unusual 5′ characteristics that allow their translation to be sustained in the face of global reductions in protein synthesis (Sendoel et al., 2017). Intriguingly, the EIF2A locus is frequently amplified in human SCCs, and although the locus contains other genes, when levels of EIF2A mRNAs are analyzed, patients with the highest levels show the poorest prognosis (Sendoel et al., 2017). These findings offer another potential therapeutic target for SCCs, which we will pursue in the future.
In summary, our studies began with the basic characterization of skin stem cells, first in vitro and then in vivo. We then turned to analyzing how K5/K14 progenitors change as they find themselves in different niches. Whether normal or cancerous, and epidermal, sweat gland, or HF, the progenitors are marked by these keratins and reside at the interface between the epithelium and mesenchyme (Fuchs and Green, 1980, Horsley et al., 2006, Lu et al., 2012, Schober and Fuchs, 2011, Tumbar et al., 2004) Their distinct microenvironments have a profound impact on their behavior, and markedly change their transcriptional and chromatin landscape (Gonzales and Fuchs, 2017). When removed from their homeostatic microenvironment, stem cells adopt plasticity, enabling them to select a broader repertoire of tissue regeneration than they normally would do (Blanpain and Fuchs, 2014). Now comfortable with being uncomfortable, I have learned that in stem cell biology, even though experiments rarely if ever deliver unequivocal answers, they almost always lead to new exciting questions. The next decade in stem cell research is bound to be even more exciting, with new insights and clinical translations.
In closing, I was fortunate as a graduate student at Princeton University to have professors of cell biology who have had a lasting impact on my own career. They taught me the importance of rigorous science and of taking a multidisciplinary and molecular approach to tackling scientific questions. At MIT with Howard Green, I applied these teachings to my passion for a mammalian cell culture system where I could examine living, normal diploid human cells under conditions where they could endlessly divide but also be induced to differentiate and make epidermis. Looking back, I see the threads of influence from all my professors woven into my career. This has always been a guide for me in my own career that education and mentorship are as important to our profession as being passionate about the science we do.
In many respects, I was destined to investigate stem cell biology, wrapped into the complex problem of tissue biology. In the now over three decades of my career, I cannot imagine focusing on any other science. I have been fortunate to witness the emergence of our ever-growing community of stem cell researchers. I feel privileged to be a part of the ISSCR community, and in 2010, it was a pleasure to serve as its President. My friendships and colleagues in this community will always be precious to me and as long lasting as epidermal stem cells. I am grateful for having been chosen to receive the McEwen Award for Innovative Stem Cell Research in 2017, as there is no greater honor than to be recognized by those for whom I have enormous respect and regard. But this honor would not have been possible were it not for the numerous students, postdocs, and staff who I have had the pleasure to mentor and guide over these years. To name but a few who have trained in my lab at Rockefeller and remain in the stem cell community: Doina Tumbar, now Professor at Cornell University; Cedric Blanpain, now Professor at the University of Brussels; Bill Lowry, Associate Professor at UCLA; Valerie Horsley and Valentina Greco, Associate Professors at Yale; Michael Rendl and Elena Ezhkova, Associate Professors at Mt Sinai Stem Cell Institute; Terry Lechler, Associate Professor at Duke; Colin Jamora and Srikala Raghavan, Associate Professors at the Stem Cell Institute in Bangalore; Ramanuj DasGupta, Professor at the Biopolis in Singapore; Mirna Perez-Moreno, Associate Professor at U Copenhagen; Kris and Agnes Kobielak, Professors at U Medical Sciences Poznan; Wen-Hui Lien (de Duve Institute); Rui Yi, Associate Professor at U Colorado Boulder Danelle Devenport, Associate Professor at Princeton, and Assistant Professors: Marcus Schober (NYU), Xiaoyang Wu (U Chicago), Ya-Chieh Hsu (Harvard Stem Cell Institute), Slobodan Beronja (Fred Hutchison Cancer Institute), Hoang Nguyen, (Baylor); Naoki Oshimori (Oregon Health Science Center); Daniel Schramek (University of Toronto), Scott Williams (University of North Carolina), Ting Chen (NIBS, Beijing), Brice Keyes (Calico), Maria Genander (Karolinska Institute), Ellen Ezratty (Columbia University), Chuck Kaufman (Washington University) and Jonathan Nowak (Harvard). Every time one of my students or postdocs departs, I am always bittersweet---sorry to see them go, but joyful to see them advance in their careers. It was the same with my very first and will be the same with those currently in my lab, who I have had the pleasure to mentor. Their natural sense of collegiality and interactiveness has been extraordinary and passes from generation to generation. This recognition is as much about them as me.

Elaine Fuchs received her undergraduate degree in Physical Chemistry from The University of Illinois, where she graduated with the highest distinction. She went on to earn her DPhil in Biochemistry from Princeton University. As a Damon Runyon postdoctoral fellow under the tutelage of Professor Howard Green, Elaine began to tackle the molecular biology of epidermal stem cells as a tool to understand how tissues balance growth and differentiation. Elaine’s early studies on the faculty at the University of Chicago culminated in several landmark papers in Cell in which she pioneered reverse genetics, starting with biochemical and molecular dissection of the stem cell’s structural proteins, keratins, and then turning to mice and final human patients to guide her to the basis of blistering skin disorders. Elaine’s group has gone on to elucidate how epidermal stem cells utilize cellular and cytoskeletal interactions to generate and replenish the skin’s barrier. Upon her move to Rockefeller University in 2002, Elaine began to focus on other skin stem cells, including those of the hair follicle and sweat glands, to illuminate how these very different tissue structures are generated and replenished. Her current interests are in the realm of how stem cells cope with stress, from wound repair to inflammation to cancer. Elaine is a Howard Hughes Medical Institute Investigator, an elected member of the National Academy of Sciences, National Academy of Medicine and the American Philosophical Society. She has received numerous awards, including the United States’ highest scientific honor, the National Medal of Science from President Obama, the Albany Prize in Medicine, the L’Oreal-UNESCO Women in Science Award, the EB Wilson Award in Cell Biology, the International Pezcoller Award for Cancer Research and the McEwen Award for Innovation in Stem Cell Research. She has also received honorary doctorates, most recently from Harvard University.
References
- Adam R.C., Yang H., Ge Y., Lien W.H., Wang P., Zhao Y., Polak L., Levorse J., Baksh S.C., Zheng D. Temporal layering of signaling effectors drives chromatin remodeling during hair follicle stem cell lineage progression. Cell Stem Cell. 2018;22:398–413.e7. doi: 10.1016/j.stem.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam R.C., Yang H., Rockowitz S., Larsen S.B., Nikolova M., Oristian D.S., Polak L., Kadaja M., Asare A., Zheng D. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015;521:366–370. doi: 10.1038/nature14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asare A., Levorse J., Fuchs E. Coupling organelle inheritance with mitosis to balance growth and differentiation. Science. 2017;355 doi: 10.1126/science.aah4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beronja S., Livshits G., Williams S., Fuchs E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat. Med. 2010;16:821–827. doi: 10.1038/nm.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beronja S., Janki P., Heller E., Lien W.H., Keyes B.E., Oshimori N., Fuchs E. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature. 2013;501:185–190. doi: 10.1038/nature12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C., Fuchs E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science. 2014;344:1242281. doi: 10.1126/science.1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C., Lowry W.E., Geoghegan A., Polak L., Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Brown J.A., Yonekubo Y., Hanson N., Sastre-Perona A., Basin A., Rytlewski J.A., Dolgalev I., Meehan S., Tsirigos A., Beronja S. TGF-β-induced quiescence mediates chemoresistance of tumor-propagating cells in squamous cell carcinoma. Cell Stem Cell. 2017;21:650–664.e8. doi: 10.1016/j.stem.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Syder A.J., Yu Q.C., Letai A., Paller A.S., Fuchs E. The genetic basis of epidermolytic hyperkeratosis: a disorder of differentiation-specific epidermal keratin genes. Cell. 1992;70:811–819. doi: 10.1016/0092-8674(92)90314-3. [DOI] [PubMed] [Google Scholar]
- Cotsarelis G., Sun T.T., Lavker R.M. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- Coulombe P.A., Hutton M.E., Vassar R., Fuchs E. A function for keratins and a common thread among different types of epidermolysis bullosa simplex diseases. J. Cell Biol. 1991;115:1661–1674. doi: 10.1083/jcb.115.6.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Green H. The expression of keratin genes in epidermis and cultured epidermal cells. Cell. 1978;15:887–897. doi: 10.1016/0092-8674(78)90273-8. [DOI] [PubMed] [Google Scholar]
- Fuchs E., Green H. Changes in keratin gene expression during terminal differentiation of the keratinocyte. Cell. 1980;19:1033–1042. doi: 10.1016/0092-8674(80)90094-x. [DOI] [PubMed] [Google Scholar]
- Fuchs E.V., Coppock S.M., Green H., Cleveland D.W. Two distinct classes of keratin genes and their evolutionary significance. Cell. 1981;27:75–84. doi: 10.1016/0092-8674(81)90362-7. [DOI] [PubMed] [Google Scholar]
- Ge Y., Gomez N.C., Adam R.C., Nikolova M., Yang H., Verma A., Lu C.P., Polak L., Yuan S., Elemento O. Stem cell lineage infidelity drives wound repair and cancer. Cell. 2017;169:636–650. doi: 10.1016/j.cell.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genander M., Cook P.J., Ramskold D., Keyes B.E., Mertz A.F., Sandberg R., Fuchs E. BMP signaling and its pSMAD1/5 target genes differentially regulate hair follicle stem cell lineages. Cell Stem Cell. 2014;15:619–633. doi: 10.1016/j.stem.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco V., Chen T., Rendl M., Schober M., Pasolli H.A., Stokes N., Dela Cruz-Racelis J., Fuchs E. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H. Cultured cells for the treatment of disease. Sci Am. 1991;265:96–102. doi: 10.1038/scientificamerican1191-96. [DOI] [PubMed] [Google Scholar]
- Gonzales K.A.U., Fuchs E. Skin and its regenerative powers: an alliance between stem cells and their niche. Dev Cell. 2017;43:387–401. doi: 10.1016/j.devcel.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch G., Schober M., Pasolli H.A., Conn E.B., Polak L., Fuchs E. Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell. 2007;12:313–327. doi: 10.1016/j.ccr.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanukoglu I., Fuchs E. The cDNA sequence of a human epidermal keratin: divergence of sequence but conservation of structure among intermediate filament proteins. Cell. 1982;31:243–252. doi: 10.1016/0092-8674(82)90424-x. [DOI] [PubMed] [Google Scholar]
- Hanukoglu I., Fuchs E. The cDNA sequence of a Type II cytoskeletal keratin reveals constant and variable structural domains among keratins. Cell. 1983;33:915–924. doi: 10.1016/0092-8674(83)90034-x. [DOI] [PubMed] [Google Scholar]
- Hirsch T., Rothoeft T., Teig N., Bauer J.W., Pellegrini G., De Rosa L., Scaglione D., Reichelt J., Klausegger A., Kneisz D. Regeneration of the entire human epidermis using transgenic stem cells. Nature. 2017;551:327–332. doi: 10.1038/nature24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V., Aliprantis A.O., Polak L., Glimcher L.H., Fuchs E. NFATc1 balances quiescence and proliferation of skin stem cells. Cell. 2008;132:299–310. doi: 10.1016/j.cell.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V., O'Carroll D., Tooze R., Ohinata Y., Saitou M., Obukhanych T., Nussenzweig M., Tarakhovsky A., Fuchs E. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell. 2006;126:597–609. doi: 10.1016/j.cell.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.C., Li L., Fuchs E. Emerging interactions between skin stem cells and their niches. Nat. Med. 2014;20:847–856. doi: 10.1038/nm.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.C., Pasolli H.A., Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes B.E., Segal J.P., Heller E., Lien W.H., Chang C.Y., Guo X., Oristian D.S., Zheng D., Fuchs E. Nfatc1 orchestrates aging in hair follicle stem cells. Proc. Natl. Acad. Sci. USA. 2013;110:E4950–E4959. doi: 10.1073/pnas.1320301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay K., Kume T., Fuchs E. FOXC1 maintains the hair follicle stem cell niche and governs stem cell quiescence to preserve long-term tissue-regenerating potential. Proc. Natl. Acad. Sci. USA. 2016;113:E1506–E1515. doi: 10.1073/pnas.1601569113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien W.H., Fuchs E. Wnt some lose some: transcriptional governance of stem cells by Wnt/beta-catenin signaling. Genes Dev. 2014;28:1517–1532. doi: 10.1101/gad.244772.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien W.H., Polak L., Lin M., Lay K., Zheng D., Fuchs E. In vivo transcriptional governance of hair follicle stem cells by canonical Wnt regulators. Nat. Cell Biol. 2014;16:179–190. doi: 10.1038/ncb2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.P., Polak L., Rocha A.S., Pasolli H.A., Chen S.C., Sharma N., Blanpain C., Fuchs E. Identification of stem cell populations in sweat glands and ducts reveals roles in homeostasis and wound repair. Cell. 2012;150:136–150. doi: 10.1016/j.cell.2012.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchuk D., McCrohon S., Fuchs E. Remarkable conservation of structure among intermediate filament genes. Cell. 1984;39:491–498. doi: 10.1016/0092-8674(84)90456-2. [DOI] [PubMed] [Google Scholar]
- Naik S., Larsen S.B., Gomez N.C., Alaverdyan K., Sendoel A., Yuan S., Polak L., Kulukian A., Chai S., Fuchs E. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature. 2017;550:475–480. doi: 10.1038/nature24271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H., Merrill B.J., Polak L., Nikolova M., Rendl M., Shaver T.M., Pasolli H.A., Fuchs E. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat. Genet. 2009;41:1068–1075. doi: 10.1038/ng.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak J.A., Polak L., Pasolli H.A., Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N., Oristian D., Fuchs E. TGF-beta promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell. 2015;160:963–976. doi: 10.1016/j.cell.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H., Polak L., Fuchs E. Lhx2 maintains stem cell character in hair follicles. Science. 2006;312:1946–1949. doi: 10.1126/science.1128004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinwald J.G., Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–343. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- Schober M., Fuchs E. Tumor-initiating stem cells of squamous cell carcinomas and their control by TGF-beta and integrin/focal adhesion kinase (FAK) signaling. Proc. Natl. Acad. Sci. USA. 2011;108:10544–10549. doi: 10.1073/pnas.1107807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramek D., Sendoel A., Segal J.P., Beronja S., Heller E., Oristian D., Reva B., Fuchs E. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science. 2014;343:309–313. doi: 10.1126/science.1248627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendoel A., Dunn J.G., Rodriguez E.H., Naik S., Gomez N.C., Hurwitz B., Levorse J., Dill B.D., Schramek D., Molina H. Translation from unconventional 5’ start sites drives tumour initiation. Nature. 2017;541:494–499. doi: 10.1038/nature21036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T., Guasch G., Greco V., Blanpain C., Lowry W.E., Rendl M., Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R., Coulombe P.A., Degenstein L., Albers K., Fuchs E. Mutant keratin expression in transgenic mice causes marked abnormalities resembling a human genetic skin disease. Cell. 1991;64:365–380. doi: 10.1016/0092-8674(91)90645-f. [DOI] [PubMed] [Google Scholar]
- Yang H., Schramek D., Adam R.C., Keyes B.E., Wang P., Zheng D., Fuchs E. ETS family transcriptional regulators drive chromatin dynamics and malignancy in squamous cell carcinomas. eLife. 2015;4:e10870. doi: 10.7554/eLife.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Adam R.C., Ge Y., Hua Z.L., Fuchs E. Epithelial-mesenchymal micro-niches govern stem cell lineage choices. Cell. 2017;169:483–496.e13. doi: 10.1016/j.cell.2017.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]