Abstract
Circadian locomotor output cycles kaput (CLOCK) is a transcription factor that activates transcription of clock-controlled genes by heterodimerizing with BMAL1 and binding to E-box elements on DNA. Although several phosphorylation sites on CLOCK have already been identified, this study characterizes a novel phosphorylation site at serine 845 (Ser-836 in humans). Here, we show that CLOCK is a novel AKT substrate in vitro and in cells, and this phosphorylation site is a negative regulator of CLOCK nuclear localization by acting as a binding site for 14-3-3 proteins. To examine the role of CLOCK phosphorylation in vivo, ClockS845A knockin mice were generated using CRISPR/Cas9 technology. ClockS845A mice are essentially normal with normal central circadian rhythms and hemodynamics. However, examination of core circadian gene expression from peripheral tissues demonstrated that ClockS845A mice have diminished expression of Per2, Reverba, Dbp, and Npas2 in skeletal muscle and Per2, Reverba, Dbp, Per1, Rora, and Npas2 in the liver during the circadian cycle. The reduction in Dbp levels is associated with reduced H3K9ac at E-boxes where CLOCK binds despite no change in total CLOCK levels. Thus, CLOCK phosphorylation by AKT on Ser-845 regulates its nuclear translocation and the expression levels of certain core circadian genes in insulin-sensitive tissues.
Keywords: Akt PKB, circadian rhythm, substrate specificity, phosphorylation, mouse genetics
Introduction
The core positive activators of the circadian transcriptional feedback loop are BMAL1, CLOCK, and NPAS2. To activate circadian transcription, BMAL1 heterodimerizes with either CLOCK or NPAS2 through their basic helix-loop-helix–per-arnt-sim domains (1). Together, these transcription factors bind to E-box DNA-response elements where they affect transcription of a number of clock-controlled genes. In fact, 5–10% of transcripts in any given tissue are estimated to undergo circadian rhythms (2, 3). Hence, the activity of these transcription factors must be intricately controlled to maintain the circadian rhythmicity of transcripts.
Circadian rhythms are regulated stringently by post-translational modifications that often follow circadian patterns. Multiple kinases have been shown to phosphorylate circadian transcription factors. For example, GSK3β phosphorylates both positive regulators CLOCK and BMAL1 (4), which reduces their stability and increases transcriptional activity. Additionally, AKT2 phosphorylates BMAL1 at Ser-42 in hepatocytes (5) promoting its cytoplasmic localization. The model states that phospho-BMAL1 either binds the adaptor protein 14-3-3 in the nucleus, which exports BMAL1 to the cytosol, and/or this interaction delays BMAL1 re-entry to the nucleus by masking its nuclear localization sequence (5).
CLOCK was the first circadian protein discovered in mammals as an effector of circadian locomotor behavior (6). Neither Clock transcripts nor CLOCK protein levels cycle in all tissues, unlike the other circadian feedback loop regulators (7, 8), suggesting that CLOCK activity is more likely regulated post-translationally. Three independent phosphoproteomic screens examining substrates specific to AKT isoform deletion (9) or insulin signaling (10, 11) found CLOCK phosphorylation on Ser-845. The role of CLOCK Ser-845 has not been confirmed or tested in vitro or in vivo.
Here, we show that CLOCK Ser-845 is phosphorylated in vitro and in cells in an AKT-dependent manner. AKT phosphorylation of CLOCK does not influence CLOCK turnover but generates a 14-3-3 binding site that influences CLOCK nuclear/cytoplasmic shuttling. To study the role of CLOCK Ser-845 phosphorylation in vivo, a CRISPR/Cas9 knockin mouse where the phospho-site serine is mutated to an alanine (CLOCKS845A) was generated. CLOCKS845A mice exhibit decreased hepatic and skeletal muscle circadian gene transcription mid-way through the light period, which correlates to the time CLOCK/BMAL1 activation occurs without differences in feeding or activity consistent with data in Clock mutant and null mice (12–15). Therefore, CLOCK Ser-845 phosphorylation site (Ser-845) impacts CLOCK activity and may link nutrient sensing to circadian transcription.
Results
AKT phosphorylates CLOCK Ser-845
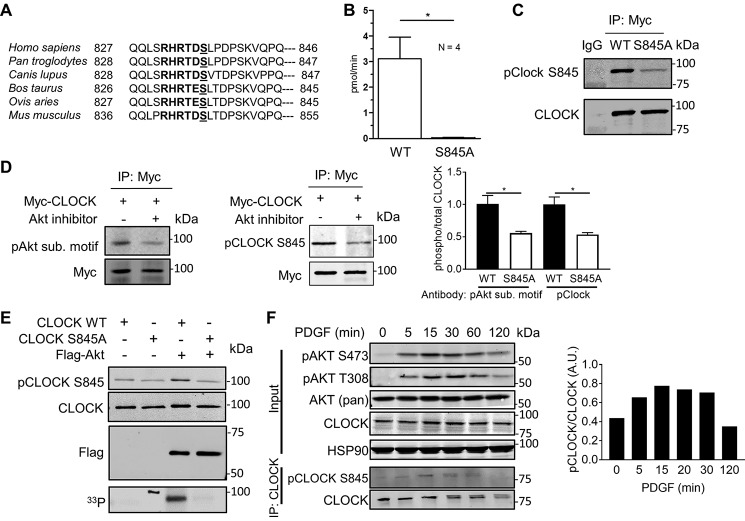
A phosphoproteomics screen examining AKT-specific substrates in mouse endothelial cells determined that endogenous CLOCK was phosphorylated at Ser-845 (9). This site of CLOCK phosphorylation was also identified by MS in two additional phospho-screens investigating insulin-responsive phosphorylation sites (10) and PI3K-Akt–sensitive sites (11) in NIH3T3-L1 adipocytes. Upon further examination, this site has an AKT substrate motif, RXRXXpS, where X represents any amino acid, and pS is a phosphoserine acceptor site. This motif in CLOCK is conserved among mammals (Fig. 1A), although it is not conserved in lower organisms such as flies, implying that this phosphorylation site is evolved in complex organisms to regulate intricate aspects of circadian rhythms.
Figure 1.
AKT phosphorylates CLOCK in vitro. A, C terminus of CLOCK has a conserved RXRXXS phosphorylation motif in mammals. B, 16-amino acid peptide starting with the −5 arginine (Arg-840 in mouse) is phosphorylated by AKT in vitro. C, Myc-CLOCK WT or Myc-CLOCK S845A was immunoprecipitated from transfected HEK293T cells. Immunoblotting with the pCLOCK Ser-845 antibody demonstrates specificity of the antibody. There was minor binding with CLOCK S845A, but the antibody binds more strongly with phosphorylated CLOCK Ser-845. D, Myc-CLOCK WT was transfected into HEK293T cells and either treated with vehicle (DMSO) or Akt inhibitor MK2206 (5 μm, 2 h). The pAkt substrate motif antibody and the pClock Ser-845 antibody bind strongly with the untreated Myc-CLOCK as quantified to the right (mean ± S.E. n = 3. *, p < 0.05). E, in vitro kinase assay with immunoisolated CLOCK and AKT shown by Western blotting (top panel) and autoradiography with [γ-33P]ATP (bottom panel). F, stimulation of NIH3T3 fibroblasts with PDGF-BB (50 ng/ml) induces phosphorylation of CLOCK Ser-845, shown using a phospho-site-specific antibody. This experiment is representative of at least three independent experiments and is quantified (pClock/total Clock) to the right. D and F, immunoprecipitated CLOCK was first blotted for p-CLOCK and then re-probed for total CLOCK levels. Unless noted, data are presented as mean ± S.E. n = 3. *, p < 0.05.
To experimentally validate CLOCK Ser-845 phosphorylation by AKT, several independent approaches were used. Initially, in vitro kinase assays were performed using immunoisolated, activated FLAG-AKT, a 16-amino acid CLOCK peptide surrounding the phospho-site or immunoprecipitated wild type (WT) CLOCK or CLOCK S845A proteins as substrates. FLAG-AKT phosphorylated the CLOCK peptide, and phosphorylation was eliminated in the peptide containing the S845A mutation (Fig. 1B). Using either a pCLOCK Ser-845 antibody (validated in Fig. 1C) or [33P]ATP incorporation into immunoisolated CLOCK (Fig. 1E, lane 3), WT CLOCK was phosphorylated by AKT, and phosphorylation was reduced in the CLOCK S845A mutant. The faint recognition of S845A CLOCK with the pCLOCK Ser-845 antibody but the elimination of [33P]ATP incorporation imply that this is due to contamination of the antibody with total anti-CLOCK antibody during the negative selection process. Next, HEK293T cells were transfected with Myc-CLOCK and treated with an allosteric inhibitor of AKT, MK2206, and CLOCK Ser-845 phosphorylation of immunoprecipitated CLOCK was examined. Inhibition of AKT reduced CLOCK phosphorylation on Ser-845 as assessed using a p-AKT substrate motif antibody (Fig. 1D, left) or the pCLOCK Ser-845 antibody (Fig. 1D, middle and quantified at right). Finally, stimulation of the PI3K-Akt signaling axis with platelet-derived growth factor (PDGF)2 in NIH3T3 cells induced time-dependent CLOCK phosphorylation after immunoprecipitation of CLOCK, as detected with the pCLOCK Ser-845 antibody (Fig. 1F). pCLOCK Ser-845 levels were not detectable in whole-cell lysates implying that the level of CLOCK phosphorylation is low or that the antibody has a low titer and/or affinity for this site. Collectively, these data demonstrate that CLOCK Ser-845 is a novel AKT substrate in vitro and in cells.
CLOCK Ser-845 phosphorylation does not influence its stability but regulates nuclear translocation via binding to 14-3-3
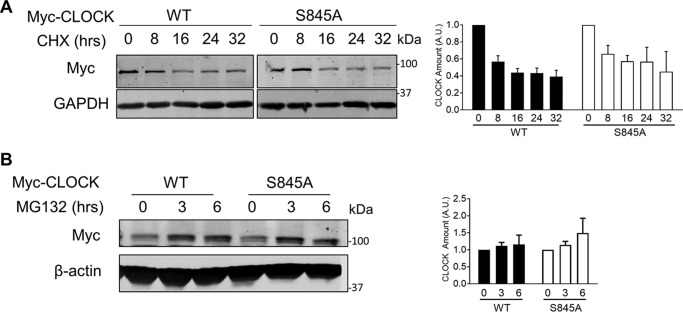
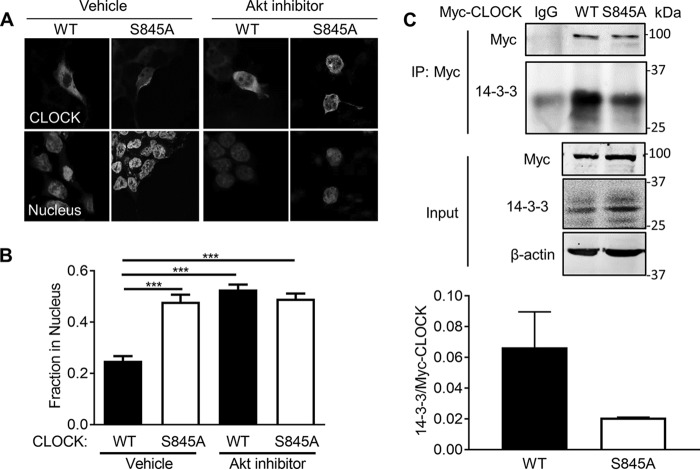
To investigate the role for CLOCK Ser-845 phosphorylation in cells, several experiments were conducted. Because phosphorylation can regulate protein stability and turnover, CLOCK stability (Myc-tagged WT and S845A CLOCK) after transfection into HEK293T cells was examined after blocking protein synthesis with cycloheximide (CHX) or proteasomal degradation with MG132. Neither CHX nor MG132 treatment had a differential effect on WT or S845A CLOCK levels after transfection (Fig. 2, A and B). Because AKT phosphorylation of transcription factors such as FOXO1 regulates its subcellular distribution, we tested whether phosphorylation of CLOCK impacted its nuclear localization. Expression of CLOCK in HEK293T cells, followed by quantitative imaging, demonstrated that ∼25% of WT CLOCK is localized to the nucleus as compared with 50% of CLOCK S845A (Fig. 3, A and quantified in B; p < 0.001). Inhibition of AKT with MK2206 increased nuclear localization of WT CLOCK to similar levels as CLOCK S845A, but it did not impact the levels of nuclear CLOCK S845A (p < 0.001). These data imply that phosphorylation of Ser-845 is important for the cytoplasmic/nuclear shuttling of CLOCK.
Figure 2.
CLOCK stability and degradation are not affected by phosphorylation at Ser-845. A, HEK293T cells were transfected with Myc-CLOCK and Myc-CLOCK S845A and treated with CHX. Quantification of four experiments, where total CLOCK was normalized to GAPDH loading control, is shown on the right. Graph is presented as mean ± S.E., n = 4 experiments. B, HEK293T cells were transfected with Myc-CLOCK WT and Myc-CLOCK S845A and treated with MG132. Graph is presented as mean ± S.E., n = 3 experiments.
Figure 3.
Phosphorylation of Ser-845 regulates nuclear CLOCK localization and its interaction with 14-3-3. A, immunofluorescence of Myc-CLOCK WT or Myc-CLOCK S845A transfected into HEK293T cells treated with either vehicle or Akt inhibitor MK2206 (5 μm). B, quantification with ImageJ of nuclear localization calculated by the percentage of CLOCK in the nucleus (co-localization with nuclear stain ToPRO3) compared with the rest of the CLOCK staining within the cell, averaged from greater than 20 cells per experiment, n = 5 individual experiments. C, co-immunoprecipitation of either Myc-CLOCK WT or S845A with endogenous 14-3-3 proteins (pan-14-3-3 antibody) from lysates of HEK293T cells. n = 3. Data are presented at mean ± S.E. ***, p < 0.001.
Many AKT substrates, especially transcription factors such as those of the FOXO family (16) and BMAL1 (5), bind to 14-3-3 proteins, thereby preventing their nuclear localization, altering their conformation, or increasing their ability to bind with other proteins. The 14-3-3 family preferentially binds to one of two canonical phosphorylated motifs or a less defined noncanonical motif. The region surrounding CLOCK Ser-845 (RHRTDSLTDP) is predicted to be a canonical Arg-containing phospho-motif for 14-3-3 binding by protein prediction programs such as the eukaryotic linear motif resource. To test this possibility, HEK293T cells were transfected with Myc-tagged WT or S845A CLOCK and Myc-tagged proteins immunoprecipitated and then blotted for associated 14-3-3. Immunoprecipitation of Myc-tagged WT CLOCK resulted in greater co-precipitation of 14-3-3 than did Myc-tagged CLOCK S845A (Fig. 3C for blots and quantification below). Residual binding of 14-3-3 to CLOCK S845A may be due to 14-3-3 binding to one of the other five predicted 14-3-3-binding motifs or through BMAL1, which also binds 14-3-3 proteins after AKT phosphorylation (5).
CLOCK S845A mice are viable and healthy
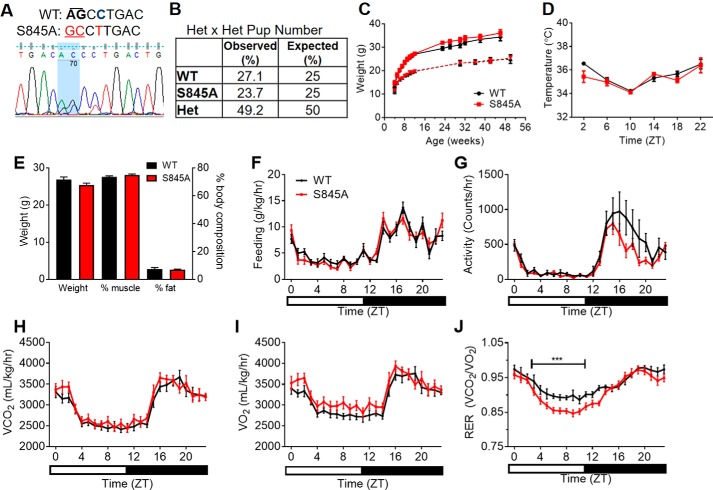
To study the importance of CLOCK Ser-845 in vivo, CRISPR/Cas9 technology was employed to generate CLOCK S845A knockin mice. Mutant mice were generated with a single allele where the codon for serine 845 (AGC) was mutated to alanine (GCC) (Fig. 4A). Additionally, to prevent the guide RNA recognizing the donor template, a single base pair synonymous substitution (cytosine to thymine) was engineered into the corresponding PAM sequence, resulting in an unchanged leucine residue just after the mutation site. The top three most likely off-target mutations were sequenced from genomic DNA template, and only those mice without off-target mutations were used for breeding. To generate the mice used for experiments, the ClockS845A heterozygous mice were backcrossed 1–2 generations to WT C57Bl/6J mice. After backcrossing, ClockS845A heterozygotes were crossed to produce heterozygotes for breeding, and F2 WT and ClockS845A homozygote (CLOCK S845A) mice were used for experiments. Upon genotyping greater than 100 pups, the observed frequencies of each genotype were as expected (Fig. 4B). Thus, this mutation does not affect development or viability.
Figure 4.
CLOCK S845A knockin mice are viable and healthy. A, sequencing demonstrating a mouse heterozygous for the CLOCK S845A mutation. The wild-type (WT) sequence has an AGC codon for a serine, whereas the S845A allele has a GCC encoding an alanine. The highlighted portion of the sequencing data shows two visible peaks for the AG → GC mutation of one allele. Just after this codon, there is silent mutation of a cytosine (bold, blue) to a thymine (red) in the mutant, which still encodes for leucine. B, percentage of each genotype from over 100 pups of heterozygous (HET) mouse mating pairs; the percentages of observed to expected mice were similar. C, weight of WT (black) and S845A (red) mice over time. Males are represented by a solid line; females are the dotted line. n = 4–5 mice per time point. D, body temperature in 12–16-week-old male mice throughout the daily cycle, where ZT0 represents lights on. n = 3–4 mice per time point. E–J, 16-week-old S845A male mice and WT littermates were used for metabolic cage experiments. E, body weight and body composition of WT and S845A mice. F, feeding patterns for WT (black) and S845A (red) over the course of a day. G, activity patterns throughout the day. H, volume of CO2 emitted; I, volume of O2 consumed. J, respiratory exchange ratio (VCO2/VO2) throughout the day. Data are represented as mean ± S.E. ZT, zeitgeber time, or time after lights on. The bar at the bottom represents the light and dark cycle.
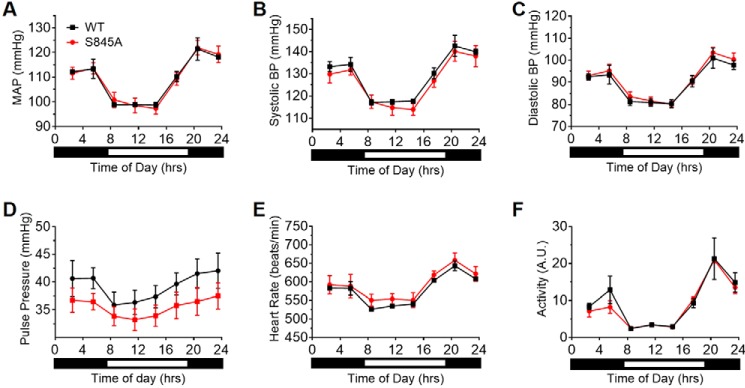
Over the course of a year, both male and female (Fig. 4C) mice were identical in body weight. In addition, the following parameters were not different in littermate WT mice versus CLOCK S845A mice: basal body temperature (Fig. 4D); body composition (Fig. 4E); food consumption (Fig. 4F); and activity (Fig. 4G). There was no change in VCO2 (Fig. 4H) or VO2 (Fig. 4I); however, there was lower respiratory exchange ratio (Fig. 4J, p < 0.001) in CLOCK S845A mice from approximately ZT5 to ZT12 (1 p.m. to 7 p.m.) suggesting a metabolic preference for fatty acid utilization over carbohydrates. Cardiovascular hemodynamics were also examined, and no differences were found in mean arterial pressure, systolic blood pressure, diastolic blood pressure, pulse pressure, heart rate, and activity when the data were analyzed as 3-h averages throughout the day (Fig. 5, A–F, respectively).
Figure 5.
CLOCK S845A mice do not have cardiovascular defects. No significant differences were found between WT (black) and S845A (red) mice in mean arterial pressure (A), systolic blood pressure (B), diastolic blood pressure (C), pulse pressure (D), heart rate (E), and activity (F). n = 3 mice per genotype. Data are represented as mean of 3 h of measurements ± S.E.
CLOCK S845A mice do not have central circadian rhythm locomotor or behavioral defects but have altered circadian gene expression in peripheral tissues
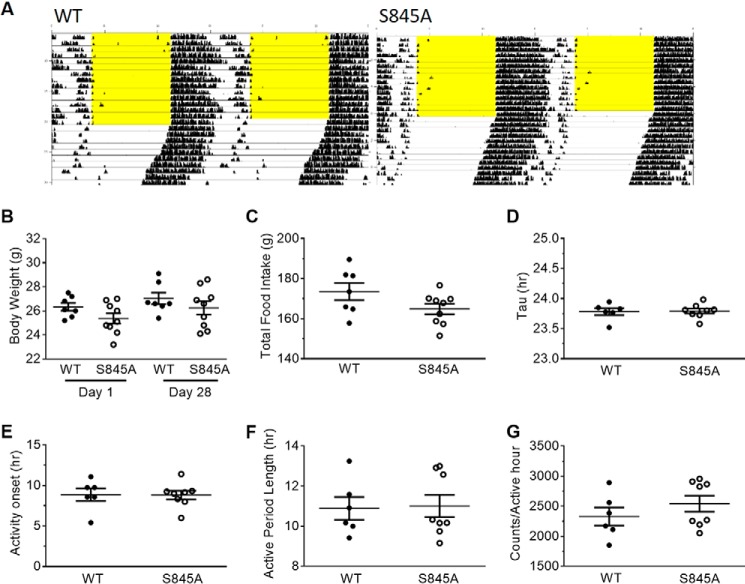
There is conflicting evidence whether CLOCK plays a role in the regulation of central circadian rhythms as measured by locomotor activity throughout the day. Clock homozygous mutant mice with a deletion of exon 19 are arrhythmic and do not maintain behavioral circadian rhythms (6, 17), but Clock KO mice do not have a locomotor rhythm defect (18). Thus, to examine the impact of Ser-845 CLOCK phosphorylation on locomotor rhythms, central circadian rhythms were examined. A representative actogram from a WT and an S845A mouse are shown in Fig. 6A. Overall there were no differences in the body weight (Fig. 6B), food intake (Fig. 6C), τ (period) (Fig. 6D), activity onset (Fig. 6E), length of active phase (α) (Fig. 6F), or activity levels/h (Fig. 6G) of S845A mice compared with WT littermate mice. Thus, CLOCK S845A mice are similar to CLOCK KO mice and do not show a locomotor phenotype (18).
Figure 6.
CLOCK S845A mice do not have a circadian locomotor phenotype. A, double-plotted representative actogram for wheel-running activity. Each horizontal line represents 24 h. The light/dark schedule is shown by 12-h lights on with yellow background and 12-h lights off with white background. Where the yellow background ends represents when the mice were subjected to complete darkness (DD). There are no differences in behavior under DD in S845A mice as compared with WT mice. B, body weight at the beginning of the study (day 1) and at the end (day 28). C, total food intake over the course of 28 days of the study. D, τ, or period, of activity under DD. E, average time of activity onset measured in hours after subjective lights on. F, average active period length. G, average amount of activity during the active phase. n = 6 WT, 8 S845A. Data are represented as individual points for each mouse with a line at the mean ± S.E.
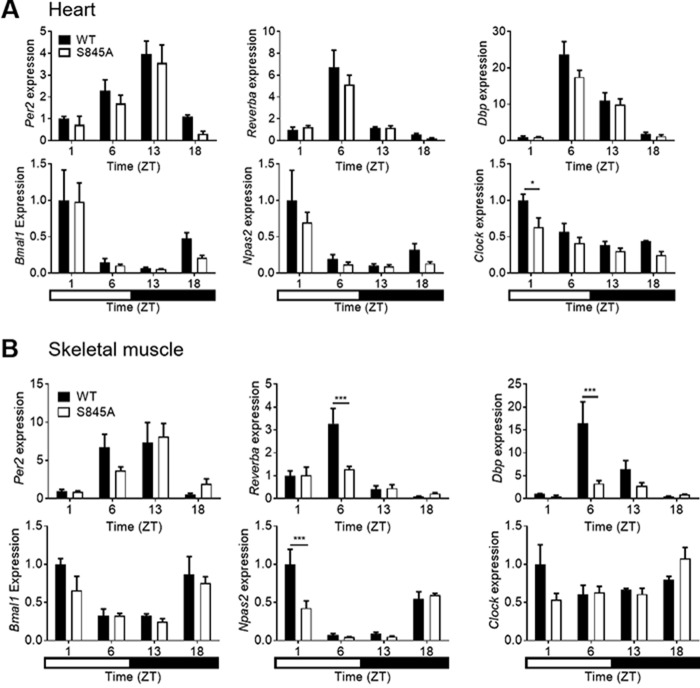
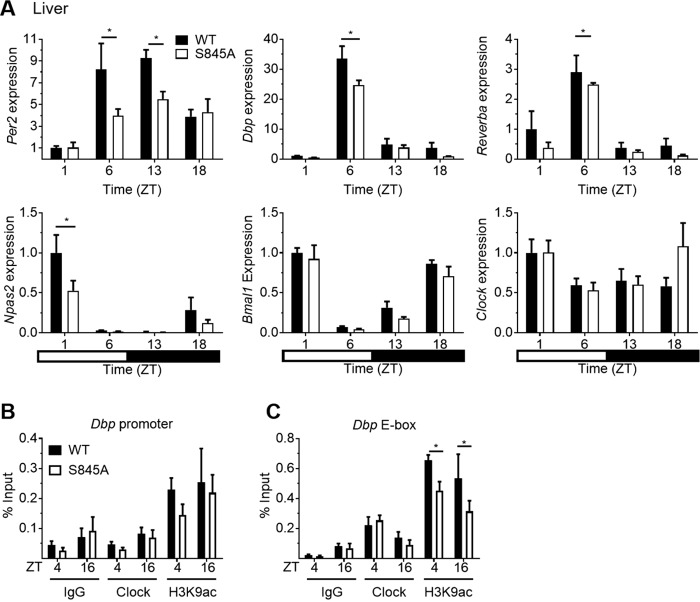
Although there was no defect in central locomotor rhythms, CLOCK activity was assessed in peripheral tissues that undergo rhythmic circadian gene expression (13). Thus, transcriptional rhythms of core circadian genes were investigated in the heart, skeletal muscle, and liver. WT and CLOCK S845A mice were fed ad libitum and sacrificed at four time points throughout the day: ZT1 (8 a.m.), ZT5 (12 p.m.), ZT13 (8 p.m.), and ZT18 (1 a.m.). The transcription of E-Box-controlled circadian genes, such as Per2, Reverbα, and Dbp, and of REV and ROR controlled genes, such as Bmal1 and Npas2, was not different in heart extracts from the two strains; however, Clock expression was lower in CLOCK S845A mice (Fig. 7A). In skeletal muscle, the expressions of Reverbα and Dbp (at ZT6) and Npas2 (at ZT1) were reduced (Fig. 7B). Interestingly, the most well-characterized organ influenced by peripheral circadian rhythms, the liver (19, 20), demonstrated a marked reduction in E-box (CLOCK)-regulated genes Per2, Reverbα, and Dbp in CLOCK S845A mice (Fig. 8A). Circadian genes controlled mainly by REV- and ROR-responsive elements, such as Npas2, Bmal1, and Clock, did not show a trend in expression differences between the two genotypes, and only Npas2 was reduced at ZT1 in CLOCK S845A mice.
Figure 7.
Circadian gene expression over a 24-h period in heart (A) and skeletal muscle (gastrocnemius) (B) from WT and CLOCK S845A mice. The top row of genes in each section (Per2, Reverba, and Dbp) shows those genes those that are mainly controlled by E-box elements, and the bottom row of each section (Bmal1, Npas2, and Clock) shows those genes those that are largely controlled by Rev and Ror response elements. Time on the x axis is given as zeitgeber time (ZT) (time after lights on). The rectangle at the bottom of the figure represents the timing of lights on (white) or lights off (black). n = 3 mice per time point. Data are represented as mean ± S.E. Statistics were calculated using a two-way analysis of variance with Bonferroni post hoc test. *, p < 0.05; ***, p < 0.001.
Figure 8.
Circadian genes controlled by E-boxes have lower expression levels in livers at the middle of the light period (Per2, Dbp, Reverba, and Per1). A, circadian gene expression in WT and S845A livers is shown in each bar graph for the genes indicated on the y axis of each graph. The black and white bar at the bottom represents the light and dark schedule over the course of a day. Data are represented as mean ± S.E. *, p < 0.05. n = 3 mice per time point. B and C, ChIP of liver chromatin in mice sacrificed at ZT4 and ZT16 with either Clock or H3K9ac antibody. IgG was used as a background control. B, ChIP using a primer pair for a Dbp promotor region without an E-box. C, a primer pair surrounding the Dbp intronic E-boxes. H3K9ac is decreased at Dbp E-boxes in S845A livers as compared with WT livers. Data are presented as mean ± S.E. *, p < 0.05, n ≥3.
Because CLOCK S845A mice had reduced levels of Dbp in skeletal muscle and liver, we examined the ability of CLOCK Ser-845 to bind with two regions of the Dbp gene, one promoter region lacking an E-Box and one intronic E-Box element in Dbp regulated by CLOCK/BMAL1 (21, 22). Chromatin immunoprecipitation (ChIP) of CLOCK or H3K9ac (a marker of active chromatin) on the Dbp promoter region without E-boxes was not different (Fig. 8B) nor was binding to Dbp E-boxes in WT or CLOCK S845A mice (Fig. 8C). However, ChIP of H3K9ac on the Dbp E-boxes (where CLOCK binds) showed significantly decreased H3K9ac at both ZT4 and ZT16 (Fig. 8C, p < 0.05). Therefore, the loss of CLOCK phosphorylation of Ser-845 reduces Dbp expression through decreased acetylation of chromatin but not via differences in CLOCK binding with DNA.
Discussion
The central finding of this study is that phosphorylation of CLOCK at Ser-845 can regulate CLOCK function in vitro and in vivo. These results are concordant with recent studies showing that BMAL1 can be similarly phosphorylated by AKT (5), demonstrating that the AKT signaling pathway has multiple mechanisms to regulate the activity of central components of the circadian clock. The present data show that AKT inhibits CLOCK function in cells directly through phosphorylation on Ser-845, which diminishes its nuclear localization. This new mechanism is complementary with AKT phosphorylation of GSK3β indirectly regulating CLOCK function (4, 23). In vivo, knockin mutation of Ser-845 to eliminate phosphorylation at this site does not produce a central circadian rhythm phenotype but influences the expression of circadian-controlled genes in peripheral tissues. The lack of a central circadian rhythm phenotype in CLOCK S845A mice is consistent with data in Clock KO mice that also do not have a locomotor rhythm defect (18), presumably due to the presence of NPAS2 as a redundant binding partner for BMAL1.
Previous work using endothelial cells isolated from AKT1- or AKT2-deficient mice demonstrated that AKT1, to a greater extent than AKT2, can phosphorylate endogenous CLOCK at Ser-845. However, because AKT2 is not as highly expressed or functionally important in endothelial cells compared with AKT1, the phosphorylation by AKT2 was not detectable in this experimental setup. Work by others in adipocytes using targeted proteomics have shown that insulin induces CLOCK Ser-845 phosphorylation (11). This result in conjunction with our data showing that circadian gene expression is altered in insulin-sensitive tissues (skeletal muscle and liver) in CLOCK S845A mice implies that regulation of CLOCK phosphorylation of Ser-845 in vivo is most likely via AKT2, the dominant isoform found in insulin-sensitive tissues (24). In addition to multiple AKT isoforms, other AGC-family kinases that phosphorylate a similar motif, such as PKA, may phosphorylate CLOCK as well. PKA is activated by forskolin, one of the key stimuli that trigger rhythmic core circadian gene transcription in cell culture (25); thus, it may play a role in CLOCK Ser-845 phosphorylation. PKA is also activated by glucagon, which is secreted during fasting at the beginning part of the light phase, around the time when there is decreased transcription of CLOCK-controlled genes in the liver. Clearly, additional experiments are needed to dissect the relationship between these kinases, CLOCK Ser-845 phosphorylation, and circadian gene expression.
To validate CLOCK as an AKT substrate, many of the above experiments were completed in HEK293T cells and NIH3T3 cells; however, the interactions described are not cell autonomous. Mechanistically, when AKT phosphorylates CLOCK at Ser-845, approximately half of the total CLOCK shifts its localization from the nucleus to the cytoplasm, similar to that seen when BMAL1 and FOXO transcription factors are phosphorylated by AKT. pCLOCK binds to 14-3-3 proteins and is prevented from dephosphorylation and re-entering the nucleus. Thus, by altering subcellular localization, the phosphorylation of CLOCK at Ser-845 can regulate the expression of clock-controlled genes as demonstrated in CLOCK 845A mutant mice.
Paradoxically, S845A mice have reduced transcription of multiple E-box–regulated genes during the day in livers and skeletal muscle, although phosphorylation of AKT substrates is likely not maximal at these time points. This may be due to its phosphorylation in vivo by other kinases, such as PKA, as mentioned above. In attempts to monitor Ser-845 phosphorylation in vivo using multiple approaches, we were unable to detect the peptide containing CLOCK Ser-845 by MS due to technical issues, and therefore, we do not have a clear time course of pCLOCK Ser-845 in tissue throughout a circadian cycle. However, this phosphopeptide has been detected in previous studies in vivo (26, 27) lending confidence that it is physiologically important.
One question that remains is why CLOCK S845A causes reduced histone acetylation of Dbp at the intronic E-box element. There is evidence that CLOCK itself has intrinsic histone acetyltransferase (HAT) activity (28); however, we were unable to detect HAT activity in recombinant CLOCK to measure whether its enzymatic activity is altered in the CLOCK S845A mutant protein. Also, it is possible that phosphorylation of CLOCK Ser-845 alters the recruitment or activity of canonical HATs or deacetylases to chromatin. Perhaps the decreased histone acetylation precedes and causes reduced gene expression. If this was the case, there would be lower levels of nuclear CLOCK over the dark cycle.
In summary, we have characterized CLOCK as a novel AKT substrate that can regulate CLOCK localization in vitro and core circadian gene expression in vivo. Additional work examining metabolic control of this pathway in disease models influenced by circadian rhythms will further enable our understanding of the importance of CLOCK Ser-845 phosphorylation.
Experimental procedures
Materials
CLOCK (5157), MYC (2276), AKT (pan) (2920), pAKT T308 (9275), pAKT S473 (9271), 14-3-3 (8312), pAkt substrate motif (9614), and GAPDH (5174) antibodies were purchased from Cell Signaling Technologies. The pCLOCK Ser-845 antibody was made for our lab by the Product Development Group, Cell Signaling Technologies. Briefly, rabbits were injected with a keyhole limpet hemocyanin-conjugated phosphopeptide (PRHRTDSLTDPSKV, mouse sequence, with the underlined S delineating the phospho-site) and antibody purified on columns with an immobilized immunogen and a nonphosphopeptide of the same motif. The HSP90 (sc-69703) antibodies were from Santa Cruz Biotechnologies (Dallas, TX). FLAG-M2 antibody (F1804) was from Sigma. β-Actin (ab8226), CLOCK (ChIP grade, ab3517), and H3K9ac (ChIP grade, ab3441) antibodies were purchased from Abcam. Secondary antibodies were fluorescent-conjugated donkey anti-mouse, -rabbit, or -goat from Rockland Immunochemicals (Pottstown, PA) (800 nm) or Life Technologies, Inc. (680 nm).
Cell culture
All HEK293T and NIH3T3 cells were cultured in 10% fetal bovine serum, 100 units/ml penicillin, and 100 g/ml streptomycin in Dulbecco's modified Eagle's medium (Gibco). All cells were grown in cell culture dishes at 37 °C and 5% CO2. The cells were then genotyped, and WT and CLOCK S845A cells passage 4–20 were used in experiments. MK2206 (Selleck Chem) was used at a 5 μm.
Plasmids
The Myc-CLOCK constructs were gifts from Dr. John Hogenesch. The phospho-site mutation in Myc-CLOCK was made using the TagMaster mutagenesis system (GM Bioscience, Frederick, MD). FLAG-Akt1 was created in a pFLAG-CMV2 backbone.
Western blotting
Western blottings were done following a standard protocol and imaged on the Odyssey imaging system by LI-COR (Lincoln, NE). Because of low level immunoreactivity of the pCLOCK antibody, blots were first probed with the pCLOCK antibody and then re-probed afterward with the total CLOCK antibody. Western blottings were quantified using the Odyssey Imaging Software to measure band intensity of the protein of interest over total protein or loading control protein (GAPDH or β-actin).
Immunoprecipitation (IP)
Cell lysates were incubated with antibodies overnight at 4 °C in IP buffer (20 mm Tris-HCl, pH 7.5, 0.1% SDS, 1% Triton X-100, 10 mm β-mercaptoethanol, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, and 1% sodium deoxycholate) with protease and phosphatase inhibitors. The next day, the appropriate protein A- or G-Sepharose beads (Millipore, Billerica, MA) were rotated in each IP for 2 h at 4 °C; then the beads were washed at least three times in IP buffer and boiled in 2× SDS loading buffer to dissociate the protein from the beads.
Immunofluorescence
HEK293T cells were transfected with Myc-CLOCK WT or Myc-CLOCK S845A. Cells were blocked with 5% BSA and stained with CLOCK antibody (1:500) overnight. AlexaFluor goat anti-rabbit 568 nm at a 1:2000 dilution was used for the secondary antibody (Thermo Fisher Scientific, Waltham, MA). Nuclei were stained with ToPRO3 (Thermo Fisher Scientific). Images were taken on the Leica SP5 confocal microscope (Wetzlar, Germany) and analyzed using ImageJ. Nuclei were outlined and measured in ImageJ by hand tracing. CLOCK staining in the nuclei was traced and measured by ImageJ. Then CLOCK staining in the whole cell was traced and measured. The percentage in the nucleus was calculated as the CLOCK staining area in the nucleus divided by total CLOCK staining area multiplied by 100.
In vitro kinase assays
Immunoisolated FLAG-AKT1 was added at 5 nm to reactions containing 5–20 μm peptide (United Peptide, Herndon, VA) or protein immunoprecipitated from in vitro translation reactions. The reaction contained 0.3 μCi/μl ATP (γ-33P) and 100 μm cold ATP. Reactions were carried out up to 30 min at 30 °C. To quench the peptide reaction, peptides were put on P81 filter paper and dropped into phosphoric acid solution. Protein substrate reactions were put in 2× SDS loading buffer and were boiled. Proteins were then separated by SDS-PAGE. Radioactivity was visualized with Phosphor Screen and Phosphor Imager. Western blotting was used in nonradioactive samples to visualize CLOCK and pCLOCK Ser-845.
Animals
Clock S845A knockin mice were made on a C57Bl/6 background using CRISPR/Cas9 technology with the help of the Yale Animal Genomics Facility. The guide RNA had sequence 5′-TGTAATACGACTCACTATAGGGGACCTTGGAAGGGTCAGTCGTTTTAGAGCTAGAAATAGC-3′. The DNA template strand had sequence 5′-TCGCACCACCAGCAACACCAGCCTCAGCAGCAACAGCAGCTTCCTCGGCACAGGACTGACGCCTTGACTGACCCTTCCAAGGTCCAGCCACAGTAGCACACACACTTCCTCTCTGACATGCGAG-3′. Mouse genomic DNA was checked for the top three most probable off-target mutations, and only those mice without detected off-target mutations were used for breeding. Mice were genotyped using two-step amplification qPCR and either the WT forward primer, 5′-GGCACAGGACTGACAG-3′, or the S845A forward primer, 5′-GCACAGGACTGACGC-3′, and the same reverse primer, 5′-CTCATCAAGGGACTGAAC-3′. The threshold cycle values were at least six cycles different. Mice were housed in the Yale Animal Facility, fed normal chow, and were kept at 25 °C on a 12-h light/dark cycle. All mice used in experiments were at least the F2 generation and were males between 12 and 20 weeks old, unless otherwise noted. All procedures were approved by the Institutional Animal Care and Use Committee of Yale University.
Quantitative RT-PCR
RNA was extracted using RNeasy kit (Qiagen). Reverse transcription was completed using the TaqMan reverse transcription reagents (Roche Applied Science). Primers were ordered from the Keck Oligonucleotide Facility (Yale University) from sequences found on PrimerBank (Harvard University) or published literature as indicated in Table 1. Gapdh was used as the reference gene to normalize RNA quantity.
Table 1.
qPCR primer sequences (mouse)
F is forward, and R is reverse.
| qPCR primer | Sequence |
|---|---|
| Bmal1 F | TGACCCTCATGGAAGGTTAGAA |
| Bmal1 R | GGACATTGCATTGCATGTTGG |
| Clock F | AGAACTTGGCATTGAAGAGTCTC |
| Clock R | GTCAGACCCAGAATCTTGGCT |
| Dbp F | CTGGCCCGAGTCTTTTTGC |
| Dbp R | CCAGGTCCACGTATTCCACG |
| Gapdh F | AATGTGTCCGTCGTGGATCTGA |
| Gapdh R | AGTGTAGCCCAAGATGCCCTTC |
| Npas2 F | AAGGATAGAGCAAAGAGAGCCT |
| Npas2 R | CATTTTCCGAGTGTTACCAGGG |
| Per2 F | AAAGCTGACGCACACAAAGAA |
| Per2 R | ACTCCTCATTAGCCTTCACCT |
| RevErbα F | TACATTGGCTCTAGTGGCTCC |
| RevErbα R | CAGTAGGTGATGGTGGGAAGTA |
| Dbp non E-box F | CCTCCTTCTCCACGTCCTGAT |
| Dbp non E-box R | GGTGAGAAAGGACAAGGGATGT |
| Dbp E-box F | TGGGACGCCTGGGTACAC |
| Dbp E-box R | GGGAATGTGCAGCACTGGTT |
Telemetry
Telemeter implantation and data collection were completed by Dr. Heino Velazquez as described previously (29). Briefly, heart rate, blood pressure, and activity measurements were collected every minute for at least 5 days. Data were analyzed by first averaging the data into 3-h bins.
Circadian wheel running assay
Mice 12–16 weeks of age were transferred to individual cages with running wheels attached to monitors (Actimetrics), and data were recorded by ClockLab software (Coulbourn Instruments). After 14 days of acclimation to 12 h of light followed by 12 h of dark (LD), the mice were kept in complete darkness (DD) for 14 days. The rpm data were collected, and all data were analyzed using the ClockLab software.
Body temperature measurements
Mouse body temperature was measured by anal probe until the electronic thermometer read an equilibrated temperature. The mice were restrained by scruffing during this process. Temperature readings taken during the dark phase of the cycle were done under red light.
Metabolic measurements
12–16-Week-old male mice were housed individually in metabolic cages for 3 days during which VO2, VCO2, activity, energy expenditure, respiratory exchange ratio, food intake, and water intake were measured. Their weight was taken, and body composition (% fat/% muscle) was measured using a TD-NMR minispec (Bruker).
ChIP
Mice were sacrificed at ZT4 and ZT16; livers were snap-frozen and stored at −80 °C. ChIP was carried out as described previously (30). Chromatin was incubated overnight, rotating at 4 °C with the following ChIP grade antibodies: CLOCK, H3K9ac, or IgG control antibody (Abcam). The IP was completed and washed with five buffers of varying salt concentrations to enhance specificity. Finally, DNA–protein complexes were eluted; samples were reverse cross-linked, and DNA was purified. qPCR was carried out on different areas of the circadian gene Dbp using the primers in Table 1. Data were expressed as “% input” by comparing the maximum amount of input DNA to the levels in the IP samples.
Data analysis
All experiments were performed at least three times; numbers of mice used in each experiment are described in the figure legends. Immunofluorescence was analyzed by ImageJ. mRNA cycling data were analyzed using JTK_Cycle (31). Wheel running data were quantified and analyzed using ClockLab. All other data were analyzed using GraphPad Prism (La Jolla, CA). Graphs comparing only two columns of data were compared using an unpaired Student's t test. Graphs with more than two columns were compared using a two-way analysis of variance. Data are presented as mean ± S.E.
Author contributions
A. K. L., W. Z., and W. C. S. conceptualization; A. K. L., W. Z., J. M. S., C. K., H. V., and W. C. S. data curation; A. K. L., W. Z., J. M. S., C. K., H. V., and W. C. S. formal analysis; A. K. L. and W. C. S. supervision; A. K. L., W. Z., and W. C. S. funding acquisition; A. K. L. and W. Z. validation; A. K. L., W. Z., H. V., and W. C. S. investigation; A. K. L. and W. Z. visualization; A. K. L., W. Z., J. M. S., C. K., and H. V. methodology; A. K. L., W. Z., and W. C. S. writing-original draft; A. K. L. and W. C. S. project administration.
Acknowledgments
We thank Roger Babbitt for invaluable technical assistance, Carlos Fernandez-Hernando and Nathan Price for helpful discussions, and Elisa Araldi, Leigh Goedeke, Nina Kristofik, and Katrina Meeth for experimental assistance. The O'Brien Kidney Center and their National Institutes of Health grant provided access to the wheel running cages and ClockLab software.
Note added in proof
In the version of this article that was published as a Paper in Press on March 27, 2018, Table 1 was inadvertently omitted. This error has now been corrected.
This work was supported by National Institutes of Health Grants R01 HL64793, R01 HL61371, HL133018, and R35 HL139945, American Heart Association MERIT and the Leducq Foundation, MIRVAD network (to W. C. S.), American Heart Association Pre-doctoral Grant 15PRE21670000, National Institutes of Health T32 Training Grant T32 GM007324 (to A. K. L.), an American Heart Association Sarnoff fellowship (to J. M. S.), and George M. O'Brien Kidney Center Grant from National Institutes of Health P30 DK079310 (to H. V.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
- PDGF
- platelet-derived growth factor
- qPCR
- quantitative PCR
- CHX
- cycloheximide
- PKA
- protein kinase A
- HAT
- histone acetyltransferase
- IP
- immunoprecipitation.
References
- 1. Huang N., Chelliah Y., Shan Y., Taylor C. A., Yoo S. H., Partch C., Green C. B., Zhang H., and Takahashi J. S. (2012) Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science 337, 189–194 10.1126/science.1222804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Panda S., Hogenesch J. B., and Kay S. A. (2002) Circadian rhythms from flies to human. Nature 417, 329–335 10.1038/417329a [DOI] [PubMed] [Google Scholar]
- 3. Rudic R. D., McNamara P., Reilly D., Grosser T., Curtis A. M., Price T. S., Panda S., Hogenesch J. B., and FitzGerald G. A. (2005) Bioinformatic analysis of circadian gene oscillation in mouse aorta. Circulation 112, 2716–2724 10.1161/CIRCULATIONAHA.105.568626 [DOI] [PubMed] [Google Scholar]
- 4. Spengler M. L., Kuropatwinski K. K., Schumer M., and Antoch M. P. (2009) A serine cluster mediates BMAL1-dependent CLOCK phosphorylation and degradation. Cell Cycle 8, 4138–4146 10.4161/cc.8.24.10273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dang F., Sun X., Ma X., Wu R., Zhang D., Chen Y., Xu Q., Wu Y., and Liu Y. (2016) Insulin post-transcriptionally modulates Bmal1 protein to affect the hepatic circadian clock. Nat. Commun. 7, 12696 10.1038/ncomms12696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vitaterna M. H., King D. P., Chang A. M., Kornhauser J. M., Lowrey P. L., McDonald J. D., Dove W. F., Pinto L. H., Turek F. W., and Takahashi J. S. (1994) Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264, 719–725 10.1126/science.8171325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. King D. P., Zhao Y., Sangoram A. M., Wilsbacher L. D., Tanaka M., Antoch M. P., Steeves T. D., Vitaterna M. H., Kornhauser J. M., Lowrey P. L., Turek F. W., and Takahashi J. S. (1997) Positional cloning of the mouse circadian clock gene. Cell 89, 641–653 10.1016/S0092-8674(00)80245-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun Z. S., Albrecht U., Zhuchenko O., Bailey J., Eichele G., and Lee C. C. (1997) RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell 90, 1003–1011 10.1016/S0092-8674(00)80366-9 [DOI] [PubMed] [Google Scholar]
- 9. Lee M. Y., Luciano A. K., Ackah E., Rodriguez-Vita J., Bancroft T. A., Eichmann A., Simons M., Kyriakides T. R., Morales-Ruiz M., and Sessa W. C. (2014) Endothelial Akt1 mediates angiogenesis by phosphorylating multiple angiogenic substrates. Proc. Natl. Acad. Sci. U.S.A. 111, 12865–12870 10.1073/pnas.1408472111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parker B. L., Yang G., Humphrey S. J., Chaudhuri R., Ma X., Peterman S., and James D. E. (2015) Targeted phosphoproteomics of insulin signaling using data-independent acquisition mass spectrometry. Sci. Signal. 8, rs6 10.1126/scisignal.aaa3139 [DOI] [PubMed] [Google Scholar]
- 11. Humphrey S. J., Yang G., Yang P., Fazakerley D. J., Stöckli J., Yang J. Y., and James D. E. (2013) Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17, 1009–1020 10.1016/j.cmet.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., and Weitz C. J. (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569 10.1126/science.280.5369.1564 [DOI] [PubMed] [Google Scholar]
- 13. DeBruyne J. P., Weaver D. R., and Reppert S. M. (2007) Peripheral circadian oscillators require CLOCK. Curr. Biol. 17, R538–R539 10.1016/j.cub.2007.05.067 [DOI] [PubMed] [Google Scholar]
- 14. Jin X., Shearman L. P., Weaver D. R., Zylka M. J., de Vries G. J., and Reppert S. M. (1999) A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96, 57–68 10.1016/S0092-8674(00)80959-9 [DOI] [PubMed] [Google Scholar]
- 15. Zhang R., Lahens N. F., Ballance H. I., Hughes M. E., and Hogenesch J. B. (2014) A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. U.S.A. 111, 16219–16224 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., and Greenberg M. E. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868 10.1016/S0092-8674(00)80595-4 [DOI] [PubMed] [Google Scholar]
- 17. Vitaterna M. H., Ko C. H., Chang A. M., Buhr E. D., Fruechte E. M., Schook A., Antoch M. P., Turek F. W., and Takahashi J. S. (2006) The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc. Natl. Acad. Sci. U.S.A. 103, 9327–9332 10.1073/pnas.0603601103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Debruyne J. P., Noton E., Lambert C. M., Maywood E. S., Weaver D. R., and Reppert S. M. (2006) A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron 50, 465–477 10.1016/j.neuron.2006.03.041 [DOI] [PubMed] [Google Scholar]
- 19. Koike N., Yoo S. H., Huang H. C., Kumar V., Lee C., Kim T. K., and Takahashi J. S. (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 10.1126/science.1226339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tahara Y., and Shibata S. (2016) Circadian rhythms of liver physiology and disease: experimental and clinical evidence. Nat. Rev. Gastroenterol. Hepatol. 13, 217–226 10.1038/nrgastro.2016.8 [DOI] [PubMed] [Google Scholar]
- 21. Yoshitane H., Ozaki H., Terajima H., Du N. H., Suzuki Y., Fujimori T., Kosaka N., Shimba S., Sugano S., Takagi T., Iwasaki W., and Fukada Y. (2014) CLOCK-controlled polyphonic regulation of circadian rhythms through canonical and noncanonical E-boxes. Mol. Cell. Biol. 34, 1776–1787 10.1128/MCB.01465-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ripperger J. A., and Schibler U. (2006) Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38, 369–374 10.1038/ng1738 [DOI] [PubMed] [Google Scholar]
- 23. Pap M., and Cooper G. M. (1998) Role of glycogen synthase kinase-3 in the phosphatidylinositol 3-Kinase/Akt cell survival pathway. J. Biol. Chem. 273, 19929–19932 10.1074/jbc.273.32.19929 [DOI] [PubMed] [Google Scholar]
- 24. Cho H., Mu J., Kim J. K., Thorvaldsen J. L., Chu Q., Crenshaw E. B. 3rd., Kaestner K. H., Bartolomei M. S., Shulman G. I., and Birnbaum M. J. (2001) Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKBβ). Science 292, 1728–1731 10.1126/science.292.5522.1728 [DOI] [PubMed] [Google Scholar]
- 25. Yagita K., and Okamura H. (2000) Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat-1 fibroblasts. FEBS Lett. 465, 79–82 10.1016/S0014-5793(99)01724-X [DOI] [PubMed] [Google Scholar]
- 26. Mertins P., Yang F., Liu T., Mani D. R., Petyuk V. A., Gillette M. A., Clauser K. R., Qiao J. W., Gritsenko M. A., Moore R. J., Levine D. A., Townsend R., Erdmann-Gilmore P., Snider J. E., Davies, et al. (2014) Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol. Cell. Proteomics 13, 1690–1704 10.1074/mcp.M113.036392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mertins P., Mani D. R., Ruggles K. V., Gillette M. A., Clauser K. R., Wang P., Wang X., Qiao J. W., Cao S., Petralia F., Kawaler E., Mundt F., Krug K., Tu Z., Lei J. T., et al. (2016) Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62 10.1038/nature18003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doi M., Hirayama J., and Sassone-Corsi P. (2006) Circadian regulator CLOCK is a histone acetyltransferase. Cell 125, 497–508 10.1016/j.cell.2006.03.033 [DOI] [PubMed] [Google Scholar]
- 29. Marin E. P., Jozsef L., Di Lorenzo A., Held K. F., Luciano A. K., Melendez J., Milstone L. M., Velazquez H., and Sessa W. C. (2016) The protein acyl transferase ZDHHC21 modulates α1 adrenergic receptor function and regulates hemodynamics. Arterioscler. Thromb. Vasc. Biol. 36, 370–379 10.1161/ATVBAHA.115.306942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goedeke L., Rotllan N., Canfrán-Duque A., Aranda J. F., Ramírez C. M., Araldi E., Lin C. S., Anderson N. N., Wagschal A., de Cabo R., Horton J. D., Lasunción M. A., Náár A. M., Suárez Y., and Fernández-Hernando C. (2015) MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat. Med. 21, 1280–1289 10.1038/nm.3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hughes M. E., Hogenesch J. B., and Kornacker K. (2010) JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms 25, 372–380 10.1177/0748730410379711 [DOI] [PMC free article] [PubMed] [Google Scholar]