Abstract
The positioning and movement of the nucleus has recently emerged as an important aspect of cell migration. Understanding of nuclear positioning and movement has reached an apogee in studies of fibroblast migration. Specific nuclear positioning and movements have been described in the polarization of fibroblast for cell migration and in active migration in 2D and 3D environments. Here, we review recent studies that have uncovered novel molecular mechanisms that contribute to these events in fibroblasts. Many of these involve a connection between the nucleus and the cytoskeleton through the LINC complex composed of outer nuclear membrane nesprins and inner nuclear membrane SUN proteins. We consider evidence that appropriate nuclear positioning contributes to efficient fibroblast polarization and migration and the possible mechanism through which the nucleus affects cell migration.
Keywords: fibroblast, LINC complex, nesprin, SUN protein, cytoskeleton, cell migration, nuclear envelope, nuclear movement
1. Introduction
Mesenchymal-derived fibroblasts are the most common cells in many connective tissues where they produce the extracellular matrix and other factors that are important for tissue and organ homeostasis and repair. Their role in these processes critically depends on their ability to migrate. Poor migration leads to wound healing defects, for example in aged individuals [1, 2], whereas uncontrolled migration contributes to inflammation and scarring [3]. Beyond their physiological importance, fibroblasts have provided an important test bed for exploring basic mechanisms of migration due to their robustness in culture and as well as their inherent propensity to migrate.
The nucleus has emerged as a surprisingly important factor in the migration of fibroblasts and other cell types. Depending on the environment, the nucleus contributes to migration by providing polarity, integrating intracellular forces, generating intracellular pressure for propulsive force, and impeding movement through narrow constrictions. How the nucleus is moved and positioned for these activities has received much attention over the last 10 years. These studies have revealed that distinct linker of nucleoskeleton and cytoskeleton (LINC) complexes contribute to nuclear movement and positioning in fibroblasts. LINC complexes are composed of outer nuclear membrane KASH proteins (or nesprins/Synes in vertebrates) and inner nuclear membrane SUN proteins that interact within the perinuclear space [4, 5]. Depending on the nesprin employed and the environment, the nucleus can attach to actin filaments, microtubules (MTs) or intermediate filaments (IFs) [6–8].
Advances have been made in understanding the role of LINC complexes in both 2D and 3D fibroblast migration. On one hand, nuclear movement has been extensively studied in 2D fibroblasts polarizing for migration in wounded monolayers. This has been a powerful system to identify the molecular components connecting the nucleus and cytoskeleton due to the ease of both genetic and cell biological manipulations and the high resolution imaging possible with flat, well-spread cells. Indeed, a macromolecular assembly of specific LINC complexes, actin cables and associated proteins has been observed to assemble during active nuclear translocation accompanying fibroblast polarization for migration [9–11]. Moreover, distinct adhesion-like structures beneath the nucleus have been described and reported to cause defects in nuclear positioning [12]. On the other hand, a novel mechanism dependent on nesprin-3, termed the “nuclear piston”, has been identified and contributes to lobopodial migration of fibroblasts migrating in 3D environments [13].
Here we review recent studies examining nuclear positioning and movements in fibroblasts polarizing for migration and during active fibroblast migration in 2D and 3D environments. In each case, we consider the molecular and mechanical mechanisms for these events, the pathways regulating them, and the roles played by nuclear positioning in fibroblast polarization and migration.
2. Nuclear translocation in fibroblasts polarizing for migration in 2D
In many migrating cells, including fibroblasts, the nucleus is positioned rearward of the cell centroid [8]. Fibroblasts have an intrinsic mechanism to establish this rearward position of the nucleus, independent of actual cell migration, as shown by experiments with serum-starved wounded monolayers [14]. Addition of the serum factor lysophosphatidic acid (LPA) to wounded monolayers of starved NIH3T3 fibroblasts or mouse embryo fibroblasts (MEFs) triggers rearward translocation of the nucleus without stimulating migration (Figure 1A). This nuclear translocation is not accompanied by nuclear rotation or changes in nuclear shape making its analysis less complex than nuclear positioning and movements that occur in actively migrating fibroblasts. As the centrosome is maintained at the cell centroid during rearward nuclear translocation, movement of the nucleus creates cell polarity by orienting the centrosome toward the leading edge (Figure 1A). Intriguingly, live cell recordings of starved NIH3T3 fibroblasts stimulated to migrate by serum reveal that productive migration commences when the nucleus moves rearward and the centrosome is oriented [14]. This result suggests that proper rearward positioning of the nucleus and centrosome orientation are required for productive fibroblast migration, a conclusion consistent with the inhibition of migration when the pathways regulating rearward translocation are disrupted (see below).
Figure 1. Nuclear translocation by TAN lines during fibroblast polarization.
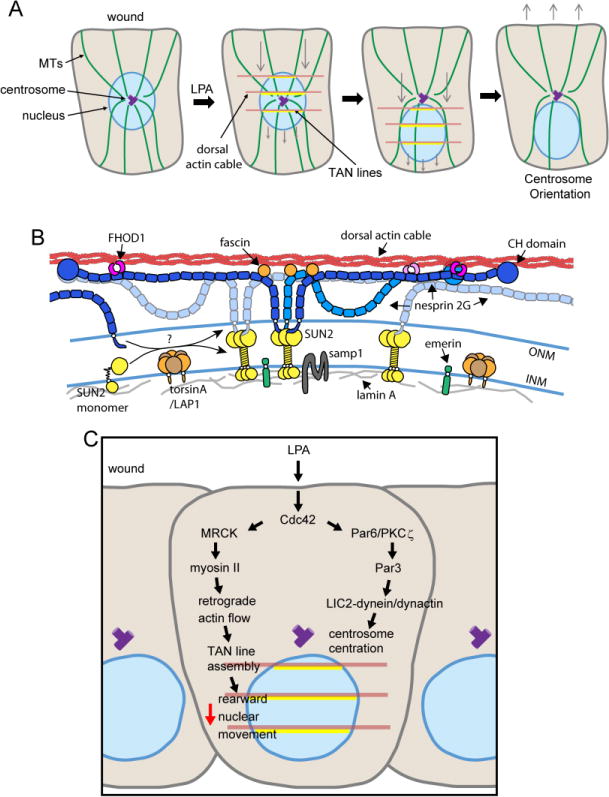
A. Model for rearward translocation of the nucleus following LPA stimulation of wound edge fibroblasts. LPA induces formation and rearward flow of dorsal actin cables (brown) that couple to the nucleus through TAN lines (yellow). Combined with centrosome centration (see panel C), nuclear translocation results in anterior orientation of the centrosome. Purple rectangles are the centrosome. Blue circle is the nucleus. Gray arrows indicate the direction of actin flow and nuclear translocation in the second and third panels and the polarity of the cell and direction of migration if the cell is stimulated in the fourth panel. Adapted from reference [83]. B. Molecular model for TAN lines. A dorsal actin cable (red) is linked to the nucleus through multiple nesprin-2G (blue) and SUN2 (yellow, shown as trimer) LINC complexes anchored by lamin A (grey). TAN line associated proteins, FHOD1 (magenta) and fascin (orange), are only depicted on the central nesprin-2G for clarity. Both are required for TAN line formation. Samp1 (dark grey) and emerin (green) contribute to TAN line anchoring. TorsinA (brown/orange) is depicted as a regulator of LINC complex formation and may also regulate TAN line assembly (see text). Adapted from reference [84]. C. Schematic of the two Cdc42 pathways activated by LPA that contribute to centrosome orientation. One is nuclear translocation mediated by dorsal actin cable (brown) flow and TAN lines (yellow); the other, centrosome centration mediated by MT anchoring at cortical sites by dynein/dynactin and Par3. Red arrow indicates the direction of nuclear translocation.
2.1 LINC complexes assemble into higher ordered TAN lines to translocate the nucleus
The rearward nuclear translocation in fibroblasts stimulated by LPA is inhibited by actin and myosin drugs and occurs at the same velocity as actin retrograde flow, indicating an actomyosin process [14]. Numerous results indicate that actin directly connects to the nucleus to move it. Dominant negative and knockdown approaches show that nesprin-2 giant (nesprin-2G) is required for nuclear movement [10, 15, 16]. Nesprin-2G is one of two giant nesprins (nesprin-1G is the other) that contain actin-binding, calponin homology (CH) domains and is the only one expressed in NIH3T3 fibroblasts [10]. Impaired nuclear movement in NIH3T3 fibroblasts depleted of nesprin-2G is rescued by expression of mini-nesprin-2G (mini-N2G), a chimeric construct containing the N-terminal CH domains and the C-terminal KASH motif [10]. Expression of mini-N2G with point mutants in the CH domains abrogating actin binding, do not rescue. Thus, nesprin-2G’s interaction with actin filaments is critical for nuclear movement in fibroblasts.
Strikingly, nesprin-2G accumulates along dorsal actin cables above the nucleus (Figure 1B) during its movement in NIH 3T3 fibroblasts and MEFs [9–11]. This result is based on colocalization of both endogenous nesprin-2G and expressed GFP-mini-N2G with dorsal actin cables [9–11]. In live cell movies, dorsal actin cables encountering the nuclear surface accumulate GFP-mini-N2G in minutes, forming linear arrays [10]. They also accumulate one of the two SUN proteins expressed in fibroblasts, SUN2, but not SUN1 or a number of other inner nuclear membrane proteins (Figure 1B). Indeed, SUN2 is detected in linear arrays in primary MEFs as well [12]. Reflecting their morphology and actin-dependence, these LINC complex arrays have been termed transmembrane actin-associated nuclear (TAN) lines [10, 11]. TAN lines form coincident with the initiation of nuclear translocation, move rearward with the nucleus and disassemble when nuclear movement ceases, providing direct correlative evidence for their involvement in the movement. This specific combination of LINC complex proteins in the nuclear envelope is required for TAN line formation and rearward nuclear translocation after LPA-stimulation and disrupting its components reduce NIH3T3 fibroblast migration speed into the wound [10, 11].
2.2 Anchorage of TAN lines by the nucleus
The coincident movement of TAN lines and the nucleus implies that TAN lines are anchored to the nucleus to transmit the force that moves it. Studies show that proteins in both nucleoplasm and inner nuclear membrane contribute to nuclear anchorage of TAN lines. Unlike factors that are required for TAN line formation, disruption of these anchorage factors causes a novel phenotype in which the TAN lines form, but slip over an immobile nucleus.
Localization and interaction studies suggest that the nuclear lamina plays a key role in anchoring the LINC complex (Figure 1B) [5, 17]. There are three lamin genes encoding lamin B1, lamin B2 and A-type lamins. A-type lamins comprise three alternatively spliced isoforms: lamin A, lamin C and lamin C2 [18]. Among these, lamin A binds to SUN proteins via its C-terminus, whereas lamin B1 and lamin C bind to SUN proteins weakly [5, 19]. MEFs null for A-type lamins, or NIH3T3 fibroblasts transiently knocked down for lamin A fail to move the nucleus or orient the centrosome after LPA stimulation [9]. While TAN lines still form in the A-type lamin disrupted cells, they slide over the nucleus instead of moving it, suggesting an anchorage defect. Consistently, diffusional mobility of mini-nesprin-2G and SUN2 measured by FRAP is increased in MEFs lacking A-type lamins [20].
Additional proteins in the nuclear envelope may assist in TAN line anchoring. One is Samp1/NET5, an inner nuclear membrane protein homologous to yeast Ima1, which is required for LINC complex stability in Schizosaccharomyces pombe [21]. Samp1 is required for nuclear movement and centrosome orientation in the LPA-stimulated NIH3T3 fibroblasts [22]. Samp1 interacts with both SUN2 and lamin A/C and is a TAN line component because it accumulates with GFP-mini-N2G along actin cables over the dorsal surface of the nucleus during rearward movement [22].
Emerin is another candidate. Emerin is a predominantly inner nuclear membrane protein that interacts directly with lamin A/C and nesprin-2 [23]. Knockdown of emerin in NIH3T3 fibroblasts results in nuclear movement and centrosome orientation defects after LPA stimulation [24]. When scrutinized for TAN line formation and motility, 80% of the emerin knockdown cells form normal TAN lines that move erratically reflecting the nonpolarized actin flow observed in emerin depleted fibroblasts. The remaining 20% of the cells phenocopy the TAN line slippage that occurs in cells lacking A-type lamins [24].
2.3 TAN Line associated proteins required for nuclear translocation
TAN lines resemble other membrane adhesion complexes, such as focal adhesions and cell-cell adhesions in that they are composed of specific membrane proteins that cluster in response to actin filaments and transmit force to a membrane. Moreover, the TAN line component nesprin-2 also accumulates at nuclear indentation sites [25] and a nesprin-2 based actin FRET tension sensor directly shows that the LINC complex is under actomyosin tension on the nuclear envelope [26]. Given the similarity to other adhesions, which are known to contain tens to hundreds of proteins, it is perhaps not surprising that additional factors have been found that contribute to TAN line formation (Figure 1B).
The formin family protein FHOD1 was identified as a putative TAN line component based upon its interaction in yeast two hybrid assays with nesprin-2G. FHOD1 has a domain structure typical of other diaphanous family formins, including FH1, FH2, DID and DAD domains, but unlike other formins, it has a second actin binding site (ABS) and caps and bundles actin filaments rather their stimulating their polymerization [27]. Biochemical studies reveal that FHOD1’s N-terminus interacts directly with spectrin repeats 11–13 of nesprin-2G near its CH domains and expression of either domain acts as a dominant negative of LPA-stimulated rearward nuclear movement [16]. FHOD1 colocalizes with actin cables in TAN lines in NIH3T3 fibroblasts and is required for nuclear movement and centrosome orientation [16]. In the absence of FHOD1, TAN lines fail to form, but there is no detectable effect on dorsal actin cables or actin retrograde flow. Both the ABS and the FH2 domain are necessary for TAN line formation and nuclear movement, suggesting that FHOD1 reinforces TAN lines by providing a cross-bridge between nesprin-2G and the actin cable (Figure 1B).
Fascin was identified as the second F-actin-bundling protein required for TAN line formation and nuclear movement. Fascin’s β-trefoil3 interacts with nesprin-2G’s spectrin repeats 51–53 near its C-terminus [15]. Similar to FHOD1, fascin colocalizes with nesprin-2G in TAN lines on the nucleus of NIH3T3 fibroblasts after LPA stimulation and is required for their formation (Figure 1B). Expressing β-trefoil3 functions as a dominant negative in LPA-stimulated rearward nuclear movement, suggesting that coupling between fascin and nesprin-2G is required for nuclear movement. The BAR domain protein amphiphysin-2 (BIN-1) may be an additional and/or alternate connection between nesprin-2G and actin cables. Amphiphysin-2 interacts with SR48-49 of nesprin-2, binds F-actin and is required for LPA-stimulated rearward translocation of the nucleus in NIH3T3 fibroblasts [28]. It has not yet been shown to localize with nesprin-2G in TAN lines.
The identification of these TAN line associated proteins leads to a new model for TAN lines (Figure 1B) that significantly modifies earlier models. First, it indicates that TAN lines are reinforced by linkages between the actin cables and nesprin-2G in addition to nesprin-2G’s intrinsic CH domains. Presumably, the multiple contacts are required to resistant the force generated to move such a large organelle. Second, it suggests that rather than projecting orthogonal to the nuclear surface, the nesprin-2G in TAN lines must instead lie along the long axis of the actin cable in order to position its CH domains and the actin bundling sites of FHOD1 and fascin (see ref. [16] for discussion). A good analogy (and one made in a previous review [29]) is that the connection between nesprin-2G and the actin cable resembles Velcro. Neither FHOD1 nor fascin depletion noticeably affected the number, apparent thickness, or movement of the dorsal actin cables, raising the possibility that their bundling capabilities strengthen dorsal actin cables so that they can resist the high load force necessary to move the nucleus. Another and not mutually exclusive possibility for FHOD1, which is homodimeric, is that it contributes to the clustering of nesprin-2G thereby increasing its ability to form TAN lines by increasing its avidity.
2.4 Regulation of nuclear movement in polarizing fibroblasts
Studies have revealed considerable information about the regulation of cytoskeletal events during nuclear translocation in polarizing fibroblasts. These studies indicate that at least one rate limiting step in nuclear translocation is the generation of the dorsal actin cables and their retrograde movement. Additionally, a recent study suggests that the assembly of TAN lines themselves may be regulated.
After LPA addition to serum-starved fibroblasts, Rho family proteins including Cdc42 are activated [30]. Microinjecting constitutively active Cdc42 constructs into starved NIH3T3 fibroblasts in wounded monolayers activates nuclear movement and centrosome orientation, whereas dominant negative Cdc42 inhibits both phenotypes in LPA-stimulated cells [14, 30], indicating that Cdc42 is both necessary and sufficient for the rearward nuclear movement as well as centrosome orientation. Cdc42 knockout MEFs also show defects in centrosome orientation [31].
Two Cdc42 effectors contribute to LPA-stimulated centrosome orientation in NIH3T3 fibroblasts (Figure 1C): myotonic dystrophy kinase-related, Cdc42 binding kinase (MRCK) and Par6 [14]. MRCK is a myosin II kinase and is responsible for activating myosin II for retrograde flow [32]. Indeed, microinjection of MRCK alone is sufficient to stimulate nuclear movement in serum-starved fibroblasts. Both major isoforms of myosin II are involved: myosin IIA for dorsal actin cable formation and myosin IIB for the directional flow of dorsal actin cables [24]. These results are consistent with previous findings that the two myosin II isoforms have unique cellular functions [33].
Par6, functions along with atypical PKCζ, Par3 and dynein to maintain the centrosome in the cell center while the nucleus moves rearward [14, 34, 35]. Par3 interacts with a specific light intermediate chain (LIC2) of dynein and together they enhance cortical interactions of MTs to maintain the centrosome position [34]. In the absence of these factors, the centrosome moves rearward with the nucleus and fails to orient (Figure 1C).
A recent study suggests that the LINC complex itself is regulated during LPA-stimulated nuclear translocation in fibroblasts [36]. TorsinA is an AAA+ ATPase of unknown function residing within the ER and perinuclear space [37–39]. It interacts with two types of activators, LAP1 in the inner nuclear membrane and LULL1 distributed throughout the ER [40] and has been reported to associate with the KASH domain from nesprin-1, -2 and -3 [41]. MEFs knocked out for torsinA as well as NIH3T3 fibroblasts depleted of torsinA by siRNAs display both nuclear movement and centrosome orientation defects [36]. LAP1, but not LULL1, is also required for the nuclear movement and centrosome orientation after LPA stimulation [36]. Both torsinA and LAP1 depletion reduce the frequency, number and persistence of TAN lines assembled after LPA stimulation, indicating that both proteins are required for the assembly and/or stability of TAN lines (Figure 1B). Wild type torsinA or a substrate locked ATPase mutant localize to TAN lines suggesting that torsinA functions to assemble TAN lines [36]. TorsinA substrates for TAN line assembly remain to be identified. Yet, GFP-mini-N2G (but not GFP-nesprin-3) exhibits reduced mobility in torsinA deficient fibroblasts [36], implying that torsinA releases nesprin-2G from a binding partner for assembly into TAN lines.
Interestingly, both torsinA and LAP1 knockdown also reduce the rate of perinuclear dorsal actin cable flow, another case where nuclear membrane proteins affect cytoplasmic actin dynamics. This result is similar to earlier work that reported that emerin knockdown alters the directionality of retrograde actin flow in NIH3T3 fibroblasts [24]. Emerin interacts with myosin IIB [24], yet how these inner nuclear membrane proteins affect cytoplasmic actin dynamics remains uncertain. One possibility suggested for emerin is that it transits between inner and outer nuclear membranes putting it in proximity to cytoplasmic actin and myosin II [24].
3. Nuclear movements and positioning in fibroblasts migrating in 2D
Nuclear movements and positioning events are considerably more complex in fibroblasts actively migrating than during the establishment of fibroblast polarity for migration described above. During 2D migration, the nucleus is actively maintained in a position just rearward of the cell centroid by multiple factors (Figure 2). In addition, a distinct movement of the nucleus has been described during fibroblast migration in 2D: nuclear rotation in the x-y axis. As TAN lines form transiently in fibroblast polarizing for migration and have not been detected in actively migrating fibroblasts, other structures and activities have been proposed to contribute to the positioning and movements of the nucleus during 2D migration and these are considered below.
Figure 2. Nuclear movements in 2D migrating fibroblasts.
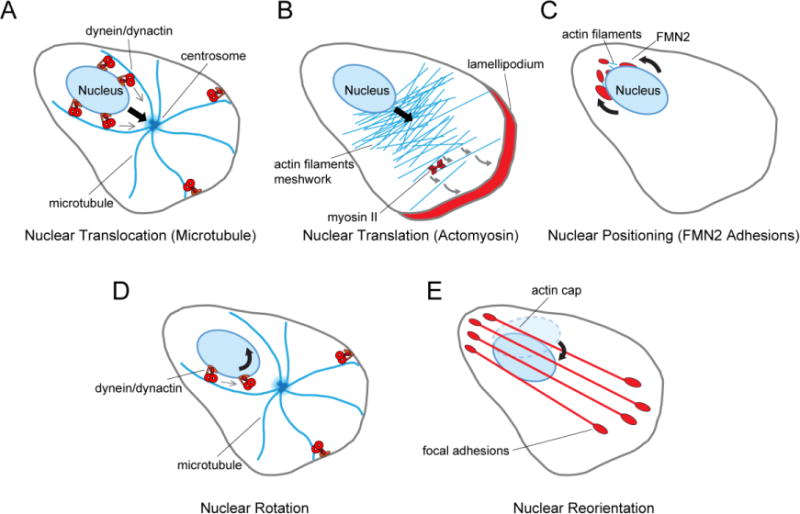
A. Nuclear translocation mediated by MTs. Nuclear dynein and dynactin engage centrosomal MTs to move the nucleus toward the centrosome. Centrosome position is maintained by cortically anchored dynein and dynactin. B: Nuclear translocation mediated by actomyosin tension. Lamellipodial protrusion generates actomyosin tension that pulls the nucleus forward. Tension is coupled to the nucleus through undefined LINC complexes (see text). C. Nuclear positioning mediated by perinuclear localized FMN2 adhesions. D. Nuclear rotation mediated dynein and dynactin. Cortically anchored dynein and dynactin maintain the position of the centrosome. E. Nuclear reorientation mediated by the actin cap. Actin fibers comprising the actin cap are anchored by focal adhesions at each end and are connected to the nucleus by undefined LINC complexes (see text). Black arrows: direction of the nuclear movement. Red molecules/structures: elements responsible for the nuclear movement.
3.1 Nuclear positioning and movement during 2D fibroblast migration
The nucleus tends to track a position just rearward of the cell centroid during 2D migration of either individual fibroblasts or fibroblasts in wounded monolayers. As the centrosome is oriented anterior to the nucleus in these cases and the centrosome tracks the cell centroid, it has long been thought that the centrosome positions the nucleus during 2D migration [42]. It has been challenging to test this hypothesis directly due to the difficulty in specifically disrupting the centrosome. Laser ablation of centrosomes has been used to understand its effect on cell migration, but the position of the nucleus was not examined [43].
Nevertheless, studies of dynein’s role in fibroblast migration provide indirect support for the idea. Dynein was initially considered to contribute to migration solely through its requirement for centrosome orientation in fibroblasts. Indeed, there is abundant evidence that dynein and its regulator dynactin contribute to centrosome orientation in fibroblasts by pulling on MTs from cortical and perhaps cytoplasmic sites [30, 34, 44, 45]. However, disrupting dynein after centrosome orientation by microinjecting inhibitory antibody or expressing dominant negative constructs still inhibits cell migration, indicating another role or roles for dynein [45]. One of these may be to actively maintain close apposition of the nucleus to the centrosome. A study of nuclear rotation during NIH3T3 fibroblast migration revealed that in addition to preventing nuclear rotation (see below), disruption of dynein or dynactin led to a more extreme position of the nucleus in the cell rear [45], indicating a role for dynein and dynactin maintaining nuclear position in migrating fibroblasts (Figure 2A). Whether dynein and dynactin function to directly move the nucleus toward the centrosome or solely function in positioning the centrosome (with the nucleus coupled to the centrosome by some other means) is not yet clear. Consistent with a direct role in moving the nucleus, a recent study showed that dynein and dynactin are recruited to NIH3T3 fibroblast nuclei by nesprin-2 and that this contributes to forward MT-dependent movement of the nucleus toward the centrosome when it is displaced by centrifugal force[46]. Thus, the role of dynein and dynactin in positioning of nuclei in migrating fibroblasts may resemble the roles they play in migrating neurons, where they are important for positioning both the nucleus and the centrosome [47, 48].
A second factor contributing to nuclear positioning during 2D fibroblast migration is the generation of pulling forces by the protruding lamellipodium (Figure 2B) [44, 49]. This has been most clearly shown by experiments in NIH3T3 fibroblasts in which photoactivatable Rac is used to create a new lamellipodium [44]. The nucleus translocates in minutes toward the direction of the new lamellipodium. This result is analogous to that in the original micropipette pulling experiment showing nuclear coupling to plasma membrane integrins [50]. Although the centrosome also moves toward the new lamellipodium, disrupting MTs has no effect on nuclear translocation, implying that the centrosome is not responsible for moving the nucleus in this case. Instead, nuclear translocation requires actomyosin and LINC complexes, although the specific nesprin and SUN proteins involved remain to be identified. Similar results were observed when forward nuclear translocation was triggered by detaching the trailing edge [44]. As specific apical or basal actin fibers did not move in concert with nucleus, the authors propose that a distributed actomyosin generated tension is transmitted to the nucleus by LINC complexes to move it forward.
A third factor that may contribute to nuclear positioning in migrating fibroblasts is a perinuclear adhesion/actin filament system that is distinct from canonical focal adhesions and stress fibers (Figure 2C) [12]. These novel adhesion-like structures are defined by their requirement for the formin FMN2 and its specific localization to these structures. The exact nature of these structures and the basis for their formation near the nucleus remain to be defined, but they are dependent on actin and independent of integrin engagement [12]. In migrating primary MEFs, these FMN2-based structures localize toward the trailing edge of moving nuclei. MEFs depleted of FMN2 loose the perinuclear adhesions, whereas other actin-dependent structures including focal adhesions and SUN2-marked TAN lines are unaffected [12]. No effect of FMN2 depletion was observed on single fibroblast velocity in 2D or the overall closure of wounded monolayers, despite observations that nuclei appeared to drift away from the cell centroid in single cells and centrosomes were not oriented in cells in wounded monolayers. A trend toward reduced wound closure was detected, suggesting a possible defect in migration persistence, but additional experiments are necessary to test this. The FMN2 perinuclear structures may play a more important role in protecting the nucleus from damage during migration through narrow constrictions in 3D matrices [12].
3.2 Nuclear rotation during fibroblast migration in 2D
It has been known for some time that nuclei rotate in the x-y plane of fibroblasts on 2D substrata [45, 51–55]. That this nuclear movement might be associated with cell migration was first suggested by experiments with wounded monolayers in which nuclear rotation was more frequent in NIH3T3 fibroblasts adjacent to or a few rows back from the wound [45]. Nuclear rotation reaches angular velocities of 8.5 degrees per minute and can result in angular changes of hundreds of degrees, although more commonly it is interrupted by pauses or reverses in direction [45]. Rotation is inhibited by MT depolymerization or disruption of dynein heavy chain or the p150Glued subunit of dynactin [45, 55]. The centrosome does not rotate with the nucleus and cortical or Golgi dynein/dynactin does not seem to be involved, leaving the only remaining model one in which dynein/dynactin from nuclear sites exerts torque on the nucleus through perinuclear microtubules (Figure 2D).
A mechanical model for how dynein exerts torque on the nucleus has been proposed [56]. The model is based on data from stationary bovine capillary endothelial cells, but is likely to apply to migrating fibroblasts as nuclear rotation in endothelial cells is also dynein-dependent, does not involve rotation with the centrosome and exhibits angular movements consistent with a persistent random walk [56]. The major prediction from the model is that rotational magnitude is dependent on the distance between the nucleus and the centrosome. Consistent with the prediction, angular displacement of the nucleus is reduced in cells on patterned islands where the centrosome is close to the nuclear centroid. As dynein is also responsible for keeping the nucleus near the centrosome, this leads to the interesting conclusion that as dynein moves the nucleus closer to the centrosome, rotations will be reduced.
A key question is whether dynein interacts with the nucleus during its rotation. Dynein and dynactin are known to interact with NIH3T3 fibroblast nuclei through a region near the C-terminus of nesprin-2 during nuclear recentration after displacement by centrifugation [46], but this has not been tested during nuclear rotation. The requirement for the LINC complex for nuclear rotation in fibroblasts is also unclear. Studies of type A lamins, which are encoded by LMNA gene and anchor the LINC complex, have yielded conflicting results. One study of migrating MEFs from LMNA−/− mice reported nuclear rotation decreased [57], whereas another reported it increased [58]. To further complicate matters, nuclear rotation is dramatically increased in MEFs knocked out for LMNB1, encoding lamin B1 [51]. Perhaps the differences in nuclear rotation in the LMNA−/− MEFs reflect differences in the levels of lamin B1 in the isolates of LMNA−/− MEFs used in the different studies.
Although nuclear rotation is MT- and dynein-dependent in migrating fibroblasts, there is evidence that it is actomyosin dependent in other contexts. The nucleus rotates in an actomyosin-dependent fashion in NIH3T3 fibroblasts subjected to cyclic stretch [59] or plated on adhesive islands that prevent migration [60]. Contrasting these results, knockdown of myosin IIB stimulates nuclear rotation in CHO cells plated on substrata supporting migration [61].
What is the function of nuclear rotation during fibroblast migration? Nuclear rotation is necessary for the alignment of the long axis of the nucleus with that of the cell, termed “nuclear reorientation” that we consider separately below. Because nuclear rotation would be expected to disrupt the transfer of actomyosin tension from the front to the back of the cell, it may also function to reset the tensional balance in the cell, perhaps to allow for establishment of a new front-back axis and direction of migration. Careful analysis of the rotational state of the nucleus during MEF migration revealed an inverse correlation between active rotation of the nucleus, nuclear shape and forward translocation of the cell [58]. Thus, during periods of slow cell translocation, the nucleus rounds-up and rotates, whereas during rapid cell translocation, the nucleus elongates, orients along the long axis of the cell and does not rotate. Nuclei also rotate in MEFs plated on microfabricated islands where they cannot migrate, but do not rotate in MEFs plated on narrow stripes where they did migrate [58]. Whether there is a causal relationship between nuclear rotation and pauses in migration requires further study.
3.3 Nuclear reorientation during fibroblast migration in 2D
The nucleus in spread or migrating fibroblasts on planar substrates tends to be a flattened ellipsoid [62–64], although as noted above, it can transition between round and ellipsoid shapes during migration [58]. That fibroblasts align the long axis of the nucleus with the long axis of the cell during 2D migration was initially noted in experiments with LMNA−/− MEFs, which fail to do this nuclear reorientation [57]. Disruption of Rho GTPase by overexpressing dominant negative constructs or C3 inhibitor reduces nuclear reorientation, suggesting that a Rho signaling pathway is required for nuclear reorientation [65]. Subsequent studies implicated an array of actin cables termed the actin cap in MEFs and Rat 2 fibroblasts (Figure 2E) [58, 66]. This actin cap consists of actin cables running parallel to the long axis of the cell and localized over the dorsal surface of the nucleus. The fibers of the actin cap terminate at focal adhesions in the front and back of the cell and may additionally attach to the nucleus because they are disrupted by expression of a dominant negative KASH construct [58, 66]. The identity of the specific LINC complex components involved in the actin cap in fibroblasts have not been identified, but nesprin-2G is required for actin cap formation in C2C12 myoblasts [58]. The forces necessary to move the nucleus during reorientation may derive from the formation of the actin cap itself [66] and/or from dynein-dependent rotation of the nucleus [58].
As noted above, the actin cap seems to constrain the rotation of the nucleus and during periods where it is not rotating and is reoriented, the cell migrates more rapidly. This suggests that the actin cap promotes rapid directed migration. Consistent with this idea, treatments that disrupt the actin cap fibers, including expression of the KASH construct and low concentration of latrunculin (60 nM), inhibit nuclear reorientation and cell migration [58, 65, 66]. However, an important caveat is that these perturbations may also affect other structures in fibroblasts important for migration. For example, 48 nM latrunculin affects lamellipodial actin dynamics in L929 mouse fibroblasts [67]. It will be important for future experiments to pin down whether the actin cap contributes to cell migration through nuclear reorientation and/or inhibition of nuclear rotation.
The actin cap has also been implicated in nuclear translocation during fibroblast migration [58]. However, it has been noted in at least one study that the nucleus translocates toward the leading edge without translocation of actin fibers in the actin cap [44]. Given the caveat with the specificity of perturbations used to disrupt the actin cap, this is still an unresolved issue.
4. Nuclear movements and positioning in fibroblasts migrating in 3D
Nuclear movement and positioning events have just begun to be explored in fibroblasts migrating in 3D environments. On top of the complexities of positioning the nucleus in fibroblasts migrating in 2D, there are many different modes of migration in 3D matrices [68]. Although some of these represent transitions of cells to a transformed phenotype, even normal fibroblasts migrate by different means depending on the type and organization of the 3D matrix [69]. Here we consider studies on nuclear positioning in 3D migrating fibroblasts with a focus on a novel form that contributes to cell migration through a mechanism called the nuclear piston. We also consider a unique challenge faced by fibroblasts migrating in 3D matrices with pore sizes smaller that the diameter of the nucleus; namely, the difficulty of squeezing the nucleus through narrow pores.
4.1 Nuclear positioning in 3D migration
A unique form of nuclear positioning has been identified in primary human foreskin fibroblasts undergoing lobopodial based migration in 3D matrices. Lobopodial migration is characterized by blunt leading edge lobopodia rather than lamellipodia and elevated Rho rather than Rac signaling [69, 70]. Whether fibroblasts migrate by one mode vs. the other depends on the mechanical properties of the 3D matrix: linearly elastic matrices (e.g., cell derived matrix) stimulate lobopodial migration [69], whereas nonlinearly elastic matrices (e.g., collagen) activate lamellipodial migration [71]. How cells sense the difference in these matrices is unknown.
Interestingly, lobopodial migration depends on the forward translocation of the nucleus which generates compartmentalized intracellular pressure anterior to the nucleus to drive lobopodia formation (Figure 3A) [13]. Two components contribute to the generation of compartmentalized pressure: a forward movement of the nucleus and a diffusion barrier between the nucleus and adjacent cell membrane. The former requires anteriorly activated myosin II and nesprin-3, which binds vimentin IFs through the adaptor plectin [72]. Vimentin immunoprecipitates contain nesprin-3, myosin II and actin suggesting a model in which nesprin-3 and vimentin IFs transmit pulling forces generated by actomyosin to the nucleus (Figure 3B) [13]. Less clearly defined is the diffusive barrier that allows pressure to build in the anterior compartment, but it may involve IFs and ER localized around the nucleus.
Figure 3. Nuclear movement by the nuclear piston in 3D lobopodial migration.
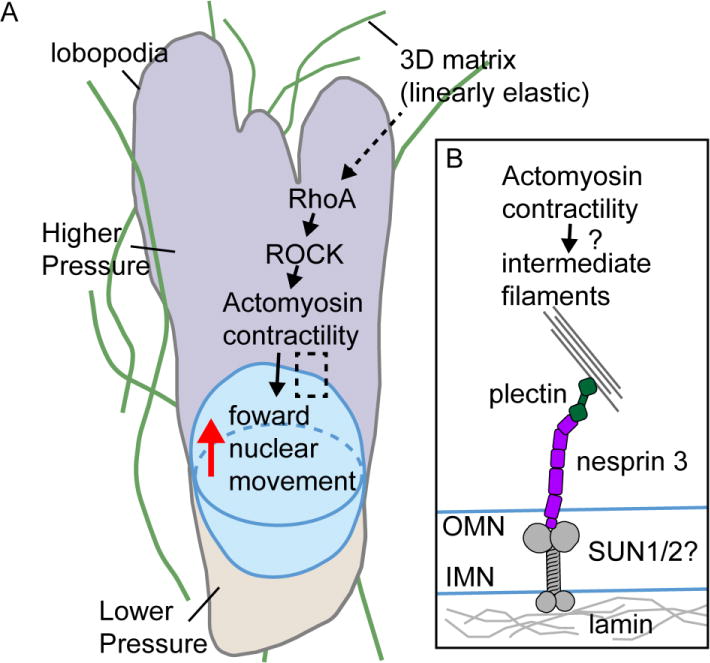
A. Linearly elastic 3D matrix triggers lobopodial migration. Rho A-activated actomyosin contractility moves the nucleus forward to generate higher pressure in the anterior compartment. B. A zoomed-in view of the boxed region in A. The nesprin-3 connection to IFs through plectin is proposed to couple the nucleus to actomyosin pulling forces.
Some important elements of the nuclear piston mechanism remain to be established, including the nature of the molecular link between contractile actin filaments and IFs that allows force transmission to the nucleus and how this resists the increasing pressure in the anterior compartment. Also, the molecular nature of the nuclear-plasma membrane “seal” that compartmentalizes the cell and allows the generation of high pressure in the anterior compartment is not yet defined. Nonetheless, the nuclear piston is an interesting new mechanism because it suggests that the nucleus is directly involved in the generation of the propulsive force for 3D cell migration. Interestingly, fibrosarcoma cells that rely on lamellipodial protrusion and matrix degradation for 3D motility, can initiate lobopodial migration if matrix metalloproteases are inhibited [73].
The factors responsible for positioning and moving the nucleus during lamellipodial based migration in 3D have not been explicitly addressed. Nonetheless, based on similarities between 2D and 3D lamellipodial migration in the positioning of the nucleus relative to the centrosome and pulling forces generated by lamellipodia formation, it seems likely at least in broad outline that similar factors will be involved.
4.2 Nuclear movement through narrow constrictions during 3D migration
The nucleus presents a unique challenge for a fibroblast migrating in a 3D matrix with a pore size is smaller than the nucleus. In fact, the passage of the nucleus through narrow constrictions (<3 μm) in dense 3D matrices is rate limiting for migration [74]. This stems from both the size of the nucleus and its inherent rigidity. Factors that decrease nuclear rigidity, such as reducing the level of A type lamins, have been shown to enhance the migration of MEFs and other cell types through narrow constrictions [75, 76]. Studies in other cell types have suggested active pulling or pushing mechanisms involving myosin II and nesprin-2G for translocating the nucleus through narrow pores [15, 77]. Whether fibroblast nuclei are actively pushed or pulled as they translocate through narrow pores and the possible molecule mechanisms involved require further study.
5. Conclusions
Studies of nuclear positioning in fibroblasts have yielded interesting new information on both the mechanisms of nuclear positioning and its functions in cell migration. Nuclear positioning involves different molecular mechanisms depending whether fibroblasts are polarizing for migration or in an active state of migration and whether they are migrating on 2D vs. 3D substratum. Importantly, the levels of complexity in nuclear positioning increase from polarizing cells to 2D migrating cells to 3D migrating cells. The multitude of mechanisms for nuclear positioning for actively migrating fibroblasts presumably reflect the need to respond to dynamic changes in intracellular forces during migration and for plasticity in the cytoskeletal-nuclear linkage.
Also striking are the unique structures that are assembled to move the nucleus in polarizing and migrating fibroblasts. The identification of adhesion-like TAN lines in polarizing fibroblasts, the actin cap and FMN2-like adhesions in 2D fibroblasts and the nuclear piston in 3D migrating cells suggest that the nucleus is a much more active participant in its positioning that previously suspected. The structures identified to date are all actin-based and most are dependent on the LINC complex, yet it is clear that MTs and IFs also contribute to nuclear positioning. It will be important to examine in more detail the molecular and structural mechanisms by which these other cytoskeletal elements are linked to the nucleus so that they can generate force to positioning it.
The mechanisms for nuclear positioning identified in migrating fibroblasts are likely to apply to other migrating cells. Nesprin-2G and SUN2 TAN lines couple the nucleus to actin cables for rearward nuclear translocation in polarizing C2C12 myoblasts [78]. Nuclear reorientation and the actin cap have been observed in C2C12 myoblasts and other cell types [58, 65]. Perinuclear FMN2 adhesion-like structures are observed in many cell types [12]. And the nuclear piston seems to operate in a variety of cells migrating in 3D elastic matrices [73]. Most of these mechanisms have been identified in cells with rigid nuclei. Whether they apply to other types of migrating cells with more pliable nuclei will be interesting to test. For example, LINC complex components are downregulated in neutrophils and so may not contribute to nuclear positioning in these cells [79]. And, the actin cap is not detected in U2OS osteosarcoma cells [66], which may have more pliable nuclei like other transformed cells.
Although it appears that the nuclear positioning mechanisms in polarizing, 2D migrating and 3D migrating fibroblasts are distinct and specific for each state, they may be more related than they currently appear. For example, both the TAN lines and the actin cap appear to prevent nuclear rotation by attaching the nucleus to actin cables. The actomyosin- and LINC complex-dependent movement of the nucleus toward lamellipodia observed in 2D migration is likely to function also in 3D lamellipodial migration. And, even the nuclear piston mechanism of forward movement in lobopodial migration in 3D requires actomyosin and the LINC complex, which have also been implicated in forward movement of the nucleus in 2D migration.
Studies of fibroblasts have also provided dramatic advances in our understanding of the functions of nuclear position. Nuclear positioning contributes to the generation of cell polarity as shown by studies of fibroblasts orienting their centrosomes [10, 11, 14]. And it contributes to the distribution of intracellular tension as shown by studies in 2D migrating fibroblasts [44, 49] and directly to the generation of cellular protrusion as shown by studies in 3D migrating fibroblasts [13, 73]. All of these functions are dependent on LINC complexes and so are not likely to be restricted to fibroblast migration, thus providing broader insight into mechanisms of nuclear positioning. Indeed, many of the genes that contribute to nuclear positioning in fibroblasts are mutated in a large number of diseases, collectively termed “nuclear envelopathies” [8, 80–82], raising the possibility that one or more of the functions of nuclear positioning described in fibroblasts may contribute to these diseases. Our prediction is that studies of nuclear positioning in fibroblasts will continue to be an important source of information for understanding both normal and disease cellular physiology.
Acknowledgments
Work in the Gundersen lab is supported by NIH grants R01 GM099481 and R01 AR068636. Ruijun Zhu was supported by an American Heart Association Predoctoral Fellowship (15PRE25400002).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ashcroft GS, Horan MA, Ferguson MW. The effects of ageing on cutaneous wound healing in mammals. Journal of Anatomy. 1995;187(Pt 1):1–26. [PMC free article] [PubMed] [Google Scholar]
- 2.Sgonc R, Gruber J. Age-Related Aspects of Cutaneous Wound Healing: A Mini-Review. Gerontology. 2013;59(2):159–164. doi: 10.1159/000342344. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Frangogiannis NG. Fibroblasts in post-infarction inflammation and cardiac repair. Biochimica et biophysica acta. 2013;1833(4):945–953. doi: 10.1016/j.bbamcr.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Padmakumar VC, et al. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. 2005;118(Pt 15):3419–30. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- 5.Crisp M, et al. Coupling of the nucleus and cytoplasm: Role of the LINC complex. The Journal of Cell Biology. 2006;172(1):41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang W, Worman HJ, Gundersen GG. Accessorizing and anchoring the LINC complex for multifunctionality. The Journal of Cell Biology. 2015;208(1):11–22. doi: 10.1083/jcb.201409047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horn HF. Chapter Six - LINC Complex Proteins in Development and Disease. In: Colin LS, editor. Current Topics in Developmental Biology. Academic Press; 2014. pp. 287–321. [DOI] [PubMed] [Google Scholar]
- 8.Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152(6):1376–89. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folker ES, et al. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proceedings of the National Academy of Sciences. 2011;108(1):131–6. doi: 10.1073/pnas.1000824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luxton GW, et al. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329(5994):956–9. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luxton GW, et al. TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus. 2011;2(3):173–81. doi: 10.4161/nucl.2.3.16243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skau CT, et al. FMN2 Makes Perinuclear Actin to Protect Nuclei during Confined Migration and Promote Metastasis. Cell. 2016;167(6):1571–1585 e18. doi: 10.1016/j.cell.2016.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Petrie RJ, Koo H, Yamada KM. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 2014;345(6200):1062–5. doi: 10.1126/science.1256965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121(3):451–63. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 15.Jayo A, et al. Fascin Regulates Nuclear Movement and Deformation in Migrating Cells. Developmental Cell. 2016;38(4):371–383. doi: 10.1016/j.devcel.2016.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kutscheidt S, et al. FHOD1 interaction with nesprin-2G mediates TAN line formation and nuclear movement. Nat Cell Biol. 2014;16(7):708–715. doi: 10.1038/ncb2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Libotte T, et al. Lamin A/C-dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol Biol Cell. 2005;16(7):3411–24. doi: 10.1091/mbc.E04-11-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Worman HJ. Nuclear lamins and laminopathies. J Pathol. 2012;226(2):316–25. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haque F, et al. SUN1 Interacts with Nuclear Lamin A and Cytoplasmic Nesprins To Provide a Physical Connection between the Nuclear Lamina and the Cytoskeleton. Molecular and Cellular Biology. 2006;26(10):3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Östlund C, et al. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. Journal of Cell Science. 2009;122(22):4099–4108. doi: 10.1242/jcs.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King MC, Drivas TG, Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134(3):427–38. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borrego-Pinto J, et al. Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci. 2012;125(Pt 5):1099–105. doi: 10.1242/jcs.087049. [DOI] [PubMed] [Google Scholar]
- 23.Clements L, et al. Direct Interaction between Emerin and Lamin A. Biochemical and Biophysical Research Communications. 2000;267(3):709–714. doi: 10.1006/bbrc.1999.2023. [DOI] [PubMed] [Google Scholar]
- 24.Chang W, et al. Emerin organizes actin flow for nuclear movement and centrosome orientation in migrating fibroblasts. Mol Biol Cell. 2013;24(24):3869–80. doi: 10.1091/mbc.E13-06-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Versaevel M, et al. Super-resolution microscopy reveals LINC complex recruitment at nuclear indentation sites. Scientific Reports. 2014;4:7362. doi: 10.1038/srep07362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arsenovic PT, et al. Nesprin-2G, a Component of the Nuclear LINC Complex, Is Subject to Myosin-Dependent Tension. Biophys J. 2016;110(1):34–43. doi: 10.1016/j.bpj.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schonichen A, et al. FHOD1 is a combined actin filament capping and bundling factor that selectively associates with actin arcs and stress fibers. J Cell Sci. 2013;126(Pt 8):1891–901. doi: 10.1242/jcs.126706. [DOI] [PubMed] [Google Scholar]
- 28.D’Alessandro M, et al. Amphiphysin 2 Orchestrates Nucleus Positioning and Shape by Linking the Nuclear Envelope to the Actin and Microtubule Cytoskeleton. Dev Cell. 2015;35(2):186–98. doi: 10.1016/j.devcel.2015.09.018. [DOI] [PubMed] [Google Scholar]
- 29.Tzur YB, Wilson KL, Gruenbaum Y. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat Rev Mol Cell Biol. 2006;7(10):782–8. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- 30.Palazzo AF, et al. Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr Biol. 2001;11(19):1536–41. doi: 10.1016/s0960-9822(01)00475-4. [DOI] [PubMed] [Google Scholar]
- 31.Czuchra A, et al. Cdc42 is not essential for filopodium formation, directed migration, cell polarization, and mitosis in fibroblastoid cells. Mol Biol Cell. 2005;16(10):4473–84. doi: 10.1091/mbc.E05-01-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung T, et al. Myotonic Dystrophy Kinase-Related Cdc42-Binding Kinase Acts as a Cdc42 Effector in Promoting Cytoskeletal Reorganization. Molecular and Cellular Biology. 1998;18(1):130–140. doi: 10.1128/mcb.18.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vicente-Manzanares M, et al. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10(11):778–90. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmoranzer J, et al. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr Biol. 2009;19(13):1065–74. doi: 10.1016/j.cub.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eng CH, Huckaba TM, Gundersen GG. The formin mDia regulates GSK3beta through novel PKCs to promote microtubule stabilization but not MTOC reorientation in migrating fibroblasts. Mol Biol Cell. 2006;17(12):5004–16. doi: 10.1091/mbc.E05-10-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saunders CA, et al. TorsinA controls TAN line assembly and the retrograde flow of dorsal perinuclear actin cables during rearward nuclear movement. J Cell Biol. 2017;216(3):657–674. doi: 10.1083/jcb.201507113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozelius LJ, et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet. 1997;17(1):40–48. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- 38.Goodchild RE, Dauer WT. Mislocalization to the nuclear envelope: An effect of the dystonia-causing torsinA mutation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(3):847–852. doi: 10.1073/pnas.0304375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naismith TV, et al. TorsinA in the nuclear envelope. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(20):7612–7617. doi: 10.1073/pnas.0308760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodchild RE, Dauer WT. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. The Journal of Cell Biology. 2005;168(6):855–862. doi: 10.1083/jcb.200411026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nery FC, et al. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. Journal of Cell Science. 2008;121(20):3476–3486. doi: 10.1242/jcs.029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luxton GW, Gundersen GG. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr Opin Cell Biol. 2011;23(5):579–88. doi: 10.1016/j.ceb.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakida NM, et al. An intact centrosome is required for the maintenance of polarization during directional cell migration. PLoS One. 2010;5(12):e15462. doi: 10.1371/journal.pone.0015462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J, et al. Actomyosin pulls to advance the nucleus in a migrating tissue cell. Biophys J. 2014;106(1):7–15. doi: 10.1016/j.bpj.2013.11.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levy JR, Holzbaur ELF. Dynein drives nuclear rotation during forward progression of motile fibroblasts. Journal of Cell Science. 2008;121(19):3187–3195. doi: 10.1242/jcs.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu R, Antoku S, Gundersen GG. Centrifugal Displacement of Nuclei Reveals Multiple LINC Complex Mechanisms for Homeostatic Nuclear Positioning. Curr Biol. 2017;27(20):3097–3110 e5. doi: 10.1016/j.cub.2017.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10(8):970–9. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- 48.Shu T, et al. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44(2):263–77. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 49.Alam SG, et al. The nucleus is an intracellular propagator of tensile forces in NIH 3T3 fibroblasts. J Cell Sci. 2015;128(10):1901–11. doi: 10.1242/jcs.161703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci U S A. 1997;94(3):849–54. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ji JY, et al. Cell nuclei spin in the absence of lamin b1. J Biol Chem. 2007;282(27):20015–26. doi: 10.1074/jbc.M611094200. [DOI] [PubMed] [Google Scholar]
- 52.Paddock SW, Albrecht-Buehler G. The degree of coupling of nuclear rotation in binucleate 3T3 cells. Exp Cell Res. 1986;166(1):113–26. doi: 10.1016/0014-4827(86)90512-4. [DOI] [PubMed] [Google Scholar]
- 53.Paddock SW, Albrecht-Buehler G. Rigidity of the nucleus during nuclear rotation in 3T3 cells. Exp Cell Res. 1988;175(2):409–13. doi: 10.1016/0014-4827(88)90205-4. [DOI] [PubMed] [Google Scholar]
- 54.Bard F, et al. Rotation of the cell nucleus in living cells: a quantitative analysis. Biol Cell. 1985;54(2):135–42. doi: 10.1111/j.1768-322x.1985.tb00388.x. [DOI] [PubMed] [Google Scholar]
- 55.Gerashchenko MV, et al. Dynein is a motor for nuclear rotation while vimentin IFs is a “brake”. Cell Biol Int. 2009;33(10):1057–64. doi: 10.1016/j.cellbi.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 56.Wu J, et al. How dynein and microtubules rotate the nucleus. J Cell Physiol. 2011;226(10):2666–74. doi: 10.1002/jcp.22616. [DOI] [PubMed] [Google Scholar]
- 57.Houben F, et al. Disturbed nuclear orientation and cellular migration in A-type lamin deficient cells. Biochim Biophys Acta. 2009;1793(2):312–24. doi: 10.1016/j.bbamcr.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 58.Kim DH, Cho S, Wirtz D. Tight coupling between nucleus and cell migration through the perinuclear actin cap. Journal of Cell Science. 2014;127(11):2528–2541. doi: 10.1242/jcs.144345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brosig M, et al. Interfering with the connection between the nucleus and the cytoskeleton affects nuclear rotation, mechanotransduction and myogenesis. The International Journal of Biochemistry & Cell Biology. 2010;42(10):1717–1728. doi: 10.1016/j.biocel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Kumar A, et al. Actomyosin contractility rotates the cell nucleus. Scientific Reports. 2014;4:3781. doi: 10.1038/srep03781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vicente-Manzanares M, et al. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol. 2007;176(5):573–80. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, et al. Moving Cell Boundaries Drive Nuclear Shaping during Cell Spreading. Biophys J. 2015;109(4):670–86. doi: 10.1016/j.bpj.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Q, et al. The regulation of dynamic mechanical coupling between actin cytoskeleton and nucleus by matrix geometry. Biomaterials. 2014;35(3):961–9. doi: 10.1016/j.biomaterials.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 64.Khatau SB, et al. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci U S A. 2009;106(45):19017–22. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maninova M, et al. The reorientation of cell nucleus promotes the establishment of front-rear polarity in migrating fibroblasts. J Mol Biol. 2013;425(11):2039–55. doi: 10.1016/j.jmb.2013.02.034. [DOI] [PubMed] [Google Scholar]
- 66.Maninova M, Vomastek T. Dorsal stress fibers, transverse actin arcs, and perinuclear actin fibers form an interconnected network that induces nuclear movement in polarizing fibroblasts. FEBS J. 2016;283(20):3676–3693. doi: 10.1111/febs.13836. [DOI] [PubMed] [Google Scholar]
- 67.Gronewold TM, et al. Effects of rhizopodin and latrunculin B on the morphology and on the actin cytoskeleton of mammalian cells. Cell Tissue Res. 1999;295(1):121–9. doi: 10.1007/s004410051218. [DOI] [PubMed] [Google Scholar]
- 68.Petrie RJ, Yamada KM. Multiple mechanisms of 3D migration: the origins of plasticity. Curr Opin Cell Biol. 2016;42:7–12. doi: 10.1016/j.ceb.2016.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petrie RJ, et al. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J Cell Biol. 2012;197(3):439–55. doi: 10.1083/jcb.201201124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petrie RJ, Yamada KM. At the leading edge of three-dimensional cell migration. Journal of Cell Science. 2012;125(24):5917–5926. doi: 10.1242/jcs.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grinnell F, et al. Dendritic Fibroblasts in Three-dimensional Collagen Matrices. Molecular Biology of the Cell. 2003;14(2):384–395. doi: 10.1091/mbc.E02-08-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilhelmsen K, et al. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171(5):799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Petrie RJ, et al. Activating the nuclear piston mechanism of 3D migration in tumor cells. J Cell Biol. 2017;216(1):93–100. doi: 10.1083/jcb.201605097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wolf K, et al. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201(7):1069–84. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davidson PM, et al. Nuclear deformability constitutes a rate-limiting step during cell migration in 3-D environments. Cell Mol Bioeng. 2014;7(3):293–306. doi: 10.1007/s12195-014-0342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harada T, et al. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J Cell Biol. 2014;204(5):669–82. doi: 10.1083/jcb.201308029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thomas DG, et al. Non-muscle myosin IIB is critical for nuclear translocation during 3D invasion. J Cell Biol. 2015;210(4):583–94. doi: 10.1083/jcb.201502039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chang W, et al. Linker of nucleoskeleton and cytoskeleton (LINC) complex-mediated actin-dependent nuclear positioning orients centrosomes in migrating myoblasts. Nucleus. 2015;6(1):77–88. doi: 10.1080/19491034.2015.1004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Olins AL, et al. The LINC-less granulocyte nucleus. Eur J Cell Biol. 2009;88(4):203–14. doi: 10.1016/j.ejcb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet. 2006;7(12):940–52. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- 81.Worman HJ, et al. Laminopathies and the long strange trip from basic cell biology to therapy. J Clin Invest. 2009;119(7):1825–36. doi: 10.1172/JCI37679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bonne G, Quijano-Roy S. Emery-Dreifuss muscular dystrophy, laminopathies, and other nuclear envelopathies. Handb Clin Neurol. 2013;113:1367–76. doi: 10.1016/B978-0-444-59565-2.00007-1. [DOI] [PubMed] [Google Scholar]
- 83.Antoku S, et al. Reinforcing the LINC complex connection to actin filaments: the role of FHOD1 in TAN line formation and nuclear movement. Cell Cycle. 2015;14(14):2200–5. doi: 10.1080/15384101.2015.1053665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Starr DA, Rose LS. TorsinA regulates the LINC to moving nuclei. J Cell Biol. 2017;216(3):543–545. doi: 10.1083/jcb.201701054. [DOI] [PMC free article] [PubMed] [Google Scholar]


