Abstract
Trauma is a major problem in the United States. Mortality from trauma is the number one cause of death under the age of 45 in the US and is the third leading cause of death for all age groups. There are nearly 200,000 deaths per year due to trauma in the US at a cost of over $671 billion in combined health care costs and lost productivity. Unsurprisingly, trauma accounts for about 30% of all life-years lost in the US. Due to immense development of trauma systems, a large majority of trauma patients survive the injury but then go on to die from complications arising from the injury. These complications are marked by early and significant metabolic changes accompanied by inflammatory responses that lead to progressive organ failure, and ultimately, death. Early resuscitative and surgical interventions followed by close monitoring to identify and rescue treatment failures are key to successful outcomes. Currently, the adequacy of resuscitation is measured using vital signs, noninvasive methods such as bedside echocardiography or stroke volume variation, and other laboratory endpoints of resuscitation, such as lactate and base deficit. However, these methods may be too crude to understand cellular and subcellular changes that may be occurring in trauma patients. Better diagnostic and therapeutic markers are needed to assess the adequacy of interventions and monitor responses at a cellular and subcellular level and inform clinical decision making before complications are clinically apparent. The developing field of metabolomics holds great promise in the identification and application of biochemical markers towards the clinical decision making process.
Keywords: metabolomics, injury, trauma, outcomes, translational
Introduction
Trauma system development has substantially decreased the mortality from trauma in the United States (1–4). However, trauma remains the number one cause of death for people under the age of 45 in the US, and is the third leading cause of death for all age groups (5). In fact, trauma accounts for about 30% of all life-years lost in the US and costs over $671 billion a year in combined health care costs and lost productivity (2013 USD) (6, 7). This is despite major advancements in damage control surgery, resuscitation and intensive care in the last fifty years.
While trauma systems have evolved to quickly identify critically injured patients and get them to major trauma centers, a substantial number of patients still deteriorate and develop systemic inflammatory response syndromes and acute organ failure despite availability of 24-hour surgical services and sophisticated intensive care. Thus, it becomes critical to identify the injured patients who are going to deteriorate as early as possible. Traditionally, we address this risk of deterioration by close clinical monitoring (8). However, conventional labs and imaging may actually be too crude and reflect downstream events instead of identifying early biochemical changes that precede physiologic changes. These methods may be too little too late to allow for effective interventions (9–11). Thus, there is a huge need to identify a set of markers that can be used to rapidly and accurately identify patients who are starting to clinically deteriorate.
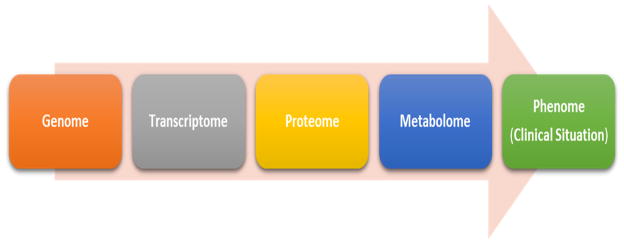
The recent development of the various “-omics” fields represent an opportune moment for the science behind trauma care (12–16). Genomics, proteomics, metabolomics are fields of increasing proximity to the clinical situation (or phenome) (Figure 1). Within the field of -omics, metabolomics holds great promise as it can provide details about the current biochemical status of a patient. Trauma is known to trigger biochemical responses resulting in altered metabolism of proteins, carbohydrates, nucleic acids, amino acids and lipids and result in a metabolic signature that represents the body’s final integrated response to the pathophysiological insults (17–19). Thus changes in the metabolome precede clinical deterioration and the ability to identify these critical metabolic changes can enable early and proactive management of clinical conditions (20). The Human Metabolome Database by the Metabolomics Innovation Centre, a Canadian publicly funded research and core facility tracks nearly 42,000 such metabolites.
Figure 1. Metabolomics and its relationship to the Clinical Situation.
Flow of biochemical information proceeds from the genome through the transcriptome, to the proteome and finally to the metabolome before presenting as the phenome (clinical situation). The metabolome thus constitutes the final integrated response from the genome, transcriptome and the proteome. It is also the last of the biochemical hierarchies before the phenome that manifests as the clinical situation. Clinical situation is in a large part a reflection of the accumulated changes of the metabolome. As such, presentation of the clinical situation is preceded by marked and quantitatively detectable changes in the metabolome which can act as early biomarkers for the clinical trajectory of trauma patients.
In this review, we discuss the state of the field of metabolomics, the process of conducting metabolomics investigation including data acquisition, processing and abstraction, and the clinical applications of metabolomics to trauma research.
Metabolomics and Data Acquisition
The metabolome constitutes a large number of highly diverse group of biochemicals which can be assayed from diverse body fluids including plasma, serum, cerebrospinal fluid and urine. The study of the metabolome requires high-resolution analytical instruments to acquire data as well specialized techniques and skills for data analysis and interpretation. There are two possible analytical methods for investigational metabolomics – nuclear magnetic resonance (NMR) and high-resolution tandem mass spectrometry with front-end ultra-performance chromatographic separation (LC-HRMS). Both NMR and LC-HRMS based analytical services are available commercially (e.g. West Coast Metabolomics Center, Metabolon, etc.) which negates the need for developing high cost in-house infrastructure.
Traditionally, high-field NMR has been the method of choice in metabolomic investigations (21–23). In fact, most of the published investigations involving the metabolic studies of trauma have relied on 1H NMR (21–24). NMR is highly quantitative which allows the acquired data to be extremely reproducible. NMR also allows unbiased identification of metabolites of interest. However, NMR has significantly poor sensitivity compared to mass spectrometry. While current NMR instruments can detect metabolites at a concentration of 1–2 μM range in volumes of 0.5 ml, early metabolic changes have to be detected at nM to pM levels which is currently impractical through NMR analysis. The second option, LC-HRMS, provides many advantages over NMR including very high sensitivity, high resolution, and the ability to detect and quantify a wider range of metabolites. As such, LC-HRMS is rapidly becoming the analytical technique of choice for metabolomic studies, and is the focus of this review (25–28).
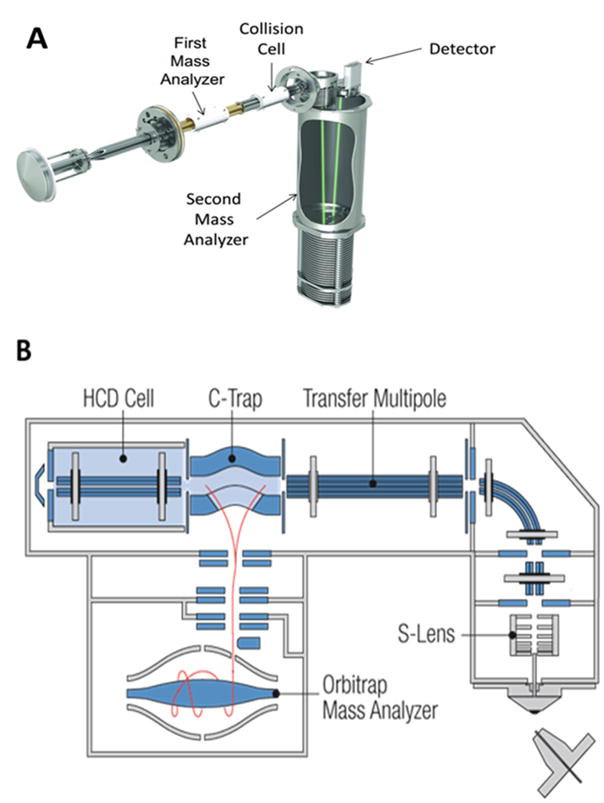
LC-HRMS instrumentation consist of a mass spectrometer, also called a mass analyzer, which converts metabolites to gas phase ions and then separates them by mass to charge (m/z) ratio (Figure 2). Nearly all current mass spectrometers consist of at least two mass analyzers operating in tandem (tandem mass spectrometry) which are separated by a collision cell. Following ionization of the metabolites at the source, the first mass analyzer selects and accelerates a beam of ions with a specific mass/charge ratio (m/z) into the collision cell. These ions are forced to collide in a controlled fashion with an inert gas, usually either nitrogen or argon, within the collision cell. A part of the kinetic energy from the collision is internalized. The resultant high energetic state leads to dissociation of bonds giving rise to molecular fragments. Neutral species are lost from the ion beam and the remaining species retaining the ionic charge are transmitted to the second mass analyzer where they are separated according to their new mass to charge ratios and recorded as counts per minute. These product ions are often characteristic of the precursor ion and aid in the identification of the individual metabolites.
Figure 2. Schematic diagrams of the standard mass analyzer configuration used in metabolomic investigations.
(A) The schematic diagram of a Sciex TripleTOF quadrupole time of flight mass analyzer (QTOF) is depicted with a quadrupole mass filter capable of 1 Dalton resolution as the first mass analyzer and a time of flight mass analyzer as the second high resolution mass analyzer. (B) The schematic diagram of a Thermo QExactive Orbitrap mass spectrometer is depicted with a quadrupole mass filter capable of 0.4 Dalton mass resolution as the first mass analyzer and an Orbitrap as the second high resolution mass analyzer. Both types of mass analyzers are commonly employed in metabolomic research.
The most versatile tandem mass analyzer combination for metabolomic investigations are the quadrupole/time of flight (QTOF) and Orbitrap families of mass analyzers (Figure 2). These provide highly accurate methods of detecting molecules and have now become the standard for metabolomics studies (25, 26, 28). These mass analyzers consist of two separate but tandemly operating quadrupole and time of flight mass analyzers (Figure 2A) or quadrupole and Orbitrap mass analyzers (Figure 2B) separated by a collision cell. A quadrupole mass analyzer consists of four parallel rods and operates by superimposing a radio frequency (RF) and a direct current (DC) voltage between opposing rods. At a given RF/DC ratio, only ions of specific mass/charge ratios will retain an oscillation path with constant amplitude that allows them to move across the length of the quadrupole and into the collision cell. Thus, although, a group of metabolite ions enters the quadrupole analyzer at the same time, by cycling through a range of RF/DC ratios different ions with different m/z ratios are forced to enter the collision cell resulting in a separation of the precursor metabolite ions. These precursor metabolite ions collide with gas molecules in the collision cell and as a result undergo fragmentation. Those fragment ions that still retain a charge, are transmitted into either the time of flight (TOF) or Orbitrap mass analyzer (Figure 2A and 2B). The TOF mass analyzer operates by accelerating pulses of ions through a very low pressure flight tube towards a detection terminal. Equicharged ions entering the time of flight analyzer will possess the same kinetic energy but will have mass dependent velocities (higher the mass, the lower the velocity). This effectively causes a temporal dispersion based on the mass, leading to effective separation and detection, by mass, of similarly charged ions. As a result of this efficient separation, mass measurements obtained from current time of flight mass analyzers are within a few parts per million of a metabolite’s mass as calculated from its chemical formula (theoretical mass). Thus time of flight mass analyzers provide a very high confidence in the identity of the metabolite. Orbitrap mass analyzers, on the other hand, force ions to move in an orbital motion around a central spindle and the resultant ion current is converted to a mass spectrum via Fourier transformation of the frequency signal. As a result, the Orbitrap m/z resolution tends to be superior to TOF mass analyzers for low molecular weight compounds, although they take longer for data acquisition compared to TOF mass analyzers.
The human metabolome and lipidome is postulated to contain more than 300,000 molecular species with a majority having masses between 50 and 1500 Daltons. This makes mass spectrometry alone insufficient for unambiguous identification. To overcome this challenge, an additional orthogonal separation method such as ultra-performance liquid chromatography (UPLC) is needed. This chromatographic method enable the separation of metabolites by their chemical properties such as hydrophobicity and greatly help to resolve different molecular species that have the same mass to charge ratio. When UPLC is employed with HRMS techniques, a minimum of three degrees of confidence (chromatographic retention time, metabolite precursor mass and metabolite product ion masses) are employed in identifying the metabolite of interest. Additional orthogonal separation techniques such as gas phase differential mobility are also available in newer mass analyzers. In recent years, the combination of these orthogonal modes of separation has led to an unparalleled degree of specificity in metabolomics studies, thus greatly increasing their relevance for research in the clinical setting.
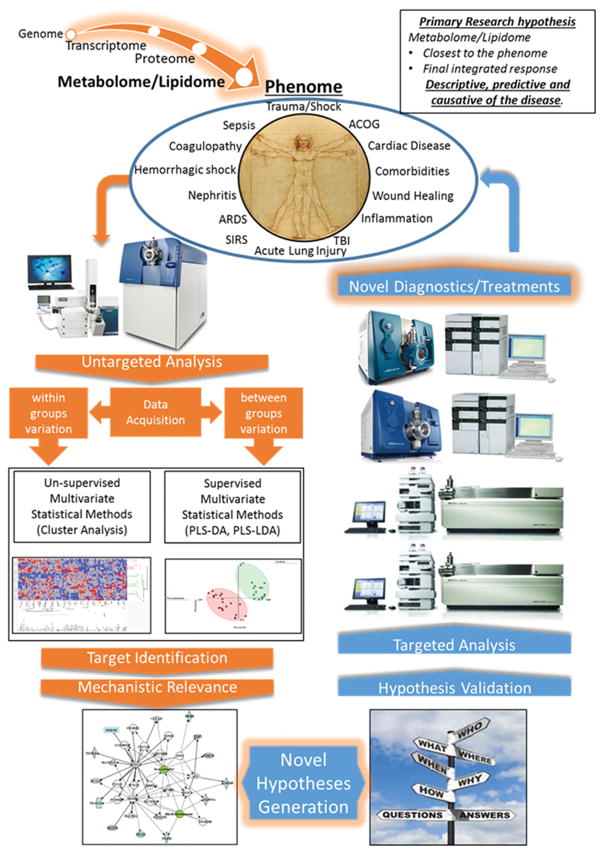
Figure 3 demonstrates a complete metabolomic workflow from sample collection, data acquisition via mass spectrometry, statistical analysis, abstraction and application to address clinical concerns.
Figure 3. The metabolome provides a window to the biochemical dysregulation that precedes poor clinical outcomes.
This figure depicts a metabolomic workflow that begins with sample collection from trauma patients which progresses to untargeted analysis to identify changes in markers and pathways. This drives hypothesis generation regarding specific markers, which are then validated through targeted analysis to form the basis of diagnostic or therapeutic interventions that enable better clinical outcomes.
Metabolomics and Data Processing
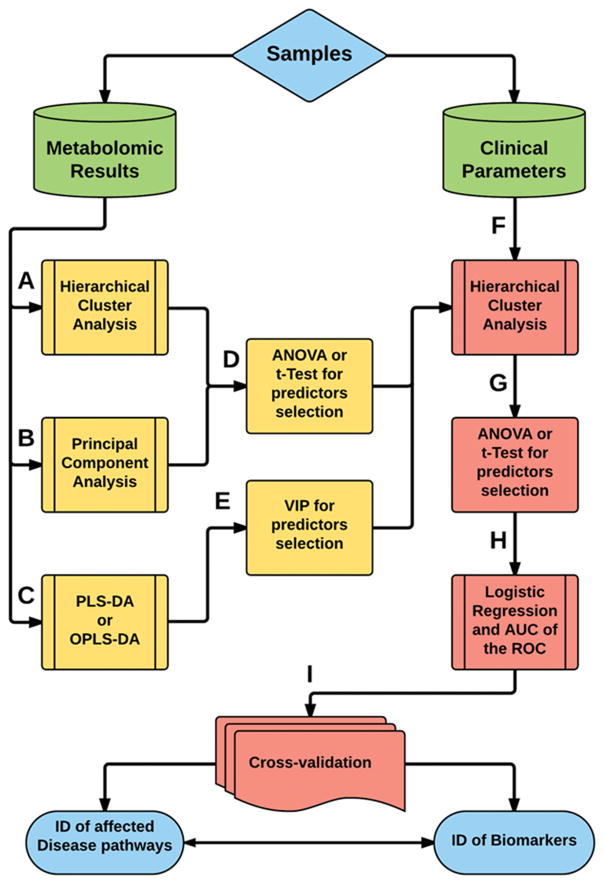
The diversity of the metabolome and the analytical capacity of the instruments generate large quantities of data per sample (often in the range of hundreds of megabytes). This data first needs to be processed using specific techniques prior to analysis including. These include data filtering, peak detection, deconvolution, peak alignment and normalization (29). The processed data is then analyzed to identify similarities and differences within the data and to assess how those differences correlate to clinical outcomes. Due to the volume of data, traditional univariate analysis comparing a single metabolite to a single outcome is often too time consuming and unwieldy in the analysis of metabolomic data. The preferred method of initial statistical investigations for metabolomic studies are unsupervised multi-parametric techniques implemented via cluster analysis or principal component analysis (Figure 4) (30). This statistical approach summarizes tendencies within the data such as clustering or outliers. Furthermore, Cluster Analysis can reveal patterns of correlation between clinical symptoms and metabolomic data that otherwise would be assessed as outliers or not noticed at all.
Figure 4. A standard statistical analysis workflow for metabolomic data.
Due to the complexity of metabolomic data, a combination of supervised and unsupervised analytical methods are employed in the statistical analysis and validation. Principal component analysis (PCA) and cluster analysis are the primary unsupervised analytical strategies (processes A and B), whereas partial least squares discriminant analysis (PLS-DA) and orthogonal partial least squared discriminant analysis (OPLS-DA) are the primary supervised statistical analysis approaches (processes C and D). Additional analysis such as 2D clustering can be incorporated to further investigate the relevance of the early statistical findings to clinical variables. Finally, validation and correctional statistics are employed to allow multiple hypothesis testing. Process A –Hierarchical Cluster Analysis: To search for cluster of patients when there is no class assignment in the study design. Process B – Principal Component Analysis (PCA): Useful to visualize clusters’ separation (when the groups are not previously assigned) or classes’ separation and determine metabolites that better explain the data variability. Process C – Partial Least Squares Discriminant Analysis (PLS-DA) or Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA): PLS-DA to visualize class-discrimination based in the identified metabolites when groups where previously assigned in the study design and OPLS-DA to separate predictor’s variables from non-correlated variables to maximize class-discrimination when there is high within-class variation. Process D – ANOVA or t-Test: Helps to select the more important candidates’ metabolites as predictors of cluster separation. Process E – Variable Importance in Projection (VIP): Helps to select the variables with more contribution to classes’ separation. Process F – Combining predictor metabolites with classical clinical variables using Hierarchical Cluster Analysis helps to associate metabolic findings to physiological states and lead to discovery of within-group pathophenotypes. Process G – ANOVA or t-Test: Application to outcomes of process F helps to select the more important predictors of pathophenotypes. Process H – Logistic Regression to test which of these predictors of pathophenotypes can be used to set a model of the pathology to select the more significant ones and Area under the Curve (AUC) of the Receiver Operating Characteristic to determine the accuracy of the selected predictors in the model determined by the predictors. Process I – Cross-Validation: To test the model with its predictors’ variables using either with previous separated test using leave-one-out cross-validation or a bootstrap approach. The final outcome of the statistical workflow are the identities of the metabolites that can be used as valid biomarkers and also the validated identities of the disease networks associated with the trauma injury.
A more powerful discriminant technique involves supervised data analysis, using tools such as Partial Least Squares Discriminant Analysis (PLS-DA) and Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA) (Figure 4) (17). However, these supervised statistical analysis methods are more susceptible to false positives compared to unsupervised techniques and need to be used with caution. Leave-One-Out-Cross-Validation approaches, or bootstrap techniques can be used to test a subset of samples and validate findings. A schematic of data processing in metabolomics is illustrated in Figure 4. These steps have to be customized for different study designs.
The discovery of statistically significant metabolic and clinical variables is the first step towards biomarker identification from metabolomic data. Logistic Regression and Area under the Curve of the Receiver Operating Characteristic are useful tools to test the accuracy of these variables in predicting outcomes. Implementation of such stringent statistical methods increases confidence in the clinical relevance of any biomarkers identified using metabolomics. Metabolites selected from these techniques are often important predictors of metabolic disturbances in the patient and have a high tendency to correlate with physiological events. Thus, they can be used to understand the underlying biochemical changes that result in the clinical state (phenome). Thus a metabolomics workflow can lead to identification of biomarkers as well as metabolic pathways involved in various clinical situations.
Metabolomics and Data Abstraction
Lastly, metabolic changes have to be understood as they relate to affected metabolic networks to be useful for creating diagnostic or treatment strategies. There are a large number of metabolomic databases that display metabolites in the context of their metabolic pathways (31–35). These databases are useful to manually verify and understand the effects of one or a few metabolic changes. However, they cannot practically be used for high throughput analysis of large datasets with thousands of data points as currently produced by sophisticated mass analyzers. Several software tools can manipulate metabolic data and visualize relationships within them using described relationships from existing databases (36). However, due to the relative novelty of the field of metabolomics, many pathways are not well characterized or are simply unavailable within existing databases (37, 38). Some options do allow customization of paths that do not fit standard/known pathways (39, 40). Often, it is up to the investigators to interpret the data to determine links to known biochemical networks.
Clinical Applications of Metabolomics in Trauma Research
The Inflammation and the Host Response Large Scale Collaborative Research Program (Glue Grant) is the pre-eminent research collaborative that has led genomic and proteomics research in trauma since its inception in 2001 with support from the National Institutes of General Medical Sciences (41–43). The investigators of this program have refined techniques to identify biomarkers that may correlate with survival(44), develop genomic score predicting clinical trajectories in trauma patients(45–47), create a method for isolating neutrophils to identify immune response to severe trauma and burn injury(48) and determine key proteins involved in acute phase response signaling, the complement system, and coagulation system pathways(49). The program has resulted in extensive knowledge and insights into the metabolic stress response to burn injuries and has outlined potential research opportunities using metabolomics, proteomics and genomics (50, 51).
Metabolomics is being increasingly studied in traumatic brain injury (TBI), severe trauma/shock and burns in an attempt to create a metabolic fingerprint for each condition that can be tracked prospectively. A review of the literature across these three areas demonstrates the enormous potential of metabolomics for trauma research. The disruption of physiologic homeostasis in TBI is thought to result in a ‘metabolic crisis’ that is reflected in the metabolome (52–59). There may be nearly 2500 molecules that are affected by TBI and can be identified in urine (60). Cerebrospinal fluid has also been studied as a source of metabolomic data in TBI patients especially since intracranial monitoring and extraventricular drains are often necessary for clinical care. Derangements of glycolysis and the tricarboxylic acid cycle as well as changes in amino acids, and phospholipid and pyrimidine metabolism have been shown in TBI patients (52, 53, 61, 62). Sphingolipids ratios have been shown to correlate with traumatic brain injury (63). Significant differences in levels of lactate, propylene glycol and glutamine have been noted compared to non-injured controls (64). Several protease substrates such as cytoskeletal proteins, transcription factors, cell cycle regulatory proteins, synaptic proteins, and cell junction proteins have been found to be constantly exposed to activated proteases in TBI patients (65). A mouse model of TBI examined via LC-MS approaches has demonstrated the negative effects on major cellular processes and lipidomic changes in the visual system following repetitive mild TBI (66). Markers of systemic metabolic derangement such as succinate, oxoproline, urate and fatty acids might contribute to coagulopathies of trauma and neutrophil priming associated with acute lung injury (18). Succinate and hypoxanthine levels may be even more sensitive biomarkers of post-shock metabolic derangement than lactate (67, 68). LC-MS lipidomic approaches and genotyping for apolipoprotein E ε 4 have shown decreased levels of several major phospholipid classes in TBI, PTSD, and TBI with PTSD patients compared to controls suggesting that phospholipid profiling may distinguish TBI and PTSD (69–71).
Several papers have described the potential impact of metabolic derangements on clinical outcomes. Medium-chain fatty acids (decanoic and octanoic acids) and sugar derivatives including 2,3-bisphosphoglyceric acid have been associated with the severity of TBI and predicted patient outcomes in some studies (72). Decreased levels of methionine and its metabolites in the plasma samples appear to correlate with severity in TBI patients (67). Yi and colleagues have identified a biomarker panel using GC-MS of nine serum metabolites (serine, pyroglutamic acid, phenylalanine, galactose, palmitic acid, arachidonic acid, linoleic acid, citric acid, and 2,3,4-trihydroxybutyrate) which might be useful to discriminate between TBI patients with and without cognitive impairment and healthy controls (73). Metabolically targeted hypothermia seems to result in differences in brain metabolites and reduction in mortality compared to body temperature targeted hypothermia treatment (74). Other studies have addressed therapies that target lipid peroxidation in TBI (75–77). Lusczek and colleagues using proton NMR have identified novel potential metabolomic biomarkers associated with mortality (i.e. succinate, malonate), injury severity (i.e. succinate, hypoxanthine) and the presence of trauma (i.e. hypoxanthine, 5-aminolevulinate) with performance comparable to lactate (78). Spinal cord injury and the resultant effects on metabolomics, lipidomics and targeted therapies have also been described by several publications (79–82).
Hemorrhagic shock and organ failure as a result of trauma can have substantial metabolomic underpinnings (18, 22, 83). This metabolic response has been demonstrated in several animal models (21, 22, 84–87). Profound metabolic changes have been shown in small but detailed analysis of critically injured adult trauma patients in the prehospital setting as well as in refractory shock. Byproducts of glycolysis, lipolysis and proteolysis were substantially elevated in both of these patient populations suggesting that resuscitation using these early metabolic signals as guidelines could be a valuable area of investigation (83). An evaluation of oxidation products and protein catabolites in trauma patients and healthy volunteers has demonstrated that amino acid and nucleotide metabolism were initially suppressed but gradually rose to catabolic range within 24 hours (88). Early changes in carbohydrate and amino acid levels correlate with systemic inflammatory response syndrome followed by later disturbances in fat metabolism that correlate with multi-organ dysfunction syndrome (18, 83, 89). Changes in tricarboxylic acid intermediates, glycolytic-gluconeogenic byproducts, purine-pyrimidine catabolism and fatty acid oxidation are hallmarks of hemorrhagic shock in a swine model (22). Furthermore, decreased lipid synthesis and urea cycle activity in the liver as well as increased hyperglycemia and lactic- and keto-acidosis have been shown to occur after injury (21, 90). These have been noted in fasting and fed states (24, 91, 92). Clinical care can both affect the metabolic response after trauma and be informed by changes in metabolic profiles. Several papers have studied resuscitation, coagulopathy and nutrition and their metabolomic impact in trauma patients. Hypothermic resuscitation can result in metabolic suppression in muscle (84). Some metabolic phenotypes may in fact predict sepsis risk in trauma patients (93). Fibrinolysis and associated proteins in trauma have also been evaluated with LC-MS(94). LC-MS has also been used to study the metabolomics of enteral and parenteral nutrition in critically ill, intensive care unit patients. Enteral nutrition increases amino acid and antioxidants production, upregulates the urea cycle and ribonucleic acid synthesis whereas parenteral nutrition only contributes to increases in amino acid production (95).
In burn injuries, elevations in cytokines, intracellular and secretory proteins, and fatty acid metabolites as measured through metabolomics and proteomics may have prognostic value (15, 96–99). NMR spectra has been used to identify significantly increased levels of 12 metabolites in patients with severe thermal injury. These metabolites, regulated by over a 100 enzymes, including α-ketoisovaleric acid, 3-methylhistidine, and β-hydroxybutyric acid, suggest that mitochondrial damage and carbohydrate, protein and fatty acid metabolism disturbances are occurring during the early stage of burn injury (100). Qi and colleagues showed that there is a significant increase in free fatty acids (FFAs) during the acute phase after a burn, and that burn severity and age correlated with an impaired response in unsaturated free fatty acids and pro-inflammatory cytokines. They found that elevated levels of saturated and mono-unsaturated FFA are correlated with increased mortality. In non-survivors, there were limited pro-inflammatory cytokines, despite high levels of saturated FFA, suggesting that these patients lacked the appropriate physiologic inflammatory response needed to react to the insult (101). Other markers of oxidative stress such as hypoxanthine, indoxyl sulfate, glucuronic acid, gluconic acid, proline, uracil, nitrotyrosine, uric acid, and trihydroxy cholanoic acid have been identified in rat models of burns suggesting potential for distinguishing between septic and non-septic burn patients (102). In addition, proteomics and metabolomics was used to profile the blister fluid from various severity burns in pediatric populations (103).
These studies scratch the surface of metabolomics as it applies to trauma research but uniformly suggest the utility of metabolomics for identifying specific biomarkers to predict clinical outcomes in injured patients and allow for earlier, and therefore potentially more effective, interventions. Other applications of metabolomics in injury care could involve determination of the presence of medications – both prescription and illicit substances, in the blood or urine (104). In an era where huge demographic shifts and medications such as novel anticoagulants are becoming a standard part of clinical care, it is becoming common to see injured elderly patients who have been on anticoagulation or numerous high-risk medications (105–107). Metabolomic changes, determined at the point-of-care, can rapidly identify critical medications and may prove to be a vital resource to the clinical team.
Limitations and Future Directions
The metabolome is constantly changing. To obtain the most accurate picture of a trauma patient’s metabolism, the acquisition of the samples has to be done as close as possible to the time of injury and processed immediately. However, several key limitations need to be solved before metabolomic analyses can be transferred from the bench to bedside to support clinical decisions in trauma care.
Historically, LC-MS instruments as well as standard workflows have been aimed at basic research and pharmaceutical applications and are extremely flexible and geared toward achieving the highest levels of sensitivity and accuracy possible. However, this does not allow for process efficiency, ease of use or instrument robustness, which limit applications in a high-volume clinical lab. Currently, there are no automated and integrated LC-MS workflows. Instrument components and testing workflows currently require continuous manual interventions which take time and risk human error. In a typical LC-MS workflow approach, a laborious manual centrifugation process, or a stand-alone, automated liquid handler, is needed to perform the sample extraction. The sample identity data must be manually written on the sample tubes or a worklist of barcode identification data must be transferred to the liquid handler. After extraction, the sample must be manually transferred to vials and the sample identity data again must be transferred. Once a sample has been analyzed by the mass spectrometer, the results must be paired with the sample identity data and all of the data must be uploaded manually into the patient medical record. Manufacturers of LC-MS instruments are starting to recognize the value of automating these workflows.
Batch-mode processing is another challenge. Currently, LC-MS processing occurs in batches of samples, which while acceptable for research and pharmaceutical applications, is not acceptable for clinical care where the results are needed as quickly as possible after each sample is collected from an individual patient. Other limitations for clinical applicability are large footprint and extensive operator training requirements. Staff performing the testing must have in-depth knowledge of instrumentation and analytical chemistry techniques, and must be proficient in the use of statistical data processing programs and to troubleshoot clogs, leaks, improperly set fittings, pump failures and column performance degradation etc.
These limitations need to be addressed before the technology can be successful implemented within a busy hospital where timely results are crucial for patient care. However, they are likely to be transient challenges as rapid technological advances are taking place that will improve system reliability, ease of use and throughput within the next few years. Manufacturers of automated liquid handlers such as Hamilton are already producing systems that process samples one at a time and directly inject them into the LC-MS systems.
Conclusion
Trauma initiates a series of events that pushes patients down a clinical trajectory that can end in death or permanent disability. This begins at the time of the event but is currently not recognized until the patient begins to have physiologic signs and symptoms from injury. Clinical criteria are downstream events and interventions based on such downstream triggers may not be early enough to be successful. Markers that plot the patient’s clinical trajectory as early and with as few time points as possible can have great potential for expediting interventions and saving lives. Metabolomics has an enormous role to play in this area. The technology necessary to use metabolomics to personalize care of trauma patients is at an exciting stage and available for such innovative uses. Since trauma continues to be a major cause of death in the US, such cutting-edge technology may be useful to solve the associated clinical problems for decades to come.
Acknowledgments
Research reported in this publication was supported by research grants from National Institutes of Health under grant numbers HD087198 (to DSW), faculty development funds from 2P60MD002256-10 (to SJ) and also received support via a Young Investigator Award from SCIEX for clinical lipidomic research (DSW). Additional funds were provided by the Clinical Center for Translational Research from VCU. Funds were also provided by CTSA UL1TR000058 from the National center for advancing translational sciences and the CCTR endowment fund of VCU (to SJ and DSW).
Footnotes
Conflict of Interests: none
References
- 1.Nathens AB, Jurkovich GJ, Rivara FP, Maier RV. Effectiveness of state trauma systems in reducing injury-related mortality: a national evaluation. Journal of Trauma. 2000;48(1):25–30. doi: 10.1097/00005373-200001000-00005. discussion -1. [DOI] [PubMed] [Google Scholar]
- 2.Mann NC, Mullins RJ, MacKenzie EJ, Jurkovich GJ, Mock CN. Systematic review of published evidence regarding trauma system effectiveness. The Journal of trauma. 1999;47(3 Suppl):S25–33. doi: 10.1097/00005373-199909001-00007. [DOI] [PubMed] [Google Scholar]
- 3.MacKenzie EJ, Rivara FP, Jurkovich GJ, Nathens AB, Frey KP, Egleston BL, Salkever DS, Scharfstein DO. A national evaluation of the effect of trauma-center care on mortality. The New England journal of medicine. 2006;354(4):366–78. doi: 10.1056/NEJMsa052049. [DOI] [PubMed] [Google Scholar]
- 4.Papa L, Langland-Orban B, Kallenborn C, Tepas JJ, 3rd, Lottenberg L, Celso B, Durham R, Flint L. Assessing effectiveness of a mature trauma system: Association of trauma center presence with lower injury mortality rate. The Journal of trauma. 2006;61(2):261–6. doi: 10.1097/01.ta.0000221789.53864.ba. discussion 6–7. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention National Center for Injury Prevention and Control. Web-based Injury Statistics Query and Reporting System (WISQARS) Centers for Disease Control and Prevention, National Center for Injury Prevention and Control. Web-based Injury Statistics Query and Reporting System (WISQARS); [Available from: http://www.cdc.gov/injury/wisqars/overview/key_data.html. [Google Scholar]
- 6.Finkelstein EACP, Miller TR, et al. Incidence and Economic Burden of Injuries in the United States. New York, NY: Oxford University Press; 2006. [Google Scholar]
- 7.Centers for Disease Control and Prevention’s National Center for Injury Prevention and Control. Web-based Injury Statistics Query and Reporting System (WISQARS): Centers for Disease Control and Prevention, National Center for Injury Prevention and Control. Web-based Injury Statistics Query and Reporting System (WISQARS) [cited 2017 1/24/17]. Available from: https://www.cdc.gov/injury/wisqars/overview/cost_of_injury.html.
- 8.Vogel JA, Liao MM, Hopkins E, Seleno N, Byyny RL, Moore EE, Gravitz C, Haukoos JS. Prediction of postinjury multiple-organ failure in the emergency department: development of the Denver Emergency Department Trauma Organ Failure score. The journal of trauma and acute care surgery. 2014;76(1):140–5. doi: 10.1097/TA.0b013e3182a99da4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang SY, Lee JH, Lee YH, Hong CK, Sung AJ, Choi YC. Comparison of the Sequential Organ Failure Assessment, Acute Physiology and Chronic Health Evaluation II scoring system, and Trauma and Injury Severity Score method for predicting the outcomes of intensive care unit trauma patients. The American journal of emergency medicine. 2012;30(5):749–53. doi: 10.1016/j.ajem.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 10.Hensler T, Sauerland S, Lefering R, Nagelschmidt M, Bouillon B, Andermahr J, Neugebauer EA. The clinical value of procalcitonin and neopterin in predicting sepsis and organ failure after major trauma. Shock. 2003;20(5):420–6. doi: 10.1097/01.shk.0000093541.78705.38. [DOI] [PubMed] [Google Scholar]
- 11.Lausevic Z, Lausevic M, Trbojevic-Stankovic J, Krstic S, Stojimirovic B. Predicting multiple organ failure in patients with severe trauma. Canadian journal of surgery Journal canadien de chirurgie. 2008;51(2):97–102. [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver SG, Winson MK, Kell DB, Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16(9):373–8. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 13.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17(7):451–9. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–9. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finnerty CC, Jeschke MG, Qian WJ, Kaushal A, Xiao W, Liu T, Gritsenko MA, Moore RJ, Camp DG, 2nd, Moldawer LL, et al. Determination of burn patient outcome by large-scale quantitative discovery proteomics. Critical care medicine. 2013;41(6):1421–34. doi: 10.1097/CCM.0b013e31827c072e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmelzer K, Fahy E, Subramaniam S, Dennis EA. The lipid maps initiative in lipidomics. Methods Enzymol. 2007;432:171–83. doi: 10.1016/S0076-6879(07)32007-7. [DOI] [PubMed] [Google Scholar]
- 17.Ramadan Z, Jacobs D, Grigorov M, Kochhar S. Metabolic profiling using principal component analysis, discriminant partial least squares, and genetic algorithms. Talanta. 2006;68(5):1683–91. doi: 10.1016/j.talanta.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 18.D’Alessandro A, Nemkov T, Moore HB, Moore EE, Wither M, Nydam T, Slaughter A, Silliman CC, Banerjee A, Hansen KC. Metabolomics of trauma-associated death: shared and fluid-specific features of human plasma vs lymph. Blood Transfusion. 2016;14(2):185–94. doi: 10.2450/2016.0208-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–83. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 20.Gowda GA, Zhang S, Gu H, Asiago V, Shanaiah N, Raftery D. Metabolomics-based methods for early disease diagnostics. Expert Rev Mol Diagn. 2008;8(5):617–33. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen MJ, Serkova NJ, Wiener-Kronish J, Pittet J-F, Niemann CU. 1H-NMR-Based Metabolic Signatures of Clinical Outcomes in Trauma Patients—Beyond Lactate and Base Deficit. The Journal of Trauma: Injury, Infection, and Critical Care. 2010;69(1):31–40. doi: 10.1097/TA.0b013e3181e043fe. [DOI] [PubMed] [Google Scholar]
- 22.Lexcen DR, Lusczek ER, Witowski NE, Mulier KE, Beilman GJ. Metabolomics classifies phase of care and identifies risk for mortality in a porcine model of multiple injuries and hemorrhagic shock. The journal of trauma and acute care surgery. 2012;73(2 Suppl 1):S147–55. doi: 10.1097/TA.0b013e3182609821. [DOI] [PubMed] [Google Scholar]
- 23.Lusczek ER, Paulo JA, Saltzman JR, Kadiyala V, Banks PA, Beilman G, Conwell DL. Urinary 1H-NMR metabolomics can distinguish pancreatitis patients from healthy controls. JOP. 2013;14(2):161–70. doi: 10.6092/1590-8577/1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witowski NE, Lusczek ER, Determan CE, Lexcen DR, Mulier KE, Wolf A, Ostrowski BG, Beilman GJ. Metabolomic analysis of survival in carbohydrate pre-fed pigs subjected to shock and polytrauma. Mol Biosyst. 2016;12(5):1638–52. doi: 10.1039/c5mb00637f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu X, Zhao X, Bai C, Zhao C, Lu G, Xu G. LC–MS-based metabonomics analysis. Journal of Chromatography B. 2008;866(1–2):64–76. doi: 10.1016/j.jchromb.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 26.Katajamaa M, Orešič M. Data processing for mass spectrometry-based metabolomics. Journal of Chromatography A. 2007;1158(1–2):318–28. doi: 10.1016/j.chroma.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 27.Spagou K, Tsoukali H, Raikos N, Gika H, Wilson ID, Theodoridis G. Hydrophilic interaction chromatography coupled to MS for metabonomic/metabolomic studies. J Sep Sci. 2010;33(6–7):716–27. doi: 10.1002/jssc.200900803. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Sun H, Zhang A, Wang P, Han Y. Ultra-performance liquid chromatography coupled to mass spectrometry as a sensitive and powerful technology for metabolomic studies. J Sep Science. 2011;34(24):3451–9. doi: 10.1002/jssc.201100333. [DOI] [PubMed] [Google Scholar]
- 29.Hendriks MMWB, Van Eeuwijk FA, Jellema RH, Westerhuis JA, Reijmers TH, Hoefsloot HCJ, Smilde AK. Data-processing strategies for metabolomics studies. TrAC Trends in Analytical Chemistry. 2011;30(10):1685–98. [Google Scholar]
- 30.Worley B, Powers R. Multivariate Analysis in Metabolomics. Curr Metabolomics. 2013;1(1):92–107. doi: 10.2174/2213235X11301010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jewison T, Su Y, Disfany FM, Liang Y, Knox C, Maciejewski A, Poelzer J, Huynh J, Zhou Y, Arndt D, et al. SMPDB 2.0: big improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 2014;42(Database issue):D478–84. doi: 10.1093/nar/gkt1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frolkis A, Knox C, Lim E, Jewison T, Law V, Hau DD, Liu P, Gautam B, Ly S, Guo AC, et al. SMPDB: The Small Molecule Pathway Database. Nucleic Acids Res. 2010;38(Database issue):D480–7. doi: 10.1093/nar/gkp1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caspi R, Karp PD. Using the MetaCyc pathway database and the BioCyc database collection. Curr Protoc Bioinformatics. 2007;Chapter 1(Unit1):17. doi: 10.1002/0471250953.bi0117s20. [DOI] [PubMed] [Google Scholar]
- 35.Karp PD, Billington R, Holland TA, Kothari A, Krummenacker M, Weaver D, Latendresse M, Paley S. Computational Metabolomics Operations at BioCyc.org. Metabolites. 2015;5(2):291–310. doi: 10.3390/metabo5020291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karnovsky A, Weymouth T, Hull T, Tarcea VG, Scardoni G, Laudanna C, Sartor MA, Stringer KA, Jagadish HV, Burant C, et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28(3):373–80. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao J, Tarcea VG, Karnovsky A, Mirel BR, Weymouth TE, Beecher CW, Cavalcoli JD, Athey BD, Omenn GS, Burant CF, et al. Metscape: a Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. Bioinformatics. 2010;26(7):971–3. doi: 10.1093/bioinformatics/btq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Junker BH, Klukas C, Schreiber F. VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics. 2006;7:109. doi: 10.1186/1471-2105-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohn H, Junker A, Hartmann A, Grafahrend-Belau E, Treutler H, Klapperstuck M, Czauderna T, Klukas C, Schreiber F. VANTED v2: a framework for systems biology applications. BMC Syst Biol. 2012;6:139. doi: 10.1186/1752-0509-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuenca AG, Maier RV, Cuschieri J, Moore EE, Moldawer LL, Tompkins RG. Inflammation Host Response to Injury LSCRP. The Glue Grant experience: characterizing the post injury genomic response. Eur J Trauma Emerg Surg. 2011;37(6):549–58. doi: 10.1007/s00068-011-0148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao W, Mindrinos MN, Seok J, Cuschieri J, Cuenca AG, Gao H, Hayden DL, Hennessy L, Moore EE, Minei JP, et al. A genomic storm in critically injured humans. J Exp Med. 2011;208(13):2581–90. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tompkins RG. Genomics of injury: The Glue Grant experience. The journal of trauma and acute care surgery. 2015;78(4):671–86. doi: 10.1097/TA.0000000000000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finnerty CC, Jeschke MG, Qian WJ, Kaushal A, Xiao W, Liu T, Gritsenko MA, Moore RJ, Camp DG, 2nd, Moldawer LL, et al. Determination of burn patient outcome by large-scale quantitative discovery proteomics. Critical care medicine. 2013;41(6):1421–34. doi: 10.1097/CCM.0b013e31827c072e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cuenca AG, Gentile LF, Lopez MC, Ungaro R, Liu H, Xiao W, Seok J, Mindrinos MN, Ang D, Baslanti TO, et al. Development of a genomic metric that can be rapidly used to predict clinical outcome in severely injured trauma patients. Critical care medicine. 2013;41(5):1175–85. doi: 10.1097/CCM.0b013e318277131c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warren HS, Elson CM, Hayden DL, Schoenfeld DA, Cobb JP, Maier RV, Moldawer LL, Moore EE, Harbrecht BG, Pelak K, et al. A genomic score prognostic of outcome in trauma patients. Mol Med. 2009;15(7–8):220–7. doi: 10.2119/molmed.2009.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orr SK, Butler KL, Hayden D, Tompkins RG, Serhan CN, Irimia D. Gene Expression of Proresolving Lipid Mediator Pathways Is Associated With Clinical Outcomes in Trauma Patients. Critical care medicine. 2015;43(12):2642–50. doi: 10.1097/CCM.0000000000001312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kotz KT, Xiao W, Miller-Graziano C, Qian WJ, Russom A, Warner EA, Moldawer LL, De A, Bankey PE, Petritis BO, et al. Clinical microfluidics for neutrophil genomics and proteomics. Nat Med. 2010;16(9):1042–7. doi: 10.1038/nm.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qian WJ, Petritis BO, Kaushal A, Finnerty CC, Jeschke MG, Monroe ME, Moore RJ, Schepmoes AA, Xiao W, Moldawer LL, et al. Plasma proteome response to severe burn injury revealed by 18O-labeled “universal” reference-based quantitative proteomics. J Proteome Res. 2010;9(9):4779–89. doi: 10.1021/pr1005026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porter C, Tompkins RG, Finnerty CC, Sidossis LS, Suman OE, Herndon DN. The metabolic stress response to burn trauma: current understanding and therapies. Lancet. 2016;388(10052):1417–26. doi: 10.1016/S0140-6736(16)31469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolf SE, Tompkins RG, Herndon DN. On the horizon: research priorities in burns for the next decade. The Surgical clinics of North America. 2014;94(4):917–30. doi: 10.1016/j.suc.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 52.Wolahan SM, Hirt D, Braas D, Glenn TC. Role of Metabolomics in Traumatic Brain Injury Research. Neurosurg Clin N Am. 2016;27(4):465–72. doi: 10.1016/j.nec.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stein NR, McArthur DL, Etchepare M, Vespa PM. Early cerebral metabolic crisis after TBI influences outcome despite adequate hemodynamic resuscitation. Neurocrit Care. 2012;17(1):49–57. doi: 10.1007/s12028-012-9708-y. [DOI] [PubMed] [Google Scholar]
- 54.Viant MR, Lyeth BG, Miller MG, Berman RF. An NMR metabolomic investigation of early metabolic disturbances following traumatic brain injury in a mammalian model. NMR Biomed. 2005;18(8):507–16. doi: 10.1002/nbm.980. [DOI] [PubMed] [Google Scholar]
- 55.Adibhatla RM, Hatcher JF. Role of Lipids in Brain Injury and Diseases. Future Lipidol. 2007;2(4):403–22. doi: 10.2217/17460875.2.4.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kochanek PM, Berger RP, Bayir H, Wagner AK, Jenkins LW, Clark RS. Biomarkers of primary and evolving damage in traumatic and ischemic brain injury: diagnosis, prognosis, probing mechanisms, and therapeutic decision making. Current opinion in critical care. 2008;14(2):135–41. doi: 10.1097/MCC.0b013e3282f57564. [DOI] [PubMed] [Google Scholar]
- 57.Manley GT, Diaz-Arrastia R, Brophy M, Engel D, Goodman C, Gwinn K, Veenstra TD, Ling G, Ottens AK, Tortella F, et al. Common data elements for traumatic brain injury: recommendations from the biospecimens and biomarkers working group. Archives of physical medicine and rehabilitation. 2010;91(11):1667–72. doi: 10.1016/j.apmr.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 58.Pitkanen A, Lukasiuk K. Molecular biomarkers of epileptogenesis. Biomark Med. 2011;5(5):629–33. doi: 10.2217/bmm.11.67. [DOI] [PubMed] [Google Scholar]
- 59.Sparvero LJ, Amoscato AA, Kochanek PM, Pitt BR, Kagan VE, Bayir H. Mass-spectrometry based oxidative lipidomics and lipid imaging: applications in traumatic brain injury. J Neurochem. 2010;115(6):1322–36. doi: 10.1111/j.1471-4159.2010.07055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ottens AK, Stafflinger JE, Griffin HE, Kunz RD, Cifu DX, Niemeier JP. Post-acute brain injury urinary signature: a new resource for molecular diagnostics. Journal of neurotrauma. 2014;31(8):782–8. doi: 10.1089/neu.2013.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolahan SM, Hirt D, Glenn TC. Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Frontiers in Neuroengineering. Boca Raton (FL): 2015. [PubMed] [Google Scholar]
- 62.Yang R, Fredman G, Krishnamoorthy S, Agrawal N, Irimia D, Piomelli D, Serhan CN. Decoding functional metabolomics with docosahexaenoyl ethanolamide (DHEA) identifies novel bioactive signals. The Journal of biological chemistry. 2011;286(36):31532–41. doi: 10.1074/jbc.M111.237990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sheth SA, Iavarone AT, Liebeskind DS, Won SJ, Swanson RA. Targeted Lipid Profiling Discovers Plasma Biomarkers of Acute Brain Injury. PloS one. 2015;10(6):e0129735. doi: 10.1371/journal.pone.0129735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glenn TC, Hirt D, Mendez G, McArthur DL, Sturtevant R, Wolahan S, Fazlollahi F, Ordon M, Bilgin-Freiert A, Ellingson B, et al. Metabolomic analysis of cerebral spinal fluid from patients with severe brain injury. Acta Neurochir Suppl. 2013;118:115–9. doi: 10.1007/978-3-7091-1434-6_20. [DOI] [PubMed] [Google Scholar]
- 65.Abou-El-Hassan H, Sukhon F, Assaf EJ, Bahmad H, Abou-Abbass H, Jourdi H, Kobeissy FH. Degradomics in Neurotrauma: Profiling Traumatic Brain Injury. Methods Mol Biol. 2017;1598:65–99. doi: 10.1007/978-1-4939-6952-4_4. [DOI] [PubMed] [Google Scholar]
- 66.Tzekov R, Dawson C, Orlando M, Mouzon B, Reed J, Evans J, Crynen G, Mullan M, Crawford F. Sub-Chronic Neuropathological and Biochemical Changes in Mouse Visual System after Repetitive Mild Traumatic Brain Injury. PloS one. 2016;11(4):e0153608. doi: 10.1371/journal.pone.0153608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dash PK, Hergenroeder GW, Jeter CB, Choi HA, Kobori N, Moore AN. Traumatic Brain Injury Alters Methionine Metabolism: Implications for Pathophysiology. Front Syst Neurosci. 2016;10:36. doi: 10.3389/fnsys.2016.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.D’Alessandro A, Moore HB, Moore EE, Reisz JA, Wither MJ, Ghasabyan A, Chandler J, Silliman CC, Hansen KC, Banerjee A. Plasma succinate is a predictor of mortality in critically injured patients. The journal of trauma and acute care surgery. 2017 doi: 10.1097/TA.0000000000001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emmerich T, Abdullah L, Crynen G, Dretsch M, Evans J, Ait-Ghezala G, Reed J, Montague H, Chaytow H, Mathura V, et al. Plasma Lipidomic Profiling in a Military Population of Mild Traumatic Brain Injury and Post-Traumatic Stress Disorder with Apolipoprotein E varepsilon4-Dependent Effect. Journal of neurotrauma. 2016;33(14):1331–48. doi: 10.1089/neu.2015.4061. [DOI] [PubMed] [Google Scholar]
- 70.Abdullah L, Evans JE, Ferguson S, Mouzon B, Montague H, Reed J, Crynen G, Emmerich T, Crocker M, Pelot R, et al. Lipidomic analyses identify injury-specific phospholipid changes 3 mo after traumatic brain injury. FASEB J. 2014;28(12):5311–21. doi: 10.1096/fj.14-258228. [DOI] [PubMed] [Google Scholar]
- 71.Emmerich T, Abdullah L, Ojo J, Mouzon B, Nguyen T, Laco GS, Crynen G, Evans JE, Reed J, Mullan M, et al. Mild TBI Results in a Long-Term Decrease in Circulating Phospholipids in a Mouse Model of Injury. Neuromolecular Med. 2017;19(1):122–35. doi: 10.1007/s12017-016-8436-4. [DOI] [PubMed] [Google Scholar]
- 72.Oresic M, Posti JP, Kamstrup-Nielsen MH, Takala RS, Lingsma HF, Mattila I, Jantti S, Katila AJ, Carpenter KL, Ala-Seppala H, et al. Human Serum Metabolites Associate With Severity and Patient Outcomes in Traumatic Brain Injury. EBioMedicine. 2016;12:118–26. doi: 10.1016/j.ebiom.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yi L, Shi S, Wang Y, Huang W, Xia ZA, Xing Z, Peng W, Wang Z. Serum Metabolic Profiling Reveals Altered Metabolic Pathways in Patients with Post-traumatic Cognitive Impairments. Sci Rep. 2016;6:21320. doi: 10.1038/srep21320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng JZ, Wang WY, Zeng J, Zhou ZY, Peng J, Yang H, Deng PC, Li SJ, Lu CD, Jiang H. Optimization of brain metabolism using metabolic-targeted therapeutic hypothermia can reduce mortality from traumatic brain injury. The journal of trauma and acute care surgery. 2017;83(2):296–304. doi: 10.1097/TA.0000000000001522. [DOI] [PubMed] [Google Scholar]
- 75.Anthonymuthu TS, Kenny EM, Bayir H. Therapies targeting lipid peroxidation in traumatic brain injury. Brain Res. 2016;1640(Pt A):57–76. doi: 10.1016/j.brainres.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bayir H, Tyurin VA, Tyurina YY, Viner R, Ritov V, Amoscato AA, Zhao Q, Zhang XJ, Janesko-Feldman KL, Alexander H, et al. Selective early cardiolipin peroxidation after traumatic brain injury: an oxidative lipidomics analysis. Ann Neurol. 2007;62(2):154–69. doi: 10.1002/ana.21168. [DOI] [PubMed] [Google Scholar]
- 77.Ji J, Kline AE, Amoscato A, Samhan-Arias AK, Sparvero LJ, Tyurin VA, Tyurina YY, Fink B, Manole MD, Puccio AM, et al. Lipidomics identifies cardiolipin oxidation as a mitochondrial target for redox therapy of brain injury. Nat Neurosci. 2012;15(10):1407–13. doi: 10.1038/nn.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lusczek ER, Muratore SL, Dubick MA, Beilman GJ. Assessment of key plasma metabolites in combat casualties. The journal of trauma and acute care surgery. 2017;82(2):309–16. doi: 10.1097/TA.0000000000001277. [DOI] [PubMed] [Google Scholar]
- 79.Dulin JN, Karoly ED, Wang Y, Strobel HW, Grill RJ. Licofelone modulates neuroinflammation and attenuates mechanical hypersensitivity in the chronic phase of spinal cord injury. J Neurosci. 2013;33(2):652–64. doi: 10.1523/JNEUROSCI.6128-11.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sauerbeck AD, Laws JL, Bandaru VV, Popovich PG, Haughey NJ, McTigue DM. Spinal cord injury causes chronic liver pathology in rats. Journal of neurotrauma. 2015;32(3):159–69. doi: 10.1089/neu.2014.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fujieda Y, Ueno S, Ogino R, Kuroda M, Jonsson TJ, Guo L, Bamba T, Fukusaki E. Metabolite profiles correlate closely with neurobehavioral function in experimental spinal cord injury in rats. PloS one. 2012;7(8):e43152. doi: 10.1371/journal.pone.0043152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Figueroa JD, Cordero K, Llan MS, De Leon M. Dietary omega-3 polyunsaturated fatty acids improve the neurolipidome and restore the DHA status while promoting functional recovery after experimental spinal cord injury. Journal of neurotrauma. 2013;30(10):853–68. doi: 10.1089/neu.2012.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peltz ED, D’Alessandro A, Moore EE, Chin T, Silliman CC, Sauaia A, Hansen KC, Banerjee A. Pathologic metabolism: an exploratory study of the plasma metabolome of critical injury. The journal of trauma and acute care surgery. 2015;78(4):742–51. doi: 10.1097/TA.0000000000000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lusczek ER, Lexcen DR, Witowski NE, Determan C, Jr, Mulier KE, Beilman G. Prolonged induced hypothermia in hemorrhagic shock is associated with decreased muscle metabolism: a nuclear magnetic resonance-based metabolomics study. Shock. 2014;41(1):79–84. doi: 10.1097/SHK.0000000000000061. [DOI] [PubMed] [Google Scholar]
- 85.D’Alessandro A, Moore HB, Moore EE, Wither M, Nemkov T, Gonzalez E, Slaughter A, Fragoso M, Hansen KC, Silliman CC, et al. Early hemorrhage triggers metabolic responses that build up during prolonged shock. Am J Physiol Regul Integr Comp Physiol. 2015;308(12):R1034–44. doi: 10.1152/ajpregu.00030.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.D’Alessandro A, Moore HB, Moore EE, Wither MJ, Nemkov T, Morton AP, Gonzalez E, Chapman MP, Fragoso M, Slaughter A, et al. Plasma First Resuscitation Reduces Lactate Acidosis, Enhances Redox Homeostasis, Amino Acid and Purine Catabolism in a Rat Model of Profound Hemorrhagic Shock. Shock. 2016;46(2):173–82. doi: 10.1097/SHK.0000000000000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morishita K, Aiboshi J, Kobayashi T, Mikami S, Yokoyama Y, Ogawa K, Yokota H, Otomo Y. Lipidomics analysis of mesenteric lymph after trauma and hemorrhagic shock. The journal of trauma and acute care surgery. 2012;72(6):1541–7. doi: 10.1097/TA.0b013e318256df15. [DOI] [PubMed] [Google Scholar]
- 88.Parent BA, Seaton M, Sood RF, Gu H, Djukovic D, Raftery D, O’Keefe GE. Use of Metabolomics to Trend Recovery and Therapy After Injury in Critically Ill Trauma Patients. JAMA Surg. 2016;151(7):e160853. doi: 10.1001/jamasurg.2016.0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mao H, Wang H, Wang B, Liu X, Gao H, Xu M, Zhao H, Deng X, Lin D. Systemic Metabolic Changes of Traumatic Critically Ill Patients Revealed by an NMR-Based Metabonomic Approach. J Proteome Res. 2009;8(12):5423–30. doi: 10.1021/pr900576y. [DOI] [PubMed] [Google Scholar]
- 90.D’Alessandro A, Slaughter AL, Peltz ED, Moore EE, Silliman CC, Wither M, Nemkov T, Bacon AW, Fragoso M, Banerjee A, et al. Trauma/hemorrhagic shock instigates aberrant metabolic flux through glycolytic pathways, as revealed by preliminary (13)C-glucose labeling metabolomics. J Transl Med. 2015;13:253. doi: 10.1186/s12967-015-0612-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Witowski N, Lusczek E, Determan C, Jr, Lexcen D, Mulier K, Ostrowski B, Beilman G. A four-compartment metabolomics analysis of the liver, muscle, serum, and urine response to polytrauma with hemorrhagic shock following carbohydrate prefeed. PloS one. 2015;10(4):e0124467. doi: 10.1371/journal.pone.0124467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Determan C, Jr, Anderson R, Becker A, Witowski N, Lusczek E, Mulier K, Beilman GJ. Fed state prior to hemorrhagic shock and polytrauma in a porcine model results in altered liver transcriptomic response. PloS one. 2014;9(6):e100088. doi: 10.1371/journal.pone.0100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blaise BJ, Gouel-Cheron A, Floccard B, Monneret G, Allaouchiche B. Metabolic phenotyping of traumatized patients reveals a susceptibility to sepsis. Anal Chem. 2013;85(22):10850–5. doi: 10.1021/ac402235q. [DOI] [PubMed] [Google Scholar]
- 94.Moore HB, Moore EE, Gonzalez E, Hansen KC, Dzieciatkowska M, Chapman MP, Sauaia A, West B, Banerjee A, Silliman CC. Hemolysis exacerbates hyperfibrinolysis, whereas platelolysis shuts down fibrinolysis: evolving concepts of the spectrum of fibrinolysis in response to severe injury. Shock. 2015;43(1):39–46. doi: 10.1097/SHK.0000000000000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Parent BA, Seaton M, Djukovic D, Gu H, Wheelock B, Navarro SL, Raftery D, O’Keefe GE. Parenteral and enteral nutrition in surgical critical care: Plasma metabolomics demonstrates divergent effects on nitrogen, fatty-acid, ribonucleotide, and oxidative metabolism. The journal of trauma and acute care surgery. 2017;82(4):704–13. doi: 10.1097/TA.0000000000001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hazeldine J, Hampson P, Lord JM. The diagnostic and prognostic value of systems biology research in major traumatic and thermal injury: a review. Burns Trauma. 2016;4:33. doi: 10.1186/s41038-016-0059-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Davis CS, Janus SE, Mosier MJ, Carter SR, Gibbs JT, Ramirez L, Gamelli RL, Kovacs EJ. Inhalation injury severity and systemic immune perturbations in burned adults. Annals of surgery. 2013;257(6):1137–46. doi: 10.1097/SLA.0b013e318275f424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Finnerty CC, Ju H, Spratt H, Victor S, Jeschke MG, Hegde S, Bhavnani SK, Luxon BA, Brasier AR, Herndon DN. Proteomics improves the prediction of burns mortality: results from regression spline modeling. Clin Transl Sci. 2012;5(3):243–9. doi: 10.1111/j.1752-8062.2012.00412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shelhamer MC, Rowan MP, Cancio LC, Aden JK, Rhie RY, Merrill GA, Wolf SE, Renz EM, Chung KK. Elevations in inflammatory cytokines are associated with poor outcomes in mechanically ventilated burn patients. The journal of trauma and acute care surgery. 2015;79(3):431–6. doi: 10.1097/TA.0000000000000786. [DOI] [PubMed] [Google Scholar]
- 100.Zhang Y, Cai B, Jiang H, Yan H, Yang H, Peng J, Wang W, Ma S, Wu X, Peng X. Use of 1H-nuclear magnetic resonance to screen a set of biomarkers for monitoring metabolic disturbances in severe burn patients. Critical care. 2014;18(4):R159. doi: 10.1186/cc13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qi P, Abdullahi A, Stanojcic M, Patsouris D, Jeschke MG. Lipidomic analysis enables prediction of clinical outcomes in burn patients. Sci Rep. 2016;6:38707. doi: 10.1038/srep38707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu XR, Zheng XF, Ji SZ, Lv YH, Zheng DY, Xia ZF, Zhang WD. Metabolomic analysis of thermally injured and/or septic rats. Burns: journal of the International Society for Burn Injuries. 2010;36(7):992–8. doi: 10.1016/j.burns.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 103.Zang T, Broszczak DA, Broadbent JA, Cuttle L, Lu H, Parker TJ. The biochemistry of blister fluid from pediatric burn injuries: proteomics and metabolomics aspects. Expert Rev Proteomics. 2016;13(1):35–53. doi: 10.1586/14789450.2016.1122528. [DOI] [PubMed] [Google Scholar]
- 104.Kreshak AA, Wardi G, Tomaszewski CA. The accuracy of emergency department medication history as determined by mass spectrometry analysis of urine: a pilot study. J Emerg Med. 2015;48(3):382–6. doi: 10.1016/j.jemermed.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 105.Pinho-Gomes AC, Hague A, Ghosh J. Management of novel oral anticoagulants in emergency and trauma surgery. The surgeon: journal of the Royal Colleges of Surgeons of Edinburgh and Ireland. 2016;14(4):234–9. doi: 10.1016/j.surge.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 106.Bauersachs RM. Managing venous thromboembolism with novel oral anticoagulants in the elderly and other high-risk patient groups. Eur J Intern Med. 2014;25(7):600–6. doi: 10.1016/j.ejim.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 107.Liu X, Baumgarten M, Smith G, Gambert S, Gottlieb S, Rattinger G, Albrecht J, Langenberg P, Zuckerman I. Warfarin usage among elderly atrial fibrillation patients with traumatic injury, an analysis of United States Medicare fee-for-service enrollees. J Clin Pharmacol. 2015;55(1):25–32. doi: 10.1002/jcph.375. [DOI] [PMC free article] [PubMed] [Google Scholar]