Abstract
The highly multiplexed polymerase chain reaction (PCR) assays used for forensic human identification perform best when used with an accurately determined quantity of input DNA. To help ensure the reliable performance of these assays, we are developing a certified reference material (CRM) for calibrating human genomic DNA working standards. To enable sharing information over time and place, CRMs must provide accurate and stable values that are metrologically traceable to a common reference. We have shown that droplet digital PCR (ddPCR) limiting dilution end-point measurements of the concentration of DNA copies per volume of sample can be traceably linked to the International System of Units (SI). Unlike values assigned using conventional relationships between ultraviolet absorbance and DNA mass concentration, entity-based ddPCR measurements are expected to be stable over time. However, the forensic community expects DNA quantity to be stated in terms of mass concentration rather than entity concentration. The transformation can be accomplished given SI-traceable values and uncertainties for the number of nucleotide bases per human haploid genome equivalent (HHGE) and the average molar mass of a nucleotide monomer in the DNA polymer. This report presents the considerations required to establish the metrological traceability of ddPCR-based mass concentration estimates of human nuclear DNA.
Keywords: Certified Reference Material (CRM), Droplet digital polymerase chain reaction (ddPCR), Human nuclear DNA, Metrological traceability
Introduction
Polymerase chain reaction (PCR) assays perform best when used with an accurately determined quantity of input DNA. This is particularly true for highly multiplexed assays, such as those used for forensic human identification [1].
We have shown that droplet digital PCR (ddPCR) limiting dilution end-point [2] measurements of the concentration of DNA copies per volume sample can be traceably linked to the International System of Units (SI) when several basic assumptions are shown to be justified [3]. Unlike values assigned using a conventional relationship between ultraviolet absorbance at 260 nm and DNA mass concentration [4], entity-based ddPCR values can be expected to be stable over time and provide results that are traceable [5] to the International System of Units (SI).
The linkage between ddPCR measurements and the SI is through the calibrated measurements of mean droplet volume (V), volume fraction of sample in the reaction mixture (F), and counts of the number of total and negative droplets (Ntot and Nneg) where negative droplets are defined as those that do not provide an above-threshold fluorescence signal at the endpoint. Poisson transformation of the ratio of negative to total droplets, λ = –ln(Nneg/Ntot), yields the mean number of copies of the PCR target in the ddPCR reaction mixture per droplet. Dividing by the total volumetric dilution factor yields the mean number of copies in the sample per droplet. The concentration of copies in the sample is then λ/(FV). The sources of bias and imprecision that contribute to the uncertainty in λ and F, u(λ) and u(F), have been discussed by others [6–9], as have the those associated with measurement of V, u(V) [9–13].
We are using ddPCR measurements to certify the concentrations of human nuclear DNA (nDNA) in a certified reference material (CRM) designed primarily for use by the forensic community. However, this community expects DNA quantity to be stated in terms of mass concentration rather than entity concentration. The entity concentration, λ/(FV) copies per nanoliter, can be transformed to mass concentration, [nDNA] nanogram per microliter, via the relationship:
| (1) |
where r is the number of assay targets per human haploid genome equivalents (HHGE), n is the number of nucleotide base pairs (bp) per double-stranded HHGE, and w̄ is the average molar mass of a bp in the DNA polymer.
For independent multiplicative factors such as these, the combined relative uncertainty of their product can be estimated from the square root of the sum-of-squares of the individual relative uncertainties [14, Section 5.1.6]:
| (2) |
The essential elements of these relationships are provided in Griffiths et al. [6, Table 1], but without addressing all assumptions, values, and uncertainties. This report presents the considerations required to establish the metrological traceability of ddPCR-based mass concentration estimates of nDNA.
Table 1.
NIST-Developed human genomic assays
| Assay Target | Chromosome Band Accession # | Primers and Probe a | Amplicon Length, bp |
|---|---|---|---|
| NEIF | Chr 2 | Fgccaaacttcagccttctcttc | 67 |
| Gene EIF5B | p11.1-q11.1 | Rctctggcaacatttcacactaca | |
| NC_000002.12 | PB+tcatgcagttgtcagaagctg | ||
| 2PR4 | Chr 2 | Fcgggtttgggttcaggtctt | 97 |
| Gene RPS27A | p16 | Rtgctacaatgaaaacattcagaagtct | |
| NC_000002.12 | PBtttgtctaccacttgcaaagctggccttt | ||
| POTP | Chr 2 | Fccaccttcctctgcttcacttt | 60 |
| STR TPOX | p25.3 | Racatgggtttttgcctttgg | |
| NC_000002.12 | PTcaccaactgaaatatg | ||
| NR4Q | Chr 4 | Ftggtgggaatgttcttcagatga | 83 |
| Gene DCK | q13.3-q21.1 | Rtcgactgagacaggcatatgtt | |
| NC_000004.12 | PB+tgtatgagaaacctgaacgatggt | ||
| D5 | Chr 5 | Fttcatacaggcaagcaatgcat | 75 |
| STR D5S2500 | q11.2 | Rcttaaagggtaaatgtttgcagtaatagat | |
| NC_000005.10 | PTataatatcagggtaaacaggg | ||
| ND6 | Chr 6 | Fgcatggctgagtctaaagttcaaag | 82 |
| STR D6S474 | q21–22 | Rgcagcctcagggttctcaa | |
| NC_000006.12 | PB+cccagaaccaaggaagatggt | ||
| D9 | Chr 9 | Fggctttgctgggtactgctt | 60 |
| STR D9S2157 | q34.2 | Rggaccacagcacatcagtcact | |
| NC_000009.12 | PTcagggcacatgaat | ||
| HBB1 | Chr 11 | Fgctgagggtttgaagtccaactc | 76 |
| Gene HBB | p15.5 | Rggtctaagtgatgacagccgtacct | |
| NC_000011.10 | PTagccagtgccagaagagccaagga | ||
| ND14 | Chr 14 | Ftccaccactgggttctatagttc | 109 |
| STR D14S1434 | q32.13C | Rggctgggaagtcccacaatc | |
| NC_000014.9 | PB+tcagactgaatcacaccatcag | ||
| 22C3 | Chr 22 | Fcccctaagaggtctgttgtgttg | 78 |
| Gene PMM1 | q13.2 | Raggtctggtggcttctccaat | |
| NC_000022.10 | PBcaaatcacctgaggtcaaggccagaaca |
F: Forward primer, R: Reverse primer,
PB : Blackhole quencher probe, PB+ : Blackhole Plus quencher probe, PT : Taqman MGB probe
Materials and Methods
ddPCR System
The measurements discussed here were performed using a Bio-Rad QX200 Droplet Digital PCR System (Hercules, CA) system. The manufacturer’s QuantaSoft version 1.7.4.0917 software was used to determine the number of positive and negative droplets at the end of 60 cycles using assay-specific intensity thresholds. These results were exported into a spreadsheet for further manipulation.
Sample Materials
Three human genomic DNA extracts were investigated, all components of a candidate CRM. These extracts were prepared from the buffy coat fraction of anticoagulated blood from anonymous donors. The component labeled “A” was derived from a single-source male, “B” from a single-source female, and “C” a mixture of a single-source male and a single-source female.
Each donor buffy coat bag was aliquoted (5 mL per aliquot) into sterile 50 mL conical tubes then stored at 4 °C prior to extraction. A modified salt-out manual extraction protocol was performed for each of the individual buffy coat samples [15], with rehydration of the DNA in 10 mmol/L tris(hydroxymethyl)aminomethane HCl, 0.1 mmol/L disodium ethylenediaminetetraacetic acid, pH 8.0 buffer (TE−4). All components were solubilized from the air-dried state in TE−4 buffer and diluted to a working concentration of ≈50 ng/μL based upon double-stranded DNA absorbance at 260 nm [4].
Since there is no consensus on the infectious status of extracted DNA, all solutions were handled as biosafety level 1 materials capable of transmitting disease [16].
ddPCR Measurements
Ten PCR assays developed at the National Institute of Standards and Technology (NIST) were used in this study. Each assay was confirmed to target one locus per HHGE using the National Center for Biotechnology Information’s BLAST/ blastn system [17]. The targets for these assays are located on eight chromosomes; three of the assays target widely separated loci on chromosome 2. Table 1 lists the genomic targets, primers and probes for these assays.
Measurements with all ten assays were made on five independently prepared aliquots of each of the three components. Each set of three aliquots was processed on one 96-well plate. The five sets were processed over four days by the same analyst using the same equipment and materials. Two technical replicates were collected for each assay for each component along with non-template controls (NTCs) for each assay. On average 17,000 droplets were counted per sample (technical replicates and NTCs). There was a maximum of three positive droplets in any of the NTCs, averaging less than one positive per NTC. Of the (3 × 5 × 10 × 2 sample + 5 × 10 × 2 NTC) = 400 ddPCR measurements, three were rejected as technical failures based on the manufacturer’s software diagnostics.
The independent aliquots were prepared as 1 volume of the ≈50 ng/μL CRM material to 3 volumes of TE−4 buffer. For each 25 μL reaction, 2.5 μL DNA of this 1:4 diluted solution was added to 22.5 μL of PCR mastermix for a total dilution factor of 1/40. For all three materials, this dilution yielded λ ≈ 0.3 copies per droplet and dilution-adjusted λ/F ≈ 12 copies per droplet. Use of this rather low λ value with unfragmented human nDNA helps to minimize assay bias at the cost of a modest increase in technical replicate variability: see the Electronic Supplementary Material (ESM) section on Poisson Sampling. The 22.5 μL of mastermix consisted of: 12.5 μL of Supermix for Probes (no dUTPs), 1.88 μL each of 5 μmol forward and reverse primers, 1.25 μL of 5 μmol probe, and 5.0 μL nuclease-free water.
For each replicate assessment, 22 μL of the mastermix solution were loaded into a 96-well plate, heat-sealed with foil, and placed on Bio-Rad Automated Droplet Generator (AutoDG). The AutoDG generated droplets and all formed droplets were loaded into a new 96-well plate. That plate was heat-sealed with foil and PCR amplified on a Pro-Flex PCR system (Applied Biosystems, Foster City CA). The amplification protocol was: 95 °C for 10 min, followed by 60 cycles of 15 s at 95 °C and 1 min at 61 °C with ramp rate set to 2.5 °C/s between temperatures. After the 60th cycle there was a 98 °C hold for 10 min followed by a 4 °C hold until the samples were removed from the thermal cycler and put onto the QX200 droplet reader.
Results and Discussion
There are two autosomal targets per diploid genome, one from each chromosome pair. Since all ten of the assays we use target autosomal loci, for simplicity the following discussions focus on the 1-to-1 relationship between a single ddPCR target and a HHGE. Except for the minor complication of X and Y sex chromosomes in males, this is identical to the 2-to-2 relationship between two ddPCR targets and the full diploid genome.
Copies per Nanoliter
Because measurements cannot be made on undiluted sample, in practice ddPCR results expressed in terms of copies per sample combine the λ and F factors. The λ/F ± u(λ/F) for a given analysis system can be characterized through repeated independent measurements of samples prepared from the same stock material. In a series of five independent determinations, three different DNA extracts were evaluated in duplicate with the ten human genomic assays listed in Table 1. For these measurements the expected relative standard uncertainty, u(λ/F)/(λ/F), for a single estimate of λ/F with a single assay is about 7.64%; see ESM Table S3 for details. This estimate is strictly appropriate only for these experiments, but may be indicative of similar processes.
Noting that the relative uncertainty for the mean, x̄, of a series of m independent determinations of some measurand X can be estimated as [14, Section 4.2.3]
| (3) |
we estimate the relative uncertainty of the mean λ/F from mrep = 5 single-assay measurements of independently prepared replicates as . The λ/F values are metrologically traceable to the derived SI unit for volume, nL, and natural unit count-one (1) [5].
The mean droplet volume was measured for droplets made using our equipment with the same lot of mastermix used for the ddPCR measurements. Using the concentrated method described in Dagata et al. [12], V for this lot was 0.7349 nL with a standard relative uncertainty, u(V)/V, of 1.15%. For a given lot of mastermix and using our equipment and supplies, the value of V remains constant well within this uncertainty over at least six months (data not shown) and is metrologically traceable to the derived SI unit for volume, nL. The u(V)/V is characteristic of the volume measurement process and is not a function of the number or nature of ddPCR measurements.
Human Genomes per Target
The concentration of a given target is not necessarily the same as the concentration of HHGEs containing that target. Fragmentation, either intentional or during sample preparation, can reduce the number of amplifiable assay-specific targets in the reaction mixture and/or increase their accessibility [18]. While the ten assays used in this study are designed to amplify one and only one site per HHGE (i.e., r = 1), sample-specific mutations can reduce or prevent amplification or introduce additional binding sites. We therefore advocate using multiple assays, designed to amplify widely separated autosomal targets, to confirm that target measurements imply genome measurements.
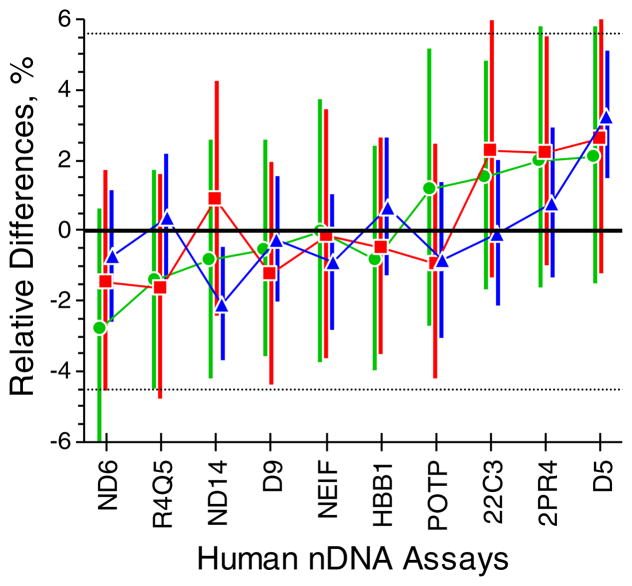
Figure 1 displays the relative performance of the assays for three human genomic extracts in TE−4 buffer. The agreement among the assays confirms that for these assays r = 1. The relative between-assay standard uncertainty, u(r)/r, is 2.17%; see ESM Table S4 for details. This value is strictly appropriate only for these experiments, but may be indicative of similar processes.
Fig. 1.
Human Genomes per Target. The “dot&bar” represent the mean relative differences among the ten assays for the three components, where the individual differences are , i indexes samples, j assays, and the “bar” above the symbol denotes averaging across replicates, plates, and (for ) assays. The solid “dots” mark the median of the posterior distribution determined by an empirical Bayesian evaluation; the vertical “bars” span the central 50%of the distribution. The green circles and lines represent the results for component “A” of the candidate CRM, red squares and lines for component “B”, and blue triangles and lines for component “C”. The thin black horizontal lines bound an approximate 95%confidence interval around the zero-difference line. See ESM Table S5 for the numerical values
By Eq. 3, the corresponding relative standard uncertainty for the mean of our massay = 10 independent assays, r̄, is . The r values are metrologically traceable to the natural unit ratio-one (1) [5].
Number of Nucleotide Bases per Haploid Human Genome Equivalent
The nucleotide base composition of the human genome is known to differ among individuals [19], but this variability is likely to be small except for rare cases (e.g., trisomies). Given current technology, determining the “exact” number of bases, n, for a personal genome is impractical. However, the Human Assembly Data web-resource maintained by The Genome Reference Consortium (GRC) provides the lengths in bp of the 22 autosomal and the X and Y sex chromosomes of the Reference Genome as estimated in the 2006 NCBI36, 2009 GRCh37, and 2013 GRCh38 assemblies (https://www.ncbi.nlm.nih.gov/grc/human/data). These chromosome lengths are identical in all “patch” releases (periodic minor updates) of these assemblies. Table 2 lists these values.
Table 2.
Lengths of chromosomes in the human reference genome, bp a
| Chromosome | Assembly
|
||
|---|---|---|---|
| GRCh38 | GRCh37 | NCBI36 | |
| 1 | 248,956,422 | 249,250,621 | 247,249,719 |
| 2 | 242,193,529 | 243,199,373 | 242,951,149 |
| 3 | 198,295,559 | 198,022,430 | 199,501,827 |
| 4 | 190,214,555 | 191,154,276 | 191,273,063 |
| 5 | 181,538,259 | 180,915,260 | 180,857,866 |
| 6 | 170,805,979 | 171,115,067 | 170,899,992 |
| 7 | 159,345,973 | 159,138,663 | 158,821,424 |
| 8 | 145,138,636 | 146,364,022 | 146,274,826 |
| 9 | 138,394,717 | 141,213,431 | 140,273,252 |
| 10 | 133,797,422 | 135,534,747 | 135,374,737 |
| 11 | 135,086,622 | 135,006,516 | 134,452,384 |
| 12 | 133,275,309 | 133,851,895 | 132,349,534 |
| 13 | 114,364,328 | 115,169,878 | 114,142,980 |
| 14 | 107,043,718 | 107,349,540 | 106,368,585 |
| 15 | 101,991,189 | 102,531,392 | 100,338,915 |
| 16 | 90,338,345 | 90,354,753 | 88,827,254 |
| 17 | 83,257,441 | 81,195,210 | 78,774,742 |
| 18 | 80,373,285 | 78,077,248 | 76,117,153 |
| 19 | 58,617,616 | 59,128,983 | 63,811,651 |
| 20 | 64,444,167 | 63,025,520 | 62,435,964 |
| 21 | 46,709,983 | 48,129,895 | 46,944,323 |
| 22 | 50,818,468 | 51,304,566 | 49,691,432 |
| X | 156,040,895 | 155,270,560 | 154,913,754 |
| Y | 57,227,415 | 59,373,566 | 57,772,954 |
as listed by The Genome Reference Consortium (GRC) (https://www.ncbi.nlm.nih.gov/grc/human/data)
The Human Assembly Data resource also provides a “Total bases” summary for every patch release of the GRCh37 and GRCh38 assemblies. These totals include bp from alternate loci (large polymorphisms in bp composition but generally not large differences in bp number) as well as un-placed and un-localized scaffolds (known bp sequences not yet positioned within the assembly) [20]. These scaffolds likely fit within the known gaps (difficult-to-sequence stretches of approximately known length) in the Reference Genome sequence and so are unlikely to have much impact on the Reference Genome bp number. The gradual increase in the number of “Total bases” thus reflects increasing knowledge of the human genome and its variability but not the bp size of the Reference genome.
Based on the GRCh38 assignment, the total length of the autosomes plus the X chromosome is 3.031 109 bp and that for the autosomes plus the Y chromosome is 2.932 109 bp. Without determining the sex chromosome ratio in a sample, an average gender-neutral HHGE then has ((3 × 3.031 + 1 × 2.932)/4) 109 = 3.006 109 bp. Using the same calculations, the GRCh37 and NCBI36 estimates are 3.012 109 bp and 2.998 109 bp. The standard deviation (and thus an estimate of the standard uncertainty) of the three values is 7.024 106 bp.
Given the upper and lower bounds, a+ and a−, of a rectangular (uniform) distribution of X, in the absence of other information the usual estimate of the expected value, x, and it standard uncertainty, u(x), is [14, Section 4.3.7]:
| (4) |
The GRCh38 value for the n of a XX female is 3.031 109 bp while for a XY male it is 2.982 109 bp. Treating these values as the bounds on the bp of a sample with an undetermined sex chromosome ratio, the estimated standard uncertainty is .
Since the measurement technologies used to construct the reference tend to contract the length of tandem repeats [21, 22], we estimate the number of bp in the reference HHGE as the largest of the currently defensible values, the GRCh37 value of 3.012 109 bp.
While we are only concerned with nDNA, the widely used spectroscopic methods are insensitive to the DNA source [4]. When comparing target-specific ddPCR and non-specific spectroscopic results, the possible influence of mitochondrial DNA (mtDNA) should be considered. While highly variable, the ratio of mtDNA to nDNA copies in whole blood is about 100-to-1 [23]. Since the reference human mitochondrial genome contains 16,569 bp [24], the impact per 100 mtDNA/ nDNA is (100 × 16,569) = 1.657 106 bp.
Noting that for independent additive factors, xi, the combined standard uncertainty of their sum, y, can be estimated as the square root of the sum-of-squares of the individual standard uncertainties, u(xi) [14, Section 5.1.2]:
| (5) |
An estimate of the combined standard uncertainty of n is then for a relative standard uncertainty, u(n)/n, of (1.58 107)/(3.012 109) = 0.525%.
While the chromosome bp lengths are not provided as certified values, the GRC is an internationally recognized authority on the reference genome. The data it provides are adequate to establish n ± u(n) as metrologically traceable to the natural unit count-one (1).
Mean Molar Mass of DNA Nucleotides
The value given in relevant literature as the mean molar mass of the nucleotide bases that comprise DNA (A: deoxyadenosine monophosphate, T: deoxythymidine monophosphate, G: deoxyguanosine monophosphate, and C: deoxycytidine monophosphate) is surprisingly variable, ranging from (308 to 330) g/mol or, expressed as A-T and G-C bp, (616 to 660) g/mol [25, 26]. Other values provided by on-line resources in response to the query “What is the average molar mass of a DNA base pair?” include (649, 650, and 654) g/mol. On evaluation, the (649 and 650) g/mol values result from averaging the molar masses of the deprotonated nucleotide monomers (loss of 2 H+ per base), with and without rounding to integer values and ignoring the loss of water during polymerization. The 654 g/mol value is the mean of the protonated monomers and ignores the loss of water. The 616 g/mol value reflects the mean mass of the deprotonated polymeric bases (loss of 1 H+ per base). The 660 g/mol value is the mean mass of the sodium salt of the polymer. Table 3 details these calculations using the estimated atomic masses, uncertainties, and ranges provided in Meija et al. [27]. The atomic mass uncertainties have been estimated following Possolo et al. [28].
Table 3.
| Base d | Formula | Nucleotide Monomer
|
Polynucleotide Polymer c
|
|||
|---|---|---|---|---|---|---|
| Acid e | Base−2 f | Acid e | Base−1 f | Na+ Salt | ||
| A | C10H14O6N5P | 331.222(1) | 329.206(1) | 313.207(1) | 312.199(1) | 334.181(1) |
| T | C10H15O8N2P | 322.208(1) | 320.192(1) | 304.193(1) | 303.185(1) | 325.167(1) |
| G | C10H14O7N5P | 347.221(1) | 345.206(1) | 329.206(1) | 328.198(1) | 350.180(1) |
| C | C9H14O7N3P | 307.197(1) | 305.181(1) | 289.182(1) | 288.174(1) | 310.156(1) |
| Mean | 326.962(2) | 324.946(2) | 308.947(2) | 307.939(2) | 329.921(2) | |
| 30:30:20:20 Mean h | 326.913(2) | 324.897(2) | 308.898(2) | 307.890(2) | 329.872(2) | |
| AT g | C20H29O14N7P2 | 653.430(2) | 649.398(2) | 617.400(2) | 615.384(2) | 659.347(2) |
| GC g | C19H28O14N8P2 | 654.419(2) | 650.387(2) | 618.388(2) | 616.372(2) | 660.336(2) |
| Mean | 653.925(3) | 649.893(3) | 617.894(3) | 615.878(3) | 659.842(3) | |
| 60:40 Mean i | 653.826(3) | 649.794(3) | 617.795(3) | 615.779(3) | 659.743(3) | |
Ignoring end-groups and methylation
Molar masses are calculated from the atomic masses, uncertainties, and ranges listed in (Meija et al., 2016) [27]. Standard uncertainties, estimated following (Possolo et al., 2017) [28], are enclosed by parentheses
Condensation polymer; n-1 waters are lost for every n condensed monomers
A: deoxyadenosine monophosphate, T: deoxythymidine monophosphate, G: deoxyguanosine monophosphate, C: deoxycytidine monophosphate
Conjugate acid, phosphate oxygens fully protonated
Conjugate base, phosphate oxygens fully deprotonated
Base pairs (bp) associated via hydrogen bonding
Calculated as (30A + 30 T + 20G + 20C)/100
Calculated as (60AT + 40GC)/100
The values based upon averaging the monomers are inappropriate for polymerized fragments, as are values that ignore counter ions when the polymers are in neutral to basic solution. The 660 g/mol estimate for the sodium salt more correctly reflects the chemical composition in TE−4 pH 8.0 buffer. Treating the “molar mass of DNA” as that of the negatively charged polymerized bases is analogous to treating “the molar mass of table salt” as that of chloride.
The mean (659.841 ± 0.003) g/mol value of the “Na+ Salt” in Table 3 is for equal proportions of AT and GC pairs. AT pairs typically outnumber GC pairs in a ratio of 60-to-40 [29]. The (659.743 ± 0.003) g/mol of the 60:40 weighted mean is a more appropriate estimate for genomic DNA.
Two factors can increase the effective mean bp mass: the addition of one water per fragment when DNA is fragmented and the variable methylation of A and C bases. The extent of fragmentation depends upon the treatment history of the material. Since the typical fragment size of the DNA in our sample materials exceeds 48,502 bp [3], the impact of adding two waters (molar mass 18.015 g/mol) per double-strand break in our materials can be conservatively estimated as 2 × 18.015/ (659.743 × 48,502) = 1.126 10−6 g/mol. Taking the maximum number of breaks to be 3.012 109/48502 = 6.210 104, the upper bound on water addition is (1.126 10−6)(6.210 104) = 0.070 g/mol. Taking no fragmentation as the lower bound and assuming fragmentation is rectangularly distributed, by Eq.4 the additional mass due to fragmentation is 0.070/2 = 0.035 g/mol and a conservative estimate of the standard uncertainty is .
Adenosine methylation of nDNA in eukaryotes is known but is infrequent [30]. About 1% of human DNA nucleotides are 5-methylcystine monophosphate [31], although this percentage is known to vary among individuals as well as tissue types [32]. Taking no methylation as the lower bound, 2% methylation as an upper bound, and assuming the methylation is rectangularly distributed between these limits, the upper bound on the additional mass per average bp due to methylation (molar mass 15.034 g/mol per methyl group) is (0.02 × 15.034) = 0.301 g/mol. By Eq. 4, the expected increase is 0.301/2 = 0.150 g/mol and a conservative estimate of the standard uncertainty is .
Combining the (659.743 ± 0.002) g/mol of the weighted mean, the (0.035 ± 0.020) from fragmentation, and the (0.150 ± 0.087) from methylation, we estimate w̄ to be (659.743 + 0.035 + 0.150) = 659.928 g/mol and from Eq. 5 . The relative standard uncertainty, u(w̄ )/w̄, is then 0.089/659.928 = 0.013%. While there is much confusion regarding the definition of what is meant by “the mean molarmass”, there is very little uncertainty once a definition is adopted.
Analogous with the GRC value for n, the International Union of Pure and Applied Chemistry’s Inorganic Chemistry Division Committee is the recognized authority on atomic mass. The data it provides, in conjunction with literature best-estimates of minor influences, establish w̄ ± u(w̄) as metrologically traceable to the derived SI unit of mass, g, and the SI unit for amount of substance, mol.
Conclusions
Segregating the experimental λ (copies per droplet), F (dilution factor), V (droplet volume), and r (targets per HHGE) factors in Eq. 1 from the constants and our estimates for n (number of bp per HHGE) and w̄ (mean molar mass per bp), the mass concentration of nDNA per microliter sample solution can be estimated as:
| (6) |
From Eq. 2, the associated relative standard uncertainty is:
| (7) |
For this series of measurements, . Note that the u(n)/n value of 0.525% completely swamps the u(w̄ )/w̄ of 0.013% but barely registers against the 3.67% of the combined uncertainties from the experimental factors.
A 95% confidence relative uncertainty, U95([nDNA]), can be estimated as:
| (8) |
where k95 is the appropriate expansion factor for the combined degrees of freedom, v([nDNA]). When all standard uncertainties are associated with large degrees of freedom, k95 = 2; otherwise v([nDNA]) can be estimated using the Welch-Satterthwaite approximation and k95 from the Student’s t distribution [14, Section G4.1]. When V is determined with the method used here, v(V) is “large”. Being based on literature data, v(n) and v(w̄) can also be considered “large”. Given the number of components, assays, and independent determinations used in this study, v(λ/F) and v(r̄) are “large enough” to justify using the k95 = 2 approximation.
The 95% confidence relative expanded uncertainty of the ddPCR assignment of [nDNA] for this series of measurements is then 2 × 3.71 ≅ 7.4%.
Since all factors are metrologically traceable to natural and/ or SI units, ddPCR-based values for the mass concentration of human nuclear DNA can be metrologically traceable to the SI.
Supplementary Material
Acknowledgments
We thank our NIST colleague Justin Zook for interpreting the information provided by The Genome Reference Consortium and an anonymous Referee for insightful suggestions and corrections. This work was supported in part by the NIST Special Programs Office project Forensic DNA.
Biography

From left to right: Blaza Toman, mathematical statistician with the Statistical Engineering Division; Margaret Kline, research biologist in the Applied Genetics Group of the Biomolecular Measurement Division; David Duewer, research chemist in the Chemical Sciences Division; and Erica Romsos, research biologist in the Applied Genetics Group. The team combines interests in forensic human identification, the use of digital PCR, metrology, and Bayesian Markov Chain Monte Carlo methods
Footnotes
Compliance with ethical standards
Human and animal rights All work presented has been reviewed and approved by the National Institute of Standards and Technology Human Subjects Protections Office. This study was determined to be “not human subjects research” (often referred to as research not involving human subjects) as defined in U. S. Department of Commerce Regulations, 15 CFR 27, also known as the Common Rule (45 CFR 46, Subpart A), for the Protection of Human Subjects by the NIST Human Subjects Protection Office and therefore not subject to oversight by the NIST Institutional Review Board.
Conflict of interest The authors declare that they have no conflict of interest.
Disclaimer Certain commercial equipment, instruments, or materials are identified in this report to specify adequately experimental conditions or reported results. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the equipment, instruments, or materials identified are necessarily the best available for the purpose.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00216-018-0982-1) contains supplementary material, which is available to authorized users.
References
- 1.Kline MC, Duewer DL, Redman JW, Butler JM. NIST Mixed Stain Study #3: DNA quantitation accuracy and its influence on short tandem repeat multiplex signal intensity. Anal Chem. 2003;75(10):2463–9. doi: 10.1021/ac026410i. [DOI] [PubMed] [Google Scholar]
- 2.Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. BioTechniques. 1992;13(3):444–9. [PubMed] [Google Scholar]
- 3.Kline MC, Duewer DL. Evaluating Droplet Digital Polymerase Chain Reaction for the Quantification of Human Genomic DNA: Lifting the Traceability Fog. Anal Chem. 2017;89(8):4648–54. doi: 10.1021/acs.analchem.7b00240. https://doi.org/10.1021/acs.analchem.7b00240. [DOI] [PubMed] [Google Scholar]
- 4.Green MR, Sambrook J. Molecular Cloning: A Laboratory Manual. 4. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- 5.De Bièvre P, Dybkaer R, Fajgelj A, Hibbert DB. Metrological traceability of measurement results in chemistry: Concepts and implementation. Pure Appl Chem. 2011;83(10):1873–935. https://doi.org/10.1351/PAC-REP-07-09-39. [Google Scholar]
- 6.Griffiths KR, Burke DG, Emslie KR. Quantitative polymerase chain reaction: a framework for improving the quality of results and estimating uncertainty of measurement. Anal Methods. 2011;3:2201–11. https://doi.org/10.1039/c1ay05069a. [Google Scholar]
- 7.Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, et al. Evaluation of a Droplet Digital Polymerase Chain Reaction Format for DNA Copy Number Quantification. Anal Chem. 2012;84:1003–11. doi: 10.1021/ac202578x. https://doi.org/10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs BKM, Goetghebeur E, Clement L. Impact of variance components on reliability of absolute quantification using digital PCR. BMC Bioinf. 2014;15:283. doi: 10.1186/1471-2105-15-283. https://doi.org/10.1186/1471-2105-15-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deprez L, Corbisier P, Kortekaas A-M, Mazoua S, Hidalgo RB, Trapmann S, et al. Validation of a digital PCR method for quantification of DNA copy number concentrations by using a certified reference material. Biomol Detect Quantif. 2016;9:29–39. doi: 10.1016/j.bdq.2016.08.002. https://doi.org/10.1016/j.bdq.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbisier P, Pinheiro L, Mazoua S, Kortekaas A-M, Chung PYJ, Gerganova T, et al. DNA copy number concentration measured by digital and droplet digital quantitative PCR using certified reference materials. Anal Bioanal Chem. 2015;407(7):1831–40. doi: 10.1007/s00216-015-8458-z. https://doi.org/10.1007/s00216-015-8458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong LH, Meng Y, Sui ZW, Wang J, Wu LQ, Fu BQ. Comparison of four digital PCR platforms for accurate quantification of DNA copy number of a certified plasmid DNA reference material. Sci Rep. 2015;5:13174. doi: 10.1038/srep13174. https://doi.org/10.1038/srep13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dagata JA, Farkas N, Kramer JA. Method for measuring the volume of nominally 100 μm diameter spherical water-in-oil emulsion droplets. 2016 NIST Special Publication 260–184. https://doi.org/10.6028/NIST.SP.260-184.
- 13.Košir AB, Divieto C, Pavšič J, Pavarelli S, Dobnik D, Dreo T, et al. Droplet volume variability as a critical factor for accuracy of absolute quantification using droplet digital PCR. Anal Bioanal Chem. 2017 doi: 10.1007/s00216-017-0625-y. https://doi.org/10.1007/s00216-017-0625-y. [DOI] [PMC free article] [PubMed]
- 14.JCGM 100:2008. Joint Committee for Guides in Metrology. Sevres, France: 2008. Evaluation of measurement data—Guide to the expression of uncertainty in measurement (GUM) See Section 4.3.7. https://www.bipm.org/en/publications/guides/#gum. [Google Scholar]
- 15.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. https://doi.org/10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chosewood LC, Wilson DE, editors. CDC/NIH. Biosafety in Microbiological and Biomedical Laboratories. 5. US Government Printing Office; Washington, DC: 2009. HHS publication No. (CDC) 21–1112. http://www.cdc.gov/biosafety/publications/bmbl5/index.htm. [Google Scholar]
- 17.NCBI. Standard Nucleotide BLAST. https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch.
- 18.Kline MC, Romsos EL, Duewer DL. Evaluating Digital PCR for the Quantification of Human Genomic DNA: Accessible Amplifiable Targets. Anal Chem. 2016;88(4):2132–9. doi: 10.1021/acs.analchem.5b03692. https://doi.org/10.1021/acs.analchem.5b03692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. https://doi.org/10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen HC, Kitts PA, et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017;27:849–64. doi: 10.1101/gr.213611.116. https://doi.org/10.1101/gr.213611.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nature. 2012;13:36–46. doi: 10.1038/nrg3117. https://doi.org/10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaisson MJP, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–11. doi: 10.1038/nature13907. https://doi.org/10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Memon AA, Zöller B, Hedelius A, Wang X, Stenman E, Sundquist J, et al. Quantification of mitochondrial DNA copy number in suspected cancer patients by a well optimized ddPCR method. Biomol Detect Quantif. 2017;13:32–9. doi: 10.1016/j.bdq.2017.08.001. https://doi.org/10.1016/j.bdq.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. doi: 10.1038/13779. https://doi.org/10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 25.Thomas RA, Krishan A, Robinson DM, Sams C, Costa F. NASA/ American Cancer Society High-Resolution Flow Cytometry Project—I. Cytometry. 2001;43:2–11. doi: 10.1002/1097-0320(20010101)43:1<2::aid-cyto1012>3.0.co;2-j. https://doi.org/10.1002/1097-0320(20010101)43:1<2::AID-CYTO1012>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 26.Doležel J, Bartoš J, Voglmayr H, Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytometry. 2003;51A:127–8. doi: 10.1002/cyto.a.10013. https://doi.org/10.1002/cyto.a.10013. [DOI] [PubMed] [Google Scholar]
- 27.Meija J, Coplen TB, Berglund M, Brand WA, De Bièvre P, Gröning M, et al. Atomic weights of the elements 2013. Pure Appl Chem. 2016;88:265–91. https://doi.org/10.1515/pac-2015-0305. [Google Scholar]
- 28.Possolo A, van der Veen AMH, Meija J, Hibbert DB. Interpreting and propagating the uncertainty of the standard atomic weights (IUPAC Technical Report) Pure Appl Chem. 2017 aop https://doi.org/10.1515/pac-2016-0402.
- 29.Vinogradov AE. Measurement by flow cytometry of genomic AT/GC ratio and genome size. Cytometry. 1994;16:34–40. doi: 10.1002/cyto.990160106. https://doi.org/10.1002/cyto.990160106. [DOI] [PubMed] [Google Scholar]
- 30.Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, et al. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–33. doi: 10.1038/nature17640. https://doi.org/10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–21. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén S-E, Greco D, et al. Differential DNA Methylation in Purified Human Blood Cells: Implications for Cell Lineage and Studies on Disease Susceptibility. PLoS One. 2012;7(7):e41361. doi: 10.1371/journal.pone.0041361. https://doi.org/10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.