Abstract
For drug delivery purposes, the ability to conveniently attach a targeting moiety that will deliver drugs to cells and then enable controlled release of the active molecule after localization is desirable. Towards this end, we designed and synthesized clickable and photocleavable lipid analog 1 to maximize the efficiency of bioconjugation and triggered release. This compound contains a dibenzocyclooctyne group for bioorthogonal derivatization linked via a photocleavable 2-nitrobenzyl moiety at the headgroup of a synthetic lipid backbone for targeting to cell membranes. To assess delivery and release using this system, we report fluorescence-based assays for liposomal modification and photocleavage in solution as well as through surface immobilization to demonstrate successful liposome functionalization and photoinduced release. In addition, fluorophore delivery to and release from live cells was confirmed and characterized using fluorescence microscopy and flow cytometry analysis in which 1 was delivered to cells, derivatized and photocleaved. Finally, drug delivery studies were performed using an azide-tagged analog of camptothecin, a potent anti-cancer drug that is challenging to deliver due to poor solubility. In this case, the ester attachment of the azide tag acted as a caging group for release by intracellular esterases rather than through photocleavage. This resulted in a dose-dependent response in the presence of liposomes containing delivery agent 1, confirming the ability of this compound to stimulate delivery to the cytoplasm of cells.
Graphical Abstract
Introduction
For the purposes of drug delivery, vehicles and targeting groups including lipids and liposomes, polymers, micro-spheres, and antibodies have received considerable attention with the goal of enhancing therapeutic properties.1 Of these, lipids represent effective agents for the delivery of drugs and imaging agents due to their biocompatibility and their ability to target molecules to cells through actions including insertion and fusion involving cellular membranes.1–5 For this reason, the development of facile methods for covalent conjugation of active molecules onto lipids and liposomal surfaces has been of considerable interest in recent years. For such bioconjugation approaches, it is beneficial to invoke bioorthogonal methods to enable the selective attachment of molecules within the complex environments of biological samples.6,7 Furthermore, if molecules are covalently attached to lipids that anchor them to the membrane, following delivery to cells, it is advantageous to utilize a controllable means to trigger the release of the active molecule from the membrane surface.
In the use of bioorthogonal derivatization for the attachment of molecules to lipids and membranes, click chemistry reactions have recently been pursued due to benefits including selectivity and fast kinetics under ambient conditions.6–11 While initial approaches focused on the copper-catalyzed azide–alkyne cycloaddition,12–19 this reaction suffers from some disadvantages as the required copper catalyst can promote the decomposition of lipid membranes20 and complicate delivery in vivo. As a result, we and others have explored the application of copper-free click chemistry for lipid and membrane derivatization.21–23 Examples of applications of this approach have included the attachment of thrombomodulin proteins to liposomes to enhance activity,22 as well as the targeting of liposomes bearing trans-cyclooctene moieties to cancer cells via the pHLIP peptide for PET imaging studies.23
With regard to the triggered release of molecules, external stimuli that have been used for this purpose include light,24–27 redox conditions,28 enzymes,29 temperature,30–33 and pH.34–37 However, most of these processes are limited by the restrictive conditions required for stimuli-responsive release as they rely on differences in properties between cell types. Of these, light-initiated release is favorable as it is a general approach that can be controlled in a spatially resolved manner to stimulate release at desired target sites, and the 2-nitrobenzyl groups is among the best studied for photoinitiated bond cleavage.38 Prior work involving the incorporation of photocleavable groups into lipids has focused on light-initiated release of molecules from liposomes.39–45 In these cases, molecules such as drugs have been non-covalently encapsulated in the liposome and release was stimulated by decomposition or disruption of the membrane. However, noncovalent molecular encapsulation can suffer from background leakage, and thus it may be preferable to covalently attach target molecules to lipids prior to triggering the release through bond cleavage.
Results and Discussion
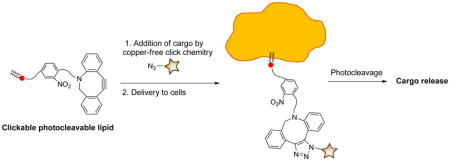
In this work, we set out to develop a facile and effective system that enables lipid conjugation for delivery purposes followed by photocleavage for the release of active molecules after targeting. To do so, we designed copper-free clickable and photocleavable lipid analog 1. Included at the headgroup of this lipid is an azodibenzocyclooctyne group (termed ADIBO or DIBAC), which benefits from fast kinetics in modification through the copper-free azide-alkyne cycloaddition reaction with azides.50–52 This group is linked to the lipid backbone via a 2-nitrobenzyl moiety to enable release of molecules attached via click chemistry through photolysis.
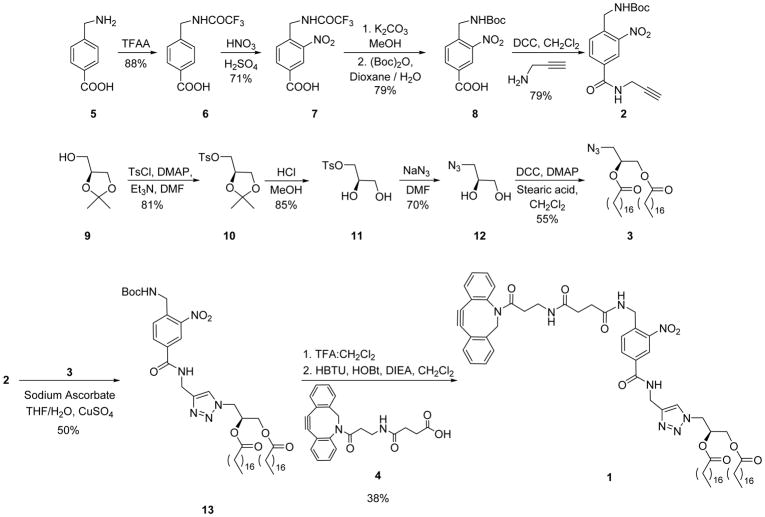
The synthetic route to clickable and photocleavable lipid 1 entailed coupling of alkyne-bearing 2-nitrobenzene 2 to azido-lipid scaffold 3 via click chemistry, followed by reaction with ADIBO-carboxylic acid 4 (Scheme 1). Alkynyl 2-nitrobenzene 2 was synthesized from 4-aminomethylbenzoic acid (5) via amine protection to 6, nitration to 7, and protecting group exchange to 8 using previously reported procedures.41 Carboxylic acid 8 was then coupled to propargylamine to generate 2. While we have previously reported azide-tagged lipid 3,47 a modified synthesis was developed to enhance yields by avoiding volatile intermediates. For the current route, S-glycerol acetonide (9) was converted to tosylate 10, as previously reported, followed by acetonide deprotection to diol 11, azide displacement to 12 and acylation with two stearic acid moieties to produce 3. Compounds 2 and 3 were next linked to form 13 via click chemistry, which was exploited due to synthetic facility and functional group tolerance. Furthermore, prior evidence has suggested that the polarity of the triazole headgroup may make this moiety an effective polar lipid headgroup mimic that lacks the charge that makes phospholipids difficult to purify and characterize.47,53 This medium polarity headgroup was seen as being beneficial for potentially enabling the ability to flip across cell membranes. Finally, the Boc protecting group of 13 was deprotected and the resulting amine was coupled to ADIBO-carboxylic acid 4, synthesized as previously described,49 to access 1.
Scheme 1.
Synthetic route to clickable and photocleavable lipid delivery agent 1.
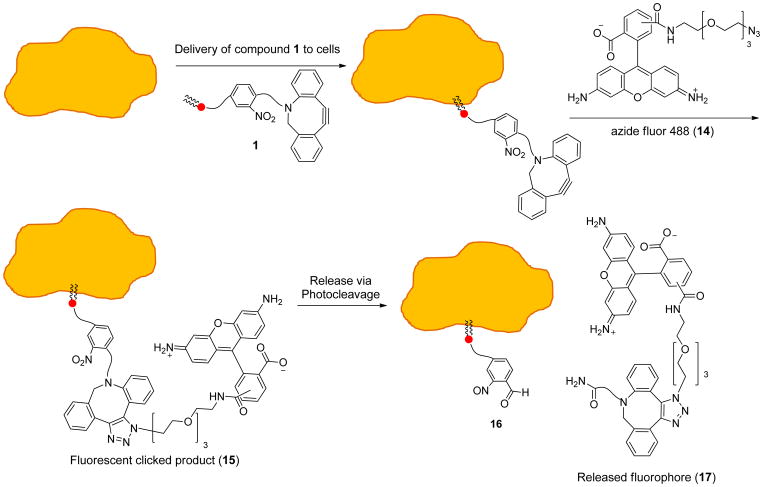
The initial thought process for delivery to and release from cells using compound 1 is depicted in Scheme 2, as evidenced by fluorescence-based derivatization, which was initially employed to characterize the system. First, compound 1 is incubated with cells leading to insertion into cell membranes due to the hydrophobic lipid backbone of 1. Following this, azide-tagged molecules can be targeted to cells through bioorthogonal copper-free click reaction with the ADIBO unit of 1. Alternatively, targeting groups could be clicked onto 1 prior to cell delivery or conjugation could be performed using liposomes containing 1 for subsequent delivery and fusion with cell membranes. In the example depicted in Scheme 2, derivatization with Azide-Fluor 488 (14) leading to triazole-linked product 15 is exploited for fluorescence-based analysis. Subsequently, irradiation with 350 nm light is performed to achieve photocleavage and thus release of the clicked molecule from cellular membranes, either into the cytoplasm or the periphery of the outside of the cell. This occurs through the traditional cleavage of the benzylic position to produce aldehyde-lipid byproduct 16 as well as the released triazole-linked conjugated molecules, in this case 17.
Scheme 2.
Approach to analysis of cell membrane delivery and release using compound 1.
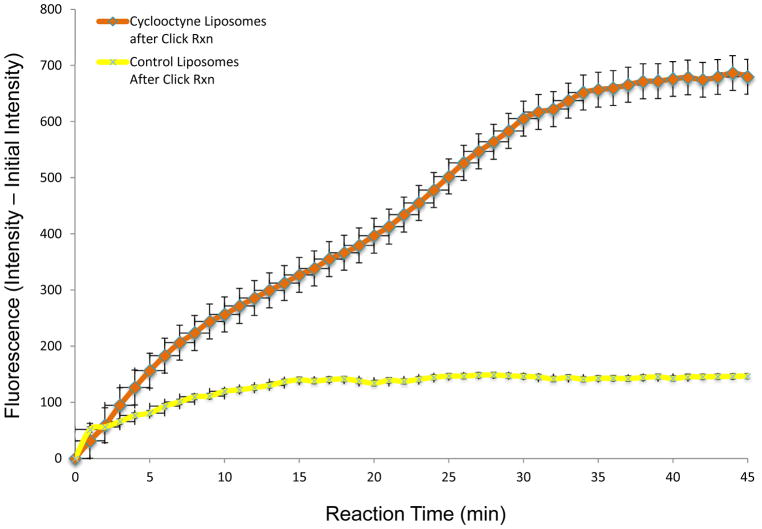
To characterize the efficacy of compound 1 for membrane delivery and release, we initiated studies using liposomes as model membranes.54–57 This was initially performed in solution through reaction of liposomes with 3-azidocoumarin (18, Scheme 3), a fluorogenic click reagent.58 To do so, unilamellar liposomes (~ 200 nm) composed primarily of phosphatidylcholine (96%) and doped with compound 1 (4%) were generated using standard procedures including hydration and extrusion as well as dynamic light scattering to characterize liposome size. Next, derivatization of liposomes with 18 was assessed in solution using a fluorimeter to track enhanced fluorescence resulting from click chemistry.58 As can be seen in Figure 1, addition of 18 led to increasing fluorescence that plateaued after ~ 40 minutes, indicating a successful click reaction. Additionally, a control was run in which liposomes lacking 1 were incubated with 18, also depicted in Figure 1. In this case, only a minor increase in fluorescence was observed, likely attributable to a slight enhancement of the background fluorescence properties of 18 caused by some insertion into the hydrophobic membrane environment. This change in fluorescence of 18 in the presence of membranes explains the double curve character of the results seen from the study sample.
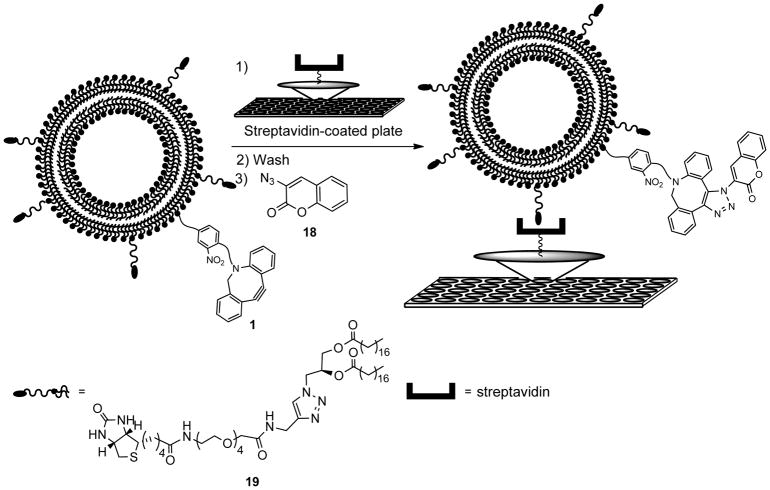
Scheme 3.
Liposome immobilization assay using reaction with 3-azidocoumarin (18) to characterize membrane derivatization.
Figure 1.
Change in fluorescence of azido-coumarin 18 in the presence of liposomes containing and lacking cyclooctyne-lipid 1.
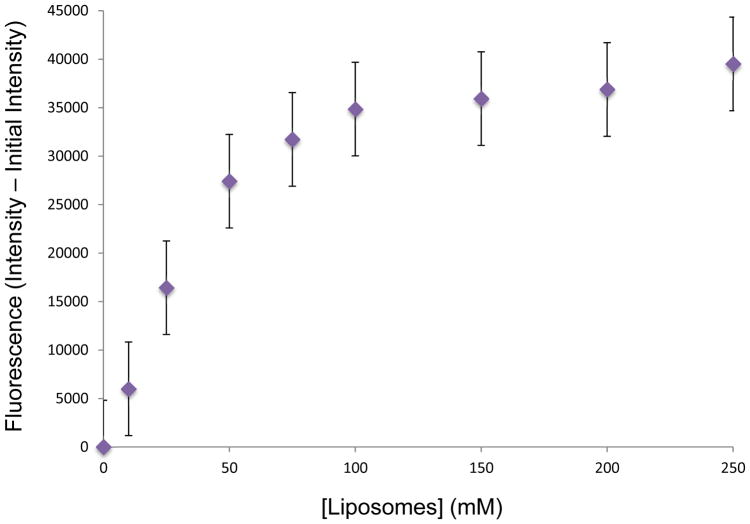
While the solution liposome assay allows for detection as a function of time, this study is complicated by the background increase in fluorescence of 18 in the presence of membranes lacking 1. Thus, for an alternative method for detecting liposome conjugation via 1, we turned to a surface immobilization assay that we have previously utilized (Scheme 3).47,59 In this assay, liposomes are immobilized onto streptavidin-coated 96-well microplates through incorporation of biotin–lipid analog 19. Following incubation of liposomes containing biotin anchor lipid 19 with azido-coumarin 18, the immobilization step enables the removal of unreacted dye from the surface-bound liposomes. Liposome derivatization was assessed using a fluorescence microplate reader to detect the enhanced fluorescence upon triazole formation with 1 incorporated into liposomes. As can be seen in Figure 2, this experiment yielded a dose-dependent response, further confirming successful liposome conjugation via copper-free click chemistry with lipid 1. In addition, the removal of unreacted dye yields a more traditional curve for product formation compared to the results obtained from the prior solution studies of liposome modification.
Figure 2.
Change in fluorescence of azido-coumarin 14 in the presence of microplate-immobilized liposomes following wash off of unreacted dye.
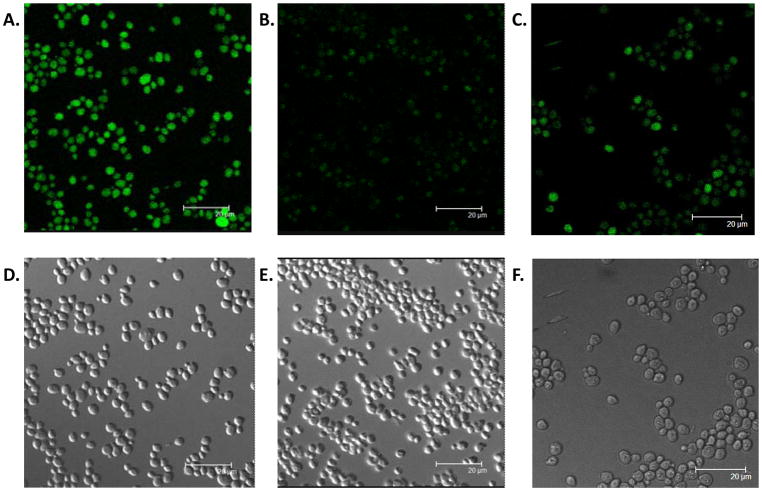
With successful results from liposome studies, we next set out to assess delivery and release using live Saccharomyces cerevisiae cells (Scheme 2). To do so, cells were grown in the presence and absence of 1 in the YPD medium. Following this, cells were incubated with commercially available Azide-Fluor 488 (14) to assess delivery of the fluorophore to cells via fluorescence microscopy, and the representative images are shown in Figure 3. Here, cells grown in the presence of lipid 1 (Figure 3A) result in significant fluorescence compared to control studies lacking 1 (Figure 3B), both of which were incubated with Azide-Fluor 488 and washed with 25% DMSO/water to remove unreacted dye. To assess release via photocleavage, cells were photolyzed for different periods of time, followed by washes to remove dye that is no longer attached to the membrane and detection through fluorescence microscopy. A representative image in Figure 3C taken after 1 hour of photocleavage demonstrates that fluorescence is significantly decreased following photocleavage. Control studies in which the labeling dye was subjected to irradiation conditions used for photolysis indicated that photobleaching is not occurring as no decrease in fluorescence was observed (data not shown). In addition, transmission images (Figures 3D–F) are included for the three described images, respectively, showing the presence of cells in each sample. Finally, released fluorophore 17 was detected by mass spectrometry, with the spectrum included in the supplementary information.
Figure 3.
Fluorescence images of A. cells grown in the presence of 1, B. cells grown in the absence of 1, and C. cells grown in the presence of 1 and irradiated for 1 hour, each of which was washed to remove unbound dye prior to imaging. D–F. Transmission images showing cell population for the three experiments, respectively.
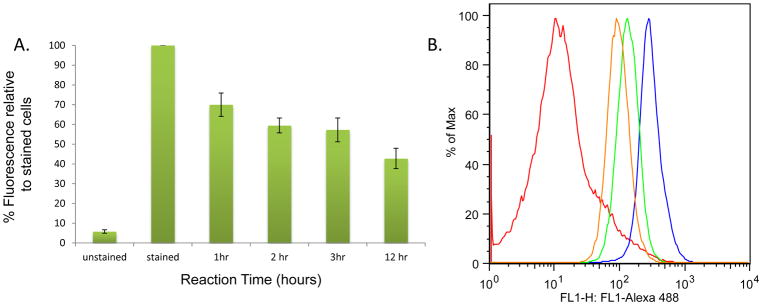
To obtain a more quantitative understanding of delivery and release in these studies, we next turned to flow cytometry-based analysis, with results shown in Figure 4. Figure 4A consists of a bar graph showing the fluorescence of control cells that were not treated with 1 or 14 (unstained), study cells after click and washing (stained), and stained cells after photocleavage for 1, 2, 3, and 12 hours followed by washing. Here, photocleavage yielded a decrease to 70%, 60%, 57%, and 42% of the original fluorescence of the unstained sample after irradiation for 1, 2, 3, and 12 hours, respectively. In addition, representative plots of fluorescence observed from flow cytometry analysis of the different described samples are shown in Figure 4B.
Figure 4.
Results from flow cytometry of labeled cells. A. Bar graph depicting fluorescence detected for cells not treated with 1 (unstained), cells treated with 1 (stained), and treated cells irradiated for 1, 2, 3, and 12 hours, each of which was washed prior to fluorescence imaging. Error bars show standard error from at least three replicate studies. B. Flow cytometry results for cells that were unstained (red), after click with 14 (blue), and after photocleavage for 3 (green) and 12 (orange) hours.
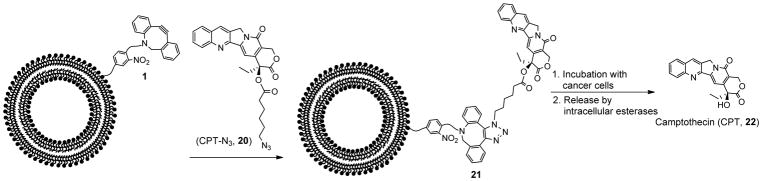
Finally, we set out to evaluate compound 1 as a tool for copper-free click-chemistry based delivery and release of an anti-cancer therapeutic using human cancer cells. For this purpose, we selected camptothecin (CPT, 22), a potent anti-cancer drug that is challenging to deliver due to its poor solubility.60 Here, we were also interested in determining whether compound 1 would allow clicked cargo to be released inside the cell, which is difficult to unambiguously determine through fluorescence analysis. Therefore, in order to deliver camptothecin via click chemistry, we switched to a release mechanism that requires delivery into cells by designing clickable analog 20 containing an azido-hexanoyl chain esterified to the free hydroxyl group of the drug (Scheme 4). This compound was expected to undergo release upon cell internalization without requiring photocleavage since the ester linker can be hydrolyzed following cell entry by intracellular esterases, thereby releasing camptothecin itself. Thus, successful release would provide evidence of the ability of lipid 1 to deliver cargo into cells and additionally validate release by mechanisms other than photocleavage.
Scheme 4.
Delivery of camptothecin to cancer cells. PC liposomes doped with 1 were reacted with azido-camptothecin 20 to produce 21 through click chemistry. The liposomes were then incubated with cancer cells to deliver camptothecin (22) through ester hydrolysis within cells.
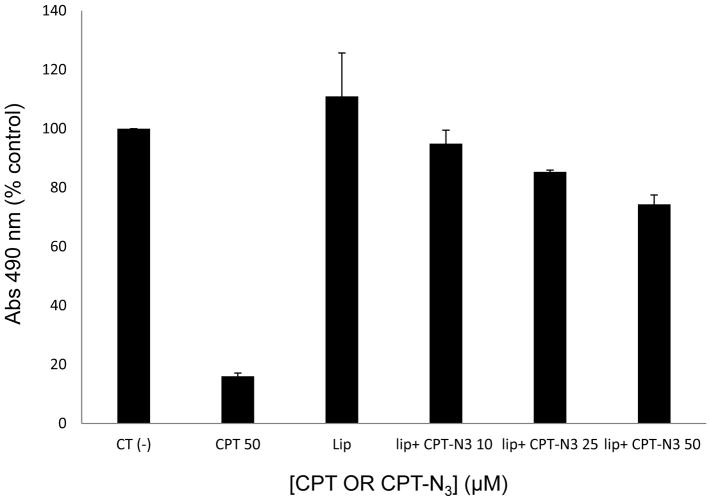
These studies were performed by incubating HeLa cells with PC liposomes doped with 6% of lipid 1 in the presence and absence of camptothecin-azide 20 (CPT-N3). This produced liposome-immobilized conjugate 21, which was confirmed by mass spectrometry (see supplementary information). Camptothecin itself was studied using a DMSO vehicle as a positive control, and all cell viability studies were performed using a standard MTS assay. In these studies, liposomes containing 1 alone did not influence cell viability, but those that were pre-incubated with added 20 ranging from 10–50 μM led to a dose-dependent decrease in living cells (Figure 5). Released camptothecin was detected by mass spectrometry as the dehydration product following extraction of cells under acidic conditions. A standard solution of camptothecin was subjected to the same extraction conditions and yielded the same peak, and both spectra are included in the supplementary information. Compound 20 alone was not viable for studies since it is not soluble in aqueous solution. These results demonstrate the delivery of camptothecin to cancer cells using compound 1.
Figure 5.
MTS assay in human cancer cell line indicates that CPT-N3 20 reduces cell proliferation in a dose-dependent manner. HeLa cells were treated with vehicle (Ct (−)- DMSO 0.01%), CPT (22, 50 μM), Liposomes control (Lip) and liposomes preincubated with CPT-N3 22 (Lip+CPT-N3) at different CPT-N3 concentrations (10 μM, 25 μM and 50 μM) for 48 hrs. Cell proliferation was measured by MTS assay and the graph represents the percentage relative to the control CT(−). The results represent the mean of 3 independent experiments. Statics t-test: * p<0.05 and ** p<0.001 two population relative to CT (−). The error bars shown correspond to the standard deviation.
These studies provide insight into the question of whether lipid 1 is effective at entering cells prior to release. For release via photocleavage, it is difficult to conclude whether this compound enters cells since lipid 1 bearing cargo could insert into the outer leaflet of cell membranes, and release could then result in a high local concentration of the drug around the periphery of the cell leading to activity. Alternatively, the derivatized lipid could initially insert into the outer leaflet of cell membranes and flip across the membrane into the inner leaflet prior to release. Finally, it could be engulfed into an endosome to stimulate cell entry. The two latter modes of action would result in the complex gaining access to the cytoplasm of the cells. Cell entry would be ideal for delivery purposes, although the likelihood of either cell penetration process would depend upon the properties of the specific drug that is being delivered. We envision that lipid 1 would be particularly effective for cell penetration and/or flipping across the membrane since it does not possess a particularly polar headgroup in the triazole. The camptothecin esterase release results provide evidence to support that lipid 1-mediated internalization does occur, since the compound would need to be exposed to the cytoplasm in order for the release by esterase hydrolysis to be activated.
In conclusion, we have designed, synthesized and evaluated compound 1 as a copper-free clickable lipid analog that enables delivery of molecular agents to cell membranes and subsequent release through photocleavage of the 2-nitrobenzyl linker connecting the introduced cargo to the membrane anchor. Through fluorogenic liposome modification and fluorescent liposome immobilization studies, we have confirmed the efficacy of lipid modification in a membrane environment for delivery purposes. We performed validation studies using two different types of live cells from different organisms, first exploiting microscopy to demonstrate that fluorescence was activated by delivery of fluorophores to S. cerevisiae cell membranes and then deactivated by photo-triggered release from the lipid backbone. Next, we probed the efficacy of delivery to human cancer cells to evaluate the prospects for future medicinal applications. In this case an alternate drug release mechanism hinging upon ester cleavage by esterase enzymes using azido-camptothecin 20 was used in part to determine if this would enable cell entry of the drug. These studies yielded a dose-dependent response of cell viability validating drug delivery to cells and also provided evidence that lipid 1 can stimulate drug entry into cells for release triggered by intracellular esterase enzymes. Therefore, this work presents a promising strategy for convenient bioconjugation and concentration of molecular cargo at cell membranes with subsequent triggered release of active molecules.
Experimental Procedures
General Experimental
Reagents and solvents were generally purchased from Acros, Aldrich or Fisher Scientific and used as received. PC (L-α-Phosphatidylcholine from chicken egg) was purchased from Avanti Polar Lipids, Inc. and 4(aminomethyl)benzoic acid was purchased from Chem Impex International. Dry solvents were obtained from a Pure solvent delivery system purchased from Innovative Technology, Inc. Column chromatography was performed using 230–400 mesh silica gel purchased from Sorbent Technologies. NMR spectra were obtained using Varian Mercury 500 and 600 spectrometers. Mass spectra were obtained with JEOL DART-AccuTOF and ABI Voyager DE Pro MALDI mass spectrometers with high-resolution capabilities. Optical rotation values were obtained using a Perkin-Elmer 241 polarimeter. Liposome extruder and polycarbonate membranes were obtained from Avestin (Ottawa, Canada). Ultrapure water was purified via a Millipore water system (≥ 18 MΩ·cm triple water purification system). 4-(((Tert-butoxycarbonyl)amino)methyl)-3-nitrobenzoic acid (8),41 (R)-(2,2-Dimethyl-1,3-dioxolan-4-yl)methyl 4-methylbenzenesulfonate (10),46,47 Biotin- PE 19,47 6-azidohexanoic acid (23)48 and ADIBO-carboxylic acid 449 were synthesized according to prior literature procedures. Solution fluorescence studies were performed using a Perkin Elmer LS55 fluorimeter. For photocleavage, samples were irradiated using a Rayonet Preparative Type RS photoreactor. Disposable polystyrene cuvettes were purchased from Fisher scientific. Microplate-based fluorescence measurements were performed using a BioTek synergy 2 multi-detection microplate reader. Black reacti-bind streptavidin high binding capacity (HBC) coated 96-well microplates were purchased from Pierce Biotechnology (Rockford, IL). Cell images were captured using a Leica SP2 laser scanning confocal microscope. msFluorescent cell counting was performed using a BD FACS (Fluorescence Activated Cell Sorting) Calibur flow cytometer.
tert-Butyl-2-nitro-4-(prop-2-yn-1-ylcarbamoyl)benzylcarbamate (2)
To 4-(((Tert-butoxycarbonyl) amino)methyl)-3-nitrobenzoic acid (8, 0.718 g, 2.40 mmol) dissolved in 200 mL of dichloromethane in a 500 mL round-bottomed flask was added dicyclohexylcarbodiimide (DCC, 0.743, 3.60 mmol), and N,N-dimethylaminopyridine (DMAP, 0.147 g, 1.20 mmol,). After 30 min, propargylamine (185 μL, 2.88 mmol) was added. The reaction was then allowed to stir overnight, after which it was washed with water, and the aqueous portion was extracted with dichloromethane (3 × 25 mL). The organic layers were then combined and dried with magnesium sulfate, filtered and concentrated by rotary evaporation. Column chromatography using gradient elution with 30–50% ethyl acetate-hexanes gave 2 as a yellow solid (638 mg, 79% yield). 1H NMR (500 MHz, CD3OD): δ 8.45 (s, 1H), 8.06 (d, J = 3.3 Hz, 1H), 7.63 (d, J = 3.6 Hz, 1H), 4.55 (s, 2H), 4.13 (d, J = 2.5 Hz, 2H), 2.58 (t, J = 2.5 Hz, 1H), 1.41 (s, 9H)). 13C NMR (126 MHz, CD3OD): δ 165.46, 156.59, 147.87, 138.06, 134.03, 132.15, 130.23, 124.01, 79.98, 79.05, 71.18, 33.57, 29.29, 28.02, 25.48, 24.79. HRMS-DART [M-H]− calcd for C16H19N3O5, 332.1246; found 332.1243.
(R)-2,3-dihydroxypropyl-4-methylbenzenesulfonate (11)
To (R)-(2,2-Dimethyl-1,3- dioxolan-4-yl)methyl 4-methylbenzenesulfonate (10, 1.00 g, 3.49 mmol)46,47 dissolved in 50 mL of methanol in a 500 mL round-bottomed flask was added 125 mL of 0.50 N hydrochloric acid, and the reaction mixture was allowed to stir for 12 h. The excess HCl was neutralized by adding solid sodium bicarbonate until carbon dioxide gas release was complete. Next, the crude mixture was passed through celite, filtered off, and the methanol was removed by rotary evaporation. Compound 11 was then eluted with 100% ethyl acetate using normal phase silica gel column chromatography and obtained as a colorless oil (816 mg, 95% yield). 1H NMR (500 MHz, CDCl3): δ 7.77 (d, J = 7.8 Hz, 2H), 7.33 (d, J = 7.8 Hz, 2H), 4.03 (s, 2H), 3.92 (s, 1H), 3.57 (m, 2H), 2.42 (s, 3H). 13C NMR (151 MHz, CDCl3): δ 145.22, 132.21, 130.02, 127.95, 70.94, 69.68, 62.77, 21.62. HRMS-DART [M+H]+ calcd for C10H14O5S, 247.0640, found 247.0644.
(S)-3-azidopropane-1,2-diol (12)
In a 250 mL round bottom flask, diol 11 (0.50 g, 2.03 mmol) and sodium azide (0.39 g, 6.09 mmol) were dissolved in N,N-dimethylformamide and the reaction was refluxed for 24 h. Next, ice water was added to the reaction mixture and the resulting mixture was extracted with dichloromethane (2 × 50 mL), dried over magnesium sulfate and the solvent was removed by rotary evaporation yielding 12 as a white solid (0.17g, 70% yield). NMR and mass spectra data matched with literature values for compound 12.47
(S)-3-(4-((4-(((tert-butoxycarbonyl)amino)methyl)-3-nitrobenzamido)methyl)-1H- 1,2,3-triazol-1-yl)propane-1,2-diyl distearate (13)
To alkyne 2 (0.400 g, 1.20 mmol) dissolved in 125 mL tetrahydrofuran-water (1:1) in a 250 mL round-bottomed flask was added azide 3 (0.780 g, 1.20 mmol). Next, copper sulfate pentahydrate (0.390 g, 1.56 mmol) and lastly sodium ascorbate (0.475 g, 2.40 mmol) were added. After overnight stirring, the solution was then concentrated and dried under high vacuum. Column chromatography with 5% methanol/dichloromethane gave product 13 as a light yellow solid (575 mg, 50% yield). 1H NMR (500 MHz, 85% CDCl3-CD3OD): δ 9.06 (s, NH), 8.54 (s, 1H), 8.13 (d, J = 8.5 Hz, 1H), 7.85 (s, 1H), 7.70 (m, 1H), 6.54 (s, NH), 5.49 – 5.37 (m, 1H), 4.71 – 4.58 (m, 4H), 4.38 (dd, J = 12.0, 3.8 Hz, 2H), 4.10 (dd, J = 7.1, 4.8 Hz, 2H), 2.41 – 2.24 (m, 4H), 1.69 – 1.53 (m, 4H), 1.45 (s, 9H), 1.27 (s, 56H), 0.89 (t, J = 6.5 Hz, 6H). 13C NMR (126 MHz, 85% CDCl3-CD3OD): δ 173.85, 173.20, 165.83, 156.71, 154.58, 148.24, 145.05, 138.24, 134.53, 132.39, 131.00, 124.46, 77.82, 69.68, 62.41, 50.48, 35.15, 34.23, 33.94, 32.62, 32.16, 31.07, 29.92, 29.72, 29.59, 29.35, 29.25, 28.43, 26.25, 25.84, 25.07, 24.96, 22.90, 14.19. HRMS-DART [M+H]+ calcd for C55H94N6O9, 983.72, found 983.73. −3.33° (c 1, CHCl3).
ADIBO–Nitrophenyl–Lipid (1)
In a 50 mL round bottom flask, compound 13 (42.6 mg, 0.113 mmol) was dissolved in 15 mL dichloromethane. To this solution was added hydroxybenzotriazole (HOBt, 15.3 mg, 0.113 mmol) and O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU, 42.8 mg, 0.113 mmol). After 30 min, a mixture containing ADIBO 449 dissolved in 10 mL dichloromethane with diisopropylethylamine (92.6 μL, 0.566 mmol) was added to the reaction mixture, which was then allowed to stir at rt under nitrogen atmosphere overnight. The next day, the crude mixture was passed through celite and then washed with 50 mL water. The collected aqueous phase was next washed with dichloromethane (3 × 20 mL). The organic layers were then combined and dried with magnesium sulfate. After filtration and concentration, column chromatography was carried out through gradient elution with 7–10% methanol-dichloromethane, which provided lipid 1 as a yellow-white solid (53.4 mg, 38% yield). 1H NMR (500 MHz, 85% CDCl3-CD3OD): δ 8.79 (t, J = 5.7 Hz, 1H), 8.54 (d, J = 1.9 Hz, 1H), 8.05 (d, J = 12.7 Hz, 1H), 7.79 (s, 1H), 7.67 (d, J = 7.6 Hz, 1H), 7.61 (d, J = 8.1 Hz, 1H), 7.42 – 7.37 (m, 3H), 7.34 – 7.26 (m, 3H), 6.89 – 6.85 (m, 1H), 5.43 – 5.37 (m, 1H), 5.16 – 5.11 (m, 1H), 4.73 – 4.68 (m, 2H), 4.65 – 4.57 (m, 4H), 4.33 (dd, J = 12.2, 4.2 Hz, 1H), 4.08 (dd, J = 12.2, 5.4 Hz, 1H), 3.37 (dq, J = 4.2, 1.4, 0.9 Hz, 1H), 3.26 – 3.21 (m, 1H), 3.15 (m, 2H), 2.36 – 2.28 (m, 2H), 2.05 – 1.98 (m, 4H), 1.58 (m, 4H), 1.41 (dd, J = 10.1, 3.7 Hz, 4H), 1.26 (s, 56H), 0.88 (t, J = 6.8 Hz, 6H). 13C NMR (151 MHz, 85% CDCl3-CD3OD): δ 173.64, 173.28, 172.97, 172.60, 172.25, 171.95, 165.55, 151.05, 148.17, 147.95, 144.92, 137.24, 134.35, 132.25, 132.13, 130.82, 129.15, 128.84, 128.67, 128.40, 128.06, 127.33, 125.74, 124.33, 124.25, 123.03, 122.59, 114.87, 107.80, 69.51, 62.24, 60.77, 55.74, 54.87, 50.35, 35.55, 35.08, 34.52, 34.12, 32.05, 31.71, 31.25, 29.83, 29.62, 24.91, 22.80, 14.16. MALDI-MS: [M+Na]+ calcd for C72H104N8O10, 1263.78; found 1263.77. +1.2° (c 1, CHCl3).
(S)-4-ethyl-3,14-dioxo-3,4,12,14-tetrahydro-1H-pyrano[3′,4′:6,7]indolizino[1,2- b]quinolin-4-yl 6-azidohexanoate) (20)
To 6-azidohexanoic acid (23) (1.13 g,7.20 mmol) dissolved in 30 mL dry dichloromethane in a 500 mL round-bottomed flask was added dimethylaminopyridine (0.88 g, 7.20 mmol), N,N′-diisopropylcarbodiimide (1.13 mL, 7.20 mmol) and Scandium(III) triflate (0.71 g, 1.44 mol). The reaction mixture was cooled to −8 °C. Lastly, (S)-(+)-Camptothecin (22) (0.83 g, 2.38 mmol) was added and the resulting mixture was allowed to stir for 18 h. The reaction mixture was washed with 0.1 N HCl and 1% (v/v) sodium bicarbonate solution. Residual water was dried over magnesium sulfate and then was passed through celite and filtered off. Compound 20 was then eluted with 100% ethyl acetate using normal phase silica gel column chromatography and obtained as a white solid (712 mg, 61% yield). 1H NMR (500 MHz, CDCl3): δ 8.40 (s, 1H), 8.22 (dd, J = 8.5, 0.8 Hz, 1H), 7.96 – 7.93 (m, 1H), 7.86 – 7.83 (m, 1H), 7.68 (m, 1H), 7.21 (s, 1H), 5.68 (d, J = 17.2 Hz, 1H), 5.41 (d, J = 17.2 Hz, 1H), 5.29 (dd, J = 3.5, 1.2 Hz, 2H), 3.22 (dt, J = 6.9, 2.7 Hz, 2H), 2.52 (m, 2H), 2.29 – 2.13 (m, 2H), 1.72 – 1.67 (m, 2H), 1.60 (m, 2H), 1.46 – 1.40 (m, 2H), 0.98 (t, J = 7.5 Hz, 3H). 13C NMR (151 MHz, CDCl3) δ 172.39, 167.52, 157.33, 152.34, 148.84, 146.21, 131.21, 129.52, 128.45, 128.22, 128.17, 128.04, 120.28, 95.89, 75.82, 67.10, 51.12, 49.91, 33.57, 31.84, 28.49, 26.03, 24.17. HRMS-DART [M+H]+ calcd for C26H25N5O5, 488.1933, found 488.1935.
Liposome preparation
Stock solutions were made of synthetic lipid 1, PC and biotin lipid 19 for microplate studies. Examples of each stock solution used are as follows: 80.0 mg lipid 1, 17.2 mg PC, and 3.90 mg 19 were each weighed into separate vials and dissolved in 500 μL chloroform to form stock solutions of 1.29 mM lipid 1, 1.67mM biotin-PE, and 44.7 mM PC. Using these stock solutions, 55.9 μL lipid 1 stock (2%), 5.40 μL biotin-PE stock (1%), and 78.2 μL PC stock (97%) were combined in a glass vial. For solution-phase fluorimeter studies, a representative sample included 31.0 μL of lipid 1 stock (4%) and 21.5 μL of PC stock (96%), while a control sample contained 22.4 μL of PC stock (100%). In each case, the chloroform solvent was removed under a stream of nitrogen and the resulting mixture was placed under vacuum overnight. The dried lipids were then hydrated by adding 500 μL of HEPES buffer (20 mM HEPES, 150 mM NaCl, pH 7.4), and rotated on a rotary evaporator for 60 min at 40 oC. The liposomes were then subjected to 10 freeze-thaw cycles and extruded at rt (21 times) to obtain uniform size and lamellarity using an extruder containing a 200 nm polycarbonate filter.
Solution phase fluorescence detection of liposome derivatization
A 2.40 mM solution of azido-coumarin 18 in dimethylsulfoxide was prepared. 4.00 μL of this solution were then diluted with 1.88 mL of HEPES buffer (20 mM HEPES, 150 mM NaCl, pH 7.4), placed into a cuvette and sealed with parafilm to minimize atmospheric exposure. After an initial fluorescence scan (λex = 350 nm; λem = 461 nm), 120 μL of control liposomal solution (2 mM, 100 % PC) was added and fluorescence readings were recorded for 45 min. The same procedure was followed for the study sample, except 120 μL of liposomal solution (2 mM, 96% PC and 4% lipid 1) were added, followed by fluorescence scanning over time.
Microplate fluorescence liposome derivatization studies
A 400 μM solution of azido-coumarin 18 in dimethylsulfoxide was prepared. In a 96-well streptavidin-coated microplate, 200 μL of wash buffer (0.5X PBS, pH 7.4, 0.56 mM phosphates) was added to each row to be used. The plates were then shaken for 30 min, after which the wash buffer was removed. Next, 0, 1, 2.5, 5.00, 7.50, 10.0, 15.0, 20.0, and 25.0 μL of liposome solution (2 mM, 97% PC, 2% lipid 1 and 1% 19) and 187.5, 198.5, 169.3, 192.3, 188.8, 177.5, 170, 162.5 μL HEPES buffer (20 mM HEPES, 150 mM NaCl, pH 7.4) were added to the appropriate wells to produce final liposome concentrations of 0, 10, 25, 50, 75, 100, 150, 200 and 250 μM in separate wells. In addition, 12.5 (control), 0.50, 1.25, 2.50, 3.75, 5.00, 7.50, 10.0, 12.5 μL azido-coumarin 18 solution (5 equivalents, 400 μM solution) were added to each well to a total volume of 200 μL. These solutions were incubated, with shaking, for 3 h at rt. Following incubation, the solutions were removed and each well was washed with 3 × 200 μL wash buffer and 200 μL HEPES buffer (20 mM HEPES, 150 mM NaCl, pH 7.4) was then added. Lastly, fluorescence was measured using a microplate reader with a 360 nm (± 40 nm) excitation filter and 460 nm (± 40 nm) emission filters.
Saccharomyces cerevisiae cells
A YPD (yeast extract peptone dextrose) agar plate was inoculated with wild type S. cerevisiae strain S 288 and incubated at 30 °C for 24 h. Next, 5 mL of YPD media were inoculated with a single colony obtained from the agar plate and incubated on a shaker at 300 rpm at 30 °C with good aeration for 15 h. Next, 500 μL aliquots from this culture were transferred into two detergent-free 250 mL Erlenmeyer flask containing 25 mL of fresh YPD medium with (study) and without (control) lipid analog 1 (100 μM in dimethylsulfoxide, 300 μL), both of which were incubated on a shaker at 300 rpm at 30 °C for 16 to 17 h. For good aeration, the medium constituted of no more than one-fifth of the total flask volume. Thereafter, the cell concentration was adjusted spectrophotometrically to approximately 106 CFU/mL. Finally, 1 mL of the cell solution was transferred into eppendorf tube, centrifuged at 9000 rpm for 4 min, and then stored at −80 °C.
Fluorescence Imaging and Flow Cytometry Analysis of Fluorescence Labeling and Photocleavage using Saccharomyces cerevisiae cells
Two 1 mL samples of S. cerevisiae cells, one grown with compound 1 (study) and one untreated (control), were incubated with 100 μM Azide-Fluor 488 (14) in 300 μL of dimethylsulfoxide (DMSO) solution overnight at rt. Next, the cell samples were centrifuged for 3 min at 2500 rpm, the dye solutions were removed, and the cells were washed once with 5% DMSO in water and twice with 20% DMSO in water, with vortexing and centrifugation at 2500 rpm for 3 min performed in between each washing step. Next, 1 mL ice-cold methanol was added to each tube, and after incubating for 10 min at 0 °C, the sample was centrifuged. Next, 1 mL of Phosphate Buffered Saline solution (PBS, pH 7.4, 1.19 mM phosphates) was used to wash the fixed cell samples, which were then centrifuged at 2500 rpm for 3 min. Cells were then subjected to fluorescence microscopy. To assess photocleavage, 200 μL of clicked cell sample were added into a disposable cuvette and 800 μL HEPES buffer (20 mM HEPES, 150 mM NaCl, pH 7.4), were added. For photocleavage, cells were irradiated with 350 nm light while suspended in a Pyrex beaker between four 350 nm bulbs in a Rayonet Preparative Type RS photoreactor. This was done for different time periods (1, 2, 3, and 12 h) to assess photocleavage over time. After photoirradiation, the cell samples in buffer were transferred into eppendorf tubes and centrifuged at 2500 rpm for 3 min, and then washed 7 times with 1 mL of 20 % DMSO/water. Each time they were vortexed and centrifuged at 9000 rpm for 4 min. Lastly, the cells were washed with 1 mL of 1X PBS buffer (pH 7.4, 1.19 mM phosphate buffer), and fluorescence microscope images after photocleavage were then obtained. For flow cytometry analysis, cell samples were analyzed using a BD FACS (Fluorescence Activated Cell Sorting) Calibur flow cytometer (Becton, Dickinson and Company, New Jersey, US) equipped with two lasers (488 nm and 635 nm). Green fluorescence (FL-1) was measured for all cell samples. Populations of the yeast cells were identified and gated according to the fluorescent intensity. Data acquisition was performed and analyzed using FlowJo software. For each sample analyzed in the flow cytometer, at least 10,000 events were acquired.
Cell proliferation assay (MTS)
HeLa cells were cultured in a humidified incubator under 5% CO2 in DMEM medium (Invitrogen) supplemented with 10% of Fetal Bovine Serum (Invitrogen). The cell viability was measured using the CellTiter 96 ®Aqueous One Solution (Promega) according to the manufacturer’s protocol. Briefly, cells were seeded (104 cells per well) 2 days prior to the experiments to a 96 well plate, and exposed to vehicle (DMSO and liposomes), CPT, CPT-N3 or CPT-N3 + Liposomes mix at different concentrations of CPT, CPT-N3 (10 μM, 25 μM and 50 μM) and incubated for 48 hrs. The liposomal solution consisted of 50 μM lipid analog 1 formed as 8.5 mM liposomes (94% PC, 6% lipid analog 1). The MTS assay was performed in 100 μL of DMEM phenol red free medium (Invitrogen) in each well and 20 μL of the CellTiter solution was added to the samples, after which the plate was placed in a 37 °C incubator with 5% CO2 until it reached the desired color. The absorbance at 490 nm was measured in a plate reader (Synergy 2, Biotek). The results are representative of 3 independent experiments, performed in quadruplicate. The inhibition of cell proliferation was expressed as the percentage of vehicle control (0.01% DMSO in the culture medium for CPT).
Supplementary Material
Acknowledgments
This material is based upon work supported by the National Science Foundation under Grant Number CHE-0954297. We acknowledge assistance from Dr. Shawn Campagna with the growing of cells, Dr. Jon Dunlap with fluorescence imaging experiments, and Dr. Tim Sparer for flow cytometry analysis.
Footnotes
Supporting Information Available: Spectra for characterizing synthetic compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
Literature Cited
- 1.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 2.Brochu H, Polidori A, Pucci B, Vermette P. Drug delivery systems using immobilized intact liposomes: A comparative and critical review. Curr Drug Deliv. 2004;1:299–312. doi: 10.2174/1567201043334678. [DOI] [PubMed] [Google Scholar]
- 3.Ferrara KW, Borden MA, Zhang H. Lipid-shelled vehicles: Engineering for ultrasound molecular imaging and drug delivery. Acc Chem Res. 2009;42:881–892. doi: 10.1021/ar8002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vermette P, Meagher L, Gagnon E, Griesser HJ, Doillon CJ. Immobilized liposome layers for drug delivery applications: inhibition of angiogenesis. J Cont Rel. 2002;80:179–195. doi: 10.1016/s0168-3659(02)00023-8. [DOI] [PubMed] [Google Scholar]
- 5.Asai T, Tsuzuku T, Takahashi S, Okamoto A, Dewa T, Nango M, Hyodo K, Ishihara H, Kikuchi H, Oku N. Cell-penetrating peptide-conjugated lipid nanoparticles for siRNA delivery. Biochem Biophys Res Commun. 2014;444:599–604. doi: 10.1016/j.bbrc.2014.01.107. [DOI] [PubMed] [Google Scholar]
- 6.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: Fishing for selectivity in a sea of functionality. Angew Chem, Int Edit. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Best MD. Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry. 2009;48:6571–6584. doi: 10.1021/bi9007726. [DOI] [PubMed] [Google Scholar]
- 8.Debets MF, van der Doelen CWJ, Rutjes F, van Delft FL. Azide: A unique dipole for metal-free bioorthogonal ligations. Chembiochem. 2010;11:1168–1184. doi: 10.1002/cbic.201000064. [DOI] [PubMed] [Google Scholar]
- 9.Mamidyala SK, Finn MG. In situ click chemistry: probing the binding landscapes of biological molecules. Chem Soc Rev. 2010;39:1252–1261. doi: 10.1039/b901969n. [DOI] [PubMed] [Google Scholar]
- 10.Moses JE, Moorhouse AD. The growing applications of click chemistry. Chem Soc Rev. 2007;36:1249–1262. doi: 10.1039/b613014n. [DOI] [PubMed] [Google Scholar]
- 11.Salisbury CM, Cravatt BF. Click chemistry-led advances in high content functional proteomics. QSAR Comb Sci. 2007;26:1229–1238. [Google Scholar]
- 12.Best MD. The Development and Application of Clickable Lipid Analogs for Elucidating and Harnessing Lipid Function. In: Bielski ZJWaR., editor. Click Chemistry in Glycoscience: New Developments and Strategies. John Wiley & Sons; Hoboken, NJ: 2013. pp. 79–105. [Google Scholar]
- 13.Best MD, Rowland MM, Bostic HE. Exploiting bioorthogonal chemistry to elucidate protein–lipid binding interactions and other biological roles of phospholipids. Acc Chem Res. 2011;44:686–698. doi: 10.1021/ar200060y. [DOI] [PubMed] [Google Scholar]
- 14.Hassane FS, Frisch B, Schuber F. Targeted liposomes: Convenient coupling of ligands to preformed vesicles using “click chemistry”. Bioconjugate Chem. 2006;17:849–854. doi: 10.1021/bc050308l. [DOI] [PubMed] [Google Scholar]
- 15.Cavalli S, Tipton AR, Overhand M, Kros A. The chemical modification of liposome surfaces via a copper-mediated 3+2 azide-alkyne cycloaddition monitored by a colorimetric assay. Chem Commun. 2006;30:3193–3195. doi: 10.1039/b606930d. [DOI] [PubMed] [Google Scholar]
- 16.van Lengerich B, Rawle RJ, Boxer SG. Covalent attachment of lipid vesicles to a fluid-supported bilayer allows observation of DNA-mediated vesicle interactions. Langmuir. 2010;26:8666–8672. doi: 10.1021/la904822f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loosli F, Doval DA, Grassi D, Zaffalon PL, Favarger F, Zumbuehl A. Clickosomes—using triazole-linked phospholipid connectors to fuse vesicles. Chem Commun. 2012;48:1604–1606. doi: 10.1039/c2cc16827h. [DOI] [PubMed] [Google Scholar]
- 18.Ma Y, Zhang HL, Sun XL. Surface-bound cytomimetic assembly based on chemoselective and biocompatible immobilization and further modification of intact liposome. Bioconjugate Chem. 2010;21:1994–1999. doi: 10.1021/bc100220j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheng RL, Luo T, Li H, Sun JJ, Wang Z, Cao A. ‘Click’ synthesized sterol-based cationic lipids as gene carriers, and the effect of skeletons and headgroups on gene delivery. Bioorg Med Chem. 2013;21:6366–6377. doi: 10.1016/j.bmc.2013.08.047. [DOI] [PubMed] [Google Scholar]
- 20.Kumar A, Erasquin UJ, Qin GT, Li K, Cai CZ. “Clickable”, polymerized liposomes as a versatile and stable platform for rapid optimization of their peripheral compositions. Chem Commun. 2010;46:5746–5748. doi: 10.1039/c0cc00784f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bostic HE, Smith MD, Poloukhtine AA, Popik VV, Best MD. Membrane labeling and immobilization via copper-free click chemistry. Chem Commun. 2012;48:1431–1433. doi: 10.1039/c1cc14415d. [DOI] [PubMed] [Google Scholar]
- 22.Zhang HL, Weingart J, Jiang R, Peng JH, Wu QY, Sun XL. Bio-inspired liposomal thrombomodulin conjugate through bio-orthogonal chemistry. Bioconjugate Chem. 2013;24:550–559. doi: 10.1021/bc300399f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emmetiere F, Irwin C, Viola-Villegas NT, Longo V, Cheal SM, Zanzonico P, Pillarsetty N, Weber WA, Lewis JS, Reiner T. F-18-Labeled-bioorthogonal liposomes for in vivo targeting. Bioconjugate Chem. 2013;24:1784–1789. doi: 10.1021/bc400322h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alvarez-Lorenzo C, Bromberg L, Concheiro A. Light-sensitive Intelligent drug delivery systems. Photochem Photobiol. 2009;85:848–860. doi: 10.1111/j.1751-1097.2008.00530.x. [DOI] [PubMed] [Google Scholar]
- 25.Yavlovich A, Smith B, Gupta K, Blumenthal R, Puri A. Light-sensitive lipid-based nanoparticles for drug delivery: design principles and future considerations for biological applications. Mol Mem Biol. 2010;27:364–381. doi: 10.3109/09687688.2010.507788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Randles EG, Bergethon PR. A Photodependent Switch of Liposome Stability and Permeability. Langmuir. 2013;29:1490–1497. doi: 10.1021/la303526k. [DOI] [PubMed] [Google Scholar]
- 27.Leung SJ, Romanowski M. Light-Activated Content Release from Liposomes. Theranostics. 2012;2:1020–1036. doi: 10.7150/thno.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ong W, Yang YM, Cruciano AC, McCarley RL. Redox-Triggered Contents Release from Liposomes. J AM Chem Soc. 2008;130:14739–14744. doi: 10.1021/ja8050469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andresen TL, Davidsen J, Begtrup M, Mouritsen OG, Jorgensen K. Enzymatic release of antitumor ether lipids by specific phospholipase A2 activation of liposome-forming prodrugs. J Med CHem. 2004;47:1694–1703. doi: 10.1021/jm031029r. [DOI] [PubMed] [Google Scholar]
- 30.Koning GA, Eggermont AMM, Lindner LH, ten Hagen TLM. Hyperthermia and Thermosensitive Liposomes for Improved Delivery of Chemotherapeutic Drugs to Solid Tumors. Pharm Res. 2010;27:1750–1754. doi: 10.1007/s11095-010-0154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pradhan P, Giri J, Rieken F, Koch C, Mykhaylyk O, Doeblinger M, Banerjee R, Bahadur D, Plank C. Targeted temperature sensitive magnetic liposomes for thermo-chemotherapy. J Cont Rel. 2010;142:108–121. doi: 10.1016/j.jconrel.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Kono K, Ozawa T, Yoshida T, Ozaki F, Ishizaka Y, Maruyama K, Kojima C, Harada A, Aoshima S. Highly temperature-sensitive liposomes based on a thermosensitive block copolymer for tumor-specific chemotherapy. Biomaterials. 2010;31:7096–7105. doi: 10.1016/j.biomaterials.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 33.Ranjan A, Jacobs GC, Woods DL, Negussie AH, Partanen A, Yarmolenko PS, Gacchina CE, Sharma KV, Frenkel V, Wood BJ, et al. Image-guided drug delivery with magnetic resonance guided high intensity focused ultrasound and temperature sensitive liposomes in a rabbit Vx2 tumor model. J Cont Rel. 2012;158:487–494. doi: 10.1016/j.jconrel.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kale AA, Torchilin VP. Design, synthesis, and characterization of pH-sensitive PEG-PE conjugates for stimuli-sensitive pharmaceutical nanocarriers: The effect of substitutes at the hydrazone linkage on the pH stability of PEG-PE conjugates. Bioconjugate Chem. 2007;18:363–370. doi: 10.1021/bc060228x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim IY, Kang YS, Lee DS, Park HJ, Choi EK, Oh YK, Son HJ, Kim JS. Antitumor activity of EGFR targeted pH-sensitive immunoliposomes encapsulating gemcitabine in A549 xenograft nude mice. J Cont Rel. 2009;140:55–60. doi: 10.1016/j.jconrel.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 36.Koren E, Apte A, Jani A, Torchilin VP. Multifunctional PEGylated 2C5- immunoliposomes containing pH-sensitive bonds and TAT peptide for enhanced tumor cell internalization and cytotoxicity. J Cont Rel. 2012;160:264–273. doi: 10.1016/j.jconrel.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Y, Chen CK, Law WC, Weinheimer E, Sengupta S, Prasad PN, Cheng C. Polylactide-graft-doxorubicin nanoparticles with precisely controlled drug loading for pH-triggered drug delivery. Biomacromolecules. 2014;15:524–532. doi: 10.1021/bm401471p. [DOI] [PubMed] [Google Scholar]
- 38.Pelliccioli AP, Wirz J. Photoremovable protecting groups: reaction mechanisms and applications. Photochem Photobiol Sci. 2002;1:441–458. doi: 10.1039/b200777k. [DOI] [PubMed] [Google Scholar]
- 39.Zhang ZY, Smith BD. Synthesis and characterization of NVOC-DOPE, a caged photoactivatable derivative of dioleoylphosphatidylethanolamine. Bioconjugate Chem. 1999;10:1150–1152. doi: 10.1021/bc990087h. [DOI] [PubMed] [Google Scholar]
- 40.Nagasaki T, Taniguchi A, Tamagaki S. Photoenhancement of transfection efficiency using novel cationic lipids having a photocleavable spacer. Bioconjugate Chem. 2003;14:513–516. doi: 10.1021/bc0256603. [DOI] [PubMed] [Google Scholar]
- 41.Chandra B, Subramaniam R, Mallik S, Srivastava DK. Formulation of photocleavable liposomes and the mechanism of their content release. Org Biomol Chem. 2006;4:1730–1740. doi: 10.1039/b518359f. [DOI] [PubMed] [Google Scholar]
- 42.Chandra B, Mallik S, Srivastava DK. Design of photocleavable lipids and their application in liposomal “uncorking”. Chem Commun. 2005:3021–3023. doi: 10.1039/b503423j. [DOI] [PubMed] [Google Scholar]
- 43.Subramaniam R, Xioa Y, Li Y, Qian SY, Sun W, Mallik S. Light-mediated and H-bond facilitated liposomal release: the role of lipid head groups in release efficiency. Tetrahedron Lett. 2010;51:529–532. [Google Scholar]
- 44.Wan YQ, Angleson JK, Kutateladze AG. Liposomes from novel photolabile phospholipids: Light-induced unloading of small molecules as monitored by PFG NMR. J Am Chem Soc. 2002;124:5610–5611. doi: 10.1021/ja016874i. [DOI] [PubMed] [Google Scholar]
- 45.Bayer AM, Alam S, Mattern-Schain SI, Best MD. Triggered liposomal release through a synthetic phosphatidylcholine analogue bearing a photocleavable moiety embedded within the sn-2 acyl chain. Chem Eur J. 2014;20:3350–3357. doi: 10.1002/chem.201304094. [DOI] [PubMed] [Google Scholar]
- 46.Nelson WL, Wennerstrom JE, Sankar SR. Absolute-configuration of glycerol derivatives. 3 Synthesis and cupra a circular-dichroism spectra of some chiral 3- aryloxy-1,2-propanediols and 3-aryloxy-1-amino-2-propanols. J Org Chem. 1977;42:1006– 1012. doi: 10.1021/jo00426a016. [DOI] [PubMed] [Google Scholar]
- 47.Losey EA, Smith MD, Meng M, Best MD. Microplate-based analysis of protein-membrane interactions via immobilization of whole liposomes containing a biotinylated anchor. Bioconjugate Chem. 2009;20:376–383. doi: 10.1021/bc800414k. [DOI] [PubMed] [Google Scholar]
- 48.Grandjean C, Boutonnier A, Guerreiro C, Fournier JM, Mulard LA. On the preparation of carbohydrate-protein conjugates using the traceless Staudinger ligation. J Org Chem. 2005;70:7123–7132. doi: 10.1021/jo0505472. [DOI] [PubMed] [Google Scholar]
- 49.Pathak RK, McKnitt CD, Popik VV, Dhar S. Copper-free click-chemistry platform to functionalize cisplatin prodrugs. Chem Eur J. 2014;20:6861–6865. doi: 10.1002/chem.201402573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuzmin A, Poloukhtine A, Wolfert MA, Popik VV. Surface functionalization using catalyst-free azide-alkyne cycloaddition. Bioconjugate Chem. 2010;21:2076–2085. doi: 10.1021/bc100306u. [DOI] [PubMed] [Google Scholar]
- 51.Debets MF, van Berkel SS, Schoffelen S, Rutjes F, van Hest JCM, van Delft FL. Aza-dibenzocyclooctynes for fast and efficient enzyme PEGylation via copper-free (3+2) cycloaddition. Chem Commun. 2010;46:97–99. doi: 10.1039/b917797c. [DOI] [PubMed] [Google Scholar]
- 52.Ning XH, Guo J, Wolfert MA, Boons GJ. Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast huisgen cycloadditions. Angew Chem, Int Edit. 2008;47:2253–2255. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Neil EJ, DiVittorio KM, Smith BD. Phosphatidylcholine-derived bolaamphiphiles via click chemistry. Org Lett. 2007;9:199–202. doi: 10.1021/ol062557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cho WW, Bittova L, Stahelin RV. Membrane binding assays for peripheral proteins. Anal Biochem. 2001;296:153–161. doi: 10.1006/abio.2001.5225. [DOI] [PubMed] [Google Scholar]
- 55.Narayan K, Lemmon MA. Determining selectivity of phosphoinositide-binding domains. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan YHM, Boxer SG. Model membrane systems and their applications. Curr Opin Chem Biol. 2007;11:581–587. doi: 10.1016/j.cbpa.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jelinek R, Silbert L. Biomimetic approaches for studying membrane processes. Mol Biosyst. 2009;5:811–818. doi: 10.1039/b907223n. [DOI] [PubMed] [Google Scholar]
- 58.Sivakumar K, Xie F, Cash BM, Long S, Barnhill HN, Wang Q. A fluorogenic 1,3-dipolar cycloaddition reaction of 3-azidocoumarins and acetylenes. Org Lett. 2004;6:4603–4606. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]
- 59.Smith MD, Best MD. Characterization of protein-membrane binding interactions via a microplate assay employing whole liposome immobilization. In: Mark SS, editor. Bioconjugation Protocols: Strategies and Methods. 2. Humana Press Inc; Totowa: 2011. pp. 477–489. [DOI] [PubMed] [Google Scholar]
- 60.O’Leary J, Muggia FM. Camptothecins: a review of their development and schedules of administration. Eur J Cancer. 1998;34:1500–1508. doi: 10.1016/s0959-8049(98)00229-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.