Abstract
Detoxicating enzymes NAD(P)H:quinone oxidoreductase 1 (NQO1) and NRH:quinone oxidoreductase 2 (NQO2) catalyze the two-electron reduction of quinone-like compounds. The protective role of the polymorphic NQO1 and NQO2 enzymes is especially of interest in the liver as the major site of drug bioactivation to chemically reactive drug metabolites. In the current study, we quantified the concentrations of NQO1 and NQO2 in 20 human liver donors and NQO1 and NQO2 activities with quinone-like drug metabolites. Hepatic NQO1 concentrations ranged from 8 to 213 nM. Using recombinant NQO1, we showed that low nM concentrations of NQO1 are sufficient to reduce synthetic amodiaquine and carbamazepine quinone-like metabolites in vitro. Hepatic NQO2 concentrations ranged from 2 to 31 μM. NQO2 catalyzed the reduction of quinone-like metabolites derived from acetaminophen, clozapine, 4′-hydroxydiclofenac, mefenamic acid, amodiaquine, and carbamazepine. The reduction of the clozapine nitrenium ion supports association studies showing that NQO2 is a genetic risk factor for clozapine-induced agranulocytosis. The 5-hydroxydiclofenac quinone imine, which was previously shown to be reduced by NQO1, was not reduced by NQO2. Tacrine was identified as a potent NQO2 inhibitor and was applied to further confirm the catalytic activity of NQO2 in these assays. While the in vivo relevance of NQO2-catalyzed reduction of quinone-like metabolites remains to be established by identification of the physiologically relevant co-substrates, our results suggest an additional protective role of the NQO2 protein by non-enzymatic scavenging of quinone-like metabolites. Hepatic NQO1 activity in detoxication of quinone-like metabolites becomes especially important when other detoxication pathways are exhausted and NQO1 levels are induced.
Introduction
The human quinone reductase family consists of two members, NAD(P)H:quinone oxidoreductase 1 (NQO1, QR1, or DT diaphorase) and NRH:quinone oxidoreductase 2 (NQO2 or QR2).1 NQO1 and NQO2 are FAD bound proteins and share significant structural similarity.2 Both enzymes catalyze the two-electron reduction of quinone-like species to facilitate subsequent enzymatic conjugation and cellular excretion, thereby surpassing the one-electron reduction to toxic semiquinone-like radicals.3,4 Reduction of substrates is enabled by a so-called ping-pong mechanism in which both the co-substrate and substrate sequentially bind to the active site resulting in FAD molecule reduction and subsequent electron transfer to the substrate.5,6 Both NQO1 and NQO2 are involved in the bioactivation of antitumor drugs, like mitomycin C and CB1954, to reactive hydroquinones.7,8 A major difference between NQO1 and NQO2 is the preference for co-substrates. In contrast to NOQ1, which utilizes NADH or NADPH as electron donor, NQO2 requires dihydro-nicotinamide riboside (NRH).4 In addition, the potent NQO1 inhibitors dicoumarol and ES936 only weakly inhibit NQO2, while potent NQO2 inhibitors, such as resveratrol and quercetin, have significantly less affinity toward NQO1.4,9−11
NQO1 is generally considered to be a detoxicating enzyme because of its activity in reduction of drug-derived quinones imines.12−14 It has been shown that NQO1 protects against menadione- and benzo[a]pyrene-3,6-quinone-induced (cyto)toxicity in NQO1-overexpressing Chinese hamster ovary cells and in transgenic mice.15,16 However, recently it was shown that menadione and aminochrome cytotoxicity was augmented in NQO1-overexpressing rat neural cells.17 The role of NQO2 as a protective enzyme has been less well studied. It was shown in vitro that NQO2 generates superoxide anions following acetaminophen binding.18 However, while menadione toxicity is decreased in NQO2-null mice, in vitro experiments indicated that NQO2 reduced reactive oxygen species formation by reduction of menadione to the more stable hydroquinone.19−21 Furthermore, two groups reported an association between a mutation in the NQO2 gene, resulting in lower levels of enzyme, and clozapine-induced agranulocytosis.22,23 Therefore, as for many drug metabolizing enzymes, available data suggests both toxifying and detoxicating properties for NQO1 and NQO2. It has recently been hypothesized that the balance between toxicity of and protection against quinones is largely determined by the intrinsic reactivity or the cellular capacity of enzymatic conjugation of the resulting hydroquinone.21
Formation of quinone-like species may result from metabolism of endogenous products such as vitamin K and xenobiotics, including drugs.24,25 A major site of exposure to drug-derived quinone-like species is the liver, since xenobiotics can be bioactivated by cytochromes P450 (CYPs) to chemically reactive metabolites including quinone-like metabolites. Basal levels of NQO1 in human liver are low, however, expression is increased in liver cancers and is strongly inducible via Nrf2 regulation.26−28 NQO2, which is independently regulated, is predominantly expressed in human liver, and Nrf2-mediated induction is weaker.18,28−30
In the current study, we have used recombinant enzymes to translate specific NQO1 and NQO2 activities in human liver cytosol from 20 donors into expression levels. Enzymatic activity and enzyme concentration dependency of NQO2 in reduction of quinone-like metabolites which are generated by bioactivation of acetaminophen, clozapine, diclofenac, and mefenamic acid was examined. NQO1- and NQO2-mediated reduction of synthetically prepared amodiaquine- and carbamazepine-derived reactive metabolites is also investigated with a range of enzyme concentrations. Conventional NQO2 inhibitors could not be applied in the current incubations because of interference with CYP activity. Instead tacrine, which was identified as a potent NQO2 inhibitor in in-house screening assays, was used to further confirm the enzymatic activity of NQO2. Together, these results demonstrate the potential of NQO1 and NQO2 as detoxicating enzymes against drug-induced liver injury.
Materials and Methods
Materials
Pooled human liver microsomes (200 donors, lot no. 1210347) were purchased from Xenotech (Lenexa, USA). Bovine serum albumin (BSA), 2,6-dichlorophenolindophenol (DCPIP), dicoumarol, glutathione (GSH), acetaminophen, mefenamic acid, resorufin, tacrine, and iminostilbene were from Sigma-Aldrich (Steinheim, Germany). 4′- and 5-Hydroxy diclofenac were obtained from Toronto Research Chemicals (North York, Canada. Amodiaquine dihydrochloride was obtained from INC Biomedicals (Aurora, OH, USA), and N-desethylamodiaquine was obtained from BD Biosciences (Franklin Lakes, NJ, USA). Clozapine was obtained from Duchefa Farma (Haarlem, The Netherlands). 1-Benzyl-1,4-dihydronicotinamide (BNAH) was obtained from TCI Europe (Zwijndrecht, Belgium). All other reagents and chemicals were of analytical grade and purchased from standard commercial suppliers.
Expression and Purification of Recombinant Human NQO2
NQO2 DNA was obtained by PCR amplification from the human cDNA ORF clone (SC319150, obtained from OriGene Technologies, Rockville, MD) using the forward primer 5′-GCAACATATGGCAGGTAAGAAAGTAC-3′ and the reverse primer 5′-GTAGAATTCATGCCCACGTGCCACAGAG-3′ to add an EcoRI site and a NdeI site at the 5′ and 3′ end of the cDNA, respectively. The amplified NQO2 fragment was gel-purified, cut with restriction enzymes EcoRI and NdeI, and inserted into the 6His-encoding pET28(a)+ vector cleaved with the same enzymes. The final construct encoding His-tagged human NQO2 was monitored by sequencing and subsequently transformed into competent E. coli BL21 cells. Expression and purification of recombinant human NQO1 and NQO2 were performed as described previously.12
Specific activities of NQO1 and NQO2 were determined using 6.25 nM NQO1 or 100 nM NQO2, 40 μM DCPIP, and 1 mM NADPH (NQO1) or 250 μM NRH (NQO2, synthesized as described below). Incubations were performed in 100 mM KPi buffer containing 0.18 mg/mL BSA, 2 mM EDTA, and 5 mM MgCl2 (pH 7.4). Reactions were carried out at 24 °C, and DCPIP reduction was monitored at 600 nm. For NQO2, activities were corrected for chemical reduction of DCPIP by NRH. Specific activities for NQO1 and NQO2 were 295 ± 18 and 7.6 ± 0.6 μmol DCPIP/min/mg protein, respectively. NQO1-specific activity was in similar range of previous reports.31,32
Synthesis of NRH
NRH was synthesized as described previously with some modifications.33 NADH (1 g, 1.5 mmol) was incubated with 0.2 units of phosphodiesterase 1 (Sigma-Aldrich, Steinheim, Germany) and 1000 units of alkaline phosphatase (Sigma-Aldrich, Steinheim, Germany) in 40 mL of 0.4 M sodium carbonate/bicarbonate buffer (pH 10.0) for 16 h at 37 °C. The incubation was then freeze-dried, and the powder was extracted five times with 12 mL methanol. The combined methanol fractions were dried by rotary evaporation, and the products were dissolved in 10 mL of water. The solution was loaded in fractions of 0.25 mL into a preparative HPLC equipped with a Waters XBridge prep 5 μm C18 (50 × 10 mm) column and eluted with 55% MeOH at a flow rate of 4.5 mL/min. NRH was detected at 335 nm and eluted at 13.6 min. The NRH peak was collected, freeze-dried, and stored at −80 °C until use.
Preparation of Liver Cytosol
Cytosol was prepared from healthy liver tissue from 20 donors (Den Braver-Sewradj et al., submitted) as follows: Tissue samples were suspended in two volumes ice-cold 100 mM KPi buffer (pH 7.4) containing 125 mM NaCl and 1 mM EDTA and homogenized using an Omni Mixer (Soryall, Inc.), followed by further homogenization by a Potter-Elvehjem (Sigma-Aldrich, Steinheim, Germany) homogenizer. The homogenate was centrifuged for 20 min at 9000g. The supernatant was subsequently centrifuged for 1 h at 100,000g. The cytosolic fraction (supernatant) was collected and dialyzed overnight at 4 °C against 100 mM KPi buffer (containing 125 mM NaCl and 1 mM EDTA, pH 7.4) using a 12–14 kDa exclusion size tubing (Spectrum Europe, Breda, The Netherlands) and stored at −80 °C until use. The protein contents in cytosolic fractions were determined using the bicinchoninic acid method with BSA as standard (ThermoFisher Scientific, Bleiswijk, The Netherlands).
Determination of Human Hepatic NQO1 Concentrations
NQO1 activity in cytosol was measured as described before with minor modifications.34 Resorufin reduction was analyzed in 96-well format at 37 °C. 500 nM of resorufin (0.1% DMSO final) was pre-incubated with 100 μM of NADH in 100 mM KPi buffer containing 0.18 mg/mL BSA, 2 mM EDTA, and 5 mM MgCl2 (pH 7.4). Reactions were started by addition of cytosol (5% v/v), and the decrease of resorufin was measured over time (λem 530 nm, λex 572 nm, PerkinElmer Victor3 1420 multilabel counter). Contribution of NQO1 to resorufin reduction was quantified by determining resofurin reduction in absence and presence of 10 μM of the specific NQO1 inhibitor dicoumarol. NQO1 activity was converted to NQO1 protein concentrations using a calibration curve constructed of purified recombinant NQO1 protein, which was incubated and analyzed under identical conditions except for the presence of cytosol (Figure S1A and B). Next, individual cytosolic protein contents (Table S1) and an intracellular hepatocyte volume of 0.55 mL/g liver were used as scaling factors for extrapolation of cytosolic to hepatic NQO1 concentrations.35
Determination of Human Hepatic NQO2 Concentrations
NQO2 activity in cytosol was measured by exploiting the selective ability of human NQO2 to reduce the drug CB1954. Measurements were based on previously described methods.36,37 Experimental conditions were pre-optimized with respect to linearity in time and with protein concentration. 100 μM of CB1954 was pre-incubated for 4 min at 37 °C with 5% cytosol in 100 mM KPi buffer containing 0.18 mg/mL BSA, 2 mM EDTA, and 5 mM MgCl2 (pH 7.4). Reactions were started by addition of 500 μM NRH and terminated after 10 min by addition of an equal volume ice-cold acetonitrile. Precipitated protein was removed by centrifugation (20 min 14,000 rpm), and parent consumption was quantified by HPLC. Fifteen μL of sample was injected on a Shimadzu Prominence LC-20 HPLC system, equipped with a Shimadzu SPD-20A UV–vis detector set at 325 nm. The analytes were separated on an Agilent XDB-C18 1.8 μm column (4.6 × 50 mm) protected by a Phenomenex security guard column (C18, 4.0 × 3.0 mm) as stationary phase and a binary gradient composed of solvent A (0.1% formic acid in water) and B (0.1% formic acid in methanol). The gradient was programmed as follows: 0 min, 20% B; 1 min 20% B; 8 min 99% B; 8.5 min, 20% B; 18 min, 20% B. The flow rate was 0.4 mL/min. Separate control incubations with NADPH excluded participation of NQO1 and nonspecific activity in CB1954 reduction. NQO2 activity was converted to NQO2 protein concentrations using a calibration curve based on recombinant NQO2 protein, which was incubated and analyzed under identical conditions except for the presence of cytosol (Figure S1C and D). Next, individual cytosolic protein contents (Table S1) and an intracellular hepatocyte volume of 0.55 mL/g liver were used as scaling factors for extrapolation of cytosolic to hepatic NQO2 concentrations.35
Reduction of Acetaminophen-, Diclofenac-, Mefenamic Acid, and Clozapine-Derived Reactive Metabolites by NQO2
Reduction of reactive metabolites by NQO2 was determined indirectly by quantifying the decrease of GSH conjugation. Reactive metabolites were generated by CYP-catalyzed oxidation using human liver microsomes as bioactivating system. Human liver microsomes (2 mg/mL) were pre-incubated with parent drug (250 μM acetaminophen or mefenamic acid, 100 μM clozapine, 50 μM 4′-hydroxy- or 5-hydroxydiclofenac, 0.5% DMSO final), GSH (500 μM), NQO2 (0–20 μM), and NRH (250 μM, when applicable). Incubations were performed in 100 mM KPi buffer containing 2 mM EDTA, 5 mM MgCl2, and 0.18 mg/mL BSA (pH 7.4) for 4 min at 37 °C. Reactions were started by addition of NADPH regenerating system (100 μM NADPH, 10 mM glucose-6-phosphate, and 0.5 U/mL glucose-6-phosphate dehydrogenase, final concentrations) and incubated for 30 min. Incubations with mefenamic acid and hydroxydiclofenac were stopped by addition of an equal volume of ice-cold methanol containing 1 mM ascorbic acid and 10 μM carbamazepine as analytical internal standard. Incubations with clozapine were terminated by addition of an equal volume ice-cold methanol containing 2% perchloric acid and 10 μM carbamazepine as analytical internal standard. Acetaminophen incubations were terminated with an equal volume ice-cold methanol. Precipitated protein was removed by centrifugation at 14,000 rpm for 15 min, and supernatants were analyzed using HPLC or LC/MS. Acetaminophen, clozapine, and their metabolites were separated on a Phenomenex Luna 5 μm C18 column (4.6 × 150 mm) protected by a Phenomenex security guard (5 μm, 4.0 × 3.0 mm). Binary gradients were used mixing solvent A (1% acetonitrile, 0.2% formic acid) and solvent B (99% acetonitrile, 0.2% formic acid) at 0.5 mL/min. Acetaminophen incubations were analyzed on HPLC (Shimadzu Prominence LC-20 coupled to a Shimadzu SPD-20A UV–vis detector set at 254 nm) using the following gradient: 0 min, 20% B; 5 min, 20% B; 30 min, 99% B; 30.5 min, 20% B; 40 min, 20% B. Clozapine was analyzed by LC/MS using an Agilent 1200 Series Rapid Resolution LC system connected to a Agilent 6230 time-of-flight (TOF) mass spectrometer. The TOF was operating in positive ion electrospray mode, and MS conditions were as follows: capillary voltage, 3500 V; drying gas (N2), 10 l/min; desolvation gas (N2), 50 psig; source temperature, 350 °C; 2 GHz, extended dynamic range mode. The following gradient was used to separate clozapine and metabolites: 0 min, 1% B; 5 min, 1% B; 30 min, 80% B; 30.5 min, 1% B; 40 min, 1% B. Hydroxydiclofenac and mefenamic acid incubations were analyzed on LC-MS as described previously.38,39 Quantification of GSH conjugates was performed with authentic references which were synthesized by the method of den Braver et al.39,40
Synthesis of Amodiaquine-Derived Quinone Imines
Synthesis of amodiaquine quinone imines was adapted from Harrison et al.41 Briefly, 5 equiv of freshly prepared silver oxide was added to 1 equiv of amodiaquine or desethylamodiaquine in anhydrous chloroform, and the mixture was stirred for 1 h at room temperature. Amodiaquine quinone imines were concentrated in vacuo and then applied to a silica-60 column to remove the residual amodiaquine or desethylamodiaquine. The identity and purity of synthetic amodiaquine quinone imines were verified by mass spectrometry and HPLC-UV.
Synthesis of Carbamazepine-Derived Iminoquinone
The iminoquinone of carbamazepine was synthesized using the method adapted from Ju et al.42 In short, 1 g of iminostilbene in 260 mL acetone was added to 2.6 g of Fremy’s salt in 260 mL 100 mM KPi buffer (pH 7.0). The mixture was stirred on ice for 10 min and slowly allowed to warm to room temperature. The resulting iminoquinone was extracted 3 times with 200 mL chloroform, washed 3 times with 100 mL water, and dried over magnesium sulfate. The resulting extract was purified using silica-60 column chromatography (hexane/ethyl acetate 7:3 v/v) and evaporated to dryness. The identity and purity were confirmed by 1H NMR.
Reduction of Synthesized Amodiaquine-Derived Quinone Imines and Carbamazepine-Derived Iminoquinone by NQO1 and NQO2
Synthetically prepared amodiaquine quinone imines and carbamazepine iminoquinone were directly incubated with NQO1 or NQO2. Incubations consisted of NQO1 (0–5 μM) or NQO2 (0–10 μM) with cofactor (250 μM NADPH or NRH, respectively) and GSH (50 μM) in 100 mM KPi buffer containing 2 mM EDTA, 5 mM MgCl2, and 0.18 mg/mL BSA (pH 7.4). After a pre-incubation for 4 min at 37 °C, reactions were started by addition of 50 μM of reactive metabolite (amodiaquine or desethylamodiaquine quinone imine, 0.02% DMF final; carbamazepine iminoquinone 0.02% DMSO final) and incubated for 1 min. Reactions were stopped by addition of 10% (v/v) of stop solution (10% perchloric acid containing 10 mM ascorbic acid) and centrifuged for 15 min at 14,000 rpm to remove precipitated protein. Amodiaquine and metabolites were analyzed on HPLC as described by Zhang et al.43 The GSH conjugate of carbamazepine iminoquinone was analyzed by LC-MS (Agilent 1200 Series Rapid Resolution LC system connected to a TOF Agilent 6230 mass spectrometer). Mobile phases consisted of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). Metabolites were separated on a Phenomenex Kinetex C18 column (5 μm, 4.6 × 100 mm) protected by a Phenomenex security guard (5 μm, 4.0 × 3.0 mm) using the following binary gradient: 0 min, 20% B; 2 min, 20% B; 12 min, 60% B; 15 min, 99% B; 15.5 min, 20% B; 25 min, 20% B. The flow rate was 0.5 mL/min, and analytes were detected with the TOF operating in positive ion electrospray mode. MS conditions were as follows: capillary voltage, 3500 V; drying gas (N2), 10 l/min; desolvation gas (N2), 50 psig; source temperature, 350 °C; 2 GHz, extended dynamic range mode.
Inhibition of NQO2 Activity by Tacrine
Tacrine was applied as a NQO2 inhibitor to determine the enzymatic activity of NQO2. Ki values of tacrine and resveratrol (positive control) were determined in 96-well format by incubating 50 nM NQO2 with 100 μM of BNAH with inhibitor (0–200 μM tacrine, 0–250 μM resveratrol) in 100 mM KPi buffer containing 2 mM EDTA, 5 mM MgCl2, and 0.18 mg/mL BSA (pH 7.4). After a pre-incubation of 15 min, reactions were started by addition of DCPIP at a concentration corresponding to its Km (25 μM, Figure S4A). DCPIP reduction was monitored in time by recording the absorbance at 600 nm using a Bio-Tek Powerwave X 340 plate reader. Ki values were determined by fitting the data to log(inhibitor) vs normalized response equation implemented in GraphPad Prism (version 5.0).
Inhibition of NQO2-mediated reduction of N-acetyl-p-benzoquinone imine (NAPQI), the acetaminophen-derived quinone imine, was investigated by incubating 2 mg/mL human liver microsomes, 500 μM GSH, 250 μM acetaminophen, 2.5 μM NQO2, and 250 μM NRH in 100 mM KPI BUFFER containing 2 mM EDTA, 5 mM MgCl2, and 0.18 mg/mL BSA (pH 7.4). Tacrine (0, 2.5, 5, or 10 μM) was included if applicable. After a pre-incubation of 4 min at 37 °C, reactions were started by addition of NADPH regenerating system (100 μM NADPH, 10 mM glucose-6-phosphate, and 0.5 U/mL glucose-6-phosphate dehydrogenase, final concentrations) and incubated for 30 min. Samples were stopped by addition of an equal volume ice-cold methanol and analyzed as described above. Inhibition of NQO2-mediated reduction of amodiaquine quinone imines was investigated by incubating 0.25 μM NQO2, 50 μM GSH, and 250 μM NRH in 100 mM KPi buffer containing 2 mM EDTA, 5 mM MgCl2, and 0.18 mg/mL BSA (pH 7.4). Tacrine (0, 5, 10, or 20 μM) was included if applicable. After a pre-incubation of 4 min at 37 °C, reactions were started by addition of 50 μM amodiaquine quinone imine and incubated for 1 min. Samples were stopped, prepared for HPLC analysis, and analyzed as described above.
Results
Concentrations of NQO1 and NQO2 in Human Liver
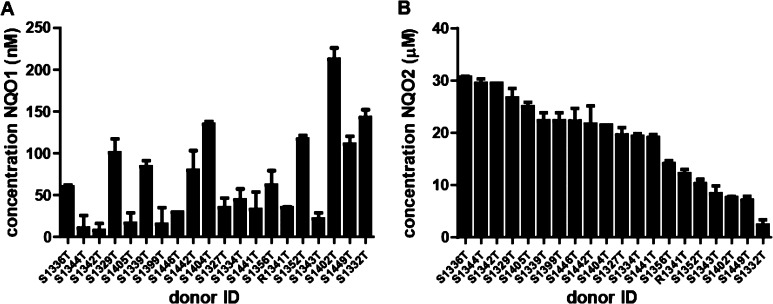
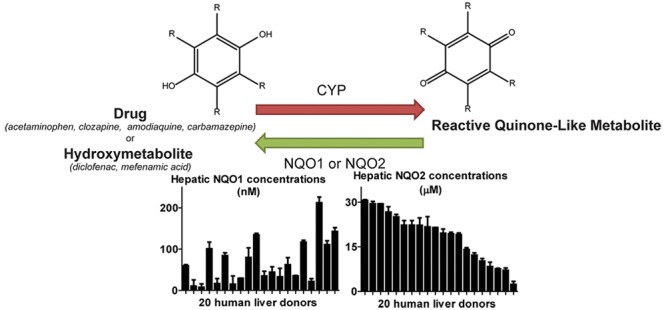
Previously, we have reaction phenotyped human liver cytosols from 20 donor livers for NQO1 and NQO2 activity using NQO1-specific resorufin reduction and CB1954 as selective NQO2 substrate (den Braver-Sewradj et al., under revision). In the current work, these specific activities are related to hepatic concentrations by using standard curves of recombinant NQO1 and NQO2 activities. Hepatic NQO1 levels ranged from 10 nM (donor S1344T) to 213 nM (donor S1402T) and NQO2 levels from 2.4 μM (donor S1332T) to 30.6 μM (donor S1336T) (Figure 2). The high variation of NQO1 levels (coefficient of variation, CV 89%) is in line with the fact that NQO1 expression is both inducible and genetically determined.26,44,45 These estimated hepatic concentrations of NQO1 and NQO2 were used in the subsequent in vitro incubations to study their activity in inactivating reactive drug metabolites.
Figure 2.
Hepatic expression levels of NQO1 (A) and NQO2 (B) in 20 donors. Liver concentrations were determined by analysis of enzyme-specific reaction velocities and comparison with calibration curves made from recombinant enzymes. Cytosolic concentrations are extrapolated to hepatic concentrations using cytosolic liver contents and intracellular hepatocytes volumes. Donors are sorted by NQO2 concentrations. Data represent average and range of duplicates.
NQO1 and NQO2 inhibition by parent drugs
NQO2 binds diverse drugs like imatinib, chloroquine, and acetaminophen.18,46,47 Therefore, prior to the bioactivation experiment, we investigated the potential inhibition of NQO1 and NQO2 activities by the parent drugs. Acetaminophen did not inhibit NQO1 and NQO2 activities up to 1 mM. However, using DCPIP as substrate, NQO1 was inhibited by diclofenac and mefenamic acid (Ki of 13 and 40 μM, respectively). Inhibition of NQO2 was only found for mefenamic acid when using menadione as substrate (Ki of 66 μM). A complete overview is given in Table S1.
NQO2-Mediated Reduction of Acetaminophen-, Clozapine-, Diclofenac-, and Mefenamic Acid-Derived Quinone-Like Metabolites Generated with Human Liver Microsomes
The quinone imines formed by CYP-catalyzed bioactivation of acetaminophen, diclofenac or mefenamic acid results were previously shown to be substrates for NQO1.12,13 No NQO1 activity was found in reduction of the reactive nitrenium ion of clozapine (unpublished data). NQO2 is not active in the presence of NADPH, but requires NRH as co-substrate.4 Since at high NRH concentrations (1 mM) significant chemical reduction of acetaminophen quinone-imine was observed (data not shown), the NRH concentration was selected at the lowest concentration for which NQO2 activity with menadione was still maximal (250 μM, Figure S2). Chemical reduction of reactive metabolites by NRH was determined in incubations in absence of NQO2 and in absence or presence of NRH. To further distinguish between enzymatic reduction and non-enzymatic scavenging of quinone-like metabolites by NQO2, incubations were performed in the presence and absence of NRH with increasing NQO2 concentrations.
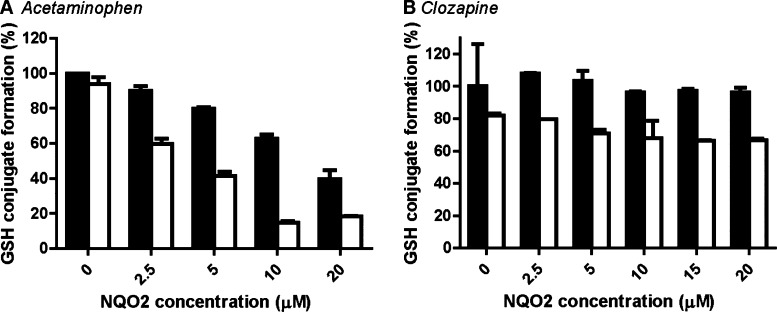
Acetaminophen oxidation by CYP results in NAPQI (Figure 1).48 Chemical reduction of NAPQI by NRH was minimal (Figure 3A, 0 μM NQO2). NQO2 was highly active with a maximal enzymatic reduction of quinone-imine at 10 μM of NQO2 (Figure 3A). In the absence of co-substrate NRH (Figure 3A, black bars), also a NQO2 concentration-dependent decrease in GSH-conjugate was observed, likely due to non-enzymatic NAPQI scavenging by NQO2.
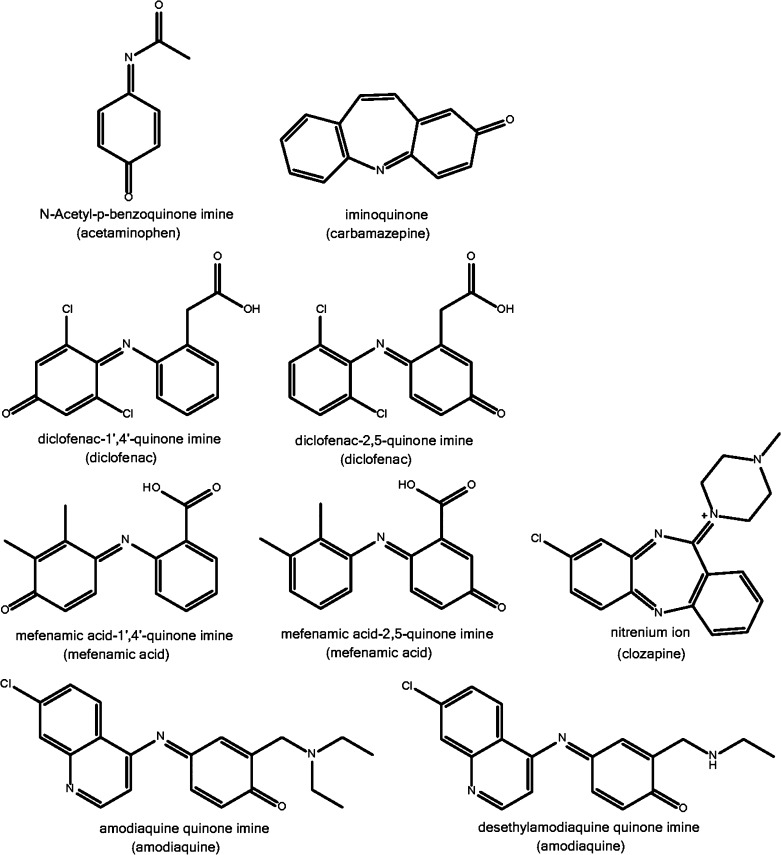
Figure 1.
Structures of reactive quinone metabolites investigated in this study. Parent drugs are indicated between brackets.
Figure 3.
NQO2-catalyzed reduction of the acetaminophen-derived quinone imine NAPQI (A) and the clozapine-derived nitrenium ion (B). CYP-catalyzed oxidation to quinones and subsequent enzymatic reduction or chemical GSH conjugation occurred simultaneously in a 30 min incubation containing acetaminophen (250 μM) or clozapine (100 μM), human liver microsomes, recombinant NQO2, and GSH (500 μM) in absence (black) or presence (white) of NRH (250 μM). Data are presented relative to the incubation without NQO2 and NRH and represent the average and range of duplicates.
Clozapine is bioactivated by CYP or myeloperoxidase to a reactive nitrenium ion, which contains a quinone-diimine moiety (Figure 1).49,50 Chemical reduction of the nitrenium ion by NRH was significant with a 20% decrease of the total GSH-conjugate formation (Figure 3B, 0 μM NQO2). An additional 15% decrease upon addition of 20 μM of NQO2 indicated that the clozapine nitrenium ion was also reduced enzymatically by NQO2 (Figure 3B).
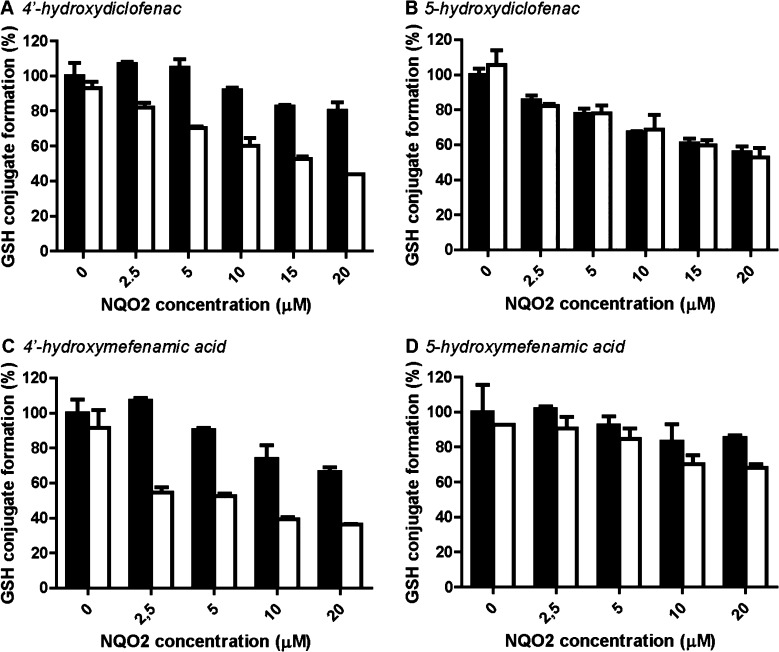
Diclofenac and mefenamic acid are bioactivated via a two-step oxidation catalyzed by CYPs, the first step being formation of 4′- and 5-hydroxymetabolites, which can be further oxidized to the respective quinone imines (Figure 1).38,39 Addition of only NRH did not decrease the formation of GSH conjugation, indicating that the chemical reduction of these quinone imines by NRH was minimal (Figure 4, 0 μM NQO2). For both diclofenac and mefenamic acid, NQO2 was clearly enzymatically active in reduction of the quinone imines derived from the 4′-hydroxy metabolites (Figure 4A and C). In contrast, the quinone imines derived from the 5-hydroxymetabolites were not (diclofenac) or barely (mefenamic acid) enzymatically reduced by NQO2 (Figure 4B and D). For both diclofenac- and mefenamic acid-derived quinone imines, significant non-enzymatic scavenging by the NQO2 protein was observed (Figure 4, black bars).
Figure 4.
NQO2-catalyzed reduction of diclofenac- and mefenamic acid-derived quinones. CYP-catalyzed formation of quinone imines from 4′-hydroxydiclofenac (50 μM) (A), 5-hydroxydiclofenac (50 μM) (B), 4′-hydroxymefenamic acid (250 μM mefenamic acid) (C), or 5-hydroxymefenamic acid (250 μM mefenamic acid) (D) and subsequent enzymatic reduction or chemical GSH conjugation was simultaneous in a 30 min incubation containing human liver microsomes, recombinant NQO2, and GSH (500 μM) in absence (black) or presence (white) of NRH (250 μM). Data represent the sum of GSH conjugates derived from 4′- or 5-hydroxydiclofenac or 4′- or 5-hydroxymefenamic acid and are presented relative to the incubation without NQO2 and NRH. Data represent the average and range of duplicates.
NQO1- and NQO2-Mediated Reduction of Amodiaquine-Derived Quinone Imines and Carbamazepine-Derived Iminoquinone
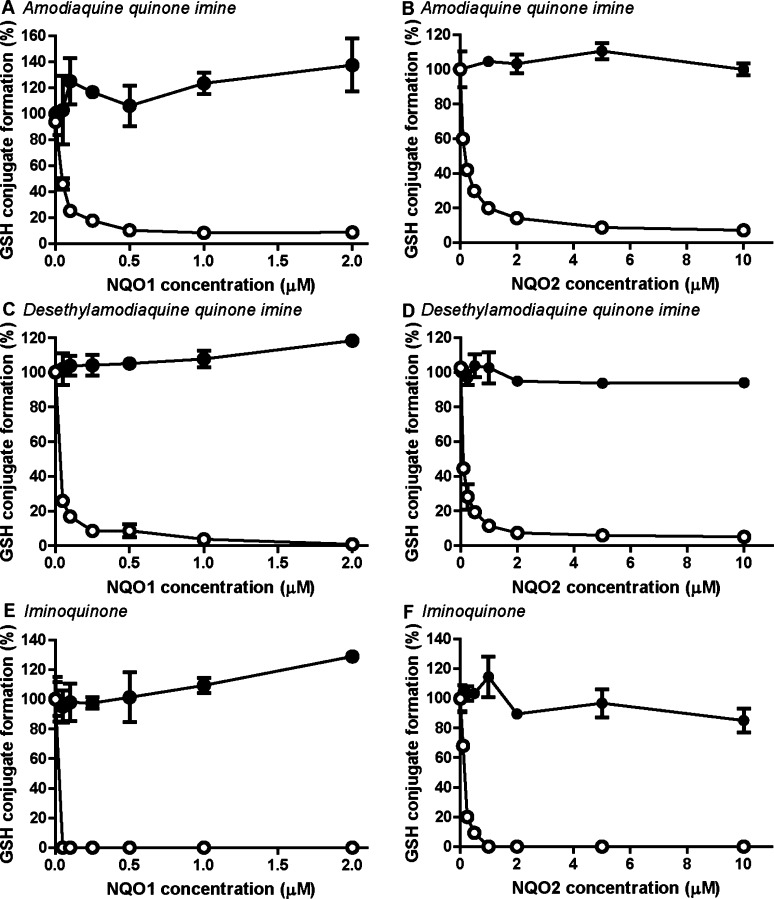
Amodiaquine can be directly bioactivated by CYPs to amodiaquine quinone imine or can be first metabolized to desethylamodiaquine which can be further oxidized to desethylamodiaquine quinone imine (Figure 1).43 Carbamazepine is oxidized by CYPs to 2-hydroxycarbamazepine which is either directly oxidized to the reactive carbamazepine iminoquinone (Figure 1) or via a secondary oxidation to 2-hydroxyiminostilbene.51 The relative stable amodiaquine-derived quinone imines and carbamazepine iminoquinone allowed direct assessment of NQO1 and NQO2 activities using synthesized metabolites. Because of the rapid enzymatic reduction and chemical GSH conjugation, incubations were stopped after 1 min. For all three reactive metabolites, both NQO1 and NQO2 were active, and complete reduction of all three reactive metabolites was already achieved at low nM (NQO1) to low μM (NQO2) concentrations of enzyme (Figure 5). For none of the reactive metabolites, chemical reduction by NRH at the chosen conditions was observed, and all were scavenged spontaneously by GSH rather than non-enzymatically by the NQO1 or NQO2 protein (Figure 5, closed circles).
Figure 5.
NQO2-catalyzed reduction of amodiaquine- and carbamazepine-derived quinones. Recombinant NQO1 or NQO2 was incubated for 1 min with purified quinone imines (50 μM) and GSH (50 μM) in absence (filled circles) or presence (open circles) of 250 μM NADPH or NRH. Amodiaquione quinone imine (A and B) and desethylamodiaquine quinoneimine (C and D) are formed by CYP-catalyzed oxidation of amodiaquine, and iminoquinone is a metabolite of carbamazepine (E and F). GSH conjugation is presented relative to the incubation without NQO and cofactor. Data represent the average and range of duplicates.
Application of Tacrine as NQO2 Inhibitor
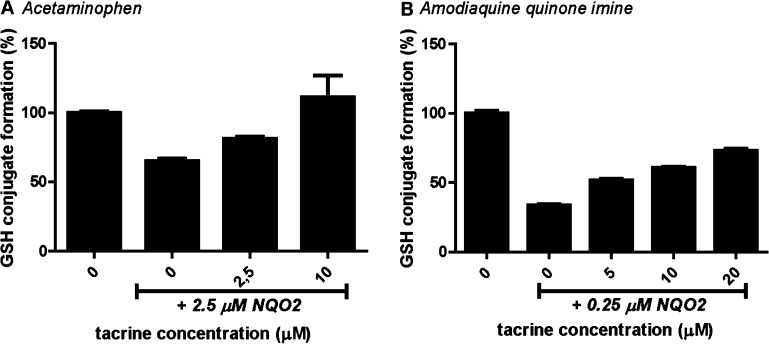
Application of an NQO2 inhibitor can further confirm enzymatic reduction, as is generally done for NQO1 using dicoumarol as specific inhibitor. However, the well-known NQO2 inhibitors resveratrol and quercetin were not suited for incubations containing human liver microsomes, because of their CYP inhibiting characteristics.52−55 Screening for NQO2 inhibition by drugs identified tacrine, which is structurally very similar to the previously reported NQO2 inhibitor 9-aminoacridine,56 as a potent NQO2 inhibitor. Inhibition of NQO2 by tacrine had a Ki of 3 and 15 uM using DCPIP or MTT as substrate, respectively (Table 1, Figure S4B). This Ki is 9-fold lower than found for NQO2 inhibition by resveratrol and 64-fold lower compared to the Ki for NQO1 (Table 1). Although tacrine is also a CYP1A2 substrate,57 no effect on CYP metabolism was observed up to 25 μM of tacrine for all four microsomal-activated drugs (data not shown). In human liver microsomes containing incubations, addition of tacrine resulted in a shift toward GSH conjugation rather than reduction of quinones for all four microsomal-activated drugs examined in this study (exemplified for acetaminophen in Figure 6A), thereby confirming a catalytic role of NQO2 in reduction of the reactive drug metabolites. Also in incubations with the synthetic quinone imine from amodiaquine, a concentration-dependent inhibition of NQO2 was observed with tacrine (Figure 6B).
Table 1. Inhibition of Recombinant NQO1 and NQO2 by Tacrine, Dicoumarol, and Resveratrol.
| enzyme | substrate | inhibitor | Kia |
|---|---|---|---|
| NQO1 | DCPIP | dicoumarol | 170 ± 1 nM |
| tacrine | 193 ± 1 μM | ||
| NQO2 | DCPIP | resveratrol | 28 ± 1 μM |
| tacrine | 3 ± 1 μM | ||
| MTT | resveratrol | 133 ± 2 μM | |
| tacrine | 15 ± 1 μM |
Values are presented as mean ± SD (n = 3).
Figure 6.
Inhibition of NQO2-mediated quinone reduction by tacrine. Acetaminophen (250 μM, A) was bioactivated by human liver microsomes to NAPQI, which was reduced by recombinant NQO2 in the presence of NRH (250 μM) and GSH (500 μM). Inhibition of NQO2 was shown over a range of tacrine concentrations. Synthetic amodiaquine quinone imine (50 μM, B) was incubated with recombinant NQO2, NRH (250 μM), and GSH (50 μM). Incubations were performed for 30 min (acetaminophen) or 1 min (amodiaquine quionone). NQO2 was pre-incubated with a range of tacrine concentrations before starting the reaction by addition of NADPH (A) or quinone imine (B). Data represents the average and range of duplicates.
Discussion
Exposure to chemically reactive drug metabolites, such as quinones-like metabolites, is postulated to contribute to the onset of many adverse drug reactions.58 Although the role of NQO1 in inactivation of reactive drug metabolites has been well studied, this capability of NQO2 had not been shown yet. In the present study, the ability of NQO2 to reduce quinone-like metabolites derived from six drugs was studied. NQO1 and NQO2 activities are especially of interest in the liver, where bioactivation of drugs and other xenobiotics by CYP enzymes is relatively high compared to other tissues. As the expression levels of quinone reductases are highly variable (and polymorphisms are known), it was of interest to further clarify the role of NQO1 and NQO2 in detoxication of quinone-like metabolites.30,45
Substrate selectivity and enzyme expression levels are a determining factor in the contribution of NQO1 or NQO2 to reduction of quinone-like metabolites. Relative activity levels of NQO1 in human liver are well investigated, and the general finding is that basal NQO1 expression in human liver is low.27,59 So far, absolute NQO1 protein concentrations have only been determined in human colon and lung.60 As abundances in colon and lung are similar to human liver, hepatic NQO1 concentrations were previously estimated to be around 300 nM.12 In the current study, we extrapolated specific NQO1 activities to NQO1 hepatic expression levels and found protein abundances in 20 human liver donors in similar levels, ranging from 8 to 213 nM (Figure 2A). These concentrations appeared sufficient to reduce quinone-like metabolites in vitro, as 50 nM of recombinant NQO1 reduced 50–100% of synthetic amodiaquine quinone imines or carbamazepine iminoquinone within a minute in the presence of 50 μM of GSH (Figure 5A, C, E). However, intact hepatocytes also contain competing enzymes such as NADPH-cytochrome P450 reductase, carbonyl reductase, and glutathione-S-transferases and up to 5–10 mM of GSH.61−63 NQO1 levels are highly inducible in human liver, especially during (drug-induced) liver injury when GSH levels usually decrease.26,45 In mice hepatocytes, which contain low basal levels of NQO1, enhanced NQO1 activities were not sufficient to protect against cytotoxicity of NAPQI.64 NQO1 KO mice studies suggested a critical (non-enzymatic) protective role of NQO1 against acetaminophen-mediated ATP-depletion instead.65 In isolated rat hepatocytes, which contain higher NQO1 levels, NQO1 inhibition by dicoumarol increased amodiaquine quinone imine cytotoxicity.14,27 In this study, using pooled primary human hepatocytes, inhibition of NQO1 did not increase cytotoxicity of amodiaquine quinone imine (Figure S5). It is important to note however that the use of pooled primary human hepatocytes neglects variability of NQO1 expression. Furthermore, it remains to be investigated whether induced NQO1 activities in human hepatocytes are sufficient to protect against drug-induced cytotoxicity.
Extrapolation of NQO2-specific activities to NQO2 hepatic expression levels showed that hepatic NQO2 concentrations ranged from 2 μM to 31 μM (Figure 2B). In subsequent human liver microsome incubations to investigate NQO2 enzymatic activity, the fact that NQO2 uses NRH instead of NADPH as co-substrate was exploited.4 Non-enzymatic quinone imine scavenging by NQO2 protein was identified by the NQO2 concentration-dependent decrease of GSH conjugation in incubations without the co-substrate NRH, while in the presence of NRH enzymatic reduction is included as well. The concentration of recombinant of NQO2 that was needed to reduce at least 50% of quinone imines generated by human liver microsomes (5 μM to 20 μM, Figure 3A and 4A and C) was in similar range as the hepatic NQO2 concentrations. Also in incubations with chemically synthesized reactive metabolites, when formation of the product is not limiting for NQO2 activity, complete reduction of amodiaquine quinone imines and carbamazepine iminoquinone was achieved at 1–5 μM of enzyme (Figure 5). With the exception of 5-hydroxydiclofenac quinone imine, NQO2-catalyzed reduction was found for all eight quinone-like metabolites tested.
Tacrine was identified as a novel and much more potent NQO2 inhibitor than resveratrol, and its utility in in vitro assays was shown here as enzymatic reduction of reactive metabolites was further confirmed by application of tacrine as NQO2 inhibitor (Table 1, Figure 6). The clinical relevance of NQO2 inhibition by tacrine is likely to be small, as the Ki value (3–15 μM, Table 1) is much higher than the plasma concentration of tacrine in humans (around 0.04 μM).66
Lowered levels of NQO2 have been correlated with an increased risk for clozapine-induced agranulocytosis.22,23 Another role in preventing agranulocytosis by NQO2, independent of non-enzymatic scavenging and enzymatic reduction, remains possible. Both NQO1 and NQO2 have been described to influence proteasomal degradation of transcripton factors.67 Thus, the high hepatic levels of NQO2 may also be related to its postulated role as a stabilizer of p53.68,69
NQO2 has been reported to reduce ortho-quinones more efficiently than NQO1.70 In line with these observations, drug-derived para-quinone imines in this and a previous study (i.e., from acetaminophen, diclofenac, mefenamic acid, amodiaquine, and carbamazepine) are more efficiently reduced by NQO1 when compared to NQO2.12 In addition, the ortho-quinone diimine (from clozapine) was reduced by NQO2 only. These results suggest a more important role of NQO1 in protection against para-quinone imines, however the role of NQO2 in human liver cannot be neglected, as expression levels are substantially higher (Figure 2).
Endogenous catalytic activity of NQO2 in humans is still unclear since its physiologically relevant co-substrate remains to be established. Although NRH has been detected in mice, cellular levels are presumably too low as co-administration of NRH significantly increased NQO2 activity in vivo.(19,71) Nevertheless, inhibition of human NQO2 catalytic activity has been suggested as one of the modes of action for the antimalarial drug primaquine.72 It has been hypothesized that levels of NRH, which can be formed by cleavage of NADH, increase during cellular stress thereby “awakening” the NQO2 enzyme.6,73 Alternatively, however, especially under acidic conditions (e.g., upon cellular ATP-depletion),74 NQO2 seems to use NADH as a co-substrate.75 According to these hypotheses, NQO2 is a latent enzyme under homeostatic conditions.
Interestingly, with the exception of the clozapine nitrenium ion, non-enzymatic scavenging of quinone-like metabolites by the NQO2 protein was observed in all incubations containing human liver microsomes as bioactivating system (Figures 3 and 4). Non-enzymatic scavenging of the 5-hydroxydiclofenac-derived quinone imine was more extensive than the 4′-hydroxydiclofenac-derived quinone imine, as was reported before, but no enzymatic reduction was observed (Figure 4B).76,77 Under the conditions used in this study, up to 5 μM of 5-hydroxydiclofenac quinone imine could be trapped with GSH combined with GST.78 Inactivation of NQO2 as a result of covalent modification by 5-hydroxydiclofenac quinone imine might therefore contribute to, but not account for, the lack of NQO2 activity. It can be argued that NQO2, in contrast to NQO1, is a preferential protein target for adduction of reactive drug metabolites and thereby further fulfills a detoxicating function.64 Indeed, NQO2 has been described to bind a number of planar aromatic compounds, thereby preventing DNA intercalation.79 Of note is that, although protein reactivity of amodiaquine quinone imine has been described, no protein scavenging was observed in incubations with chemically synthesized amodiaquine quinone imines (Figure 5).25,80 An important scavenging role of NQO2 is consistent with relatively high protein concentrations in the cell.
In conclusion, we showed that the estimated hepatic NQO1 levels in livers from human donors were sufficiently high to reduce quinone-like drug metabolites from amodiaquine and carbamazepine. We also demonstrated that NQO2 was able to catalyze reduction of quinone-like drug metabolites in vitro at physiologically relevant enzyme concentrations and was strongly inhibited by tacrine, a newly identified NQO2 inhibitor. Additionally, NQO2 acted as a non-enzymatic scavenger of quinone-like metabolites. The physiological relevance of these findings remains to be established. Nevertheless, together with their roles in cellular signaling, the present results provide further evidence that both proteins may be important factors in risk assessment of drug toxicity.
Glossary
Abbreviations
- BNAH
1-benzyl-1,4-dihydronicotinamide
- CYP
cytochrome P450
- DCPIP
2,6-dichlorophenolindophenol
- NQO1
NAD(P)H:quinone oxidoreductase 1
- NQO2
NRH:quinone oxidoreductase 2
- NRH
dihydronicotinamide riboside
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.chemrestox.7b00289.
Data on quinone reductase inhibition by the parent drugs and hydroxydiclofenac, the enzyme kinetics of NRH oxidation by NQO2, the determination of the Ki and mode of inhibition of tacrine on NQO2 activity with DCPIP, and the cytotoxicity of amodiaquine quinone in pooled primary human hepatocytes in the absence and presence of the NQO1 inhibitor dicoumarol (PDF)
Author Contributions
† These authors contributed equally to this study.
S.P.d.B.-S., M.W.d.B., S.J.D., N.P.E.V., J.N.M.C., and J.C.V. were supported by the European Community under Innovative Medicines Initiative (IMI) Program (grant 115336) (MIP-DILI). Y.Z. was funded by the China Scholarship Council.
The authors declare no competing financial interest.
Supplementary Material
References
- Vasiliou V.; Ross D.; Nebert D. W. (2006) Update of the NAD(P)H:quinone oxidoreductase (NQO) gene family. Hum. Genomics 2, 329–35. 10.1186/1479-7364-2-5-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal A. K.; Burnett P.; Adesnik M.; McBride O. W. (1990) Nucleotide and deduced amino acid sequence of a human cDNA (NQO2) corresponding to a second member of the NAD(P)H:quinone oxidoreductase gene family. Extensive polymorphism at the NQO2 gene locus on chromosome 6. Biochemistry 29, 1899–906. 10.1021/bi00459a034. [DOI] [PubMed] [Google Scholar]
- Dinkova-Kostova A. T.; Talalay P. (2010) NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch. Biochem. Biophys. 501, 116–23. 10.1016/j.abb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.; Knox R.; Sun X. Z.; Joseph P.; Jaiswal A. K.; Zhang D.; Deng P. S.; Chen S. (1997) Catalytic properties of NAD(P)H:quinone oxidoreductase-2 (NQO2), a dihydronicotinamide riboside dependent oxidoreductase. Arch. Biochem. Biophys. 347, 221–8. 10.1006/abbi.1997.0344. [DOI] [PubMed] [Google Scholar]
- Li R.; Bianchet M. A.; Talalay P.; Amzel L. M. (1995) The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: mechanism of the two-electron reduction. Proc. Natl. Acad. Sci. U. S. A. 92, 8846–50. 10.1073/pnas.92.19.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella F.; Ferry G.; Delagrange P.; Boutin J. A. (2005) NRH:quinone reductase 2: An enzyme of surprises and mysteries. Biochem. Pharmacol. 71, 1–12. 10.1016/j.bcp.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Siegel D.; Yan C.; Ross D. (2012) NAD(P)H:quinone oxidoreductase 1 (NQO1) in the sensitivity and resistance to antitumor quinones. Biochem. Pharmacol. 83, 1033–1040. 10.1016/j.bcp.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox R. J.; Burke P. J.; Chen S.; Kerr D. J. (2003) CB 1954: from the Walker tumor to NQO2 and VDEPT. Curr. Pharm. Des. 9, 2091–2104. 10.2174/1381612033454108. [DOI] [PubMed] [Google Scholar]
- Zhao Q.; Yang X. L.; Holtzclaw W. D.; Talalay P. (1997) Unexpected genetic and structural relationships of a long-forgotten flavoenzyme to NAD(P)H:quinone reductase (DT-diaphorase). Proc. Natl. Acad. Sci. U. S. A. 94, 1669–74. 10.1073/pnas.94.5.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buryanovskyy L.; Fu Y.; Boyd M.; Ma Y.; Hsieh T.; Wu J. M.; Zhang Z. (2004) Crystal structure of quinone reductase 2 in complex with resveratrol. Biochemistry 43, 11417–26. 10.1021/bi049162o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winski S. L.; Faig M.; Bianchet M. A.; Siegel D.; Swann E.; Fung K.; Duncan M. W.; Moody C. J.; Amzel L. M.; Ross D. (2001) Characterization of a mechanism-based inhibitor of NAD(P)H:Quinone oxidoreductase 1 by biochemical, x-ray crystallographic, and mass spectrometric approaches. Biochemistry 40, 15135–15142. 10.1021/bi011324i. [DOI] [PubMed] [Google Scholar]
- Vredenburg G.; Elias N. S.; Venkataraman H.; Hendriks D. F. G.; Vermeulen N. P. E.; Commandeur J. N. M.; Vos J. C. (2014) Human NAD(P)H:quinone oxidoreductase 1 (NQO1)-mediated inactivation of reactive quinoneimine metabolites of diclofenac and mefenamic acid. Chem. Res. Toxicol. 27, 576–86. 10.1021/tx400431k. [DOI] [PubMed] [Google Scholar]
- Powis G.; See K. L.; Santone K. S.; Melder D. C.; Hodnett E. M. (1987) Quinoneimines as substrates for quinone reductase (NAD(P)H: (Quinone-acceptor)oxidoreductase) and the effect of dicumarol on their cytotoxicity. Biochem. Pharmacol. 36, 2473–2479. 10.1016/0006-2952(87)90519-3. [DOI] [PubMed] [Google Scholar]
- Tafazoli S.; O’Brien P. J. (2009) Amodiaquine-induced oxidative stress in a hepatocyte inflammation model. Toxicology 256, 101–9. 10.1016/j.tox.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Joseph P.; Long D. J.; Klein-Szanto A. J. P.; Jaiswal A. K. (2000) Role of NAD(P)H:quinone oxidoreductase 1 (DT diaphorase) in protection against quinone toxicity. Biochem. Pharmacol. 60, 207–214. 10.1016/S0006-2952(00)00321-X. [DOI] [PubMed] [Google Scholar]
- Radjendirane V.; Joseph P.; Lee Y. H.; Kimura S.; Klein-Szanto A. J.; Gonzalez F. J.; Jaiswal A. K. (1998) Disruption of the DT diaphorase (NQO1) gene in mice leads to increased menadione toxicity. J. Biol. Chem. 273, 7382–9. 10.1074/jbc.273.13.7382. [DOI] [PubMed] [Google Scholar]
- Xiong R.; Siegel D.; Ross D. (2014) Quinone-induced protein handling changes: Implications for major protein handling systems in quinone-mediated toxicity. Toxicol. Appl. Pharmacol. 280, 285–295. 10.1016/j.taap.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen T. P.; Björklund M. (2014) NQO2 is a reactive oxygen species generating off-target for acetaminophen. Mol. Pharmaceutics 11, 4395–404. 10.1021/mp5004866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long D. J.; Iskander K.; Gaikwad A.; Arin M.; Roop D. R.; Knox R.; Barrios R.; Jaiswal A. K. (2002) Disruption of dihydronicotinamide riboside:quinone oxidoreductase 2 (NQO2) leads to myeloid hyperplasia of bone marrow and decreased sensitivity to menadione toxicity. J. Biol. Chem. 277, 46131–9. 10.1074/jbc.M208675200. [DOI] [PubMed] [Google Scholar]
- Gaikwad N. W.; Yang L.; Rogan E. G.; Cavalieri E. L. (2009) Evidence for NQO2-mediated reduction of the carcinogenic estrogen ortho-quinones. Free Radical Biol. Med. 46, 253–62. 10.1016/j.freeradbiomed.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassagnes L. E.; Perio P.; Ferry G.; Moulharat N.; Antoine M.; Gayon R.; Boutin J. A.; Nepveu F.; Reybier K. (2015) In cellulo monitoring of quinone reductase activity and reactive oxygen species production during the redox cycling of 1,2 and 1,4 quinones. Free Radical Biol. Med. 89, 126–134. 10.1016/j.freeradbiomed.2015.07.150. [DOI] [PubMed] [Google Scholar]
- Ostrousky O.; Meged S.; Loewenthal R.; Valevski A.; Weizman A.; Carp H.; Gazit E. (2003) NQO2 gene is associated with clozapine-induced agranulocytosis. Tissue Antigens 62, 483–91. 10.1046/j.1399-0039.2003.00133.x. [DOI] [PubMed] [Google Scholar]
- van der Weide K.; Loovers H.; Pondman K.; Bogers J.; van der Straaten T.; Langemeijer E.; Cohen D.; Commandeur J.; van der Weide J. (2017) Genetic risk factors for clozapine-induced neutropenia and agranulocytosis in a Dutch psychiatric population. Pharmacogenomics J. 17, 471–478. 10.1038/tpj.2016.32. [DOI] [PubMed] [Google Scholar]
- Gong X.; Gutala R.; Jaiswal A. K. (2008) Quinone Oxidoreductases and Vitamin K Metabolism. Vitam. Horm. 78, 85–101. 10.1016/S0083-6729(07)00005-2. [DOI] [PubMed] [Google Scholar]
- Attia S. M. (2010) Deleterious effects of reactive metabolites. Oxid. Med. Cell. Longevity 3, 238–253. 10.4161/oxim.3.4.13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksunes L. M.; Goedken M.; Manautou J. E. (2006) Up-regulation of NAD(P)H quinone oxidoreductase 1 during human liver injury. World J. Gastroenterol. 12, 1937–1940. 10.3748/wjg.v12.i12.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel D.; Ross D. (2000) Immunodetection of NAD(P)H:quinone oxidoreductase 1 (NQO1) in human tissues. Free Radical Biol. Med. 29, 246–53. 10.1016/S0891-5849(00)00310-5. [DOI] [PubMed] [Google Scholar]
- Strassburg A.; Strassburg C. P.; Manns M. P.; Tukey R. H. (2002) Differential gene expression of NAD(P)H:quinone oxidoreductase and NRH:quinone oxidoreductase in human hepatocellular and biliary tissue. Mol. Pharmacol. 61, 320–325. 10.1124/mol.61.2.320. [DOI] [PubMed] [Google Scholar]
- Wang W.; Jaiswal A. K. (2006) Nuclear factor Nrf2 and antioxidant response element regulate NRH:quinone oxidoreductase 2 (NQO2) gene expression and antioxidant induction. Free Radical Biol. Med. 40, 1119–1130. 10.1016/j.freeradbiomed.2005.10.063. [DOI] [PubMed] [Google Scholar]
- Riches Z.; Liu Y.; Berman J. M.; Walia G.; Collier A. C. (2017) The ontogeny and population variability of human hepatic dihydronicotinamide riboside:quinone oxidoreductase (NQO2). J. Biochem. Mol. Toxicol. 31, e21921. 10.1002/jbt.21921. [DOI] [PubMed] [Google Scholar]
- Siegel D.; Bolton E. M.; Burr J. a; Liebler D. C.; Ross D. (1997) The reduction of alpha-tocopherolquinone by human NAD(P)H: quinone oxidoreductase: the role of alpha-tocopherolhydroquinone as a cellular antioxidant. Mol. Pharmacol. 52, 300–305. 10.1124/mol.52.2.300. [DOI] [PubMed] [Google Scholar]
- Siegel D.; Gustafson D. L.; Dehn D. L.; Han J. Y.; Boonchoong P.; Berliner L. J.; Ross D. (2004) NAD(P)H:Quinone Oxidoreductase 1: Role as a Superoxide Scavenger. Mol. Pharmacol. 65, 1238–1247. 10.1124/mol.65.5.1238. [DOI] [PubMed] [Google Scholar]
- Megarity C. F.; Gill J. R. E.; Caraher M. C.; Stratford I. J.; Nolan K. A.; Timson D. J. (2014) The two common polymorphic forms of human NRH-quinone oxidoreductase 2 (NQO2) have different biochemical properties. FEBS Lett. 588, 1666–1672. 10.1016/j.febslet.2014.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. Y.; Westphal A. H.; de Haan L. H. J.; Aarts J. M. M. J. G.; Rietjens I. M. C. M.; van Berkel W. J. H. (2005) Human NAD(P)H:quinone oxidoreductase inhibition by flavonoids in living cells. Free Radical Biol. Med. 39, 257–65. 10.1016/j.freeradbiomed.2005.03.013. [DOI] [PubMed] [Google Scholar]
- vom Dahl S.; Hallbrucker C.; Lang F.; Gerok W.; Häussinger D. (1991) A non-invasive technique for cell volume determination in perfused rat liver. Biol. Chem. Hoppe-Seyler 372, 411–8. 10.1515/bchm3.1991.372.1.411. [DOI] [PubMed] [Google Scholar]
- Jamieson D.; Wilson K.; Pridgeon S.; Margetts J. P.; Edmondson R. J.; Leung H. Y.; Knox R.; Boddy A. V. (2007) NAD(P)H:quinone oxidoreductase 1 and nrh:quinone oxidoreductase 2 activity and expression in bladder and ovarian cancer and lower NRH:quinone oxidoreductase 2 activity associated with an NQO2 exon 3 single-nucleotide polymorphism. Clin. Cancer Res. 13, 1584–90. 10.1158/1078-0432.CCR-06-1416. [DOI] [PubMed] [Google Scholar]
- Knox R. J.; Jenkins T. C.; Hobbs S. M.; Chen S.; Melton R. G.; Burke P. J. (2000) Bioactivation of 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB 1954) by human NAD(P)H quinone oxidoreductase 2: a novel co-substrate-mediated antitumor prodrug therapy. Cancer Res. 60, 4179–86. [PubMed] [Google Scholar]
- den Braver M. W.; den Braver-Sewradj S. P.; Vermeulen N. P. E.; Commandeur J. N. M. (2016) Characterization of cytochrome P450 isoforms involved in sequential two-step bioactivation of diclofenac to reactive p-benzoquinone imines. Toxicol. Lett. 253, 46–54. 10.1016/j.toxlet.2016.04.022. [DOI] [PubMed] [Google Scholar]
- Venkataraman H.; Den Braver M. W.; Vermeulen N. P. E.; Commandeur J. N. M. (2014) Cytochrome P450-mediated bioactivation of mefenamic acid to quinoneimine intermediates and inactivation by human glutathione S-transferases. Chem. Res. Toxicol. 27, 2071–2081. 10.1021/tx500288b. [DOI] [PubMed] [Google Scholar]
- den Braver M. W.; Vermeulen N. P. E.; Commandeur J. N. M. (2017) Generic method for the absolute quantification of glutathione S-conjugates: Application to the conjugates of acetaminophen, clozapine and diclofenac. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 1046, 185–194. 10.1016/j.jchromb.2017.02.004. [DOI] [PubMed] [Google Scholar]
- Harrison A. C.; Kitteringham N. R.; Clarke J. B.; Park B. K. (1992) The mechanism of bioactivation and antigen formation of amodiaquine in the rat. Biochem. Pharmacol. 43, 1421–1430. 10.1016/0006-2952(92)90198-R. [DOI] [PubMed] [Google Scholar]
- Ju C.; Uetrecht J. P. (1999) Detection of 2-hydroxyiminostilbene in the urine of patients taking carbamazepine and its oxidation to a reactive iminoquinone intermediate. J. Pharmacol. Exp. Ther. 288, 51–56. [PubMed] [Google Scholar]
- Zhang Y.; Vermeulen N. P. E.; Commandeur J. N. M. (2017) Characterization of human cytochrome P450 mediated bioactivation of amodiaquine and its major metabolite N-desethylamodiaquine. Br. J. Clin. Pharmacol. 83, 572–583. 10.1111/bcp.13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias V. G.; Lakhman S. S.; Forrest A.; Relling M. V.; Blanco J. G. (2006) Higher activity of polymorphic NAD(P)H:quinone oxidoreductase in liver cytosols from blacks compared to whites. Toxicol. Lett. 164, 249–258. 10.1016/j.toxlet.2006.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougée L. R. A.; Riches Z.; Berman J. M.; Collier A. C. (2016) The ontogeny and population variability of human hepatic NADPH dehydrogenase quinone oxido-reductase 1 (NQO1). Drug Metab. Dispos. 44, 967–974. 10.1124/dmd.115.068650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger J. A.; Hantschel O.; Superti-Furga G.; Kuriyan J. (2009) The structure of the leukemia drug imatinib bound to human quinone reductase 2 (NQO2). BMC Struct. Biol. 9, 7. 10.1186/1472-6807-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K. K. K.; Shilton B. H. (2013) Chloroquine binding reveals flavin redox switch function of quinone reductase 2. J. Biol. Chem. 288, 11242–51. 10.1074/jbc.M113.457002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du K.; Ramachandran A.; Jaeschke H. (2016) Oxidative stress during acetaminophen hepatotoxicity: Sources, pathophysiological role and therapeutic potential. Redox Biol. 10, 148–156. 10.1016/j.redox.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovic S.; Boerma J. S.; van Bergen L.; Vermeulen N. P. E.; Commandeur J. N. M. (2010) Role of human glutathione S-transferases in the inactivation of reactive metabolites of clozapine. Chem. Res. Toxicol. 23, 1467–76. 10.1021/tx100131f. [DOI] [PubMed] [Google Scholar]
- Williams D. P.; Pirmohamed M.; Naisbitt D. J.; Maggs J. L.; Park B. K. (1997) Neutrophil cytotoxicity of the chemically reactive metabolite(s) of clozapine: possible role in agranulocytosis. J. Pharmacol. Exp. Ther. 283, 1375–82. [PubMed] [Google Scholar]
- Pearce R. E.; Uetrecht J. P.; Leeder J. S. (2005) Pathways of carbamazepine bioactivation in vitro: II. The role of human cytochrome P450 enzymes in the formation of 2-hydroxyiminostilbene. Drug Metab. Dispos. 33, 1819–1826. 10.1124/dmd.105.004861. [DOI] [PubMed] [Google Scholar]
- Chan W. K.; Delucchi A. B. (2000) Resveratrol, a red wine constituent, is a mechanism-based inactivator of cytochrome P450 3A4. Life Sci. 67, 3103–12. 10.1016/S0024-3205(00)00888-2. [DOI] [PubMed] [Google Scholar]
- Chun Y. J.; Kim M. Y.; Guengerich F. P. (1999) Resveratrol is a selective human cytochrome P450 1A1 inhibitor. Biochem. Biophys. Res. Commun. 262, 20–4. 10.1006/bbrc.1999.1152. [DOI] [PubMed] [Google Scholar]
- Muthiah Y.; Ong C.; Sulaiman S.; Ismail R. (2016) Inhibition of human cytochrome p450 2c8-catalyzed amodiaquine n-desethylation: Effect of five traditionally and commonly used herbs. Pharmacogn. Res. 8, 292. 10.4103/0974-8490.188886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar T. M.; Kumar R. M.; Agrawal A.; Dubey G. P.; Ilango K. (2015) Comparative inhibitory potential of selected dietary bioactive polyphenols, phytosterols on CYP3A4 and CYP2D6 with fluorometric high-throughput screening. J. Food Sci. Technol. 52, 4537–4543. 10.1007/s13197-014-1472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan K. A.; Doncaster J. R.; Dunstan M. S.; Scott K. A.; Frenkel A. D.; Siegel D.; Ross D.; Barnes J.; Levy C.; Leys D.; Whitehead R. C.; Stratford I. J.; Bryce R. A. (2009) Synthesis and biological evaluation of coumarin-based inhibitors of NAD(P)H: quinone oxidoreductase-1 (NQO1). J. Med. Chem. 52, 7142–7156. 10.1021/jm9011609. [DOI] [PubMed] [Google Scholar]
- Pool W. F.; Reily M. D.; Bjorge S. M.; Woolf T. F. (1997) Metabolic disposition of the cognition activator tacrine in rats, dogs, and humans. Species comparisons. Drug Metab. Dispos. 25, 590–7. [PubMed] [Google Scholar]
- Park B. K.; Boobis A.; Clarke S.; Goldring C. E. P.; Jones D.; Kenna J. G.; Lambert C.; Laverty H. G.; Naisbitt D. J.; Nelson S.; Nicoll-Griffith D. A.; Obach R. S.; Routledge P.; Smith D. A.; Tweedie D. J.; Vermeulen N.; Williams D. P.; Wilson I. D.; Baillie T. A. (2011) Managing the challenge of chemically reactive metabolites in drug development. Nat. Rev. Drug Discovery 10, 292–306. 10.1038/nrd3408. [DOI] [PubMed] [Google Scholar]
- Schlager J. J.; Powis G. (1990) Cytosolic NAD(P)H:(quinone-acceptor)oxidoreductase in human normal and tumor tissue: effects of cigarette smoking and alcohol. Int. J. Cancer 45, 403–9. 10.1002/ijc.2910450304. [DOI] [PubMed] [Google Scholar]
- Tang Z.; Wu M.; Li Y.; Zheng X.; Liu H.; Cheng X.; Xu L.; Wang G.; Hao H. (2013) Absolute quantification of NAD(P)H:quinone oxidoreductase 1 in human tumor cell lines and tissues by liquid chromatography–mass spectrometry/mass spectrometry using both isotopic and non-isotopic internal standards. Anal. Chim. Acta 772, 59–67. 10.1016/j.aca.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Di L. (2014) The role of drug metabolizing enzymes in clearance. Expert Opin. Drug Metab. Toxicol. 10, 379–393. 10.1517/17425255.2014.876006. [DOI] [PubMed] [Google Scholar]
- Hayes J. D.; Flanagan J. U.; Jowsey I. R. (2005) GLUTATHIONE TRANSFERASES. Annu. Rev. Pharmacol. Toxicol. 45, 51–88. 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- Rooseboom M.; Commandeur J. N. M.; Vermeulen N. P. E. (2004) Enzyme-catalyzed activation of anticancer prodrugs. Pharmacol. Rev. 56, 53–102. 10.1124/pr.56.1.3. [DOI] [PubMed] [Google Scholar]
- Moffit J. S.; Aleksunes L. M.; Kardas M. J.; Slitt A. L.; Klaassen C. D.; Manautou J. E. (2007) Role of NAD(P)H:quinone oxidoreductase 1 in clofibrate-mediated hepatoprotection from acetaminophen. Toxicology 230, 197–206. 10.1016/j.tox.2006.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J. H.; Kim Y.-H.; Noh J.-R.; Gang G.-T.; Kim K.-S.; Chung H. K.; Tadi S.; Yim Y.-H.; Shong M.; Lee C.-H. (2015) The protective role of NAD(P)H:quinone oxidoreductase 1 on acetaminophen-induced liver injury is associated with prevention of adenosine triphosphate depletion and improvement of mitochondrial dysfunction. Arch. Toxicol. 89, 2159–2166. 10.1007/s00204-014-1340-5. [DOI] [PubMed] [Google Scholar]
- Jann M. W.; Shirley K. L.; Small G. W. (2002) Clinical pharmacokinetics and pharmacodynamics of cholinesterase inhibitors. Clin. Pharmacokinet. 41, 719–739. 10.2165/00003088-200241100-00003. [DOI] [PubMed] [Google Scholar]
- Sollner S.; Macheroux P. (2009) New roles of flavoproteins in molecular cell biology: an unexpected role for quinone reductases as regulators of proteasomal degradation. FEBS J. 276, 4313–24. 10.1111/j.1742-4658.2009.07143.x. [DOI] [PubMed] [Google Scholar]
- Iskander K.; Paquet M.; Brayton C.; Jaiswal A. K. (2004) Deficiency of NRH:Quinone oxidoreductase 2 increases susceptibility to 7,12-dimethylbenz(a)anthracene and benzo(a)pyrene-induced skin carcinogenesis. Cancer Res. 64, 5925–5928. 10.1158/0008-5472.CAN-04-0763. [DOI] [PubMed] [Google Scholar]
- Hsieh T. C.; Wang Z.; Hamby C. V.; Wu J. M. (2005) Inhibition of melanoma cell proliferation by resveratrol is correlated with upregulation of quinone reductase 2 and p53. Biochem. Biophys. Res. Commun. 334, 223–230. 10.1016/j.bbrc.2005.06.073. [DOI] [PubMed] [Google Scholar]
- Fu Y.; Buryanovskyy L.; Zhang Z. (2008) Quinone reductase 2 is a catechol quinone reductase. J. Biol. Chem. 283, 23829–35. 10.1074/jbc.M801371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskander K.; Li J.; Han S.; Zheng B.; Jaiswal A. K. (2006) NQO1 and NQO2 regulation of humoral immunity and autoimmunity. J. Biol. Chem. 281, 30917–30924. 10.1074/jbc.M605809200. [DOI] [PubMed] [Google Scholar]
- Graves P. R.; Kwiek J. J.; Fadden P.; Ray R.; Hardeman K.; Coley A. M.; Foley M.; Haystead T. a J. (2002) Discovery of novel targets of quinoline drugs in the human purine binding proteome. Mol. Pharmacol. 62, 1364–1372. 10.1124/mol.62.6.1364. [DOI] [PubMed] [Google Scholar]
- Boutin J. A. (2016) Quinone reductase 2 as a promising target of melatonin therapeutic actions. Expert Opin. Ther. Targets 20, 303–317. 10.1517/14728222.2016.1091882. [DOI] [PubMed] [Google Scholar]
- Bronk S. F.; Gores G. J. (1991) Efflux of protons from acidic vesicles contributes to cytosolic acidification of hepatocytes during ATP depletion. Hepatology 14, 626–33. 10.1002/hep.1840140409. [DOI] [PubMed] [Google Scholar]
- Jamieson D.; Tung A. T. Y.; Knox R. J.; Boddy A. V. (2006) Reduction of mitomycin C is catalysed by human recombinant NRH:quinone oxidoreductase 2 using reduced nicotinamide adenine dinucleotide as an electron donating co-factor. Br. J. Cancer 95, 1229–33. 10.1038/sj.bjc.6603414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida T.; Onozato T.; Kanazawa T.; Tanaka S.; Kuroda J. (2012) Increase in covalent binding of 5-hydroxydiclofenac to hepatic tissues in rats co-treated with lipopolysaccharide and diclofenac: involvement in the onset of diclofenac-induced idiosyncratic hepatotoxicity. J. Toxicol. Sci. 37, 1143–1156. 10.2131/jts.37.1143. [DOI] [PubMed] [Google Scholar]
- Boerma J. S.; Dragovic S.; Vermeulen N. P. E.; Commandeur J. N. M. (2012) Mass spectrometric characterization of protein adducts of multiple P450-dependent reactive intermediates of diclofenac to human glutathione-S-transferase P1–1. Chem. Res. Toxicol. 25, 2532–2541. 10.1021/tx300334w. [DOI] [PubMed] [Google Scholar]
- den Braver M. W.; Zhang Y.; Venkataraman H.; Vermeulen N. P. E.; Commandeur J. N. M. (2016) Simulation of interindividual differences in inactivation of reactive para-benzoquinone imine metabolites of diclofenac by glutathione S-transferases in human liver cytosol. Toxicol. Lett. 255, 52–62. 10.1016/j.toxlet.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Leung K. K. K.; Shilton B. H. (2015) Binding of DNA-Intercalating Agents to Oxidized and Reduced Quinone Reductase 2. Biochemistry 54, 7438–7448. 10.1021/acs.biochem.5b00884. [DOI] [PubMed] [Google Scholar]
- Lohmann W.; Karst U. (2007) Generation and identification of reactive metabolites by electrochemistry and immobilized enzymes coupled on-line to liquid chromatography/mass spectrometry. Anal. Chem. 79, 6831–6839. 10.1021/ac071100r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.