Abstract
The past several years have seen an explosion in development of applications for the CRISPR-Cas9 system, from efficient genome editing, to high-throughput screening, to recruitment of a range of DNA and chromatin-modifying enzymes. While homology-directed repair (HDR) coupled with Cas9 nuclease cleavage has been used with great success to repair and re-write genomes, recently developed base editing systems present a useful orthogonal strategy to engineer nucleotide substitutions. Base editing relies on recruitment of cytidine deaminases to introduce changes (rather than double stranded breaks and donor templates), and offers potential improvements in efficiency while limiting damage and simplifying the delivery of editing machinery. At the same time, these systems enable novel mutagenesis strategies to introduce sequence diversity for engineering and discovery. Here, we review the different base editing platforms, including their deaminase recruitment strategies and editing outcomes, and compare them to other CRISPR genome editing technologies. Additionally, we discuss how these systems have been applied in therapeutic, engineering, and research settings. Lastly, we explore future directions of this emerging technology.
Introduction
The ability to precisely edit genomic DNA using the CRISPR-Cas9 system has revolutionized the field of genome engineering (reviewed in (Doudna and Charpentier, 2014; Hsu et al., 2014; Komor et al., 2017a; Sander and Joung, 2014; Shalem et al., 2015; Sternberg and Doudna, 2015; Wang et al., 2016)). This targeted nuclease system was discovered as a bacterial adaptive immune system where foreign DNA is incorporated into an array of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) (Barrangou, 2013; Ishino et al., 1987; Jansen et al., 2002; Mojica et al., 2000). The short segments of foreign DNA captured between repeats are then used to generate guide RNAs, composed of a targeting CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA). These complex with Cas9 endonuclease, which is targeted to the foreign DNA via the crRNA sequence and a required protospacer adjacent motif (PAM) to digest the foreign DNA (Brouns et al., 2008; Garneau et al., 2010; Marraffini and Sontheimer, 2008). Significant work has been done to reveal the underlying mechanisms of this process (Deltcheva et al., 2011; Gasiunas et al., 2012; Jiang and Doudna, 2017; Jinek et al., 2012; Nishimasu et al., 2014) and enable its transfer to eukaryotic systems (Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013). In most applications, single guide RNAs (sgRNAs) (Jinek et al., 2012), which combine the crRNA and tracrRNA, are used to target the Cas9 nuclease to a specific site in the genome, where it introduces a double strand break (DSB).
Once a DSB is formed by Cas9 and an sgRNA, it can be repaired through non-homologous end-joining (NHEJ) or homology directed repair (HDR) (Jasin and Haber, 2016). NHEJ introduces insertions or deletions at the damage site (Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013), and is useful for generating frameshifts in coding regions (causing effective gene knockout (Shalem et al., 2014; Shalem et al., 2015; Wang et al., 2014) or disrupting transcription factor binding motifs (Canver et al., 2015; Ernst et al., 2016; Rajagopal et al., 2016; Sanjana et al., 2016) and splicing elements (Charton et al., 2016; Kapahnke et al., 2016; Mou et al., 2017). Alternatively, templated repair of the DSB can be used to introduce specific changes through HDR in the presence of either single or double-stranded DNA donor templates that encode the desired sequence. Cas9-directed HDR has enabled dramatic improvements in endogenous tagging of genes (Bak and Porteus, 2017; Lackner et al., 2015; Leonetti et al., 2016), generation of in-frame protein variants (Findlay et al., 2014; Garst et al., 2017; Ryan et al., 2014), as well as in clinical correction of deleterious mutations (Dever et al., 2016; DeWitt et al., 2016; Yang et al., 2016b). Significant optimization of Cas9-mediated genome-editing has allowed for single nucleotide editing efficiencies reaching 60% or more in cell culture systems (Richardson et al., 2016). However, this strategy generally depends on inducing a double strand break; therefore, small indels are an unintended by-product, which can have major negative functional consequences if the targeted gene is knocked out.
In order to limit DNA damage during editing and eliminate the need to deliver an HDR donor template, a number of groups have recently developed CRISPR base editing systems. These make use of Cas9 variants, cytidine deaminases, and manipulation of DNA repair pathways to achieve specific editing outcomes. Importantly, base editing systems enable both highly targeted single base changes (see Precision Base Editing) and local sequence diversification (see Localized Sequence Diversification). Here we review the different base editor systems, including their components, and how they affect the repair of the deaminated base with respect to efficiency and possible base substitutions. Given the therapeutic potential for base editing, current approaches for identifying off-targets of the system are discussed in detail. Lastly, we review the diverse applications of the base editors and highlight future directions for the field.
Section 1: Repurposing Deaminases for Base Editing and Diversification
Natural Roles of Deaminases
Deaminases play important roles in diverse processes including musculoskeletal myogenesis (Hsieh et al., 2014), central nervous system development and function (Li and Church, 2013), and in innate and adaptive immune pathways (Hamilton et al., 2010; Keegan et al., 2001; Samuel, 2012; Vieira and Soares, 2013). The Apolipoprotein B mRNA Editing Enzyme, Catalytic Polypeptide-Like (APOBEC) family of cytidine deaminases (Chiu and Greene, 2006; Navaratnam and Sarwar, 2006) restrict viral infection by editing viral genomes. For example, APOBEC3G was identified as a cellular factor capable of restricting HIV infection (Sheehy et al., 2002; Yu, 2006); it deaminates HIV viral cDNA (Harris et al., 2003; Lecossier et al., 2003), rendering it incapable of replication. APOBEC3 proteins have also been shown to restrict the activity of endogenous retroelements (Esnault et al., 2005; Koito and Ikeda, 2013; Muckenfuss et al., 2006). Deaminases also enable functional diversification: APOBEC1 was the first RNA editing enzyme identified (Powell et al., 1987), and functions in lipid homeostasis by editing apolipoprotein B (apoB) mRNA, converting a glutamine to a stop codon. This creates a truncated APOB48 protein specifically in the small intestine that transports dietary lipids, while the full-length APOB100 form retains the ability to bind the LDL receptor and performs a more general role in lipid transport around the body (Chan, 1992).
Activation-induced cytidine deaminase (AID) is responsible for antibody diversification through somatic hypermutation and class switch recombination. During somatic hypermutation, AID is specifically targeted to the Ig locus, where it creates diverse mutations which are then subjected to selection through antigen binding; this allows for maturation of both affinity and specificity. Loss of AID function prevents class switch recombination and somatic hypermutation, which causes a form of hyper-immunoglobulin M syndrome where low affinity antibodies are generated (Revy et al., 2000; Xu et al., 2012). Promiscuous targeting of AID outside the Ig locus has been associated with B cell lymphomas (Kuppers et al., 1999; Xu et al., 2012) and has been shown to be required for development of germinal-center derived B cell lymphomas in several mouse models (Pasqualucci et al., 2008; Ramiro et al., 2006). Intriguingly, the process of somatic hypermutation mediated by AID biases repair pathways to error-prone outcomes through mechanisms which are incompletely understood (Larson et al., 2005; Odegard and Schatz, 2006; Reynaud et al., 2003).
These deaminases share a common mechanism by which they modify cytidine to uridine. RNA targets of cytidine deamination do not undergo further processing, but for DNA, the conversion generates a DNA lesion that can be repaired through several pathways: (1) uridine can be replaced by a thymidine during DNA replication, (2) base excision repair (BER) can excise the uridine allowing the insertion of an alternative nucleotide, or (3) mismatch repair (MMR) by way of translesion synthesis, where an error-prone polymerase goes through the lesion, increasing the possibility of mutations at nearby nucleotides. Incorporation of uracil is common, and its repair is often performed in an error-free manner; however, the potential for error-prone repair facilitates antiviral defense and functional diversification.
Precision Base Editing with BE3/4 and Target-AID
The ability to manipulate the damage and repair pathways utilized by cytidine deaminases, combined with the precise targeting of Cas9, is a powerful strategy to introduce specific point mutations. Cas9-targeted deamination was first demonstrated in the pioneering work of the Liu group to develop the Base Editor (BE) system (Komor et al., 2016). Since a major goal of this work was to induce base changes without double strand DNA breaks, the initial BE1 system included only rat deaminase APOBEC1 (rAPOBEC1) fused to deactivated Cas9 (dCas9), in which the two nuclease domains of Cas9 are inactivated by mutation of D10A and either H840A or N863A (Jinek et al., 2012; Qi et al., 2013). Transfection of BE1 and a targeted sgRNA successfully converted cytidines to thymidines within a window of −16 to −12 bases from the PAM of the sgRNA (Figure 1A).
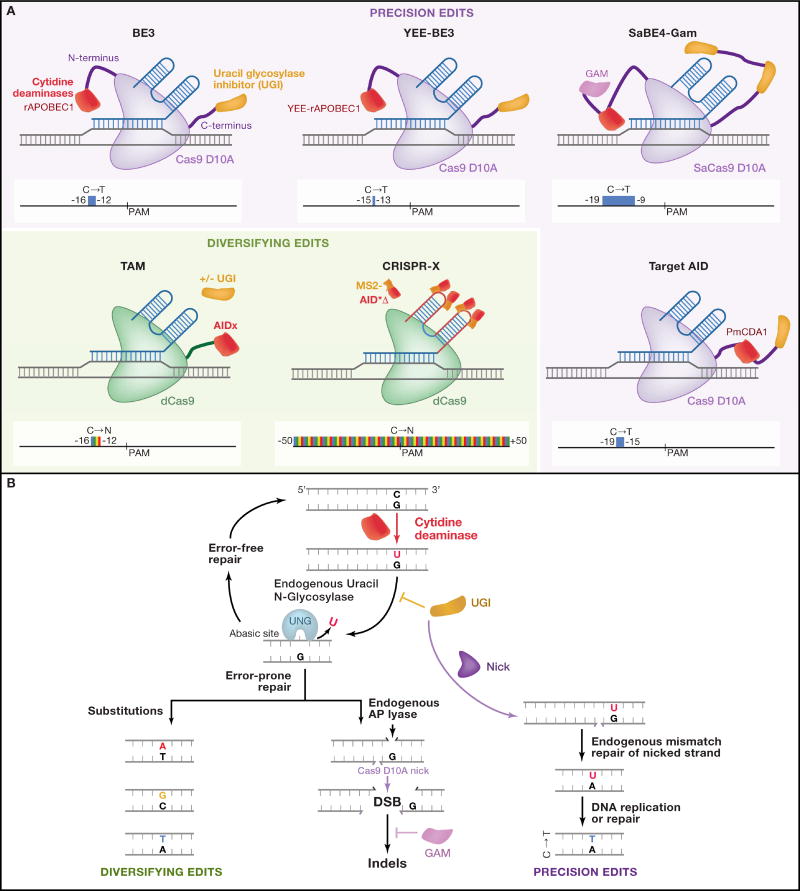
Figure 1. Molecular components and mechanisms for base editing.
A) The molecular components of the Base editor (BE), Target-AID, Targeted AID-mediated Mutagenesis (TAM), and CRISPR-X technologies are shown. The BE3, YEE-BE3, and Target-AID systems employ the Cas9 D10A (purple) nickase along with a cytidine deaminase (red) and a uracil DNA glycosylase inhibitor (yellow). The BE4 systems also uses the Gam protein (pink). The TAM and CRISPR-X systems used dCas9 (green) to recruit variants of the deaminase AID (AIDx or MS2-AID*Δ). Below each technology, the targeting window is indicated. The increased spectrum of base substitutions that can be achieved with TAM and CRISPR-X is represented by the multi-color editing windows.
B) For base editors, the mechanisms of DNA repair are critical for the eventual editing outcome. After the initial deamination (C to U) conversion, uracil DNA glycosylase can remove the base and perform error-free (correctly replacing U with C) or error-prone repair generating different substitutions (left side). Alternatively a uracil DNA glycosylase inhibitor (UGI) can be used to block base excision by Uracil N-Glycosidase (UNG). Nicking via a Cas9 nickase of the non-deaminated strand promotes long patch base excision repair where the deaminated strand is preferred for templated repair generating a U:A base pair that is then resolved as a T:A base pair. Upon formation of the abasic site, an AP lyase can remove the site forming a ssDNA break. This excision in conjunction with the Cas9 D10A can create a DSB. Addition of the bacteriophage protein Gam reduces the frequency of indel formation by directly binding the double strand break, which may select against cells with DSBs. Endogenous repair machinery is labeled in black and represented with black triangles. Exogenous components of the base editing tools are illustrated and labeled in color.
An optimized BE2 system resulted from the addition of the uracil DNA glycosylase inhibitor (UGI) (Figure 1A) from the Bacillus subtilis bacteriophage PBS1 (Mol et al., 1995). UGI directly binds and inhibits uracil DNA glycosylase, thus blocking uridine excision and the ensuing BER pathway. This improvement increased base editing efficiency of the C>T substitution three-fold by effectively disfavoring error-free repair (Figure 1B). Further improvement was realized by changing the dCas9 to a ‘nickase’ Cas9 D10A, which nicks the strand opposite the deaminated cytidine. In principle this should initiate long-patch base excision repair, where the deaminated strand is preferentially used to template the repair to produce a U:A base pair. This intermediate is then converted to T:A during DNA replication (Heller and Marians, 2006; Robertson et al., 2009; Zharkov, 2008) (Figure 1B). The optimized BE3 editor consists of rAPOBEC1 fused to the N-terminus of nickase Cas9 D10A and a UGI fused to the C-terminus. This iteration, BE3, achieved a six-fold increase in efficiency over BE2, with up to 37% of targeted alleles edited. It is worth noting that the optimized BE3 system also exhibited a slightly increased indel frequency of 1.1%, as compared to <0.1% for the BE2 system, since nicks can lead to NHEJ at a low rate (Certo et al., 2011).
A similar system, Target-AID (Nishida et al., 2016), uses nickase Cas9 D10A to recruit the cytidine deaminase PmCDA1 (from sea lamprey) to a gene of interest. Using this system to target the ADE1 gene in yeast, ~15–50% of cells could be induced to exhibit the red colony phenotype for the mutated ADE1 gene (Box 1). When this system was transported into mammalian cells using a dCas9-PmCDA1 fusion to target the HPRT gene, a smaller fraction (~2%) of cells exhibited resistance to 6-Thioguanine (6TG). Introducing the Cas9 D10A nickase increased the frequency of resistant cells to 35%, but subsequent targeted sequencing revealed that many of these mutations were deletions. To address this, the authors added UGI to block the removal of the uracil base, and observed a two- to three-fold increase of mutation frequency and reduction in deletions. Further characterization through deep sequencing at other targeted loci indicated mutation frequencies of 15–55% localized to −19 to −15 bases with respect to the PAM, similar to those observed in yeast. Thus, while BE3 and Target-AID use different cytidine deaminases and protein fusion arrangement (Figure 1A), both demonstrate the utility of an opposite strand nick and UGI to increase C>T base editing efficiency.
Box 1. Quantifying Mutation Frequency.
Quantifying base editing mutation rates is a challenging problem, especially at the low frequencies induced over large sequence space generated by diversifying base editors (analogous to the problems encountered when sequencing the B-cell receptor repertoire). In addition to sequencing assays, fluorescent and drug resistance assays have been used to estimate the rate of mutations.
Measurement by Phenotype
In a number of studies, change in a gene’s phenotype has been used as a proxy for mutation rate. For example, both the TAM and CRISPR-X systems were initially quantitated by following GFP fluorescence (by repair of a stop codon or loss of fluorescence, respectively). Similarly, on-target editing by the Target-AID system has been measured in S. cerevisiae by following loss of function in the CAN1 gene (causing resistance to canavanine) or ADE1 gene (red pigment upon loss of function). In this study, loss of LYP1, whose knockout provides protection against S-aminoethyl-L-cysteine, was used as a separate marker for non-specific mutations in the genome. In mammalian cells, the HPRT gene was targeted, where its loss of function conferred resistance to 6TG (Nishida et al., 2016). Although these various phenotypic measurements provide a rapid method for assessing mutation frequency, it is important to note that this may be inaccurate in some cases because not all mutations lead to a measurable phenotype; conversely, the phenotype may require all alleles to be mutated simultaneously. As an example of this discrepancy, the rescue of GFP fluorescence using the TAM system was observed in 4–5% of cells, but sequencing of the GFP loci revealed more than 20% of the loci contained at least one mutation (Ma et al., 2016).
Measurement by Sequencing
High-throughput sequencing of the locus can address these issues and has been used to measure the frequency of mutations induced by base editors. Sequencing data is typically filtered for read quality and overall alignment to the regions targeted for mutagenesis before quantifying the frequency of mutation at each base position in the targeting window, when compared to the unmutated gene or parent population.
One key limitation to high-throughput sequencing approaches for detecting mutations is the inherent error rate. Errors can be introduced in a number of ways: (1) SNPs or sequence heterogeneity may already exist in some cultured cell lines, (2) PCR error can occur during the preparation of sequencing libraries, or (3) bases can be incorrectly read during sequencing. Error rates for high-throughput sequencing systems are typically well below 1% (Reuter et al., 2015), but still limit the detection frequency of mutations. These errors can be estimated and accounted for by preparing and sequencing unmutated samples in parallel. For example, in the CRISPR-X system, mutation frequencies for individual bases were calculated by normalizing to the average frequency of mutations in non-mutated samples. The standard deviation of the mutation frequency for non-mutated samples was used to esitmate the noise in sequencing. The calculated standard deviation of the mutation frequency at a single base was 0.05%, allowing for the detection of mutations as low as 0.05%. Using this background, mutation frequency was quantified at each base position in the 100bp targeting window for the 30 sgRNAs tested, and ranged as high as 20% (Hess et al., 2016). While it may be possible to detect even more rare mutations, these may require sequencing samples with high coverage over the targeted area in order to confidently distinguish them from errors in sequencing.
A second limitation is the ability to detect co-occurring mutations when multiple sgRNAs are introduced over a large region (>600bp). While long read technologies exist, they are limited in throughput, and for highly diverse populations, the lack of sequencing depth can misrepresent the population. Alternatively, by measuring the relative frequencies of mutations in the population after each round of selection, it may be possible to determine which mutations co-occur (Sakoparnig et al., 2015; Wang et al., 2012; Yeang et al., 2008; Zhang et al., 2014).
A final consideration, especially for experiments with diversifying base edits, is the number of cells that should be maintained for the experiment. This number plays a critical role in experimental design to avoid bottlenecking the population and to determine the sequencing depth needed to assay the population. Estimating diversity through sequencing is a complex problem due to sequencing errors, as described above. Significant work has been done on this problem when estimating the diversity of the antibody repertoire (reviewed in (Benichou et al., 2012; Robins, 2013)). This undertaking has proved challenging due to the complexity of the library and the need to separate the mutation and selection processes. However, through a number of statistical models (Elhanati et al., 2015; Greiff et al., 2015; Mora et al., 2010) and other strategies (reviewed in (Hoehn et al., 2016)), the potential diversity of the antibody repertoire has been estimated to have a lower bound of 1018 (Elhanati et al., 2015), though in fact there may only be 3–9 × 106 different antibodies among ~109 B-cells in the human body (Arnaout et al., 2011). While quantifying mutation frequency over a single sgRNA-targeted region is somewhat simpler, significant variability has been observed in terms of the range of mutations that can be induced with diversifying base editors. For example, for the TAM and CRISPR-X systems, mutation frequencies at single bases for a single sgRNA targeting an integrated GFP locus were found to range between ~1 and 25% (Hess et al., 2016; Ma et al., 2016). Given the challenge of quantifying diversity in a starting population of mutated cells for diversifying base editors, it is helpful to estimate the total diversity created by a single sgRNA. For example, between 20–80% of all possible single base mutations were detectable above background sequencing noise within the +20 to +40bp mutation hotspot, measured in a population (0.5–1.5 million cells extracted) with an average of 4.5 million sequencing reads using the CRISPR-X system (Hess et al., 2016). This calculation provides an estimate of the number of possible variants made by a single sgRNA. In this way, mutation frequency, size of the mutagenized region, population size, and sequencing read depth can be taken into account when designing selection experiments.
Within a year of its publication, BE3 has been widely applied to edit diverse cell, plant, and animal genomes (see Applications of Precision Editing), but further development of base editor tools has continued in parallel. The Liu group has now introduced BE4, with optimized longer linkers and two fused copies of UGI to further reduce (uracil N-glycosylase) UNG-mediated base excision repair (Komor et al., 2017b) (Fig. 1A). This design decision is well-supported by experiments in UNG knockout cells that clearly demonstrate UNG is necessary for generating most undesirable substitutions and indels. The additional fusion of the DNA end-binding Gam protein (from Mu bacteriophage) (d'Adda di Fagagna et al., 2003) to BE4 helped moderately reduce undesirable indels. In theory, this works by Gam binding the double-stranded breaks that occasionally result from the combination of an endogenous DNA-(apurinic or apyrimidinic site) lyase (AP lyase) nick on the target strand and a Cas9 D10A nick on the non-target strand (Fig. 1B). The authors speculate that Gam competes with error-prone DSB repair processes and induces cell death, thus removing indels from the surviving population of cells. In a head-to-head experiment with 6 sites in HEK293T cells, BE4 generally outperformed BE3 and Target-AID, although Target-AID is optimal at more PAM-distal C’s because it has a different targeting window. When compared with BE4 containing SpCas9 D10A, BE4 with SaCas9 D10A appeared to maintain consistently high editing efficiency at cytosines within −19 to −9 bases with respect to the SaCas9 PAM. Therefore, the choice of optimal precision base editor for a given target site will likely depend on PAM availability, position of target C relative to PAM, acceptability of indels in population, and delivery modality constraints.
Localized Sequence Diversification with CRISPR-X and TAM
While BE4 and Target-AID perform a specific C>T conversion, an alternative approach is to use deamination as a means of creating a diverse library of point mutations localized to a targeted region of the genome. This is similar to the process of somatic hypermutation during antibody affinity maturation, normally performed by AID. Two technologies, Targeted AID-mediated Mutagenesis (TAM) (Ma et al., 2016) and CRISPR-X (Hess et al., 2016), use this strategy to generate localized sequence diversity through base editing.
In the TAM system (Ma et al., 2016), human AID is fused to the C-terminus of dCas9 (Figure 1A). When this system was targeted to a copy of GFP containing a premature stop codon with a pool of sgRNAs, 2% of cells exhibited GFP fluorescence, indicating successful editing. This efficiency could be doubled by removing the C-terminus of AID (AIDx), which includes a nuclear export signal (NES) that normally limits AID activity. Sequencing of the targeted GFP locus revealed mutation frequencies >20%, with transitions and transversions from cytidine and guanine to the other 3 bases with only a slight bias for C>T and G>A transitions. Coexpression of UGI resulted in an increase in frequency of mutation when using single sgRNAs, but limited mutations to C>T transitions within a window of −16 to −12 bases with respect to the PAM of the sgRNA.
In the CRISPR-X system (Hess et al., 2016) dCas9 is used to recruit a hyperactive variant of the AID enzyme. Three mutations (K10E, T82I, and E156G) were previously identified in a bacterial screen that increased the deamination activity of AID (Wang et al., 2009); this variant was combined with a deletion of the NES to generate AID*Δ. Instead of fusing the enzyme to dCas9, AID*Δ was fused to MS2 protein and recruited to the locus via two MS2 RNA hairpins inserted into the sgRNA backbone (Konermann et al., 2015) (Figure 1A). Compared to native AID without an NES (AIDΔ), which induced mutations within a targeting window of +12bp to +32bp from the PAM site, CRISPR-X using AID*Δ induced a ~2–6 fold increased frequency of mutations (>20%) over a larger window of −50bp to +50bp. Similar to the TAM system, the conversion of C to A, G, or T and G to A, C, or T showed only a slight preference for C>T. Mutations at A and T bases were also observed, but at a much lower frequency. Interestingly, using CRISPR-X, the position of targeted mutations appears to be sensitive to the direction of transcription, with the region of highest mutation downstream of the PAM site with respect to the direction of transcription. This is consistent with previous observations that mutagenesis by AID depends on interactions with the transcription machinery (Chaudhuri et al., 2003). Nonetheless, when CRISPR-X was targeted to regions upstream of annotated transcription start sites, mutations could be detected at similar frequencies to coding regions.
Although the designs of each of these base editing systems are different, a common thread is the ability to generate point mutations at cytosine and guanine nucleotides with a low rate of indels, in contrast with HDR-based methods which often generate more indels than substitutions. This is critical when modifying protein coding genes where indels could generate frameshifts that make truncated or nonsense protein products. The diverse editing outcomes generated by these systems enable both precision editing of single C>T bases as well as sequence diversification; each technology facilitates a range of possible applications.
Section 2: Applications of base editing
Though base editing technologies are still new, there have already been many successful applications in vitro and in vivo, ranging from precise, therapeutic and agronomic editing, to broad screens for directed evolution and mapping of protein-drug interactions (Figure 2). These pioneering experiments have been conducted in a wide panel of cell lines and organisms, summarized in Tables 1 and 2. In this section, we review how current base editing systems have been applied.
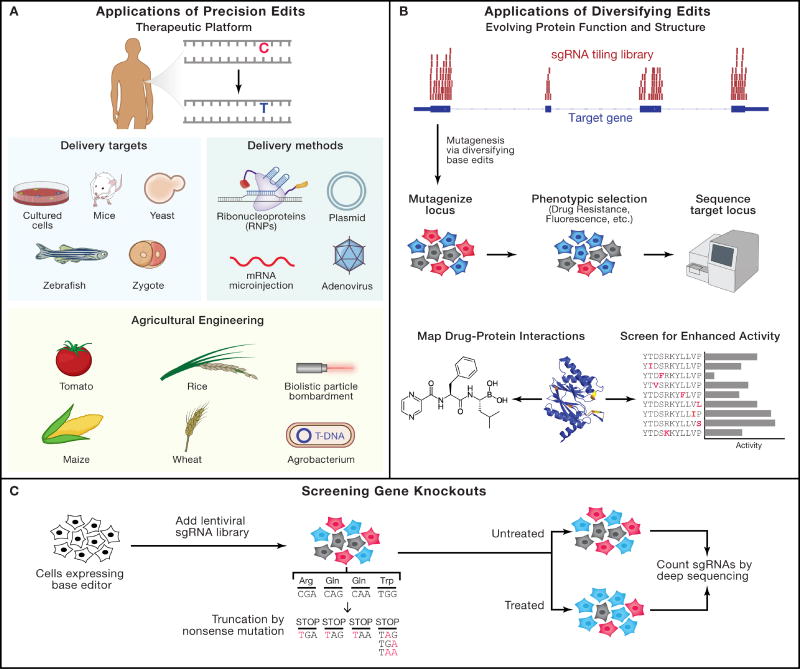
Figure 2. Applications of base editing technologies.
Base editing has been applied in two main areas: precision editing and screening applications.
A) For precision editing, corrective or beneficial conversion of cytidines to thymidines has been demonstrated in many systems. Multiple delivery targets and methods that have been used are shown (see also Table 1). Precision editing has also been used for agricultural engineering using agrobacterium or biolistic DNA particle bombardment to deliver base editors to tomato, maize, wheat, and rice plants.
B) Diversifying base editors have been used to evolve and study protein function and structure. In these experiments, sgRNAs tiling a region are used to generate targeted mutations and the subsequent population of variants is screened for specific phenotypes (i.e. drug resistance/fluorescence), followed by targeted sequencing of the locus. To validate identified mutations, enriched variants are individually installed and drug resistance or the activity of the individual clones are validated.
C) The precise base editing system can also be used for screening applications. Targeting the BE3 system to Arg, Gln, and Trp codons can edit each into stop codons, generating truncated gene products (CRISPR-STOP, iSTOP). This platform provides a potential alternative to Cas9 gene screens without inducing double strand breaks.
Table 1. Efficiency of precision base editors in diverse model systems.
A summary of base editing experiments over the past two years. Efficiency data is presented as the approximate minimum and maximum across loci and replicates in a given model system. Where data is available, base editing is compared with nuclease-active Cas9. Quantification of substitution and indel frequencies in studies using cell populations were based on high-throughput sequencing; studies using whole-organisms were based on genotyping. RNP: Ribonucleoprotein complex of base editor and sgRNA, n.r.: not reported, n/a: not applicable.
| Gene (variant) | Cell type | Delivery Method | Base Editor |
Substit ution freque ncy |
Indel freque ncy |
Comparison to Active Cas9 (with or without donor) |
Substit ution frequen cy |
Indel freque ncy |
Reference |
|---|---|---|---|---|---|---|---|---|---|
| APOE4 (C158R) | Mouse astrocyte | Plasmid nucleofection | BE3 | 58–75% | 4–6% | SpCas9 and donor 200-nt ssDNA | 0.1–0.3% | 26–40% | (Komor et al., 2016) |
| TP53 (Y163C) | HCC1954 human breast cancer cell line | Plasmid nucleofection | BE3 | 3–8% | <0.7% | SpCas9 and donor 200-nt ssDNA | 0% | 6–8% | (Komor et al., 2016) |
| 6 loci (incl. EMX1, FANCF, RNF2) | HEK293T cells | Plasmid lipofection | BE3 | 5–37% | 1–6% | SpCas9 and donor 200-nt ssDNA | <2% | 2–7% | (Komor et al., 2016) |
| 6 loci (incl. EMX1, FANCF, RNF2) | HEK293T cells | Plasmid lipofection | BE4 | 20–50% | 0.5–3% | (Komor et al., 2017b) | |||
| 6 loci (incl. EMX1, FANCF, RNF2) | HEK293T cells | Plasmid lipofection | Target-AID | 5–50% | 0.5–3% | (Komor et al., 2017b) | |||
| 4 loci (incl. FANCF) | HEK293T cells | Plasmid lipofection | SaBE4-Gam | 40–65% | 0.5–4% | (Komor et al., 2017b) | |||
| 4 loci (incl. EMX1, FANCF, VEGFA) | HEK293T cells | RNP lipofection | BE3 | 5–65% | <5% | (Rees et al.,2017) | |||
| TYR1, TYR2, TYR3 | Zebrafish embryo | RNP lipid nanoparticle injection | BE3 | 0–5% | <2% | (Rees et al.,2017) | |||
| VEGFA site 2 | Murine NIH/3T3 cells | Plasmid lipofection | BE3 | 10–85% | n.r. | (Rees et al.,2017) | |||
| VEGFA site 2 | Murine NIH/3T3 cells | RNP lipofection | BE3 | 5–45% | n.r. | (Rees et al.,2017) | |||
| VEGFA site 2 | Mouse inner ear | RNP lipid nanoparticle injection | BE3 | <1.5% | <0.1% | (Rees et al.,2017) | |||
| Dmd, Tyr | Mouse Embryo | RNP electroporation | BE3 | 56–69% | n.r. | (Kim et al., 2017b) | |||
| Dmd, Tyr | Mouse Embryo | mRNA microinjection | BE3 | 44–100% | n.r. | (Kim et al., 2017b) | |||
| Tyr | Mouse Embryo | mRNA microinjection | HF-BE2 | 11.6–50% | 4.5% | (Liang et al., 2017) | |||
| HPRT, EMX1, Efempl, MGAT1 | CHO cells | Plasmid lipofection and G418 selection | Target-AID | 17–55% | 4–12% | SpCas9 without donor | 0.1–0.6% | 71–93% | (Nishida et al., 2016) |
| Can1 | Saccharomyces cerevisiae BY4741 | Transformation and selection for mutations | Target-AID | 96% | 0.2% | SpCas9 without donor | <5% | 5–40% | (Nishida et al., 2016) |
| PCSK9 | Adult mouse liver | Adenoviral vector | BE3 | 19–34% | 1–2% | SpCas9 without donor | n/a | 50% | (Chadwick et al., 2017), (Ding et al., 2014) |
| FTIPIe and ALS | Rice calli | Agrobacterium transfection and selection for mutations | Target-AID | 34–89% | 11–66% | (Shimatani et al.,2017) | |||
| NRT1.1B and SLR1 | Rice calli | Agrobacterium transformation and selection for delivery | BE3 without UGI | 2.7–13.3% | 8.9–10.8% | (Lu and Zhu, 2017) | |||
| DELLA and ETR1 | Tomato | Agrobacterium transformation and selection for delivery | Target-AID | 33–85% | 14–69% | SpCas9 without donor | 0.5–2% | 25–44% | (Shimatani et al., 2017) |
| OSCDC48 | Rice calli | Agrobacterium transformation and selection for delivery | BE3 | 43.5% | n.r. | (Zong et al., 2017) | |||
| TaLOX2 | Wheat immature embryo cells | Biolistic gold-DNA particle bombardment without selection | BE3 | 1.3% | n.r. | (Zong et al., 2017) | |||
| ZmCENH3 | Maize immature embryo cells | Agrobacterium transformation and selection for delivery | BE3 | 10.1% | n.r. | (Zong et al., 2017) |
RNP: Ribonucleoprotein complex of base editor and sgRNA, n.r.: not reported, n/a: not applicable.
Table 2. Comparison of diversifying base edits in screening applications.
A summary of base editing screens. Selection strategy, cell type, delivery method and variant of base editor used are shown. The composition of the libraries and control libraries are described along with the number of identified variants or sgRNAs with phenotypes. A short summary of results for each screen is included.
| Gene | Selection strategy |
Cell type | Delivery | Base editor |
Number of sgRNA |
Number of significant hits/variants |
Result | Reference |
|---|---|---|---|---|---|---|---|---|
| PSMB5 | Bortezomib resistance | K562 cells | Lentiviral vector | CRISPR-X | 143 targeting exons 705 safe-targeting controls | 11 variants | Discovery of 2 known and 9 novel cancer drug resistance mutations | (Hess et al., 2016) |
| wtGFP | Fluorescence shift; cell sorting | K562 Cells | Plasmid nucleofection or Lentiviral Vector | CRISPR-X | 4 targeting wtGFP 4 safe-targeting controls | 1 variant | Recovery of known S65T substitution that changes fluorescence spectra to EGFP | (Hess et al., 2016) |
| BCR-ABL Kinase Domain | Imatinib resistance | K562 Cells | Plasmid nucleofection | TAM | 16 targeting exon 6 3 targeting AAVS1 | 6 variants | Discovery of 1 known and 5 novel cancer drug resistance mutations | (Ma et al., 2016) |
| HPRT Puromycin mCherry | 6TG selection, Puromycin selection, and mCherry FACS | HEK293T cells | Lentiviral vector | BE3 | 3 targeting HPRT 4 targeting Puromycin 4 targeting mCherry 16 controls | 7 sgRNA | Demonstration of CRISPR-STOP approach in a functional screen | (Kuscu et al., 2017)( |
Applications of Precision Editing
As a therapeutic platform
The precise conversion of cytidine to thymidine by the BE system lends itself as a potential therapeutic approach for the targeted insertion of beneficial mutations. Komor et al. first demonstrated the potential use of this system by correcting Alzheimer’s Disease-associated mutations in APOE4 in mouse astrocytes, and TP53 mutations in a mouse breast cancer line (Komor et al., 2016). Since this initial demonstration, there have been many successful examples of base editing to substitute a desired mutation (Table 1). Base editing systems, most notably BE3, have been used in a wide range of mammalian cell culture models including HEK293Ts (Chadwick et al., 2017; Kim et al., 2017a; Kim et al., 2017c; Komor et al., 2016), human iPSCs (Chadwick et al., 2017), CHO cells (Nishida et al., 2016), U2OS cells (Kim et al., 2017c; Komor et al., 2016), HCC1954 cells (Komor et al., 2016), mouse Neuro-2a cells, (Chadwick et al., 2017), and mouse astrocytes (Komor et al., 2016). There have also been successful demonstrations of base editing in vivo via delivery by adenoviral vector into mouse liver (Chadwick et al., 2017) and by RNP delivery into the mouse inner ear (Rees et al., 2017). Zebrafish (Rees et al., 2017; Zhang et al., 2017) and mouse embryos (Kim et al., 2017b; Liang et al., 2017) have also been successfully base edited, with the latter being reintroduced into surrogate mothers and giving rise to properly-edited offspring.
Komor et al. estimated that 3000 genetic variants listed in ClinVar would be correctable with a C>T substitution. However, given PAM sequence requirements, narrow base editing windows relative to the guide-targeted PAM sequence, and the potential need to avoid editing multiple cytidines in the editing window, they further estimated that only 300–900 would be targetable (Komor et al., 2016). To address these limitations, the BE system has been fused to an array of Cas9 variants with varied PAM-sequence specificities derived from S. pyogenes (‘NGG’), S. aureus (‘NNGRRT’), or evolved artificially (‘NGA’, VQR-Cas9; ‘NGAG’, EQR-Cas9; ‘NGCG’, VRER-Cas9; ‘NNNRRT’, SaKKH-Cas9), thus allowing the BE system to target 2200 of these C:T or G:A point mutations (Kim et al., 2017c; Komor et al., 2017b). The space of targetable mutations has been further expanded by the fusion of cytidine deaminases to other programmable DNA binding proteins such as zinc-fingers and TALENs, which are not restricted by PAM sites (Yang et al., 2016a).
Base editing systems may offer several advantages over traditional HDR-based CRISPR-Cas9 genome editing methods as a therapeutic tool. Importantly, these systems exhibit significantly lower indel rates, which decreases the chance of disrupting the function of the target gene through NHEJ (Komor et al., 2016). More recently, base editing constructs (e.g. YEE-BE3) have been engineered to decrease the number of undesirable substitutions generated proximal to the target site by narrowing the editing window to 1–2 bases (Kim et al., 2017c) (Box 2). Comparisons between the frequency of off-target mutations generated using base editing versus cleavage with active Cas9 nuclease suggest that base editing may have greater specificity (Kim et al., 2017a; Kim et al., 2017c). To further reduce non-specific mutations, BE3 has been fused to a high-fidelity SpCas9 (HF-Cas9) (Kleinstiver et al., 2016), which has reduced affinity to off-target mismatch sites, resulting in the creation of a HF-BE3 with lower non-specific editing rates (Rees et al., 2017). However, this benefit came at the cost of lower on-target editing rates.
Box 2. Specificity and Off-target base editing.
Unintended off-target mutations pose a challenge to the application of any genome editing technology. Researchers measuring and improving base-editing specificity can employ some of the same tools developed for preceding technologies, including computational prediction of off-targets and targeted deep sequencing to detect off-target mutations (reviewed in (Hsu et al., 2014; Tsai and Joung, 2016; Tycko et al., 2016)). However, distinct properties of base editors also create a need for distinct specificity methods and tools.
Proximal off-targets
Base editing at additional cytosines proximal to the targeted base may be unintended and considered as off-target mutations. One solution is to more tightly restrict the targetable window. Optimizing the rAPOBEC1 with a mutant deaminase domain effectively reduced the window from 4–6 bases to as little as 1–2 bases (Kim et al., 2017c). A triple mutant rAPOBEC1 (YEE-BE3) achieves this extreme precision at the cost of 2.9-fold lower average maximal editing than BE3, while double mutant YE1-BE3 retained maximal editing with a targeting window of 2–3 bases. Notably, the restricted targeting window decreases the likelihood of finding an appropriate NGG PAM site for a given target. However, base editors can be built with a variety of engineered S. pyogenes Cas9 PAM variants (Kleinstiver et al., 2016; Kleinstiver et al., 2015b) or S. aureus Cas9 and its engineered PAM variants (Kleinstiver et al., 2015a) to widen the number of available targeting sites (Kim et al., 2017c; Komor et al., 2017b). Careful experimental design can sometimes obviate the need for these engineered variants as proximal off-target mutations are relatively inconsequential in coding regions if the off-target mutation creates a synonymous codon (Komor et al., 2016).
Genome-wide off-targets
Base editors can also induce distal off-target mutagenesis at other binding sites across the genome. In fact, as Cas9 binding is more promiscuous than Cas9 cleavage (Wu et al., 2014), one might expect less specificity from base editors than active Cas9 nuclease. However, head-to-head comparisons across a range of mismatched target sites show generally similar specificity rules, with notable exceptions in which their off-targets do not coincide (Kim et al., 2017a). For example, BE3 appears more tolerant of DNA-sgRNA mismatch bulges than Cas9. Unsurprisingly, Cas9 is more prone to off-target effects at sites without editable cytosines in the BE3-targetable window as the base editor induces minimal mutagenesis of non-cytosine bases.
These exceptions, along with the fact that most off-target detection methods rely on DSBs and would not be applicable to base editors, motivate the development of new methods for the unbiased detection of base editing genome-wide. The developers of Digenome-seq, an in vitro genome-wide detection method (Kim et al., 2015), recently modified the protocol so it could be applied to base editors (Kim et al., 2017a). Their modified protocol took advantage of the deaminated uracil intermediate, which rarely occurs in the genome. They applied the USER enzymes, (a uracil DNA glycosylase and DNA glycosylase-lyase endonuclease VIII), to cleave off the base-edited uracils on one strand while the Cas9 D10A nicks the opposite strand. This generates an enrichment of free DNA ends at the targeted site which can be computationally identified in aligned whole genome sequencing data. Given that this process requires uracil DNA glycosylase activity to cleave the DNA, BE3 was modified so that it no longer included the uracil glycosylase inhibitor. Across 7 different guide RNAs, they found C to U conversions at only 18 ± 9 sites in the human genome. These off-targets were often different from those of active Cas9.
Standard whole genome sequencing (WGS) can also be used to measure editing genome-wide in cells. WGS was applied in the smaller yeast genome and detected a slight increase in single nucleotide variants which could be attributed to the deaminase but not the Cas9 (Nishida et al., 2016). When WGS was applied in a F0 base-edited mouse, no off-target mutations were discovered, demonstrating the high specificity of base editing in some applications (Kim et al., 2017b). While WGS is appropriate for clonal or largely homogenous genome mixtures, the modified Digenome-seq in vitro method is more sensitive to rare off-targets, detecting sites with substitution frequencies as low as 0.1% of alleles. Nonetheless, there remains a need for improved methods for unbiased off-target detection in cells and tissues with increased sensitivity to detect variants in heterogenous populations.
Minimizing off-target editing
Optimizing sgRNA design can also lower the frequency of off-targets at mismatched sites. While there are a number of active nuclease Cas9 sgRNA design tools that can predict off-targets, most do not explicitly consider the targetable windows of base editors as a design constraint. At least one tool (www.benchling.com) allows users to rank base editing sgRNAs by their predicted off target effects, but there remains a need for data-driven models of base editing specificity. It is important to consider not only mismatched sites, but also sites with small indels or non-canonical PAMs (Leenay et al., 2016). Ideally, sgRNAs should also be designed to be specific in the actual genome under experimentation, and not merely in the reference genome (Yang et al., 2014). Beyond careful sgRNA selection, specificity may be improved by modifying the sgRNA construct. For example, extending the sgRNA with two additional guanines (ggX20) maintained on-target efficiency while improving specificity in one study (Kim et al., 2017a).
Cas9 fusion proteins can also be optimized for improved specificity. The engineered rAPOBEC1 variants with restricted targeting windows decrease off-target editing at mismatched sites (Kim et al., 2017a). Alternatively, installing the BE3 fusion onto an engineered ‘high fidelity’ HF-Cas9 (Kleinstiver et al., 2016) reduces off-target effects, though this results in somewhat reduced on-target efficiency (Rees et al., 2017). However, delivering BE3 as a purified ribonucleoprotein (RNP) resulted in even greater specificity than HF-BE3 plasmid delivery while maintaining on-target efficiency (Rees et al., 2017), which is consistent with previous demonstrations of Cas9 RNP specificity (Kim et al., 2014; Zuris et al., 2015).
Overall, off-targets can be minimized by iterating a process of: 1) selecting specific sgRNAs (Doench et al., 2016; Perez et al., 2017; Scott and Zhang, 2017), 2) employing optimized Cas9 variants and delivery methods, 3) detecting mutations with unbiased genome-wide methods, and 4) validating off-target edits with sensitive targeted sequencing.
Additional engineering has been done to control the dynamics of base editing. The Liu group embedded self-cleaving catalytic RNA aptazymes in the sgRNA, which block the sgRNA spacer sequence until exposure to a ligand (Tang et al., 2017). This makes base editing responsive to a ligand, although the system exhibited some activity without ligand and was somewhat less efficient than standard sgRNAs. Additional means of inducing Cas9 activity, such as light (Nihongaki et al., 2015; Polstein and Gersbach, 2015), cell surface interactions (Dingal et al., 2017), and other small molecules (Gao et al., 2016; Senturk et al., 2017; Zetsche et al., 2015b), should be transferrable to base editors and enable fine control of editing dynamics.
Notably, the unbiased, sensitive, genome-wide off-target detection methods in cells (i.e. GUIDE-seq (Tsai et al., 2015)) and tissues (i.e. BLESS (Ran et al., 2015)) rely on double-stranded breaks and are not compatible with base editors in their current form. While base editor off-targets can be characterized with in vitro assays (i.e. Digenome-seq (Kim et al., 2017a)), whole genome sequencing, and targeted sequencing; unbiased methods have not been developed for detecting off-target base edits in cells and tissues. Additional improvements to specificity and off-target detection would accelerate translation of safe and effective base editing therapeutics.
Engineering Agricultural Traits
The recent demonstrations of efficient base editing in crops (Lu and Zhu, 2017; Shimatani et al., 2017; Zong et al., 2017) (Figure 2A) provide a new method to select for and fine-tune desirable crop traits which are attributable to point mutations, such as 14 key agronomic traits in rice (Huang et al., 2010) and the flavor of tomatoes (Tieman et al., 2017). This can be further extended to other agricultural demands including improving nutrition and production of crops (reviewed in (Baltes et al., 2017)). In rice, multiplex base editing of ALS and FTIP1e generated double mutants resistant to two herbicides (Shimatani et al., 2017). Interestingly, despite using similar Target-AID constructs, the rate of indels has been significantly higher in crops (Lu and Zhu, 2017; Shimatani et al., 2017) (Table 1) than in yeast or mammalian cells (Nishida et al., 2016). Nonetheless, indel rates are still lower than using traditional HDR-based CRISPR-Cas9 gene editing (Li et al., 2013; Mao et al., 2013). Base editing tools have also been successfully used in wheat, maize (Zong et al., 2017), and Arabidopsis (Chen et al., 2017). Delivery of base editing systems to plants has been performed using either transformed agrobacterium (Hooykaas and Schilperoort, 1992; Voytas, 2013) or biolistic particle bombardment (Sanford, 1988). Although delivery of DNA via agrobacterium is more controllable for single copy insertions, the particle bombardment strategy can be used to deliver the system to a wider range of organisms, including those that are not compatible with agrobacterium colonization (Nam et al., 1997). Both methods have been effective (Table 1) in a variety of plants. Given these initial successes, it is likely that base editing may be broadly useful in other varieties of crops, plants (Baltes et al., 2017) and animals (Petersen, 2017) as well.
Applications of Diversifying Editing
Evolving protein function and structure
Base editing systems have been used for protein evolution and saturation mutagenesis (Table 2 and Figure 2B), and present a number of useful features when compared to traditional methods such as error-prone PCR, recombination, and DNA synthesis (reviewed in (Packer and Liu, 2015)). More recently, active Cas9 has been used together with a synthetic library of oligonucleotide HDR templates (Findlay et al., 2014; Garst et al., 2017; Ryan et al., 2014) to maximize variant sequence space by allowing complete control of all base substitutions at a specified locus. However, these techniques require laborious synthesis of donor templates for each region, and carry the risk of DNA damage and indel generation by active Cas9. The ability to efficiently create diverse populations of mutations at precise regions within endogenous genes using diversifying base editors eliminates the need to separately synthesize and deliver a donor library -- which may be constrained by donor length, sequence complexity, and library size. Modifying the endogenous gene obviates the need to use artificial promoters to express a variant library, which may change the native regulation of protein function and localization. Proteins evolved in bacteria or yeast may also not function appropriately when transferred back into mammalian systems (Way et al., 2014; Yuen et al., 2006). At the same time, the targeted nature of the base editing approach decreases the number of cells and reduces sequencing cost that would otherwise be needed to analyze a mutagenized genome generated with radiation or chemical mutagenesis.
AID has evolved as an efficient generator of genetic diversity. B cell lymphoma lines with misregulated AID-dependent somatic hypermutation have been used to evolve antibodies or fluorescent proteins (Arakawa et al., 2008; Lim et al., 2016; Wang et al., 2004), but the promiscuous mutagenesis in this system is relatively inefficient. In contrast, by directly recruiting AID, diversifying base editing systems (CRISPR-X and TAM) can facilitate efficient, targeted mutagenesis of key structural or functional domains of endogenous targets through carefully designed sgRNA pools. For example, the CRISPR-X platform has been used to evolve wtGFP to EGFP using a targeted pool of sgRNAs. In this proof-of-principle study, dCas9-expressing K562 cells were electroporated with MS2-AID*Δ and sgRNAs targeting wtGFP near known spectrum-shifting mutation sites; cells with shifted emission spectra were sorted by FACS and the GFP locus was sequenced. The identified residue change (S65T) matched the previously known mutation to confer the wtGFP to EGFP spectral shift (Heim and Tsien, 1996). The broad editing window of CRISPR-X and its ability to generate diverse base transitions and transversions are well-suited for the directed evolution of proteins (Hess et al., 2016).
In addition to the evolution of protein function, base editing systems can also be used to map key structural features required for protein-protein and protein-drug interactions. For example, base editing across full protein coding regions performed using the TAM and CRISPR-X platforms has enabled the identification of known and novel drug-resistant gene variants, such as mutations in PSMB5 that confer bortezomib resistance (Hess et al., 2016), and mutations in the BCR-ABL kinase domain that confer imatinib resistance (Ma et al., 2016). Identifying variant-drug interactions can improve molecular diagnosis to inform clinical treatment, lend insight towards a mechanistic understanding of drug resistance, and may facilitate drug design. Other groups have successfully used active Cas9 to mutagenize protein coding regions, wherein error-prone NHEJ mechanisms create gene variants that confer novel phenotypes such as drug resistance (Donovan et al., 2017; Ipsaro et al., 2017). While using active Cas9 for mutagenesis is straightforward, a large portion of NHEJ-generated mutations are frameshifts and multiple base indels, which may be either highly useful or undesirable depending on the application (Shalem et al., 2014; van Overbeek et al., 2016; Wang et al., 2014). Therefore, this approach may require a larger starting cell population as well as a strong selectable phenotype to enrich for single point mutants. Thus, diversifying base editors present a useful alternative for broad mutagenesis screens by minimizing indels and creating more varied point mutations.
Screening Gene Knockouts with CRISPR-STOP and iSTOP
Precision base editing can also be used to screen the effects of gene knockouts. The CRISPR-STOP and iSTOP systems use BE3 to efficiently generate early stop codons at arginine, glutamine, and tryptophan residues (Figure 2C), effectively causing gene inactivation (Billon et al., 2017; Kuscu et al., 2017). In addition to transcriptional repression using CRISPRi (Gilbert et al., 2014; Gilbert et al., 2013), CRISPR-STOP and iSTOP may be an attractive alternative approach for disrupting the function of genes present at a high copy number, where active Cas9 nuclease can sometimes create heterogenous repair outcomes and trigger excessive DNA damage and cell death (Aguirre et al., 2016; Morgens et al., 2017; Munoz et al., 2016; Wang et al., 2015). In contrast with CRISPRi screens, CRISPR-STOP and iSTOP generate gene knockouts via early truncation at targeted loci rather than transcriptional knockdown, with potentially different phenotypic consequences. While genome-wide screens with CRISPR-STOP and iSTOP have not yet been conducted, these tools may have advantages where a defined protein truncation product is desired and it is necessary to avoid DSBs.
Section 3: Future Directions and Areas for Improvement
While base editing has already been successful in initial studies, several technological improvements would enable additional applications: increased editing specificity, decreased PAM site dependencies, and an expansion of control over the types and frequencies of base transitions and transversions. Nevertheless, current tools enable significant extension of current applications and exploration of new ground. In this final section, we briefly discuss how base editing systems might be improved, current limitations for their delivery in vivo, and future possibilities for base editing in therapy and engineering.
Future Improvements to Base Editing Tools
Additional engineering to the enzymes recruited by dCas9 may increase the versatility of base editing technology. Specifically, engineering the deaminase or DNA repair machinery may make it possible to direct different specific conversion events (e.g. convert C>A instead of C>T). It should also be noted that the deaminases comprise a large family of diverse enzymes. In addition to the APOBEC family and AID, these include adenosine (Alseth et al., 2014; Bass, 2002) and guanine deaminases (Roy and Roy, 1967; Yuan et al., 1999). For example, ADA is an adenosine deaminase involved in the purine salvage process that converts adenosine to inosine; loss of this enzyme causes autosomal recessive Severe Combined Immunodeficiency (SCID) (Hershfield, 1998; Hirschhorn, 1990). Recruitment of adenosine deaminases would expand the range of targetable bases to allow conversion of adenine (and thymidine as its reverse complement) to alternative bases. Furthermore, ADAR enzymes (Li and Church, 2013; Nishikura, 2010), adenosine deaminases able to act on RNA, open up the potential for post-transcriptional editing of mRNA (Hanswillemenke et al., 2015; McMahon et al., 2016; Montiel-Gonzalez et al., 2013; Schneider et al., 2014) using a modified Cas9 system capable of targeting RNA (Nelles et al., 2016) or the C2c2 CRISPR effector (Abudayyeh et al., 2016; East-Seletsky et al., 2016), which can be targeted to RNA.
New base editing Cas9 variants (alternate PAM sites (Kim et al., 2017c), Cpf1 (Zetsche et al., 2015a), high-fidelity variants (Rees et al., 2017)) may each have different susceptibilities to off-target effects, and will need to be carefully characterized. Although significant work has been done to quantify the on-target and off-target effects for active Cas9 (reviewed in (Hsu et al., 2014; Tsai and Joung, 2016; Tycko et al., 2016)), similar methods for base editing are only emerging (Box 2). These new methods will be critical to benchmark improvements in base editing platforms.
Therapeutic delivery of CRISPR base editors
Therapeutic base editing will depend on efficient delivery. RNP lipofection (Rees et al., 2017), mRNA microinjection (Kim et al., 2017b), and administration of adenoviral vectors (Chadwick et al., 2017) have been used for making therapeutic precision edits (Table 1). A comparison of the plasmid and RNP delivery methods found that RNPs had higher specificity (Rees et al., 2017). Another study that compared mRNA injections and RNPs found that mutant ratios among blastocysts and transferred embryos that gave rise to offspring were both efficient (Kim et al., 2017b). Given that the choice of delivery method may influence editing efficiency and specificity, it will be important to systematically and directly compare delivery methods for each application. Notably, adenovirus has been superseded by adeno-associated virus (AAV) in in vivo gene therapy pipelines because of AAV’s favorable efficiency and immunogenicity profile (Valdmanis and Kay, 2017). However, base editors are large fusion proteins that exceed the packaging capacity of AAV. Further efforts to reduce the size (Friedland et al., 2015; Zetsche et al., 2015b) and limit immunogenicity of base editing components or vehicles (Abudayyeh et al., 2016; Bessis et al., 2004; Chew et al., 2016; Hirsch et al., 2016; Wang et al., 2017; Yin et al., 2016; Zuris et al., 2015) may also improve their clinical utility.
Identifying and characterizing pathogenic variants
Base editing can efficiently install mutations and generate variant libraries to screen for specific phenotypes; combining these two current applications may facilitate high throughput characterization of known and putatively pathogenic SNPs/SNVs. While the majority of known disease-associated gene variants are point mutations (Landrum et al., 2016), there is often insufficient statistical power from human subject studies to establish a causal relationship between specific variants and their putative phenotypes (Strande et al., 2017). In cases where base editors can target and correct the mutations, such edits can rescue the wild type phenotype in vitro or in animal models, validating the mutation. Recently, active Cas9 has also been used to validate possible causal variants affecting MYB expression through saturation mutagenesis (Canver et al., 2017). In addition to characterizing variants, base editors with broad, diversifying properties can be used to identify known and novel variants that affect measurable phenotypes (e.g. alternative splicing behaviors or gene expression levels). The information generated from these approaches could be used to support a causal variant-phenotype relationship, or improve our understanding of disease mechanisms and identify key functional residues.
Investigating the non-coding genome
Base editing systems may also be well-suited to investigate sequence-function relationships in enhancers and other non-coding regions. Previously, active Cas9 and CRISPRi have been used to tile enhancer regions to identify important regulatory elements that control gene expression (Canver et al., 2017; Canver et al., 2015; Diao et al., 2017; Fulco et al., 2016; Gasperini et al., 2017; Korkmaz et al., 2016; Sanjana et al., 2016; Xie et al., 2017). Precise base editing systems, and those with broader editing windows like CRISPR-X, may be similarly applied to interrogate regulatory elements through saturation mutagenesis. While base editors used in such a way may identify the sequence determinants of a regulatory element’s function at single-nucleotide resolution, the narrower functional and mutagenic windows may practically constrain the amount of sequence space that can be screened. Mutations generated by current base editors may not necessarily disrupt regulatory function, whereas active Cas9 and CRISPRi may be more readily able to inhibit activity -- although repressing enhancers and other regulatory elements with CRISPRi may also vary in efficiency depending on the chosen repressor and surrounding epigenetic signature.
Mapping Protein-Protein and Protein-Drug Interactions
The CRISPR-X and TAM base editing systems have been used to identify mutations that confer drug resistance. Both systems may be extended to map and characterize protein-protein and protein-ligand interactions to determine biological function. As with the identification of bortezomib- and imatinib-resistant mutants, an efficient interaction screen would likely require a selectable phenotype amenable to high-throughput screening, such as cell growth and survival, reporter expression, or two-component reporter systems such as the KInase Substrate Sensor (KISS) (Lievens et al., 2014).
One of the benefits of Cas9 based technologies is the straightforward path to simultaneously target multiple sites in the genome. Indeed, both Target-AID and CRISPR-X systems have been used to mutagenize two loci within a cell (Hess et al., 2016; Nishida et al., 2016). This capacity opens up a number of possibilities for future applications. Because the repair mechanism for base editing does not involve double strand breaks -- a requirement for HDR-mediated editing -- multiple edits can be made without risk of translocations or paired deletions between the cut sites. Simultaneous mutagenesis of two sites should make it possible to study protein-protein interactions through co-evolution.
Base Editing for Directed Evolution and Synthetic Biology
While base editing can be used to investigate molecular interactions, it can also be used for the directed evolution of improved biomolecules. As an initial proof-of-principle, wild-type GFP was evolved to EGFP using CRISPR-X (Hess et al., 2016). This strategy can be used in both industrial engineering and synthetic biology applications (Cobb et al., 2013). For example, metabolic engineering (Jakociunas et al., 2017) of microbes or yeast can be accomplished by generating focused libraries of mutants in situ to refine the function of a desired enzyme or pathway. The base editing diversification strategies provide a straightforward means of generating new libraries without requiring the library to be removed from the organism, randomized through gene shuffling or error-prone PCR, and replaced. The endeavor to engineer promoters, sensors, or other regulatory elements and functional circuits may also benefit from base editing tools, which can mutate these components within mammalian cells in their native genomic context (Lienert et al., 2014; Mathur et al., 2017).
Conclusion
In summary, base editing presents a useful alternative to HDR-mediated genome editing with active Cas9 nuclease, avoiding double strand breaks and the resulting indels. Capable of generating either precise C>T edits or targeted diversification of a genomic region, base editing systems have been applied to therapeutic correction of deleterious mutations, engineering of agricultural crops, and the study and evolution of protein structure and function. As with any emerging technology, base editing systems will require additional fine tuning and characterization, especially for therapeutic applications of precise base editing. Nonetheless, base editing is an exciting new addition to the CRISPR toolbox, and its potential in genome engineering and biology is just beginning to be realized.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang CZ, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, et al. Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer Discov. 2016;6:914–929. doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseth I, Dalhus B, Bjoras M. Inosine in DNA and RNA. Curr Opin Genet Dev. 2014;26:116–123. doi: 10.1016/j.gde.2014.07.008. [DOI] [PubMed] [Google Scholar]
- Arakawa H, Kudo H, Batrak V, Caldwell RB, Rieger MA, Ellwart JW, Buerstedde JM. Protein evolution by hypermutation and selection in the B cell line DT40. Nucleic Acids Res. 2008;36:e1. doi: 10.1093/nar/gkm616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout R, Lee W, Cahill P, Honan T, Sparrow T, Weiand M, Nusbaum C, Rajewsky K, Koralov SB. High-resolution description of antibody heavy-chain repertoires in humans. PLoS One. 2011;6:e22365. doi: 10.1371/journal.pone.0022365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak RO, Porteus MH. CRISPR-Mediated Integration of Large Gene Cassettes Using AAV Donor Vectors. Cell Rep. 2017;20:750–756. doi: 10.1016/j.celrep.2017.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes NJ, Gil-Humanes J, Voytas DF. Genome Engineering and Agriculture: Opportunities and Challenges. Prog Mol Biol Transl Sci. 2017;149:1–26. doi: 10.1016/bs.pmbts.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R. CRISPR-Cas systems and RNA-guided interference. Wiley Interdisciplinary Reviews: RNA. 2013;4:267–278. doi: 10.1002/wrna.1159. [DOI] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benichou J, Ben-Hamo R, Louzoun Y, Efroni S. Rep-Seq: uncovering the immunological repertoire through next-generation sequencing. Immunology. 2012;135:183–191. doi: 10.1111/j.1365-2567.2011.03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessis N, GarciaCozar FJ, Boissier MC. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 2004;11(Suppl 1):S10–17. doi: 10.1038/sj.gt.3302364. [DOI] [PubMed] [Google Scholar]
- Billon P, Bryant EE, Joseph SA, Nambiar TS, Hayward SB, Rothstein R, Ciccia A. CRISPR-Mediated Base Editing Enables Efficient Disruption of Eukaryotic Genes through Induction of STOP Codons. Mol Cell. 2017 doi: 10.1016/j.molcel.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, Van Der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Lessard S, Pinello L, Wu Y, Ilboudo Y, Stern EN, Needleman AJ, Galacteros F, Brugnara C, Kutlar A, et al. Variant-aware saturating mutagenesis using multiple Cas9 nucleases identifies regulatory elements at trait-associated loci. Nat Genet. 2017;49:625–634. doi: 10.1038/ng.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certo MT, Ryu BY, Annis JE, Garibov M, Jarjour J, Rawlings DJ, Scharenberg AM. Tracking genome engineering outcome at individual DNA breakpoints. Nat Methods. 2011;8:671–676. doi: 10.1038/nmeth.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick AC, Wang X, Musunuru K. In Vivo Base Editing of PCSK9 (Proprotein Convertase Subtilisin/Kexin Type 9) as a Therapeutic Alternative to Genome Editing. Arterioscler Thromb Vasc Biol. 2017 doi: 10.1161/ATVBAHA.117.309881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L. Apolipoprotein B, the major protein component of triglyceride-rich and low density lipoproteins. J Biol Chem. 1992;267:25621–25624. [PubMed] [Google Scholar]
- Charton K, Suel L, Henriques SF, Moussu JP, Bovolenta M, Taillepierre M, Becker C, Lipson K, Richard I. Exploiting the CRISPR/Cas9 system to study alternative splicing in vivo: application to titin. Hum Mol Genet. 2016;25:4518–4532. doi: 10.1093/hmg/ddw280. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wang Z, Ni H, Xu Y, Chen Q, Jiang L. CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in Arabidopsis. Sci China Life Sci. 2017;60:520–523. doi: 10.1007/s11427-017-9021-5. [DOI] [PubMed] [Google Scholar]
- Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, Church GM. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods. 2016;13:868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Greene WC. Multifaceted antiviral actions of APOBEC3 cytidine deaminases. Trends Immunol. 2006;27:291–297. doi: 10.1016/j.it.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Cobb RE, Sun N, Zhao H. Directed evolution as a powerful synthetic biology tool. Methods. 2013;60:81–90. doi: 10.1016/j.ymeth.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Weller GR, Doherty AJ, Jackson SP. The Gam protein of bacteriophage Mu is an orthologue of eukaryotic Ku. EMBO Rep. 2003;4:47–52. doi: 10.1038/sj.embor.embor709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, Pavel-Dinu M, Saxena N, Wilkens AB, Mantri S, et al. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt MA, Magis W, Bray NL, Wang T, Berman JR, Urbinati F, Heo SJ, Mitros T, Munoz DP, Boffelli D, et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci Transl Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y, Fang R, Li B, Meng Z, Yu J, Qiu Y, Lin KC, Huang H, Liu T, Marina RJ, et al. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat Methods. 2017;14:629–635. doi: 10.1038/nmeth.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, Cowan CA, Rader DJ, Musunuru K. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingal PDP, Kipniss NH, Labanieh L, Gao Y, Qi LS. Engineering Cell Sensing and Responses Using a GPCR-Coupled CRISPR-Cas System. bioRxiv. 2017:152496. doi: 10.1038/s41467-017-02075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan KF, Hegde M, Sullender M, Vaimberg EW, Johannessen CM, Root DE, Doench JG. Creation of Novel Protein Variants with CRISPR/Cas9-Mediated Mutagenesis: Turning a Screening By-Product into a Discovery Tool. PLoS One. 2017;12:e0170445. doi: 10.1371/journal.pone.0170445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JH, Tjian R, Doudna JA. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhanati Y, Sethna Z, Marcou Q, Callan CG, Jr, Mora T, Walczak AM. Inferring processes underlying B-cell repertoire diversity. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2014.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Melnikov A, Zhang X, Wang L, Rogov P, Mikkelsen TS, Kellis M. Genome-scale high-resolution mapping of activating and repressive nucleotides in regulatory regions. Nat Biotechnol. 2016;34:1180–1190. doi: 10.1038/nbt.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, Marco E, Gotta GM, Maeder ML, Kennedy EM, et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, Engreitz JM. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 2016;354:769–773. doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Xiong X, Wong S, Charles EJ, Lim WA, Qi LS. Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat Methods. 2016;13:1043–1049. doi: 10.1038/nmeth.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis M-E, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Garst AD, Bassalo MC, Pines G, Lynch SA, Halweg-Edwards AL, Liu R, Liang L, Wang Z, Zeitoun R, Alexander WG, et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat Biotechnol. 2017;35:48–55. doi: 10.1038/nbt.3718. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini M, Findlay GM, McKenna A, Milbank JH, Lee C, Zhang MD, Cusanovich DA, Shendure J. CRISPR/Cas9-Mediated Scanning for Regulatory Elements Required for HPRT1 Expression via Thousands of Large, Programmed Genomic Deletions. Am J Hum Genet. 2017;101:192–205. doi: 10.1016/j.ajhg.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiff V, Bhat P, Cook SC, Menzel U, Kang W, Reddy ST. A bioinformatic framework for immune repertoire diversity profiling enables detection of immunological status. Genome Med. 2015;7:49. doi: 10.1186/s13073-015-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton CE, Papavasiliou FN, Rosenberg BR. Diverse functions for DNA and RNA editing in the immune system. RNA Biol. 2010;7:220–228. doi: 10.4161/rna.7.2.11344. [DOI] [PubMed] [Google Scholar]
- Hanswillemenke A, Kuzdere T, Vogel P, Jekely G, Stafforst T. Site-Directed RNA Editing in Vivo Can Be Triggered by the Light-Driven Assembly of an Artificial Riboprotein. J Am Chem Soc. 2015;137:15875–15881. doi: 10.1021/jacs.5b10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7:932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- Hershfield MS. Adenosine deaminase deficiency: clinical expression, molecular basis, and therapy. Semin Hematol. 1998;35:291–298. [PubMed] [Google Scholar]
- Hess GT, Fresard L, Han K, Lee CH, Li A, Cimprich KA, Montgomery SB, Bassik MC. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods. 2016;13:1036–1042. doi: 10.1038/nmeth.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch ML, Wolf SJ, Samulski RJ. Delivering Transgenic DNA Exceeding the Carrying Capacity of AAV Vectors. Methods Mol Biol. 2016;1382:21–39. doi: 10.1007/978-1-4939-3271-9_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn R. Adenosine deaminase deficiency. Immunodefic Rev. 1990;2:175–198. [PubMed] [Google Scholar]
- Hoehn KB, Fowler A, Lunter G, Pybus OG. The Diversity and Molecular Evolution of B-Cell Receptors during Infection. Mol Biol Evol. 2016;33:1147–1157. doi: 10.1093/molbev/msw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooykaas PJ, Schilperoort RA. Agrobacterium and plant genetic engineering. Plant Mol Biol. 1992;19:15–38. doi: 10.1007/BF00015604. [DOI] [PubMed] [Google Scholar]
- Hsieh CL, Liu H, Huang Y, Kang L, Chen HW, Chen YT, Wee YR, Chen SJ, Tan BC. ADAR1 deaminase contributes to scheduled skeletal myogenesis progression via stage-specific functions. Cell Death Differ. 2014;21:707–719. doi: 10.1038/cdd.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- Ipsaro JJ, Shen C, Arai E, Xu Y, Kinney JB, Joshua-Tor L, Vakoc CR, Shi J. Rapid generation of drug-resistance alleles at endogenous loci using CRISPR-Cas9 indel mutagenesis. PLoS One. 2017;12:e0172177. doi: 10.1371/journal.pone.0172177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino M, Sawada Y, Yaegashi T, Demura M, Fujinaga K. Nucleotide sequence of the adenovirus type 40 inverted terminal repeat: close relation to that of adenovirus type 5. Virology. 1987;156:414–416. doi: 10.1016/0042-6822(87)90421-1. [DOI] [PubMed] [Google Scholar]
- Jakociunas T, Jensen MK, Keasling JD. System-level perturbations of cell metabolism using CRISPR/Cas9. Curr Opin Biotechnol. 2017;46:134–140. doi: 10.1016/j.copbio.2017.03.014. [DOI] [PubMed] [Google Scholar]
- Jansen R, Embden J, Gaastra W, Schouls L. Identification of genes that are associated with DNA repeats in prokaryotes. Molecular microbiology. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Jasin M, Haber JE. The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair (Amst) 2016;44:6–16. doi: 10.1016/j.dnarep.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Doudna JA. CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahnke M, Banning A, Tikkanen R. Random Splicing of Several Exons Caused by a Single Base Change in the Target Exon of CRISPR/Cas9 Mediated Gene Knockout. Cells. 2016;5 doi: 10.3390/cells5040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan LP, Gallo A, O’Connell MA. The many roles of an RNA editor. Nat Rev Genet. 2001;2:869–878. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim JI, Kim JS. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. 231 p following 243. [DOI] [PubMed] [Google Scholar]
- Kim D, Lim K, Kim ST, Yoon SH, Kim K, Ryu SM, Kim JS. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat Biotechnol. 2017a;35:475–480. doi: 10.1038/nbt.3852. [DOI] [PubMed] [Google Scholar]
- Kim K, Ryu SM, Kim ST, Baek G, Kim D, Lim K, Chung E, Kim S, Kim JS. Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol. 2017b;35:435–437. doi: 10.1038/nbt.3816. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol. 2017c;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, Joung JK. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015a;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015b;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koito A, Ikeda T. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front Microbiol. 2013;4:28. doi: 10.3389/fmicb.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017a;169:559. doi: 10.1016/j.cell.2017.04.005. [DOI] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Zhao KT, Packer MS, Gaudelli NM, Waterbury AL, Koblan LW, Kim YB, Badran AH, Liu DR. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv. 2017b;3:eaao4774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz G, Lopes R, Ugalde AP, Nevedomskaya E, Han R, Myacheva K, Zwart W, Elkon R, Agami R. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol. 2016;34:192–198. doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- Kuppers R, Klein U, Hansmann ML, Rajewsky K. Cellular origin of human B-cell lymphomas. N Engl J Med. 1999;341:1520–1529. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- Kuscu C, Parlak M, Tufan T, Yang J, Szlachta K, Wei X, Mammadov R, Adli M. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat Methods. 2017;14:710–712. doi: 10.1038/nmeth.4327. [DOI] [PubMed] [Google Scholar]
- Lackner DH, Carre A, Guzzardo PM, Banning C, Mangena R, Henley T, Oberndorfer S, Gapp BV, Nijman SM, Brummelkamp TR, et al. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat Commun. 2015;6:10237. doi: 10.1038/ncomms10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson ED, Cummings WJ, Bednarski DW, Maizels N. MRE11/RAD50 cleaves DNA in the AID/UNG-dependent pathway of immunoglobulin gene diversification. Mol Cell. 2005;20:367–375. doi: 10.1016/j.molcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- Leenay RT, Maksimchuk KR, Slotkowski RA, Agrawal RN, Gomaa AA, Briner AE, Barrangou R, Beisel CL. Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. Molecular cell. 2016;62:137–147. doi: 10.1016/j.molcel.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonetti MD, Sekine S, Kamiyama D, Weissman JS, Huang B. A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc Natl Acad Sci U S A. 2016;113:E3501–3508. doi: 10.1073/pnas.1606731113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Church GM. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat Neurosci. 2013;16:1518–1522. doi: 10.1038/nn.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Sun H, Sun Y, Zhang X, Xie X, Zhang J, Zhang Z, Chen Y, Ding C, Xiong Y, et al. Effective gene editing by high-fidelity base editor 2 in mouse zygotes. Protein Cell. 2017;8:601–611. doi: 10.1007/s13238-017-0418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Lohmueller JJ, Garg A, Silver PA. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat Rev Mol Cell Biol. 2014;15:95–107. doi: 10.1038/nrm3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens S, Gerlo S, Lemmens I, De Clercq DJ, Risseeuw MD, Vanderroost N, De Smet AS, Ruyssinck E, Chevet E, Van Calenbergh S, et al. Kinase Substrate Sensor (KISS), a mammalian in situ protein interaction sensor. Mol Cell Proteomics. 2014;13:3332–3342. doi: 10.1074/mcp.M114.041087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AW, Williams GT, Rada C, Sale JE. Directed evolution of human scFvs in DT40 cells. Protein Eng Des Sel. 2016;29:39–48. doi: 10.1093/protein/gzv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Zhu JK. Precise Editing of a Target Base in the Rice Genome Using a Modified CRISPR/Cas9 System. Mol Plant. 2017;10:523–525. doi: 10.1016/j.molp.2016.11.013. [DOI] [PubMed] [Google Scholar]
- Ma Y, Zhang J, Yin W, Zhang Z, Song Y, Chang X. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods. 2016;13:1029–1035. doi: 10.1038/nmeth.4027. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant. 2013;6:2008–2011. doi: 10.1093/mp/sst121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur M, Xiang JS, Smolke CD. Mammalian synthetic biology for studying the cell. J Cell Biol. 2017;216:73–82. doi: 10.1083/jcb.201611002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AC, Rahman R, Jin H, Shen JL, Fieldsend A, Luo W, Rosbash M. TRIBE: Hijacking an RNA-Editing Enzyme to Identify Cell-Specific Targets of RNA-Binding Proteins. Cell. 2016;165:742–753. doi: 10.1016/j.cell.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Molecular microbiology. 2000;36:244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, Mosbaugh DW, Tainer JA. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell. 1995;82:701–708. doi: 10.1016/0092-8674(95)90467-0. [DOI] [PubMed] [Google Scholar]
- Montiel-Gonzalez MF, Vallecillo-Viejo I, Yudowski GA, Rosenthal JJ. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc Natl Acad Sci U S A. 2013;110:18285–18290. doi: 10.1073/pnas.1306243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora T, Walczak AM, Bialek W, Callan CG., Jr Maximum entropy models for antibody diversity. Proc Natl Acad Sci U S A. 2010;107:5405–5410. doi: 10.1073/pnas.1001705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens DW, Wainberg M, Boyle EA, Ursu O, Araya CL, Tsui CK, Haney MS, Hess GT, Han K, Jeng EE, et al. Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat Commun. 2017;8:15178. doi: 10.1038/ncomms15178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H, Smith JL, Peng L, Yin H, Moore J, Zhang XO, Song CQ, Sheel A, Wu Q, Ozata DM, et al. CRISPR/Cas9-mediated genome editing induces exon skipping by alternative splicing or exon deletion. Genome Biol. 2017;18:108. doi: 10.1186/s13059-017-1237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Munoz DM, Cassiani PJ, Li L, Billy E, Korn JM, Jones MD, Golji J, Ruddy DA, Yu K, McAllister G, et al. CRISPR Screens Provide a Comprehensive Assessment of Cancer Vulnerabilities but Generate False-Positive Hits for Highly Amplified Genomic Regions. Cancer Discov. 2016;6:900–913. doi: 10.1158/2159-8290.CD-16-0178. [DOI] [PubMed] [Google Scholar]
- Nam J, Matthysse AG, Gelvin SB. Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell. 1997;9:317–333. doi: 10.1105/tpc.9.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaratnam N, Sarwar R. An overview of cytidine deaminases. Int J Hematol. 2006;83:195–200. doi: 10.1532/IJH97.06032. [DOI] [PubMed] [Google Scholar]
- Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell. 2016;165:488–496. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M. CRISPR-Cas9-based photoactivatable transcription system. Chem Biol. 2015;22:169–174. doi: 10.1016/j.chembiol.2014.12.011. [DOI] [PubMed] [Google Scholar]
- Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353 doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- Packer MS, Liu DR. Methods for the directed evolution of proteins. Nat Rev Genet. 2015;16:379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC, 3rd, Nussenzweig MC, Dalla-Favera R. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- Perez AR, Pritykin Y, Vidigal JA, Chhangawala S, Zamparo L, Leslie CS, Ventura A. GuideScan software for improved single and paired CRISPR guide RNA design. Nat Biotechnol. 2017;35:347–349. doi: 10.1038/nbt.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen B. Basics of genome editing technology and its application in livestock species. Reprod Domest Anim. 2017;52(Suppl 3):4–13. doi: 10.1111/rda.13012. [DOI] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal N, Srinivasan S, Kooshesh K, Guo Y, Edwards MD, Banerjee B, Syed T, Emons BJ, Gifford DK, Sherwood RI. High-throughput mapping of regulatory DNA. Nat Biotechnol. 2016;34:167–174. doi: 10.1038/nbt.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HA, Komor AC, Yeh WH, Caetano-Lopes J, Warman M, Edge ASB, Liu DR. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun. 2017;8:15790. doi: 10.1038/ncomms15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell. 2015;58:586–597. doi: 10.1016/j.molcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Reynaud CA, Aoufouchi S, Faili A, Weill JC. What role for AID: mutator, or assembler of the immunoglobulin mutasome? Nat Immunol. 2003;4:631–638. doi: 10.1038/ni0703-631. [DOI] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, Bray NL, Corn JE. Non-homologous DNA increases gene disruption efficiency by altering DNA repair outcomes. Nat Commun. 2016;7:12463. doi: 10.1038/ncomms12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins H. Immunosequencing: applications of immune repertoire deep sequencing. Curr Opin Immunol. 2013;25:646–652. doi: 10.1016/j.coi.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Roy JE, Roy KL. The mechanism and specificity of guanine deaminase. Can J Biochem. 1967;45:1263–1269. doi: 10.1139/o67-147. [DOI] [PubMed] [Google Scholar]
- Ryan OW, Skerker JM, Maurer MJ, Li X, Tsai JC, Poddar S, Lee ME, DeLoache W, Dueber JE, Arkin AP, et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife. 2014;3 doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoparnig T, Fried P, Beerenwinkel N. Identification of constrained cancer driver genes based on mutation timing. PLoS Comput Biol. 2015;11:e1004027. doi: 10.1371/journal.pcbi.1004027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE. ADARs: viruses and innate immunity. Curr Top Microbiol Immunol. 2012;353:163–195. doi: 10.1007/82_2011_148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JC. The biolistic process. Trends in Biotechnology. 1988;6:299–302. [Google Scholar]
- Sanjana NE, Wright J, Zheng K, Shalem O, Fontanillas P, Joung J, Cheng C, Regev A, Zhang F. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353:1545–1549. doi: 10.1126/science.aaf7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MF, Wettengel J, Hoffmann PC, Stafforst T. Optimal guideRNAs for re-directing deaminase activity of hADAR1 and hADAR2 in trans. Nucleic Acids Res. 2014;42:e87. doi: 10.1093/nar/gku272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DA, Zhang F. Implications of human genetic variation in CRISPR-based therapeutic genome editing. Nat Med. 2017 doi: 10.1038/nm.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senturk S, Shirole NH, Nowak DG, Corbo V, Pal D, Vaughan A, Tuveson DA, Trotman LC, Kinney JB, Sordella R. Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nat Commun. 2017;8:14370. doi: 10.1038/ncomms14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol. 2017;35:441–443. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, Doudna JA. Expanding the Biologist’s Toolkit with CRISPR-Cas9. Mol Cell. 2015;58:568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- Strande NT, Riggs ER, Buchanan AH, Ceyhan-Birsoy O, DiStefano M, Dwight SS, Goldstein J, Ghosh R, Seifert BA, Sneddon TP, et al. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am J Hum Genet. 2017;100:895–906. doi: 10.1016/j.ajhg.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Hu JH, Liu DR. Aptazyme-embedded guide RNAs enable ligand-responsive genome editing and transcriptional activation. Nat Commun. 2017;8:15939. doi: 10.1038/ncomms15939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D, Zhu G, Resende MF, Jr, Lin T, Nguyen C, Bies D, Rambla JL, Beltran KS, Taylor M, Zhang B, et al. A chemical genetic roadmap to improved tomato flavor. Science. 2017;355:391–394. doi: 10.1126/science.aal1556. [DOI] [PubMed] [Google Scholar]
- Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet. 2016;17:300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nature biotechnology. 2015;33:187. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko J, Myer VE, Hsu PD. Methods for Optimizing CRISPR-Cas9 Genome Editing Specificity. Mol Cell. 2016;63:355–370. doi: 10.1016/j.molcel.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdmanis PN, Kay MA. Future of rAAV Gene Therapy: Platform for RNAi, Gene Editing, and Beyond. Hum Gene Ther. 2017;28:361–372. doi: 10.1089/hum.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek M, Capurso D, Carter MM, Thompson MS, Frias E, Russ C, Reece-Hoyes JS, Nye C, Gradia S, Vidal B, et al. DNA Repair Profiling Reveals Nonrandom Outcomes at Cas9-Mediated Breaks. Mol Cell. 2016;63:633–646. doi: 10.1016/j.molcel.2016.06.037. [DOI] [PubMed] [Google Scholar]
- Vieira VC, Soares MA. The role of cytidine deaminases on innate immune responses against human viral infections. Biomed Res Int. 2013;2013:683095. doi: 10.1155/2013/683095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas DF. Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol. 2013;64:327–350. doi: 10.1146/annurev-arplant-042811-105552. [DOI] [PubMed] [Google Scholar]
- Wang C, Huang R, He B, Du Q. Improving the thermostability of alpha-amylase by combinatorial coevolving-site saturation mutagenesis. BMC Bioinformatics. 2012;13:263. doi: 10.1186/1471-2105-13-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, La Russa M, Qi LS. CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- Wang HX, Li M, Lee CM, Chakraborty S, Kim HW, Bao G, Leong KW. CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery. Chem Rev. 2017;117:9874–9906. doi: 10.1021/acs.chemrev.6b00799. [DOI] [PubMed] [Google Scholar]
- Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci U S A. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Yang Z, Rada C, Neuberger MS. AID upmutants isolated using a high-throughput screen highlight the immunity/cancer balance limiting DNA deaminase activity. Nat Struct Mol Biol. 2009;16:769–776. doi: 10.1038/nsmb.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way JC, Collins JJ, Keasling JD, Silver PA. Integrating biological redesign: where synthetic biology came from and where it needs to go. Cell. 2014;157:151–161. doi: 10.1016/j.cell.2014.02.039. [DOI] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Duan J, Li B, Zhou P, Hon GC. Multiplexed Engineering and Analysis of Combinatorial Enhancer Activity in Single Cells. Molecular Cell. 2017;66:285–299. e285. doi: 10.1016/j.molcel.2017.03.007. [DOI] [PubMed] [Google Scholar]
- Xu Z, Zan H, Pone EJ, Mai T, Casali P. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat Rev Immunol. 2012;12:517–531. doi: 10.1038/nri3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Briggs AW, Chew WL, Mali P, Guell M, Aach J, Goodman DB, Cox D, Kan Y, Lesha E, et al. Engineering and optimising deaminase fusions for genome editing. Nat Commun. 2016a;7:13330. doi: 10.1038/ncomms13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Grishin D, Wang G, Aach J, Zhang C-Z, Chari R, Homsy J, Cai X, Zhao Y, Fan J-B. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nature communications. 2014;5 doi: 10.1038/ncomms6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016b;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeang CH, McCormick F, Levine A. Combinatorial patterns of somatic gene mutations in cancer. FASEB J. 2008;22:2605–2622. doi: 10.1096/fj.08-108985. [DOI] [PubMed] [Google Scholar]
- Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, Park A, Yang J, Suresh S, Bizhanova A, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XF. Innate cellular defenses of APOBEC3 cytidine deaminases and viral counter-defenses. Curr Opin HIV AIDS. 2006;1:187–193. doi: 10.1097/01.COH.0000221590.03670.32. [DOI] [PubMed] [Google Scholar]
- Yuan G, Bin JC, McKay DJ, Snyder FF. Cloning and characterization of human guanine deaminase. Purification and partial amino acid sequence of the mouse protein. J Biol Chem. 1999;274:8175–8180. doi: 10.1074/jbc.274.12.8175. [DOI] [PubMed] [Google Scholar]
- Yuen CM, Rodda SJ, Vokes SA, McMahon AP, Liu DR. Control of transcription factor activity and osteoblast differentiation in mammalian cells using an evolved small-molecule-dependent intein. J Am Chem Soc. 2006;128:8939–8946. doi: 10.1021/ja062980e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015a;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol. 2015b;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wu LY, Zhang XS, Zhang S. Discovery of co-occurring driver pathways in cancer. BMC Bioinformatics. 2014;15:271. doi: 10.1186/1471-2105-15-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qin W, Lu X, Xu J, Huang H, Bai H, Li S, Lin S. Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nat Commun. 2017;8:118. doi: 10.1038/s41467-017-00175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zharkov DO. Base excision DNA repair. Cell Mol Life Sci. 2008;65:1544–1565. doi: 10.1007/s00018-008-7543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu JL, Wang D, Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol. 2017;35:438–440. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]
- Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]