Dear Editor,
Highly pathogenic avian influenza (HPAI) viruses of the H5 subtype remain a serious concern for both poultry and human health. The clade 2.3.4.4 H5 viruses have evolved into four genetic groups (nominally designated A, B, C, and D). Clade 2.3.4.4.A and B H5N8 viruses appear to have been reintroduced from poultry into wild bird populations, likely in Asia, and then spread globally through wild bird migration during 2014–2015 (2.3.4.4.A) and 2016–2017 (2.3.4.4.B)1, 2.
Clade 2.3.4.4.C and D H5 HPAI viruses have circulated exclusively in China and a few other countries in Southeast Asia. Indeed, clade 2.3.4.4.C H5N6 HPAI viruses have been the predominant clade detected in poultry populations in China3. Recently, clade 2.3.4.4 H5N6 subtypes have also been associated with human infection in China (http://www.wpro.who.int/emerging_diseases/AvianInfluenza/en/).
The clade 2.3.4.4.A H5N8 viruses were identified in China in 2013 and were subsequently introduced into Korea in early 20144, 5. Since then, the spread of clade 2.3.4.4.A viruses mediated by wild bird migration has resulted in intra- and intercontinental transmission. These viruses then reassorted with low pathogenic avian influenza (LPAI) of the Eurasian or North American lineage to result in novel H5Nx subtypes (such as N1, N2, N3, N5, and N8), which subsequently spread broadly throughout Eurasia and North America6, 7.
In mid-2016, a novel clade of 2.3.4.4.B H5N8 viruses was detected in wild birds in Uvs-Nuur Lake in Siberia, Russia, near the border with Mongolia and in Qinghai Lake in China. These H5N8 viruses were novel reassortants consisting of five segments (polymerase basic-2 (PB2), polymerase basic-1 (PB1), polymerase acidic (PA), nucleoprotein (NP), and matrix (MP)) from Eurasian LPAI viruses most similar to H5N8 viruses isolated in China in 20138, 9. These Russia–Mongolia H5N8 viruses then further reassorted with the NA genes from Eurasian LPAI viruses to result in additional H5N5 and H5N6 subtypes. This 2.3.4.4.B lineage has been detected in most European countries, Central Asia, the Middle East, and Africa during 2016–201710.
In Korea, there have been four outbreaks of H5N1 HPAI from 2003 to 2011. However, since 2014, large epizootics in poultry have been caused by clade 2.3.4.4.A H5N8 viruses. Then, in late 2016, outbreaks in poultry resulted from clade 2.3.4.4.C H5N6 and clade 2.3.4.4.B H5N8 HPAI viruses, demonstrating the expanding diversity of the threat to the poultry industry in Korea11, 12.
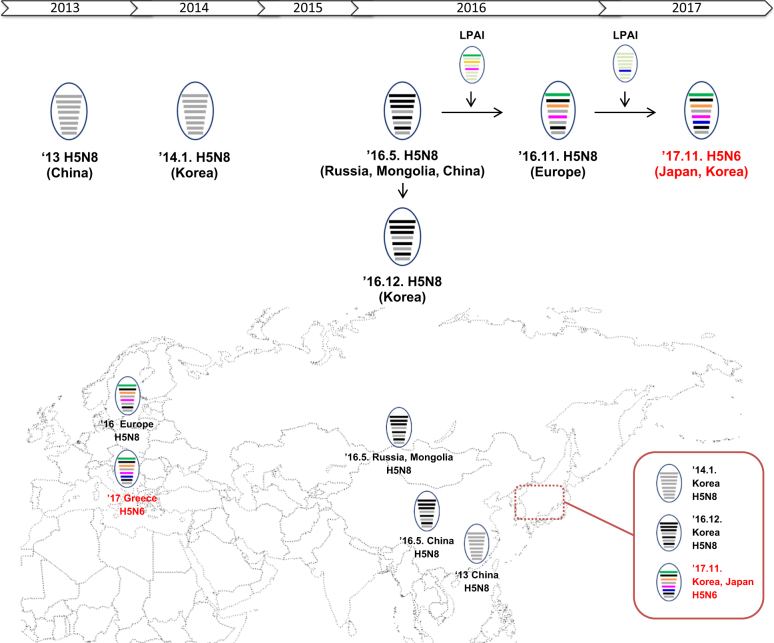
In November 2017, we detected two novel H5N6 HPAI viruses, A/duck/Korea/HD1/2017(H5N6) (HD1) and A/mallard/Korea/Jeju-H24/2017(H5N6) (Jeju-H24), through our active surveillance program for avian influenza. On November 17, an H5N6 HPAI virus was detected in a broiler duck farm in Gochang of Jeonbuk Province (nominated HD1). The second H5N6 virus was isolated from fecal samples taken from a wild mallard (Anas platyrhynchos) collected in Jeju on November 27 (nominated Jeju-H24). Virus isolation, sequencing, and phylogenetic analysis were performed as described (Supplementary Materials and Methods 1). The two isolates shared 99.69–100% nucleotide homology across all eight genes. A BLAST search (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) revealed that these novel H5N6 viruses were reassortants with H5 genes most genetically similar to clade 2.3.4.4.B viruses derived from European H5N8 HPAI viruses circulating during the 2016–2017 epizootic and N6 genes of a Eurasian LPAI virus lineage circulating in wild birds (Fig. 1, Supplementary Figure S1 and Table S1).
Fig. 1. Evolutionary timeline of the genetic cassette of the current Korean 2.3.4.4B H5Nx highly pathogenic avian influenza viruses, generated through reassortment.
Current reassortant H5N6 viruses consist of PB2, PB1, PA, HA, NP, M, and NS segments from European clade 2.3.4.4.B H5N8 viruses isolated in 2016 and N6 gene segments from low pathogenic avian influenza viruses. Gray colors represent the lineage of the segments originating from viruses of clade 2.3.4.4.B in 2013–2014, and the other colors denote the segments derived from low-pathogenic avian influenza virus through reassortment events. The eight horizontal bars indicate the gene segments in the order (top to bottom of the virion) PB2, PB1, PA, HA, NP, NA, M, and NS. HA hemagglutinin, M matrix, NA neuraminidase, NP nucleoprotein, NS nonstructural, PA polymerase acidic, PB1 polymerase basic 1, PB2 polymerase basic 2
Phylogenetic analyses of all genes except for the NA gene showed that the closest genetic relatives to the two viruses isolated in Korea were isolated from both wild and domestic birds sampled in a number of European countries in 2016/17. Furthermore, both the NP and NA genes were most similar to the H5N6 reassortant virus isolated from a chicken in Greece during this epizootic (Supplementary Figure S1 and S2).
The H5N6 viruses had multiple polybasic amino acid motifs at the HA cleavage site, characteristic of highly pathogenic influenza viruses (PLRERRRKR/GLF). Interestingly, the HPAI cleavage site motif differed between the Greek and the Korean clade 2.3.4.4.B strains. However, it is identical to the Japanese H5N6 HPAI virus (A/mute swan/Shimane/3211A001/2017) isolated from a wild bird on 5 November 2017, perhaps indicating heterogeneity in the emerging 2.3.4.4.B H5N6 lineage that is potentially being maintained in wild birds.
The clade 2.3.4.4.B viruses have consistently evolved into novel H5Nx subtypes with various gene constellations by reassortment with LPAI viruses since 2013. In particular, the internal gene segments derived from Eurasian LPAI viruses in wild birds that have reassorted with the clade 2.3.4.4 ‘H5N8 backbone’ and the H5N5 and H5N8 HPAI subtypes of various genotypes were detected in Germany, Italy, the Netherlands, Croatia and the Republic of Georgia13–15. Reassortant H5Nx viruses might diffuse among wild bird species in Eurasian breeding or staging sites, such as Central Asia and Siberia, and these viruses then disseminate into Eastern Asia (Korea and Japan). Conversely, there might be some degree of temporo-spatial segregation across Eurasian wild bird populations, resulting in maintenance of distinct variants and, therefore, heterogeneity in emerging strains in different geographic regions. These hypotheses are, as yet, difficult to test owing to significant sampling biases in wild bird influenza monitoring and would require more strategically targeted surveillance in ecologically relevant populations.
The novel clade 2.3.4.4.B H5N6 viruses isolated in November 2017 were phylogenetically distinguishable from the clade 2.3.4.4 viruses (H5N6 and H5N8) previously isolated in Korea during 2016–2017. In late 2016, clade 2.3.4.4.C H5N6 HPAI viruses that originated from China were introduced to Korea and Japan by wild birds and caused enormous economic losses to the poultry industry, particularly in Korea12. Recent H5N6 viruses from 2017 showed <95% homologies in eight gene segments with previous Korean H5N6 viruses isolated in 2016. In addition, novel H5N6 viruses showed <97% in nucleotide identities with three genes (PB2, PA, and NP) with previous Korean H5N8 viruses of clade 2.3.4.4.B in the 2016/2017 winter season. These results indicate that new H5N6 viruses are unlikely to have been circulating undetected in Korea; therefore, they are more likely to represent a new introduction mediated by migratory birds.
In this study, we reported novel reassortant H5N6 HPAI viruses isolated from wild birds and poultry in November 2017. Genetic analysis indicated that these isolates had been generated through reassortment events between European H5N8 HPAI viruses in 2016/2017, which were descendants of Russia–Mongolia H5N8 viruses, and Eurasian lineage LPAI viruses. Novel H5N6 viruses with similar gene constellations were founded in both Greece and Korea, potentially derived by new reassortant events in Eurasian wild bird populations at breeding sites or staging sites and then spread to multiple geographic regions through different migratory subpopulations. More detailed information on HPAI viruses identified through active surveillance programs throughout Eurasia is needed to better evaluate the ecology, evolution, dynamics and diffusion of these viruses in wild birds, particularly in breeding sites in Mongolia and Siberia where these species likely converge in the summer with the potential for reassortment-driven expansion in diversity, and to assess the potential risks to animal and public health.
Electronic supplementary material
Acknowledgements
We thank Jung-Eui Lee, In-Kyung Kim, and Byung-suk Jeon for their excellent technical assistance. We also thank the Animal and Plant Quarantine Agency (APQA), Ministry of Agriculture, Food and Rural Affairs (MAFRA) and the Regional Office for Animal Disease Control for their efforts to control HPAI. This study was supported by a grant from the Animal and Plant Quarantine Agency (APQA) (no. B-1541781-2008-2019-01).
Contributor Information
Yong-Joo Kim, Email: kyjvet@korea.kr.
Youn-Jeong Lee, Email: leeyj700@korea.kr.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41426-018-0104-3).
References
- 1.Global Consortium for H5N8 and Related Influenza Viruses. Role for migratory wild birds in the global spread of avian influenza H5N8. Science. 2016;354:213–7. doi: 10.1126/science.aaf8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee DH, Bertran K, Kwon JH, et al. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J. Vet. Sci. 2017;18(S1):269–80. doi: 10.4142/jvs.2017.18.S1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi Y, Chen Q, Wang Q, et al. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe. 2016;20:810–21. doi: 10.1016/j.chom.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 4.Wu H, Peng X, Xu L, et al. Novel reassortant influenza A(H5N8) viruses in domestic ducks, eastern China. Emerg. Infect. Dis. 2014;20:1315–8. doi: 10.3201/eid2008.140339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee YJ, Kang HM, Lee EK, et al. Novel reassortant influenza A(H5N8) viruses, South Korea, 2014. Emerg. Infect. Dis. 2014;20:1087–9. doi: 10.3201/eid2006.140233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee DH, Bahl J, Torchetti MK, et al. Highly pathogenic avian influenza viruses and generation of novel reassortants, United States, 2014-5. Emerg. Infect. Dis. 2016;22:1283–5. doi: 10.3201/eid2207.160048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee MS, Chen LH, Chen YP, et al. Highly pathogenic avian influenza viruses H5N2, H5N3, and H5N8 in Taiwan in 2015. Vet. Microbiol. 2016;187:50–57. doi: 10.1016/j.vetmic.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Lee DH, Sharshov K, Swayne DE, et al. Novel reassortant clade 2.3.4.4 avian influenza A(H5N8) virus in wild aquatic birds, Russia, 2016. Emerg. Infect. Dis. 2017;23:359–60. doi: 10.3201/eid2302.161252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, Liu H, Bi Y, et al. Highly pathogenic avian influenza A(H5N8) virus in wild migratory birds, Qinghai Lake, China. Emerg. Infect. Dis. 2017;23:637–41. doi: 10.3201/eid2304.161866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Organization for Animal Health. Update on avian influenza in animals (type H5 and H7). www.oie.int/animal-health-in-the-world/update-on-avian-influenza/
- 11.Kim YI, Park SJ, Kwon HI, et al. Genetic and phylogenetic characterization of a novel genotype of highly pathogenic avian influenza (HPAI) H5N8 viruses in 2016/2017 in South Korea. Infect. Genet. Evol. 2017;53:56–67. doi: 10.1016/j.meegid.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Lee EK, Song BM, Lee YN, et al. Multiple novel H5N6 highly pathogenic avian influenza viruses, South Korea, 2016. Infect. Genet. Evol. 2017;51:21–23. doi: 10.1016/j.meegid.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Beerens N, Heutink R, Bergervoet SA, et al. Multiple reassorted viruses as cause of highly pathogenic avian influenza A(H5N8) virus epidemic, the Netherlands, 2016. Emerg. Infect. Dis. 2017;23:1974–81. doi: 10.3201/eid2312.171062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pohlmann A, Starick E, Harder T, et al. Outbreaks among wild birds and domestic poultry caused by reassorted influenza A(H5N8) clade 2.3.4.4 viruses, Germany, 2016. Emerg. Infect. Dis. 2017;23:633–6. doi: 10.3201/eid2304.161949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fusaro A, Monne I, Mulatti P, et al. Genetic diversity of highly pathogenic avian influenza A (H5N8/H5N5) viruses in Italy, 2016-17. Emerg. Infect. Dis. 2017;23:1543–7. doi: 10.3201/eid2309.170539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.