Abstract
Prognosis of triple-negative breast cancer (TNBC) remains poor. To identify shared and selective vulnerabilities of basal-like TNBC, the most common TNBC subtype, a directed siRNA lethality screen was performed in 7 human breast cancer cell lines, focusing on 154 previously identified dependency genes of one TNBC line. Thirty common dependency genes were identified, including multiple proteasome and RNA splicing genes, especially those associated with the U4/U6.U5 tri-snRNP complex (e.g., PRPF8, PRPF38A). PRPF8 or PRPF38A knockdown or the splicing modulator E7107 led to widespread intronic retention and altered splicing of transcripts involved in multiple basal-like TNBC dependencies, including protein homeostasis, mitosis and apoptosis. E7107 treatment suppressed the growth of basal-A TNBC cell line and patient-derived basal-like TNBC xenografts at a well-tolerated dose. The anti-tumor response was enhanced by adding the proteasome inhibitor bortezomib. Thus, inhibiting both splicing and the proteasome might be an effective approach for treating basal-like TNBC.
Keywords: Triple-negative breast cancer, lethality screen, RNA splicing, MCL1, E7107, targeted therapy
INTRODUCTION
Triple negative breast cancers (TNBCs), defined by their lack of estrogen (ER), progesterone and HER2 receptors, are a heterogeneous group of poorly differentiated and aggressive tumors that disproportionately affect younger and African American women (1–3). TNBCs are classically defined as “basal-like” or “claudin-low” based on gene expression. Basal-like tumors (the most common variant) display a luminal/myoepithelial or progenitor-like phenotype, whereas claudinlow tumors express predominantly mesenchymal traits. mRNA expression analysis further divides TNBC into six molecular subtypes, which are basal-like (BL-1, BL-2, IM), claudin-low (M, MSL) or molecular apocrine tumors (LAR) (4). Basal-A and basal-B breast cancer cell lines resemble basal-like and claudin-low tumors, respectively (5). Genome sequencing corroborates TNBC diversity - apart from TP53 mutations, which occur in >80% of TNBCs, no recurrent genetic lesions that could serve as drug targets have been identified (6–8).
To identify novel TNBC therapeutic targets, we previously performed a genome-wide siRNA lethality screen to identify selective basal-like TNBC genetic dependencies by comparing a pair of isogenic, genetically well-defined TNBC cell lines, basal-A BPLER and myoepithelial HMLER (9). The screen identified 154 genes selectively needed for BPLER survival. MCL1 and multiple proteasome genes scored as top BPLER dependency genes and as shared dependencies of basal-A cell lines. Basal-A cell lines were selectively sensitive to the proteasome inhibitor drug bortezomib. Proteasome addiction was linked to Mcl-1 dependence. However, the maximum tolerated dose (MTD) of bortezomib was needed for an effective anti-tumor response, which may explain the failure of bortezomib in clinical trials in TNBC.
Because of TNBC heterogeneity and the inadequacy of current treatment, here we sought to identify additional shared genetic dependencies that might be useful TNBC drug targets. To this end, we performed a targeted siRNA lethality screen using 4 basal-A and 3 luminal human cell lines, focusing on the 154 BPLER dependency genes. Knockdown of only 30 genes resulted in at least a mean 2-fold loss in viability of the 4 TNBC lines. These included MCL1 and 5 genes linked to the proteasome, confirming our earlier work (9). mRNA splicing genes were prominent basal-A shared dependencies, comprising 12 of the remaining 24 hits. These results confirm other studies that identified splicing as a selective TNBC dependency (10–12). Over 200 proteins assemble with small nuclear RNAs (snRNAs) to form multiple small nuclear ribonucleoprotein (snRNP) complexes that control distinct steps of splicing (13). Four of the 12 splicing gene hits are either components of the U4/U6.U5 tri-snRNP complex (PRPF6, PRPF8, PRPF38A) or associated with it (USP39). The tri-snRNP complex plays a key early role in splicing - it associates with the initial spliceosome A complex to form the B complex to promote the catalytic activation of the spliceosome (14–17). More differentiated luminal breast cancer cell lines were not as dependent on these splicing genes, suggesting that transient inhibition of splicing might be well-tolerated by normal cells. Because of the over-representation of hits in this functionally important complex, we focused on these splicing hits in this study.
MATERIALS AND METHODS
Cell lines
HCC70, HCC1143, HCC1187, HCC1937, HCC1954, HCC1806, AU565, BT474, MCF7, MDAMB231, MDAMB436 and BT549 cell lines were purchased directly from ATCC. The MB468 cell line was engineered to express luciferase and was obtained from Dr. A. Kung (Memorial Sloan Kettering Cancer Center). All cell lines were tested for Mycoplasma infection by PCR every 3 months. Only early-passage cell lines were used and cells were kept in culture no longer than 21 days. Cell lines were obtained in 2012 with the exception of MDAMB231 that was obtained in 2016. Cell lines were authenticated by short tandem repeat (STR) analysis. Bortezomib was purchased from LC Laboratories. E7107 (18) was kindly provided by H3 Biomedicine, Inc.
High throughput screening (HTS)
Automated siRNA screening was performed using robotics at the ICCB-L Screening Facility at Harvard Medical School, as previously described (9). Briefly, HTS conditions optimized in our previous study were tested in 22 human breast cancer cell lines, and cell lines showing a Z’ factor >0.6 were selected for targeted screening using a customized 252 siRNA library of validated siRNAs (siGenome, Dharmacon) against the 154 BPLER dependency genes identified in (9). Cells (1000/well) were reverse-transfected with individual siRNAs (50 nM) in triplicate in 384-well flat clear bottom white polystyrene microplates (Corning) using Dharmafect 1 (0.04 µl/well) and a 1:1 mixture of Optimem/10% FBS RPMI 1640 medium (final volume: 25 µl/well). Fresh 10% FBS RPMI 1640 (15 µl/well) was added 24 hr after transfection and cell viability was measured by CellTiterGlo chemoluminescence assay (Promega) 72 hr after transfection using an Infinite M100 Pro high throughput plate reader (Tecan), after verifying that the detected signal was within a linear range. For each siRNA, the viability scores of the 3 replicates were averaged and compared to the average score of cells transfected with a control siRNA with no detectable effect on viability (Non-targeting Control #4, Dharmacon). The heatmap of cell viability ratios was generated using the Gplot2 package within R.
RNA library preparation and sequencing
Libraries were prepared using Illumina TruSeq Stranded mRNA sample preparation kits from 500 ng of purified total RNA according to the manufacturer’s protocol. The finished dsDNA libraries were quantified by Qubit fluorometer, Agilent TapeStation 2200, and qRT-PCR using the Kapa Biosystems library quantification kit according to the manufacturer’s protocols. Uniquely indexed libraries were pooled in equimolar ratios and sequenced on two Illumina NextSeq500 runs with paired-end 75bp reads at the Dana-Farber Cancer Institute Molecular Biology Core Facilities.
RNA-seq data analysis
Raw RNA-seq data were deposited in the GEO database (GSE90519). All of the analysis scripts to reproduce the analysis are stored in the Github repository and are accessible online (19). A step-by-step description of data analysis procedures is also available online (20). Pathway analyses were based on the Reactome web-based tool within the Molecular Signatures Database (MSigDB) of the Broad Institute (21) using a cut-off of q<0.05. Gene networks were generated using GeneMania (22) focusing on physical interactions, pathway and shared protein domain interactions.
Protein analysis
Immunoblot was performed as previously described (9). Briefly, cells were lysed in 1X Cell Lysis Buffer (Cell Signaling) and 30–50 µg of total protein were boiled for 5 min in 1X NuPage buffer + DTT (Life Technologies), resolved on a 4–20% Tris-HCl polyacrylamide gradient Criterion gel (Biorad) and transferred to a PVDF membrane using a Criterion wet-transfer system (Biorad) for 3 hr at 0.35A. A list of antibodies used is shown in supplementary Materials and Methods.
qRT-PCR and RT-PCR
Total RNA was extracted from pelleted cells using the RNeasy Plus Mini Kit (Qiagen, Cat#74134). First strand cDNA synthesis was performed using the iScript cDNA synthesis kit (BioRad, Cat#1708891) using 1 µg of total RNA and following the manufacturer’s protocol.
Mouse studies
Animal experiments involving cell-line-derived xenografts were performed in the AAALAC-accredited Laboratory Animal Science Center (LASC) at Boston University Medical Campus. All experiments were conducted in accordance with Boston University Institutional Animal Care and Use Committee (IACUC). Approximately 6-week-old, female, nude mice (NU/J; Jackson Laboratory) were used. Briefly, for each mouse 3×106 or 1×107 HCC1187 cells or 2×106 MB468 cells were injected subcutaneously in the right flank in a 1:1 solution of Matrigel (BDBioscience) and 10% FBS RPMI 1640 using 26G needles, in a final volume of 100 µl. When tumors were palpable (usually within ~ 4–6 weeks), mice were randomized into groups with tumors of similar size (50 +/− 10 mm3) and treated by tail-vein injection with 5 mg/kg E7107, 0.4–1.2 mg/kg bortezomib or vehicle (DMSO) in 100 µl volume in PBS once a day for 4 or 5 consecutive days. Tumor volume was measured every 2 or 3 days by caliper and statistical significance determined by one-sided t-test. PDX-based experiments were conducted in collaboration with Champions Oncology Inc based on a comparable protocol, with the exception that Ncr nude mice (Taconic) were used instead of NU/J. Each of the models was previously validated by histopathology and characterized by RNA-seq and whole-exome sequencing. Data are available on the Champions Oncology website (23).
RESULTS
A targeted siRNA screen identifies core survival networks in basal-A breast cancer cell lines
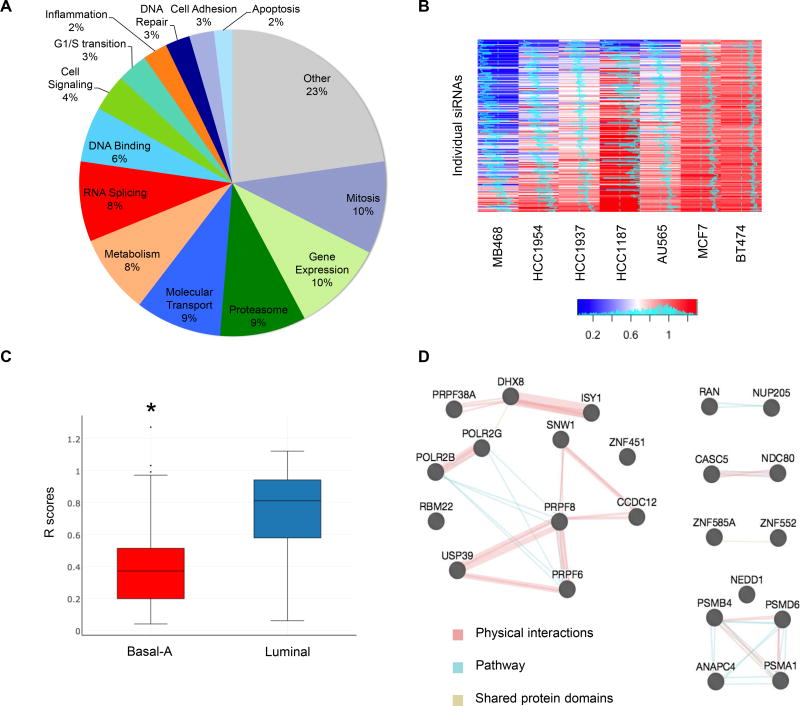
77% of the 154 BPLER dependency genes (9) clustered within 13 well-defined gene networks that control key cellular functions (Figure 1A). To identify common basal-A TNBC dependencies, a directed siRNA screen was performed on 4 other human basal-A cell lines and 3 luminal cell lines as controls using a customized 252 siRNA library (siGenome, Dharmacon) that targeted each of the 154 BPLER dependency genes (Figure 1A). Transfection of each of these siRNAs individually reduced BPLER viability, suggesting that this library is enriched for functional siRNAs. To focus on genes that are not essential for viability of more differentiated cells, we excluded from consideration genes whose knockdown was lethal for the luminal cell lines. All seven cell lines used in the screen were efficiently transfected (>90%) and gave reproducible results under high throughput screening conditions (Z’ factor >0.6). For each network, we assessed between 3 and 25 distinct siRNAs. The readout was the ratio in cell viability (R) between cells transfected with individual siRNAs vs. non-targeting control, 72 hr after transfection. siRNAs returning an R value < 0.75 were considered hits. Each basal-A cell line displayed a unique vulnerability profile, not surprisingly given the genetic and epigenetic heterogeneity of these tumors (Figure 1B,Table S1). MB468 had the highest number of hits with 132 of 154 genes (213 of 252 siRNAs) scoring positive; HCC1954, HCC1937 and HCC1187 had 102, 91 and 44 gene hits (or 167, 122 and 65 siRNAs), respectively. 30 genes scored with a ≥2-fold reduction in basal-A median cell viability. These were selected as high-confidence hits for further analysis (Table S2). Similar results were obtained based on mean cell viability. The screen was not powered to systematically identify differential vulnerabilities between basal-A and luminal cell lines. Nonetheless, basal-A cell lines were generally more sensitive to knockdown of these genes than luminal cell lines (median R = 0.39 vs. 0.86; p = 2.22E-28, onesided t-test; Figure 1C). RNA splicing (12 genes or 25 siRNAs; q=1.6E-07) and ubiquitinproteasome (5 genes or 9 siRNAs; q=0.007) Reactome modules were over-represented among these hits. The list also included the Bcl2-family gene MCL1, the mitotic spindle checkpoint genes NDC80 and CASC5, and the nuclear transport genes RAN (also involved in mitosis) and NUP205. We previously showed that survival of basal-A cell lines depends on MCL1 and the proteasome (9). TNBC are well known to depend on mitosis. Both proteasome and RNA splicing hits formed a dense protein-protein interaction network based on GeneMania (19) (Figure 1D). NDC80 and CASC5 proteins were also predicted to interact. Thus, our siRNA screen reidentified known basal-like TNBC vulnerabilities and pointed to novel candidate targets, including specific RNA splicing and nuclear export genes.
Figure 1. A targeted siRNA screen identifies shared vulnerabilities of basal-A TNBC vs luminal breast cancer cell lines.
(A) Distribution of pathway assignments of the 252 siRNAs used in this screen.
(B) Viability heat map of the indicated cell lines 72 hr after transfection with individual siRNAs targeting the 154 BPLER dependency genes, relative to transfection with a non-targeting siRNA. Histograms in heatmap indicate score distance from 0.5 (midline). Histogram in color key indicates counts of specific scores across all cell lines. MB468, HCC1954, HCC1937 and HCC1187 cell lines are basal-A; AU565, MCF7 and BT474 are luminal
(C) Box-whisker plot showing the distribution of R scores associated with the genes in Table S2 in basal-A vs. luminal cell lines. * indicates p = 2.22E-28, one-sided t-test.
(D) Functional interaction map of the genes in Table S2 generated using GeneMania (19). Nodes represent proteins. Edges indicate physical (red), pathway (blue) or shared domain (yellow) interactions. Nodes are grouped based on functional similarity. Edge thickness indicates number of studies in the literature supporting the interaction.
Basal-A cell lines selectively depend on specific RNA splicing, mitosis and nuclear export genes
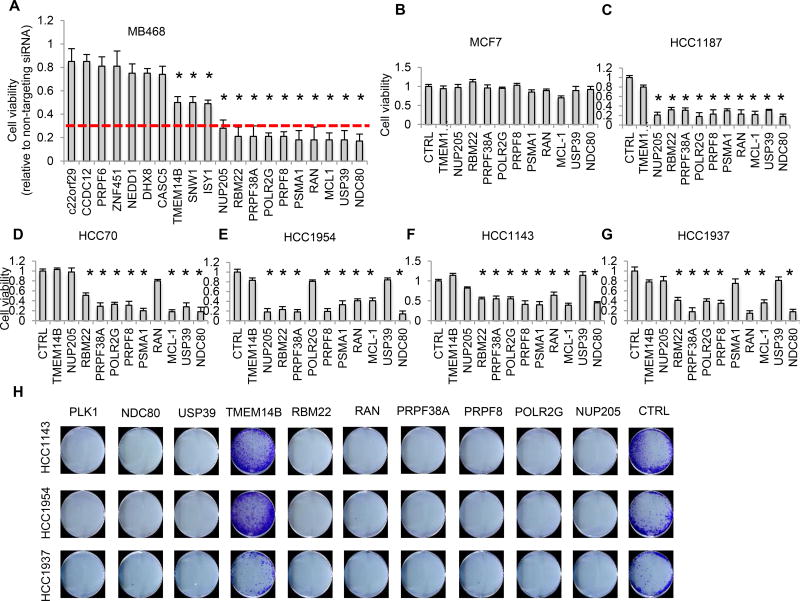
To confirm the results of the screen, we selected 20 of the 30 high confidence shared dependency genes for further experimental validation in MB468 cells (which scored positive for each of these hits in the primary screen) using a different siRNA (Silencer, Ambion; Figure 2A), focusing on the most enriched functional pathways. Thirteen hits (65%) reconfirmed as MB468 dependency genes, based on the criteria used for the primary screen. Ten hits, whose knockdown reduced MB468 viability ≥3-fold, were examined in 5 basal-A and 1 luminal cell line. These hits included genes involved in proteasome degradation (PSMA1), splicing (PRPF8, PRPF38A, RBM22, POLR2G, USP39), mitosis (NDC80), nuclear transport (RAN, NUP205) and apoptosis (MCL1). Knockdown of these genes had no significant effect on luminal MCF7 cell viability (Figure 2B), but silencing PSMA1, PRPF8, PRPF38A or NDC80 (but not the non-validated hit TMEM14B used as control) reduced cell viability 72–96 hr after transfection by ≥3-fold in 4 of 5 basal-A cell lines (Figures 2C–2G). To further gauge the usefulness of these hits as candidate TNBC targets, we assessed the effect of gene knockdown on colony formation in 3 basal-A cell lines (HCC1143, HCC1954 and HCC1937) (Figure 2H). Knockdown of all 8 selected hits (but not TMEM14B) abrogated colony formation after 10–14 days, including hits that only weakly inhibited the short-term viability of these cell lines in bulk (e.g. NUP205, USP39).
Figure 2. Experimental validation using independent siRNAs of the top secondary screen hits involved in proteasome degradation, RNA splicing, mitosis and nuclear transport.
(A) Viability of MB468 cells 72 hr after transfection with Silencer siRNAs targeting the indicated genes, relative to cells transfected with the non-targeting siRNA. Red line indicates the cut-off used for prioritizing genes for further validation (<0.3).
(B–G) Viability of MCF7 (B), HCC1187 (C), HCC70 (D), HCC1954 (E), HCC1143 (F) or HCC1937 (G) cells after transfection with Silencer siRNAs against the indicated genes or with a non-targeting siRNA (CTRL). All cell lines were assessed at 72 hr post-transfection except for HCC1187 that were assessed at 96 hr.
(H) Colony assay of indicated cell lines after transfection with Silencer siRNAs against the indicated genes or non-targeting siRNA (CTRL). Cells were plated at clonal density 48 hr post-transfection and grown for an additional 10–14 days. Colonies were visualized by crystal violet staining. PLK1 is shown as positive control.
Data in (A–G) represent the mean ± S.D. of at least 3 independent experiments. * indicates p<0.05 (one-sided t-test).
RNA splicing genes are commonly up-regulated in basal-like primary tumors
Many of the shared basal-A dependency genes are involved in splicing. To explore splicing as a potential TNBC drug target, we examined expression of 72 genes associated with different steps of RNA splicing (based on the MSigDB database) in 476 human breast specimens in the TCGA database, including 387 primary tumors (109 basal-like, 268 luminal) and 99 normal breast tissues. Hierarchical clustering based on these genes separated basal-like and luminal tumors and normal breast tissue. Basal-like tumors showed the highest spliceosome gene expression (Figure S1A). Splicing genes, as a group, were modestly, but significantly, up-regulated in basal-like tumors relative to luminal tumors and normal breast (mean fold-change: 1.2 and 1.4, respectively; p=0.007 and 1.8E-9 by two-sided t-test; Figure S1B). Ten genes were up-regulated by ≥2-fold - SF3B4 (in the SF3b complex), LSM2, LSM4 and LSM7 (in the tri-snRNP complex) and Sm proteins associated with snRNP biogenesis (SNRPA, SNRPB, SNRPC, SNRPD1, SNRPE, SNRPG; Figure S1C). None of the RNA splicing dependency gene mRNAs in our screen were up-regulated by ≥1.5-fold in basal-like TNBCs compared to luminal or normal tissues and thus would have been difficult to identify by mRNA expression. However, PRPF8, PRPF38A and RBM22 proteins were generally more abundant in luminal and TNBC (both basal-A and basal-B) cell line lysates than in BPE, a non-transformed breast epithelial cell line (Figure S1D). Moreover, knockdown of each of these three genes selectively triggered caspase-3 cleavage in MB468, but not BPE, confirming the functional relevance of these hits (Figure S1E).
PRPF8 and PRPF38A are both required for optimal RNA splicing
PRPF8 is a core component of the S. cerevisiae and human U4/U6.U5 tri-snRNP complexes (15–17). Prp38, the yeast homolog of PRPF38A, is required for catalytic activation of the tri-snRNP complex (24,25). Thus, PRPF8 and PRPF38A are both important for tri-snRNP function. To study early events associated with inhibiting this complex, we first validated 2 sets of siGenome siRNAs (4 individual siRNAs each) against PRPF8 (PRPF8-01, -02, -04, -05) and PRPF38A (PRPF38A-01, 02, 03, 04). All 8 siRNAs induced > 90% target mRNA silencing in MB468 cells 48 hr after transfection as determined by qRT-PCR (Figure S2A). PRPF8 and PRPF38A protein knockdown was confirmed by immunoblot for 2 of 2 PRPF8 (PRPF8-01, -02) and 2 of 2 PRPF38A siRNAs (PRPF38A-02, -03) tested (Figure S2B). Unlike PRPF38A Silencer siRNA (Figure S1E), neither PRPF38A-02 nor PRPF38A-03 siGenome siRNAs suppressed PRPF8 proteins levels.
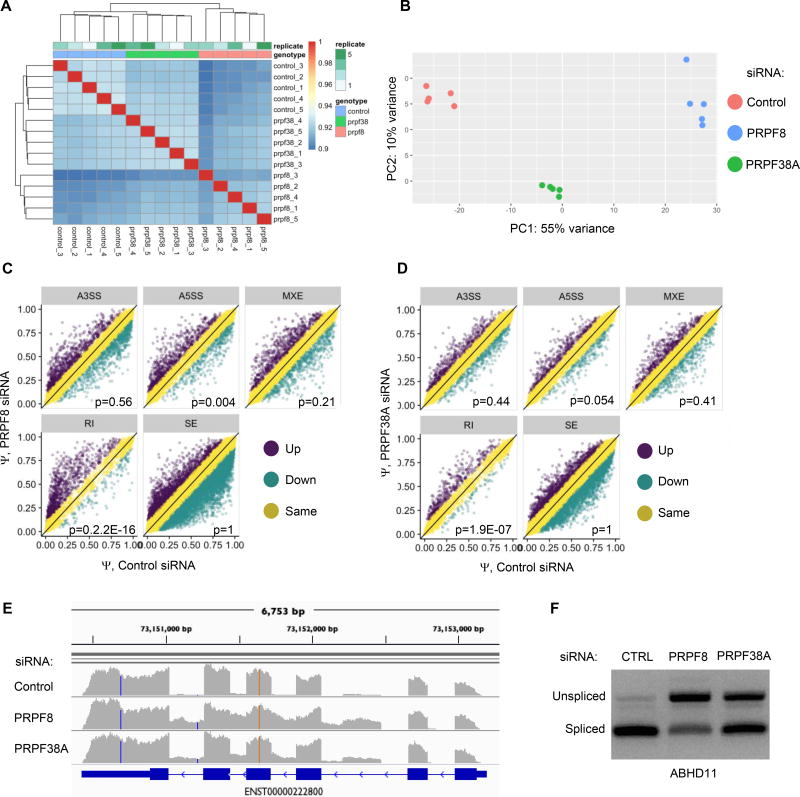
Next, we performed RNA-seq in quintuplicate in MB468 cells knocked down for PRPF8 (PRPF8-KD) or PRPF38A (PRPF38A-KD) for 48 hr using PRPF8-01 and PRPF38A-03 siRNAs, respectively. 50–70% of cells were viable, and only live cells were analyzed. The dominant PRPF8 isoform was knocked down by >98%, although a minor isoform, comprising ~5% of PRPF8 transcripts, persisted based on the estimated TPM values from Sailfish (26). All 3 known splicing variants of PRPF38A were silenced by >95%. Replicate samples clustered based on Spearman correlation of TMM-normalized counts (Figure 3A). Principal component analysis (PCA) on the 500 most variable genes clearly separated PRPF8A-KD, PRPF38A-KD and control samples (Figure 3B; QC data available on line).
Figure 3. PRPF8 or PRPF38A knockdown triggers intronic retention.
(A) Correlation (Spearman) of gene expression of TMM-normalized RNA-seq counts from 5 independent replicates of viable control (transfected with non-targeting siRNA), PRPF8-KD, PRPF38A-KD MB468 cells. Heatmap shows clustering of replicate samples.
(B) Principal component analysis of RNA-seq gene-level expression data in the same samples as in (A), using the 500 most variable genes.
(C,D) MISO analysis of specific classes of RNA splicing alterations within differentially expressed transcripts in PRPF8-KD (C) and PRPF38A-KD (D) samples relative to control samples transfected with non-targeting siRNA. p <0.05 indicates significant enrichment for alterations of the indicated class (Wilcoxon Test).
(E) RNA-seq read coverage for ABHD11 in PRPF8-KD and PRPR38A-KD and control cells. Blue boxes and lines indicate exonic and intronic regions, respectively.
(F) RT-PCR, using intron-spanning primers for ABHD11, of total RNA from MB468 cells 48 hr after transfection with the indicated siRNAs. Intron-retaining transcripts migrate more slowly.
To determine how PRPF8 or PRPF38A affect RNA splicing, we first assessed the relative representation of specific classes of splicing events in PRPF8-KD and PRPF38A-KD - alternative 5’ splice sites (A5SS), alternative 3’ splice sites (A3SS), mutually exclusive exons (MXE), retained introns (RI) and skipped exons (SE), based on MISO analysis (27) (Figures 3C and 3D). Both PRPF8-KD and PRPF38A-KD had significantly higher RIs than control cells (p=2.2E-16 and p=1.9E-07, respectively; Wilcoxon test). PRPF8-KD also showed greater A5SS (p=0.004) use, but this association was weaker in PRPF38A-KD (p=0.054). There were no significant differences in A3SS, MXE or SE with knockdown of either gene. Among 5,231 introns considered in MISO, 1,261 (24.1%) and 535 (10.2%) showed ≥10% differential retention in PRPF8-KD and PRPF38A-KD, respectively (Table S3). Of note, 83% of retained introns in PRPF38A-KD were also found in PRPF8-KD, suggesting that the two proteins affected RNA splicing through a similar mechanism. RT-PCR verified that both PRPF8 and PRPF38A knockdown caused intronic retention in ABHD11 transcripts, randomly chosen among the top shared differential RI events (Figures 3E and 3F). Thus, both PRPF8 and PRPF38A were required for optimal RNA splicing, although PRPF8 more than PRPF38A.
PRPF8 recapitulates the functional effect of PRPF38A on gene expression
We next used DESeq2 software (28) to examine whether PRPF8 and PRPF38A silencing selectively affects the expression of certain types of genes (Tables S4 and S5). 1,293 and 2,261 mRNAs were up- and down-regulated by ≥2-fold, respectively, in PRPF8-KD relative to control cells with q-value <0.01 (excluding genes with <20 normalized counts). PRPF38A-KD showed a more restricted effect with 579 up- and 610 down-regulated genes based on the same criteria (Figures S2C and S2D). Amongst the genes up- and down-regulated in PRPF38A-KD, 70% and 75% were similarly changed in PRPF8-KD (Figure S2E). To estimate the reproducibility of the RNA-seq data, we selected a set of 12 genes and measured their expression by qRT-PCR. Ten of 12 and all 12 genes reconfirmed as differentially expressed in PRPF8-KD and PRPF38A-KD cells, respectively, with fold-changes between 2 and 33 (Figure S2F). 80% of the expressed genes, validated as differentially expressed in PRPF8-KD or PRPF38A-KD, showed similar changes in HCC1187 knocked down for PRPF8 or PRPF38A (Figure S2G). Thus, much of the effect of PRPF38A on gene expression was recapitulated by PRPF8, but not vice versa.
PRPF8 and PRPF38A knockdown affects the pre-mRNA splicing of well-defined classes of genes required for TNBC survival
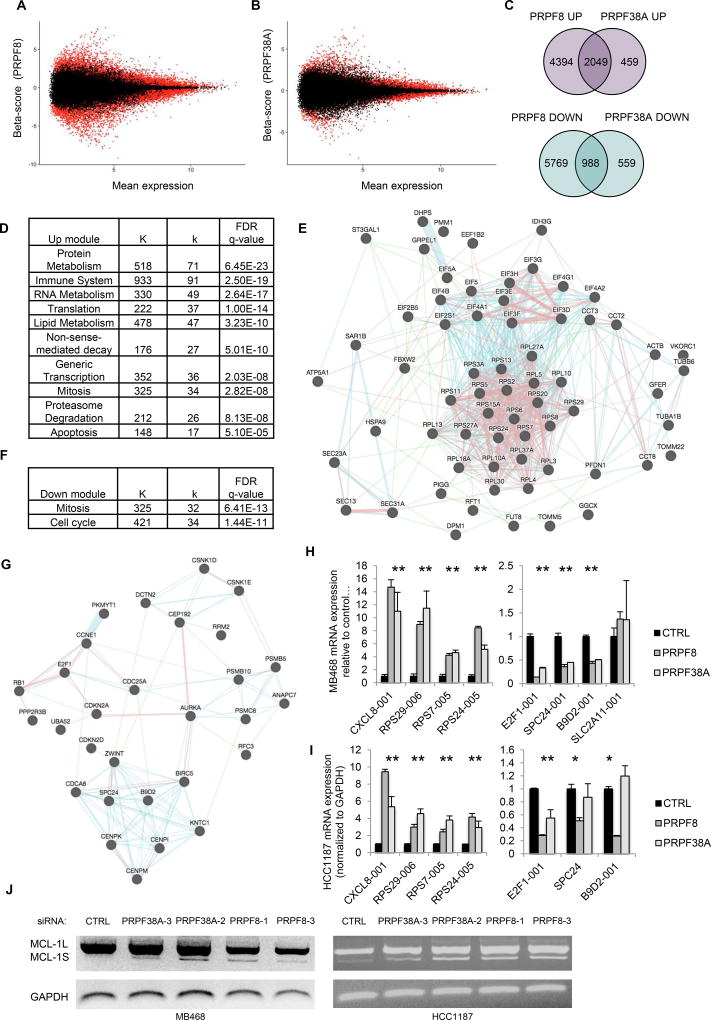
We next used Sleuth software to evaluate the expression of specific splice variants in PRPF8-KD and PRPF38A-KD. Of 196,501 transcripts, 29,980 (15.2%) and 16,955 (8.6%) transcripts were differentially expressed in PRPF8-KD and PRPF38A-KD, respectively (Figure 4A and 4B; Tables S6 and S7). We further analyzed transcripts with a β-score ≥ 0.69 or ≤ −0.69 (approximately 3-fold change) and q ≤0.01. Based on these criteria, 6,443 transcripts were upregulated and 6,767 were down-regulated in PRPF8-KD cells, while 2,508 were up-regulated and 1,557 were down-regulated in PRPF38A-KD cells (Figure 4C). 82% and 64% of up- and down-regulated transcripts in PRPF38A-KD cells were similarly altered in PRPF8-KD cells, corroborating the link between PRPF8 and PRPF38A.
Figure 4. PRPF8 or PRPF38A knockdown regulate splice variants of genes implicated in protein metabolism, mitosis, proteasome function and apoptosis.
(A–B) Bland-Altman (MA) plots of transcript-level mean expression by RNA-seq in viable MB468 cells 48 hr after transfection with PRPF8 (A) or PRPF38A (B) siRNAs, relative to nontargeting siRNA. B score is the beta coefficient from the Sleuth analysis. Red indicates q <0.05.
(C) Venn diagrams showing the number of shared up-regulated (top) or down-regulated (bottom) transcripts in MB468 cells transfected with PRPF8 or PRPF38A siRNAs relative to nontargeting siRNA.
(D) Significantly over-represented Reactome modules (q<0.01) among up-regulated transcripts in PRPF8-KD and PRPF38A-KD cells. Full list in Table S8. K indicates the total number of genes in each module; k indicates the number of up-regulated transcripts assigned to each module.
(E) Functional interaction map of protein metabolism proteins corresponding to transcripts upregulated in viable PRPF8-KD and PRPF38A-KD. Map generation and interaction key as in Fig. 1D.
(F) Significantly over-represented Reactome modules (q<0.01) among down-regulated transcripts in MB468 cells knocked down for PRPF38A, as in (D). Full list in Table S9.
(G) Functional interaction map of mitosis proteins (based on MSigDB) corresponding to transcripts down-regulated in viable PRPF8-KD and PRPF38A-KD cells as in (E).
(H,I) Levels of the indicated transcripts in MB468 (H) or HCC1187 (I) 48 hr after transfection with siRNAs against PRPF8 or PRPF38A relative to non-targeting siRNA, as determined by qRT-PCR using isoform-specific primers. Data represent the mean ± S.D. of at least 2 independent experiments (*, p <0.05; t-test relative to control).
(J) RT-PCR analysis of MCL1 splicing using primers that distinguish between different MCL1 isoforms in MB468 and HCC1187 cells 48 hr after transfection with indicated siRNAs. Top and bottom bands correspond to MCL1-001 and MCL1-002 transcripts, respectively.
Reactome pathway analysis (29) of shared PRPF8 and PRPF38A dependent transcripts identified at least 100 over-represented modules (q<0.01; Figure 4D and Table S8). Up-regulated transcripts were particularly enriched for immune system protein genes (e.g. IL-8, IL-6, TNF), ribosomal protein genes (e.g. RPS5, RPS6, RPS24) and translation initiation factors (e.g. EIF4A1, EIF4A2, EIF4G1). Genes involved in mitosis (e.g. AURKB, CENPA, CENPN), the proteasome (e.g. PSMA3, PSMB4, PSMD2) and apoptosis (e.g. MCL1, TP53, DIABLO, BAX, CASP7, TNF) were also highly enriched (Figure 4E). These transcripts were frequently RI or nonsense variants encoding for non-functional proteins. Specifically, 41 of 56 (73%) of translation initiation, 63 of 98 (64%) of protein metabolism (mainly ribosomal protein transcripts), 18 of 38 (47%) of mitosis and 16 of 34 (47%) of proteasome related transcripts were inactive splice variants. In contrast, key immune system transcripts were protein-coding variants. Therefore, both PRPF8 and PRPF38A knockdown altered the pre-mRNA splicing of well-defined classes of genes, some of which directly control apoptosis or are known to be selectively required for TNBC survival (e.g. mitosis and proteasome genes). Conversely, down-regulated transcripts in PRPF8-KD and PRPF38A-KD were mostly enriched for genes implicated in mitosis and the mitotic spindle checkpoint (Figure 4F and 4G; Table S9). Unlike the upregulated mitosis transcripts, 30 of 35 (86%) of down-regulated mitosis transcripts were protein-coding variants. These data were validated by flow cytometry and immunoblot experiments, which showed that PRPF8 knockdown increased by 45% the number of cells in G2/M phase (from 29% to 42%; Figure S3A) and increased phosphorylated histone H3 protein levels (Figure S3B), confirming that mitosis was disrupted.
To further corroborate these data, we measured the expression of 8 deregulated transcripts (4 up and 4 down) that could be unequivocally distinguished by qRT-PCR using isoform-specific primers, focusing on immune (up), ribosomal (up) and mitosis (down) related transcripts (Figure 4H). Seven of 8 transcripts were differentially expressed by ~2–12-fold in both PRPF8-KD and PRPF38A-KD. Of these, 7 and 5 were also differentially expressed by >2-fold in HCC1187 cells knocked down for PRPF8 or PRPF38A, respectively (Figure 4I).
We next asked whether any of the top basal-A dependency genes (Table S2) were affected by PRPF8 and PRPF38A knockdown. MCL1 (ranked #2 in our screen), PSMB4, POLR2G, C22orf29, ZNF451, ZNF552 and ZNF585A each had at least one transcript with β-score ≤ −0.69 or ≥0.69 and q ≤0.01 in both PRPF8-KD and PRPF38A-KD. In particular, the MCL1-002 transcript that encodes for the pro-apoptotic protein Mcl-1S was up-regulated in both PRPF8-KD and PRPF38A-KD, while the MCL1-001 transcript that encodes for anti-apoptotic Mcl-1L was down-regulated in PRPF8-KD. These results were confirmed by RT-PCR using 2 different siRNAs for PRPF8 and PRPF38A (Figure 4J, left). In HCC1187 cells, both Mcl-1S and Mcl-1L transcripts were induced by knockdown of PRPF8 or PRPF38A (Figure 4J, right). Thus, PRPF8 and PRPF38A knockdown enhanced expression of pro-apoptotic Mcl-1S.
To exclude potential off-target effects, we selected a subset of transcripts implicated in immune response (CXCL8-001), protein translation (RPS29-006, RPS7-005, RPS24-005), cell cycle (E2F1-001), mitosis (SPC4-001, B9D2-001), the proteasome (PSMB10-003) and apoptosis (MCL1-002) and reassessed each of them separately in non-transformed BPE cells and 5 breast cancer cell lines of different subtypes, using 2 to 4 pre-validated PRPF8 siRNAs (Figures S4 and S5). Changes in splicing of all these transcripts, except CXCL8 and PSMB10, were found using at least 2 distinct siRNAs in 3 or more cell lines (Table S10). Taken together, these data indicate that certain classes of transcripts, including multiple TNBC dependency genes, are altered by PRPF8 or PRPF38A knockdown.
The splicing modulator E7107 affects multiple TNBC survival networks and is cytotoxic against basal-A cells
E7107 is a clinical-stage small molecule that perturbs the activity of the core splicing factor SF3B1. Its effect on PRPF8 or PRPF38A function is unknown. Because SF3B1 is essential for splicing and operates upstream of the tri-snRNP complex, we postulated that E7107 may affect PRPF8 and PRPF38A sensitive transcripts and thus basal-like TNBC survival networks. To explore this hypothesis, we assessed genome-wide mRNA expression changes associated with exposure to E7107 for 24 hr using MB468 as a model. A total of 11,217 and 15,869 transcripts were up-regulated and down-regulated, respectively, based on the same sleuth criteria used for PRPF8 and PRPF38A RNA-seq analysis (Table S11). The majority of up-regulated transcripts (58%) were non-coding variants (mostly intron-retaining), similar to what observed after PRPF8 or PRPF38A knockdown. To define functional pathways most impacted by E7107, we focused on the top 1,000 up- and down-regulated transcripts based on b-value. Both genesets were enriched for genes involved in mitosis, RNA splicing, DNA repair, proteasome degradation and nuclear transport (q<0.01 for each module; Tables S12 and S13). Among PRPF8/PRPF38A-sensitive mRNAs, 42.5% of up-regulated and 28.2% of down-regulated transcripts were similarly affected by E7107 in the same direction, which is more than expected by chance (p=9.6E-68 and p=0.04, respectively, by hypergeometric distribution).
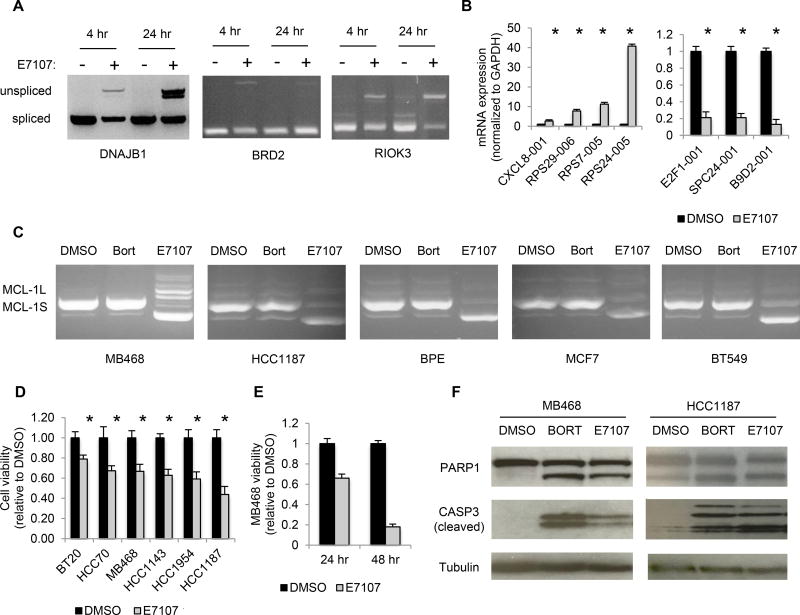
To begin to validate these data, we assessed by qRT-PCR or RT-PCR the expression of 11 of these transcripts in MB468 cells 24 hr after treatment with E7107. E7107 caused marked intronic retention based on the accumulation of unspliced transcripts of DNAJB1, BRD2 and RIOK3 (Figure 5A). Like PRPF8 and PRPF38A knockdown (Figures 4H–4I), E7107 activated the protein-coding isoform of CXCL8, induced intron-retaining transcripts of ribosomal proteins RPS7, RPS24 and RPS29, and suppressed protein coding variants of E2F1 and spindle checkpoint SPC24 and B9D2 transcripts (Figure 5B). Moreover, E7107 altered MCL1 splicing to the pro-apoptotic splice variant in 5 breast cell lines, including non-transformed BPE cells (Figure 5C). The effect of E7107 on MCL1 splicing was not secondary to cell death, since MCL1 splicing was not affected by bortezomib, which is cytotoxic to basal-A TNBC cell lines (9) (Figure 5C). Thus, E7107 recapitulated key splicing effects of PRPF8 and PRPF38A knockdown and affected similar processes implicated in basal-like TNBC survival.
Figure 5. The splicing modulator drug E7107 causes intron retention, affects PRPF8- and PRPF38A-sensitive splice variants and promotes apoptosis in basal-A cell lines in vitro.
(A) RT-PCR using intron-spanning primers for the indicated genes of total RNA from MB468 cells treated with 100 nM E7107 (+) or DMSO (−) for the indicated time.
(B) Transcript mRNA levels, determined by qRT-PCR of the indicated genes using isoformspecific primers, in MB468 cells treated with 100 nM E7107 or DMSO for 24 hr. Data represent the mean ± S.D. of at least 3 independent experiments (*, p <0.05; t-test relative to control).
(C) RT-PCR analysis of MCL1 splicing as in (Fig. 4J) in the indicated cell lines treated with E7107, bortezomib (Bort; used as control) or DMSO for 24 hr.
(D–E) Viability of indicated cell lines after treatment with E7107 or DMSO after 24 hr (D) or after culture for an additional 24 hr in drug-free medium (E), relative to DMSO-treated cells. Data represent the mean ± S.D. of at least 3 independent experiments (*, p <0.05; t-test relative to control).
(F) Immunoblot probed for the indicated proteins of lysates of MB468 and HCC1187 cells treated with 100 nM E7107, 12.5 nM Bortezomib (Bort) or DMSO for 24 hr.
We next tested whether E7107 treatment was cytotoxic against basal-like TNBC cells in vitro. Among 6 basal-A lines tested, 5 showed <66% viability 24 hr after treatment compared to vehicle-treated cells (Figure 5D). In MB468, viability declined to <20% after an additional 24 hr in drug-free medium, suggesting that the cells had become committed to death (Figure 5E). The sensitivity of other cell lines at 48 hr was not tested. In both HCC1187 and MB468, E7107 activated PARP1 and caspase-3 cleavage, two indicators of apoptosis, within 24 hr (Figure 5F). SF3B1 knockdown also killed both cell lines (Figure S6). These data suggest that SF3B1 modulators might be harnessed therapeutically to target multiple basal-like TNBC dependencies at once.
E7107 impedes TNBC growth in both cell-line- and patient-derived xenograft models
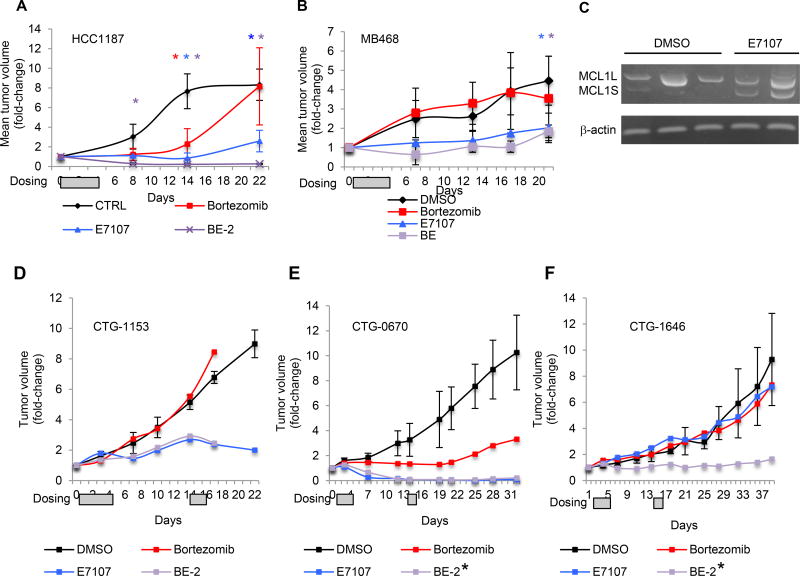
We next evaluated E7107 in 5 mouse TNBC models - 2 basal-A cell line xenografts (HCC1187 and MB468 in Nu/J mice) and 3 basal-like TNBC patient-derived xenografts (PDXs; CTG0670, CTG1153 and CTG1646 in NCr mice). E7107 (5 mg/kg) was administered when tumor volume reached ~100 mm3 (Day 0) by tail-vein injection for 4 days (1–4) or 4+2 days (1–4,15,16) in the cell-line and PDX models, respectively. In the HCC1187 model, E7107 decreased mean primary tumor volume by 80% after 22 days, compared to vehicle (p=0.002, one-sided t-test; Figure 6A). Comparable results were obtained in a replicate experiment (Figure S7A). MB468 xenografts were also sensitive to E7107 (55% mean tumor volume reduction after 21 days; p=0.01, onesided t-test; Figure 6B). Within MB468 xenografts, E7107 induced MCL1S expression as early as 6 hr after a single dose, although MCL1L expression persisted (Figure 6C).
Figure 6. E7107 impedes in vivo growth of cell-line- and patient-derived TNBC xenografts, and its activity is augmented in combination with bortezomib.
(A–B) Mean tumor volume fold-change (relative to Day 0) +/− SD in mice bearing palpable HCC1187 (n=16) (A) or MB468 (n=16) (B) xenografts in the right flank treated with intravenous bortezomib (0.4 mg/kg; red), E7107 (5 mg/kg; blue), both (purple) or DMSO (black) on days 1–4, according to the BE-2 dosing schedule, as shown in Table S14. * indicates significant difference (p <0.05; one-sided t-test) for bortezomib (red), E7107 (blue) or both (purple) relative to DMSO.
(C) RT-PCR analysis of MCL1 splicing as in (Fig. 4J) in pre-established MB468 xenografts 6 hr after treatment with one E7107 dose (5mg/kg) or DMSO.
(D–E) Tumor volume in mice bearing palpable CTG1153 (n=4) (D), CTG0670 (n=4) (E) or CTG1646 (n=4) (F) TNBC patient-derived xenografts in the right flank treated with bortezomib (0.4 mg/kg; red), E7107 (5 mg/kg; blue), both (purple) or DMSO (black) on days 1–4, 15, 16, according to the BE-2 (CTG1153) or BE-2* (CTG0670 and CTG1646) dosing schedules, as shown in Table S14. Data represent individual tumor volumes (relative to pre-treatment volume) for each mouse. Gray boxes in A,B,D-F indicate treatment days.
The PDX models originate from heavily pre-treated basal-like TNBC patients, who were refractory to an anthracycline, cyclophosphamide and paclitaxel. To assess the therapeutic effect of E7107, we adopted a “one animal per model per treatment” (1x1x1) study design (30), but included 3 animals in the control group to estimate the variability of each model. For each PDX, all vehicle-treated animals developed tumors that expanded over 22–38 days (Figures 6D–F). E7107 induced a partial response in CTG1153 (74% tumor volume reduction compared to control mean after 22 days; Figure 6D) and complete response in CTG0670 (>99% tumor volume reduction relative to control mean after 12 days; Figure 6E), while it was ineffective against CTG1646 (Figure 6F).
The combination of E7107 and bortezomib improves tumor response
We previously showed that bortezomib is effective against basal-A TNBC xenografts, but only at the MTD (9). To test whether bortezomib could augment E7107 antitumor activity, we first conducted a tolerability study in HCC1187 tumor-bearing Nu/J mice using increasing doses of bortezomib and a fixed E7107 dose (125 µg, ~5 mg/kg), co-administered intravenously. We assessed 5 bortezomib/E7107 (BE) regimens (Table S14). BE-3 was the maximum tolerated dose (MTD). BE-2 (50% the MTD), which was well tolerated, was used for all efficacy studies. This low dose of bortezomib (10 µg, ~0.4 mg/kg) alone has no therapeutic effect in these models. As above, cell line xenografts were treated for 4 days (1–4) and PDXs for 4+2 days (1–4,15,16). BE-2 induced nearly complete response against pre-established HCC1187 xenografts after 8 days (92% mean tumor volume reduction; p=0.01, one-sided t-test; Figure 6A), whereas neither bortezomib nor E7107 alone did. BE-2 somewhat improved the response to E7107 in MB468 xenografts but not significantly (Figure 6B).
We next assessed the combination in PDX models. In CTG1153, BE-2 was lethal after 16 days, suggesting that BE-2 is less well tolerated in NCr than Nu/J mice. BE-2 did not improve the CTG1153 response to E7107 over this time (Figure 6D). We next tested reducing the number of injections of bortezomib from 3 to 2 without changing E7017 dosing (BE-2*; Table S14) in CTG1646 and CTG0670. BE-2* was well tolerated in both models for over 30 days. In CTG0670, E7107 with or without bortezomib induced a complete response (Figure 6E). In CTG1646, where neither E7107 nor bortezomib had a significant effect on its own, the combination reduced tumor volume by 85% compared to control mice (Figure 6F). Overall, a bortezomib/E7107 combination reduced tumor volume in 5 of 5 TNBC models, inducing a complete or nearly complete response in 2 models.
DISCUSSION
Here we used a previous siRNA genetic dependency screen in one basal-A TNBC cell line (BPLER) as a starting point to identify common gene dependencies of basal-like TNBC as potential drug targets. Only 30 of 154 BPLER dependency genes were shared with other basal-A TNBC cell lines, consistent with this subtype’s heterogeneity. These 30 genes included 5 proteasome genes and MCL1, which we previously identified as shared dependencies of basal-A TNBC. Another prominent set of genes (12 of the remaining 24) function in RNA splicing. Although splicing involves hundreds of genes, these hits concentrated in a particular splicing complex, the U4/U6.U5 tri-snRNP complex, which joins the spliceosome complexes at the 5’ and 3’ ends of introns at an early step of intron excision. Indeed, TNBC cells knocked down for either of two tri-snRNP genes, PRPF8 and PRPF38A, showed widespread intron retention. Splicing alterations were not randomly distributed, but were concentrated in mRNAs participating in vital processes on which TNBC are known to depend, including protein translation (especially ribosomal proteins and translation initiation factors), protein degradation, mitosis, cell cycle progression, and the immune response. Abnormal splicing of MCL1 increased expression of its proapoptotic splicing variant and reduced expression of its antiapoptotic variant. Treatment with the SF3B1 modulator E7107 similarly affected MCL1, suggesting that MCL1 splicing is sensitive to RNA splicing perturbation, confirming previous reports (31,32), and not uniquely dependent on the tri-snRNP complex. E7107 also affected different gene networks critically implicated in TNBC survival, including mitosis, proteasome degradation, RNA splicing, nuclear export and DNA repair. Thus, multiple mechanisms, beside MCL1 inhibition, are likely involved in TNBC response to E7107. Additional studies will be needed to dissect the precise contribution of each of these mechanisms, particularly in vivo.
We first investigated the effect of knocking down tri-snRNP genes, which were high-confidence hits in our screen. No prior studies have looked at how these splicing factors affect pre-mRNA expression or splicing on a genome-wide scale. PRPF8 acts as a scaffold to assemble the complex. The function of PRPF38A is less clear. PRPF38A interacts with 28 other RNA splicing proteins, including tri-snRNP proteins, A complex proteins, B complex proteins, proteins involved in B complex activation, and U2 snRNA-interacting proteins. As such, PRPF38A is one of the largest protein-protein-interaction hubs of the human spliceosome (25).
RNA sequencing of cells knocked down for PRPF8 or PRPF38A showed that certain groups of functionally related transcripts are selectively sensitive to tri-snRNP disruption. Ribosomal protein transcripts comprised the most over-represented class of differentially spliced transcripts. This makes sense, since these mRNAs have short half-lives (~8–15 min). Mitosis and apoptosis transcripts (also enriched) also have short half-lives (33). Together, these data suggest that transcripts that turnover rapidly may be most affected by a dysfunctional spliceosome. The fact that the same transcripts were similarly affected in 6 cell lines (including non-transformed cells) suggests that certain genes may be intrinsically susceptible to splicing inhibition. Another key finding is that new transcripts are frequently intron-retaining or nonsense splice variants. Knockdown of these splicing genes selectively affected genes/pathways that are specific dependencies of basal-A TNBC cells, including mitosis, the ubiquitin-proteasome system and MCL1. Thus, suppressing splicing could be a way to target multiple TNBC vulnerabilities at once.
Splicing is emerging as an attractive therapeutic target for both hematological and solid malignancies (34). Human cancers frequently coopt the RNA splicing machinery to reprogram gene expression to their advantage. Although several splicing modulator natural products have been identified, only one splicing modulator drug, E7107, a derivative of pladienolide D, which perturbs SF3B1 activity (12,18,35), has been tested in the clinic (36,37). A 2nd-generation modulator of the SF3b complex, dubbed H3B-8800, is currently in phase 1 trials but no data have been released yet. To our knowledge, no selective modulators of the tri-snRNP complex are currently available for clinical testing. Although our study suggests that key TNBC survival networks are equally sensitive to inhibition of the SF3b and tri-snRNP complexes in vitro, it will be interesting to define whether specific modulation of either complex is associated with distinct safety and efficacy profiles in vivo. Here we showed, for the first time, that E7107 monotherapy induced a complete response in a TNBC PDX model. Our data also suggest that combining splicing modulation with proteasome inhibition should enhance anti-tumor efficacy and broaden responses.
Supplementary Material
Acknowledgments
We thank Tony Godfrey of Boston University for useful discussions, Arya Ashok and Maria Mancini of Champions Oncology for PDX-related studies; Caroline Shamu, Jennifer Smith and Sean Johnston of ICCB-L Screening Facility for help with siRNA screening; all members of the Lieberman and Petrocca laboratories for helpful suggestions.
Additional information: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number 5K22CA184244 to F. Petrocca, and by a Boston University/Boston Medical Center Startup Fund to F. Petrocca and a Breast Cancer Alliance Exceptional Project Grant to J. Lieberman. Approximately 50% percent of the project costs were financed by federal money and the remaining 50% by nongovernmental sources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Gusterson B. Do “basal-like” breast cancers really exist? Nat Rev Cancer. 2009;9:128–34. doi: 10.1038/nrc2571. [DOI] [PubMed] [Google Scholar]
- 3.Metzger-Filho O, Tutt A, de Azambuja E, Saini KS, Viale G, Loi S, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012;30:1879–87. doi: 10.1200/JCO.2011.38.2010. [DOI] [PubMed] [Google Scholar]
- 4.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nat Cell Biol. 2012;486:395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–9. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrocca F, Altschuler G, Tan SM, Mendillo ML, Yan H, Jerry DJ, et al. A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell. 2013;24:182–96. doi: 10.1016/j.ccr.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcotte R, Sayad A, Brown KR, Sanchez-Garcia F, Reimand J, Haider M, et al. Functional Genomic Landscape of Human Breast Cancer Drivers, Vulnerabilities, and Resistance. Cell. 2016;164:293–309. doi: 10.1016/j.cell.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu TYT, Simon LM, Neill NJ, Marcotte R, Sayad A, Bland CS, et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525:384–8. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwata M, Ozawa Y, Uenaka T, Shimizu H, Niijima J, Kanada RM, et al. E7107, a new 7-urethane derivative of pladienolide D, displays curative effect against several human tumor xenografts. Cancer Res. 2004;64:691–1. [Google Scholar]
- 13.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108–21. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA. 2006;12:1418–30. doi: 10.1261/rna.55406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen THD, Galej WP, Bai X-C, Savva CG, Newman AJ, Scheres SHW, et al. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature. 2015;523:47–52. doi: 10.1038/nature14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agafonov DE, Kastner B, Dybkov O, Hofele RV, Liu WT, Urlaub H, et al. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science. 2016;351:1416–20. doi: 10.1126/science.aad2085. [DOI] [PubMed] [Google Scholar]
- 17.Wan R, Yan C, Bai R, Wang L, Huang M, Wong CCL, et al. The 3.8 Å structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science. 2016;351:466–75. doi: 10.1126/science.aad6466. [DOI] [PubMed] [Google Scholar]
- 18.Fan L, Lagisetti C, Edwards CC, Webb TR, Potter PM. Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing. ACS Chem Biol. 2011;6(6):582–9. doi: 10.1021/cb100356k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.https://github.com/hbc/petrocca-tnbc-splicing
- 20.http://bioinformatics.sph.harvard.edu/petrocca-tnbc-splicing
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–20. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.http://championsoncology.com
- 24.Xie J, Beickman K, Otte E, Rymond BC. Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. The EMBO Journal. 1998;17:2938–46. doi: 10.1093/emboj/17.10.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schütze T, Ulrich AKC, Apelt L, Will CL, Bartlick N, Seeger M, et al. Multiple protein–protein interactions converging on the Prp38 protein during activation of the human spliceosome. RNA. 2016;22:265–77. doi: 10.1261/rna.054296.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patro R, Mount SM, Kingsford C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol. 2014;32(5):462–4. doi: 10.1038/nbt.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Meth. 2010;7:1009–15. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:31. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–7. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21:1318–25. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 31.Laetsch TW, Liu X, Vu A, Sliozberg M, Vido M, Elci OU, et al. Multiple components of the spliceosome regulate Mcl1 activity in neuroblastoma. Cell Death Dis. 2014;5:e1072. doi: 10.1038/cddis.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore MJ, Wang Q, Kennedy CJ, Silver PA. An Alternative Splicing Network Links Cell-Cycle Control to Apoptosis. Cell. 2010;142:625–36. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MSH. Database for mRNA Half-Life of 19 977 Genes Obtained by DNA Microarray Analysis of Pluripotent and Differentiating Mouse Embryonic Stem Cells. DNA Research. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonnal S, Vigevani L, Valcárcel J, rcel JVA. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11:847–59. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 35.Yokoi A, Kotake Y, Takahashi K, Kadowaki T, Matsumoto Y, Minoshima Y, et al. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J. 2011;278:4870–80. doi: 10.1111/j.1742-4658.2011.08387.x. [DOI] [PubMed] [Google Scholar]
- 36.Hong DS, Kurzrock R, Naing A, Wheler JJ, Falchook GS, Schiffman JS, et al. A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest New Drugs. 2013;32:436–44. doi: 10.1007/s10637-013-0046-5. [DOI] [PubMed] [Google Scholar]
- 37.Eskens FALM, Ramos FJ, Burger H, O'Brien JP, Piera A, de Jonge MJA, et al. Phase I Pharmacokinetic and Pharmacodynamic Study of the First-in-Class Spliceosome Inhibitor E7107 in Patients with Advanced Solid Tumors. Clinical Cancer Research. Clinical Cancer Research. 2013;19:6296–304. doi: 10.1158/1078-0432.CCR-13-0485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.