Abstract
Quantitative and qualitative analysis of amino acids in biofluids offers relevant information in diagnosis of diseases, evaluation of nutritional state and in elucidating metabolic influences on physiology. A simple, rapid and robust procedure based on CE-LIF has been optimised for human plasma samples in terms of sample treatment, separation and quantitation. Time required for derivatization was 15 min and analysis time 35 min. 4-Fluoro-7-nitro-2,1,3-benzoxadiazole (NBD-F) was the labeling agent used for obtaining fluorescence derivatives. Electrophoretic conditions were: 175 mM borate buffer at pH 10.25 prepared with 12.5 mM β-cyclodextrin. The voltage applied was +21 kV. Fourteen amino acids could be quantified: L-proline, L-phenylalanine, L-leucine, L-isoleucine, L-ornithine, D-ornithine, L-glutamine, L-alanine, L-threonine, glycine, L-serine, D-serine, taurine and L-glutamate. With this chiral CE-LIF method, Land D-amino acids are adequately separated. The method was validated for a representative group of amino acids in human plasma: L-proline, L-isoleucine, L-ornithine, L-glutamine, L-alanine L-threonine, glycine, L-serine, D-serine, and glutamate. The method has been successfully applied to human plasma from patients with bipolar disorder, all of which were taking lithium as a mood stabilizer. Eleven amino acids were quantified in plasma from nine patients, aged 24-55 years. The results were in accordance to published values for the bipolar patients. The method is useful particularly in studies where plasma amino acid levels can be used as biomarkers for diagnosis of diseases, evaluating the disease progression, and monitoring response to drug therapy.
Keywords: amino acids, biofluid, bipolar disorder, fluorescence, plasma
1 Introduction
Amino acids are organic compounds that play a major role in a number of important biological processes including energy metabolism, neurotransmission, lipid transport, etc. Quantitative and qualitative analysis of amino acids has been applied in the diagnosis of diseases (e.g., inborn errors of metabolism), evaluation of nutritional state and in elucidating metabolic influences on physiology [1-3]. Plasma levels of amino acids have been shown to be associated with symptoms and severity of a number of neuropsychiatric diseases such as schizophrenia, depression and bipolar disorder (BPD) [3, 4]. BPD, formerly known as manic-depressive psychosis, is one of the most debilitating and common psychiatric disorders worldwide. BPD is well distinguished by flashing emotional and behavioral disruptions [5]. As free amino acids are important for neurotransmission and receptor function, changes in their metabolism can be used not only for disease diagnosis, but also for monitoring treatment outcomes. Over or under expression of specific amino acids have been observed in patients of neuropsychiatric conditions when compared to controls: for example, some authors have found higher concentrations of glutamate, glutamine and glycine in BPD patients [4, 6]; depresed patients had a significant increase in plasma levels of glutamate, glycine, glutamine and taurine when compared to controls [3]. Studies have suggested that higher plasma serine concentration is a possible biomarker for schizophrenia, mania, paranoia, psychotic depression and unipolar depression [7-9]. Thus, these neuroactive amino acids (NAA) can be utilized as biomarkers in the diagnosis and prognosis of a number of neurological disorders. Some of these amino acids are present in trace levels in biological fluids. Increased complexity of the biological matrices makes it even more difficult to quantify these amino acids [10]. Thus, it is necessary to develop a highly sensitive analytical methodology for the determination of these amino acids. As was described elsewhere [11], quantitative measurement of the complete range of amino acids in biological samples is an important challenge in clinical biochemistry for several reasons: (1) amino acids do not have a chromophore; (2) most of them are highly hydrophilic and therefore are difficult to extract using organic solvents for gas chromatography (GC), show poor retention in reverse phase liquid chromatography (RP-LC) and are difficult to separate from the solvent peak. The chromatographic separation using RP needs analyte derivatization or the use of ion-paring agents to increase the chromatographic retention of analytes and to preclude their co-elution with the void volume; (3) in GC, the derivatizated compounds are very volatile, usually resulting in more than one derivative per analyte and some of these are lost during the sample treatment. Although, GC-MS involves time-consuming derivatization or complicated extraction procedures, it is the gold standard technique for the diagnosis of inborn errors of metabolism.
Different analytical techniques based on chromatographic and electrophoretic methodologies have been reported in the literature for analyzing amino acids from different biological matrices. Among the LC techniques, both ion-exchange, reverse phase high-performance liquid chromatography (RP-HPLC) and reverse phase high-performance liquid chromatography coupled with mass spectrometry (RP-HPLC-MS) are used extensively. Other common LC based methods include hydrophilic interaction chromatography (HILIC) [12]. Different detectors can been used with LC including UV [13, 14], fluorescence [15, 16], and mass spectrometry (MS) [17]. With UV-VIS and fluorescence detection, derivatization (pre- or post- separation) is often necessary due to the lack of a chromophore [18, 19]. Ion exchange separation usually requires a post-column derivatization with ninhydrin [18]. O-phthaldialdehyde (OPA) in combination with 2-mercaptoethanol (2-ME) is used to produce fluorescent isoindole derivatives which can be separated by HPLC and detected fluorimetrically. However, this reagent reacts only with primary amines in the presence of thiol and generates unstable derivatives [20]. For chiral analysis, by substituting a chiral thiol reagent for 2-ME, diasteromeric derivatives are produced and can be separated by HPLC [16].
Another technique that is routinely used to detect changes in the metabolic profiles is capillary electrophoresis (CE). Some reviews described advances in amino acid analysis by CE [21-23]; according to some of them, LIF provides the lowest reported limits of detection among the detection methods available for use with CE [24]. Several labelling techniques can be used with CE-LIF; each of these has their own strength and drawbacks. As reviewed previously [25], six different labelling agents are commonly used: NDA (naphthalene dicarboxaldehyde) [26], OPA [27], FQ (3-(2-Furoyl)quinoline-2-carboxaldehyde) [28-30] and fluorescein derivatives such as FITC (Fluorescein-5-isothiocyanate) [31, 32] or CFSE (Carboxyfluorescein succinimidyl ester) [33]. The main drawbacks are the use of toxic chemicals such as sodium cyanide (NDA, FQ, CBQCA), the expensive UV laser (OPA, NDA) [34], and the generation of a large number of fluorescein related labelling by-products. Moreover, FQ derivatives are not soluble enough in aqueous buffers [35]. Also, NDA, OPA, CBQCA and FQ only react with primary amines; proline, a secondary amine cannot be labelled with these reagents. 4-Fluoro-7-nitro-2,1,3-benzoxadiazole (NBD-F) is a labelling agent that can be used for both primary and secondary amines. Sensitive detection of NBD-derivatives can be performed using HPLC and CE coupled to common argon ion laser [36]. When compared to fluorescein based dyes, the advantages of using NBD-F are the short time required to achieve derivatization and the cleaner electropherograms due to low number of by-products. Tseng [37] described a MEKC method based on NBD-F with a background electrolyte containing sodium cholate, β-cyclodextrin, Brij 35 in aqueous borate buffer, pH 9.3 containing 7% methanol; they applied this method to quantify amino acids in biofluids, and thisincludes a complex extraction procedure and complicated electropherograms. We recently validated a bioanalytical method based on capillary electrophoresis lased induced fluorescence (CE-LIF) using NBD-F as a derivatizing agent for the analysis of amino acids in urine and hippocampus tissue [11]. To our knowledge, there are no other published methods for the analysis of amino acids in plasma or serum by CE-LIF using NBD-F. We have also quantified the L-and D-serine enantiomers using the current methodology using the labeling agent, NBD-F. A previous study by Singh and colleagues [32] has used FITC to quantify selected amino acid enantiomers including, serine. However, none of the reported capillary electrophoresis based methods on have been applied to study concentrations of amino acids using plasma samples from psychiatric patients.
The goal of this work was to optimize and validate a method for analyzing amino acids in plasma by a CE method combining chiral selector and LIF detection. This validated method was applied to measure selected amino acids (L-proline, L-phenylalanine, L-leucine, L-isoleucine, L-ornithine, L-glutamine, L-alanine, L-threonine, glycine, L-serine, D-serine, taurine and glutamate) using plasma samples of patients with bipolar disorder.
2. Materials and methods
2.1. Chemicals and reagents
Analytical grade standards of L- and D-amino acids: phenylalanine 99%, glutamine 98%, serine 99%, aspartic acid 98%, glutamic acid 99%, valine, alanine, threonine, proline 99%, isoleucine 98%, leucine, ornithine hydrochloride 99%, 2-aminoadipic acid (internal standard - IS), glycine 99%, taurine 99%, β- cyclodextrin 97%, and boric acid 99.5% were purchased from Sigma–Aldrich (Steinheim, Germany). Hydrochloric acid was from Fluka (Buchs, Switzerland) and sodium hydroxide from Panreac Química S.A.U. (Barcelona, Spain). NBD-F 99% was from TCI (Tokyo, Japan). All solutions and dilutions were prepared with purified water from a Milli-Qplus185 system (Millipore, Billerica, MA, USA). Individual 25 mM stock solution of each amino acid was prepared in purified water and stored at -20°C. From this, a dilution of 1 mM of each amino acid was prepared and stored at +4°C during the working week. These solutions were diluted as required on the day of the analysis. The derivatization solution of NBD-F was prepared by dissolving 80 mM NBD-F in methanol and mixing with equal parts of 500 μM HCl to give a final concentration of 40 mM NBD-F/250 μM HCl in 50% methanol.
2.2. Instrumentation
CE experiments were carried out on a P/ACE MDQ system (Beckman-Coulter, Fullerton, CA, USA) with LIF detector, an argon source operating at λexc: 488 nm and λem: 522 nm, a capillary silica column (Beckman Coulter, Madrid, Spain) 60 cm in total length, and 75μm of I.D. All solutions were kept refrigerated at 7 ± 2°C in the CE autosampler. Data acquisition and instrument control were carried out using 32 Karat™ system software version 7.0 (Beckman Coulter, Fullerton, CA, USA).
At the beginning of its use the capillary was conditioned by flushing with 1 M NaOH (15 min) and water (15 min). Between the runs, the capillary was flushed with HCl 0.1 M (3 min), water (5 min) and background electrolyte (BGE) (5 min). Injections were made at the anodic end with a pressure of 0.5 psi (33 mbar) for 10 s. After optimization, running buffer (BGE) consisted of 175 mM borate buffer at pH 10.25 (pH adjusted with 2M NaOH) and 12.5mM β-cyclodextrin. The voltage applied was +21 kV and the current observed under these conditions was 140 μA. The capillary thermostat was set at 17°C. The buffer vials were refreshed every six analysis to maintain consistency.
2.3. Plasma Samples
Study population
The study was approved by the Combined Neuroscience Institutional Review Board of the National Institutes of Health. All subjects provided written informed consent. All subjects were studied as inpatients at the National Institute of Mental Health Clinical Research Center, Mood Disorders Research Unit in Bethesda, Maryland. As described previously [38], patients corresponded to bipolar I or II disorder and currently experiencing a major depressive episode without psychotic features were enrolled in the study. This study included nine subjects; one male and eight females, with an age range of 24 to 55 years (mean age=39.2 [S.D. 10.5] years). These patients were also taking the mood stabilizer, lithium (0.6–1.2 mEq/L) for 4 weeks before and during the study, no other psychotropic medications were taken. Pool of plasma for method development and validation was from healthy volunteers.
All samples were stored at −80°C until the day of the assay.
Ultrafiltration
Plasma proteins were removed by filtration using the Centrifree® ultrafiltration devices from Millipore (Ireland Ltd., Ireland; 30kDa cut off). A volume of 150μL of plasma was placed into a single Centrifree® device and centrifuged at 2,000 × g, 4°C for 60 min. The filtered plasma was then used for amino acid analysis.
Derivatization procedure
In a glass tube, 20μL of filtered plasma, or the working standard solution was mixed with 20μL of 200 mM aminoadipic acid (IS), 25μL of 40mM NBD-F (derivatization solution) and 150μL of borate buffer 10 mM, pH 10. The resultant mixture was vortex for 10 seconds and metabolite derivatization was performed at 60°C for 15 min in an oven. Immediately after derivatization, samples were kept refrigerated in the CE autosampler at 7°C for at least 30 min before CE-LIF analysis.
Identification of amino acids in the plasma
Selected amino acids (L-proline, L-phenylalanine, L-leucine, L-isoleucine, L-ornithine, L-glutamine, L-alanine, L-threonine, glycine, L-serine, D-serine, taurine and glutamate) were analyzed in this study. In order to identify and quantify these amino acids in plasma, the standard solution of the pure amino acid was injected alone, followed by the sample, and then the sample that was spiked with the standard. Moreover, L- and D- pure enantiomers for each amino acid were injected to check whether the final analytical method was capable of separating these isomers.
2.4. Validation study
The method was validated for selected amino acids for selectivity, linearity, accuracy, instrumental precision, method precision (both with standards and samples), limit of quantification (LOQ) and limit of detection (LOD). Selectivity was checked by (1) analyzing the profile for ultrafiltered and non derivatized plasma pool without the labelling agent; no peak was observed in this case; (2), analyzing a blank containing the derivatizing reagent to distinguish the reagent peaks; and (3) comparing the electropherograms of derivatized samples with and without the IS to check that there were no others peaks at IS migration time. Linearity was estimated by assaying at least five levels of concentrations of the standards in triplicate, covering all the expected values ranging from 25 to 200% or 300% of mean values found in a preliminary assay. The individual ranges are described in table 1. Recovery was estimated by comparing in triplicate, the values of spiked samples prepared in a linear range (taking into account the endogenous concentrations which had been previously measured in the samples). Instrumental precision was evaluated by multiple injections (n=10) of a homogeneous derivatized standard solution. Within-day precision of the method was checked by injecting individual preparations of standards and samples in the mid-range of the calibration curve. Intermediate precision was tested in the same way, but on a different day, with freshly prepared buffers and reagents. LOQ for the selected amino acids were estimated using the Eurachem method [39] by injecting six replicates of each standard at least four levels of concentration ranging from 0.02-5.0 μM for D-serine and 0.1-20 μM for the rest of the amino acids. LOQ was established by representing R.S.D. of the six replicates versus concentration and interpolating the concentration corresponding to 10%. LOD was calculated by means of the relation LOD: (3/10)×LOQ and checked experimentally.
Table 1. Validation data for selected amino acids in human plasma with the optimized CE-LIF method.
| L-proline | L-isoleucine | L-ornithine | L-glutamine | L-alanine | L-threonine | glycine | L-serine | D-serine | L-glutamate | |
|---|---|---|---|---|---|---|---|---|---|---|
| Linearity (N=20) | ||||||||||
| Slope | 0.0078 ± 0.0003 | 0.0059 ± 0.0004 | 0.0017 ± 0.0002 | 0.0048± 0.0003 | 0.00382 ± 8.4E-05 | 0.0052 ± 0.0003 | 0.0059 ±0.0004 | 0.0055 ± 0.0003 | 0.0047 ± 0.0002 | 0.00481 ± 0.00003 |
| Intercept | −0.075 ± 0.099 | −0.053 ± 0.067 | −0.020 ± 0.028 | −0.37 ± 0.27 | −0.028 ± 0.038 | −0.19 ± 0.12 | −0.012 ± 0.0179 | −0.21 ± 0.13 | 0.0004 ± 0.003 | 0.012 ± 0.012 |
| r | 0.997 | 0.992 | 0.991 | 0.997 | 0.999 | 0.997 | 0.995 | 0.996 | 0.998 | 0.9997 |
| Range (μM) | 25-300 | 25-300 | 25-300 | 125-1500 | 25-750 | 25-600 | 25-750 | 25-750 | 0.5-25 | 25-300 |
| Accuracy | ||||||||||
| % | 97.1 | 101.5 | 97.1 | 101.8 | 94.0 | 96.0 | 96.7 | 103.7 | 100.9 | 92.0 |
| RSD(%) | 3.7 | 5.2 | 6.9 | 4.5 | 7.76 | 7.3 | 6.0. | 6.2 | 4.5 | 9.8 |
| Instrumental precisión (N=12) RSD(%) Method Precision Standard | 4.3 | 2.2 | 4.3 | 5.2 | 1.4 | 5.9 | 4.3 | 5.6 | 4.8 | 1.6 |
| Inter-assay (N=12) RSD (%) | 6.5 | 4.2 | 6.8 | 6.2 | 3.8 | 5.5 | 5.2 | 6.7 | 4.4 | 2.8 |
| Intra-assay (N=6) RSD (%) Method Precision Sample | 3.1 | 3.5 | 2.9 | 3.2 | 3.6 | 3.7 | 3.7 | 3.3 | 2.7 | 3.0 |
| Inter-assay (N=12) RSD (%) | 6.0 | 5.3 | 6.0 | 5.7 | 5.8 | 5.5 | 4.4 | 5.3 | 6.8 | 3.4 |
| Intra-assay (N=6) RSD (%) | 5.4 | 5.2 | 4.0 | 5.6 | 2.0 | 2.9 | 2.8 | 5.1 | 5.0 | 2.0 |
| LOQ (EURACHEM)(nM) | 738 | 630 | 1.44 μM | 146 | 889 | 714 | 128 | 174 | 231 | 203 |
| LOD(nM) | 221 | 189 | 433 | 43.7 | 267 | 214 | 38.3 | 52.2 | 69.5 | 61.0 |
3. Results and Discussion
3.1. Method optimization
The principal goal of this study was to develop and validate a robust and rapid CE-LIF method for the determination of amino acid profile in human plasma. The method was based on our previous CE-LIF method for the analysis of amino acids in urine and hippocampus tissue samples [11], with some optimizations for this new biological matrix, human plasma. This validated method was used to measure selected amino acids in patients with BPD.
In order to analyze amino acid concentrations in human plasma by CE-LIF, several optimizations were performed. Plasma proteins were first removed as amino acid profiling requires initial plasma deproteinization in order to avoid capillary clogging and changes due to absorptions in the capillary wall. These procedures involve the use of organic solvents or strong organic acids. The use of acid is also associated with some disadvantages; the labeling reaction (before derivatization with AQC, FITC or NBD-F etc.) is performed in an alkaline medium by the addition of borate buffer. The acidified sample requires much more buffer for pH switching, thus increasing the ionic strength of the sample. The advantages to be gained from adopting the strategy of ultrafiltration are mainly introducing neither any effect on the physico-chemical status of the free amino acid fraction in the plasma nor any substances that could affect derivatization. Because of the reasons cited above and based on the data available in the literature [14, 40], ultrafiltration was selected as the method for sample deproteinization. Plasma diluted (1:1) with pure water was compared with undiluted plasma sample. Better sensitivity without signal saturation was observed with the undiluted sample (data not shown). As an initial screen, previously described CE-LIF method [11] was applied to the filtered plasma after derivatization, but an unknown compound co-eluted with L-serine (which was not observed in our previous method). Lower concentrations of borate buffer were tested (90, 70, 50 mM); decreasing the buffer concentration, analysis time was lowered but resolution equal to zero was achieved, nevertheless L and D-serine were already resolved. Higher concentrations of borate buffers were assayed, 110 mM, 125 mM, 150mM and 175 mM after 30 seconds of sample injection (figure 1). Different sample injection times ranging from 10, 20, 30, 40, 50 seconds were also tested, large differences in sensitivity were found. At more than 30 s of sample injection, peaks such as D-serine had higher signal and lower deviation, but a higher risk of peak co-migration was observed near the derivatizing peak. Thus the 30 s injection time was applied for D-serine, and 10 s sample injection was applied all other amino acids analyzed. With the use of 110 mM borate buffer (figure 1A), L-serine showed a single peak suggesting that the co-eluting compound migrated within at the same time. As can be seen from figure 1, L-serine was fully resolved with higher concentrations of buffer (175mM; figure 1D) and the peak shape was good. Moreover, both glutamine and alanine were fully resolved when compared to 125 mM and 150 mM concentration (figure 1B, 1C). Migration time, number of theoretical plates and resolution were calculated for these two critical pair of peaks (table 2); best results for resolution >1.5 were obtained with 175 mM BGE.
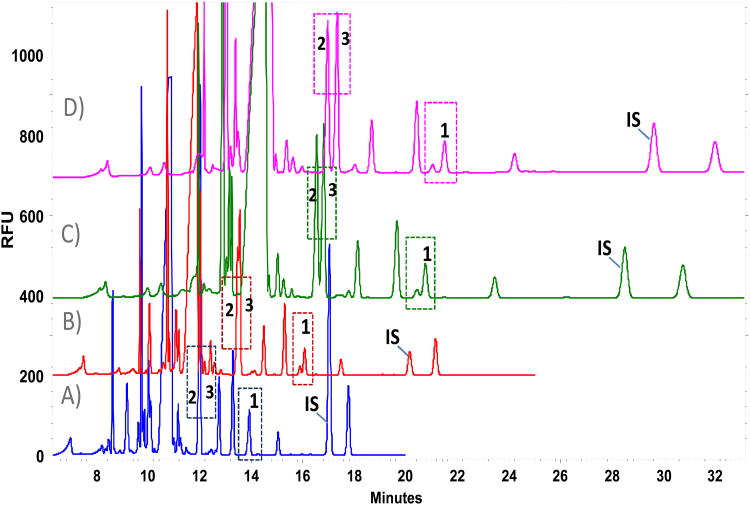
Figure 1.
Influence of buffer concentration over amino acid profile; borate buffer, pH 10.25 and 12.5 mM β-cyclodextrin, 30 s of sample injection. A) 110mM, [c.o.= 88μA]; B) 125mM, [c.o.= 100μA] C) 150mM, [c.o.= 119μA]; D) 175mM, [c.o.= 136μA]. Key. [c.o. ]= Current; [1]=L-serine; [2]=L-glutamine; [3]=L-alanine; [IS] = L-2-aminoadipic acid (A. 500 μM, B. 100 μM, C-D. 250 μM).
Table 2.
Performance parameters on critical separations of glutamine/alanine and Unknown/L-serine through the increase of running buffer concentration.
| Buffer 110mM [c.o.= 88μA] | Buffer 125mM [c.o.= 100μA] | Buffer 150mM [co.= 119μA] | Buffer 175mM [co.= 136μA] | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||
| Analyte | tm (min) | N(USP) | R(USP) | tm (min) | N(USP) | R(USP) | tm (min) | N(USP) | R(USP) | tm (min) | N(USP) | R(USP) |
| Gln | 12.0 | 139113 | 0.0 | 13.5 | 29080 | 0.3 | 15.5 | 160923 | 1.1 | 17.0 | 101646 | 1.7 |
| Ala | 12.0 | 13.6 | 104822 | 15.7 | 161776 | 17.3 | 98086 | |||||
|
| ||||||||||||
| Uknown | 13.9 | 110615 | 0.0 | 15.9 | 148677 | 1.1 | 18.9 | 140663 | 1.1 | 21.0 | 94944 | 1.7 |
| L-Ser | 13.9 | 16.1 | 155873 | 19.1 | 151890 | 21.5 | 95162 | |||||
Abbreviations: Gln: glutamine; Ala: alanine; L-Ser: L-serine; tm: migration time; N: theoretical plates; R: resolution; [c.o.]: current observed.
Addition of methanol or acetonitrile to the BGE (up to 20% (v/v) was studied as the fluorescence of NBD derivatives is very sensitive to the hydrophobicity of the BGE, but higher migration time in the whole profile and wider peaks were obtained. The final optimized conditions for the optimized CE-LIF method were: running buffer 175mM borate buffer at pH 10.25, 12.5 mM β-cyclodextrin, 10 s of sample injection (33mbar), L-2-aminoadipic acid (IS) using ultrafiltered, undiluted plasma samples.
The last step before validation was the identification of the selected amino acids in the plasma. As described in the method section, migration times and peak areas were compared between the ultrafiltered sample, pure standard and spiked sample (figure 2). Fourteen amino acids that were identified in the plasma profile include, L-proline, L-phenylalanine, L-leucine, L-isoleucine, L-ornithine, D-ornithine, L-glutamine, L-alanine, L-threonine, glycine, L-serine, D-serine, taurine and glutamate. Finally, for the identified amino acids pure L- and D- enantiomers were injected to ensure that the method could separate the enantiomers. The enantiomers of all these amino acids were satisfactorily separated. As a chiral selector was included in the BGE, chiral separation of the standards with resolution≥ 1.5 was achieved (data not shown).
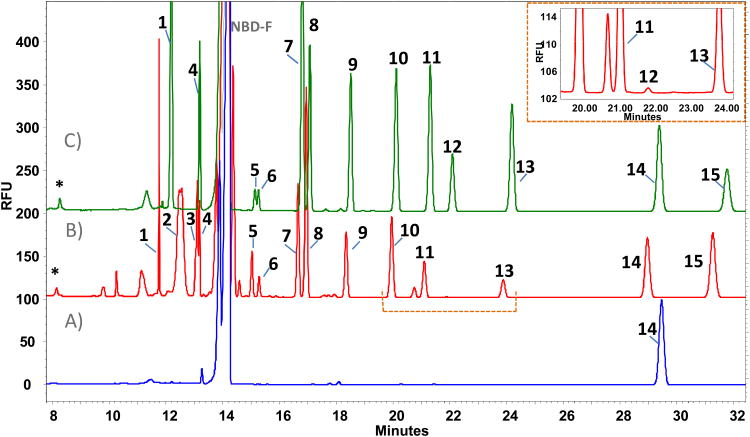
Figure 2.
A) Blank of sample (NBD-F). B) Plasma profile. C) Standards. Peak identification: 1. L-proline [200μM]; 2. L-phenylalanine; 3. L-leucine; 4. L-isoleucine [100μM]; 5. L-ornithine [100μM]; 6. D-ornithine [100μM]; 7. L-glutamine [500μM]; 8. L-alanine [250μM]; 9. L-threonine [200μM]; 10. glycine [200μM]; 11. L-serine [250μM]; 12. D-serine [100μM]; 13. taurine [150μM]; 14. L-2-aminoadipic acid [IS=200 μM]; 15. L-glutamate [100μM]; D. NBD-F hydrolysis products. Conditions: 175mM borate buffer, pH 10.25 and 12.5 mM β-cyclodextrin, 10 s of sample injection.
3.2. Validation
A complete validation was performed for only a representative group of amino acids including, L-proline, L-isoleucine, L-ornithine, L-glutamine, L-alanine, L-threonine, glycine, L- and D- serine, and L-glutamate. A summary of the validation parameters for the selected amino acids are shown in table 1.
During validation, standards fit the linear model (r > 0.99) for all amino acids and no bias was found for most of them excluding, L-glutamine, L-threonine and L-serine. However, in spite of the bias, no practical consequence was seen in the recovery. The recoveries ranged from 92.0 to 103.7.1 % and the differences were not statistically significant. For standards (n=12), the instrumental precision ranged from 1.4% to 5.9%. Intra-assay precision for standards ranged from 2.9% to 3.7% (n=6) and inter-assay precision from 2.8% to 6.8% (n=12). Intra-assay precision for samples: six samples prepared from the same pool were treated from the beginning and run in the same assay, the daily RSDs ranged from 2.0-5.6% and from 3.4% to 6.8% in different days (n=12). Theoretical LOQ calculated by the Eurachem method for these amino acids ranged from 128 nM for glycine to 1.44 μM for L-ornithine which were lower than the values observed in the samples.
3.3. Quantitation of amino acids in plasma from BD patients
The validated CE-LIF method was applied to study plasma samples from nine patients diagnosed with BD. Eleven amino acids were determined in all the profiles: L-proline, L-isoleucine, L-ornithine, L-glutamine, L-alanine, L-threonine, glycine, L- and D- serine, taurine and L-glutamate. All the results were in μM range and are summarized in table 3. Interestingly, five patients whose samples were collected from 2007 to 2009 showed lower concentration of L-glutamine and higher concentration of glutamate than the more recent samples. A possible explanation for this is that serum hydrolases may increase the free glutamic acid by breaking down glutamine [41] during storage. Although these samples were storage at −80 °C, the length of storage (3-5 years) could have contributed to this effect in both amino acids. Therefore, only the most recent samples were considered for statistical analysis of L-glutamate and L-glutamine. All the obtained results and other values from literature are presented in table 4. Major amino acids with concentrations from 220-590 μM were L-glutamine, L-alanine and glycine, amino acids at middle concentrations from 50 μM to 120 μM were L-threonine, L-serine, L-ornithine, L-proline, L-isoleucine, taurine and L-glutamate, and minor amino acids such as D-serine had a concentration of around 2 μM. Even though other previous studies analyzed plasma samples not only from bipolar but also from depressed patients under lithium or different treatments, our results are concordant with the literature. The glutamate and glycine results were very similar to the previous values reported for depressed patients [42] and major depressed BP patients [43]. Taurine values were lower compared with previously studied depressed patients [3, 44] but comparable levels were observed with BP patients [43]. For this instance antidepressant treatment may significantly decrease plasma taurine levels. Amino acid values obtained for L-alanine, L-glutamine, L-ornithine, L-isoleucine, L-threonine and L-serine, reported here are in agreement with the literature [3.4,42,43,44]. Interestingly most previously published articles reported total serine instead of the separate enantiomers.
Table 3.
Quantitation of amino acids on plasma from BPD patients.
| Plasma concentrations (μM) | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Patient | Year | Gender | Age | L-proline | L-isoleucine | L-ornithine | L-glutamine | L-alanine |
|
| ||||||||
| 1 | 2007 | Female | 34 | 70.9 | 60.0 | 121.8 | 476.8 | 349.2 |
| 2 | 2008 | Female | 24 | 36.1 | 38.7 | 60.5 | 403.0 | 268.1 |
| 3 | 2008 | Male | 55 | 67.1 | 63.3 | 94.6 | 560.8 | 399.9 |
| 4 | 2009 | Female | 37 | 73.1 | 75.4 | 78.9 | 471.0 | 440.1 |
| 5 | 2009 | Female | 44 | 63.7 | 68.0 | 113.5 | 354.9 | 322.5 |
| 6 | 2010 | Female | 48 | 41.8 | 46.2 | 77.4 | 633.7* | 298.3 |
| 7 | 2010 | Female | 29 | 23.0 | 43.3 | 54.8 | 531.8* | 278.8 |
| 8 | 2010 | Female | 32 | 56.9 | 45.4 | 433.7* | 214.4 | |
| 9 | 2011 | Female | 50 | 57.4 | 42.7 | 81.7 | 698.2* | 261.1 |
|
|
||||||||
| Average | 54.5 | 53.7 | 84.3 | 574.3 | 314.7 | |||
| SD | 16.2 | 12.4 | 21.0 | 116.1 | 67.4 | |||
| Plasma concentrations (μM) | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Patient | Sample year | L-threonine | glycine | L-serine | D-serine | L-glutamate | taurine |
|
| |||||||
| 1 | 2007 | 136.7 | 209.3 | 125.2 | 1.3 | 130.5 | 57.0 |
| 2 | 2008 | 93.4 | 197.1 | 93.6 | 1.7 | 123.1 | 33.3 |
| 3 | 2008 | 134.7 | 251.7 | 126.4 | 2.3 | 85.5 | 124.8 |
| 4 | 2009 | 116.0 | 192.7 | 85.1 | 4.5 | 211.5 | 55.3 |
| 5 | 2009 | 99.9 | 209.3 | 92.5 | 1.9 | 193.1 | 79.8 |
| 6 | 2010 | 177.5 | 247.5 | 109.4 | 3.5 | 22.4* | 26.5 |
| 7 | 2010 | 120.2 | 380.1 | 94.3 | 2.7 | 27.5* | 55.6 |
| 8 | 2010 | 131.7 | 145.2 | 73.4 | 1.2 | 27.7* | 21.3 |
| 9 | 2011 | 135.4 | 254.5 | 110.0 | 2.8 | 35.5* | 24.0 |
|
|
|||||||
| Average | 127.3 | 231.9 | 101.1 | 2.4 | 28.3 | 53.1 | |
| SD | 23.2 | 61.8 | 16.9 | 1.0 | 5.4 | 31.3 | |
Values of L-glutamine and L-glutamate considered for statistical analysis.
Table 4.
Amino acids quantitation in plasma from BD patients (n=9) and comparison with similar studies [3, 4, 42-44].
| Amino acid [μmol/L] | Current results (n=9) | Mitani, H. [3] (n=23) | Hoekstra, R. [4] (n=5) | Hoekstra, R. [4] (n=20) | Mayoral- Mariles, A. [42] (n=17-21) | Altamura [45] (n=25) | Mattos-Pinto, V.L. [44] (n=5) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||||
| Average | SEM | Average | SEM | Average | SEM | Average | SEM | Average | SEM | Average | SEM | Average | SEM | |
| L-glutamine | 574.3* | 50.3 | 393.4 | 40.4 | 583.4 | 28.4 | 447.0 | 65.2 | ||||||
| L-alanine | 314.7 | 23.8 | 404.4 | 42.4 | 273.0 | 11.0 | ||||||||
| glycine | 231.9 | 21.9 | 205.0 | 23.5 | 319.2 | 47.3 | 288.8 | 20.9 | 240.4 | 13.5 | 215.0 | 10.4 | 159.0 | 11.0 |
| L-threonine | 127.3 | 8.2 | 155.2 | 17.2 | 80.9 | 5.4 | 84.0 | 2.0 | ||||||
| L-serine | 101.1 | 6.0 | 107.4 | 9.6 | 93.1 | 3.6 | 108.0§ | 4.2 | 153.0§ | 9.0 | ||||
| L-ornithine | 84.3 | 7.4 | 97.0 | 2.0 | ||||||||||
| L-proline | 54.5 | 5.7 | ||||||||||||
| L-isoleucine | 53.7 | 4.4 | 44.9 | 2.5 | 71.0 | 11.0 | ||||||||
| taurine | 53.1 | 11.1 | 133.9 | 19.4 | 40.0 | 2.6 | 147.0 | 6.0 | ||||||
| L-glutamic acid | 28.3* | 2.7 | 94.0 | 10.0 | 39.4 | 8.6 | 56.8 | 6.1 | 44.1 | 3.3 | 336.0 | 31.8 | 58.0 | 2.0 |
| D-serine | 2.4 | 0.4 | 2.1 | 0.2 | ||||||||||
|
| ||||||||||||||
| Subject details | Inpatients with a diagnosis of bipolar I or II depression under lithium treatment | Patients with depression (17 out of 23 treated with imipramine drug) | Manic patients with bipolar I disorder under lithium | Manic patients with bipolar I disorder | Mild depressed eldery women | Bipolar and unipolar major depressed subjects | Patients with major depression diagnosed of anxiety disorder | |||||||
L-glutamine values from 4 patients (collected between 2010-2011). Data presented are mean ± SEM (standard error of the mean).
L-glutamate values from 4 patients (collected between 2010-2011). Data presented are mean ± SEM (standard error of the mean).
-Quantification of total serine.
4. Conclusions
We developed a sensitive CE-LIF method for the analysis of 14 amino acids in human plasma using 175mM borate running buffer at pH 10.25, 12.5 mM β-cyclodextrin, 10 s of sample injection (33mbar) and L-2-aminoadipic acid as the internal standard. The method has been validated for 10 amino acids including some D- and L-enantiomers such as D- and L-serine, which cannot be detected with other general methods based on GC and HPLC. Validation parameters are adequate for bioanalysis. Undiluted human plasma samples can be processed by ultrafiltration, derivatization and analyzed for amino acid profiles. The results obtained for representative amino acids plasma from BP patients are in agreement with literature values. The method is useful particularly in studies where plasma amino acid levels in patients are used as biomarkers for diagnosis of diseases, evaluating the disease progression, and monitoring response to drug therapy.
Acknowledgments
The authors thank Irving W. Wainer and Carlos A. Zarate, for the plasma samples and for their irreplaceable support and also Santiago Angulo for his scientific support. Alma Villaseñor acknowledges EADS-CASA for her fellowship. The authors gratefully acknowledge the financial support from Ministry of Science and Innovation MICINN CTQ2011-23562. This work was supported in part by the Intramural Research Programs of the National Institute of Aging, National Institutes of Health (NIH), and the National Institute of Mental Health, NIH.
Abbreviations
- BDP
bipolar disorder
References
- 1.He Y, Yu Z, Giegling I, Xie L, Hartmann AM, Prehn C, Adamski J, Kahn R, Li Y, Illig T, Wang-Sattler R, Rujescu D. Transl Psychiatry. 2012;2:e149. doi: 10.1038/tp.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu HB, Fang L, Hu ZC, Chen YC, Chen JJ, Li FF, Lu J, Mu J, Xie P. Psychiatry Res. 2012 doi: 10.1016/j.psychres.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 3.Mitani H, Shirayama Y, Yamada T, Maeda K, Ashby CR, Kawahara R. Prog Neuropsychopharmacol Biol Psychiatry. 2006;30:1155–1158. doi: 10.1016/j.pnpbp.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 4.Hoekstra R, Fekkes D, Loonen AJ, Pepplinkhuizen L, Tuinier S, Verhoeven WM. Eur Neuropsychopharmacol. 2006;16:71–77. doi: 10.1016/j.euroneuro.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Zarate CA, Manji HK. Mt Sinai J Med. 2008;75:226–247. doi: 10.1002/msj.20042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sussulini A, Prando A, Maretto DA, Poppi RJ, Tasic L, Banzato CE, Arruda MA. Anal Chem. 2009;81:9755–9763. doi: 10.1021/ac901502j. [DOI] [PubMed] [Google Scholar]
- 7.Waziri R, Wilson R, Sherman AD. Br J Psychiatry. 1983;143:69–73. doi: 10.1192/bjp.143.1.69. [DOI] [PubMed] [Google Scholar]
- 8.Waziri R, Wilcox J, Sherman AD, Mott J. Psychiatry Res. 1984;12:121–136. doi: 10.1016/0165-1781(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 9.Maes M, De Backer G, Suy E, Minner B. Neuropsychobiology. 1995;31:10–15. doi: 10.1159/000119166. [DOI] [PubMed] [Google Scholar]
- 10.Zinellu A, Sotgia S, Pisanu E, Scanu B, Sanna M, Usai MF, Chessa R, Deiana L, Carru C. Anal Bioanal Chem. 2010;398:1973–1978. doi: 10.1007/s00216-010-4134-5. [DOI] [PubMed] [Google Scholar]
- 11.Lorenzo MP, Navarrete A, Balderas C, Garcia A. J Pharm Biomed Anal. 2013;73:116–124. doi: 10.1016/j.jpba.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Martens-Lobenhoffer J, Bode-Böger SM. Clin Chem. 2006;52:488–493. doi: 10.1373/clinchem.2005.060152. [DOI] [PubMed] [Google Scholar]
- 13.Sethuraman R, Krishnamoorthy MG, Lee TL, Liu EH, Chiang S, Nishimura W, Sakai M, Minami T, Tachibana S. Clin Chem. 2007;53:1489–1494. doi: 10.1373/clinchem.2007.086702. [DOI] [PubMed] [Google Scholar]
- 14.Jaworska M, Stańczyk M, Wilk M, Kłaczkow G, Anuszewska E, Barzał J, Rzepecki P. Amino Acids. 2012;43:1653–1661. doi: 10.1007/s00726-012-1243-9. [DOI] [PubMed] [Google Scholar]
- 15.Frank MP, Powers RW. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;852:646–649. doi: 10.1016/j.jchromb.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant SL, Shulman Y, Tibbo P, Hampson DR, Baker GB. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;844:278–282. doi: 10.1016/j.jchromb.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 17.Shimbo K, Oonuki T, Yahashi A, Hirayama K, Miyano H. Rapid Commun Mass Spectrom. 2009;23:1483–1492. doi: 10.1002/rcm.4026. [DOI] [PubMed] [Google Scholar]
- 18.Kaspar H, Dettmer K, Chan Q, Daniels S, Nimkar S, Daviglus ML, Stamler J, Elliott P, Oefner PJ. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:1838–1846. doi: 10.1016/j.jchromb.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fekkes D. J Chromatogr B Biomed Appl. 1996;682:3–22. doi: 10.1016/0378-4347(96)00057-6. [DOI] [PubMed] [Google Scholar]
- 20.Devall AJ, Blake R, Langman N, Smith CG, Richards DA, Whitehead KJ. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;848:323–328. doi: 10.1016/j.jchromb.2006.10.049. [DOI] [PubMed] [Google Scholar]
- 21.Poinsot V, Gavard P, Feurer B, Couderc F. Electrophoresis. 2010;31:105–121. doi: 10.1002/elps.200900399. [DOI] [PubMed] [Google Scholar]
- 22.Poinsot V, Rodat A, Gavard P, Feurer B, Couderc F. Electrophoresis. 2008;29:207–223. doi: 10.1002/elps.200700482. [DOI] [PubMed] [Google Scholar]
- 23.Iadarola P, Ferrari F, Fumagalli M, Viglio S. Electrophoresis. 2008;29:224–236. doi: 10.1002/elps.200700662. [DOI] [PubMed] [Google Scholar]
- 24.Lapainis T, Sweedler JV. J Chromatogr A. 2008;1184:144–158. doi: 10.1016/j.chroma.2007.10.098. [DOI] [PubMed] [Google Scholar]
- 25.Szöko E, Tábi T. J Pharm Biomed Anal. 2010;53:1180–1192. doi: 10.1016/j.jpba.2010.07.045. [DOI] [PubMed] [Google Scholar]
- 26.Siri N, Lacroix M, Garrigues JC, Poinsot V, Couderc F. Electrophoresis. 2006;27:4446–4455. doi: 10.1002/elps.200600165. [DOI] [PubMed] [Google Scholar]
- 27.Taga A, Honda S. Journal of Chromatography A. 1996;742:8. [Google Scholar]
- 28.Veledo MT, de Frutos M, Diez-Masa JC. J Chromatogr A. 2005;1079:335–343. doi: 10.1016/j.chroma.2005.03.111. [DOI] [PubMed] [Google Scholar]
- 29.Boulat O, McLaren DG, Arriaga EA, Chen DD. J Chromatogr B Biomed Sci Appl. 2001;754:217–228. doi: 10.1016/s0378-4347(00)00611-3. [DOI] [PubMed] [Google Scholar]
- 30.Bergquist J, Vona MJ, Stiller CO, O'Connor WT, Falkenberg T, Ekman R. J Neurosci Methods. 1996;65:33–42. doi: 10.1016/0165-0270(95)00141-7. [DOI] [PubMed] [Google Scholar]
- 31.Nouadje G, Rubie H, Chatelut E, Canal P, Nertz M, Puig P, Couderc F. J Chromatogr A. 1995;717:293–298. doi: 10.1016/0021-9673(95)00747-3. [DOI] [PubMed] [Google Scholar]
- 32.Singh NS, Paul RK, Sichler M, Moaddel R, Bernier M, Wainer IW. Anal Biochem. 2012;421:460–466. doi: 10.1016/j.ab.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen HL, Zhang XJ, Qi SD, Xu HX, Sung JJ, Bian ZX. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3248–3252. doi: 10.1016/j.jchromb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Le Potier I, Smadja C, Zhang J, Taverna M. Anal Bioanal Chem. 2006;386:1387–1394. doi: 10.1007/s00216-006-0709-6. [DOI] [PubMed] [Google Scholar]
- 35.Klinker CC, Bowser MT. Anal Chem. 2007;79:8747–8754. doi: 10.1021/ac071433o. [DOI] [PubMed] [Google Scholar]
- 36.Hu S, Li PC. J Chromatogr A. 2000;876:183–191. doi: 10.1016/s0021-9673(00)00163-1. [DOI] [PubMed] [Google Scholar]
- 37.Tseng HM, Li Y, Barrett DA. Anal Bioanal Chem. 2007;388:433–439. doi: 10.1007/s00216-007-1239-6. [DOI] [PubMed] [Google Scholar]
- 38.Diazgranados N, Ibrahim L, Brutsche NE, Newberg A, Kronstein P, Khalife S, Kammerer WA, Quezado Z, Luckenbaugh DA, Salvadore G, Machado-Vieira R, Manji HK, Zarate CA. Arch Gen Psychiatry. 2010;67:793–802. doi: 10.1001/archgenpsychiatry.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Group EW. Available from: http://www.eurachem.org/guides/pdf/valid.pdf.
- 40.Jensen BP, Chin PK, Begg EJ. Anal Bioanal Chem. 2011;401:2187–2193. doi: 10.1007/s00216-011-5303-x. [DOI] [PubMed] [Google Scholar]
- 41.Deyl Z, Hyanek J, Horakova M. J Chromatogr. 1986;379:177–250. doi: 10.1016/s0378-4347(00)80685-4. [DOI] [PubMed] [Google Scholar]
- 42.Mayoral-Mariles A, Cruz-Revilla C, Vega-Manriquez X, Aguirre-Hernández R, Severiano-Pérez P, Aburto-Arciniega E, Jiménez-Mendoza A, Guevara-Guzmán R. Arch Med Res. 2012;43:375–382. doi: 10.1016/j.arcmed.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 43.Altamura C, Maes M, Dai J, Meltzer HY. Eur Neuropsychopharmacol. 1995;5(1):71–75. doi: 10.1016/0924-977x(95)00033-l. [DOI] [PubMed] [Google Scholar]
- 44.Pinto VL, de Souza PF, Brunini TM, Oliveira MB, Moss MB, Siqueira MA, Ferraz MR, Mendes-Ribeiro AC. J Affect Disord. 2012;140:187–192. doi: 10.1016/j.jad.2012.02.008. [DOI] [PubMed] [Google Scholar]