mRNA live-cell imaging reveals that plasmodesmata targeting is crucial for determining mobile mRNA movement.
Abstract
Many plant mRNAs move from cell to cell or long distance to execute non-cell-autonomous functions. These mobile mRNAs traffic through the phloem to regulate many developmental processes, but despite the burgeoning discovery of mobile mRNAs, little is known about the mechanism underlying the intracellular sorting of these mRNAs. Here, we exploited a fluorescence-based mRNA labeling system, using the bacteriophage coat protein MS2, fused to GFP (MS2-GFP) and an MS2 recognition site in the RNA of interest, to visualize the intracellular trafficking of mobile mRNAs in living plant cells of Nicotiana benthamiana. We first improved this system by using the nuclear localization sequence from FD, which substantially reduced the fluorescent background of MS2-GFP in the cytoplasm. The modified system allowed us to observe the cytoplasmic fluorescent foci dependent on MS2-binding sites. Coexpressing the MS2-GFP system with a virus movement protein, which is a plasmodesmata (PD)-localized nonspecific RNA-binding protein, targeted cytoplasmic fluorescent foci to the PD, suggesting that the cytoplasmic fluorescent foci contain mRNA and MS2-GFP. Our ex vivo RNA imaging revealed that mobile but not nonmobile mRNAs were selectively targeted to PD. Real-time images of intracellular translocation revealed that the translocation of mRNA and organelles in the transvacuolar strands may be governed by the same mechanism. Our study suggests that PD targeting of mRNA is a selective step in determining mRNA cell-to-cell movement of mRNAs.
Many mRNAs can move cell to cell or long distance to non-cell-autonomously regulate developmental processes. In response to environmental stimuli, these mobile mRNAs are transcribed in distal tissues, then translocate through the phloem and unload to the sink tissues (Banerjee et al., 2006; Lu et al., 2012; Huang et al., 2012), where they are believed to be translated to exert their functions (Lough and Lucas, 2006). Because a small amount of mobile mRNA is sufficient to generate multiple copies of a protein, this mRNA-based regulatory network provides an efficient way to incorporate spatial stimuli into development. Analysis of mRNA composition in phloem exudates and in grafted plants has revealed several thousands of mobile mRNAs (Guo et al., 2013; Thieme et al., 2015; Yang et al., 2015; Zhang et al., 2016), suggesting that long-distance transported mRNAs may be widely used in plants as a signaling mechanism.
Despite the increasing evidence of long-distance trafficking of mobile mRNAs, our knowledge of the mechanisms underlying mobile mRNA trafficking is limited. Simulation by use of a computational model suggests that the movement of mobile mRNAs may be non-sequence specific, and the mobility of mRNA is determined by transcript abundance (Calderwood et al., 2016). In contrast, analysis of mRNA movement by grafting experiments has shown that high accumulation of a nonmobile mRNA such as GFP in the cytosol is not sufficient to target GFP RNA for long-distance movement, but insertion of an RNA fragment from a mobile GA-INSENSITIVE or FLOWERING LOCUS T (FT) mRNA is sufficient to target GFP mRNA movement (Huang and Yu, 2009; Lu et al., 2012). In addition, recent evidence indicates that the tRNA-like structure on a subset of mobile mRNAs is a signal triggering mRNA movement (Zhang et al., 2016). Thus, how mobile mRNA is translocated cell to cell remains to be elucidated.
Until recently, our detection of mobile mRNA largely relied on reverse transcription quantitative PCR (RT-qPCR) or RNA-sequencing analyses of grafted plants (Banerjee et al., 2006; Huang et al., 2012; Lu et al., 2012; Thieme et al., 2015; Yang et al., 2015). However, the long-distance trafficking of mobile mRNAs in grafted plants consists of multiple translocation steps, including intracellular trafficking of mobile mRNAs from the nucleus to cytosol, cell-to-cell movement in mesophyll cells and the companion cell/sieve element complex, translocation through phloem, and unloading into and cell-to-cell movement in the destination tissues. Different RNA transport sequences may participate in various steps to determine RNA translocation. Therefore, an approach is needed to effectively investigate the individual translocation steps of mobile mRNA.
Among the RNA live-imaging systems, indirect RNA labeling by fluorescent-tagged RNA-binding proteins (RBPs) is well developed (Keryer-Bibens et al., 2008; Urbanek et al., 2014). The general principles of in vivo mRNA labeling are to exploit a GFP-fused RBP, such as an MS2 bacteriophage coat protein (MS2-GFP), with the target RNA fused to a tandem repeat of the MS2 recognition RNA stem loop (SL) and the interaction between MS2-GFP and RNA-SL allowing the visualization of mRNA in the living cells. To reduce the cytosolic fluorescent background derived from MS2-GFP, the nuclear localization signal (NLS) derived from Simian Virus 40 (SV40) is used to restrict MS2-GFP to the nucleus (Bertrand et al., 1998). When coexpressed with an mRNA containing SL, MS2SV40-GFP tethers to its RNA recognition site, and the RNA-protein complexes are retained in the cytosol to form concentrated fluorescent foci. This system has been successfully used to visualize mRNA in yeast, Drosophila melanogaster, mammals, and plants (Bertrand et al., 1998; Rook et al., 2000; Forrest and Gavis, 2003; Hamada et al., 2003; Sambade et al., 2008; Park et al., 2014). One drawback of the MS2 system is that the NLS from SV40 is inefficient in restricting small GFP fusion proteins to the nucleus (Köhler, 1998), and the small amount of MS2SV40-GFP proteins accumulates in the cytosol, thereby producing a low level of cytosolic fluorescent background (Wu et al., 2012). However, this system usually produces substantial fluorescent background in the cytosol when used in plants, possibly because the large vacuoles occupy most of the volume in plant cells, which compresses the cytosol in a thin layer around the cells. Thus, even a small amount of cytosolic fluorescence background is concentrated at the cell periphery and greatly interferes in the detection of mRNA signals.
In this study, we adopted an MS2-GFP-based mRNA labeling system to visualize real-time trafficking of mRNA in living cells. We improved the MS2 system and substantially reduced the cytoplasmic fluorescent background. By using this modified system, we showed that the detection of cytoplasmic green fluorescent foci depended on the binding sites of MS2. Ex vivo RNA imaging differentiated the localization of mobile and nonmobile mRNAs, which suggests that the intracellular targeting of mRNA may be determined by intrinsic mRNA localization signals. Further colocalization studies identified mobile mRNAs targeted to plasmodesmata (PD), which supports that the cell-to-cell movement of mobile mRNA is mediated through PD.
RESULTS
NLSs from Arabidopsis thaliana FD Effectively Reduced the Cytoplasmic Background of the mRNA Labeling System
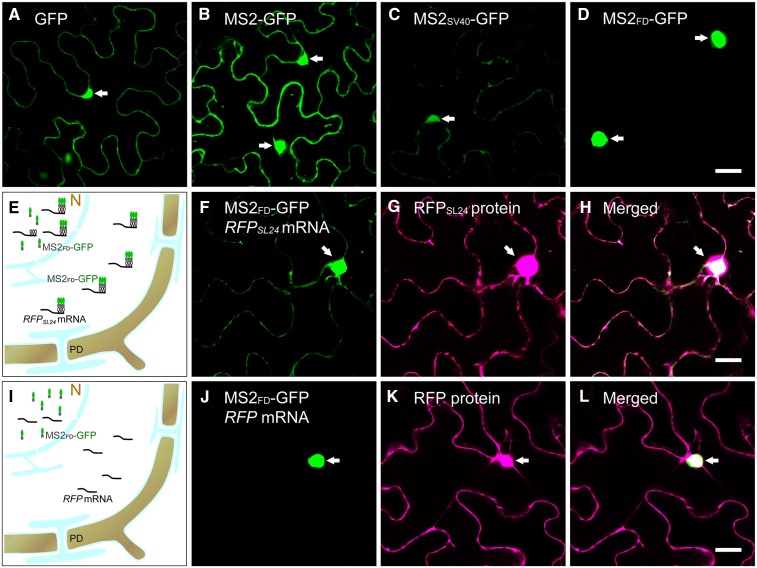
The MS2-based RNA live imaging system has been widely used in microbial and animal systems to visualize the subcellular localization of mRNAs. Transient expression of GFP or MS2-GFP by agroinfiltration showed GFP or MS2-GFP localized in both the nucleus and cytosol (Fig. 1, A and B). Although the insertion of SV40 NLS into MS2-GFP (MS2SV40-GFP) greatly reduced cytosolic MS2-GFP, we detected substantial cytoplasmic MS2-GFP background (Fig. 1C), which greatly interferes with the detection of signals. To improve the nuclear retention of MS2SV40-GFP in living plant cells, we selected the plant endogenous NLS to limit MS2-GFP to the nucleus. Arabidopsis FD is a transcription factor that exclusively localizes in the nucleus of cells in the apex (Abe et al., 2005; Wigge et al., 2005). Use of the online NLS prediction tool SeqNLS predicted that the potential NLS of FD is located at 203 to 235 amino acids. Consistent with this prediction, confocal microscopy revealed that the C-terminal fragment of FD (211–240 amino acids) was sufficient to target MS2-GFP to the nucleus (Fig. 1D). This fusion construct (MS2FD-GFP) was driven by a CaMV35S promoter for our mRNA live imaging system.
Figure 1.
Improvement of the MS2-GFP RNA labeling system in plants. A to D, Agroinfiltration of GFP (A), MS2-GFP (B), MS2SV40-GFP (C), and MS2FD-GFP (D) in a N. benthamiana leaf. E and I, Illustrations of the MS2-GFP RNA labeling system. N, Nucleus. F to H, Coexpression of MS2FD-GFP with RFPSL24 mRNA. F, Green channel; G, red channel; H is merged from F and G. J to L, Coexpression of MS2FD-GFP with RFP mRNA. J, Green channel; K, red channel; L is merged from J and K. The nucleus is indicated by arrows. Bar = 20 µm.
In previous mammalian studies, a minimum of 24 repeats of SL sequences was required to detect a fluorescent GFP signal in the cytosol (Fusco et al., 2003). To determine the specificity of MS2FD-GFP in living plant cells, we generated the cDNA of RFP containing 24 repeats of MS2 recognition SL sequences (RFPSL24) driven by a CaMV35S promoter. With MS2FD-GFP coexpressed with RFPSL24 by agroinfiltration in Nicotiana benthamiana leaves, MS2FD-GFP could complex with RFPSL24 mRNA, and we detected GFP signals in both the nucleus and cytosol (Fig. 1, E–H). In the control experiment, coexpression of MS2FD-GFP with RFP without SL sequences restricted MS2FD-GFP to the nucleus (Fig. 1, I–L). These findings suggest that the detection of GFP fluorescent signals in the cytosol depended on MS2 recognition of SL sequences. In addition, the insertion of 24 repeats of SL sequences in RFP mRNA did not interrupt the transcription or translation of mRNA because our real-time RT-qPCR revealed comparable mRNA levels in RFP- or RFPSL24-expressing leaves (Supplemental Fig. S1), and RFP fluorescent signals were detected in leaves coexpressed with RFP or RFPSL24 mRNA, so RFP proteins were translated from both mRNAs (Fig. 1, G and K).
Movement Protein of Tobacco mosaic virus Targets mRNAs to PD
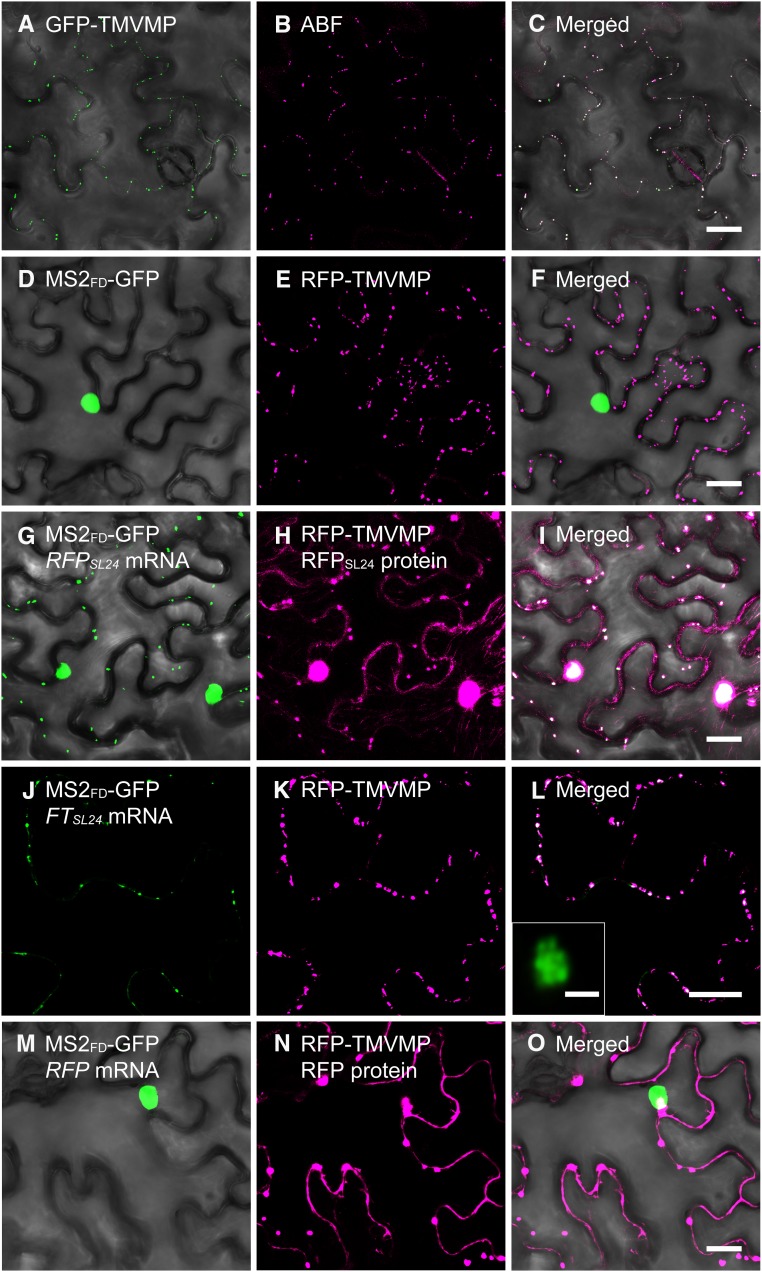
The TMVMP (movement protein of Tobacco mosaic virus) is a PD-localized protein with non-sequence-specific RNA-binding activity (Citovsky et al., 1990). To further verify our MS2 system, we used PD-localized TMVMP (Fig. 2, A–C) to target RFPSL24 mRNA to PD. On coexpression of RFP-TMVMP with MS2FD-GFP, TMVMP did not interfere in the nuclear localization of MS2FD-GFP (Fig. 2, D–F). Remarkably, the punctate appearance of green fluorescent foci, which were colocalized with RFP-TMVMP (Fig. 2, G–L; Supplemental Fig. S2), was observed in cells with coexpression of RFP-TMVMP and RFPSL24 (Fig. 2, G–I) or FTSL24 mRNA (Fig. 2, J–L) but not RFP mRNA (Fig. 2, M–O). Further Airyscan confocal microscopy revealed aggregation of multiple fluorescent dots in fluorescent foci (Fig. 2L, inset). These results suggest that TMVMP binds with target mRNA and relocates the mRNA to PD. Thus, the cytosolic GFP in our images were derived from ribonucleoprotein complexes containing MS2FD-GFP and RFPSL24 or FTSL24 mRNA.
Figure 2.
Targeting of mRNA to PD by TMVMP. A and B, Expression of GFP-TMVMP (A) in an ABF-stained N. benthamiana leaf (B). C, Merged image of GFP-TMVMP and ABF-stained PD. D to O, Coexpression of MS2FD-GFP with RFP-TMVMP (D–F), MS2FD-GFP with RFP-TMVMP and RFPSL24 mRNA (G–I), MS2FD-GFP with RFP-TMVMP and FTSL24 mRNA (J–L), or MS2FD-GFP with RFP-TMVMP and RFP mRNA (M–O). L, inset, Airyscan confocal microscopy of a single GFP foci in J. A, D, G, J, and M, Green channel; B, E, H, K, and N, red channel; C, F, I, L, and O, merged images of green and corresponding red channels. Bar = 20 µm in all panels, except L inset, in which bar = 1 µm.
Selective Subcellular Distribution of Mobile and Nonmobile mRNA in Living Plant Cells
Spatial and temporal mRNA localization to specific subcellular compartments is crucial for physiological function and developmental regulation (Buxbaum et al., 2015). We hypothesized that mobile mRNA is selectively targeted to specific subcellular compartments, such as PD, for cell-to-cell movement. To test this hypothesis, we examined the subcellular distribution of a nonmobile mRNA, RFPSL24, and a mobile mRNA, FTSL24, in living plant cells. To verify that FTSL24 mRNA remains functional after conjugation with 24 repeats of SL sequences, FTSL24 was driven by a CaMV35S promoter and introduced into the Arabidopsis ft-10 mutant, a late-flowering mutant (Yoo et al., 2005). The flowering time was earlier for ft-10 transformants harboring 35Spro:FTSL24 than the ft-10 mutant (Supplemental Fig. S3A), so 35Spro:FTSL24 complemented the ft-10 mutant. In addition, Arabidopsis grafting experiments showed that FTSL24 mRNA moved long distance from the 35Spro:FTSL24 transformant stocks to the wild-type scions (Supplemental Fig. S3B), so FTSL24 mRNA remained a mobile mRNA.
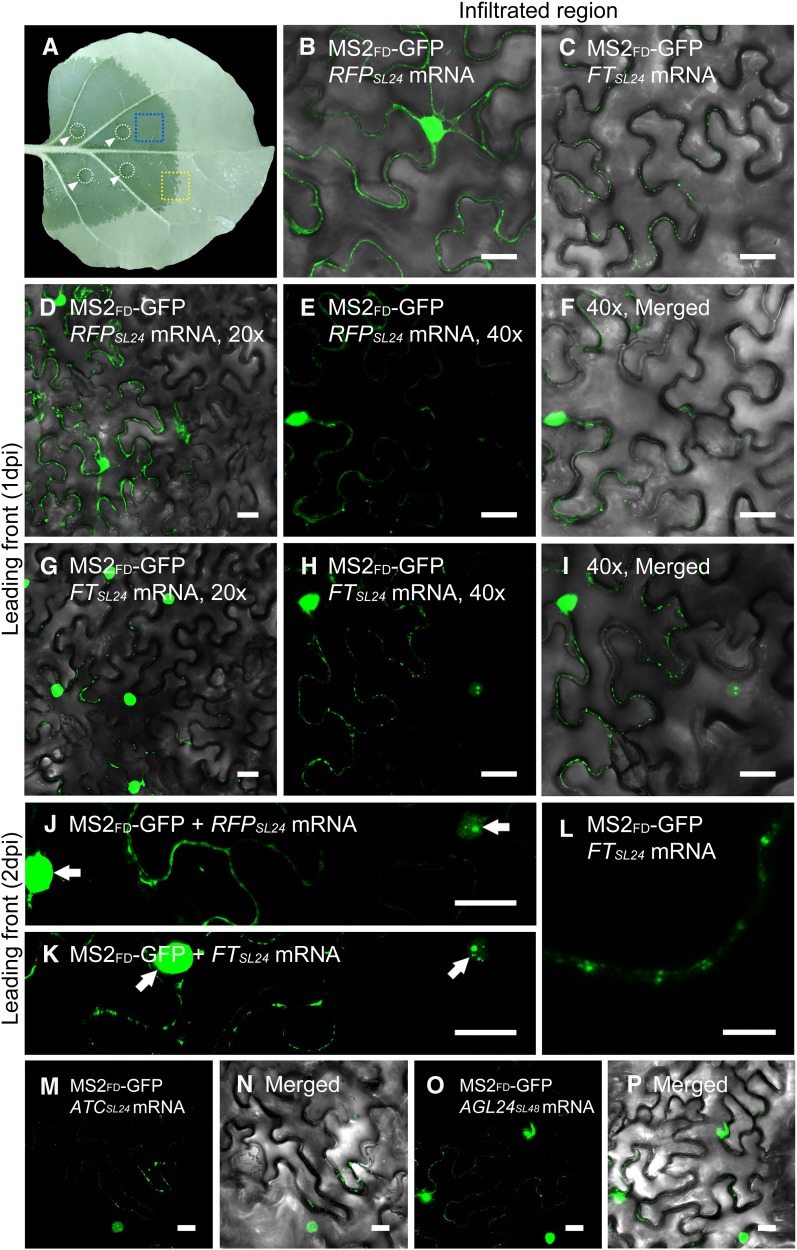
We transiently expressed FTSL24 or RFPSL24 mRNA in N. benthamiana leaves by agroinfiltration (Fig. 3A). At 1 d postinfiltration (dpi), green fluorescent signals of RFPSL24 were evenly distributed at the cell periphery (Fig. 3B), whereas FTSL24 signals were accumulated in individual foci at the cell periphery (Fig. 3C). However, at later stages (2 dpi), RFPSL24 or FTSL24 mRNAs were highly accumulated in the cytosol, with less differential accumulation between RFPSL24 and FTSL24 mRNA (Supplemental Fig. S4). However, in some cases, FTSL24 mRNA signals were still distributed throughout the cell periphery, with a few dispersed punctate foci (Supplemental Fig. S4B), which contrasted with the even distribution of RFPSL24 mRNA (Supplemental Fig. S4A).
Figure 3.
Differential subcellular distribution of mobile mRNA and nonmobile mRNA in living plant cells. A, Image of an agroinfiltrated N. benthamiana leaf. The infiltrated positions are indicated by circles and arrowheads. The infiltrated area is visible by the dark color. Leaf sections of the infiltrated region (blue rectangle) or leading front (yellow rectangle) were cut out for confocal analyses. B and C, The infiltrated region of a N. benthamiana leaf coexpressing MS2FD-GFP and RFPSL24 (B) or FTSL24 mRNA (C) at 1 dpi. Bar = 20 μm. D to K, The leading front tissues of a N. benthamiana leaf coexpressing with MS2FD-GFP and RFPSL24 (D–F and J) or FTSL24 mRNA (G–I and K). Bar = 20 µm. F and I are the merged image of E and H with its corresponding bright-field image. L, Airyscan analysis of N. benthamiana leaf coexpressing MS2FD-GFP and FTSL24 mRNA. Bar = 5 µm. M to P, The leading front of N. benthamiana leaf coexpressing MS2FD-GFP and ATCSL24 (M and N) or AGL24SL48 mRNA (O and P). Bar = 20 µm. N and P are the merged image of M and O with its corresponding bright field image. Arrows in J and K indicate nucleus.
The CaMV35S promoter is a strong, constitutive promoter, and vigorous expression of delivered constructs may overload the RNA mobile machineries and result in nonspecific localization of mRNA, which results in a narrow time frame for observing the subcellular distribution of mRNA. Cells in the leading front region of the leaves usually begin to express delivered constructs. The low expression of FTSL24 and RFPSL24 mRNA in the leading front cells allows us to visualize intracellular targeting of FTSL24 and RFPSL24 mRNA in these cells. To minimize the nonspecific targets of overaccumulated mRNA, we used these leading front regions for analysis (Fig. 3A). By confocal microscopy, the leading front cells were distinguished by a clear boundary between GFP-expressing and non-GFP-expressing cells (Fig. 3, D–I). In these boundary cells, fluorescent signals with a few speckles were first observed in the nucleus (Fig. 3, J and K). In cells neighboring these early expressed cells, the signals in the nucleus rapidly reached saturation, with few fluorescent signals observed in the cytosol (Fig. 3, J and K). The cytoplasmic fluorescent signals in the cells expressing RFPSL24 or FTSL24 exhibited distinct distribution patterns: FTSL24 signals were concentrated in individual foci located at the cell periphery, whereas RFPSL24 signals were evenly distributed at the cell periphery (Fig. 3, E and H). High-resolution images with Airyscan showed that the FTSL24 signals displayed a puncta pattern, probably recognized as the patterns of PD (Fig. 3L). To further validate the subcellular distribution patterns of mobile and nonmobile mRNAs, MS2FD-GFP and RFPSL24 or FTSL24 mRNA were cobombarded to N. benthamiana leaves. In agreement with the results of agroinfiltration, FTSL24 mRNA displayed punctate patterns, whereas RFPSL24 mRNA were distributed at the cell periphery (Supplemental Fig. S5).
To further examine whether the punctate distribution of mRNA also occurs with other mobile mRNAs, we selected two mobile mRNAs, Arabidopsis thaliana CENTRORADIALIS homolog (ATC) and AGAMOUS-LIKE24 (AGL24), for analysis (Yang and Yu, 2010; Huang et al., 2012). On coexpressing ATCSL24 or AGL24SL24 mRNA with MS2FD-GFP in N. benthamiana cells, the two mRNAs also showed a punctate distribution on the plasma membrane (Fig. 3, M–P). Thus, the three mobile mRNAs were targeted to a PD-like structure when transiently expressed in N. benthamiana cells.
Mobile FTSL24 mRNA Colocalizes with PD
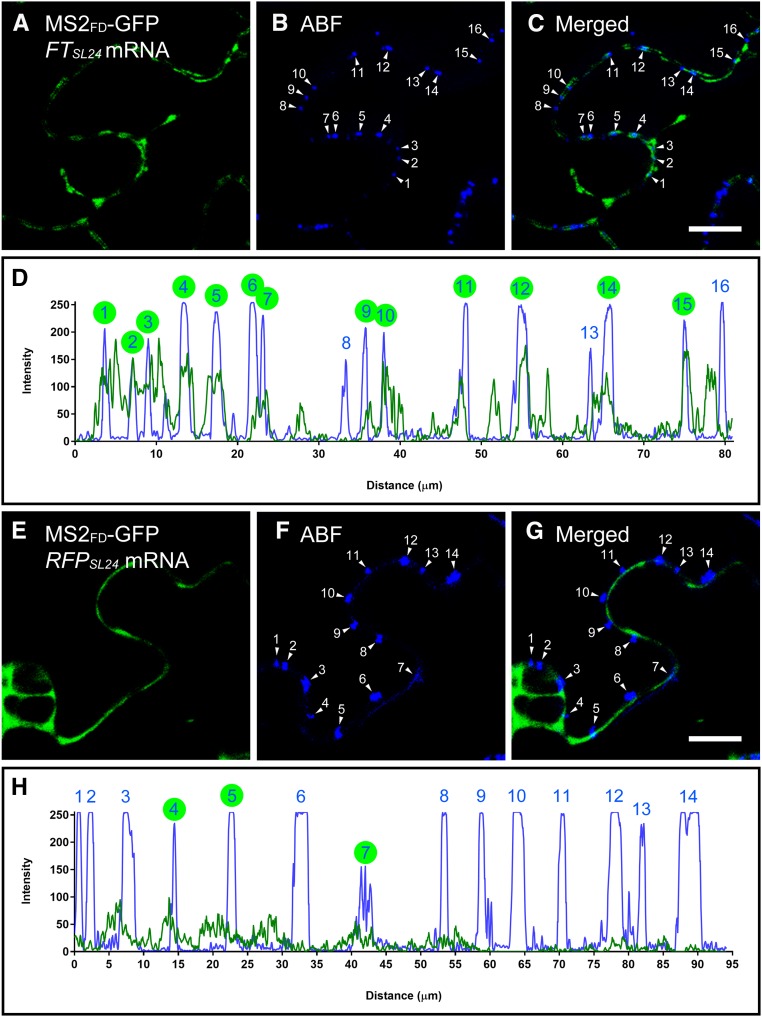
In plants, PD are the channels mediating cell-to-cell trafficking of many macromolecules including proteins and microRNA (Vatén et al., 2011). To verify that the punctate foci of FTSL24 mRNA were PD, we examined colocalization of FTSL24 mRNA and PD markers. Aniline blue fluorophore (ABF) is a widely used PD marker that stains callose located at the PD neck (Vatén et al., 2011). In ABF-stained N. benthamiana leaves expressing MS2FD-GFP and FTSL24 or RFPSL24 mRNA, FTSL24 signals colocalized with PD (Fig. 4, A–D). In contrast, RFPSL24 signals mainly distributed in the cytoplasmic space adjacent to the plasma membrane (Fig. 4, E–H). Fluorescence overlapping spectra showed the concordant distribution of ABF with FTSL24 but not RFPSL24 mRNA. In 16 ABF-stained PD analyzed, FTSL24 signals were detected in 13 (Fig. 4D), whereas RFPSL24 signals were detected in only 3 of 14 stained PD (Fig. 4H). Consistent with these results, colocalization analyses with FM4-64-stained plasma membrane showed FTSL24 mRNA distributed as puncta on the plasma membrane (Supplemental Figure S6, A–C), whereas RFPSL24 mRNA was distributed in the cytoplasmic space adjacent to the plasma membrane (Supplemental Fig. S6, D–F). Thus, our results suggest that FTSL24 but not RFPSL24 mRNA was targeted to PD.
Figure 4.
Mobile FTSL24 mRNA is targeted to PD. A to C, Colocalization of FTSL24 mRNA and PD. PD was stained with ABF. The numbers in B and C represent individual PD stained with ABF. C is merged from A and B. Bar = 10 µm. D, Quantitative analysis of colocalization by overlapping fluorescence spectra. Blue, PD stained with ABF; green, FTSL24 mRNA. E to G, Colocalization analysis of RFPSL24 mRNA and PD. The numbers in F and G represent individual PD stained with ABF. G is merged from E and F. Bar = 10 μm. H, Quantitative analysis of colocalization by overlapping fluorescence spectra. Blue, PD stained with ABF; green, FTSL24 mRNA. In D and H, PD with colocalization of FTSL24 or RFPSL24 mRNA is indicated by green circles.
Dynamic Translocation of RFPSL24 or FTSL24 mRNA to PD
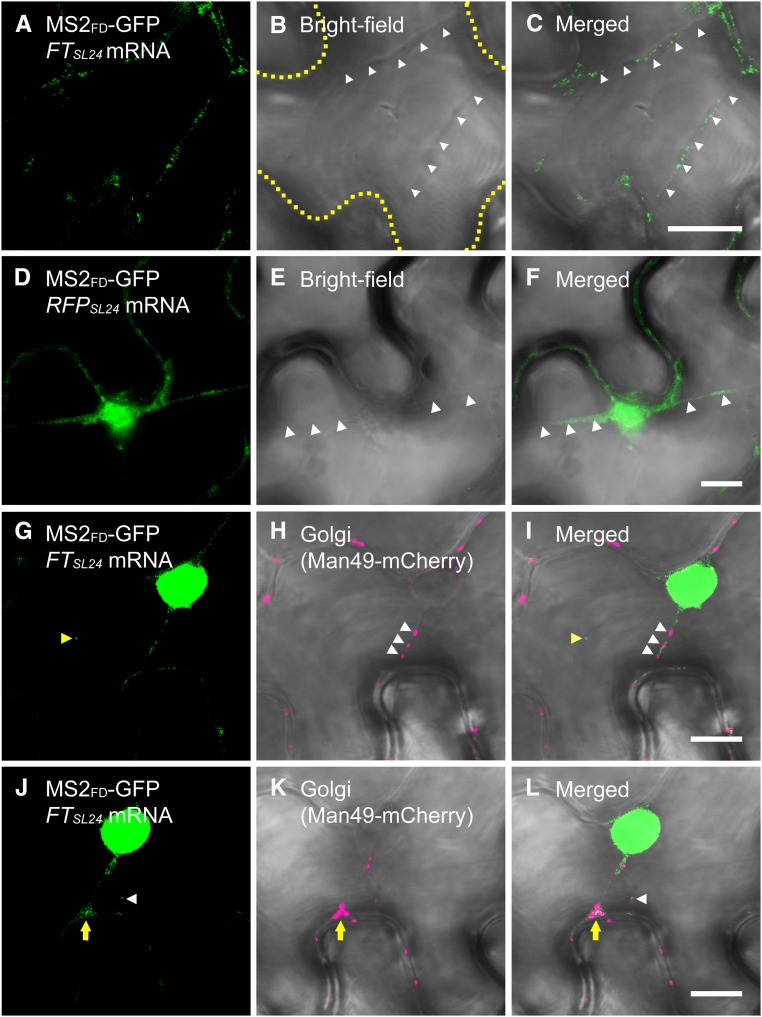
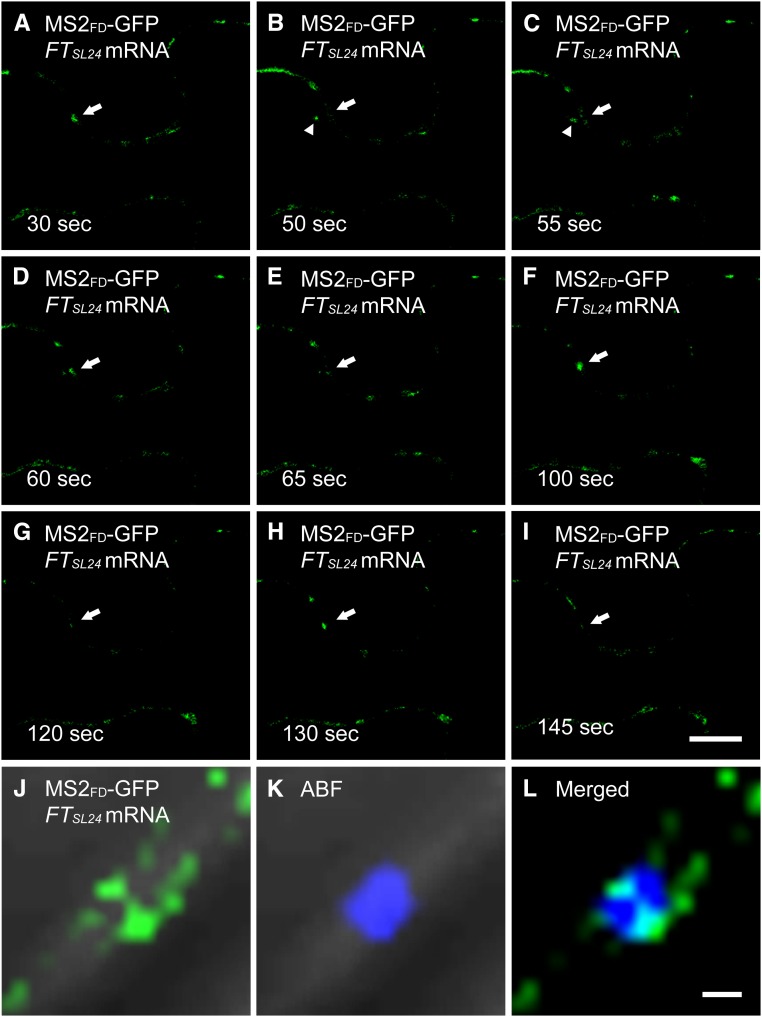
In plant cells, different parts of the cytoplasm are connected by transvacuolar strands that may serve as a route for intracellular transport of organelles and metabolites (Hoffmann and Nebenführ, 2004). To further investigate the dynamic translocation of RFPSL24 or FTSL24 mRNA to PD, we used real-time images of FTSL24 mRNA recorded at a rate of 1 frame per second. Live-cell images showed that both FTSL24 and RFPSL24 mRNA trafficked along the transvacuolar strands to the cell periphery (Fig. 5, A–F; Supplemental Movie S1). Previous results show that Golgi is transported through transvacuolar strands to the cell periphery (Nebenführ et al., 1999). Coexpression of FTSL24 mRNA with Golgi marker Man49-mCherry, the first 49 amino acids of α-1,2 mammosidase I (Saint-Jore-Dupas et al., 2006), revealed that the transport of Golgi and FTSL24 mRNA was mediated along the same transvacuolar strands (Fig. 5, G–I), which suggests that the transport of FTSL24 mRNA and Golgi in transvacuolar strands may be governed by the same mechanism. In addition, analysis of trajectories revealed the bidirectional transport of FTSL24 mRNA and Golgi in transvacuolar strands (Supplemental Movie S1). At the contact sites where transvacuolar strands connect with cell membranes, FTSL24 mRNA or Golgi temporarily accumulated before further transport along the cell periphery (Fig. 5, J–L). At the cell periphery, FTSL24 mRNA was not constantly located at the same foci; instead, the signals were intermittently detected at the same spot (Fig. 6, A–I; Supplemental Movie S2). This finding is consistent with the hypothesis that mobile mRNA is targeted to PD and then moves through the PD into neighboring cells. In contrast, RFPSL24 mRNA distributed in the cell periphery and exhibited no preference for specific subcellular targeting (Supplemental Movie S3). In agreement with this notion, in ABF-stained cells coexpressing MS2FD-GFP and FTSL24 mRNA, FTSL24 mRNA located oppositely on two distinct, parallel plasma membranes, colocalized with PD bridging these two contact cells (Fig. 6, J–L), which suggests that mobile FTSL24 RNA was selectively targeted to PD and translocated through PD for cell-to-cell movement.
Figure 5.
Dynamic translocation of RFPSL24 or FTSL24 mRNA in living plant cells. Translocation of FTSL24 mRNA (A–C) or RFPSL24 mRNA (D–F) through a cytoplasmic strand-like structure. Strands are indicated by white arrowheads. The outline of the cell is indicated by a yellow dashed line in B. C is merged from A and B, and F is merged from D and E. G to I, FTSL24 mRNA and Golgi (Man49-mCherry) associated with cytoplasmic strands. Yellow arrowheads indicate single FTSL24 mRNA particle identified in the cytosol, and white arrowheads indicate Golgi traffic through cytoplasmic strands. I is merged from G and H. J to L, Postponement of transport of FTSL24 mRNA and Golgi (Man49-mCherry) at the contact site of the cytoplasmic strand and cell membrane (indicated by yellow arrows). White arrowheads indicate single FTSL24 mRNA particle identified in the cytosol. L is merged from J and K. Bar = 10 µm.
Figure 6.
Intermittent detection of FTSL24 mRNA in PD. A to I, Time-lapse images of an N. benthamiana leaf coexpressing MS2FD-GFP and FTSL24 mRNA. The GFP signal in specific punctate spots is indicated by arrows. The trafficking of FTSL24 mRNA is indicated by arrowheads in B and C. Note that the GFP signal in specific punctate spots is only detected in A, D, F, and H. Bar = 10 µm. J to L, Localization of FTSL24 mRNA in PD stained with ABF. L is merged from J and K. Bar = 1 μm.
DISCUSSION
The visualization of real-time RNA trafficking in living cells is a powerful approach to understand the cellular distribution of specific mRNA. In the past few years, many methods for RNA visualization have been developed (Urbanek et al., 2014). Among these methods, indirect RNA labeling by genetically encoded fluorescent RBPs has allowed for visualizing mRNA in specific tissues (Keryer-Bibens et al., 2008; Tyagi, 2009; Urbanek et al., 2014). Despite the significant advances in mRNA live imaging in many species, studies in plants are relatively limited (Urbanek et al., 2014; Tilsner, 2015). One possible reason is that the inefficient nuclear targeting of the MS2SV40-GFP system used in plants usually produces significant fluorescent background in the cytosol, which greatly interferes with the detection of mRNA signals. By substituting the NLS of SV40 with Arabidopsis FD in this study, we substantially reduced the cytoplasmic fluorescent background. The high signal-to-noise ratio allowed us to detect the GFP signal in the cytosol dependent on MS2 recognition of SL sequences (Fig. 1). In addition, high-resolution Airyscan microscopy revealed individual dotted fluorescent signals gathered by TMVMP in PD-localized foci (Fig. 2L, inset). In mammalian cells, each fluorescent particle represents a single mRNA molecule labeled with multiple MS2-GFP (Fusco et al., 2003). Thus, it is likely that the dotted GFP signals we found may represent a single mRNA molecule. However, further quantitative analysis of fluorescent signals is required to verify whether these fluorescent particles represent a single mRNA.
The targeting of mRNA to subcellular compartments is an efficient mechanism to spatially locate signals and determinants for cellular differentiation (Bertrand et al., 1998; Schnorrer et al., 2000; Park et al., 2014). To asymmetrically target mRNA, the RNA zip codes (cis-acting elements on mRNA) are required to address subcellular mRNA targeting (Long et al., 1997; Bertrand et al., 1998; Chartrand et al., 1999). In plants, this intracellular mRNA targeting is extended to the intercellular level where many mRNAs can move cell to cell or long distance to exert their functions. Similarly, analysis of long-distance movement of mobile RNAs identified putative RNA transport sequences that are necessary and sufficient to target mobile mRNAs for long-distance movement (Banerjee et al., 2009; Huang and Yu. 2009; Lu et al., 2012; Zhang et al., 2016). By using an mRNA visualization system, we differentiated the subcellular distribution of mobile and nonmobile mRNAs: The intracellular targeting of nonmobile RFP mRNA displayed a cytosolic distribution, whereas mobile FT, ATC, and AGL24 mRNAs were targeted to PD (Figs. 3 and 4), which suggests that intrinsic signals on mobile mRNAs may determine their subcellular trafficking. In plants, PD are important intercellular cytoplasmic connecting channels for exchange of various macro- and micromolecules (Turnbull and Lopez-Cobollo, 2013). The selective targeting of mobile mRNA to PD (Figs. 3 and 4) further supports that the trafficking of mobile mRNA is triggered by specific RNA sequences rather than transcript abundance (Huang and Yu, 2009; Zhang et al., 2016). The differential targeting of mobile and nonmobile mRNAs may be a determining step for RNA movement. However, the long-distance movement of phloem-mediated mobile mRNAs involves multiple steps, including (1) intracellular targeting of mobile mRNAs to PD, (2) cell-to-cell movement of mobile mRNAs from companion cells to sieve elements, (3) translocation of mobile mRNAs in sieve elements, and (4) unloading to and cell-to-cell movement of mobile mRNAs in the sink tissues. Whether multiple RNA transport sequences are required for successful long-distance trafficking remains to be investigated.
The selective PD targeting of mobile but not nonmobile mRNAs (Fig. 3) implies the sequences or RNA structure on mobile mRNA may determine subcellular localization of mRNA. In yeast ASH1 mRNA, sequences required for mRNA asymmetric distribution have been located in the 3′ untranslated region and coding sequence (Long et al., 1997; Bertrand et al., 1998; Chartrand et al., 1999). Computational prediction and x-ray crystallography revealed that the sequences involved in ASH1 mRNA localization form secondary and tertiary structures (Chartrand et al., 1999; Edelmann et al., 2017). Whether similar RNA structures determine PD targeting of mobile mRNAs remains to be investigated.
One of the drawbacks of the MS2-dependent RNA visualization system is the requirement for multiple repeats of an MS2-recognized SL structure on target mRNA. The insertion of a large fragment of the MS2 recognition SL sequences (24 repeats of SL sequences is 870 bp) on target mRNA may interfere in the biological function of target mRNA. Our real-time RT-qPCR and confocal microscopy analyses showed that the insertion of SL sequences on target mRNA did not greatly interfere in the transcription and translation of RFPSL24 mRNA (Supplemental Fig. S1; Fig. 1G) or long-distance movement of FTSL24 RNA (Supplemental Fig. S3). However, we cannot rule out that the insertion of SL sequences on other mRNAs may disrupt their function. In our experiments, a minimum of 24 repeats of SL sequences is required for detecting the fluorescent signal in living plant cells, which agrees with results in mammalian cells (Fusco et al., 2003). Given that the intensity of the fluorescent signal is proportional to the number of GFP copies, the number of SL repeats can be reduced by fusing multiple GFP copies in one MS2 protein.
Virus MPs are RNA-binding proteins required for intercellular movement and systemic infection of viruses (Citovsky et al., 1990). Our coexpression of TMVMP with RFP RNA showed that TMVMP can target RFP RNA to PD (Fig. 2), whereas expression of FT RNA alone was sufficient for PD localization (Fig. 4, A–D), which suggests that plant endogenous RBPs may target mobile mRNAs to PD. These RBPs may recognize a specific structure on mobile mRNAs (e.g. the tRNA-like structures) to direct the movement of mobile mRNAs (Zhang et al., 2016). The different structure motifs on mobile mRNAs may recruit specific RBPs at particular stages, or reciprocally, the enrolled RBPs may induce a conformational change in mRNA structure for further transport (Edelmann et al., 2017). The crucial roles of RBPs in the subcellular localization of mobile mRNAs may be mediated by bridging mobile mRNAs to endomembrane or cytoskeleton systems or be associated with motor proteins (Takizawa et al., 1997; Schnorrer et al., 2000; Pohlmann et al., 2015). Indeed, analysis of RBPs in phloem exudates identified multiple putative systemic RBPs involved in phloem-mediated mRNA transport (Ham et al., 2009; Li et al., 2011). Whether these systemic RBPs are involved in PD targeting of mobile mRNAs remains to be investigated.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Nicotiana benthamiana plants were grown under a 16-h/8-h day/night cycle, under white fluorescent light, at an intensity of 100 μmol m−2 s−1; temperature and humidity of the growth chamber were set to 27°C and 60%, respectively.
The Arabidopsis thaliana ft-10 mutant was transformed by the floral-dip method (Clough and Bent, 1998). The transformants were selected on Murashige and Skoog medium with 40 µg mL−1 hygromycin.
Plasmid Construction and Organelle Markers
The online prediction tool SeqNLS (http://mleg.cse.sc.edu/seqNLS/; Lin and Hu, 2013) was used for in silico prediction of the FD (At4G35900) protein NLS.
For constructing MS2FD-GFP, the C-terminal fragment of Arabidopsis FD (211–240 amino acids) that contains a predicted NLS, was PCR amplified. The SV40 NLS in pG14-MS2-GFP (Addgene plasmid #27117) was replaced with the PCR-amplified FD fragment and subcloned into the pCAMBIA1390 vector to give p1390-35S-MS2FD-GFP.
For constructing 35Spro:FTSL24, 35Spro:ATCSL24, 35Spro:AGL24SL24, or 35Spro:RFPSL24, 24 copies of a tandem repeat SL sequence (SL24) were generated and inserted into the 5′ end of FT, ATC, AGL24, or RFP coding sequences. The resulting constructs were driven by a CaMV35S promoter.
For Arabidopsis grafting experiments, inflorescence grafting was used to examine the long-distance movement of FTSL24 mRNA (Huang and Yu, 2009).
Transient Expression and ABF Treatment in N. benthamiana Leaves
Before transient expression, Agrobacterium tumefaciens strain AGL1 carrying individual constructs was cultured in media containing 50 µg mL−1 kanamycin, 10 mm MES, pH 5.7, and 20 µm acetosyringone at 28°C overnight. Subsequently, A. tumefaciens cells were pelleted and resuspended in infiltration solution (10 mm MgCl2, 10 mm MES, pH 5.7, and 200 µm acetosyringone) to OD600 1.0 and left at room temperature for 1 h. Coinfiltration was conducted with bacteria solutions prepared as equivalent volume mixtures infiltrated into the underside of 3-week-old N. benthamiana leaves by using a syringe (with needle removed).
ABF (Biosupplies) was stocked as a 0.1-mg/mL solution in distilled water and stored at 4°C. Before detecting callose deposits, the stock solution was diluted to 1:3 with infiltration buffer and infiltrated into N. benthamiana leaves.
Confocal Laser Scanning Microscopy, Airyscan High-Resolution Imaging, and Colocalization Assays
At 1 or 2 dpi with A. tumefaciens or 10 min after ABF infiltration in N. benthamiana leaves, an ∼0.5-cm2 sample was removed from infiltrated leaves or the leading front region of infiltrated leaves, covered with glass slides, and observed under the confocal laser scanning microscope (LSM880; Carl Zeiss). The settings for excitation laser/detection filters (in nm) were GFP, argon 488/band-pass 510 to 550; RFP, DP SS561/band-pass 590 to 650; and ABF, diode 405/band-pass 430 to 500. High-resolution images were acquired with the Airyscan module mounted on the LSM880 system. Colocalization was analyzed with the Profile assay included in the ZEN software accompanying the LSM880.
RNA Extraction, RT-qPCR, and RT-PCR Analysis
To validate the transient expression of RFPSL24 mRNA (Supplemental Fig. S1), total RNA from A. tumefaciens-infiltrated N. benthamiana leaves was extracted by the Trizol reagent method (Invitrogen). After DNase I (Invitrogen) treatment, 5 μg total RNA was used to synthesize first-strand cDNA with Superscript III reverse transcriptase (Invitrogen) with a reaction volume of 20 μL. The volume was then brought up to 500 μL, and an aliquot of 5 μL was used for real-time PCR. For each reaction, 200 nm gene-specific primers were used with the AB 9500 Real-Time PCR System (Applied Biosystems). PCR parameters were 95°C for 10 min, followed by 40 cycles of two steps (95°C for 15 s and 60°C for 1 min). RT-qPCR was conducted with two independent experimental replicates, and triplicate reactions were conducted for each sample. The expression of ACTIN was used as a normalization control. One representative result is shown in Supplemental Figure S1. The primers used in RT-PCR and RT-qPCR analysis are in Supplemental Table S1.
To examine long-distance movement of FTSL24 mRNA in grafting experiments, 5 μg total RNA was used to synthesize the first-strand cDNA with oligo(dT)20 and Superscript III reverse transcriptase (Invitrogen). To detect the FTSL24 transgene, FT-For and NOSter-Rev were used as primers in the PCR and 2 μL first-strand cDNA served as a template. PCR conditions were 94°C for 1 min; 35 cycles for three steps (94°C for 30 s, 60°C for 30 s, and 68°C for 1 min); and 68°C for 7 min. An aliquot of 5 μL PCR products was separated on 0.8% agarose gels to visualize the amplified DNA fragments.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_119756 (FD), AB027504 (FT), AB024715 (ATC), AF005158 (AGL24).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Primers used in RT-PCR and RT-qPCR analysis.
Supplemental Figure S1. RT-qPCR analysis of N. benthamiana leaves expressing RFPSL24 or RFP mRNA.
Supplemental Figure S2. Colocalization spectroscopy of RFPSL24 mRNA and RFP-TMVMP.
Supplemental Figure S3. FTSL24 mRNA remains a mobile mRNA.
Supplemental Figure S4. Subcellular distribution pattern of labeled mRNAs.
Supplemental Figure S5. Intracellular distribution of RFPSL24 and FTSL24 mRNA in cobombardment experiments.
Supplemental Figure S6. Colocalization of mRNA and the plasma membrane marker FM4-64.
Supplemental Movie 1. Bidirectional transport of FTSL24 mRNA and Golgi in transvacuolar strands of N. benthamiana leaves.
Supplemental Movie 2. Intermittent detection of FTSL24 mRNA at the same punctate foci.
Supplemental Movie 3. Transport of RFPSL24 mRNA and Golgi in N. benthamiana cells.
Acknowledgments
We thank Miss Y.S. Chang for assistance in Arabidopsis grafting, Dr. N.S. Lin for providing TMVMP-RFP and TMVMP-GFP, and Dr. R.H. Singer for providing MS2SV40-GFP clones. We thank Dr. Marjori Matzke for critical reading of the manuscript.
Footnotes
Articles can be viewed without a subscription.
This work was supported by grants from Academia Sinica, Taiwan (AS-105-TP-B03).
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Banerjee AK, Chatterjee M, Yu Y, Suh S-G, Miller WA, Hannapel DJ (2006) Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18: 3443–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Lin T, Hannapel DJ (2009) Untranslated regions of a mobile transcript mediate RNA metabolism. Plant Physiol 151: 1831–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM (1998) Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445 [DOI] [PubMed] [Google Scholar]
- Buxbaum AR, Haimovich G, Singer RH (2015) In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol 16: 95–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood A, Kopriva S, Morris RJ (2016) Transcript abundance explains mRNA mobility data in Arabidopsis thaliana. Plant Cell 28: 610–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P, Meng XH, Singer RH, Long RM (1999) Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol 9: 333–336 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Knorr D, Schuster G, Zambryski P (1990) The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60: 637–647 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Edelmann FT, Schlundt A, Heym RG, Jenner A, Niedner-Boblenz A, Syed MI, Paillart JC, Stehle R, Janowski R, Sattler M, Jansen RP, Niessing D (2017) Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA-transport complex. Nat Struct Mol Biol 24: 152–161 [DOI] [PubMed] [Google Scholar]
- Forrest KM, Gavis ER (2003) Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol 13: 1159–1168 [DOI] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E (2003) Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol 13: 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Zhang J, Sun H, Salse J, Lucas WJ, Zhang H, Zheng Y, Mao L, Ren Y, Wang Z, et al. (2013) The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet 45: 51–58 [DOI] [PubMed] [Google Scholar]
- Ham BK, Brandom JL, Xoconostle-Cázares B, Ringgold V, Lough TJ, Lucas WJ (2009) A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21: 197–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Ishiyama K, Choi S-B, Wang C, Singh S, Kawai N, Franceschi VR, Okita TW (2003) The transport of prolamine RNAs to prolamine protein bodies in living rice endosperm cells. Plant Cell 15: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Nebenführ A (2004) Dynamic rearrangements of transvacuolar strands in BY-2 cells imply a role of myosin in remodeling the plant actin cytoskeleton. Protoplasma 224: 201–210 [DOI] [PubMed] [Google Scholar]
- Huang N-C, Yu T-S (2009) The sequences of Arabidopsis GA-INSENSITIVE RNA constitute the motifs that are necessary and sufficient for RNA long-distance trafficking. Plant J 59: 921–929 [DOI] [PubMed] [Google Scholar]
- Huang N-C, Jane W-N, Chen J, Yu T-S (2012) Arabidopsis thaliana CENTRORADIALIS homologue (ATC) acts systemically to inhibit floral initiation in Arabidopsis. Plant J 72: 175–184 [DOI] [PubMed] [Google Scholar]
- Keryer-Bibens C, Barreau C, Osborne HB (2008) Tethering of proteins to RNAs by bacteriophage proteins. Biol Cell 100: 125–138 [DOI] [PubMed] [Google Scholar]
- Köhler RH. (1998) GFP for in vivo imaging of subcellular structures in plant cells. Trends Plant Sci 3: 317–320 [Google Scholar]
- Li P, Ham BK, Lucas WJ (2011) CmRBP50 protein phosphorylation is essential for assembly of a stable phloem-mobile high-affinity ribonucleoprotein complex. J Biol Chem 286: 23142–23149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Hu J (2013) SeqNLS: nuclear localization signal prediction based on frequent pattern mining and linear motif scoring. PLoS One 8: e76864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP (1997) Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277: 383–387 [DOI] [PubMed] [Google Scholar]
- Lough TJ, Lucas WJ (2006) Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu Rev Plant Biol 57: 203–232 [DOI] [PubMed] [Google Scholar]
- Lu K-J, Huang N-C, Liu Y-S, Lu C-A, Yu T-S (2012) Long-distance movement of Arabidopsis FLOWERING LOCUS T RNA participates in systemic floral regulation. RNA Biol 9: 653–662 [DOI] [PubMed] [Google Scholar]
- Nebenführ A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin LA (1999) Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol 121: 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HY, Lim H, Yoon YJ, Follenzi A, Nwokafor C, Lopez-Jones M, Meng X, Singer RH (2014) Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science 343: 422–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmann T, Baumann S, Haag C, Albrecht M, Feldbrügge M (2015) A FYVE zinc finger domain protein specifically links mRNA transport to endosome trafficking. eLife 4: 06041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook MS, Lu M, Kosik KS (2000) CaMKIIalpha 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci 20: 6385–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambade A, Brandner K, Hofmann C, Seemanpillai M, Mutterer J, Heinlein M (2008) Transport of TMV movement protein particles associated with the targeting of RNA to plasmodesmata. Traffic 9: 2073–2088 [DOI] [PubMed] [Google Scholar]
- Schnorrer F, Bohmann K, Nüsslein-Volhard C (2000) The molecular motor dynein is involved in targeting swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat Cell Biol 2: 185–190 [DOI] [PubMed] [Google Scholar]
- Saint-Jore-Dupas C, Nebenführ A, Boulaflous A, Follet-Gueye M-L, Plasson C, Hawes C, Driouich A, Faye L, Gomord V (2006) Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18: 3182–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD (1997) Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 389: 90–93 [DOI] [PubMed] [Google Scholar]
- Thieme CJ, Rojas-Triana M, Stecyk E, Schudoma C, Zhang W, Yang L, Miñambres M, Walther D, Schulze WX, Paz-Ares J, Scheible W-R, Kragler F (2015) Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat Plants 1: 15025. [DOI] [PubMed] [Google Scholar]
- Tilsner J. (2015) Techniques for RNA in vivo imaging in plants. J Microsc 258: 1–5 [DOI] [PubMed] [Google Scholar]
- Tyagi S. (2009) Imaging intracellular RNA distribution and dynamics in living cells. Nat Methods 6: 331–338 [DOI] [PubMed] [Google Scholar]
- Turnbull CG, Lopez-Cobollo RM (2013) Heavy traffic in the fast lane: long-distance signalling by macromolecules. New Phytol 198: 33–51 [DOI] [PubMed] [Google Scholar]
- Urbanek MO, Galka-Marciniak P, Olejniczak M, Krzyzosiak WJ (2014) RNA imaging in living cells - methods and applications. RNA Biol 11: 1083–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatén A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, et al. (2011) Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 21: 1144–1155 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Wu B, Chao JA, Singer RH (2012) Fluorescence fluctuation spectroscopy enables quantitative imaging of single mRNAs in living cells. Biophys J 102: 2936–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H-W, Yu T-S (2010) Arabidopsis floral regulators FVE and AGL24 are phloem-mobile RNAs. Bot Stu 51: 17–26 [Google Scholar]
- Yang Y, Mao L, Jittayasothorn Y, Kang Y, Jiao C, Fei Z, Zhong GY (2015) Messenger RNA exchange between scions and rootstocks in grafted grapevines. BMC Plant Biol 15: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH (2005) CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol 139: 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Thieme CJ, Kollwig G, Apelt F, Yang L, Winter N, Andresen N, Walther D, Kragler F (2016) tRNA-related sequences trigger systemic mRNA transport in plants. Plant Cell 28: 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]