Messenger RNA is degraded as it moves through the tomato phloem, and the mobility of a transcript cannot be reliably predicted based on its abundance in the Nicotiana benthamiana leaf or on whether it harbors a tRNA-like structural motif.
Abstract
Recent heterograft analyses showed that large-scale messenger RNA (mRNA) movement takes place in the phloem, but the number of mobile transcripts reported varies widely. However, our knowledge of the mechanisms underlying large-scale mRNA movement remains limited. In this study, using a Nicotiana benthamiana/tomato (Solanum lycopersicum) heterograft system and a transgenic approach involving potato (Solanum tuberosum), we found that: (1) the overall mRNA abundance in the leaf is not a good indicator of transcript mobility to the root; (2) increasing the expression levels of nonmobile mRNAs in the companion cells does not promote their mobility; (3) mobile mRNAs undergo degradation during their movement; and (4) some mRNAs arriving in roots move back to shoots. These results indicate that mRNA movement has both regulated and unregulated components. The cellular origins of mobile mRNAs may differ between herbaceous and woody species. Taken together, these findings suggest that the long-distance movement of mRNAs is a complex process and that elucidating the physiological roles associated with this movement is challenging but remains an important task for future research.
Higher plants have evolved a communication system that enables the coordination of developmental and environmental cues between different organs (Spiegelman et al., 2013; Turnbull and Lopez-Cobollo, 2013). This communication is achieved by long-distance signaling that takes place in the vasculature tissue (Banerjee et al., 2006). Phloem, one of the major components in the vasculature system, has long been recognized as a tissue that transports carbohydrates and amino acids. In recent years, it has been found that this tissue also is a conduit for signals (e.g. mRNAs, small RNAs, proteins, small peptides, and hormones; Turgeon and Wolf, 2009).
Although mRNAs have been traditionally viewed as local intermediate components between the genomic DNA and the protein in a cell, a handful of classical studies have shown that some mRNAs are able to traffic from source to sink tissues via the phloem and exert important physiological functions in the distal recipient organs. For example, the development of sink tissues, such as young leaves (Kim et al., 2001), tubers (Banerjee et al., 2006), and roots (Notaguchi et al., 2012), is partially controlled by the long-distance mobile mRNAs generated in the source tissues.
A recent study on the interaction between the parasitic Cuscuta pentagona and its host indicated that a large amount of bidirectional mRNA exchange occurred via the vasculature between the two species (Kim et al., 2014). In recent years, a few heterograft systems, in which one species/genotype was used as a scion and another as a rootstock, were used to demonstrate that large-scale migration of mRNAs occurs from leaf to root, or vice versa. Notaguchi et al. (2015) determined that 138 transcripts moved from the rootstock of Arabidopsis (Arabidopsis thaliana) to the Nicotiana benthamiana scion via the phloem. Using two different ecotypes of Arabidopsis, Thieme et al. (2015) determined that 2,006 mRNAs could move from the shoot to the root, or vice versa, under either normal or mineral-deficient conditions. In a grapevine (Vitis vinifera) heterograft system, 3,333 mRNAs were found to be transmissible (Yang et al., 2015). In another heterograft system, an analysis of the response to early phosphate deficiency revealed that 3,546 mRNAs moved from cucumber (Cucumis sativus) source leaves to watermelon (Citrullus lanatus) sink tissues via the phloem (Zhang et al., 2016b).
This variability, with over 1 order of magnitude difference in the number of mRNAs transported, suggests a great deal of species specificity and that not all the mobile mRNAs are functional. Zhang et al., 2016a, discovered that, among the large number of mobile mRNAs identified in the Arabidopsis heterografts, 11.4% of them harbored a tRNA-like structure (TLS). Further experimental evidence supported the hypothesis that not only does this motif impart stability and mobility to mRNAs but also that mRNAs with this structure can be translated into proteins after transport (Zhang et al., 2016a). However, how this mechanism can be applied to other plant heterograft systems remains unknown. A computational analysis conducted by Calderwood et al. (2016) on the same set of mobile mRNAs suggested that most of the identified mRNA species are mobile as a consequence of local abundance in companion cells. A similar correlation between abundance and long-distance mobility has been noted by other researchers as well (Kim et al., 2014; Yang et al., 2015). The extent to which this correlation, drawn from either mathematical modeling methods or computational analysis, applies to natural plant systems needs further experimental verification.
In this study, we developed a heterograft system using N. benthamiana and tomato (Solanum lycopersicum) to investigate shoot-to-root mRNA movement. Compared with most published heterograft systems, the phylogenetic distance between these two species is relatively large, which allowed for more accurate distinction of scion mRNAs from those in the rootstock. In addition, the ease of transformation of the two partners is advantageous in future molecular and physiological studies. We used this system to test if: (1) the abundance of mRNAs in the leaf is a determining factor for mRNA mobility; (2) the movement of mRNAs is a regulated or unregulated process; (3) mobile mRNAs undergo degradation during their movement from shoots to roots; and (4) shoot-to-root-to-shoot cycling movement occurs. In addition, a transgenic approach involving potato (Solanum tuberosum) was used to test the relationship between the abundance of companion cell transcripts and their shoot-to-root mobility.
RESULTS
mRNA Migration from Shoot to Root
A heterograft system, in which N. benthamiana was used as the scion and tomato as the rootstock, was established to identify the shoot-to-root long-distance movement of mRNAs through the phloem (Fig. 1A). We compared the morphology of leaves and roots in both the heterografts and the nongrafted counterparts. No visible phenotypic differences were observed, indicating that the heterografts had largely maintained the endogenous physiology of the nongrafted plants (data not shown).
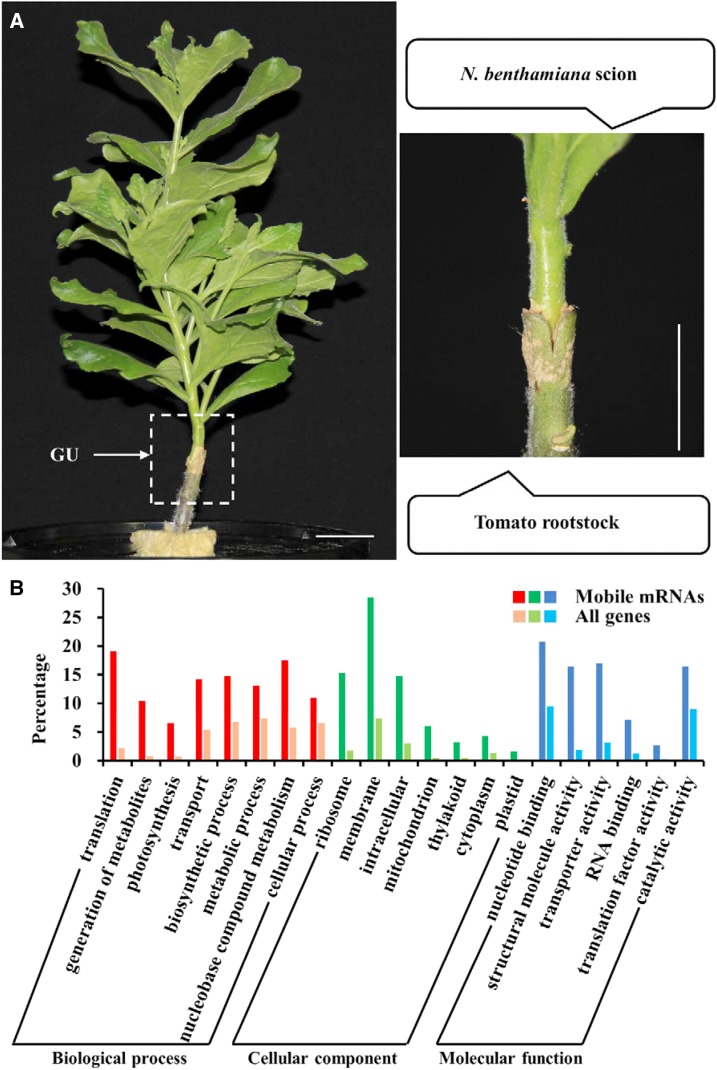
Figure 1.
Shoot-to-root mobile mRNAs identified from the N. benthamiana/tomato heterografts grown in a hydroponic condition. A, Representative heterograft established between N. benthamiana and tomato. The white box indicates the graft union (GU). The image at right shows an enlargement of the region boxed at left. Bars = 2 cm. B, GO terms enriched in the 183 mobile mRNAs. Columns indicate the percentages of genes that were enriched in the GO terms. All genes refers to the whole-genome background.
A total of 183 N. benthamiana mRNAs were determined to move into the tomato root in heterografts grown in a hydroponic system (Supplemental Table S1). Gene Ontology (GO) analysis of these mobile mRNAs indicated that certain biological processes, such as translation (19.1%), transport (14.2%), and biosynthetic process (14.8%), were overrepresented. Within the cellular component category, ribosome (15.3%), membrane (28.4%), and intracellular (14.8%) were overrepresented. Moreover, nucleotide binding (20.8%), transporter activity (16.9%), and RNA binding (7.1%) were overrepresented in the molecular function category (Fig. 1B; Supplemental Table S2).
Orthologs of some of the mRNAs identified from this work also were reported in previous studies in which traditional phloem collection methods were used (Ivashikina et al., 2003; Doering-Saad et al., 2006; Omid et al., 2007; Deeken et al., 2008; Kanehira et al., 2010). For example, IAA1, the mRNA encoding an auxin-responsive protein, was identified as mobile in our system. A previous analysis discovered a member of the Aux/IAA family encoding a transcriptional regulator in auxin signaling in the phloem sap of melon (Cucumis melo; Omid et al., 2007). IAA18 and IAA28, two additional auxin signaling-related transcripts, were found to be expressed in the vasculature tissue of mature Arabidopsis leaves and transported to the root tip to regulate lateral root formation (Notaguchi et al., 2012). In addition, a number of mRNAs encoding essential translation machinery components (e.g. ribosomal protein, elongation factor, and eukaryotic translation initiation factor) were found to be mobile. Previous proteome studies have discovered that many proteins related to translation exist in the phloem (Lin et al., 2009; Ma et al., 2010; Ham and Lucas, 2014) even though the translational machinery is absent in sieve elements.
The Abundance of mRNA Is Not Associated with Mobility
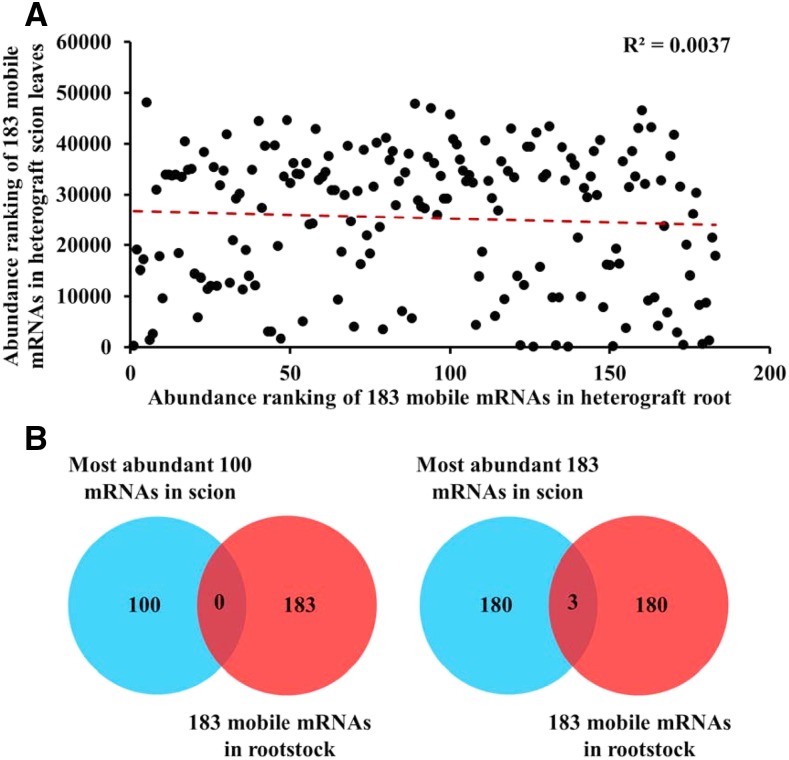
To determine whether more abundant leaf mRNAs are prone to move, we collected mature leaves from the N. benthamiana scions in the N. benthamiana/tomato heterograft and sequenced the mRNAs. The abundances of the transcripts in leaves corresponding to the 183 mobile mRNAs were compared with the abundances of the same 183 mRNAs in the heterograft roots. No correlation (R2 = 0.0037) was found between the shoot-to-root mobility of the mRNAs and their abundances in the leaves, the origin of these mobile mRNAs (Fig. 2A; Supplemental Table S3). Further detailed analysis showed that none of the 100 most abundant leaf mRNAs moved from shoots to roots; among the 183 mobile mRNAs, only three transcripts were ranked among the 183 most abundant leaf mRNAs (Fig. 2B). On the contrary, some mRNAs with low abundance in the leaf were able to move to the rootstock. The abundances of approximately half (91 out of 183) of the mobile mRNAs were ranked lower than 30,000th among the entire leaf transcriptome. For example, the abundance of Niben101Scf00291g02025.1, an mRNA ranked the eighth most mobile, was ranked 30,982th in abundance in the leaf; Niben101Scf05425g00016.1 was ranked 16th in the mobile list but was 33,512th most abundant in the leaf transcriptome. Although most of the shoot-to-root mobile mRNAs had higher abundances in the scion than in the rootstock, it is noteworthy that the abundances of 27 mobile mRNAs were higher in the rootstock than in the leaf. For example, Niben101Scf01796g01005.1, an mRNA that was ranked 17th most mobile, had a reads per kilobase of exon per million fragments mapped value of 1.14 in the rootstock but only 0.2 in the leaf (the 40,445th most abundant in the leaf transcriptome; Supplemental Table S4).
Figure 2.
Correlation analysis of the abundances of leaf mRNAs and their mobility. Expression levels of the graft-transmissible mRNAs in leaf and root tissues were ranked by mean reads per kilobase of exon per million fragments mapped values derived from heterografts (n = 3) grown with full-strength nutrients. A, Correlation of abundances for the 183 mobile mRNAs in the scion and rootstock. The red dashed line is the trend line. B, Venn diagram showing the number of mobile mRNAs among the most abundant 100 or 183 mRNAs in the scion. Blue represents the number of the most abundant mRNAs in the N. benthamiana scion, and red represents the number of the mobile mRNAs in the tomato rootstock.
Overexpressing Phloem-Immobile mRNAs in the Companion Cells Does Not Promote Mobility
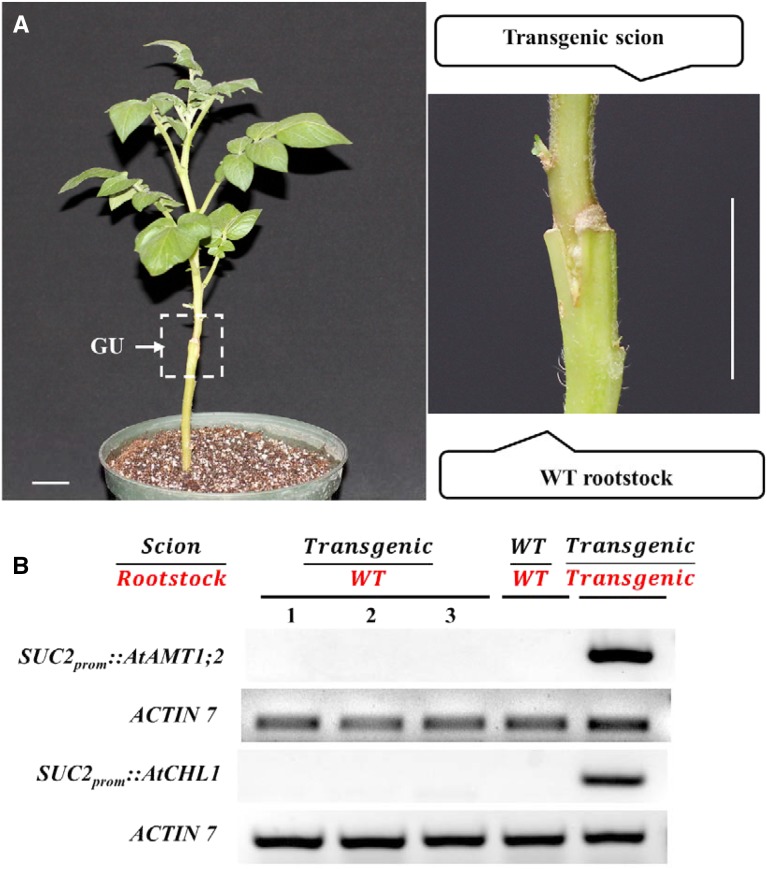
The lack of correlation between the mobility of mRNAs and their abundances in the leaf indicated that using whole leaf tissues is not informative enough to predict the mobility of mRNAs. One possibility is that only cells directly connected to the sieve tubes act as sources of mobile mRNAs. In apoplastic loading species, such as N. benthamiana, the phloem is isolated from surrounding cells, and it is generally believed that the components moving in the sieve tubes originate in the associated companion cells (King and Zeevaart, 1974; Stadler et al., 2005). Using a simple computational model, Calderwood et al. (2016) analyzed a series of mobile RNA data sets associated with Arabidopsis and postulated that mRNAs with higher abundance in the companion cells are more prone to move (Mustroph et al., 2009; Calderwood et al., 2016). To test this hypothesis, we investigated whether increasing the abundance of a phloem-immobile mRNA will make it mobile. We overexpressed the Arabidopsis dual-affinity nitrate transporter gene (AtCHL1) and the Arabidopsis ammonium transporter gene (AtAMT1;2) in the phloem of potato driven by the strong companion cell-specific sucrose transporter2 (SUC2) promoter. Neither of the homologous mRNA transcripts of these two genes were detected to be mobile in our heterograft system. In addition, the phloem immobility of homologs of these two transcripts in previously published heterograft systems was further confirmed by exploring the PlaMoM database (Guan et al., 2017). Previous analyses had shown that the size of the shoot-to-root mobile mRNAs can be as large as 4 kb (Notaguchi et al., 2015; Thieme et al., 2015; Zhang W et al., 2016). Therefore, the length (i.e. 1,545 and 1,773 bp, respectively) of AtAMT1;2 and AtCHL1 should not act as a negative barrier to the movement of the individual mRNAs from the companion cell to the sieve tube. Multiple transgenic lines were generated, and the transgenic potato plant with the highest expression of either AtCHL1 or AtAMT1;2 in the companion cells was used as the scion that was grafted onto the wild-type potato rootstock (Fig. 3A; Supplemental Fig. S1). The lack of amplification via PCR of these genes in the rootstock of the heterografts indicated that the increased abundances of these phloem-immobile mRNAs transcribed in the companion cells did not promote shoot-to-root movement (Fig. 3B).
Figure 3.
Mobility of nonmobile mRNAs overexpressed in transgenic potato phloem companion cells. A, Representative SUC2prom::AtCHL1 transgenic potato/wild-type (WT) potato heterograft. The white box indicates the graft union (GU). The image at right shows an enlargement of the region boxed at left. Bars = 2 cm. B, Reverse transcription (RT)-PCR analysis of AtAMT1;2 and AtCHL1 in transgenic potato grafts. Both AtAMT1;2 and AtCHL1 are phloem-immobile mRNAs. Transgenic potato plants with overexpressed AtAMT1;2 or AtCHL1 were grafted onto the wild type, and the expression of these genes in the rootstock was compared with that in the homografts in which the scion and the rootstock were the same genotype (wild type or transgenic). For the transgenic/wild-type grafts, three shoots excised from the same transgenic potato plant with either overexpressed AtAMT1;2 or AtCHL1 were grafted on the tomato rootstock.
Mobile mRNAs Differentially Accumulate along the Movement Path
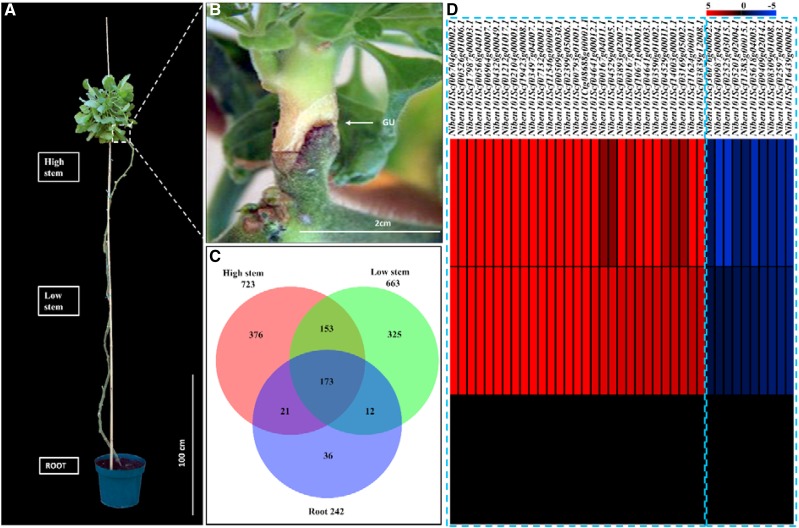
In addition to the cell specificity that might confer the mobility of an mRNA, transport from shoot to root could depend on other factors, such as sequence structure and half-life (Calderwood et al., 2016; Zhang et al., 2016a). In the Arabidopsis/C. pentagona system, three host-born mRNAs were chosen as representatives to investigate the fate of mobile mRNAs once entering the parasitic plant. It was found that the accumulation patterns (i.e. increased or decreased abundance) of these mobile mRNAs differed during their transport path (LeBlanc et al., 2013). To understand whether mobile mRNAs accumulate differently once passing the graft joint on a more global scale, N. benthamiana scions were grafted onto the tops of 2.5-m-tall tomato plants (Fig. 4, A and B). RNAs were extracted from three locations, high stem (10–20 cm below the grafting joint), low stem (120–130 cm below the grafting joint), and root, and sequenced. The analysis revealed that 1,096 mRNAs from the N. benthamiana scion passed the grafting joint and moved downward to the tomato rootstock. However, 78% (854 transcripts) of the mobile mRNAs did not arrive in the root. The total number of mobile mRNAs decreased from the high stem (723) to the low stem (663) and the root (242; Fig. 4C; Supplemental Tables S5–S7). Although the abundance of most mRNAs decreased during the shoot-to-root transport, 10 mRNAs showed higher abundance in the root than in the high stem and low stem (Fig. 4D; Table I; Supplemental Table S8). It is noteworthy that two of these root-accumulated mobile mRNAs were annotated as casein kinase I, which has been reported to be involved in plant hormonal metabolism and the establishment of root architecture (Liu et al., 2003). Another interesting observation is that some of the mobile mRNAs identified in low stem or root were not detected in tissues located above it. For example, 325 mRNAs detected in the low stem were not detectable in the high stem site; similarly, 36 mRNAs detected in root were not detectable in either location in the stem. A similar phenomenon was observed in the Arabidopsis heterograft, where some of the rootstock mRNAs identified in the scion flowers were not detectable in the inflorescence stem, a tissue mediating the mRNA transport (Thieme et al., 2015). One possible explanation for this is that the abundances of these mobile mRNAs are so low that they cannot be readily detected in those tissues using RNA sequencing (RNA-seq) analysis.
Figure 4.
Differentially accumulated mobile mRNAs along the movement path in a long-stem heterograft. A, Representative long-stem heterograft. An actively growing N. benthamiana scion was grafted on the top of a 2.5-m-tall tomato rootstock. B, Magnification of the dashed box in A. GU, Graft union. C, Venn diagram indicating the number of mobile mRNAs identified at different locations on the tomato rootstock. D, Heat map showing the abundance profiles of the 39 differentially accumulated mobile mRNAs. Colors indicate different log2 (fold change) values, which are related to the mRNA abundance in the root (0 in the heat map). The left dashed box indicates the 29 mRNAs decreased in abundance during the transport, and the right dashed box indicates the 10 mobile mRNAs overaccumulated in the heterograft root.
Table I. Top 10 most highly accumulated mobile mRNAs in the roots of long-stem heterografts.
| mRNA Accession | Annotation |
|---|---|
| Niben101Scf36076g00002.1 | Elongation factor 1-α1 |
| Niben101Scf00987g00004.1 | Casein kinase I |
| Niben101Scf02525g03015.1 | Casein kinase I |
| Niben101Scf05201g02004.1 | Magnesium transporter NIPA2 |
| Niben101Scf11383g00015.1 | 40S ribosomal protein S24-2 |
| Niben101Scf05618g04003.1 | tRNA [guanine(26)-N(2)]-dimethyltransferase |
| Niben101Scf09409g02014.1 | Choline/ethanolaminephosphotransferase1 |
| Niben101Scf08309g01008.1 | Homogentisate 1,2-dioxygenase |
| Niben101Scf02597g00003.1 | 30S ribosomal protein S8 |
| Niben101Scf16439g00004.1 | 30S ribosomal protein S8 |
mRNAs Move from Scion to Rootstock to Scion
In addition to the shoot-to-root movement of mRNAs (Thieme et al., 2015; Yang et al., 2015; Zhang et al., 2016b), Arabidopsis and grapevine heterograft systems indicate that mRNAs transcribed in roots also can move from roots to shoots (Thieme et al., 2015; Yang et al., 2015). In these previous studies, two separate groups of mRNAs, originating either from leaves or roots, were demonstrated to be mobile. We asked whether mRNAs in the rootstock that had arrived from the scion can be transported back to the shoot. In contrast to previous analyses, our interest was only focused on the same set of mRNAs that are transcribed in the leaf. To test this, we developed a system in which potato and N. benthamiana were individually grafted onto two separate stems emerging from the same tomato rootstock (Fig. 5A). The potato scion was first grafted onto one of the two stems on the tomato rootstock (Fig. 5B). Once the first heterograft was established and the potato scion was actively growing, the young sink leaves on the potato shoot were removed and an N. benthamiana scion was grafted onto the second stem of the tomato rootstock (Fig. 5C). Since only mature source leaves of the potato scion remained, N. benthamiana mRNAs identified in the potato leaves would have to be imported either via the phloem, against source-to-sink bulk flow, or via the nonvasculature cell-to-cell-based rootstock-to-scion transport approach mediated by plasmodesmata (Lucas et al., 1995). A total of 58 N. benthamiana mobile mRNAs were detected in the mature leaves of the potato scions (Supplemental Table S9).
Figure 5.
Scion-to-rootstock-to-scion mRNA cycling revealed by a split-shoot heterograft system. A, Representative split-shoot heterograft in which the scion on the right is N. benthamiana, the scion on the left is potato, and the rootstock is tomato. The white boxes indicate the graft union (GU). B, Closeup view of the heterograft union between potato and tomato. C, Closeup view of the heterograft union between N. benthamiana and tomato. Bars = 2 cm.
Functional analysis of the 58 scion-to-rootstock-to-scion mobile mRNAs revealed that diverse biological processes were overrepresented, including those related to translation, transport, and biosynthetic processes (Supplemental Table S10). Within the cellular component category, intracellular, membrane, and thylakoid were overrepresented. Moreover, structural molecule activity, transporter activity, and hydrolase activity were overrepresented in the molecular function category.
Mobile mRNAs Are Highly System Specific
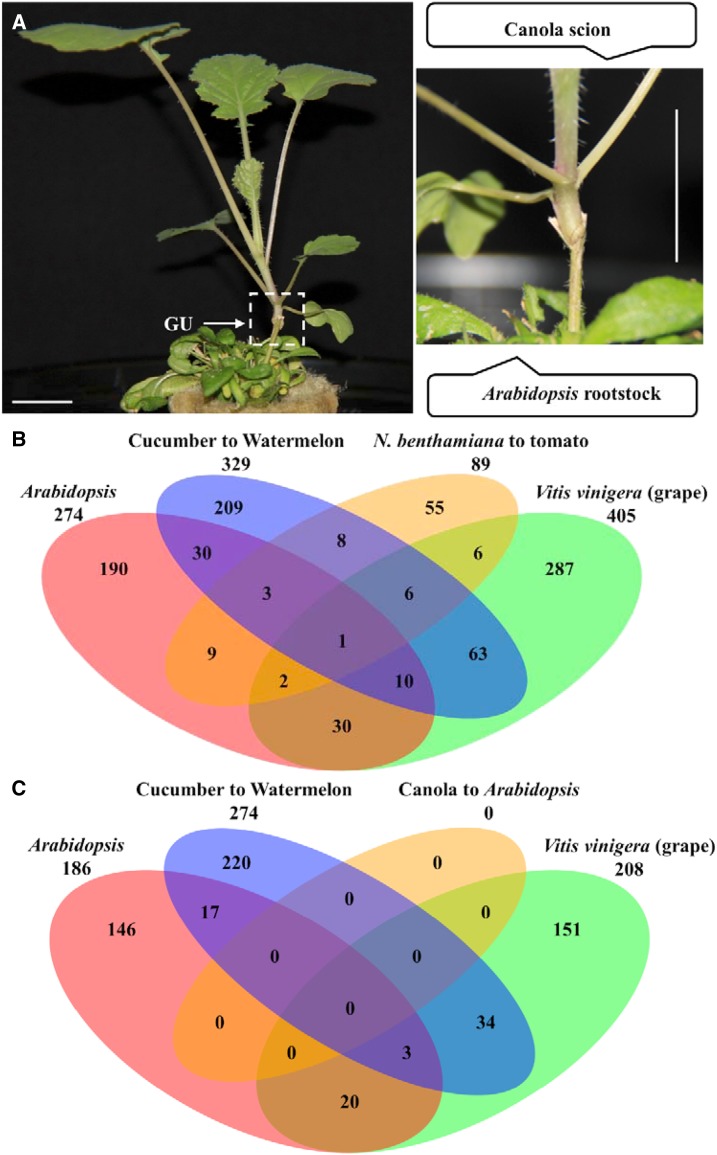
Although large-scale movement of mRNAs has been demonstrated, it is not clear how many of the mobile sequences are functional. If the mRNAs do play physiological roles, one would predict sequence conservation between different systems. Identification of the core mRNAs shared by these heterograft systems and functional characterization of these mRNAs will shed light on their physiological roles. However, most species involved in these previous systems are not easy to transform, which is a prerequisite for future in-depth functional analyses of the mobile mRNAs. To address this issue, we developed another heterograft system using canola (Brassica napus) and Arabidopsis, both of which are transformable, as scion and rootstock, respectively (Fig. 6A). Although canola and Arabidopsis are both Brassicaceae species, there is considerable variation in their genome sequences, which facilitates the accurate identification of species-specific mRNAs. Surprisingly, RNA-seq analysis of the Arabidopsis roots only led to the identification of 23 shoot-to-root mobile mRNAs derived from canola (Supplemental Table S11). In previous heterograft studies, 1,698, 1,963, and 1,593 shoot-to-root mobile mRNAs were detected in Arabidopsis, grapevine, and cucumber systems, respectively. In our N. benthamiana/tomato system, 1,163 mRNAs, when counts from all experimental systems were combined, were detected to be mobile from shoot to root (Supplemental Table S12).
Figure 6.
Heterograft established between canola and Arabidopsis and comparison of mobile mRNAs identified from different heterograft systems. A, Representative canola/Arabidopsis heterograft grown in a hydroponic solution. The white box indicates the graft union (GU). Bars = 2 cm. B, Venn diagram of orthologous mobile mRNAs identified in the N. benthamiana/tomato system and other heterograft systems. C, Venn diagram of orthologous mobile mRNAs identified in the canola/Arabidopsis system and other heterograft systems. Mobile mRNAs having orthologs in the proteomes of the other three systems based on reciprocal BLASTP hits were included in the comparison.
To explore whether growth condition and physiological status can cause the movement of different sets of mRNAs, we compared the 183 mobile mRNAs identified in the regular stem N. benthamiana/tomato heterograft grown in a hydroponic condition with the 242 mobile mRNAs identified in the long-stem N. benthamiana/tomato heterograft grown in soil. Forty-one and 100 mRNAs were found to be short stem + hydroponic and long stem + soil specific, respectively (Supplemental Table S13). The large difference in the number and identity of mobile mRNAs also was observed in the C. pentagona system, where 9,518 or 347 host mRNAs were detected in C. pentagona when the host species was either Arabidopsis or tomato, respectively (Kim et al., 2014). Together, these findings suggest that the movement of mobile mRNAs could be determined by the species involved in the system as well as other factors such as the growth environment.
To detect whether a core group of shoot-to-root mobile mRNAs exists in various systems, we compared the identified mobile mRNAs from the N. benthamiana/tomato heterografts with those identified in Arabidopsis, grapevine, and cucumber heterograft systems (Thieme et al., 2015; Yang et al., 2015; Zhang et al., 2016b). Through orthology analysis, we found that only one mobile mRNA, chlorophyll a/b binding protein8 (Niben101Scf01328g01012), was shared among these four systems (Fig. 6B). A physiological role for the chlorophyll a/b-binding protein in the stem and root has not been reported, although previous localization studies have shown that this gene is lowly expressed in the stem and root (Sakamoto et al., 1991; Tada et al., 1991; Song et al., 2007). Future functional study specifically targeted to the movement of this transcript will provide more insight into the regulation of the transport of this transcript. A similar analysis was carried out with the canola/Arabidopsis heterograft system and those in Arabidopsis, grape, and cucumber, and no mRNA was found to be shared (Fig. 6C).
Harboring the TLS Motif Does Not Necessarily Lead to High Mobility
Recently, Zhang et al., 2016a demonstrated that a 70- to 73-nucleotide-long TLS motif confers shoot-to-root or root-to-shoot mobility of mRNAs and that approximately 11.4% and 7.5% of the mobile mRNAs identified from the Arabidopsis Columbia/Arabidopsis PED and the grapevine heterografts, respectively, harbor this motif. We used TLSfinder in the PlaMoM database to scan the mobile transcripts identified in the N. benthamiana/tomato system (Guan et al., 2017). Among the 1,163 mobile mRNAs, 126 (10.8%) harbor at least one TLS motif in the coding sequence or untranslated region, which was significantly enriched (P < 0.05, hypergeometric test) when compared with the genome background (5,066 TLS-containing genes out of 57,140 total genes; 8.9%). We then asked to what extent this positive relationship is reflected in individual mRNAs. To our surprise, among the top 100 most abundant mRNAs in leaves of our N. benthamiana/tomato heterograft system, 18 harbored the TLS motif but none of them were mobile; among the top 2,000 most abundant mRNAs, 174 harbored the TLS motif but only 11 were mobile. In addition, in the long-stem N. benthamiana/tomato heterograft system, we found that the majority of the mobile mRNAs harboring the TLS motif did not arrive in the tomato root. Only 23 of the 122 mobile transcripts having this motif that successfully passed the graft junction were detected in the roots. It is important to note that our discovery on the lack of correlation between mobility and the harboring of the TLS motif does not negate the importance of this motif because it is possible that, under certain circumstances (e.g. low minerals), some of the immobile mRNAs containing this motif could become mobile (Thieme et al., 2015; Zhang et al., 2016b).
DISCUSSION
Previous studies using EDTA-facilitated exudation, laser-capture microdissection, fluorescence-activated cell sorting, or cucurbit exudation identified large numbers of mRNAs in the phloem translocation stream (Ivashikina et al., 2003; Doering-Saad et al., 2006; Omid et al., 2007; Deeken et al., 2008; Kanehira et al., 2010). However, the authenticity of these mRNAs was often questioned (Turgeon and Wolf, 2009; Liu et al., 2012; Zhang et al., 2012, 2014). Recent studies using different heterograft systems strongly suggest the existence of large-scale, long-distance movement of mRNAs via the phloem (Notaguchi et al., 2015; Thieme et al., 2015; Yang et al., 2015; Zhang et al., 2016b). In addition, several earlier studies on the interaction of the host plants Arabidopsis and tomato with the parasite C. pentagona have demonstrated that large numbers of host plant mRNAs move to the parasitic species via the phloem (LeBlanc et al., 2013; Kim et al., 2014). These discoveries collectively indicate that movement of mRNAs via the phloem is a common physiological process. In this study, we established an N. benthamiana/tomato heterograft system to identify shoot-to-root phloem-mobile mRNAs. The relatively large differences in genome sequence between the two species allowed confident and exhaustive identification of mobile mRNAs. In addition, we took advantage of this system to study the accumulation pattern of shoot-born mRNAs during their movement to the root.
Regulated and Unregulated Movement of mRNAs
It has been demonstrated that some phloem-mobile mRNAs play physiological roles in recipient sink organs such as young leaves, roots, and tubers (Kim et al., 2001; Banerjee et al., 2006; Notaguchi et al., 2012). However, the identification of large numbers of mobile mRNAs (hundreds to thousands) from recent plant-parasitic or heterograft experimental systems and the high variation across different systems have raised concerns as to the percentage of these mobile mRNAs associated with physiological functions. Is mRNA transport a regulated physiological and developmental process or due primarily to the leakage of leaf mRNAs into the sieve tubes? In the grapevine study, the number of mobile mRNAs identified from the field-grown heterografts was much smaller than that in the in vitro-grown grafts (Yang et al., 2015). One explanation is that more mobile mRNAs were degraded before reaching the recipient organs in the field-grown grafts due to the longer distance than in the in vitro-grown grafts (Yang et al., 2015). Similar tendencies for RNA movement were reported for both the Arabidopsis grafts and the parasitic plant and its host plant (LeBlanc et al., 2013; Kim et al., 2014; Thieme et al., 2015). Our long-stem N. benthamiana/tomato experiments showed that a total of 1,096 mRNAs passed the graft joint, but 854 of them disappeared, including some of those with the TLS motif, during their movement from shoot to root. The disappearance of mRNAs during their movement can be attributed to two reasons. One is that companion cells along the transport phloem in the stem could specifically arrest certain types of mRNAs from the sieve tube for translation or degradation. The other is that there is an RNase in the sieve tube that can degrade mRNAs lacking protection. Although it is generally believed that the phloem is devoid of RNase and, therefore, RNA degradation is unlikely (Morris, 2017), the small molecular masses (e.g. 25–29 kD; Bariola et al., 1999) of some of the RNase enzymes are certainly an advantage for their movement from the companion cell to the sieve element. Previous studies have shown that the size-exclusion limit between companion cells and sieve elements is large enough to allow the transit of a 67-kD GFP in Arabidopsis (Stadler et al., 2005). The lack of detection of the RNase activity in the phloem may be due to inadequate sensitivity of the chemical kits used for the assay (Sasaki et al., 1998; Doering-Saad et al., 2002). It is reasonable to assume that the movement of mRNAs undergoing degradation in the phloem is not regulated; therefore, it is less likely that there are physiological functions associated with them.
However, evidence of a regulated process for mRNA movement exists. It was demonstrated that the long-distance trafficking of StBEL5 in potato is regulated and plays a physiological role in tuberization (Banerjee et al., 2006). A recent study by Zhang et al. (2016a) demonstrated that a TLS motif confers mobility for mRNAs. In the grapevine study, some mRNAs were transmitted at rates higher than random transmission (Yang et al., 2015). In our N. benthamiana/tomato grafts, a number of mRNAs had higher abundances in the rootstock than in the leaf, which is the origin of the synthesis of these mobile mRNAs (Table I; Supplemental Table S4). An mRNA encoding casein kinase I was discovered to be mobile and accumulated in both the regular and the long-stem N. benthamiana/tomato heterografts. In rice (Oryza sativa), casein kinase I is involved in auxin metabolism and root development. Arabidopsis plants with down-regulated casein kinase I have fewer lateral and adventitious roots as well as shortened primary roots (Liu et al., 2003). It is intriguing to study whether the long-distance movement of the casein kinase mRNA plays a role in the establishment of root architecture. Transcript abundance in the companion cells was proposed as one of the mechanisms related to mRNA mobility (Calderwood et al., 2016). However, concerns associated with the mathematical model used to draw the conclusion have been raised (Morris, 2017). Our transgenic potato experiment showed that the abundance itself is not enough to lead to the movement of an mRNA because overexpressing two phloem-immobile mRNAs, AtCHL1 or AtAMT1;2, in the companion cells did not promote their shoot-to-root movements (Fig. 3). This result is in agreement with a recent study in which the overexpression of GFP in the companion cells did not lead to movement of the transcript from the leaf to the root (Paultre et al., 2016). It is important to note that the results from the transgenic potato experiment do not negate the importance of transcript abundance and cell specificity to phloem mobility. Rather, they indicate that other unidentified regulatory mechanisms (e.g. motif structure, length, interaction with RNA-binding proteins or the plasmodesmata) between the companion cell and the sieve element, etc., in addition to transcript abundance, also may be involved in conferring mobility.
Cell Origin: Apoplastic and Symplastic Loading Species May Differ
When we compared transcript abundance in the entire leaf versus phloem mobility in the N. benthamiana/tomato heterografts, we did not observe any correlation (Fig. 2A), indicating that more abundant transcripts in the leaf do not necessarily lead to higher probability of shoot-to-root movement. This lack of correlation is in conflict with the grapevine graft system, in which 17 of the highly abundant 33 leaf mRNAs were mobile (Yang et al., 2015). This large difference between the two systems led us to hypothesize whether leaf ultrastructure could play a role in this physiological process. N. benthamiana, the scion in our heterograft system, is an apoplastic loading species. The phloem companion cell-sieve element (CC-SE) complex is almost entirely isolated from its surrounding cell types, including the mesophyll, while in grapevine, a woody species and a symplastic loader, the plasmodesmatal connections between the mesophyll cells and the CC-SE complex are very abundant. Therefore, when mRNAs extracted from the entire leaf are used for abundance analysis, the most abundant mRNAs in an apoplastic loader may not have the highest mobility unless they are transcribed in the phloem companion cells. Transcripts generated in the mesophyll cells cannot move to the CC-SE due to the lack of plasmodesmata between them. In contrast, in a symplastic loading species, such as grapevine, mRNAs can potentially move directly to the translocation stream sieve element from surrounding cells such as mesophyll. To explore the origin of the mobile mRNAs identified from our heterograft system in which the scion, N. benthamiana, is an apoplastic species, we compared the 1,163 mobile mRNAs derived from the three types of N. benthamiana/tomato heterografts (Supplemental Table S12) with the phloem mRNAs identified with either phloem sap collection or the laser-capture microdissection method (Ivashikina et al., 2003; Doering-Saad et al., 2006; Omid et al., 2007; Deeken et al., 2008; Kanehira et al., 2010). A total of 128 mobile mRNAs were found to be detected in previous studies. A hypergeometric test showed that these mobile mRNAs were overrepresented in the mRNAs identified from previous studies (P < 0.05; Supplemental Table S14). Previous studies have shown that mRNAs can move from cell to cell via the plasmodesmata (Lucas et al., 1995; Roberts and Oparka, 2003). Hence, the abundance of mRNAs in the leaf could be a better indicator of mRNA mobility in symplastic loading species than in apoplastic species. This hypothesis also could be used to explain why a large number of transcripts harboring the TLS motif in the leaf of the N. benthamiana/tomato heterograft did not move to the root. It is highly possible that this motif could confer mobility only when the mRNAs are transcribed in the companion cells.
Scion-to-Rootstock-to-Scion mRNA Cycling
While the xylem mainly serves to conduct water and minerals from roots to shoots, the phloem serves as a conduit to transport carbohydrates, amino acids, and a number of signaling molecules. Previous studies in different heterograft systems indicate that mRNAs not only move from shoots to roots via the phloem but also move from roots to shoots (Thieme et al., 2015; Yang et al., 2015). It has been suggested that the root-to-shoot movement of mobile mRNAs can take place either in the phloem against the source-to-sink bulk flow or via the cell-to-cell-based root-to-shoot transport system (Thieme et al., 2015; Ham and Lucas, 2017). The attentions of these prior studies were either on shoot- or root-derived mRNAs. To find out whether root-arriving, shoot-born mRNAs can move back to the shoot, we designed a split-shoot system involving tripartite grafting. To our surprise, 58 N. benthamiana mRNAs were found in the potato shoot. To fulfill this scion-to-rootstock-to-scion movement of mRNAs, the scion-to-rootstock mobile mRNAs from N. benthamiana need to be unloaded from the phloem to the apoplast/xylem area in tomato and then are loaded to either an adjacent shoot-ward transport phloem or to other root cells, as discussed above, before the rootstock-to-scion movement takes place. How the scion-to-rootstock-transmitted mRNAs are released from the phloem to the apoplast/xylem area and exchanged to other cells is a fascinating question to be addressed in the future. It has been known for a long time that the exchange of carbon-containing compounds and minerals between the apoplast/xylem and phloem happens in plants, and membrane transporters play important roles in this process (van Bel, 1990; Zhang and Turgeon, 2009; White, 2012). However, there are no mRNA transporters that have been discovered to efflux mRNAs. An alternative explanation is that the scion-to-rootstock-arriving mRNAs are initially unloaded from the phloem in the stele region via plasmodesmata, as indicated by a recent study in which the shoot-to-root-arriving proteins can be unloaded symplastically (Paultre et al., 2016), and then, by an unknown mechanism, the unloaded mRNAs are transferred to cells (e.g. phloem and/or cortical) and move to the shoot.
CONCLUSION
Heterografting is a powerful system to identify the long-distance transport of mobile mRNAs. However, the large variation in the number and identity of the mobile mRNAs derived from different systems indicates that a more general understanding of the mechanisms of the movement of these transcripts is needed before large-scale functional characterizations on individual mRNAs is pursued. Endogenous physiological differences among plant species may be one of the reasons for the large diversity of identified mobile mRNAs. Different heterograft combinations and the degree of the distortion associated with the involved species may be another reason leading to the large variation of identified mobile transcripts. Identifying the core mobile mRNAs across species and dissecting the physiological functions associated with them are challenging but remain important tasks for future research.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seedlings of Nicotiana benthamiana, tomato (Solanum lycopersicum ‘Heinz 1706-BG’), and potato (Solanum tuberosum ‘Désirée’) were grown in Propagation mix (Sun Gro Horticulture) in a greenhouse with a day/night temperature of 28°C/25°C and a light intensity of approximately 400 to 600 µmol m−2 s−1 on a 14-h/10-h light regime. Canola (Brassica napus ‘Westar’) and Arabidopsis (Arabidopsis thaliana ecotype Columbia) were grown in a growth chamber with a day/night temperature of 26°C/23°C and a light intensity of 250 µmol m−2 s−1 on a 12-h/12-h light regime.
Grafting and Tissue Sampling
To produce heterografts between N. benthamiana and tomato, 3-week-old tomato and N. benthamiana seedlings were used as rootstock and scion, respectively. A V-shaped wedge of an N. benthamiana stem was inserted into a slit in a tomato stem. Both the scion and the graft joint were kept in a transparent plastic bag, and the heterografts were placed beneath a bench with dim light. After 1 week, the plastic bag was gradually opened over a period of 3 d and the established heterografts were transferred to full-strength hydroponic growth solution and grown in a growth chamber with a day/night temperature of 28°C/25°C and a light intensity of 400 to 600 µmol m−2 s−1 on a 14-h/10-h regime. At this stage, visible flower buds from the N. benthamiana scion were pinched off to avoid contaminating the tomato root tissue with pollen.
To overexpress nonmobile mRNAs in the phloem companion cells of potato, two representative Arabidopsis genes, AtCHL1 and AtAMT1;2, were cloned and individually fused behind the promoter of AtSUC2, a phloem companion cell-specific gene. Primers flanked with restriction sites used to amplify both promoters and genes are listed in Supplemental Table S15. pRI 101-AN was used as the binary vector. Transgenic potato plants were produced by Agrobacterium tumefaciens-mediated transformation following the protocol described by Zhou et al. (2017). Grafting transgenic potato onto wild-type potato followed the same procedure described above. These plants were grown in a greenhouse under the aforementioned conditions.
To study whether the accumulation pattern of mobile mRNAs differs during their movement, the shoot tips of 2.5-m-tall (5-month-old) tomato plants were severed and N. benthamiana scions were grafted onto the tops of these tomato plants. The established heterografts were allowed to grow for 6 weeks in a greenhouse in the conditions described above.
To study whether scion-to-rootstock mobile mRNAs can move back to the scion after they arrive in the root, tomato seedlings were trained to have two major stems during their first 3 weeks of growth. A potato scion was grafted on one of the two stems to form the first heterograft. After 3 weeks, the young sink leaves on the actively growing potato scion were pinched off and an N. benthamiana scion was grafted onto the second stem of the tomato to create a split-shoot plant. These tripartite heterografts were grown in the same greenhouse in the conditions described above for another 3 weeks before the mature leaves on the potato scion were harvested.
To produce the canola/Arabidopsis heterograft, shoots of 2-week-old canola plants were grafted onto the severed inflorescence stems of 4-week-old Arabidopsis. The procedure was similar to that used for the N. benthamiana/tomato system. The heterografts were allowed to grow for 6 weeks in a chamber with a day/night temperature of 26°C/23°C and a light intensity of 250 µmol m−2 s−1 in a 12-h/12-h regime to produce several actively growing mature canola leaves. At 5 d prior to harvesting the Arabidopsis roots, the Arabidopsis leaves were covered by aluminum foil to facilitate the source-to-sink bulk flow from the canola leaves to the Arabidopsis roots.
RNA Isolation and RT-PCR
Total RNA was isolated from tissues using the E.Z.N.A. Total RNA Kit I (Omega Bio-tek) following the manufacturer’s instructions. One microgram of total RNA was treated with RQ1 DNase (Promega) for 30 min to remove genomic DNA and then converted into cDNA using iScript Reverse Transcription Supermix (Bio-Rad). Gene-specific primers used in RT-PCR are listed in Supplemental Table S15. ExTaq DNA Polymerase (Takara Bio) was used to amplify the PCR products. The PCR cycle consisted of 95°C for 3 min, followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s.
RNA-Seq and RNA-Seq Data Processing
Strand-specific RNA-seq libraries were constructed from 4 μg of total RNA using the TruSeq Stranded mRNA Library Prep Kit (Illumina) following the manufacturer’s protocol. Three independent biological replicates for either leaf or root from the different heterograft combinations described above were prepared and sequenced on an Illumina NextSeq 500 system at the Genomics Resources Core Facility of Weill Cornell Medical College. Libraries from different species were sequenced on different lanes of the NextSeq 500 system to avoid cross-contaminations between samples.
Raw RNA-seq reads were first processed to trim the adaptor and low-quality sequences using Trimmomatic (Bolger et al., 2014), and trimmed reads shorter than 40 bp were discarded. The remaining high-quality reads were first aligned to an rRNA database using Bowtie (Langmead et al., 2009), allowing up to three mismatches. These rRNA-mapped reads were excluded from subsequent analyses. The remaining cleaned RNA-seq reads were mapped to the corresponding genomes using HISAT (Kim et al., 2015). Following alignments, raw counts for each gene model were derived and then normalized using the reads per kilobase of exon per million fragments mapped method (Mortazavi et al., 2008).
Graft-Transmissible mRNA Identification
To identify mRNAs transmitted from the N. benthamiana scion to the tomato rootstock, the cleaned reads from the tomato rootstock were first mapped to the tomato genome (Tomato Genome Consortium, 2012) using HISAT, allowing up to two edit distances. The unmapped reads were compared with the RNA-seq reads from the nongrafted tomato plants, and those having perfect matches were discarded. The remaining reads were further mapped to the N. benthamiana genome (Bombarely et al., 2012) using HISAT, allowing up to one edit distance. Reads mapped to the N. benthamiana genome were regarded as transmitted. The graft-transmissible mRNAs were identified if the corresponding reads were detected in at least two out of the three biological replicates. Similar procedures were followed to identify mobile mRNAs for the tripartite N. benthamiana/tomato/potato system and the canola/Arabidopsis system. GO terms enriched in the mobile mRNAs were identified using GO::TermFinder (Boyle et al., 2004).
Differential Accumulation Analysis in Long-Stem N. benthamiana/Tomato Heterografts
Plant tissues from high stem (a region approximately 10–20 cm below the grafting joint), low stem (a region approximately 120–130 cm below the grafting joint), and root were collected for RNA-seq analysis. Identification of the mobile mRNAs followed the same procedure as described above. Raw counts were then fed to edgeR (Robinson et al., 2010) to identify differentially accumulated genes. The method of Benjamini and Hochberg (1995) was used to adjust raw P values with a confidence threshold of false discovery rate < 0.01. The abundance of each mRNA in high stem, low stem, and root was divided by that in the root to obtain the relative fold changes along the movement path.
TLS Analysis
TLSs for either the entire N. benthamiana leaf transcriptome or all of the identified mobile mRNAs were scanned using TLSfinder in the PlaMoM database (Guan et al., 2017) with default parameter settings.
Orthology Analysis
Complete proteome sequences of N. benthamiana were compared with those of Arabidopsis, grapevine (Vitis vinifera), and cucumber (Cucumis sativus) using BLASTP with an e-value cutoff of 1e-05. Orthologous pairs between N. benthamiana and Arabidopsis, grapevine, or cucumber were identified based on the reciprocal best hits of BLASTP. The orthologous pairs that shared the same N. benthamiana genes were identified as those present in all four species. The same procedure was used to identify orthologous pairs between canola and Arabidopsis, grapevine, or cucumber. Mobile mRNAs identified from the different heterograft systems and presented in the four-way orthology were used for the comparisons.
Accession Number
The sequence data were deposited in the National Center for Biotechnology Information Sequence Read Archive under accession number SRP111187.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Companion cell-specific overexpression of two phloem-immobile mRNAs in potato.
Supplemental Table S1. N. benthamiana mRNAs detected in the tomato root samples in the N. benthamiana/tomato heterografts.
Supplemental Table S2. Enrichment of GO slim terms in the 183 mobile mRNAs detected from the N. benthamiana/tomato heterografts.
Supplemental Table S3. Expression of genes in the leaf of N. benthamiana collected from the N. benthamiana/tomato heterografts.
Supplemental Table S4. Abundance of mobile mRNAs in the scion or rootstock in the N. benthamiana/tomato heterografts.
Supplemental Table S5. Mobile mRNAs detected from the root samples of tomato in the long-stem N. benthamiana/tomato heterografts.
Supplemental Table S6. Mobile mRNAs detected from the low-stem region of the tomato rootstock in the long-stem N. benthamiana/tomato heterografts.
Supplemental Table S7. Mobile mRNAs detected from the high-stem region of the tomato rootstock in the N. benthamiana/tomato heterografts.
Supplemental Table S8. Differentially expressed mobile mRNAs detected in the high-stem, low-stem, and root regions in the long-stem N. benthamiana/tomato heterografts.
Supplemental Table S9. Mobile mRNAs detected from mature potato leaves in the split-shoot grafting system.
Supplemental Table S10. GO terms enriched in the shoot-to-root-to-shoot mobile mRNAs.
Supplemental Table S11. Mobile mRNAs detected in the Arabidopsis root collected from the canola/Arabidopsis heterografts.
Supplemental Table S12. Combined list of shoot-to-root mobile mRNAs in the different heterograft systems.
Supplemental Table S13. Mobile mRNAs identified in the roots from different N. benthamiana/tomato heterografts.
Supplemental Table S14. Comparison of the 1,163 mobile mRNAs identified from the three N. benthamiana/tomato heterografts with phloem mRNAs identified from previous studies.
Supplemental Table S15. Primers used in this study.
Acknowledgments
We thank the Tomato Genetics Resource Center at the University of California, Davis, for providing the tomato seeds and the Plant Gene Resources of Canada for supplying the canola cv Westar seeds.
Footnotes
This work was supported by funds from Purdue University as part of AgSEED Crossroads funding to support Indiana’s Agriculture and Rural Development (to C.Z.), the National Key Research and Development Project of China (2016YFD0101206), and the U.S. National Science Foundation (IOS-1339287 and IOS-1539831).
Articles can be viewed without a subscription.
References
- Banerjee AK, Chatterjee M, Yu Y, Suh SG, Miller WA, Hannapel DJ (2006) Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18: 3443–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola PA, MacIntosh GC, Green PJ (1999) Regulation of S-like ribonuclease levels in Arabidopsis: antisense inhibition of RNS1 or RNS2 elevates anthocyanin accumulation. Plant Physio1 119: 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller LA, Martin GB (2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant Microbe Interact 25: 1523–1530 [DOI] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G (2004) GO::TermFinder: open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood A, Kopriva S, Morris RJ (2016) Transcript abundance explains mRNA mobility data in Arabidopsis thaliana. Plant Cell 28: 610–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeken R, Ache P, Kajahn I, Klinkenberg J, Bringmann G, Hedrich R (2008) Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J 55: 746–759 [DOI] [PubMed] [Google Scholar]
- Doering-Saad C, Newbury HJ, Bale JS, Pritchard J (2002) Use of aphid stylectomy and RT-PCR for the detection of transporter mRNAs in sieve elements. J Exp Bot 53: 631–637 [DOI] [PubMed] [Google Scholar]
- Doering-Saad C, Newbury HJ, Couldridge CE, Bale JS, Pritchard J (2006) A phloem-enriched cDNA library from Ricinus: insights into phloem function. J Exp Bot 57: 3183–3193 [DOI] [PubMed] [Google Scholar]
- Guan D, Yan B, Thieme C, Hua J, Zhu H, Boheler KR, Zhao Z, Kragler F, Xia Y, Zhang S (2017) PlaMoM: a comprehensive database compiles plant mobile macromolecules. Nucleic Acids Res 45: D1021–D1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham BK, Lucas WJ (2014) The angiosperm phloem sieve tube system: a role in mediating traits important to modern agriculture. J Exp Bot 65: 1799–1816 [DOI] [PubMed] [Google Scholar]
- Ham BK, Lucas WJ (2017) Phloem-mobile RNAs as systemic signaling agents. Annu Rev Plant Biol 68: 173–195 [DOI] [PubMed] [Google Scholar]
- Ivashikina N, Deeken R, Ache P, Kranz E, Pommerrenig B, Sauer N, Hedrich R (2003) Isolation of AtSUC2 promoter-GFP-marked companion cells for patch-clamp studies and expression profiling. Plant J 36: 931–945 [DOI] [PubMed] [Google Scholar]
- Kanehira A, Yamada K, Iwaya T, Tsuwamoto R, Kasai A, Nakazono M, Harada T (2010) Apple phloem cells contain some mRNAs transported over long distances. Tree Genet Genomes 6: 635–642 [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G, LeBlanc ML, Wafula EK, dePamphilis CW, Westwood JH (2014) Genomic-scale exchange of mRNA between a parasitic plant and its hosts. Science 345: 808–811 [DOI] [PubMed] [Google Scholar]
- Kim M, Canio W, Kessler S, Sinha N (2001) Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293: 287–289 [DOI] [PubMed] [Google Scholar]
- King RW, Zeevaart JA (1974) Enhancement of phloem exudation from cut petioles by chelating agents. Plant Physiol 53: 96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc M, Kim G, Patel B, Stromberg V, Westwood J (2013) Quantification of tomato and Arabidopsis mobile RNAs trafficking into the parasitic plant Cuscuta pentagona. New Phytol 200: 1225–1233 [DOI] [PubMed] [Google Scholar]
- Lin MK, Lee YJ, Lough TJ, Phinney BS, Lucas WJ (2009) Analysis of the pumpkin phloem proteome provides insights into angiosperm sieve tube function. Mol Cell Proteomics 8: 343–356 [DOI] [PubMed] [Google Scholar]
- Liu DD, Chao WM, Turgeon R (2012) Transport of sucrose, not hexose, in the phloem. J Exp Bot 63: 4315–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xu ZH, Luo D, Xue HW (2003) Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J 36: 189–202 [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Bouché-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, Hake S (1995) Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270: 1980–1983 [DOI] [PubMed] [Google Scholar]
- Ma Y, Miura E, Ham BK, Cheng HW, Lee YJ, Lucas WJ (2010) Pumpkin eIF5A isoforms interact with components of the translational machinery in the cucurbit sieve tube system. Plant J 64: 536–550 [DOI] [PubMed] [Google Scholar]
- Morris RJ. (2017) On the selectivity, specificity and signalling potential of the long-distance movement of messenger RNA. Curr Opin Plant Biol 43: 1–7 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaguchi M, Wolf S, Lucas WJ (2012) Phloem-mobile Aux/IAA transcripts target to the root tip and modify root architecture. J Integr Plant Biol 54: 760–772 [DOI] [PubMed] [Google Scholar]
- Notaguchi M, Higashiyama T, Suzuki T (2015) Identification of mRNAs that move over long distances using an RNA-Seq analysis of Arabidopsis/Nicotiana benthamiana heterografts. Plant Cell Physiol 56: 311–321 [DOI] [PubMed] [Google Scholar]
- Omid A, Keilin T, Glass A, Leshkowitz D, Wolf S (2007) Characterization of phloem-sap transcription profile in melon plants. J Exp Bot 58: 3645–3656 [DOI] [PubMed] [Google Scholar]
- Paultre DS, Gustin MP, Molnar A, Oparka KJ (2016) Lost in transit: long-distance trafficking and phloem unloading of protein signals in Arabidopsis homografts. Plant Cell 28: 2016–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Oparka K (2003) Plasmodesmata and the control of symplastic transport. Plant Cell Environ 26: 103–124 [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto M, Sanada Y, Tagiri A, Murakami T, Ohashi Y, Matsuoka M (1991) Structure and characterization of a gene for light-harvesting Chl a/b binding protein from rice. Plant Cell Physiol 32: 385–393 [Google Scholar]
- Sasaki T, Chino M, Hayashi H, Fujiwara T (1998) Detection of several mRNA species in rice phloem sap. Plant Cell Physiol 39: 895–897 [DOI] [PubMed] [Google Scholar]
- Song G, Honda H, Yamaguchi K (2007) Expression of a rice chlorophyll a/b binding protein promoter in sweetpotato. J Am Soc Hortic Sci 132: 551–556 [Google Scholar]
- Spiegelman Z, Golan G, Wolf S (2013) Don’t kill the messenger: long-distance trafficking of mRNA molecules. Plant Sci 213: 1–8 [DOI] [PubMed] [Google Scholar]
- Stadler R, Wright KM, Lauterbach C, Amon G, Gahrtz M, Feuerstein A, Oparka KJ, Sauer N (2005) Expression of GFP-fusions in Arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel post-phloem domain in roots. Plant J 41: 319–331 [DOI] [PubMed] [Google Scholar]
- Tada Y, Sakamoto M, Matsuoka M, Fujimura T (1991) Expression of a monocot LHCP promoter in transgenic rice. EMBO J 10: 1803–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme CJ, Rojas-Triana M, Stecyk E, Schudoma C, Zhang W, Yang L, Miñambres M, Walther D, Schulze WX, Paz-Ares J, et al. (2015) Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat Plants 1: 15025. [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon R, Wolf S (2009) Phloem transport: cellular pathways and molecular trafficking. Annu Rev Plant Biol 60: 207–221 [DOI] [PubMed] [Google Scholar]
- Turnbull CG, Lopez-Cobollo RM (2013) Heavy traffic in the fast lane: long-distance signalling by macromolecules. New Phytol 198: 33–51 [DOI] [PubMed] [Google Scholar]
- van Bel A. (1990) Xylem-phloem exchange via the rays: the undervalued route of transport. J Exp Bot 41: 631–644 [Google Scholar]
- White PJ. (2012) Long-distance transport in the xylem and phloem. In Marschner P, ed, Marschner’s Mineral Nutrition of Higher Plants, Ed 3 Academic Press, Cambridge, MA, pp 49–70 [Google Scholar]
- Yang Y, Mao L, Jittayasothorn Y, Kang Y, Jiao C, Fei Z, Zhong GY (2015) Messenger RNA exchange between scions and rootstocks in grafted grapevines. BMC Plant Biol 15: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Turgeon R (2009) Downregulating the sucrose transporter VpSUT1 in Verbascum phoeniceum does not inhibit phloem loading. Proc Natl Acad Sci USA 106: 18849–18854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yu X, Ayre BG, Turgeon R (2012) The origin and composition of cucurbit “phloem” exudate. Plant Physiol 158: 1873–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Han L, Slewinski TL, Sun J, Zhang J, Wang ZY, Turgeon R (2014) Symplastic phloem loading in poplar. Plant Physiol 166: 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Thieme CJ, Kollwig G, Apelt F, Yang L, Winter N, Andresen N, Walther D, Kragler F (2016a) tRNA-related sequences trigger systemic mRNA transport in plants. Plant Cell 28: 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zheng Y, Ham BK, Chen J, Yoshida A, Kochian LV, Fei Z, Lucas WJ (2016b) Vascular-mediated signalling involved in early phosphate stress response in plants. Nat Plants 2: 16033. [DOI] [PubMed] [Google Scholar]
- Zhou X, Zha M, Huang J, Li L, Imran M, Zhang C (2017) StMYB44 negatively regulates phosphate transport by suppressing expression of PHOSPHATE1 in potato. J Exp Bot 68: 1265–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]