Summary
Interleukin‐1β (IL‐1β) is a potent mediator of innate immunity commonly up‐regulated in a broad spectrum of inflammatory diseases. When bound to its cell surface receptor, IL‐1β initiates a signalling cascade that cooperatively induces the expression of canonical IL‐1 target genes such as IL‐8 and IL‐6. Here, we present galectin‐3 as a novel regulator of IL‐1β responses in corneal keratinocytes. Using the SNAP‐tag system and digitonin semi‐permeabilization, we show that recombinant exogenous galectin‐3 binds to the plasma membrane of keratinocytes and is internalized into cytoplasmic compartments. We find that exogenous galectin‐3, but not a dominant negative inhibitor of galectin‐3 polymerization lacking the N‐terminal domain, exacerbates the response to IL‐1β by stimulating the secretion of inflammatory cytokines. The activity of galectin‐3 could be reduced by a novel d‐galactopyranoside derivative targeting the conserved galactoside‐binding site of galectins and did not involve interaction with IL‐1 receptor 1 or the induction of endogenous IL‐1β. Consistent with these observations, we demonstrate that small interfering RNA‐mediated suppression of endogenous galectin‐3 expression is sufficient to impair the IL‐1β‐induced secretion of IL‐8 and IL‐6 in a p38 mitogen‐activated protein kinase‐independent manner. Collectively, our findings provide a novel role for galectin‐3 as an amplifier of IL‐1β responses during epithelial inflammation through an as yet unidentified mechanism.
Keywords: corneal keratinocyte, galectin‐3, innate immunity, interleukin‐1β, p38 mitogen‐activated protein kinase
Introduction
Interleukin‐1 (IL‐1) is a multifunctional cytokine closely associated with acute and chronic inflammation and a powerful inducer of the innate immune response. It is produced by immune and non‐immune cells, and exists in two distinct forms, IL‐1α and IL‐1β, both of which elicit largely identical biological responses through interaction with the IL‐1 receptor type 1 (IL1R1) on target cells.1, 2 Binding of IL‐1 to its receptor results in the activation of protein kinase pathways that promote the expression of canonical IL‐1 target genes with pleiotropic activities, such as IL‐8 and IL‐6.2 Abnormal production of IL‐1 has been associated with a number of inflammatory, autoimmune, infectious and degenerative diseases, which has led to the development of numerous strategies to therapeutically inhibit IL‐1 activity.3, 4
Multiple mechanisms contribute to maintain a tight control over IL‐1 activity. These include processing of the IL‐1β precursor through inflammasome‐dependent and ‐independent pathways, and a number of receptor antagonists and decoy receptors that modulate the extent of the IL‐1β signalling cascade.5 Synergistic events resulting from combinatorial stimuli can also exacerbate the responses to IL‐1β, particularly those involving cytokines that share transcriptional pathways with IL‐1β, such as with IL‐17 and tumour necrosis factor‐α.6, 7 In addition to cytokines, other mediators of inflammation such as galectin‐3 are expressed and rapidly released to sites of insult; however, very little is known about their contribution to the IL‐1β inflammatory response.
Galectin‐3 is a 28 000 to 35 000 MW chimeric protein originally identified as Mac‐2 on the surface of murine macrophages.8 It is composed of a single carbohydrate‐recognition domain and a large N‐terminal domain that contributes to self‐aggregation. When secreted, galectin‐3 is able to crosslink surface glycoproteins and stimulate important pathways involved in the innate immune response and act as a regulatory molecule at various stages during acute and chronic inflammation.9 Galectin‐3 can be found at elevated levels in immune cells as well as in body fluids during cancer and inflammatory disease,10, 11, 12 and its expression appears to be dependent on cell differentiation and activation.9 Galectin‐3 is also highly expressed in epithelial cells and is involved in the pathogenesis of both skin and corneal diseases.13, 14 In the present study, we identify a novel function of galectin‐3 as an amplifier of IL‐1β responses. We find that galectin‐3 interacts with the plasma membrane of corneal keratinocytes and exacerbates the response to IL‐1β by stimulating the secretion of inflammatory cytokines. We further demonstrate that the effect of galectin‐3 involves the galactoside‐binding pocket and N‐terminal polymerization domain of the lectin, and does not require interaction of galectin‐3 with IL1R1 or the p38 mitogen‐activated protein kinase (MAPK) pathway. These data establish a critical role for galectin‐3 in promoting the pro‐inflammatory activities of IL‐1β in corneal epithelium and indicate that down‐regulation of galectin‐3 is necessary to re‐establish tissue homeostasis.
Materials and methods
Antibodies and reagents
Antibodies to human IL‐1β (1 : 1000; mAb201) and IL‐8 (1 : 500; mAb208) for use in immunoblot analyses were purchased from R&D Systems (Minneapolis, MN). Anti‐human IL‐6 (1 : 1000; ab93356) and anti‐human IL1R1 (1 : 1000; EP409Y) antibodies were purchased from Abcam (Cambridge, MA) and Novus Biologicals (Littleton, CO), respectively. Antibodies to human galectin‐3 (1 : 2000; sc20157), caveolin‐1 (1 : 2000; sc894) and glyceraldehyde 3‐phosphate dehydrogenase (GAPDH; 1 : 2000; sc25778) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX). Antibodies to p38 (1 : 500; ab7952) and p‐p38 (1 : 100; sc101758) were purchased from Abcam and Santa Cruz Biotechnology, respectively. Anti‐α‐tubulin (1 : 2000; T5168) antibody was purchased from Sigma‐Aldrich (St Louis, MO). Anti‐human IL1R1 (1 : 100; N20, sc‐688) antibody for immunostaining was purchased from Santa Cruz Biotechnology, Inc. and recombinant human IL‐1β (201‐LB) was purchased from R&D Systems.
Recombinant human galectin‐3 (rhGal3) and galectin‐3C (rhGal3C) were cloned and expressed as previously reported.15 Briefly, Rosetta Escherichia coli clones carrying the rhGal3 or rhGal3C vectors were selected and grown using Lysogeny broth medium containing ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml). Heterologous protein expression was induced by the addition of 0·3 mm of isopropyl β‐d‐1 thiogalactopyranoside and the induced cultures were incubated overnight at 15° with shaking. The rhGal3 and rhGal3C were purified from lysates by affinity chromatography using lactosyl sepharose. To eliminate contaminating bacterial endotoxins, rhGal3 and rhGal3C were further purified by polymyxinB affinity chromatography (Sigma‐Aldrich). The absence of lipopolysaccharide was confirmed using the ToxinSensor™ Chromogenic LAL Endotoxin Assay Kit (GenScript, Piscataway, NJ). Protein solutions were concentrated by centrifugal filtration (VWR, Radnor, PA), dialysed against phosphate‐buffered saline (PBS) containing 10% glycerol and stored at −20°.
Synthesis of SNAP‐galectin‐3
The coding sequence of the SNAP‐tag was amplified by PCR using the pSNAP‐tag(m) vector (New England Biolabs, Ipswich, MA) as template and the primers SNAP‐F 5′‐GGCGGCGGCCATATGGACAAAGACTGCG‐3′) and SNAP‐R (5′‐AAAAATTGTCTGCCATTACCGTTCGTATAA‐3′). The coding sequence of galectin‐3 was amplified by PCR using cDNA from telomerase‐immortalized human corneal keratinocytes as template and the primers galectin‐F (5′‐TTATACGAACGGTAATGGCAGACAATTTTT‐3′) and galectin‐R (5′‐GGCGGCGGCGGATCCTTATATCATGGTATA‐3′). The SNAP‐galectin‐3 direct fusion sequence was amplified by touchdown PCR using the SNAP and galectin‐3 amplicons and the primers SNAP‐F and galectin‐R. The obtained product was cloned into the SNAP‐tag expression vector pTWIN‐1 (New England Biolabs) using the NdeI and BamHI restriction enzymes, resulting in an N‐terminal fusion of SNAP to galectin‐3. The protein was synthesized and purified as described previously.15 The in‐gel detection of the SNAP‐tag fusion protein was performed using SNAP‐Cell 505‐Star green fluorescent substrate (New England Biolabs).
Chemical synthesis of the galectin inhibitor
Synthesis of 1,1′‐sulphanediyl‐bis‐{3‐deoxy‐3‐[4‐(5‐fluoropyrid‐2‐yl)‐1H‐1,2,3‐triazol‐1‐yl]‐β‐d‐galactopyranoside} was performed following procedures described in the literature.16 All reagents and solvents were dried before use according to standard methods. Commercial reagents were used without further purification. Purification of the compound was carried out by column chromatography on silica gel (40–60 μm, 60 Å) and preparative HPLC (Agilent 1260 infinity system, column SymmetryPrep‐C18, 17 ml/min H2O‐MeCN gradient 10–100% 15 min with 0·1% formic acid). Specific rotations were measured on a PerkinElmer Model 341 Polarimeter. NMR spectra 1H, 13C, 2D COSY and HMQC were recorded with a Bruker Avance II 400 MHz spectrometer (400 Hz for 1H, 100 Hz for 13C) at ambient temperature (see Supplementary material, Fig. S1). HRMS was determined by direct infusion on a Waters XEVO‐G2 QTOF mass spectrometer using electrospray ionization. The purity of the inhibitor was > 95% as determined by HPLC analysis (Agilent series 1100 system, column Eclipse XDB‐C18, 0·8 ml/min H2O‐MeCN gradient 5–95% 13 min with 0·1% trifluoroacetic acid). Galectin affinities were determined in a competitive fluorescence anisotropy assay as reported previously.16
Cell culture and human tissue
Telomerase‐immortalized human corneal keratinocytes were grown as previously reported. Briefly, cells were plated at a seeding density of 5 × 104 cells/cm2 and maintained in keratinocyte serum‐free medium (Thermo Fisher Scientific, Waltham, MA) until confluence. Thereafter, cells were grown in Dulbecco's modified Eagle's medium (DMEM)/F12 supplemented with 10% calf serum and 10 ng/ml epidermal growth factor for 7 days to promote stratification and differentiation, as previously reported.17 Where indicated, cells were incubated with IL‐1β (10 ng/ml) in serum‐free DMEM. Discarded healthy human corneal epithelial tissue was collected from a donor who underwent LASIK surgery and frozen in optimal cutting temperature compound for sectioning using a cryostat.
Semipermeabilization assay
Cell cultures grown in six‐well culture plates were treated with 100 μg/ml SNAP‐rhGal3. After 6 hr, cells were incubated in a shaker at 4° with NEH buffer (150 mm NaCl, 0·2 mm EDTA, 20 mm HEPES‐NaOH, pH 7·4) containing 42 μg/ml digitonin for 10 min.18 Cells were scraped and centrifuged at 12 000 g in a table‐top centrifuge for 15 min as described previously.19 Supernatants (cytosolic fraction) and pellets (homogenate) were analysed by SDS–PAGE followed by immunoblot analysis.
Immunoblot analyses
Protein from cell cultures was extracted using RIPA buffer (150 μm NaCl, 50 μm Tris–HCl, pH 8·0, 1% Nonidet P‐40, 0·5% deoxycholate, 0·1% SDS) supplemented with Complete™ Protease Inhibitor Cocktail (Roche Diagnostics, Indianapolis, IN). After homogenization with a pellet pestle, the protein cell extracts were centrifuged at 12 000 g for 45 min, and the protein concentration of the supernatant was determined using the Pierce BCA™ Protein Assay Kit (Thermo Fisher Scientific). Proteins in cell lysates (20 μg) or cell culture media (20 μl) were resolved in 15% SDS–PAGE, and electroblotted onto nitrocellulose membranes (Bio‐Rad, Hercules, CA). Membranes were then incubated with primary antibodies in Tris‐buffered saline and Tween 20 supplemented with 5% non‐fat milk overnight at 4°, followed by the appropriate secondary antibodies coupled to horseradish peroxidase (Santa Cruz Biotechnology, Inc.). Peroxidase activity was detected on HyBlot CL autoradiography film (Denville Scientific, Inc., Plainfield, NJ). Immunoblots were quantified using imagej ® software.
Immunofluorescence microscopy
Human corneal tissue sections (10 μm) and keratinocyte cultures grown in four‐well chamber slides (Thermo Fisher Scientific) were fixed for 10 min in methanol at −20° and washed with PBS. After blocking for 10 min in PBS with 3% bovine serum albumin, slides were incubated overnight with primary antibody diluted in PBS with 1% bovine serum albumin. Secondary antibodies were incubated for 1 hr at room temperature. Nuclei were counterstained using Vectashield with DAPI (Vector Laboratories, Burlingame, CA). Incubation with primary antibodies was routinely omitted in control experiments. The sections and cultures were viewed by confocal microscopy using a DM 6000 CS confocal laser scanning microscope (Leica Microsystems, Buffalo Grove, IL).
SNAP fluorescence labelling was used as described by the manufacturer (New England Biolabs). Briefly, stratified human corneal keratinocytes were incubated with 100 μg/ml of SNAP‐galectin‐3 fusion protein for 0, 2 or 6 hr and washed with DMEM supplemented with 10% serum three times for 30 min. Cultures were then labelled with 5 μm of SNAP‐Surface 549 fluorescent substrate for 30 min at 37°. Thereafter, cells were incubated with SNAP‐Surface Block, labelled with 2 μm of SNAP‐Cell 505‐Star green fluorescent substrate for 30 min at 37° and fixed in 100% methanol. Slides were mounted in VectaShield mounting medium containing DAPI, and observed on a Zeiss Axio Observer Z1 inverted fluorescence microscope (Carl Zeiss Microimaging GmbH, Jena, Germany).
Small interfering RNA knockdown
Depletion of galectin‐3 was achieved using Silencer® Select Pre‐designed small interfering RNA (siRNA) (S8149; Thermo Fisher Scientific) targeting human LGALS3 mRNA. A non‐specific scrambled siRNA (4390843; Thermo Fisher Scientific) served as the negative control. For knockdown, cells in six‐well culture plates were transfected twice – at 80% confluence and 3 days post‐confluence – by 6 hr incubation with 2 μl siRNA in 2 μl Lipofectamine 2000 (Thermo Fisher Scientific) dissolved in 800 μl Opti‐MEM plus GlutaMax reduced‐serum medium (Thermo Fisher Scientific). Following each transfection, keratinocyte serum‐free medium (for cells treated at 80% confluence) or DMEM (for stratifying cells) was added to cultures for 20 hr. Thereafter, the medium was switched to DMEM/F‐12 to promote stratification and differentiation.
Real‐time quantitative PCR
Total RNA was isolated from cell cultures using the extraction reagent TRIzol (Thermo Fisher Scientific) following the manufacturer's instructions. Residual genomic DNA was eliminated by DNase I digestion of the RNA preparation. One microgramme of total RNA was used for cDNA synthesis (iScript™ cDNA Synthesis; Bio‐Rad). Detection of gene expression was performed by quantitative polymerase chain reaction (qPCR) using primers for IL‐8 and IL‐6 as described previously.20 Gene expression was measured using the KAPA SYBR® FAST qPCR kit (Kapa Biosystems, Wilmington, MA) in a Mastercycler ep realplex thermal cycler (Eppendorf, Hauppauge, NY). The following parameters were used: 2 min at 95°, followed by 40 cycles of 5 seconds at 95° and 30 seconds at 60°. Expression values were normalized for the housekeeping gene GAPDH (PrimePCR GAPDH primers; Bio‐Rad) and fold‐changes were calculated using the comparative CT method.
Galectin‐3 affinity chromatography
A galectin‐3 affinity column was prepared by coupling 5 mg of rhGal3 to cyanogen bromide‐activated Sepharose 4B (GE Healthcare, Milwaukee, WI) following the manufacturer's instructions. Binding activity of rhGal3 conjugated to beads was assessed by incubation of 200 μg asialofetuin (Sigma‐Aldrich) with 50 μl rhGal3 beads in the presence or absence of 0·1 m lactose for 1 hr at room temperature. Asialofetuin was detected on a 10% SDS–PAGE gel by GelCode® Blue Stain (Thermo Fisher Scientific). For galectin‐3 affinity chromatography, aliquots of cell extracts (50 μg) were incubated with 100 μl rhGal3‐conjugated agarose beads in PBS, pH 7·5 for 1 hr at 37° with gentle mixing every 10 min. After washing with PBS, beads were eluted in sequence with 0·5 m sucrose and 0·5 m lactose. Eluates were then resolved on 10% SDS–PAGE and analysed by immunoblot using an IL1R1 antibody.
Cell surface biotinylation
Cells were surface labelled with the Pierce® Cell Surface Protein Isolation kit (Thermo Fisher Scientific) following the manufacturer's instructions. Briefly, cells were incubated with 0·25 mg/ml cell‐impermeable sulpho‐NHS‐SS‐Biotin. After washing with Tris‐buffered slaine, cells were lysed in the presence of Complete™ Protease Inhibitor Cocktail (Roche Diagnostics). Cell lysates were incubated with 500 μl NeutrAvidin™ for 1 hr at room temperature. Biotinylated proteins were finally eluted with dithiothreitol.
Statistics
Statistical analysis was carried out with graphpad prism 7 (GraphPad Software, San Diego, CA) for Macintosh.
Results
Exogenous galectin‐3 binds to and is internalized by corneal keratinocytes
Increased levels of galectin‐3 in biological fluids have been associated with the pathophysiology of a number of diseases. At the ocular surface, elevated levels of galectin‐3 have been found in tears of patients with immune disorders in whom the integrity of the corneal epithelium is impaired.10, 21 To test whether exogenous galectin‐3 interacts with stratified corneal keratinocytes, we generated a plasmid containing the full‐length encoding region of galectin‐3 fused at its N terminus to SNAP‐tag, a 182‐residue‐long (19 400 MW) self‐labelling polypeptide. SDS–PAGE analysis of the resulting fusion protein, termed SNAP‐rhGal3, confirmed that the product migrated as a single band with an expected size of ~55 000 (Fig. 1a). Incubation of corneal keratinocytes with increasing concentrations of SNAP‐rhGal3 resulted in the detection of exogenous galectin‐3 in conjunction with endogenous galectin‐3 in cell lysates (Fig. 1b). In these experiments, the uptake of SNAP‐rhGal3 was inhibited by 15 min pre‐incubation of the fusion protein with the galectin‐3 inhibitor lactose, demonstrating that the interaction between the corneal keratinocytes and exogenous galectin‐3 is carbohydrate‐dependent.
Figure 1.
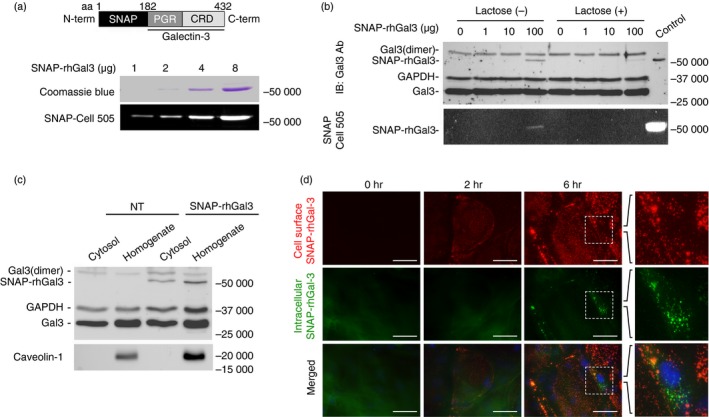
Exogenous galectin‐3 binds to and is internalized by corneal keratinocytes. (a) A SNAP‐galectin‐3 chimera, SNAP‐rhGal3, was produced by fusing the coding sequence of the SNAP‐tag (181 amino acids) to the N terminus of human galectin‐3 (amino acids 183–432). Increasing amounts of the protein were visualized on a polyacrylamide gel by Coomassie blue staining and fluorescence imaging. CRD, carbohydrate recognition domain; PGR, proline, glycine and tyrosine‐rich domain. (b) Corneal keratinocytes were treated with varying doses of SNAP‐rhGal3, pre‐incubated for 15 min with or without 0·1 m lactose, a competitive inhibitor of galectin‐3, for 6 hr at 37°. The presence of exogenous and endogenous galectin‐3 in cell lysates was determined by immunoblot. Exogenous galectin‐3 was visualized by fluorescence imaging. (c) Corneal keratinocytes were treated with or without 100 μg/ml SNAP‐rhGal3 for 6 hr at 37°. Thereafter, cells were permeabilized with digitonin for 10 min and the amount of galectin‐3 or caveolin‐1 leaked into the supernatant (cytosol), or remaining in the cells (homogenate), was analysed by immunoblot. NT, non‐treated. (d) Corneal keratinocytes were incubated with 100 μg/ml of SNAP‐rhGal3 for 0, 2 or 6 hr. Fluorescence microscopy images show surface localization (red) and internalization (green) of SNAP‐rhGal3. Nuclei were counterstained using DAPI (blue). Scale bar, 50 μm. [Colour figure can be viewed at http://wileyonlinelibrary.com]
The basis of this interaction was defined in more detail using a semi‐permeabilization assay.18 In this assay, stratified cultures of corneal keratinocytes were treated with SNAP‐rhGal3 and the cytosolic fraction was extracted using plasma membrane semi‐permeabilization with digitonin. As shown in Figure 1(c), the SNAP‐rhGal3 was detected in the cytosol with the appropriate molecular mass, suggesting internalization of the intact fusion protein into intracellular compartments. A portion of SNAP‐rhGal3 was detected in the homogenate fraction, which tested positive for the plasma membrane protein marker caveolin‐1. To further determine the cellular distribution of exogenous galectin‐3 in cell culture, the SNAP‐rhGal3 fusion protein was visualized using fluorescence microscopy. A distinct punctate staining pattern corresponding to exogenous galectin‐3 and characteristic of apically localized proteins was observed at 2 hr in the plasma membrane using the SNAP‐Surface 549 substrate (Fig. 1c). At 6 hr, the location of SNAP‐rhGal3 at the cell surface became more evident and correlated with the sporadic appearance of the protein in intracellular compartments as shown using the SNAP‐Cell 505 substrate.
Exogenous galectin‐3 amplifies the response to IL‐1β
Interleukin‐1 is a major cytokine constitutively expressed by stratified epithelia and critical to the regulation of the immune response following injury. Downstream signalling responses associated with the up‐regulation of IL‐1 include the induction of IL‐8, a strong chemotactic and angiogenic factor, and IL‐6, a potent inducer of lymphocyte differentiation.22 To examine the impact of exogenous galectin‐3 on the IL‐1 pathway, we first determined the localization and expression of its primary receptor, IL1R1, in human corneal keratinocytes. Immunofluorescence analyses revealed that IL1R1 localized throughout the stratified epithelium in donor tissue (Fig. 2a), and was detected by immunoblot analysis in cultured cells as a single band with an apparent molecular weight of ~75 000 (Fig. 2b). Consistent with previous data, we found that treatment of stratified human corneal keratinocytes with IL‐1β for 6 hr resulted in increased levels of IL‐8 and IL‐6 in the cell‐culture media (Fig. 2b,c). Further, we observed that the levels of these inflammatory cytokines were not affected by the sole addition of exogenous galectin‐3 to the media. However, when added in combination with IL‐1β, there was a threefold increase in downstream cytokine secretion compared with treatment with IL‐1β alone. At 24 hr, incubation of the stratified cultures with a combination of IL‐1β and exogenous galectin‐3 resulted in an approximately twofold increase in IL‐8 and IL‐6 secretion compared with treatment with IL‐1β alone. Under these conditions, the expression of IL1R1 remained unaffected (Fig. 2b).
Figure 2.
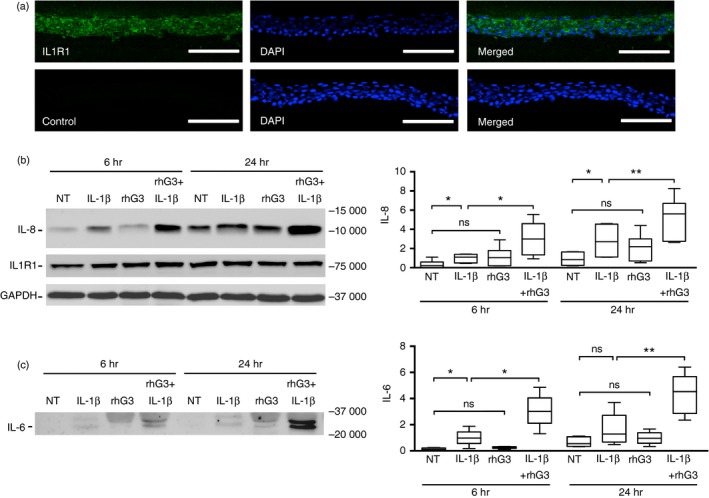
Exogenous galectin‐3 amplifies the response to interleukin‐1β (IL‐1β). (a) Immunofluorescence microscopy demonstrating the presence of IL‐1 receptor 1 (IL1R1) (green) in healthy human corneal epithelial tissue. Nuclei were counterstained using DAPI (blue). Scale bar, 100 μm. (b and c) Stratified cultures of human corneal keratinocytes were incubated with IL‐1β (10 ng/ml) or recombinant human galectin‐3 (100 μg/ml) alone or in combination for 6 and 24 hr at 37°. Levels of IL‐8 and IL‐6 in the cell culture media, and IL1R1 in cell lysates, were determined by immunoblot. NT, non‐treated. Results in (b) and (c) represent six independent experiments. The box and whisker plots show the 25th and 75th centiles (box), the median and the minimum and maximum data values (whiskers). Significance was determined using repeated measures one‐way analysis of variance with Dunnett's post hoc multiple comparison test. *P < 0·05, **P < 0·01; ns, non‐significant. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Next, we used a galectin‐3 mutant lacking the N‐terminal polymerization domain to investigate whether galectin‐3 multimerization was necessary to amplify the response to IL‐1β. We found that, in contrast to exogenous full‐length galectin‐3, addition of IL‐1β and the galectin‐3 N‐terminal deletion mutant failed to significantly amplify the levels of IL‐8 in media compared with the addition of IL‐1β alone (Fig. 3a), suggesting that N‐terminally mediated galectin‐3 aggregation is necessary to potentiate the IL‐1β response. Further, the involvement of the galactoside‐binding pocket of galectin‐3 in promoting cytokine secretion was tested using a novel synthetic galactoside derivative galectin inhibitor (Fig. 3b). We first tested the direct interaction of the synthetic molecule galectin inhibitor with a panel of galectins using fluorescence anisotropy. The data showed that the galactoside derivative was a potent inhibitor of galectin‐3 with a K d value of 20 nm. A similar observation was made for galectin‐1 and galectin‐2, which were inhibited with only nanomolar K d values, whereas other galectins required much higher concentrations to reach inhibitory activity. Importantly, treatment of corneal keratinocyes with the inhibitor was sufficient to reduce the effect of IL‐1β and exogenous galectin‐3 on IL‐8 secretion (Fig. 3b), indicating that the canonical carbohydrate‐binding site of galectin‐3 participates in the amplification of the response to IL‐1β.
Figure 3.
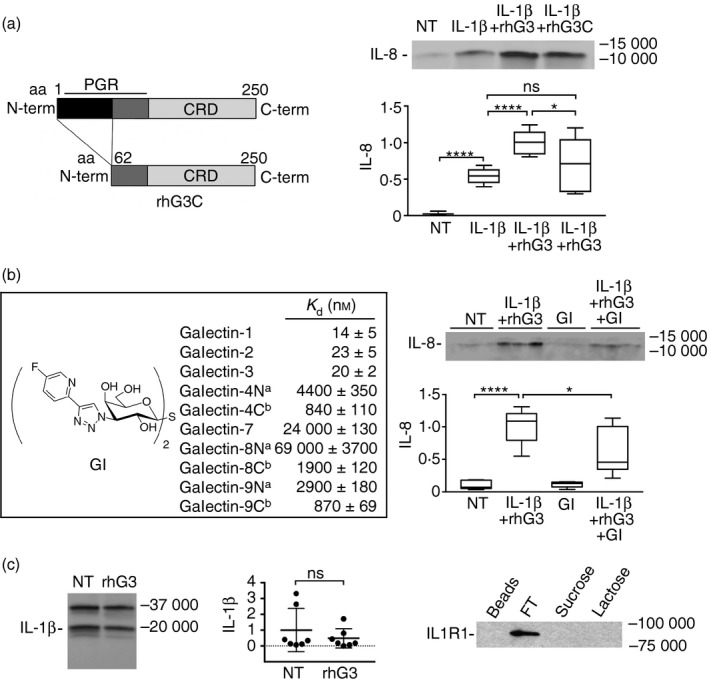
The galectin‐3 pro‐inflammatory activity is established through the N and C termini. (a) A truncated form of galectin‐3 lacking the first 62 amino acids in the N‐terminal domain (rhG3C) was obtained by site‐directed mutagenesis as previously described.15 Stratified cultures of human corneal keratinocytes were incubated with interleukin‐1β (IL‐1β) (10 ng/ml) alone or in combination with 100 μg/ml exogenous full‐length galectin‐3 (rhG3) or rhG3C for 24 hr at 37°. Levels of IL‐8 in the cell‐culture media were determined by immunoblot. CRD, carbohydrate recognition domain; PGR, proline, glycine and tyrosine‐rich domain. NT, not treated. (b) The 1H‐NMR and 13C‐NMR spectra of the galectin inhibitor (GI) are described in the Supplementary material (Figure S1). Galectin affinities were determined in a competitive fluorescence anisotropy assay. Stratified cultures of human corneal keratinocytes were incubated with 1 μm GI alone or in combination with IL‐1β and rhG3 for 24 hr at 37°. Levels of IL‐8 in the cell‐culture media were determined by immunoblot. aN, N‐terminal domain; bC, C‐terminal domain. (c) By immunoblot, addition of exogenous full‐length galectin‐3 had no effect on the levels of IL‐1β in the cell culture media. (d) Cell extracts were collected from stratified keratinocyte cell cultures and subjected to galectin‐3 affinity chromatography. The column was eluted with a non‐competing disaccharide, sucrose, before elution with the competing sugar lactose. FT, flow through. Results in (a) represent three independent experiments performed at least in duplicate, whereas results in (b) represent two independent experiments performed in triplicate. Dissociation constants (K d) in (b) were determined in at least two independent experiments each including at least four data‐points. The box and whisker plots show the 25th and 75th centiles (box), the median and the minimum and maximum data values (whiskers). Results in (c) represent seven independent experiments and data are reported as mean ± standard deviation. Significance was determined using ordinary one‐way analysis of variance with Tukey's post hoc multiple comparison test (a and b), and paired t test (c). *P < 0·05, ****P < 0·0001; ns, non‐significant.
Given that galectin‐3 has been shown to potentiate IL‐1 production by immune cells,23 we then examined whether exogenous full‐length galectin‐3 could influence IL‐1β production in our experiments. We found that corneal keratinocytes secrete both the 31 000 MW IL‐1β precursor and the 17 000 MW mature molecule (Fig. 3c). Addition of exogenous galectin‐3 had no effect on the levels of mature IL‐1β, indicating that the effect of galectin‐3 as an amplifier of the inflammatory response is not associated with the higher concentration of IL‐1β in corneal keratinocytes. Moreover, in galectin‐3 affinity chromatography experiments, we found that the activities of exogenous galectin‐3 as an amplifier of the IL‐1β‐mediated inflammatory response did not involve interaction of the lectin with IL1R1 (Fig. 3d).
Suppression of endogenous galectin‐3 impairs the IL‐1β response
Considering that galectin‐3 is one of the most highly expressed glycogenes in the human ocular surface epithelia,24 we performed loss‐of‐function experiments to investigate whether endogenous galectin‐3 plays a role in modulating the IL‐1β response. Here, the response to IL‐1β was analysed in corneal keratinocytes transfected with siRNA targeting galectin‐3. Consistent with published results, siRNA effectively silenced the expression of galectin‐3 in the stratified cultures (Fig. 4a). Transfection with galectin‐3 siRNA also effectively reduced the content of IL‐8 and IL‐6 in the media following treatment with IL‐1β for 6 hr compared with a scramble siRNA control. Surprisingly, and contrary to changes observed at the protein level, knockdown of galectin‐3 did not affect the transcription of the IL‐8 and IL‐6 genes (Fig. 4b). To verify our findings we further examined the involvement of endogenous galectin‐3 in the regulation of p38 MAPK, a common downstream kinase in the canonical IL‐1 cascade that promotes transcription of the IL‐8 and IL‐6 genes.2 In these experiments, p38 phosphorylation was induced within 15 min following the addition of IL‐1β to the cell culture and was not affected by the silencing of galectin‐3 (Fig. 4c). Further, we demonstrated that abrogation of galectin‐3 did not change the levels of total and cell surface IL1R1 (Fig. 4d). Hence, it appeared plausible that the function of galectin‐3 in amplifying the secretion of IL‐8 and IL‐6 was independent of IL1R1 signalling events.
Figure 4.
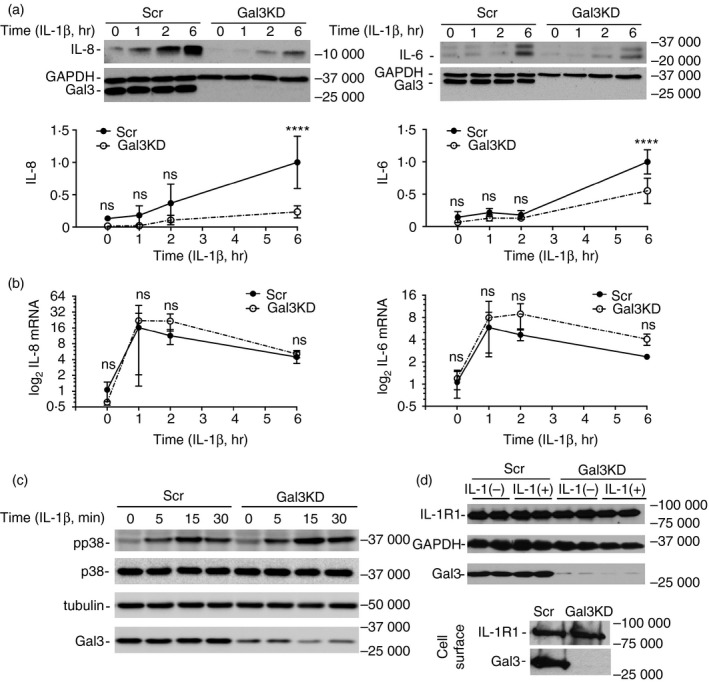
Suppression of endogenous galectin‐3 impairs the interleukin‐1β (IL‐1β) response in a p38 mitogen‐activated protein kinase (MAPK) ‐independent manner. (a) Stratified human corneal keratinocytes were transfected with galectin‐3 small interfering RNA (siRNA) (Gal3KD) or control siRNA (Scr) using Lipofectamine 2000. The cultures were then incubated with IL‐1β (10 ng/ml) for 0, 1, 2 and 6 hr at 37°. Levels of IL‐8 and IL‐6 in the cell culture media, and galectin‐3 in cell lysates, were determined by immunoblot. (b) Real‐time quantitative PCR analysis of IL‐8 and IL‐6 gene expression in stratified human corneal keratinocytes transfected with galectin‐3 or control siRNA. The cultures were incubated with IL‐1β (10 ng/ml) for 0, 1, 2 and 6 hr at 37°. (c) Cells were treated with IL‐1β (10 ng/ml) for 0, 5, 15 and 30 min at 37°. Phosphorylated p38 MAPK (pp38) and total p38 MAPK (p38) were measured by immunoblot. (d) Cells transfected with galectin‐3 or control siRNA were incubated with or without 10 ng/ml IL‐1β for 6 hr at 37°. Levels of IL1R1 in cell lysates were determined by immunoblot. Cell surface labelling of IL1R1 and galectin‐3 was carried out using biotinylation as described in Materials and methods. Results in (a) and (b) represent four independent experiments and data are reported as mean ± standard deviation. Significance was determined using two‐way analysis of variance with Sidak's post hoc multiple comparison test. ****P < 0·0001; ns, non‐significant.
Discussion
Epithelial tissues covering the exposed surfaces of the body act as a first line of defence against damage and infection both by forming a physical barrier that prevents pathogen colonization and by the release of inflammatory cytokines and chemokines that activate the innate immune system. During the past decade, galectins have emerged as an important component in the initiation of innate immunity, serving as danger signals for infective microorganisms, and by orchestrating the activities of innate immune cells such as neutrophils, mast cells, macrophages and dendritic cells, in either an extracellular or intracellular manner.25, 26, 27 The contribution of galectins to the epithelial immune response, on the other hand, has been far less explored. In this study, we have found that galectin‐3 interacts with the plasma membrane of corneal keratinocytes to augment the pro‐inflammatory activities of IL‐1β. We show that these activities are established through the N and C termini of galectin‐3, which contain functional domains associated with receptor aggregation and carbohydrate recognition, and do not involve interaction of galectin‐3 with IL1R1 or the p38 MAPK signalling pathway.
Increased concentrations of galectins have been detected during inflammatory disease in body fluids such as tears and synovia10, 21, 28 and in the bloodstream of patients with different types of cancer.29 In a secreted form, galectin‐3 elicits diverse biological responses that include adhesion of tumour cells to the vascular endothelium30 and activation of specific cell surface receptors such as Toll‐like receptor 4.31 Exposure to galectin‐3 also exerts cytokine‐like regulatory actions and, indeed, promotes secretion of IL‐1β in monocytes,23 macrophages32 and microglia.33 Interestingly, in our study, we found that exogenous galectin‐3 itself did not affect the levels of IL‐1β, instead acting as an amplifier of the IL‐1β‐mediated inflammatory response. The failure of galectin‐3 to induce IL‐1β was not entirely surprising, since the ability of galectin‐3 to promote cytokine secretion appears to be cell‐type specific.34
More intriguing was the observation that galectin‐3 promoted secretion of IL‐8 and IL‐6 in the presence of IL‐1β. The pro‐inflammatory activities of IL‐1β are intimately associated with the activation of a complex signalling system that results in the rapid induction of canonical IL‐1 target genes.2 In this context, the sole stimulation of intestinal epithelial cells and osteoblasts with IL‐1β has been shown to promote IL‐8 and IL‐6 expression, respectively, via p38 MAPK activation.35, 36 Consistent with these results, we found that stimulation of corneal keratinocytes with IL‐1β promoted IL‐8 and IL‐6 expression; however, the increase in mRNA levels for these cytokines was not affected by the targeted silencing of galectin‐3. Further, we did not observe a direct effect of galectin‐3 on p38 phosphorylation and galectin‐3 did not bind IL1R1, suggesting that the amplifying effect of galectin‐3 is independent of IL‐1β signalling events and is achieved at the post‐transcriptional/translational level.
It is possible to speculate that galectin‐3 serves as a positive‐feedback mechanism following transcription of the IL‐8 and IL‐6 genes. Although a plethora of information is available on the transcriptional regulation and extracellular actions of IL‐8 and IL‐6, the mechanism by which cytokines traffic and are released remains unclear for most cell types. In some instances, poor correlations have been found between cytokine mRNA and protein levels both in vitro and in vivo.37, 38 Cytokine secretion is a tightly controlled and cell‐type‐specific process that takes place through multiple mechanisms other than the classical secretory pathway.39, 40 For example, IL‐8 in human neutrophils localizes to organelles distinct from the classical secretory vesicles41 and IL‐6 has been found in tubulovesicular structures that bud off the Golgi and fuse with recycling endosomes in transit to the plasma membrane.42 Interleukin‐1 is also known to stimulate trafficking of IL‐6 to the cell surface in the absence of secretory granule release.43 Based on our own findings, we hypothesize that galectin‐3 in corneal keratinocytes promotes secretion of inflammatory cytokines through an as yet unidentified receptor involved in the regulation of cytokine trafficking and secretion.
Under inflammatory conditions, epithelial cells remodel their cell‐surface glycans by modifying the expression of glycosyltransferases responsible for the biosynthesis of carbohydrate chains.44 These events result in abnormalities in signalling and intracellular processes that have been linked to disease pathogenesis. In this context, galectins have emerged as important effectors that translate glycan‐containing information into a broad spectrum of cellular responses.45 To date, several cell surface receptors have been identified for galectin‐3 in human corneal keratinocytes, including transmembrane mucins,17 the matrix metalloproteinase inducer CD147,46 and integrin α 3/β 1.47 Binding to these receptors has been associated with the ability of galectin‐3 to provide barrier function in the cornea and the regulation of epithelial motility during wound repair. Understanding how the inflammatory microenvironment affects the interaction of galectin‐3 with these and other potential receptors in epithelial surfaces remains largely unknown and will be the goal of future studies.
Disclosures
The authors declare that they have no conflicts of interest with the contents of this article.
Supporting information
Figure S1. 1H‐NMR and 13C‐NMR spectra of the galectin inhibitor.
Acknowledgements
This work was supported by the National Institutes of Health, NEI Grants R01EY014847 (PA) and R01EY026147 (PA), the Japan Eye Bank Association (YU), and a postdoctoral fellowship from the Uehara Memorial Foundation in Japan (YU). The authors thank Ilene K. Gipson of the Schepens Eye Research Institute for providing the human corneal keratinocyte cell line, Hakon Leffler of the Department of Laboratory Medicine at Lund University for performing the competitive fluorescence anisotropy assay, and Hans T. Schambye of Galecto Biotech AB for additional synthesis of the galectin inhibitor.
References
- 1. Sims JE, Smith DE. The IL‐1 family: regulators of immunity. Nat Rev Immunol 2010; 10:89–102. [DOI] [PubMed] [Google Scholar]
- 2. Weber A, Wasiliew P, Kracht M. Interleukin‐1 (IL‐1) pathway. Sci Signal 2010; 3:1–6. cm1. [DOI] [PubMed] [Google Scholar]
- 3. Dinarello CA, Simon A, van der Meer JW. Treating inflammation by blocking interleukin‐1 in a broad spectrum of diseases. Nat Rev Drug Discov 2012; 11:633–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garlanda C, Dinarello CA, Mantovani A. The interleukin‐1 family: back to the future. Immunity 2013; 39:1003–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dinarello CA. Interleukin‐1 in the pathogenesis and treatment of inflammatory diseases. Blood 2011; 117:3720–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chabaud M, Page G, Miossec P. Enhancing effect of IL‐1, IL‐17, and TNF‐α on macrophage inflammatory protein‐3α production in rheumatoid arthritis: regulation by soluble receptors and Th2 cytokines. J Immunol 2001; 167:6015–20. [DOI] [PubMed] [Google Scholar]
- 7. Elias JA, Lentz V. IL‐1 and tumor necrosis factor synergistically stimulate fibroblast IL‐6 production and stabilize IL‐6 messenger RNA. J Immunol 1990; 145:161–6. [PubMed] [Google Scholar]
- 8. Ho MK, Springer TA. Mac‐2, a novel 32,000 Mr mouse macrophage subpopulation‐specific antigen defined by monoclonal antibodies. J Immunol 1982; 128:1221–8. [PubMed] [Google Scholar]
- 9. Henderson NC, Sethi T. The regulation of inflammation by galectin‐3. Immunol Rev 2009; 230:160–71. [DOI] [PubMed] [Google Scholar]
- 10. Uchino Y, Mauris J, Woodward AM, Dieckow J, Amparo F, Dana R et al Alteration of galectin‐3 in tears of patients with dry eye disease. Am J Ophthalmol 2015; 159:1027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu LG. Circulating galectin‐3 in the bloodstream: an emerging promoter of cancer metastasis. World J Gastrointest Oncol 2010; 2:177–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rabinovich GA, Toscano MA. Turning ‘sweet’ on immunity: galectin‐glycan interactions in immune tolerance and inflammation. Nat Rev Immunol 2009; 9:338–52. [DOI] [PubMed] [Google Scholar]
- 13. Larsen L, Chen HY, Saegusa J, Liu FT. Galectin‐3 and the skin. J Dermatol Sci 2011; 64:85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Argueso P. Glycobiology of the ocular surface: mucins and lectins. Jpn J Ophthalmol 2013; 57:150–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mauris J, Mantelli F, Woodward AM, Cao Z, Bertozzi CR, Panjwani N et al Modulation of ocular surface glycocalyx barrier function by a galectin‐3 N‐terminal deletion mutant and membrane‐anchored synthetic glycopolymers. PLoS ONE 2013; 8:e72304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Delaine T, Collins P, MacKinnon A, Sharma G, Stegmayr J, Rajput VK et al Galectin‐3‐binding glycomimetics that strongly reduce bleomycin‐induced lung fibrosis and modulate intracellular glycan recognition. ChemBioChem 2016; 17:1759–70. [DOI] [PubMed] [Google Scholar]
- 17. Argueso P, Guzman‐Aranguez A, Mantelli F, Cao Z, Ricciuto J, Panjwani N. Association of cell surface mucins with galectin‐3 contributes to the ocular surface epithelial barrier. J Biol Chem 2009; 284:23037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu X, Fagotto F. A method to separate nuclear, cytosolic, and membrane‐associated signaling molecules in cultured cells. Sci Signal 2011; 4:pl2. [DOI] [PubMed] [Google Scholar]
- 19. Geiger R, Andritschke D, Friebe S, Herzog F, Luisoni S, Heger T et al BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat Cell Biol 2011; 13:1305–14. [DOI] [PubMed] [Google Scholar]
- 20. Hayashi F, Means TK, Luster AD. Toll‐like receptors stimulate human neutrophil function. Blood 2003; 102:2660–9. [DOI] [PubMed] [Google Scholar]
- 21. Hrdlickova‐Cela E, Plzak J, Smetana K Jr, Melkova Z, Kaltner H, Filipec M et al Detection of galectin‐3 in tear fluid at disease states and immunohistochemical and lectin histochemical analysis in human corneal and conjunctival epithelium. Br J Ophthalmol 2001; 85:1336–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kinoshita S, Adachi W, Sotozono C, Nishida K, Yokoi N, Quantock AJ et al Characteristics of the human ocular surface epithelium. Prog Retin Eye Res 2001; 20:639–73. [DOI] [PubMed] [Google Scholar]
- 23. Jeng KC, Frigeri LG, Liu FT. An endogenous lectin, galectin‐3 (εBP/Mac‐2), potentiates IL‐1 production by human monocytes. Immunol Lett 1994; 42:113–6. [DOI] [PubMed] [Google Scholar]
- 24. Mantelli F, Schaffer L, Dana R, Head SR, Argueso P. Glycogene expression in conjunctiva of patients with dry eye: downregulation of Notch signaling. Invest Ophthalmol Vis Sci 2009; 50:2666–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Kooyk Y, Rabinovich GA. Protein–glycan interactions in the control of innate and adaptive immune responses. Nat Immunol 2008; 9:593–601. [DOI] [PubMed] [Google Scholar]
- 26. Cerliani JP, Stowell SR, Mascanfroni ID, Arthur CM, Cummings RD, Rabinovich GA. Expanding the universe of cytokines and pattern recognition receptors: galectins and glycans in innate immunity. J Clin Immunol 2011; 31:10–21. [DOI] [PubMed] [Google Scholar]
- 27. Sato S, Nieminen J. Seeing strangers or announcing “danger”: galectin‐3 in two models of innate immunity. Glycoconj J 2002; 19:583–91. [DOI] [PubMed] [Google Scholar]
- 28. Ohshima S, Kuchen S, Seemayer CA, Kyburz D, Hirt A, Klinzing S et al Galectin 3 and its binding protein in rheumatoid arthritis. Arthritis Rheum 2003; 48:2788–95. [DOI] [PubMed] [Google Scholar]
- 29. Duckworth CA, Yu L‐G. Galectins in the blood circulation: potential therapeutic targets of cancer metastasis. Galectins Dis Implicat Target Therap Am Chem Soc 2012; 18:309–22. [Google Scholar]
- 30. Zhao Q, Guo X, Nash GB, Stone PC, Hilkens J, Rhodes JM et al Circulating galectin‐3 promotes metastasis by modifying MUC1 localization on cancer cell surface. Cancer Res 2009; 69:6799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burguillos MA, Svensson M, Schulte T, Boza‐Serrano A, Garcia‐Quintanilla A, Kavanagh E et al Microglia‐secreted galectin‐3 acts as a toll‐like receptor 4 ligand and contributes to microglial activation. Cell Rep 2015; 10:1626–38. [DOI] [PubMed] [Google Scholar]
- 32. Tian J, Yang G, Chen HY, Hsu DK, Tomilov A, Olson KA et al Galectin‐3 regulates inflammasome activation in cholestatic liver injury. FASEB J 2016; 30:4202–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jeon SB, Yoon HJ, Chang CY, Koh HS, Jeon SH, Park EJ. Galectin‐3 exerts cytokine‐like regulatory actions through the JAK‐STAT pathway. J Immunol 2010; 185:7037–46. [DOI] [PubMed] [Google Scholar]
- 34. Filer A, Bik M, Parsonage GN, Fitton J, Trebilcock E, Howlett K et al Galectin 3 induces a distinctive pattern of cytokine and chemokine production in rheumatoid synovial fibroblasts via selective signaling pathways. Arthritis Rheum 2009; 60:1604–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parhar K, Ray A, Steinbrecher U, Nelson C, Salh B. The p38 mitogen‐activated protein kinase regulates interleukin‐1β‐induced IL‐8 expression via an effect on the IL‐8 promoter in intestinal epithelial cells. Immunology 2003; 108:502–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patil C, Zhu X, Rossa C Jr, Kim YJ, Kirkwood KL. p38 MAPK regulates IL‐1β induced IL‐6 expression through mRNA stability in osteoblasts. Immunol Invest 2004; 33:213–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gowrisankar YV, Clark MA. Angiotensin II induces interleukin‐6 expression in astrocytes: role of reactive oxygen species and NF‐κB. Mol Cell Endocrinol 2016; 437:130–41. [DOI] [PubMed] [Google Scholar]
- 38. Huang TJ, Li YY, Weng YJ, Cheng CC, Hsu RW. Interleukin‐6 protein expression is more important than interleukin‐6 mRNA levels in assessing surgical invasiveness. J Surg Res 2007; 142:53–8. [DOI] [PubMed] [Google Scholar]
- 39. Lacy P, Stow JL. Cytokine release from innate immune cells: association with diverse membrane trafficking pathways. Blood 2011; 118:9–18. [DOI] [PubMed] [Google Scholar]
- 40. Stanley AC, Lacy P. Pathways for cytokine secretion. Physiology (Bethesda) 2010; 25:218–29. [DOI] [PubMed] [Google Scholar]
- 41. Pellme S, Morgelin M, Tapper H, Mellqvist UH, Dahlgren C, Karlsson A. Localization of human neutrophil interleukin‐8 (CXCL‐8) to organelle(s) distinct from the classical granules and secretory vesicles. J Leukoc Biol 2006; 79:564–73. [DOI] [PubMed] [Google Scholar]
- 42. Manderson AP, Kay JG, Hammond LA, Brown DL, Stow JL. Subcompartments of the macrophage recycling endosome direct the differential secretion of IL‐6 and TNFα . J Cell Biol 2007; 178:57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kandere‐Grzybowska K, Letourneau R, Kempuraj D, Donelan J, Poplawski S, Boucher W et al IL‐1 induces vesicular secretion of IL‐6 without degranulation from human mast cells. J Immunol 2003; 171:4830–6. [DOI] [PubMed] [Google Scholar]
- 44. Dewald JH, Colomb F, Bobowski‐Gerard M, Groux‐Degroote S, Delannoy P. Role of cytokine‐induced glycosylation changes in regulating cell interactions and cell signaling in inflammatory diseases and cancer. Cells 2016; 5:pii: E43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cerliani JP, Blidner AG, Toscano MA, Croci DO, Rabinovich GA. Translating the ‘Sugar Code’ into immune and vascular signaling programs. Trends Biochem Sci 2017; 42:255–73. [DOI] [PubMed] [Google Scholar]
- 46. Mauris J, Woodward AM, Cao Z, Panjwani N, Argueso P. Molecular basis for MMP9 induction and disruption of epithelial cell–cell contacts by galectin‐3. J Cell Sci 2014; 127:3141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Saravanan C, Liu FT, Gipson IK, Panjwani N. Galectin‐3 promotes lamellipodia formation in epithelial cells by interacting with complex N‐glycans on α 3/β 1 integrin. J Cell Sci 2009; 122:3684–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. 1H‐NMR and 13C‐NMR spectra of the galectin inhibitor.


