Abstract
BACKGROUND: Barrett's esophagus metaplasia is the key precursor lesion of esophageal adenocarcinoma. The aim of this study was to find a subset of markers that may allow the identification of patients at risk for esophageal adenocarcinoma, and to determine genes differentially expressed in esophageal squamous cell carcinoma. METHODS: Laser capture microdissection technique was applied to procure cells from defined regions. Genome-wide RNA profiling was performed on esophageal adenocarcinoma (n = 21), Barrett's esophagus (n = 20), esophageal squamous carcinoma (n = 9) and healthy esophageal biopsies (n = 18) using the Affymetrix Human Genome U133plus 2.0 array. Microarray results were validated by quantitative real-time polymerase chain reaction in a second and independent cohort and by immunohistochemistry of two putative markers in a third independent cohort. RESULTS: Through unsupervised hierarchical clustering and principal component analysis, samples were separated into four distinct groups that match perfectly with histology. Many genes down-regulated in esophageal cancers belong to the epidermal differentiation complex or the related GO-group “cornified envelope” of terminally differentiated keratinocytes. Similarly, retinol metabolism was strongly down-regulated. Genes showing strong overexpression in esophageal carcinomas belong to the GO groups extracellular region /matrix such as MMP1, CTHRC1, and INHBA. According to an analysis of genes strongly up-regulated in both esophageal adenocarcinoma and Barrett's esophagus, REG4 might be of particular interest as an early marker for esophageal adenocarcinoma. CONCLUSIONS: Our study provides high quality data, which could serve for identification of potential biomarkers of Barrett's esophagus at risk of esophageal adenocarcinoma progression.
Introduction
Barrett's esophagus, a condition in which a metaplastic columnar mucosa that confers a predisposition to cancer replaces an esophageal squamous mucosa, is a major risk factor for esophageal adenocarcinoma (EAC). The frequency of this deadly tumor has increased significantly during the past four decades [1]. Barrett's esophagus (BE) is associated with a 40-fold increased risk for the development of EAC, which gradually progresses to carcinoma through a metaplasia–dysplasia–carcinoma sequence [2]. Thus, a better understanding of the pathogenesis of Barrett's associated cancer is required for risk prediction and the development of effective therapeutic strategies. Esophageal squamous cell carcinoma (ESCC) is another histological subtype which is detected mostly at advanced stages leading to a low survival of the patients [3].
In many types of carcinomas, biomarkers can enhance our ability for diagnosis, prognosis, and for therapy prediction. At present there are no clinical or histological parameters that allow the identification of patients bearing a high risk of progression of BE to EAC. One of the reasons might be that esophageal Barrett's adenocarcinomas are quite heterogeneous. A number of possible markers have been identified, e.g. aneuploidy [4], copy number alterations [5] and deletion of the p16 tumor suppressor gene [6]. Similarly, miRNAs might be potential biomarkers [7]. EGFR was found to be overexpressed in high-grade dysplasia and adenocarcinoma of the esophagus [8]. However, none of the currently known molecular biomarkers has sufficient predictive power to identify BE progression in a clinically useful manner [9].
Gene expression profiling by microarrays is a powerful technique for delineating biomarkers and pathways associated with cancer progression [10]. Here, also laser capture microdissection (LMD) was applied to collect cells from defined regions of histologic tissue. We focused on neoplastic progression and found a set of genes representing potential biomarkers which might help to create a more standardized, reproducible and reliable diagnostic procedure in order to stratify patients with BE in terms of their risk to develop cancer. Expression of these genes can be determined in a quantitative way, and thus may be able to complement conventional semi-quantitative histological analysis.
Materials and Methods
Patients and Tissue Specimens
Unselected patients seeking treatment for Barrett's esophagus or esophageal cancer at Charité-Universitätsmedizin (Berlin, Germany) were enrolled in the study. Gene expression profiling using whole-human-genome microarrays (Affymetrix U133plus2) was performed on 21 EAC, 20 BE, 9 ESCC and 18 NE biopsies (biopsies from patients without esophageal pathology), collected from 68 individuals. Patients' age ranged from 22 to 79 years, with a median age of 58 years. The validation cohort for RT-PCR verification consisted of 49 esophageal specimens, including 10 EAC, 7 ESCC, 20 BE and 12 NE. Patients' age ranged from 24 to 79 years, with a median age of 59 years. NE biopsies were collected from patients with esophageal pain but diagnosed as normal squamous without pathological changes. BE biopsies were taken during upper gastrointestinal endoscopy with biopsy sampling from year 2005 to 2010. Surgical specimens of chemotherapy-naive patients with known EAC of histological grading G1, UICC stage II and III, who had undergone esophagectomy, were obtained from the tumor bank of Charité Comprehensive Cancer Center (Charité Campus Buch, Berlin, Germany). Samples were immediately snap-frozen in liquid nitrogen and stored at −80 °C. Another independent validation cohort used for immunohistochemistry consisted of 36 formalin fixed and paraffin embedded (FFPE) specimens. In total, 12 EAC, 12 BE and 12 NE were examined. Patients' age ranged from 34 to 87 years, with a median age of 60 years. We obtained tissue specimens from all subjects with informed written consent approved by the local ethics committee of the Charité-Universitätsmedizin, Berlin. Each specimen included in this study was histopathologically approved according to grade and stage (MV, Institute of Pathology, University Hospital Bayreuth, Germany) using standard International Federation of Gastrointestinal criteria.
Laser Capture Microdissection
Laser Capture Microdissection (LMD) was performed for all cases except healthy tissue. Frozen specimens were serially sectioned in 5-μm slices with a cryostat and the first and last slide was stained with hematoxylin and eosin to define the analyzed regions. After transferring the 5 μm sections in between onto MMI membrane slides, these were fixed in 70% isopropyl alcohol and then stained with the MMI basic staining kit. LMD regions were defined by MV (Institute of Pathology, Klinikum Bayreuth, Germany). For LMD a Cellcut device equipped with a Nikon TE300 microscope (MMI AG, Switzerland) was used for isolating desired cells from sections. Then desired cells or areas were selected, cut and collected.
Microarray Analysis
Total RNA was extracted using the RNeasy Micro Kit (Qiagen) according to the manufacturer's protocol. RNA quality was analyzed by Bioanalyzer 2100 (Agilent). Only high quality RNA with RIN above 7.0 was processed further. The Affymetrix Human Genome U133plus 2.0 array (Affymetrix, Santa Clara, CA) was used for this study. Preparation of labeled cRNA and hybridization were done using the gene chip hybridization, wash, and stain kit (Affymetrix, Santa Clara, CA), as described previously [11], [12], [13]. Two cycle labeling was applied to all samples. Chip data quality was strictly controlled. Besides the common quality control provided by gene chip operating software, a scientific workflow for chip quality control was set up in our lab including pseudo image plots, scatter MA plot and relative log expression plot. Additionally, correlation analysis across gene chips was also performed by Agilent GeneSpring software GX 10.2.
Data Analysis
Probe set data were generated with GeneChip Operation Software (GCOS, Affymetrix). Raw data were further processed using tools available from Bioconductor version 2.6 [14]. Background correction and normalization were carried out using the Affymetrix package [15]. Results were filtered by the detection P-value, considering probes with P-value <0.01 as ‘present’ and all others ‘absent’. Probes ‘present’ in at least 50% of the samples of one group were examined for further analysis. Differential expression with respect to control samples was determined using the limma package in Bioconductor [16]. The False Discovery Rate (FDR) was employed using Benjamini-Hochberg procedure for multiple testing of the resulting P-value significance. Principal component analysis (PCA) and unsupervised hierarchical clustering were used to evaluate the relationships between patients' groups. Gene annotations were based on Affymetrix Human Genome U133 Plus 2.0 Array annotation data (hgu133plus2 package). Gene Ontology enrichment was carried out using the DAVID Functional Annotation Bioinformatics Microarray Analysis [17]. A list of all the genes included in these microarrays and the normalized data have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/info/linking.html) under GEO accession number GSE26886.
RNA Extraction and qRT-PCR
RNA extractions were carried out using the RNeasy Mini Kit (Qiagen, Valencia, CA). Total RNA quality and yield were assessed using a bioanalyzer system (Agilent 2100 Bioanalyzer; Agilent Technologies, Palo Alto, CA) and a spectrophotometer (NanoDrop ND-1000; NanoDrop Technologies, Wilmington, DE). Only RNA with an RNA integrity number >7.0 was used for RT-PCR analysis. For quantification, the expression levels of genes found by microarray analyses, RT-PCR was performed using pre-designed gene-specific TaqMan probes and primer sets purchased from Applied Biosystems. RT-PCR amplification was carried out using RNA UltraSense One-Step Quantitative RT-PCR System (Invitrogen) on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems), according to the manufacturer's instructions. All samples were amplified in triplicate reactions. Gene expression was quantified relative to the expression of internal control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using Sequence Detector Software (SDS 2.2, Applied Biosystems) and the relative quantification method for quantitative RT-PCR. In addition, GAPDH levels were also used to determine whether a given sample contained sufficient mRNA to be included in the study. Samples for which the Ct value for GAPDH above 22 were excluded from analysis. The relative expression of each individual gene was calculated based on the determined ∆CT values. The ∆CT value is the difference between the Ct value for an individual gene and the internal reference control gene GAPDH [18]. ∆CT values correspond to a log-2 scale. The result of quantitative gene expression was calculated as a 2−∆CT relative to GAPDH expression in the corresponding samples.
Immunohistochemistry
Immunohistochemistry was performed on 5 μm thin formalin fixed paraffin sections using the biotin blocking system (Dako, Glostrup, Denmark) and the ImmPRESS universal reagent (Vector Laboratories Inc., Burlingame, CA). Primary antibody for CTHRC1 detection was mouse monoclonal anti-human CTHRC1 antibody (Abcam ab72527, 1:1000). Primary antibody for INHBA detection was rabbit polyclonal anti-human INHBA antibody (Sigma-Aldrich Corporation, HPA020031, 1:200).
Results and Discussion
Gene Expression Profiles of Samples are Consistent with Histological Typing
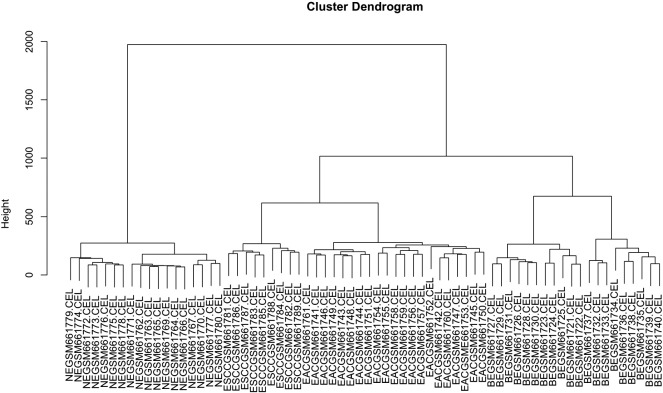
Gene expression profiling using whole-human-genome microarrays (Affymetrix U133plus2) was performed on 21 EAC, 20 BE, 9 ESCC and 18 NE biopsies. By unsupervised hierarchical clustering the tissue samples were separated into 4 distinct groups consistent with their histological subtype. Clustering of the gene expression profiles is depicted in the dendrogram (Figure 1). All 18 samples taken from normal esophageal epithelium cluster together on the left side group called CONTROL. Esophageal carcinoma samples belong to the group in the middle, but they fall into two strictly separated clusters. On the middle left, esophageal squamous cell carcinomas (ESCC) cluster together, and on the middle right esophageal adenocarcinomas (EAC) are found. Samples taken from Barrett's esophagus (BE) cluster together on the right side. Please note that for each group all samples cluster together with not one mismatch. Another approach, Principal component analysis (Supplement Figure 1) demonstrated a similar grouping. The excellent agreement of gene expression profiles with the histological subtypes might be a result of laser capture microdissection, which is likely to be the superior approach in terms of controlling tissue purity.
Figure 1.
Unsupervised hierarchical clustering of samples obtained from four different tissue types. Clustering of the gene expression profiles of the 68 cases under study produced four main clusters. Depicted is a full-length view of the cluster dendrogram with cases orientated along the horizontal axis. Hierarchical clustering analysis is called unsupervised if the relatedness of cases is determined only by the similarity of their whole human genome expression profiles, independently of clinical or pathological parameters. Here, hierarchical cluster analysis showed that the four tissues types are clearly distinct. All 18 samples taken from healthy esophageal epithelium (NE) cluster together in the group on the left side. Carcinoma samples belong to the cluster group in the middle, encompassing two strictly separated clusters of esophageal squamous cell carcinomas (ESCC) and of esophageal adenocarcinomas (EAC). Samples taken from Barrett's esophagus (BE) form another cluster group on the right side. Please note that all samples of the different tissue types cluster together with not one mismatch.
Several gene expression profiling studies of Barrett's esophagus and esophageal carcinomas have been published to date, e.g., [19]. Cellular contamination within patient samples, or a mixture of tissue types within a biopsy is a strong challenge for efforts to determine an accurate cancer–related gene expression profile because gene expression measurements can become skewed by this heterogeneity. Here, we have applied laser microdissection (LMD) in order to get access to almost “pure” samples of Barrett's epithelium, esophageal adenocarcinoma or squamous carcinoma tissue. LMD enables the isolation of specific cell populations from complex tissues under morphological control, which is likely to be the superior approach in terms of controlling tissue purity. In our study, LMD was applied not only for all esophageal tumor samples, but also for BE samples because BE biopsies often contain only few BE glands besides squamous epithelium or stromal cells. LMD enabled us to obtain purified columnar cells from each BE biopsy and use them as a source for RNA extraction. Unsupervised hierarchical clustering and principal component analysis of microarray data obtained distinguished samples into four distinct groups according to their histopathology, demonstrating the quality of the LMD approach and the microarray analysis.
Validation in an Independent Cohort
To verify our observed gene expression results, we investigated expression levels of 12 randomly chosen genes with P < .01 by real-time PCR and confirmed conclusively the microarray results generated from the training cohort. Comparison of the obtained fold-changes by array analysis versus real-time PCR confirmed our approach as conservative, revealing higher expression differences in the former (Supplement Table 2).
Examination of the Marker Expression on the Protein Level
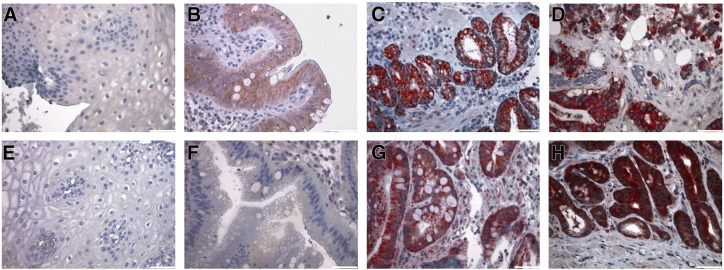
CTHRC1 and INHBA antibodies detected a specific protein band on western blots and thus were used for another validation of gene expression results. Immunohistochemical analysis demonstrated differential abundance of CTHRC1 and INHBA on the protein level in paraffin tissue specimens of a third independent patient cohort of 12 EAC, 5 intraepithelial neoplasia (IEN) specimens, and 7 BE from patients without tumor formation in the first five years after biopsy sampling representing low risk BE, and 12 NE specimens. Immunohistochemical staining showed that CTHRC1 was abundant in 10 of 12 EAC specimens and in the 5 IEN specimens, but staining was low in 7 BE specimens from patients without tumor formation. In NE biopsies no CTHRC1 staining was detected. INHBA was highly abundant in all EAC samples, highly abundant in the IEN specimens, but staining was low in the BE samples and not detected at all in NE specimens (Figure 2).
Figure 2.
Immunohistochemical detection of CTHRC1 (A-D) and INHBA (E-H) expression in healthy epithelium, BE, intraepithelial neoplasia and esophageal adenocarcinoma. Immunohistochemical detection of CTHRC1 and INHBA (red) was performed on 5 μm thin formalin fixed paraffin sections. Cell nuclei were counterstained using hematoxylin (blue). No specific CTHRC1 / INHBA expression could be found in healthy esophageal epithelium (A + E), low CTHRC1 / INHBA expression was detected in Barrett's glands (B + F). In adenocarcinomas, high CTHRC1 and INHBA expression was predominantly observed in mucosa glands (D + H), moreover, high expression of CTHRC1 / INHBA was observed in the intraepithelial neoplasia (C + G). Representative sections are shown. Negative controls were obtained by omission of primary antibody (data not shown). Pictures are 200× magnified.
Analysis of Differentially Expressed Genes
The aim of this study was the generation of a database, which allows the identification of genes or groups of genes with strong differential expression in esophageal carcinomas. To get an overview, gene ontology analysis was used to determine significantly over- or underrepresented biological processes, cellular components, and molecular functions within the entire set of differentially expressed genes. Entrez IDs were assigned to gene ontology categories according to the annotations in the Affymetrix Human Genome U133 Plus 2.0 Array annotation data (hgu133plus2 package). Then these Entrez IDs were subjected to DAVID functional annotation bioinformatics microarray analysis, using the default feature listings and algorithm settings, with the whole human genome as background. Ontology categories with FDR adjusted (Benjamini) P values <.05 were recorded (Suppl. Table 1).
This database can be queried in different ways - pursuing different aims. In order to detect cancer-associated changes but to rule out epithelium-specific changes, esophageal adenocarcinomas were compared with their precursor tissue type, BE, and squamous carcinomas were compared with healthy normal esophageal tissue, NE. In an approach to identify the most consistently changed individual genes, we searched for so-called peak genes with a fold change >6-fold increase/decrease between any two tissue groups.
On the one hand, BE but also EAC and ESCC samples showed an up-regulated representation of genes coding for proteins of the extracellular environment (Table 1) such as MMP1, SPP1, CTHRC1, INHBA and SULF1. Matrix metalloproteinases (MMPs) are a family of zinc-dependent proteases that target the degradation and proteolytic process of components of the extracellular matrix [20]. An elevated expression of MMP1 in esophageal cancers has been described [21]. Another gene, strongly up-regulated in both types of esophageal carcinomas, Activin, the disulfide-linked homodimer of inhibin-ßA (INHBA), is a ligand of the transforming growth factor-beta (TGF-ß) superfamily. INHBA overexpression promotes cell proliferation and may be epigenetically regulated in esophageal adenocarcinoma [22]. Overexpression of activin A in esophageal squamous cell carcinoma has been associated with advanced nodal status, clinical stage, and a worse overall prognosis [23]. Similarly, collagen triple helix repeat-containing 1 (CTHRC1) is known to be aberrantly up-regulated in most human solid tumors [24]. CTHRC1 drives cancer progression perhaps by increasing cancer cell migration [25]. High levels of PMEPA1/TMEPAI expression have been found in renal cell carcinoma, colon cancer, breast cancer, and ovarian cancer as well as in several cancer cell lines [26], [27]. Genome-wide studies, which compared the gene expression levels of invasive cancer tissues with normal counterpart tissues or pre-invasive cancers, suggested that TMEPAI is one of the most highly inducible genes in invasive cancers that converts TGF-β signaling from a tumor suppressor to a tumor promoter [28]. These lines of evidence suggest an oncogenic function of TMEPAI in many cancers. Genes strongly overexpressed in esophageal carcinomas might be helpful for diagnosis and future therapeutic strategies.
Table 1.
Peak Genes Up-Regulated in Esophageal Carcinomas
| Gene Symbol | Description | Entrez | BE logFC | BE adj.P | EAC logFC | EAC adj.P | ESCC logFC | ESCC adj.P |
|---|---|---|---|---|---|---|---|---|
| BGN | biglycan | 633 | 0,18 | 5,56E−01 | 2,56 | 4,84E−13 | 2,70 | 6,16E-10 |
| C11orf96 | chromosome 11 open reading frame 96 | 387,763 | 2,12 | 3,20E−08 | 4,61 | 9,83E-21 | 3,76 | 2,93E-12 |
| COCH | coagulation factor C homolog, cochlin (Limulus polyphemus) | 1690 | 0,76 | 1,03E-01 | 2,95 | 8,35E-09 | 2,77 | 1,33E-05 |
| COL8A1 | collagen, type VIII, alpha 1 | 1295 | -0,67 | 1,45E-01 | 2,56 | 1,59E-07 | 2,72 | 1,23E-05 |
| CTHRC1 | collagen triple helix repeat containing 1 | 115,908 | 3,00 | 7,35E-10 | 5,31 | 1,50E-19 | 5,50 | 5,21E-15 |
| FOXC1 | forkhead box C1 | 2296 | -1,35 | 2,50E-03 | 1,23 | 4,79E-03 | 2,57 | 1,68E-05 |
| IL8 | interleukin 8 | 3576 | 0,68 | 2,48E-01 | 2,73 | 6,77E-06 | 2,58 | 9,25E-04 |
| INHBA | inhibin, beta A | 3624 | 0,33 | 3,88E-01 | 4,26 | 1,23E-17 | 5,13 | 8,01E-16 |
| LEPREL4 | leprecan-like 4 | 10,609 | 0,99 | 5,50E-04 | 3,03 | 6,17E-17 | 3,15 | 6,56E-13 |
| MMP1 | matrix metallopeptidase 1 (interstitial collagenase) | 4312 | 2,51 | 3,07E-04 | 5,49 | 3,87E-12 | 7,12 | 6,53E-12 |
| MTHFD1L | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like | 25,902 | 0,30 | 3,51E-01 | 2,55 | 3,31E-12 | 2,94 | 2,54E-10 |
| NNMT | nicotinamide N-methyltransferase | 4837 | 2,10 | 1,08E-05 | 4,18 | 4,33E-14 | 4,06 | 1,02E-09 |
| PMEPA1 | prostate transmembrane protein, androgen induced 1 | 56,937 | 1,27 | 7,58E-04 | 4,37 | 1,38E-18 | 3,94 | 4,07E-12 |
| RGS1 | regulator of G-protein signaling 1 | 5996 | 0,05 | 9,16E-01 | 2,89 | 3,07E-09 | 2,77 | 4,90E-06 |
| SOCS3 | suppressor of cytokine signaling 3 | 9021 | 1,02 | 1,70E-02 | 3,27 | 2,75E-11 | 3,89 | 5,91E-10 |
| SPP1 | secreted phosphoprotein 1 / osteopontin | 6696 | 0,02 | 9,73E-01 | 2,89 | 1,26E-06 | 5,57 | 5,96E-11 |
| SULF1 | sulfatase 1 | 23,213 | -0,16 | 7,48E-01 | 3,05 | 1,51E-08 | 3,13 | 4,29E-06 |
| USP42 | ubiquitin specific peptidase 42 | 84,132 | 0,02 | 9,23E-01 | 2,25 | 6,15E-18 | 2,91 | 2,70E-17 |
| WDR72 | WD repeat domain 72 | 256,764 | 1,93 | 7,52E-03 | 4,68 | 3,28E-09 | 5,76 | 1,97E-08 |
| WISP1 | WNT1 inducible signaling pathway protein 1 | 8840 | 0,17 | 6,57E-01 | 2,26 | 4,30E-08 | 3,10 | 1,48E-08 |
Depicted are genes whose expression is strongly up-regulated in esophageal carcinomas in comparison to NE. From left to right: Gene symbol and description; Entrez-ID; BE fold change, fold increase of gene expression in BE vs. healthy samples; BE adjusted P-value; EAC fold change, fold increase of gene expression in esophageal adenocarcinoma vs. healthy samples; EAC adjusted P-value; ESCC fold change, fold increase of gene expression in esophageal squamous carcinoma vs. healthy samples; ESCC adjusted P-value; Note that a log2-fold change is used. P-value, significance of this difference in gene expression.
On the other hand, genes belonging to the GO group epithelium development were down-regulated in BE but also in EAC and ESCC samples when compared to healthy esophageal epithelium. Interestingly, many of the down-regulated peak genes in esophageal cancers (Table 2) belong to the so-called epidermal differentiation complex (EDC) [29]. Other genes strongly down-regulated in EAC and ESCC such as CRTC1 and SCEL belong to the GO-group cornified envelope of terminally differentiated keratinocytes. Expression of these genes is necessary for differentiation frequently turned down in carcinomas. For instance, CRNN, which was found to be significantly down-regulated in esophageal carcinomas, is a member of the EDC. The loss of CRNN expression correlates with longer tumor length, deeper tumor invasion depth, more advanced lymph node metastasis, and poorer survival [30]. Another gene belonging to the EDC is S100A9, which was also strongly down-regulated in both EAC and ESCC. In a proteomic approach, S100A9 was found to be strongly down-regulated in ESCC [31]. Interestingly, also the transglutaminase 3, which is responsible for crosslinking EDC matrix proteins was down-regulated. This means that the formation of the epidermal layer is disturbed in the carcinomas. Similarly SCEL, responsible for the assembly of proteins in the cornified envelope was strongly down-regulated. Recent results that SCEL is necessary for metastatic colorectal cancer tumor growth in the liver [32].
Table 2.
Peak Genes Down-Regulated in Esophageal Carcinomas
| Gene Symbol | Description | Entrez | BE logFC | BE adj.P | EAC logFC | EAC adj.P | ESCC logFC | ESCC adj.P |
|---|---|---|---|---|---|---|---|---|
| ADH7 | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | 131 | −4,03 | 2,26E-13 | −6,12 | 2,28E-21 | −4,24 | 1,98E-10 |
| CRNN | Cornulin | 49,860 | −6,01 | 2,99E-12 | −9,17 | 5,23E-20 | −9,22 | 6,37E− 15 |
| DHRS9 | dehydrogenase/reductase (SDR family) member 9 | 10,170 | -1,51 | 3,37E-03 | −3,84 | 4,12E-11 | −4,45 | 1,86E-09 |
| DSC2 | desmocollin 2 | 1824 | −1,48 | 7,32E-04 | −3,74 | 3,27E-13 | −4,09 | 1,51E-10 |
| DSG3 | desmoglein 3 | 1830 | −5,04 | 4,29E-13 | −7,04 | 2,27E-19 | −3,47 | 8,82E-06 |
| EMP1 | epithelial membrane protein 1 | 2012 | −3,32 | 3,56E-09 | −5,68 | 8,34E-18 | −4,67 | 2,91E-10 |
| FLG | filaggrin | 2312 | −3,94 | 2,75E-09 | −5,99 | 7,96E-16 | −5,72 | 8,26E-11 |
| GRHL3 | grainyhead-like 3 (Drosophila) | 57,822 | −4,56 | 4,93E-17 | −5,33 | 1,77E-20 | −4,17 | 1,81E-11 |
| IL1RN | interleukin 1 receptor antagonist | 3557 | −2,47 | 4,32E-06 | −5,51 | 5,58E-17 | −5,20 | 1,64E-11 |
| KRT13 | keratin 13 | 3860 | −6,47 | 1,69E-09 | −10,04 | 1,72E-16 | −6,59 | 6,13E-07 |
| KRT4 | keratin 4 | 3851 | −5,36 | 1,36E-08 | −8,57 | 1,16E-15 | −8,22 | 9,77E-11 |
| MAL | mal, T-cell differentiation protein | 4118 | −4,30 | 2,94E-09 | −8,95 | 1,01E-21 | −9,01 | 2,42E− 16 |
| MALL | mal, T-cell differentiation protein-like | 7851 | -1,70 | 1,23E-03 | −3,91 | 3,81E-11 | −4,17 | 1,70E-08 |
| S100A8 | S100 calcium binding protein A8 | 6279 | −5,18 | 1,97E-07 | −8,87 | 5,95E-15 | −3,98 | 1,11E-03 |
| S100A9 | S100 calcium binding protein A9 | 6280 | −5,13 | 4,54E-10 | −7,82 | 5,04E-17 | −4,88 | 9,69E-07 |
| SCEL | sciellin | 8796 | −5,39 | 1,46E-11 | −8,30 | 2,71E-19 | −7,72 | 4,33E-13 |
| SERPINB11 | serpin peptidase inhibitor, clade B (ovalbumin), member 11 | 89,778 | −2,69 | 8,06E-15 | −3,03 | 2,51E-17 | −2,89 | 5,76E-12 |
| SERPINB13 | serpin peptidase inhibitor, clade B (ovalbumin), member 13 | 5275 | −6,23 | 5,27E-12 | −8,59 | 9,12E-18 | −5,04 | 1,51E-06 |
| SERPINB2 | serpin peptidase inhibitor, clade B (ovalbumin), member 2 | 5055 | −5,29 | 5,33E-13 | −7,68 | 5,23E-20 | −6,32 | 7,07E-12 |
| SERPINB4 | serpin peptidase inhibitor, clade B (ovalbumin), member 4 | 6318 | −4,76 | 7,70E-08 | −6,95 | 5,53E-13 | −6,15 | 7,70E-08 |
| SPINK7 | serine peptidase inhibitor, Kazal type 7 (putative) | 84,651 | −4,18 | 8,83E-10 | −7,40 | 2,24E-19 | −7,23 | 6,51E-14 |
| SPRR1A | small proline-rich protein 1A | 6698 | −5,76 | 3,10E-08 | −9,65 | 6,86E-16 | −5,21 | 4,41E-05 |
| SPRR1B | small proline-rich protein 1B | 6699 | −5,34 | 3,36E-08 | −8,60 | 3,64E-15 | −3,78 | 1,19E-03 |
| SPRR3 | small proline-rich protein 3 | 6707 | −5,33 | 2,19E-08 | −9,49 | 3,09E-17 | −8,46 | 6,13E-11 |
| TGM3 | transglutaminase 3 | 7053 | −5,73 | 2,54E-14 | −8,39 | 8,01E-22 | −7,90 | 2,55E-15 |
Depicted are genes whose expression is strongly down-regulated in esophageal carcinomas in comparison to NE. From left to right: Gene symbol and description; Entrez-ID; BE fold change, fold decrease of gene expression in BE vs. healthy samples; BE adjusted P-value; EAC fold change, fold decrease of gene expression in esophageal adenocarcinoma vs. healthy samples; EAC adjusted P-value; ESCC fold change, fold decrease of gene expression in esophageal squamous carcinoma vs. healthy samples; ESCC adjusted P-value; Note that a log2-fold change is used. P-value, significance of this difference in gene expression.
Interestingly, also the expression of MAL (T-cell differentiation-related gene) was significantly reduced in both EAC and ESCC (Table 2). MAL promoter hypermethylation correlating with decreased mRNA expression was reported [33] MAL is thought to be a novel suppressor gene [34] with diagnostic value in colorectal and esophageal cancers [35]. The protein has been localized to the endoplasmic reticulum of T-cells and is a candidate linker protein in T-cell signal transduction. The role of MAL for cancer immunity has not been determined yet.
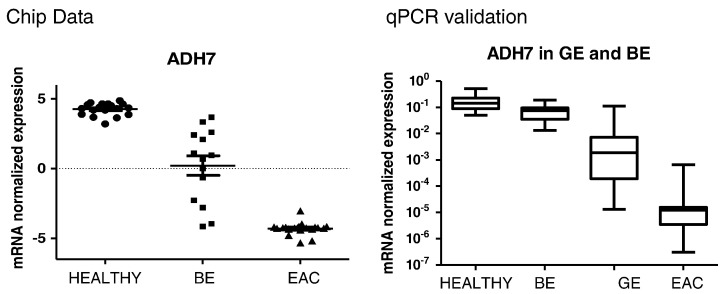
Retinol metabolism was also down-regulated in both types of carcinomas, which is reflected by a strong loss of the expression of ADH7 (Table 2, Figure 3). The enzyme encoded by ADH7 is a member of the alcohol dehydrogenase family. However, it is inefficient in ethanol oxidation, but active as a retinol dehydrogenase. Since retinoic acid regulates the proliferation, differentiation, and apoptosis of premalignant and malignant cells during carcinogenesis [36], it is conceivable that a disturbance of its formation leads to various disorders including malignancy. Moreover, we noted a reduced activity in genes that are related to retinol metabolism (UGT2B15, ADH7, ADH1C, DHRS9, CYP4B1) within EAC compared to BE. These changes may signify a kind of dedifferentiation and indicate that cancer cells are more susceptible to vitamin A insufficiency. Indeed, recent studies indicated that loss of retinoic acid signaling is an early event during esophageal carcinogenesis [37] and that administration of retinoic acid reduces growth of esophageal squamous carcinoma cell lines [38].
Figure 3.
ADH7 – marker for Barrett‘s and Gastric mucosa? Reduced expression of retinol metabolic enzyme ADH7 was found in esophageal adenocarcinoma (EAC) but also in Barrett's esophagus (BE) and already in metaplastic gastric epithelium (GE) of the cardia. Data obtained by microarray analysis (left side) are in agreement with those of RT-PCR of an independent set of samples. Left side: mRNA expression of ADH7 in the training cohort determined by microarray analysis. Gene expression is significantly differential expressed between groups (corrected P-value <0.0001). Data analysis was performed on 18 Healthy, 20 BE and 21 EAC samples. The horizontal axis depicts the three patient groups: Healthy, BE and EAC. Gene expression is presented as normalized (log2 scale) signal intensity. Right side: Relative mRNA expression of ADH7 in the validation cohort. The mRNA expression was examined in 12 samples from Healthy tissue, 20 BE, 8 GE and 10 EAC by means of quantitative real-time PCR. Quantitation is relative to the transcription of GAPDH. Significance in differential expression of individual gene between groups was calculated using Kruskal-Wallis test (P-value <0.001). The horizontal axis shows four patient groups: Healthy, BE, GE and EAC.
In an approach to identify genes, which might be helpful for early diagnosis of esophageal adenocarcinomas, we searched for genes which were strongly up-regulated in EAC but also in BE. Genes like this (Table 3) often have a role for transcriptional activation of other genes such as GATA6, TOX3, PROM or are components of the cell membrane. These genes could be helpful for early diagnosis of cancer in Barrett tissues but their expression could also be increased due to the replacement of the squamous mucosa by columnar epithelium in BE and EAC. Therefore, the expression of the encoded proteins in gastrointestinal carcinomas was checked using the protein atlas database [39]. For instance, GREM1 (gremlin 1, DNA family BMP antagonist), ITLN1 (intelectin 1) and TM4SF4 (transmembrane 4 L six family member 4) were significantly up-regulated in BE and EAC. Expression of these genes was highly homogeneous in all samples of a given tissue and therefore might be of particular value for early diagnosis. Interestingly, the encoded proteins were detected in stomach carcinomas, but not in the respective non-malignant tissues. Similarly, REG4 (Regenerating islet-derived family, member 4) was significantly up-regulated in both BE and even stronger in EAC (Table 3). Colorectal cancers along with few stomach, pancreatic and ovarian cancers showed moderate to strong cytoplasmic immunoreactivity for REG4. Aberrant expression of REG4 was associated with the growth, survival, adhesion and resistance to apoptosis of tumor cells (e.g. [40]). Moreover, REG4 is considered a novel prognostic factor in some gastroenterological carcinomas [41] and was identified as a transcriptional target of GATA6 [42]. Patients suffering from esophageal cancer with an amplification of GATA6 have a poorer survival [43]. In our study, GATA6 was found to be significantly up-regulated in both BE and EAC (Table 3).
Table 3.
Peak Genes Up-Regulated in BE and EAC
| Gene Symbol | Description | Entrez | BE logFC | BE adj.P | EAC logFC | EAC adj.P |
|---|---|---|---|---|---|---|
| AGR2 | anterior gradient 2 homolog (Xenopus laevis) | 10,551 | 7,28 | 4,72E-19 | 6,40 | 4,29E-29 |
| AGR3 | anterior gradient 3 homolog (Xenopus laevis) | 155,465 | 7,12 | 7,63E-24 | 5,44 | 7,67E-19 |
| CENPV | centromere protein V | 201,161 | 4,68 | 1,61E-22 | 5,05 | 1,49E-24 |
| CLRN3 | clarin 3 | 119,467 | 5,02 | 1,31E-10 | 5,82 | 5,09E-13 |
| CTSE | cathepsin E | 1510 | 7,35 | 1,37E-20 | 4,87 | 1,62E-13 |
| GATA6 | GATA binding protein 6 | 2627 | 7,21 | 2,07E-26 | 6,76 | 2,71E-25 |
| GDA | guanine deaminase | 9615 | 4,67 | 4,08E-15 | 4,69 | 1,33E-15 |
| GOLM1 | golgi membrane protein 1 | 51,280 | 7,37 | 1,57E-35 | 6,01 | 1,83E-30 |
| LGALS4 | lectin, galactoside-binding, soluble, 4 | 3960 | 8,14 | 2,31E-37 | 7,16 | 4,47E-34 |
| MUC13 | mucin 13, cell surface associated | 56,667 | 4,46 | 2,43E-16 | 4,88 | 1,92E-18 |
| OLFM4 | olfactomedin 4 | 10,562 | 6,74 | 2,10E-09 | 6,07 | 2,08E-08 |
| PIGR | polymeric immunoglobulin receptor | 5284 | 8,49 | 2,89E-27 | 7,14 | 8,31E-24 |
| PIP5K1B | phosphatidylinositol-4-phosphate 5-kinase, type I, beta | 8395 | 5,82 | 1,52E-25 | 5,13 | 3,55E-23 |
| POSTN | periostin, osteoblast specific factor | 10,631 | 5,95 | 2,32E-22 | 5,17 | 6,02E-20 |
| PROM1 | prominin 1 | 8842 | 6,47 | 4,89E-26 | 6,02 | 8,39E-25 |
| REG4 | regenerating islet-derived family, member 4 | 83,998 | 6,75 | 7,04E-13 | 5,10 | 2,70E-09 |
| RHPN2 | rhophilin, Rho GTPase binding protein 2 | 85,415 | 4,83 | 2,76E-25 | 5,91 | 2,79E-30 |
| SPINK1 | serine peptidase inhibitor, Kazal type 1 | 6690 | 7,11 | 2,55E-21 | 6,55 | 4,01E-20 |
| SULT1C2 | sulfotransferase family, cytosolic, 1C, member 2 | 6819 | 8,03 | 8,34E-24 | 5,67 | 2,76E-17 |
| TFF1 | trefoil factor 1 | 7031 | 8,50 | 8,34E-24 | 5,53 | 8,74E-16 |
| TMC5 | transmembrane channel-like 5 | 79,838 | 6,42 | 2,15E-28 | 6,33 | 2,20E-28 |
| TMPRSS3 | transmembrane protease, serine 3 | 64,699 | 3,75 | 4,90E-13 | 4,72 | 2,51E-17 |
| TOX3 | TOX high mobility group box family member 3 | 27,324 | 6,64 | 1,42E-34 | 6,10 | 1,83E-32 |
| TSPAN8 | tetraspanin 8 | 7103 | 8,71 | 2,36E-43 | 7,91 | 6,74E-41 |
| VCAN | versican | 1462 | 3,63 | 1,25E-14 | 4,63 | 1,59E-19 |
Depicted are the genes whose expression is strongly up-regulated in Barrett's esophagus (BE) as well as in esophageal adenocarcinoma (EAC) in comparison to NE. From left to right: Gene symbol and description; Entrez-ID; BE fold change, fold increase of gene expression in BE vs. healthy samples; BE adjusted P-value; EAC fold change, fold increase of gene expression in esophageal adenocarcinoma vs. healthy samples; EAC adjusted P-value; Note that a log2-fold change is used. P-value, significance of this difference in gene expression.
Several researchers have investigated gene expression in Barrett's esophagus. Greenawalt et al. [44] found that SOCS3, MMP3 and MMP10 were up-regulated in ESCC and EAC, and that SERPINB2, SEPINB3 and SPINK5 were down-regulated in esophageal carcinomas, which is in accordance with our data. Interestingly, they also searched for genes up-regulated in BE and found an increased expression of TFF1, LGALS4 and GATA6 (Table 3). Botelho et al. [45] searched for gene expression using paraffin-embedded Barrett's esophagus tissue samples. They also found down-regulation of ADH7 in esophageal carcinomas and an up-regulation of TSPAN8 in BE (Table 2, Table 3). Hyland et al. [46] performed gene expression analysis s of Barrett's esophagus and matched normal mucosa. Ten of the top 50 genes in their list of differentially expressed genes in BE vs. NE belong to the 25 peak genes depicted in Table 3. Warnecke-Eberz et al. [19] proposed a diagnostic signature of 19 genes for esophageal carcinomas of both subtypes that contained at least 3 genes of Table 1.
Recently, the Cancer Genome Atlas Research Network performed a comprehensive molecular analysis of esophagus carcinomas derived from Western and Eastern populations [47]. The previously detected amplifications of GATA4 and GATA6 genes were confirmed. Interestingly, an up-regulation of the p63 transcription network only in ESCC was reported. Accordingly, we found an up-regulation of TP63 in ESCC (0.67 log2fold vs. NE) in comparison to EAC (−5.6 log2fold) and BE (−4.6 log2fold). However, we noted a down-regulation of the EDC genes in esophageal carcinomas in comparison to NE (Table 2). EDC genes are thought to be target genes of p63 [44].
In summary, a database of gene expression in BE and esophageal carcinomas was generated which can be queried in different ways, e.g. in order to find carcinoma-associated markers with differential expression in both esophageal squamous cell carcinomas and adenocarcinomas. Another aim is to identify a subset of markers that may allow detecting patients at risk for esophageal adenocarcinoma at a preneoplastic stage. Of course, there are many other kinds of possible queries, e.g. related to networks of genes, which are affected by the changes in differentiation during cancer development. The examples given here are intended to show that by using this database genes or groups of related genes can be identified which might be helpful for early diagnosis of esophageal carcinomas or for treatment and monitoring of this disease. The next step is to kick off a retrospective study with more patients and long follow-up in order to evaluate the changes in gene expression identified in our experimental approach.
Conclusions
Our study provides high quality data, e.g. in order to determine a subset of markers for the identification of patients at risk for esophageal adenocarcinoma at a preneoplastic stage or markers which may lead to a better disease management.
Acknowledgements
The authors thank Mrs. Sabine Grigull, Mrs. Gudrun Koch and Mrs Claudia Roefzaad for excellent technical assistance.
Footnotes
Funding: This work was supported by grants from Charité University Hospital and Otto-von-Guericke University Magdeburg, Germany.
Disclaimers: The authors indicated no potential conflicts of interest.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2017.10.003.
Appendix A. Supplementary data
Supplementary material
References
- 1.Spechler SJ, Souza RF. Barrett's esophagus. N Engl J Med. 2014;371(9):836–845. doi: 10.1056/NEJMra1314704. [DOI] [PubMed] [Google Scholar]
- 2.Wang KK, Sampliner RE, Practice Parameters Committee of the American College of G Updated guidelines 2008 for the diagnosis, surveillance and therapy of Barrett's esophagus. Am J Gastroenterol. 2008;103(3):788–797. doi: 10.1111/j.1572-0241.2008.01835.x. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez-Plaza JJ, Hulak N, Garcia-Fuentes E, Garrido-Sanchez L, Zhumadilov Z, Akilzhanova A. Oesophageal squamous cell carcinoma (ESCC): Advances through omics technologies, towards ESCC salivaomics. Drug Discov Ther. 2015;9(4):247–257. doi: 10.5582/ddt.2015.01042. [DOI] [PubMed] [Google Scholar]
- 4.Reid BJ, Levine DS, Longton G, Blount PL, Rabinovitch PS. Predictors of progression to cancer in Barrett's esophagus: baseline histology and flow cytometry identify low- and high-risk patient subsets. Am J Gastroenterol. 2000;95(7):1669–1676. doi: 10.1111/j.1572-0241.2000.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paulson TG, Maley CC, Li X, Li H, Sanchez CA, Chao DL, Odze RD, Vaughan TL, Blount PL, Reid BJ. Chromosomal instability and copy number alterations in Barrett's esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15(10):3305–3314. doi: 10.1158/1078-0432.CCR-08-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardie LJ, Darnton SJ, Wallis YL, Chauhan A, Hainaut P, Wild CP, Casson AG. p16 expression in Barrett's esophagus and esophageal adenocarcinoma: association with genetic and epigenetic alterations. Cancer Lett. 2005;217(2):221–230. doi: 10.1016/j.canlet.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 7.Yang H, Gu J, Wang KK, Zhang W, Xing J, Chen Z, Ajani JA, Wu X. MicroRNA expression signatures in Barrett's esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15(18):5744–5752. doi: 10.1158/1078-0432.CCR-09-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cronin J, McAdam E, Danikas A, Tselepis C, Griffiths P, Baxter J, Thomas L, Manson J, Jenkins G. Epidermal growth factor receptor (EGFR) is overexpressed in high-grade dysplasia and adenocarcinoma of the esophagus and may represent a biomarker of histological progression in Barrett's esophagus (BE) Am J Gastroenterol. 2011;106(1):46–56. doi: 10.1038/ajg.2010.433. [DOI] [PubMed] [Google Scholar]
- 9.Prasad GA, Bansal A, Sharma P, Wang KK. Predictors of progression in Barrett's esophagus: current knowledge and future directions. Am J Gastroenterol. 2010;105(7):1490–1502. doi: 10.1038/ajg.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bucca G, Carruba G, Saetta A, Muti P, Castagnetta L, Smith CP. Gene expression profiling of human cancers. Ann N Y Acad Sci. 2004;1028:28–37. doi: 10.1196/annals.1322.003. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Ma C, Kemmner W. Wdr66 is a novel marker for risk stratification and involved in epithelial-mesenchymal transition of esophageal squamous cell carcinoma. BMC Cancer. 2013;13:137. doi: 10.1186/1471-2407-13-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehlker M, Huska MR, Jons T, Andrade-Navarro MA, Kemmner W. Concerted down-regulation of immune-system related genes predicts metastasis in colorectal carcinoma. BMC Cancer. 2014;14:64. doi: 10.1186/1471-2407-14-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kretschmer C, Sterner-Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Mol Cancer. 2011;10(1):15. doi: 10.1186/1476-4598-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 16.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1):1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Rubie C, Kempf K, Hans J, Su T, Tilton B, Georg T, Brittner B, Ludwig B, Schilling M. Housekeeping gene variability in normal and cancerous colorectal, pancreatic, esophageal, gastric and hepatic tissues. Mol Cell Probes. 2005;19(2):101–109. doi: 10.1016/j.mcp.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Warnecke-Eberz U, Metzger R, Holscher AH, Drebber U, Bollschweiler E. Diagnostic marker signature for esophageal cancer from transcriptome analysis. Tumour Biol. 2016;37(5):6349–6358. doi: 10.1007/s13277-015-4400-4. [DOI] [PubMed] [Google Scholar]
- 20.Liu M, Hu Y, Zhang MF, Luo KJ, Xie XY, Wen J, Fu JH, Yang H. MMP1 promotes tumor growth and metastasis in esophageal squamous cell carcinoma. Cancer Lett. 2016;377(1):97–104. doi: 10.1016/j.canlet.2016.04.034. [DOI] [PubMed] [Google Scholar]
- 21.Murray GI. Matrix metalloproteinases: a multifunctional group of molecules. J Pathol. 2001;195(2):135–137. doi: 10.1002/1096-9896(200109)195:2<135::AID-PATH939>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 22.Seder CW, Hartojo W, Lin L, Silvers AL, Wang Z, Thomas DG, Giordano TJ, Chen G, Chang AC, Orringer MB. INHBA overexpression promotes cell proliferation and may be epigenetically regulated in esophageal adenocarcinoma. J Thorac Oncol. 2009;4(4):455–462. doi: 10.1097/JTO.0b013e31819c791a. [DOI] [PubMed] [Google Scholar]
- 23.Yoshinaga K, Mimori K, Yamashita K, Utsunomiya T, Inoue H, Mori M. Clinical significance of the expression of activin A in esophageal carcinoma. Int J Oncol. 2003;22(1):75–80. [PubMed] [Google Scholar]
- 24.Kim HC, Kim YS, Oh HW, Kim K, Oh SS, Kim JT, Kim BY, Lee SJ, Choe YK, Kim DH. Collagen triple helix repeat containing 1 (CTHRC1) acts via ERK-dependent induction of MMP9 to promote invasion of colorectal cancer cells. Oncotarget. 2014;5(2):519–529. doi: 10.18632/oncotarget.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eriksson J, Le Joncour V, Nummela P, Jahkola T, Virolainen S, Laakkonen P, Saksela O, Holtta E. Gene expression analyses of primary melanomas reveal CTHRC1 as an important player in melanoma progression. Oncotarget. 2016;7(12):15065–15092. doi: 10.18632/oncotarget.7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rae FK, Hooper JD, Nicol DL, Clements JA. Characterization of a novel gene, STAG1/PMEPA1, upregulated in renal cell carcinoma and other solid tumors. Mol Carcinog. 2001;32(1):44–53. doi: 10.1002/mc.1063. [DOI] [PubMed] [Google Scholar]
- 27.Brunschwig EB, Wilson K, Mack D, Dawson D, Lawrence E, Willson JK, Lu S, Nosrati A, Rerko RM, Swinler S. PMEPA1, a transforming growth factor-beta-induced marker of terminal colonocyte differentiation whose expression is maintained in primary and metastatic colon cancer. Cancer Res. 2003;63(7):1568–1575. [PubMed] [Google Scholar]
- 28.Watanabe Y, Itoh S, Goto T, Ohnishi E, Inamitsu M, Itoh F, Satoh K, Wiercinska E, Yang W, Shi L. TMEPAI, a transmembrane TGF-beta-inducible protein, sequesters Smad proteins from active participation in TGF-beta signaling. Mol Cell. 2010;37(1):123–134. doi: 10.1016/j.molcel.2009.10.028. [DOI] [PubMed] [Google Scholar]
- 29.Kypriotou M, Huber M, Hohl D. The human epidermal differentiation complex: cornified envelope precursors, S100 proteins and the 'fused genes' family. Exp Dermatol. 2012;21(9):643–649. doi: 10.1111/j.1600-0625.2012.01472.x. [DOI] [PubMed] [Google Scholar]
- 30.Hsu PK, Kao HL, Chen HY, Yen CC, Wu YC, Hsu WH, Chou TY. Loss of CRNN expression is associated with advanced tumor stage and poor survival in patients with esophageal squamous cell carcinoma. J Thorac Cardiovasc Surg. 2014;147(5):1612–1618.e4. doi: 10.1016/j.jtcvs.2013.09.066. [DOI] [PubMed] [Google Scholar]
- 31.Pawar H, Srikanth SM, Kashyap MK, Sathe G, Chavan S, Singal M, Manju HC, Kumar KV, Vijayakumar M, Sirdeshmukh R. Downregulation of S100 Calcium Binding Protein A9 in Esophageal Squamous Cell Carcinoma. ScientificWorldJournal. 2015;2015:325721. doi: 10.1155/2015/325721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou CK, Fan CC, Lin PS, Liao PY, Tung JC, Hsieh CH, Hung MC, Chen CH, Chang WC. Sciellin mediates mesenchymal-to-epithelial transition in colorectal cancer hepatic metastasis. Oncotarget. 2016;7(18):25742–25754. doi: 10.18632/oncotarget.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Z, Wang L, Zhang Y, Cheng Y, Gao Y, Feng X, Dong M, Cao Z, Chen S, Yu H. MAL hypermethylation is a tissue-specific event that correlates with MAL mRNA expression in esophageal carcinoma. Sci Rep. 2013;3:2838. doi: 10.1038/srep02838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mimori K, Nishida K, Nakamura Y, Ieta K, Yoshikawa Y, Sasaki A, Ishii H, Alonso MA, Mori M. Loss of MAL expression in precancerous lesions of the esophagus. Ann Surg Oncol. 2007;14(5):1670–1677. doi: 10.1245/s10434-006-9064-2. [DOI] [PubMed] [Google Scholar]
- 35.Buffart TE, Overmeer RM, Steenbergen RD, Tijssen M, van Grieken NC, Snijders PJ, Grabsch HI, van de Velde CJ, Carvalho B, Meijer GA. MAL promoter hypermethylation as a novel prognostic marker in gastric cancer. Br J Cancer. 2008;99(11):1802–1807. doi: 10.1038/sj.bjc.6604777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lotan R. Retinoids and chemoprevention of aerodigestive tract cancers. Cancer Metastasis Rev. 1997;16(3-4):349–356. doi: 10.1023/a:1005808429176. [DOI] [PubMed] [Google Scholar]
- 37.Qiu H, Zhang W, El-Naggar AK, Lippman SM, Lin P, Lotan R, Xu XC. Loss of retinoic acid receptor-beta expression is an early event during esophageal carcinogenesis. Am J Pathol. 1999;155(5):1519–1523. doi: 10.1016/s0002-9440(10)65467-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller A, Nakagawa H, Rustgi AK. Retinoic acid and N-(4-hydroxy-phenyl) retinamide suppress growth of esophageal squamous carcinoma cell lines. Cancer Lett. 1997;113(1-2):95–101. doi: 10.1016/s0304-3835(97)04601-6. [DOI] [PubMed] [Google Scholar]
- 39.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 40.Ohara S, Oue N, Matsubara A, Mita K, Hasegawa Y, Hayashi T, Usui T, Amatya VJ, Takeshima Y, Kuniyasu H. Reg IV is an independent prognostic factor for relapse in patients with clinically localized prostate cancer. Cancer Sci. 2008;99(8):1570–1577. doi: 10.1111/j.1349-7006.2008.00846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Numata M, Oshima T. Significance of regenerating islet-derived type IV gene expression in gastroenterological cancers. World J Gastroenterol. 2012;18(27):3502–3510. doi: 10.3748/wjg.v18.i27.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawasaki Y, Matsumura K, Miyamoto M, Tsuji S, Okuno M, Suda S, Hiyoshi M, Kitayama J, Akiyama T. REG4 is a transcriptional target of GATA6 and is essential for colorectal tumorigenesis. Sci Rep. 2015;5:14291. doi: 10.1038/srep14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin L, Bass AJ, Lockwood WW, Wang Z, Silvers AL, Thomas DG, Chang AC, Lin J, Orringer MB, Li W. Activation of GATA binding protein 6 (GATA6) sustains oncogenic lineage-survival in esophageal adenocarcinoma. Proc Natl Acad Sci U S A. 2012;109(11):4251–4256. doi: 10.1073/pnas.1011989109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20(22):3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Botelho NK, Schneiders FI, Lord SJ, Freeman AK, Tyagi S, Nancarrow DJ, Hayward NK, Whiteman DC, Lord RV. Gene expression alterations in formalin-fixed, paraffin-embedded Barrett esophagus and esophageal adenocarcinoma tissues. Cancer Biol Ther. 2010;10(2):172–179. doi: 10.4161/cbt.10.2.12166. [DOI] [PubMed] [Google Scholar]
- 46.Hyland PL, Hu N, Rotunno M, Su H, Wang C, Wang L, Pfeiffer RM, Gherman B, Giffen C, Dykes C. Global changes in gene expression of Barrett's esophagus compared to normal squamous esophagus and gastric cardia tissues. PLoS One. 2014;9(4):e93219. doi: 10.1371/journal.pone.0093219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cancer Genome Atlas Research N, Analysis Working Group: Asan U, Agency BCC, Brigham, Women's H, Broad I, Brown U, Case Western Reserve U, Dana-Farber Cancer I, Duke U Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material