Abstract Abstract
Ganoderma is a cosmopolitan white rot fungal genus, famous for its medicinal properties. In the present study, two new Ganoderma species were collected from south-eastern China and described on the basis of morphological characters and phylogenetic analyses of sequences of the internal transcribed spacer (ITS) region, the translation elongation factor 1-α gene (EF1-α) and the second subunit of RNA polymerase II (RPB2). Specimens of both species were found on living trees of Casuarina equisetifolia. Ganoderma angustisporum sp. nov. is characterised by its sessile basidiomata and almond-shaped, slightly truncate, narrow basidiospores (9–11.3 × 4–5.2 µm). Ganoderma casuarinicola sp. nov. is characterised by its strongly laccate reddish-brown pileal surface, luminous yellow to yellowish-brown cutis and ellipsoid, truncate basidiospores (9–10.2 × 5–6 µm). The two new species are compared with their related taxa. Phylogenetic analyses confirmed that G. angustisporum and G. casuarinicola are distinct species within Ganoderma.
Keywords: Ganodermataceae, medicinal mushroom, morphology, phylogeny, taxonomy, wood-rotting fungi
Introduction
Ganoderma P. Karst. is easily recognised by its characteristic appearance, double-walled and truncate basidiospores (Karsten 1881; Moncalvo and Ryvarden 1997). According to Christian (2015), there are 428 names in the Index Fungorum (http://www.indexfungorum.org/) and 456 records of taxa (420 with status legitimate) in MycoBank (http://www.mycobank.org/). Ganoderma products as dietary supplement are very popular in Asia, especially in China and China is very rich in Ganoderma species (Zhao and Zhang 2000; Wang et al. 2009; Cao et al. 2012; Cao and Yuan 2013; Li et al. 2014b). The great variability in macroscopic characters of basidiomes has resulted in a large number of synonyms and confusions in the taxonomy of this genus. Using DNA sequence data for the identification of Ganoderma species is of greatest importance.
Some Ganoderma species are well known for causing wood decay in a wide range of tree species around the world. For example, G. boninense Pat. is a causal agent of oil palm basal stem rot and is responsible for considerable yield losses in southeast Asian oil palm plantations (Pilotti 2005). Especially in Indonesia and Malaysia, G. boninense and G. philippii (Bres. & Henn. ex Sacc.) Bres. cause great economic loss of palm oil, tea and rubber (Zakaria et al. 2009).
Casuarina equisetifolia Forst. is used as an industrial raw material and wood fuel, as well as for conservation of coastal ecosystems and for agricultural land protection against salinity intrusion (Hossain et al. 1998; Chowdhury et al. 2009). In China, Casuarina equisetifolia is widely planted in the coastal areas of Guangxi, Guangdong, Fujian, Hainan and Taiwan provinces. During collections of wood-rotting fungi in South China in recent years, two Ganoderma species growing on Casuarina equisetifolia, which could not be identified to any known species, were collected. Those two species are here described based on morphological characters and phylogenetic analyses.
Materials and methods
Morphological studies
The examined specimens were deposited in the herbarium of the Institute of Microbiology, Beijing Forestry University (BJFC). Macro-morphological descriptions were based on field notes. Special colour terms followed Petersen (1996). Micro-morphological data were obtained from the dried specimens and observed under a light microscope following Li et al. (2014a) and Han et al. (2016). Sections were studied at a magnification of up to 1000× using a Nikon E 80i microscope and phase contrast illumination. Drawings were made with the aid of a drawing tube. Microscopic features, measurements and drawings were made from slide preparations stained with Cotton Blue and Melzer’s reagent. Spores were measured from sections cut from the tubes. To represent variation in the size of basidiospores, 5% of measurements were excluded from each end of the range and extreme values are given in parentheses.
The following abbreviations are used: IKI = Melzer’s reagent, IKI– = neither amyloid nor dextrinoid, KOH = 5% potassium hydroxide, CB = Cotton Blue, CB+ = cyanophilous, Q is an average computed by dividing the length by the width of each spore separately, n (a,b) = a spores measured from b specimens.
Molecular study and phylogenetic analysis
The CTAB rapid plant genome extraction kit-DN14 (Aidlab Biotechnologies Co. Ltd., Beijing, China) was used to extract total genomic DNA from dried specimens according to the manufacturer’s instructions with some modifications (Chen et al. 2016, 2017). The genes ITS, EF1-α and RPB2 were amplified by polymerase chain reaction (PCR) technique. The ITS region was amplified with primer pair ITS5 (GGA AGT AAA AGT CGT AAC AAG G) and ITS4 (TCC TCC GCT TAT TGA TAT GC) (White et al. 1990). Part of the EF1-α gene was amplified with primer pair EF1-983F (GCY CCY GGH CAY CGT GAY TTY AT) and EF1-1567R (ACH GTR CCR ATA CCA CCR ATC TT) (Rehner and Buckley 2005). Part of the RPB2 gene was amplified with primer pairs 5F (GAY GAY MGW GAT CAY TTY GG) and 7CR (CCC ATR GCT TGY TTR CCC AT) (Liu et al. 1999). The PCR cycling for ITS was as follows: initial denaturation at 95 °C for 3 min, followed by 35 cycles at 94 °C for 40 s, 54 °C for 45 s and 72 °C for 1 min and a final extension of 72 °C for 10 min. The PCR cycling for EF1-α was as follows: initial denaturation at 95 °C for 3 min, followed by 34 cycles at 94 °C for 40 s, 56 °C for 45 s and 72 °C for 1 min and a final extension of 72 °C for 10 min. The PCR cycling for RPB2 was as follows: initial denaturation at 95 °C for 5 min, followed by 35 cycles at 95 °C for 1 min, 58 °C for 2 min and 72 °C for 1.5 min and a final extension of 72 °C for 10 min. The PCR products were purified and sequenced at Beijing Genomics Institute (China), using forward and reverse PCR primers. All newly generated sequences were deposited in GenBank (Table 1).
Table 1.
Species, specimens, geographic origin and GenBank accession numbers of sequences used in this study.
| Species name | Voucher no. | Geographic origin | GenBank accession numbers | References | ||
|---|---|---|---|---|---|---|
| ITS | EF1-α | RPB2 | ||||
| Ganoderma angustisporum | Cui 13817 (holotype) | Fujian, China | MG279170* | MG367563* | MG367507* | this study |
| G. angustisporum | Cui 14578 | Guangdong, China | MG279171* | MG367564* | – | this study |
| G. angustisporum | Cui 16340 | Guangxi, China | MG279172* | – | – | this study |
| G. aridicola | Dai 12588 (holotype) | Durban, South Africa | KU572491 | KU572502 | – | Xing et al. 2016 |
| G. boninense | WD 2028 | Japan | KJ143905 | KJ143924 | KJ143964 | Zhou et al. 2014 |
| G. boninense | WD 2085 | Japan | KJ143906 | KJ143925 | KJ143965 | Zhou et al. 2014 |
| G. casuarinicola | Dai 16336 (holotype) | Guangdong, China | MG279173* | MG367565* | MG367508* | this study |
| G. casuarinicola | Dai 16337 | Guangdong, China | MG279174* | MG367566* | MG367509* | this study |
| G. casuarinicola | Dai 16338 | Guangdong, China | MG279175* | MG367567* | MG367510* | this study |
| G. casuarinicola | Dai 16339 | Guangdong, China | MG279176* | MG367568* | MG367511* | this study |
| G. curtisii | CBS 100131 | NC, USA | JQ781848 | KJ143926 | KJ143966 | Zhou et al. 2014 |
| G. curtisii | CBS 100132 | NC, USA | JQ781849 | KJ143927 | KJ143967 | Zhou et al. 2014 |
| G. destructans | CBS 139793 (type) | Pretoria, South Africa | NR132919 | – | – | Coetzee et al. 2015 |
| G. destructans | CMW 43670 | Pretoria, South Africa | KR183856 | – | – | Coetzee et al. 2015 |
| G. destructans | Dai 16431 | South Africa | MG279177* | MG367569* | MG367512* | this study |
| G. enigmaticum | CBS 139792 (type) | Pretoria, South Africa | NR132918 | – | – | Coetzee et al. 2015 |
| G. enigmaticum | Dai 15970 | Africa | KU572486 | KU572496 | MG367513* | Xing et al. 2016; this study |
| G. enigmaticum | Dai 15971 | Africa | KU572487 | KU572497 | MG367514* | Xing et al. 2016; this study |
| G. heohnelianum | Dai 11995 | Yunnan, China | KU219988 | MG367550* | MG367497* | Song et al. 2016b; this study |
| G. heohnelianum | Yuan 6337 | Guangxi, China | MG279160* | MG367551* | MG367498* | this study |
| G. heohnelianum | Cui 13982 | Guangxi, China | MG279178* | MG367570* | MG367515* | this study |
| G. leucocontextum | GDGM 44489 | Xizang, China | KM396271 | – | – | Li et al. 2014b |
| G. leucocontextum | GDGM 44490 | Xizang, China | KM396272 | – | – | Li et al. 2014b |
| G. leucocontextum | Dai 15601 | Xizang, China | KU572485 | KU572495 | MG367516* | Xing et al. 2016; this study |
| G. lingzhi | Wu 1006-38 (holotype) | Hubei, China | JQ781858 | JX029976 | JX029980 | Cao et al. 2012 |
| G. lingzhi | Cui 14342 | Sichuan, China | MG279179* | MG367571* | MG367517* | this study |
| G. lingzhi | Cui 14375 | Sichuan, China | MG279180* | MG367572* | MG367518* | this study |
| G. lobatum | JV 1008/31 | USA | KF605671 | MG367553* | MG367499* | this study |
| G. lobatum | JV 1008/32 | USA | KF605670 | MG367554* | MG367500* | this study |
| G. lucidum | K 175217 | UK, Europe | KJ143911 | KJ143929 | KJ143971 | Zhou et al. 2014 |
| G. lucidum | Cui 14404 | Sichuan, China | MG279181* | MG367573* | MG367519* | this study |
| G. lucidum | Cui 14405 | Sichuan, China | MG279182* | MG367574* | MG367520* | this study |
| G. martinicense | LIP SW-Mart08-44 | Martinica | KF963257 | – | – | Welti and Courtecuisse 2010 |
| G. martinicense | LIP SW-Mart08-55 (type) | Martinica | KF963256 | – | – | Welti and Courtecuisse 2010 |
| G. martinicense | He 2240 | USA | MG279163* | MG367557* | MG367503* | this study |
| G. multipileum | CWN 04670 | Taiwan, China | KJ143913 | KJ143931 | KJ143972 | Zhou et al. 2014 |
| G. multipileum | Dai 9447 | Hainan, China | KJ143914 | – | KJ143973 | Zhou et al. 2014 |
| G. multipileum | Cui 14373 | Sichuan, China | MG279184* | MG367575* | MG367521* | this study |
| G. multiplicatum | SPC9 | Brazil | KU569553 | – | – | Bolaños et al. 2016 |
| G. multiplicatum | 60119011 | Brazil | MG279185* | – | – | this study |
| G. multiplicatum | URM 83346 | Brazil | JX310823 | – | – | Bolaños et al. 2016 |
| G. orbiforme | Cui 13918 | Hainan, China | MG279186* | MG367576* | MG367522* | this study |
| G. orbiforme | Cui 13880 | Hainan, China | MG279187* | MG367577* | MG367523* | this study |
| G. philippii | Cui 14443 | Hainan, China | MG279188* | MG367578* | MG367524* | this study |
| G. philippii | Cui 14444 | Hainan, China | MG279189* | MG367579* | MG367525* | this study |
| G. resinaceum | Rivoire 4150 | France, Europe | KJ143915 | – | – | Zhou et al. 2014 |
| G. resinaceum | CBS 194.76 | Netherlands, Europe | KJ143916 | KJ143934 | – | Zhou et al. 2014 |
| G. ryvardenii | HKAS 58053 (type) | Cameroon, Africa | HM138671 | – | – | Kinge and Mih 2011 |
| G. ryvardenii | HKAS 58054 | Cameroon, Africa | HM138672 | – | – | Kinge and Mih 2011 |
| G. ryvardenii | HKAS 58055 | Cameroon, Africa | HM138670 | – | – | Kinge and Mih 2011 |
| G. shandongense | Dai 15785 | Shandong, China | MG279190* | MG367580* | MG367526* | this study |
| G. shandongense | Dai 15787 | Shandong, China | MG279191* | MG367581* | MG367527* | this study |
| G. shandongense | Dai 15791 | Shandong, China | MG279192* | MG367582* | MG367528* | this study |
| G. sinense | Wei 5327 | Hainan, China | KF494998 | KF494976 | MG367529* | this study |
| G. sinense | Cui 13835 | Hainan, China | MG279193* | MG367583* | MG367530* | this study |
| G. tropicum | He 1232 | Guangxi, China | KF495000 | MG367584* | MG367531* | this study |
| G. tropicum | Yuan 3490 | Yunnan, China | JQ781880 | KJ143938 | – | Cao et al. 2012 |
| G. tropicum | Dai 16434 | Hainan, China | MG279194* | MG367585* | MG367532* | this study |
| G. tsugae | Dai 12751b | CT, USA | KJ143919 | KJ143939 | KJ143977 | Zhou et al. 2014 |
| G. tsugae | Cui 14110 | Jilin, China | MG279195* | MG367586* | MG367533* | this study |
| G. tsugae | Cui 14112 | Jilin, China | MG279196* | MG367587* | MG367534* | this study |
| G. weberianum | CBS 219.36 | Philippines | JQ520219 | – | – | Zhou et al. 2014 |
| G. williamsianum | Wei 5032 | Hainan, China | KU219994 | – | – | Song et al. 2016b |
| G. williamsianum | Dai 16809 | Thailand | MG279183* | MG367588* | MG367535* | this study |
| G. zonatum | FL-02 | FL, USA | KJ143921 | KJ143941 | KJ143979 | Zhou et al. 2014 |
| G. zonatum | FL-03 | FL, USA | KJ143922 | KJ143942 | KJ143980 | Zhou et al. 2014 |
| Outgroup | ||||||
| Amauroderma rugosum | Cui 9011 | Guangdong, China | KJ531664 | KU572504 | MG367506* | Li and Yuan 2015; this study |
| Tomophagus colossus | TC-02 | Vietnam | KJ143923 | KJ143943 | – | Zhou et al. 2014 |
* Newly generated sequences for this study.
Bold names = new species.
Besides the sequences generated from this study, other reference sequences were selected from GenBank for phylogenetic analyses. Sequences were aligned in MAFFT 6 (Katoh and Toh 2008; http://mafft.cbrc.jp/alignment/server/) using the “G-INS-I” strategy and manually adjusted in BioEdit (Hall 1999). Sequence alignment was deposited in TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S22403; submission ID 22403). Amauroderma rugosum (Blume & T. Nees) Torrend and Tomophagus colossus (Fr.) C.F. Baker were selected as outgroups.
Phylogenetic analyses in this study followed the approach of Song et al. (2016a) and Song and Cui (2017). The maximum likelihood (ML) and Bayesian inference (BI) methods were used to analyse the combined dataset of ITS, EF1-α and RPB2 sequences. ML analysis was conducted with RAxML-HPC252 on the Cipres Science Gateway (Miller and Pfeiffer 2011) involved 100 ML searches; all model parameters were estimated by the programme. The ML bootstrap values (ML-BS) were obtained with 1000 rapid bootstrapping replicates. BI was performed with MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003), with a mixed model partition. A suitable substitution model for each partition of the dataset was determined using the Akaike Information Criterion implemented in MrMODELTEST 2.3. Four Markov chains were run from the random starting tree for 1 million generations to make the average standard deviation of split deviation frequencies lower than 0.01. Trees were sampled every 100 generations. The burn-in was set to discard the first 25% of the trees. A majority rule consensus tree of all the remaining trees was used to calculate Bayesian posterior probabilities (BPP). The ML and BI algorithms generated congruent topologies in main lineages; thus, only the topology from the ML algorithm was presented along with BS and BPP greater than 75% and 0.95, respectively, at the nodes.
Results
Phylogenetic analysis
The combined ITS, EF1-α and RPB2 dataset included sequences from 66 fungal samples representing 27 taxa. The selected models were K80 for 5.8S, K80 + G for ITS1, HKY + G for ITS2, GTR + I + G for ITS1+ ITS2 + 5.8S. The best model selected and applied in the BI analysis for the combined ITS, EF1-α and RPB2 partition was a GTR+I+G model. BI analysis and ML analysis resulted in the same topology with an average standard deviation of split frequencies = 0.006025 (BI).
Taxonomy
Ganoderma angustisporum
J.H. Xing, B.K. Cui & Y.C. Dai sp. nov.
MB823320
Figure 2.
Basidiomata of Ganoderma species. a, b G. angustisporum (Cui 13817) c, d G. casuarinicola (Dai 16336). Scale bars: 2 cm.
Figure 3.
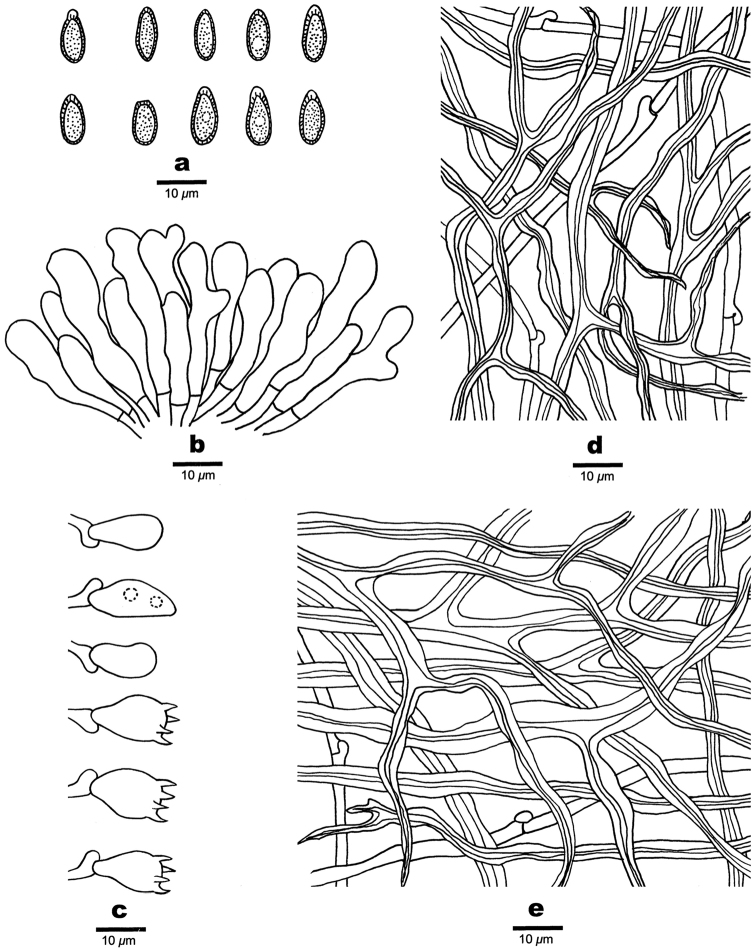
Microscopic structures of Ganoderma angustisporum (drawn from the holotype). a Basidiospores b Apical cells from the pellis c Basidia and basidioles d Hyphae from context e Hyphae from trama. Scale bars: 10 µm.
Diagnosis.
Ganoderma angustisporum is characterised by its sessile basidiomata, white pore surface, almond-shaped, slightly truncate and narrow basidiospores.
Holotype.
CHINA. Fujian Prov., Pingtan County, on living tree of Casuarina equisetifolia, 18 August 2016, Cui 13817 (BJFC!).
Etymology.
angustisporum (Lat.): referring to the narrow basidiospores.
Description.
Basidiomes annual, sessile and broadly attached, applanate, shell-shaped, projecting up to 13.5 cm, 10 cm wide and 1.1 cm thick at base, corky when fresh, becoming hard corky to woody hard upon drying. Pileal surface strongly laccate, reddish-brown to dark brown, with a thin crust, concentrically zonate or azonate; margin distinct, slightly obtuse. Pore surface white when fresh, turning light buff when dry; pores round to angular, 3–5 per mm; dissepiments slightly thick to thick, entire. Context corky, homogeneous, greyish-brown, bearing distinct concentric growth zones, black melanoid band present, up to 0.4 cm thick. Tubes woody hard, greyish-brown, up to 0.7 cm long. Hyphal system trimitic; generative hyphae bearing clamp connections; all the hyphae IKI–, CB+; tissues darkening in KOH. Pellis: pellis cells regularly arranged into a palisade; terminal cells clavate, yellowish to pale brown, thin-walled, occasionally with blunt outgrowth and protuberance in the apical or lateral parts, bearing a simple septum at base, moderately amyloid at maturity, 15–33 × 4–10 μm. Context generative hyphae colourless, thin-walled, bearing clamp connections, unbranched, 2–4.5 µm in diam; skeletal hyphae dominant, pale yellowish-brown, thick-walled to subsolid, frequently branched, interwoven, 3–6 µm in diam; binding hyphae abundant, pale yellowish-brown, thick-walled with a narrow lumen to subsolid, frequently branched, tortuous, interwoven, 1–2.5 µm in diam. Tubes generative hyphae colourless, thin-walled, bearing clamp connections, unbranched, slightly swollen at the distal end, 2–2.8 μm in diam; skeletal hyphae dominant, pale brown to distinctly brown, thick-walled with a medium or narrow lumen to subsolid, frequently branched, strongly interwoven, 3–4.5 μm in diam; binding hyphae brownish-yellow, thick-walled to almost solid, frequently branched, interwoven, 1–1.8 μm in diam. Basidia barrel-shaped, yellowish to pale brown, with a clamp connection and four sterigmata, 11–16 × 6.5–9 µm; basidioles pear-shaped to fusiform, 8–15 × 5–8 µm. Basidiospores mostly almond-shaped at maturity, slightly truncate, yellowish to pale brown, IKI–, CB+, double-walled, exospore smooth, endospore with coarse echinulate, (8–)9–10.5(–11) × (3.5–)4–5 µm, L = 8.89 μm, W = 4.27 μm, Q = 2.01–2.24 (n = 60/2, with the turgid vesicular appendix excluded); (8–)9–11.3(–12) × (3.8–)4–5.2 µm, L = 10.26 μm, W = 4.31 μm, Q = 2.36–2.4 (n = 60/2, with the turgid vesicular appendix included).
Type of rot.
A white rot.
Additional specimens examined.
CHINA. Guangdong Prov., Maoming, Dianbai, on living trees of Casuarina equisetifolia, 20 June 2017, Cui 14578, Cui 16494 and Cui 16495 (BJFC!).
Ganoderma casuarinicola
J.H. Xing, B.K. Cui & Y.C. Dai sp. nov.
MB823321
Figure 4.
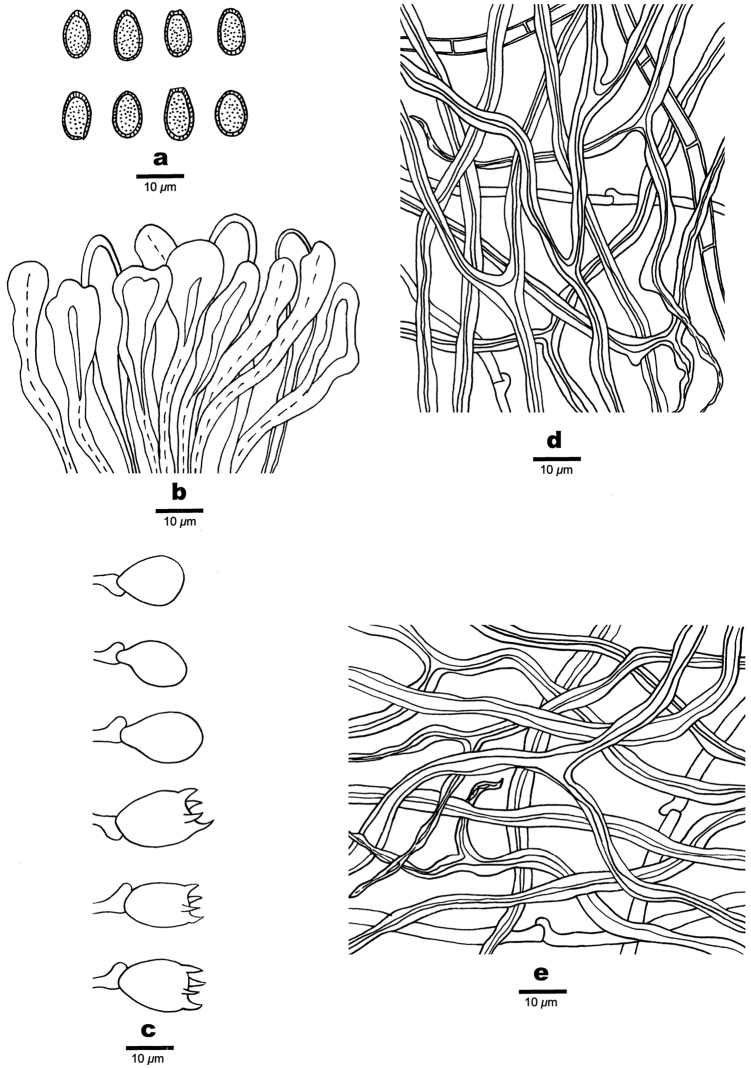
Microscopic structures of Ganoderma casuarinicola (drawn from the holotype). a Basidiospores b Apical cells from the cuticle c Basidia and basidioles d Hyphae from context e Hyphae from trama. Scale bars: 10 µm.
Diagnosis.
Ganoderma casuarinicola is characterised by its strongly laccate reddish-brown pileal surface, white pore surface, luminous yellow to yellowish-brown cutis.
Holotype.
CHINA. Guangdong Prov. Zhanjiang, Dianbai, on living tree of Casuarina equisetifolia, 4 October 2015, Dai 16336 (BJFC!).
Etymology.
casuarinicola (Lat.): referring to the host tree genus Casuarina.
Description.
Basidiomes annual, stipitate to substipitate, pileus sectorial to shell-shaped, projecting up to 10 cm, 7 cm wide and 2 cm thick at base, corky, without odour when fresh, becoming hard corky to woody hard when dry. Pileal surface strongly laccate, reddish-brown, with a thin crust; margin obtuse, cream to reddish-brown. Stipe flattened or subcylindrical, lateral, reddish-brown, up to 6 cm long and 1.7 cm in diam. Pore surface white when fresh, turning cream when dry; pores round to angular, 4–6 per mm; dissepiments thin to slightly thick, entire. Context corky, heterogeneous, the upper layer generally light yellow up to 0.1 cm thick and the lower layer generally dark brown close to the tubes up to 1 cm thick, showing distinct concentric growth zones, black melanoid band absent. Tubes woody hard, greyish-brown, up to 0.9 cm long. Hyphal system trimitic; generative hyphae bearing clamp connections, occasionally with simple septa; all the hyphae IKI–, CB+; tissues darkening in KOH. Pellis: Pellis cells regularly arranged into a palisade; terminal cells clavate, luminous yellow to yellowish-brown, thick-walled, occasionally expanded at the apex, moderately amyloid at maturity, 40–70 × 5–13 μm. Context generative hyphae colourless, thin-walled, with clamp connections, occasionally branched, 2–4 µm in diam; skeletal hyphae dominant, pale yellowish-brown, thick-walled to subsolid, frequently branched, interwoven, 3–5.5 µm in diam; binding hyphae abundant, pale yellowish-brown, thick-walled with a narrow lumen to subsolid, frequently branched, tortuous, interwoven, 1–3 µm in diam. Tubes generative hyphae colourless, thin-walled, mostly bearing clamp connections, occasionally with simple septa, occasionally branched, slightly swollen at the distal end, 1.5–3 μm in diam; skeletal hyphae dominant, pale brown to distinctly brown, thick-walled with a medium or narrow lumen to subsolid, frequently branched, strongly interwoven, 2–4.5 μm in diam; binding hyphae brownish-yellow, thick-walled to almost solid, frequently branched, interwoven, 1.5–2.5 μm in diam. Basidia barrel-shaped, yellowish to pale brown, with a clamp connection and four sterigmata, 12–18 × 9.5–13 µm; basidioles pear-shaped, 9–16 × 8–12 µm. Basidiospores mostly ellipsoid at maturity, truncate, yellowish to pale brown, IKI–, CB+, double-walled, exospore smooth, endospore with coarse echinulate, (8–)8.5–9 (–10) × (4.2–)5.5–6.5(–7) µm, L = 8.82 μm, W = 5.65 μm, Q = 1.52–1.60 (n = 60/2, with the turgid vesicular appendix excluded); (8.3–)9–10.2(–11.5) × (4.5–)5–6(–7) µm, L = 9.85 μm, W = 5.77 μm, Q = 1.68–1.72 (n = 60/2, with the turgid vesicular appendix included).
Type of rot.
a white rot.
Additional specimens examined.
CHINA. Guangdong Prov., Zhanjiang, Dianbai, on living trees of Casuarina equisetifolia, 4 October 2015, Dai 16337, Dai 16338, Dai 16339, Dai 17892, Cui 16370, Cui 16376 and Cui 16377 (BJFC!).
Discussion
The two new Ganoderma species were found on living trees of Casuarina equisetifolia from the southeast coast of China. However, they are quite different from each other in morphology. Their main morphological differences are presented in Table 2. Phylogenetically, the two new species grouped together with some other laccate Ganoderma species in a well-supported clade.
Table 2.
Morphological differences between the two new Ganoderma species collected on Casuarina from China.
| Species | Pileal surface | Context | Cuticle cells | Shape of basidiospores | Size of basidiospores |
|---|---|---|---|---|---|
| G. angustisporum | reddish brown to dark brown | homogeneous, black melanoid band present | thin-walled, septate | almond-shaped | (8–)9–10.5(–11) × (3.5–)4–5 µm (with the turgid vesicular appendix excluded) (8–)9–11.3(–12) × (3.8–)4–5.2 µm (with the turgid vesicular appendix included) |
| G. casuarinicola | reddish brown | not fully homogeneous, black melanoid band absent | thick-walled to subsolid, non-septate | ellipsoid | (8–)8.5–9 (–10) × (4.2–)5.5–6.5(–7) µm (with the turgid vesicular appendix excluded)(8.3–)9–10.2(–11.5) × (4.5–)5–6(–7) µm (with the turgid vesicular appendix included) |
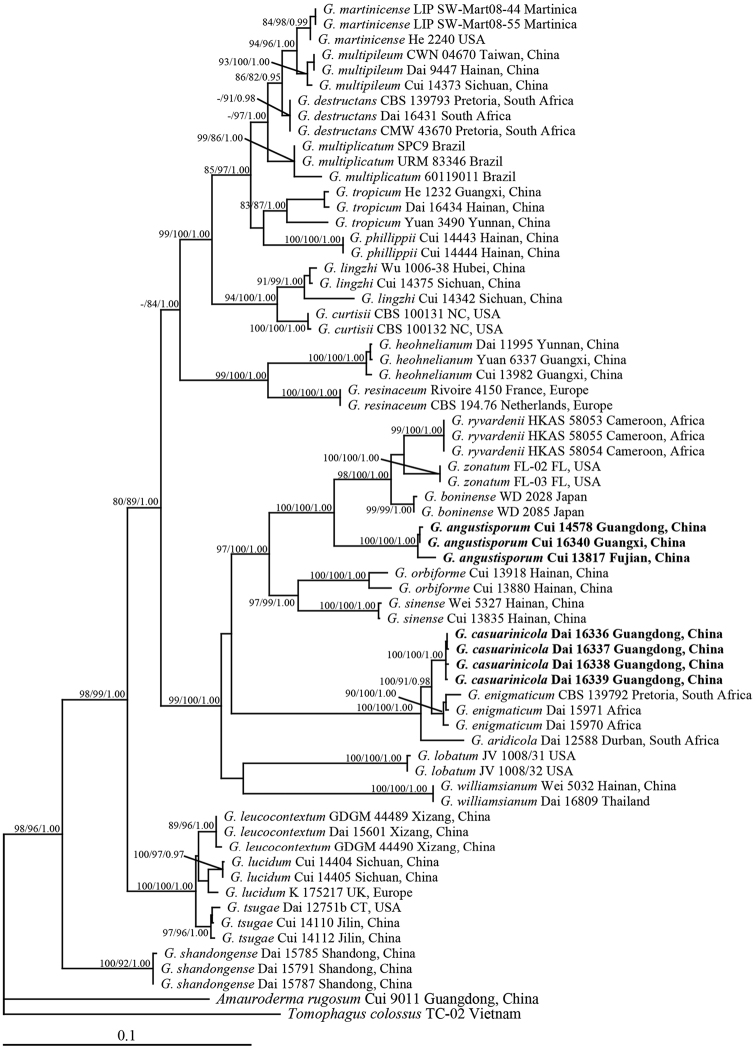
In the phylogenetic tree inferred from ITS, EF1-α and RPB2 sequences, G. angustisporum clustered together with G. boninense, G. ryvardenii Tonjock & Mih and G. zonatum Murrill, these four species forming a strong support (BS = 100%, BPP =1.00; Fig. 1) lineage and could be distinctly separated from each other in the tree. Morphologically, all these four species produce laccate and sessile basidiomata, but G. angustisporum has the narrowest basidiospores amongst so far accepted Ganoderma species. Ganoderma boninense is another species producing relatively narrow basidiospores, but its basidiospores (8.7–12.8 × 4.7–6 μm, Zhou et al. 2014) are slightly wider than G. angustisporum. Moreover, its pileal surface ranges from orange to reddish-brown, even to fuscous or almost black and it lacks concentric growth zones in the context. Ganoderma ryvardenii differs from G. angustisporum by its larger pores (2–4 per mm), reddish basidiomata with waved margin and wider basidiospores (10–13 × 6–8 μm; Kinge and Mih 2011). Ganoderma zonatum mainly differs by larger basidiospores (10–12 × 5.3–6.3 μm, Zhou et al. 2014) and the absence of black melanoid band in the context.
Figure 1.
Phylogeny of the new Ganoderma species and related taxa based on ITS+EF1-α+RPB2 sequence data. Branches are labelled with bootstrap values (ML) higher than 75%, and posterior probabilities (BI) higher than 0.95. Bold names = new species.
In the phylogenetic tree, we obtained G. casuarinicola as sister to G. enigmaticum M.P.A. Coetzee, Marinc. & M.J. Wingf., a species described from South Africa, but morphologically, G. enigmaticum can be easily distinguished from G. casuarinicola by its golden yellow pileal surface without furrows and narrower basidiospores (8–11 × 3.5–6 μm, Coetzee et al. 2015). These two species gathered together with another South Africa species G. aridicola J.H. Xing & B.K. Cui, but G. aridicola is a sessile species with dark brown to black pileal surface, while G. casuarinicola has a reddish-brown pileal surface. Besides, G. casuarinicola has smaller basidia than G. aridicola (15–25 × 8–11 μm, Xing et al. 2016). Morphologically, G. casuarinicola resembles G. tsugae Murrill in having a reddish-brown pileal surface, white pore surface, similar wide ellipsoid basidiospores and lacking the black melanoid band in the context, but G. tsugae mainly differs by the absence of a light yellow layer under the laccate crust and concentric growth zones in the context (Zhou et al. 2014) and they are distinct from each other in the phylogenetic tree (Fig. 1). Besides, G. tsugae grows exclusively on conifers, especially on Tsuga, Abies and Larix, while G. casuarinicola occurs on hardwoods.
In conclusion, both morphology and phylogeny inferred from the combined ITS, EF1-α and RPB2 sequences support that the specimens, collected on living trees of Casuarina equisetifolia from the southeast coast of China, are two new species within the Ganoderma genus.
Supplementary Material
Acknowledgments
The research was financed by the National Natural Science Foundation of China (Project No. 31670016) and the Fundamental Research Funds for the Central Universities (Project No. 2016ZCQ04).
Citation
Xing J-H, Sun Y-F, Han Y-L, Cui B-K, Dai Y-C (2018) Morphological and molecular identification of two new Ganoderma species on Casuarina equisetifolia from China. MycoKeys 34: 93–108. https://doi.org/10.3897/mycokeys.34.22593
References
- Bolaños AC, Bononi VLR, Gugliotta ADM. (2016) New records of Ganoderma multiplicatum (Mont.) Pat. (Polyporales, Basidiomycota) from Colombia and its geographic distribution in South America. Check List 12: 1948. https://doi.org/10.15560/19236
- Cao Y, Wu SH, Dai YC. (2012) Species clarification of the prize medicinal Ganoderma mushroom “Lingzhi”. Fungal Diversity 56: 49–62. https://doi.org/10.1007/s13225-012-0178-5 [Google Scholar]
- Cao Y, Yuan HS. (2013) Ganoderma mutabile sp. nov. from southwestern China based on morphological and molecular data. Mycol Prog 12: 121–126. https://doi.org/10.1007/s11557-012-0819-9 [Google Scholar]
- Chen JJ, Cui BK, Dai YC. (2016) Global diversity and molecular systematics of Wrightoporia s. l. (Russulales, Basidiomycota). Persoonia 37: 21–36. https://doi.org/10.3767/003158516X689666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, Wu F, Wang M, Cui BK. (2017) Species diversity and molecular systematics of Fibroporia (Polyporales, Basidiomycota) and its related genera. Mycol Prog 16: 521–533. https://doi.org/10.1007/s11557-017-1285-1 [Google Scholar]
- Chowdhury MQ, Ishiguri F, Iizuka K, Takashima Y, Matsumoto K, Hiraiwa T, Ishido M, Sanpe H, Yokota S, Yoshizawa N. (2009) Radial variations of wood properties in Casuarina equisetifolia growing in Bangladesh. Journal of Wood Science 55: 139–143. https://doi.org/10.1007/s10086-008-1004-2 [Google Scholar]
- Coetzee MPA, Marincowitz S, Muthelo VG, Wingfield MJ. (2015) Ganoderma species, including new taxa associated with root rot of the iconic Jacaranda mimosifolia in Pretoria, South Africa. IMA Fungus 6: 249–256. https://doi.org/10.5598/imafungus.2015.06.01.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- Han ML, Chen YY, Shen LL, Song J, Vlasák J, Dai YC, Cui BK. (2016) Taxonomy and phylogeny of the brown-rot fungi: Fomitopsis and its related genera. Fungal Divers 80: 343–373. https://doi.org/10.1007/s13225-016-0364-y [Google Scholar]
- Hossain MK, Akhter S, Riadh SM. (1998) Effect of polybag size on initial growth of Casuarina equisetifolia seedlings in the nursery. Chittagong University Journal of Science 22: 43–46. [Google Scholar]
- Karsten PA. (1881) Enumeralio boletinearum et polyporearum fennicarum, systemate novo dispositarum. Revue Mycologique 3: 16–19. [Google Scholar]
- Katoh K, Toh H. (2008) Recent developments in the MAFFT multiple sequence alignment program. Briefings In Bioinformatics 9: 286–298. https://doi.org/10.1093/bib/bbn013 [DOI] [PubMed] [Google Scholar]
- Kinge TR, Mih AM. (2011) Ganoderma ryvardense sp. nov. associated with basal stem rot (BSR) disease of oil palm in Cameroon. Mycosphere 2: 179–188. [Google Scholar]
- Li HJ, Cui BK, Dai YC. (2014a) Taxonomy and multi-gene phylogeny of Datronia (Polyporales, Basidiomycota). Persoonia 32: 170–182. https://doi.org/10.3767/003158514X681828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Yuan HS. (2015) Type studies on Amauroderma species described by J.D. Zhao et al. and the phylogeny of species in China. Mycotaxon 130: 79–89. http://dx.doi.org/10.5248/130.79 [Google Scholar]
- Li TH, Hu HP, Deng WQ, Wu SH, Wang DM, Tsering T. (2014b) https://doi.org/10.1016/j.myc.2014.03.005
- Liu YL, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol 16: 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Moncalvo JM, Ryvarden L. (1997) A nomenclatural study of the Ganodermataceae Donk. Synopsis Fungorum 11: 1–114. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2011) The CIPRES science gateway: a community resource for phylogenetic analyses. Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery. ACM, 41. https://doi.org/10.1145/2016741.2016785
- Petersen JH. (1996) Farvekort. The Danish Mycological Society´s colour-chart. Foreningen til Svampekundskabens Fremme, Greve.
- Pilotti CA. (2005) Stem rots of oil palm caused by Ganoderma boninense: pathogen biology and epidemiology. Mycopathologia 159: 129–137. https://doi.org/10.1007/s11046-004-4435-3 [DOI] [PubMed] [Google Scholar]
- Rehner SA, Buckley E. (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-a sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. https://doi.org/10.3852/mycologia.97.1.84 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. https://doi.org/10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Song J, Cui BK. (2017) Phylogeny, divergence time and historical biogeography of Laetiporus (Basidiomycota, Polyporales). BMC Evol Biol 17: 102. https://doi.org/10.1186/s12862-017-0948-5 [DOI] [PMC free article] [PubMed]
- Song J, Chen JJ, Wang M, Chen YY, Cui BK. (2016a) Phylogeny and biogeography of the remarkable genus Bondarzewia (Basidiomycota, Russulales). Scientific Reports 6: 34568. https://doi.org/10.1038/srep34568 [DOI] [PMC free article] [PubMed]
- Song J, Xing JH, Decock C, He XL, Cui BK. (2016b) Molecular phylogeny and morphology reveal a new species of Amauroderma (Basidiomycota) from China. Phytotaxa 260: 47. https://doi.org/10.11646/phytotaxa.260.1.5
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. https://doi.org/10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DM, Wu SH, Li TH. (2009) Two records of Ganoderma new to mainland China. Mycotaxon 108: 35–40. https://doi.org/10.5248/108.35 [Google Scholar]
- Welti S, Courtecuisse R. (2010) The Ganodermataceae, in the French West Indies (Guadeloupe and Martinique). Fungal Diversity 43: 103–126. https://doi.org/10.1007/s13225-010-0036-2 [Google Scholar]
- White TJ, Bruns TD, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR protocols, a guide to methods and applications. Academic Press, San Diego, 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Xing JH, Song J, Decock C, Cui BK. (2016) Morphological characters and phylogenetic analysis reveal a new species within the Ganoderma lucidum complex from South Africa. Phytotaxa 266: 115–124. https://doi.org/10.11646/phytotaxa.266.2.5 [Google Scholar]
- Zakaria L, Ali NS, Salleh B. (2009) Molecular analysis of Ganoderma species from different hosts in Peninsular Malaysia. Journal of Biological Sciences 9: 12–20. https://doi.org/10.3923/jbs.2009.12.20 [Google Scholar]
- Zhao JD, Zhang XQ. (2000) Flora Fungorum Sinicorum 18. Ganodermataceae. Science Press, Beijing. [In Chinese]
- Zhou LW, Cao Y, Wu SH, Vlasák J, Li DW, Li MJ, Dai YC. (2014) Global diversity of the Ganoderma lucidum complex (Ganodermataceae, Polyporales) inferred from morphology and multilocus phylogeny. Phytochemistry 114: 7–15. https://doi.org/10.1016/j.phytochem.2014.09.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.