ABSTRACT
Human adenovirus (HAdV) E1B-55K is a multifunctional regulator of productive viral replication and oncogenic transformation in nonpermissive mammalian cells. These functions depend on E1B-55K's posttranslational modification with the SUMO protein and its binding to HAdV E4orf6. Both early viral proteins recruit specific host factors to form an E3 ubiquitin ligase complex that targets antiviral host substrates for proteasomal degradation. Recently, we reported that the PML-NB-associated factor Daxx represses efficient HAdV productive infection and is proteasomally degraded via a SUMO-E1B-55K-dependent, E4orf6-independent pathway, the details of which remained to be established. RNF4, a cellular SUMO-targeted ubiquitin ligase (STUbL), induces ubiquitinylation of specific SUMOylated proteins and plays an essential role during DNA repair. Here, we show that E1B-55K recruits RNF4 to the insoluble nuclear matrix fraction of the infected cell to support RNF4/Daxx association, promoting Daxx PTM and thus inhibiting this antiviral factor. Removing RNF4 from infected cells using RNA interference resulted in blocking the proper establishment of viral replication centers and significantly diminished viral gene expression. These results provide a model for how HAdV antagonize the antiviral host responses by exploiting the functional capacity of cellular STUbLs. Thus, RNF4 and its STUbL function represent a positive factor during lytic infection and a novel candidate for future therapeutic antiviral intervention strategies.
IMPORTANCE Daxx is a PML-NB-associated transcription factor that was recently shown to repress efficient HAdV productive infection. To counteract this antiviral measurement during infection, Daxx is degraded via a novel pathway including viral E1B-55K and host proteasomes. This virus-mediated degradation is independent of the classical HAdV E3 ubiquitin ligase complex, which is essential during viral infection to target other host antiviral substrates. To maintain a productive viral life cycle, HAdV E1B-55K early viral protein inhibits the chromatin-remodeling factor Daxx in a SUMO-dependent manner. In addition, viral E1B-55K protein recruits the STUbL RNF4 and sequesters it into the insoluble fraction of the infected cell. E1B-55K promotes complex formation between RNF4- and E1B-55K-targeted Daxx protein, supporting Daxx posttranslational modification prior to functional inhibition. Hence, RNF4 represents a novel host factor that is beneficial for HAdV gene expression by supporting Daxx counteraction. In this regard, RNF4 and other STUbL proteins might represent novel targets for therapeutic intervention.
KEYWORDS: STUbL, Daxx, E1B-55K, HAdV, PML-NB, RNF4, SUMO, ubiquitin, human adenovirus
INTRODUCTION
PTM (posttranslational modification) of substrate proteins with ubiquitin or SUMO (small ubiquitin-like modifier) has been shown to regulate a diverse number of cellular processes, including proteasomal protein degradation, transcription factor activity, nuclear/cytoplasmic shuttling, and DDR (DNA damage response) (1, 2). Intriguingly, several pathogens have evolved strategies to take advantage of the cellular ubiquitin and SUMO machinery, either by modulating essential viral proteins or restricting cellular protein functions by PTM (3).
HAdV (human adenoviruses) counteract cellular antiviral responses by producing the E1B-55K (early region 1B, 55 kDa) protein that targets cellular proteins, such as Mre11, p53, DNA ligase IV, Tip60, integrin α3, ATRX, and SPOC1, for proteasomal degradation in cooperation with the E4orf6 (early region 4 open reading frame 6) protein. Together with a variety of host factors, such as Cullin5, Rbx1/RCO1/Hrt1, and elongins B/C, they assemble an SCF-like E3 ubiquitin ligase complex (3, 4).
We reported previously that the transcriptional repressor Daxx (death domain-associated protein) represents a negative regulator of HAdV5 gene expression during productive infection (5–10). Daxx is mainly found in the nucleus, associated with PML-NBs (PML nuclear bodies), or at heterochromatin areas in a complex with ATRX (X-linked α-thalassemia retardation syndrome protein) (2, 11). PML-NB association of Daxx was found to alleviate gene repression and activate apoptosis, while chromatin-bound Daxx acts in a transcriptionally repressive manner (12–14). Daxx association with either PML-NBs or chromatin depends on the status of the host cell and on the interaction of Daxx with other nuclear proteins (e.g., PML and ATRX), which can be regulated by PTM. It was also observed that cell cycle-dependent phosphorylation regulates the exit of Daxx from PML-NBs prior to assembly to ATRX and chromatin-associated proteins, like histone deacetylases, acetylated histone H4, and DEK, at condensed chromatin regions (15–19).
We demonstrated that the functional Daxx/ATRX chromatin-remodeling complexes in the nucleus of infected cells efficiently repress HAdV replication. These data provide evidence that chromatin-modulating proteins play a major role in host cell intrinsic defense mechanisms against HAdV. To oppose this repression, this virus antagonizes ATRX protein concentrations by proteasomal degradation via the E1B-55K/E4orf6 E3 ubiquitin ligase complex during productive infection (20). In addition, we also discovered that E1B-55K alone inhibits the innate antiviral activities of Daxx by targeting this cellular protein for proteasomal degradation via a so-far unknown proteasome-dependent pathway independent of E4orf6 (20). These findings illustrate the importance of E1B-55K in processes blocking innate antiviral activities.
The cellular RNF4 (RING-finger protein 4) protein is a member of the STUbL (SUMO-targeted ubiquitin ligases) protein family (2). STUbL proteins bind via SUMO-SIM interaction on SUMO-conjugated factors and thereby promote ubiquitinylation and proteasomal degradation of SUMO-modified target proteins (21). RNF4 contains four functional SIM regions in the N terminus and a RING domain at the C-terminal region, which is responsible for dimerization and activation of the ligase activity (22). Importantly, RNF4 mediates the ubiquitinylation, and thus proteasomal degradation, of SUMO-modified PML, the scaffolding factor of PML-NBs (21, 23–25). Since PML-NBs are involved in virus infection, it is not surprising that RNF4 severely affects viral life cycles (21).
Here, we show that RNF4 is sequestered into the insoluble nuclear matrix fraction of the host cell during HAdV infection. This relocalization is mediated by RNF4 interaction with E1B-55K, independently of the SIM and ARM regions in the host protein. Furthermore, we provide evidence that E1B-55K connects RNF4 with the anti-HAdV transcription factor Daxx to simultaneously modulate Daxx PTM. This block of antiviral capacity is supported by the finding that HAdV gene expression is reduced in RNF4-depleted cells. Taken together, our data demonstrate that E1B-55K together with RNF4 foster Daxx PTM most presumably prior to Daxx proteasomal degradation during HAdV infection; thus, RNF4 expression is favorable for viral gene expression and replication.
RESULTS
HAdV infection sequesters RNF4 STUbL into the insoluble matrix fraction of the host cell nucleus.
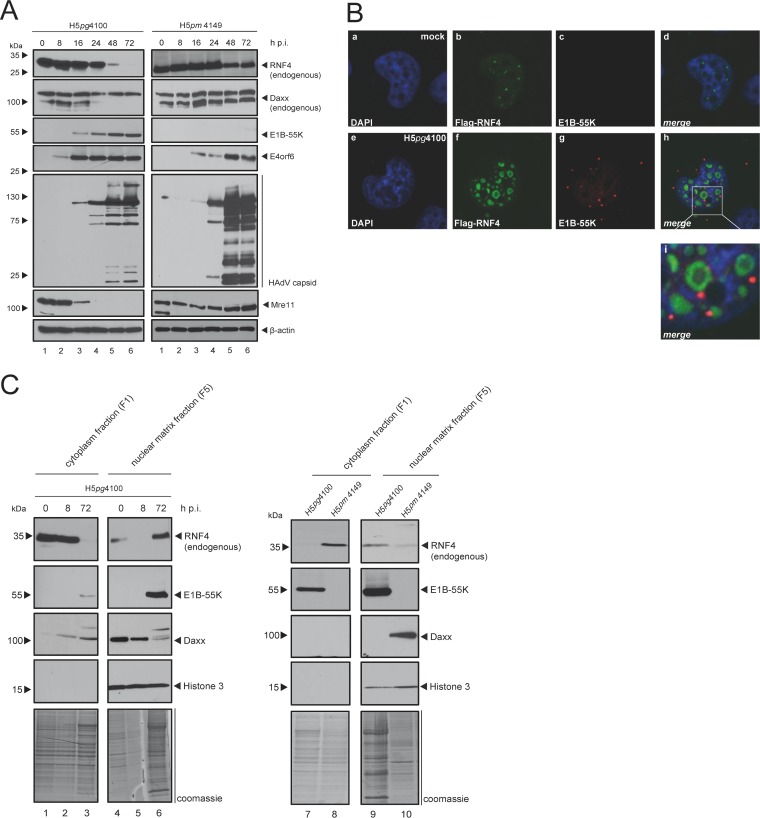
Since host DNA damage repair (DDR) impedes viability and propagation of DNA viruses, HAdV efficiently target a multitude of host cell DDR repair regulatory factors, such as Mre11 and SPOC1, in order to promote productive infection (26). RNF4 plays a critical role in the cellular response to DNA double-strand breaks (DSB), prompting us to examine RNF4 protein levels in low-salt radioimmunoprecipitation assay (RIPA) extracts from infected human cells (Fig. 1A, left). We observed that RNF4 levels are reproducibly reduced in soluble extracts 48 h after wild-type (wt) infection (H5pg4100). Moreover, we confirmed that Mre11 protein levels are also significantly decreased during HAdV infection. This protein is a component of the MRN repair complex and represents a classical target of the adenoviral E3 ubiquitin ligase complex containing E1B-55K, E4orf6, and additional host determinants (5, 7–9, 27, 28). In parallel, we also observed that the PML-NB-associated Daxx protein levels are decreased during HAdV infection. This is consistent with earlier findings demonstrating that SUMOylated E1B-55K binds and sequesters Daxx into the proteasomal pathway of the cell by a mechanism still not understood in detail (8). We note that the Daxx antibody also detects a higher migrating unspecific band, which we refer to as unspecific (see Fig. 5F).
FIG 1.
HAdV-mediated modulation of RNF4 protein during infection. (A) H1299 cells were infected with either wt virus (H5pg4100, left) or with an E1B-55K null mutant (H5pm4149, right) at a multiplicity of 50 FFU per cell and harvested after indicated time points postinfection. Total cell extracts were prepared with low-salt RIPA buffer, separated by SDS-PAGE, and subjected to immunoblotting using RNF4 mouse MAb (kindly provided by Takeshi Urano), Daxx rabbit pAb 07-471 (Upstate), mouse MAb 2A6 (α-E1B-55K), E4orf6 mouse MAb RSA3, HAdV-5 rabbit polyclonal serum L133, Mre11 rabbit pAb pNB 100-142 (Novus Biologicals, Inc.), rabbit MAb α-E2A (α-E2A/DBP), and MAb AC-15 (anti-β-actin) as a loading control. Molecular sizes, in kDa, are indicated on the left, with relevant proteins on the right. (B) H1299 cells transfected with 2 μg pFlag-RNF4-WT, infected with wt virus (H5pg4100) at a multiplicity of 20 FFU per cell, and fixed with 4% PFA after 48 h postinfection. Cells were labeled with anti-Flag mouse MAb M2 (Sigma-Aldrich, Inc.), detected with Alexa 488 (α-Flag; green) and mouse MAb 2A6 (α-E1B-55K), detected with Cy3 (α-E1B-55K; red)-conjugated secondary antibody. Nuclei are labeled with DAPI (4,6-diamidino-2-phenylindole). Representative α-Flag (green; Bb, Bf), α-E1B-55K (red; Bc, Bg), and DAPI (blue; Ba, Be) staining patterns, an overlay of the single images (merge; Bd, Bh), and an enlarged overlay (merge; Bi) are shown (magnification, ×7,600). (C) H1299 cells were infected with wt virus (H5pg4100, left and right) or an E1B-55K null mutant (H5pm4149, right) at a multiplicity of 50 FFU per cell and harvested after indicated time points postinfection (left) or after 48 h (right). Cell extracts were fractionated into cytoplasm and insoluble nuclear factions. Equivalent amounts of protein for each fraction were separated by SDS-PAGE and subjected to immunoblotting using the Ab indicated in panel A plus rabbit MAb H3 (α-histone 3). Molecular sizes, in kDa, are indicated on the left, and relevant proteins are on the right.
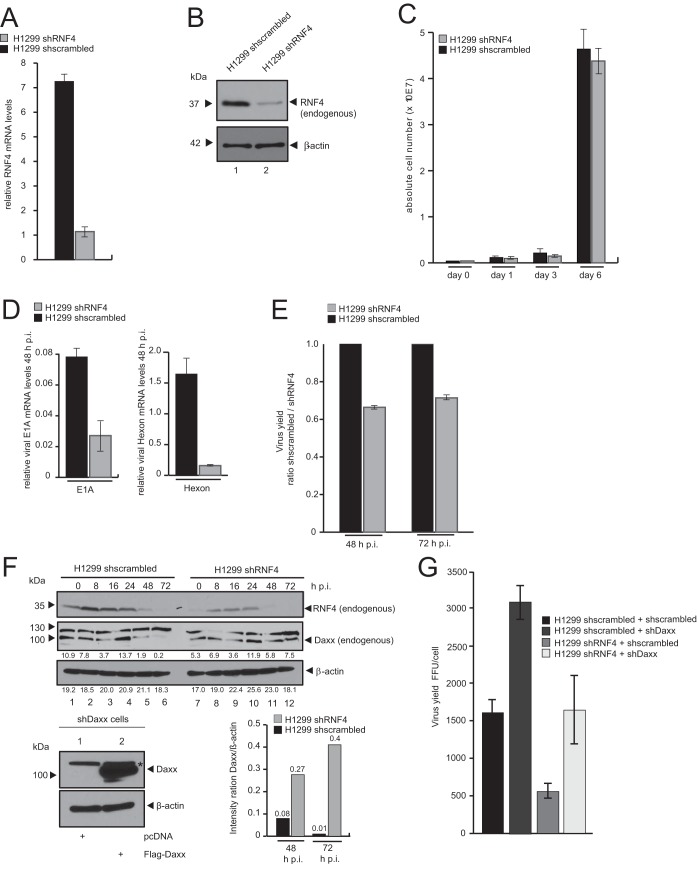
FIG 5.
RNF4 knockdown reduces HAdV viral gene expression and progeny production. (A) H1299 shscrambled and H1299 shRNF4 cells were harvested, and total RNA was extracted, reverse transcribed, and quantified by reverse transcription-PCR (RT-PCR) analysis using primers specific for RNF4. The data were normalized to 18S rRNA levels. The data are presented as relative RNF4 mRNA levels compared between H1299 shRNF4 and H1299 shscrambled control cells. (B) Endogenous RNF4 protein levels in H1299 shscrambled and H1299 shRNF4 cells were determined by preparing whole-cell extracts followed by SDS-PAGE and immunoblotting using RNF4 mouse MAb (kindly provided by T. Urano) and mouse MAb AC-15 (α-β-actin) as a loading control. Molecular sizes, in kDa, are indicated on the left, and relevant proteins are on the right. (C) A total of 1 × 105 cells (H1299 shscrambled and H1299 shRNF4) were cultivated, and absolute cell numbers were determined after the indicated time points. The means and standard deviations are presented for three independent experiments. (D) H1299 shscrambled and H1299 shRNF4 cells were infected with wt virus (H5pg4100) at a multiplicity of 20 FFU per cell. The cells were harvested 16 and 48 h postinfection, and total RNA was extracted, reverse transcribed, and quantified by RT-PCR analysis using primers specific for HAdV-C5 E1A and Hexon. The data were normalized to 18S rRNA levels, and the means and standard deviations are presented for three independent experiments. (E) H1299 shscrambled and H1299 shRNF4 cells were infected with wt virus (H5pg4100) at a multiplicity of 50 FFU per cell. Viral particles were harvested 48 and 72 h posttransfection, and virus yield was determined by quantitative E2A/DBP immunofluorescence staining on HEK293 cells. The means and standard deviations are presented for three independent experiments. Values are shown as the shscrambled/shRNF4 ratio. (F) H1299 shscrambled and H1299 shRNF4 cells were infected with wt virus (H5pg4100) at a multiplicity of 50 FFU per cell and harvested after indicated time points postinfection. Total cell extracts were prepared, separated by SDS-PAGE, and subjected to immunoblotting using RNF4 mouse MAb (kindly provided by T. Urano), Daxx rabbit pAb 07-471 (Upstate), and MAb AC-15 (α-β-actin) as a loading control. Daxx and β-actin blots were used for quantitative analysis and amount comparison for Daxx/β-actin intensity ratio at 48 and 72 h p.i. HepaRG shDaxx cells were cotransfected with 5 μg of Flag-Daxx (lower) harvested 24 h posttransfection, and total cell extracts were prepared. Proteins were separated by SDS-PAGE and subjected to immunoblotting. Input levels of total cell lysates were detected using Daxx rabbit pAb 07-471 (Upstate) and mouse MAb AC-15 (α-β-actin) as a loading control. Molecular sizes, in kDa, are indicated on the left, and relevant proteins are on the right. (G) H1299 shscrambled and H1299 shRNF4 cells cotransfected with 5 μg shDaxx construct and 24 h later superinfected with wt virus (H5pg4100) at a multiplicity of 50 FFU per cell. Viral particles were harvested 48 h posttransfection, and virus yield was determined by quantitative E2A/DBP immunofluorescence staining on HEK293 cells. The means and standard deviations are presented for three independent experiments.
We next tested RNF4 levels in mutant virus-infected cells that do not express viral E1B-55K (H5pm4149) (Fig. 1A, right). We observed that without E1B-55K present, the Daxx level is not reduced and RNF4 protein level is much less reduced compared to that of wild-type-infected cells. As expected, Mre11 also is not affected in E1B-55K-lacking infected cells due to a nonfunctional E1B-55K/E4orf6 E3 ubiquitin ligase complex.
Simultaneously, we determined RNF4 localization during HAdV wt (H5pg4100) infection. Intriguingly, we discovered that RNF4 is still detectable and thus is not degraded 48 h postinfection (h p.i.) (Fig. 1B, panel f). We even found this host STUbL juxtaposed to a specific E1B-55K fraction in round aggregates within non-4′,6-diamidino-2-phenylindole (DAPI)-stained regions in the nucleus (Fig. 1B, panels h and i), presumably representing the insoluble fraction of the infected cell. These results show that RNF4 was efficiently removed from the soluble fraction during the course of HAdV infection and relocalized adjacent to E1B-55K-containing aggregates in the host nucleus.
To verify this observation, we next performed subcellular fractionation of infected cells at 8 and 72 h postinfection, subjected these extracts to Western blotting, and analyzed them with antibodies directed against RNF4 and E1B-55K. In addition, we included antibodies that recognize human histone 3 as a control for the nuclear fraction (Fig. 1C). In accordance with previous observations (2), E1B-55K was found in all cell fractions 72 h postinfection (Fig. 1C, lanes 3, 6, 7, and 9). However, the larger SUMOylated moieties of the viral protein were mainly observed in the insoluble matrix fraction of the infected cell at 48 h postinfection (Fig. 1C, right, lane 9). As expected, we detected RNF4 in cytoplasmic and soluble nuclear fractions at time zero (Fig. 1C, left, lanes 1 and 4); however, the subcellular RNF4 distribution was clearly perturbed during HAdV infection. By 72 h postinfection, this cellular STUbL was completely sequestered into the insoluble matrix fraction (F5; Fig. 1C, left, lane 6). An additional comparison with H5pm4149-infected cells not expressing E1B-55K showed that relocalization of RNF4 into the nuclear matrix fraction is E1B-55K dependent, as the majority of RNF4 was detected in the cytoplasm fraction 72 h after H5pm4149 infection (Fig. 1C, right, lanes 8 and 10) compared to wt infection (H5pg4100).
Additionally, we tested the intracellular Daxx distribution, since this PML-NB component and anti-HAdV factor is sequestered into the host proteasomal degradation pathway during infection. Previously, we found that this process solely depends on the presence of SUMO-conjugated E1B-55K (29). Here, Daxx is mainly detectable in the nuclear matrix fraction; however, 72 h postinfection, Daxx showed an additional band with higher molecular mass, pointing to significant PTM accompanied by a severe reduction in protein levels (Fig. 1C, left, lane 6).
Hence, taken together, our immunofluorescence data and fractionation assay results reveal that E1B-55K localizes juxtaposed to the host STUbL RNF4 in the insoluble fraction of the infected cell. In parallel, we observed reduced Daxx protein levels in the same insoluble fraction during HAdV infection only when E1B-55K is expressed.
HAdV E1B-55K protein is a novel interaction partner of the host STUbL RNF4.
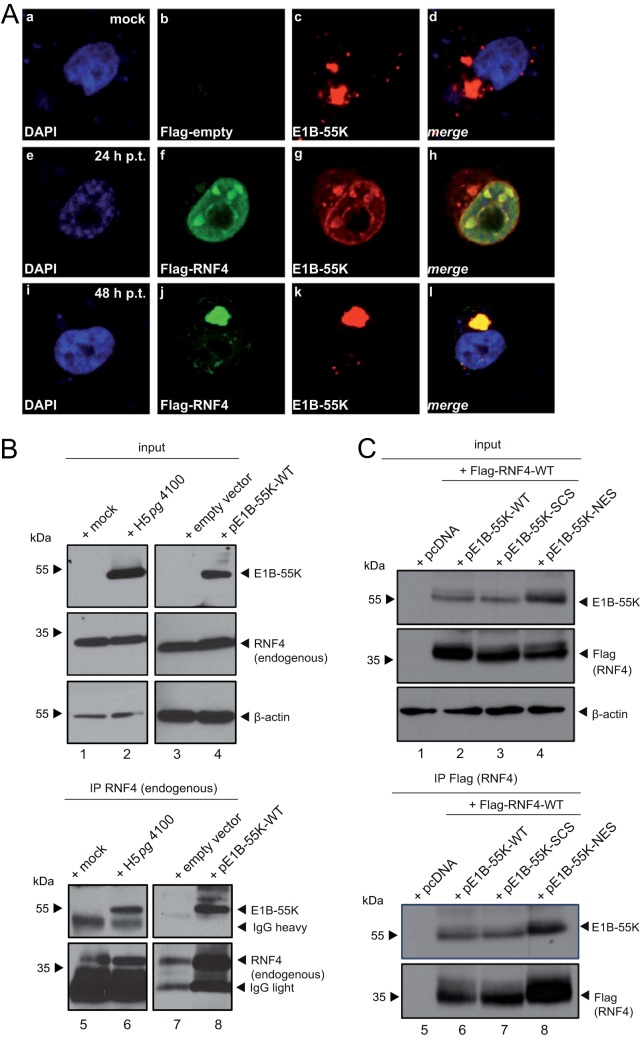
Given the above-described results, we further investigated intracellular localization of E1B-55K and RNF4 in transient-transfection experiments. Consistent with previous results, immunofluorescence analyses in E1B-55K-transfected human cells revealed that this viral protein mostly concentrates in perinuclear bodies 48 h posttransfection and infection (2, 30). In contrast to the mostly diffuse nuclear localization 24 h posttransfection (Fig. 2A, panel f), by 48 h posttransfection, RNF4 in the transiently transfected cells was completely sequestered into the E1B-55K-containing aggregates (Fig. 2A, panel l).
FIG 2.
E1B-55K interaction with RNF4 protein. (A) H1299 cells were cotransfected with 2 μg pFlag-RNF4-WT and 2 μg pE1B-55K. After 24 and 48 h posttransfection (h p.t.), cells were fixed with 4% PFA and labeled with α-Flag mouse MAb M2 (Sigma-Aldrich, Inc.), detected with Alexa 488 (α-Flag; green) and mouse MAb 2A6 (α-E1B-55K), and detected with Cy3 (α-E1B-55K; red)-conjugated secondary antibody. Mock cells are transfected but not infected. Nuclei are labeled with DAPI (4,6-diamidino-2-phenylindole). Representative α-Flag (green; Db, Df, Dj), α-E1B-55K (red; Dc, Dg, Dk), DAPI (blue; Da, De, Di) staining patterns and overlays of the single images (merge; Dd, Dh, Dl) are shown (magnification, ×7600). (B) H1299 cells were transfected with an empty vector control or a plasmid encoding E1B-55K and harvested 48 h posttransfection, or they were infected with wt virus (H5pg4100) at a multiplicity of 50 FFU per cell and harvested 24 h postinfection, and total cell extracts were prepared. Immunoprecipitation of endogenous RNF4 was performed using RNF4 mouse pAb A01 (Abnova), and proteins were separated by SDS-PAGE and subjected to immunoblotting. Input levels of total cell lysates and coprecipitated proteins were detected using mouse MAb 2A6 (α-E1B-55K), RNF4 mouse pAb A01 (Abnova), and mouse MAb AC-15 (α-β-actin) as a loading control. Note that heavy chains (IgH) are detected at 55 kDa. Molecular sizes in kDa are indicated on the left, and relevant proteins are on the right. (C) H1299 cells were cotransfected with 5 μg pFlag-RNF4-WT and 3 μg pE1B-55K-wt, 6 μg SCS (SUMO conjugation site K104R mutant), or 1.5 μg NES (nuclear export signal mutant) and harvested 48 h posttransfection, and total cell extracts were prepared. Immunoprecipitation of pFlag-RNF4 was performed using α-Flag mouse MAb M2 (Sigma-Aldrich, Inc.), and proteins were separated by SDS-PAGE and subjected to immunoblotting. Input levels of total cell lysates and coprecipitated proteins were detected using the Ab indicated in panel B. Molecular sizes, in kDa, are indicated on the left, and relevant proteins are on the right.
Since we observed recruitment of RNF4 with E1B-55K and the SUMOylated E1B-55K into the insoluble matrix fraction during infection, we next tested whether E1B-55K interacts with the endogenous RNF4 protein fraction in infected cells. As anticipated, in wt (H5pg4100)-infected cells, E1B-55K coimmunoprecipitated with RNF4-specific antibody, revealing an interaction between both factors (Fig. 2B, lane 6). No E1B-55K signal was observed in the corresponding negative controls (Fig. 2B, lane 5). Agreeing with the data obtained in infected cells, we also detected RNF4 binding to E1B-55K in the absence of any viral background (Fig. 2B, lane 8).
We next investigated the impact of E1B-55K SUMOylation on the protein interaction with the host STUbL protein RNF4. Therefore, we coexpressed RNF4 with E1B-55K wt and the SUMO-deficient mutant K104R/SCS (Fig. 2C). To control our findings, we also included the NES mutant of E1B-55K, which is even more efficiently SUMO modified (Fig. 2A, panels g and k) (31–37) than the wt protein. Our results show that loss of SUMO conjugation in pE1B-55K-SCS-transfected human cells does not significantly affect the binding ability between the viral factor and RNF4.
Together these data show that HAdV induces an altered localization of the host STUbL protein into specific insoluble E1B-55K-containing aggregates and that E1B-55K SUMOylation does not abrogate binding of the viral factor to RNF4.
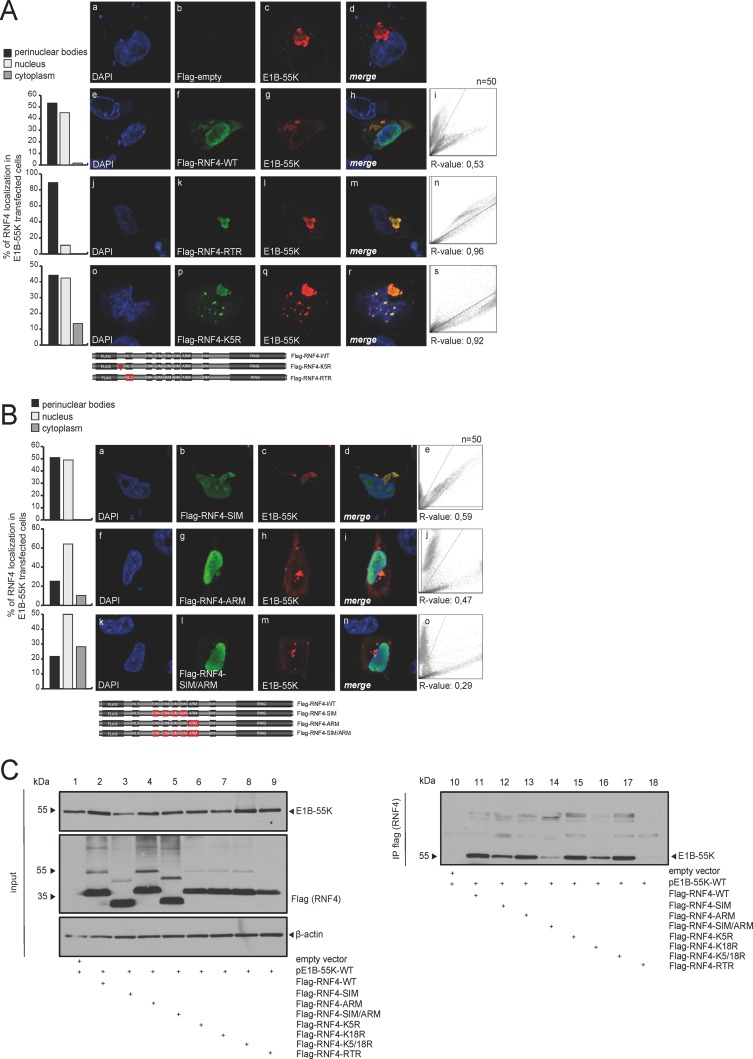
HAdV E1B-55K interaction with RNF4 is NLS, SIM, and ARM independent.
To test whether the putative RNF4 nuclear localization sequence (NLS) domain is involved in the E1B-55K-mediated relocalization of RNF4 into perinuclear aggregates, RNF4 variants with mutated NLS signals were generated. Intracellular fluorescence analyses revealed that E1B-55K-mediated relocalization of RNF4 is independent of the putative NLS in the STUbL protein, since the Flag-RNF4-RTR version with mutated NLS signal was sequestered into the E1B-55K-containing aggregates in the presence of the viral protein (Fig. 3A, panels k and l). Our quantitation revealed that the E1B-55K protein was approximately 40% more efficient in relocalizing the RTR mutant of RNF4 into perinuclear body aggregates. Similarly, a mutant with a severe defect in ubiquitin modification of the STUbL protein itself (Flag-RNF4-K5R) did not affect E1B-55K-mediated relocalization of RNF4 (Fig. 3A, panels p and q).
FIG 3.
E1B-55K binding is mediated by several regions in the RNF4 protein. (A) H1299 cells were cotransfected with 2 μg of pE1B-55K and 2 μg pFlag-empty, pFlag-RNF4-WT, RTR, or K5R. Cells were fixed with 4% PFA after 48 h posttransfection and labeled with α-Flag mouse MAb M2 (Sigma-Aldrich, Inc.), detected with Alexa 488 (α-Flag; green)-conjugated secondary antibody. Representative α-Flag (green; Bb, Bf, Bk, Bp), α-E1B-55K (red; Bc, Bg, Bl, Bq), and DAPI (blue; Ba, Be, Bj, Bo) staining patterns, overlays of the single images (merge; Bd, Bh, Bm, Br), and two-dimensional intensity histograms (Bi, Bn, Bs) are shown (n = 50 cells). Schematic representation of pFlag-RNF4-WT, the pFlag-RNF4-RTR (3-amino acid [aa] mutation in the putative NLS signal K192021R), and pFlag-RNF4-K5R construct (1-aa mutation in the putative ubiquitinylation site). Mutated regions were marked in red. (B) H1299 cells were cotransfected with 2 μg of pE1B-55K and 2 μg pFlag-RNF4-SIM, ARM, or SIM/ARM. Cells were fixed with 4% PFA after 48 h posttransfection and labeled as indicated in panel A. Representative α-Flag (green; Cb, Cg, Cl), α-E1B-55K (red; Cc, Ch, Cm), and DAPI (blue; Ca, Cf, Ck) staining patterns, overlays of the single images (merge; Cd, Ci, Cn), and 2D intensity histograms (Ce, Cj, Co) are shown (n = 50 cells). Schematic representation of the mutated pFlag-RNF4 constructs SIM (deletion of SIM1-4), ARM (deletion of ARM, positions 73 to 83), and SIM/ARM (deletion of SIM1-4 and ARM). Mutated regions were marked in red. Colocalization of Flag-RNF4 and E1B-55K was analyzed using coloc2 in Fiji (30) and calculated using Pearson's correlation coefficient (R value). (C) H1299 cells were cotransfected with a plasmid encoding E1B-55K and pFlag-RNF4-WT, SIM, ARM, SIM/ARM, K5R, K18R, K5/18R, and RTR and harvested 48 h posttransfection, and total cell extracts were prepared. Immunoprecipitation of pFlag-RNF4 was performed using α-Flag mouse MAb M2 (Sigma-Aldrich, Inc.). Proteins were separated by SDS-PAGE and subjected to immunoblotting. Input levels of total cell lysates and coprecipitated proteins were detected using mouse MAb 2A6 (α-E1B-55K), anti-Flag mouse MAb M2 (Sigma-Aldrich, Inc.), and mouse MAb AC-15 (α-β-actin) as a loading control. Molecular sizes, in kDa, are indicated on the left, and relevant proteins are on the right.
RNF4 contains tandem SUMO-interacting motifs (SIM), which have specific consensus sequences to interact with SUMO or SUMO-like domains of their ubiquitinylation targets (38). Besides the SIM, a conserved arginine-rich motif (ARM) acts as a novel recognition motif in RNF4 for selective target recruitment. Results obtained by intracellular fluorescence analyses showed that both factors still colocalize in the host nucleus as well as in perinuclear aggregates despite the SIM or ARM mutations in RNF4 (Fig. 3B, panels b, c, g, h, l, and m). Although quantitation analyses show no change in R values for RNF4 colocalization with E1B-55K between the wild type and SIM/ARM mutants, we observe differences in intracellular distributions of the protein complex. RNF4-SIM/E1B-55K complexes are distributed in accordance with RNF4-WT/E1B-55K complexes within perinuclear bodies and the nucleus. Interestingly, this changes when the ARM region of RNF4 is altered, as RNF4 shows additional cytoplasmic localization in E1B-55K-transfected cells (Fig. 3B, panels g and l), indicating that E1B-55K-mediated relocalization into the nuclear matrix is not as efficient as that with wild-type RNF4 protein.
To investigate whether the NLS, SIM, ARM, or defective ubiquitin modification mutations in RNF4 affect binding to E1B-55K, we performed additional coimmunoprecipitation studies. As anticipated, in E1B-55K-transfected human cells, E1B-55K coimmunoprecipitated with RNF4-specific antibody, confirming the interaction between both factors (Fig. 3C, lanes 11 to 18), while no E1B-55K signal was observed in the corresponding negative controls (Fig. 3C, lane 10). We observed only a minor reduction in E1B-55K binding to RNF4 without a functional SIM domain (Fig. 3C, lane 12) and therefore conclude that the viral protein is not recruited via a SIM-dependent mechanism, as shown for other SUMOylated targets of RNF4. The ARM region does not affect the RNF4 SIM-independent binding to E1B-55K; however, reduced binding was observed with the NLS mutant Flag-RNF4-RTR (Fig. 3C, lane 18). Flag-RNF4-RTR still colocalized with E1B-55K, supporting the fact that reduced binding is sufficient for both proteins to localize together in perinuclear bodies and in the nucleus.
HAdV infection promotes RNF4 interaction with Daxx during infection.
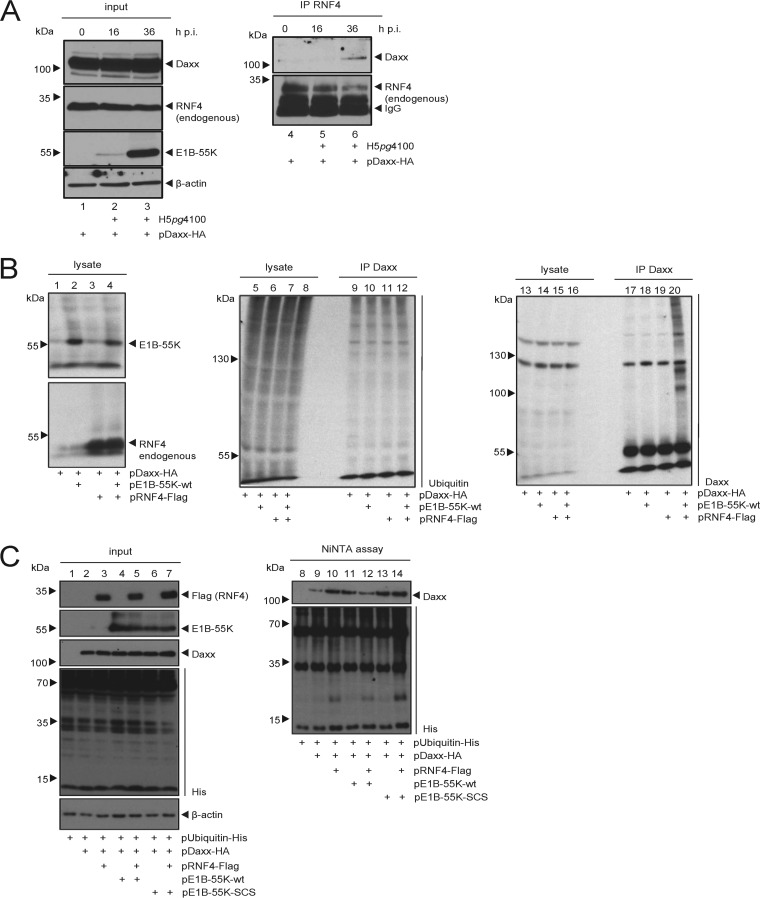
We next asked whether RNF4 binding to E1B-55K interferes with the viral factor's association with already known interaction partners, such as the PML-NB-associated HAdV restriction factor Daxx. Since we had already observed intracellular localization of Daxx within the nuclear matrix fraction together with RNF4 and E1B-55K, described above, we examined the binding between RNF4 and Daxx at different times postinfection (Fig. 4A). We cotransfected hemagglutinin (HA)-tagged Daxx and superinfected with HAdV wt virus (Fig. 4A). Our results indicate that Daxx does not show RNF4 binding in uninfected cells (Fig. 4A, lane 4). However, with an ongoing increase of E1B-55K protein expression during infection (Fig. 4A, lanes 1 to 3), we clearly detect an interaction between Daxx and RNF4 (Fig. 4A, lane 6). This supports the notion that HAdV infection and the presence of E1B-55K promotes binding between RNF4 and Daxx.
FIG 4.
HAdV infection promotes RNF4-Daxx interaction and Daxx ubiquitin PTM. (A) H1299 cells were infected with wt virus (H5pg4100) at a multiplicity of 50 FFU per cell (left), cotransfected with 2 μg of HA-Daxx prior to infection, and harvested 16 and 36 h postinfection, and then total cell extracts were prepared. Immunoprecipitation of endogenous RNF4 was performed using RNF4 mouse pAb A01 (Abnova), and proteins were separated by SDS-PAGE and subjected to immunoblotting. Input levels of total cell lysates and coprecipitated proteins were detected using mouse MAb 2A6 (α-E1B-55K), RNF4 mouse pAb A01 (Abnova), Daxx rabbit pAb 07-471 (Upstate), and mouse MAb AC-15 (α-β-actin) as a loading control. Note that light chains (IgG) are detected at 20 kDa. Molecular sizes, in kDa, are indicated on the left, and relevant proteins are on the right. (B) H1299 cells were stably transfected with 5 μg of pRNF4-Flag and pE1B-55K. Cells were treated with 25 mM NEM and 10 μM Mg132 and incubated for an additional 4 h. At 28 h posttransfection, cell pellets were resuspended in 1% SDS lysis buffer and cleared by centrifugation. Modification of Daxx was analyzed by immunoblotting after SDS-PAGE. Input levels of total cell lysates and immunoprecipitated Daxx were detected using Daxx rabbit pAb 07-471 (Upstate), MAb P4D1 (α-Ubiquitin), RNF4 mouse pAB A01 (Abnova), and MAb 2A6 (α-E1B-55K). Molecular sizes, in kDa, are indicated on the left, and relevant proteins are on the right. (C) H1299 cells were stably transfected with 10 μg pUbiquitin-His and 5 μg each of pDaxx-HA, pRNF4-Flag, and either pE1B-55K or pE1B-55K-SCM. Cells were treated with 25 mM NEM, and total cell lysates were prepared with guanidinium chloride buffer 28 h posttransfection and subjected to Ni-NTA purification of ubiquitin-His-conjugated proteins. Proteins were separated by SDS-PAGE and subjected to immunoblotting. Input levels of total cell lysates and Ni-NTA-purified proteins were detected using Daxx rabbit pAb 07-471 (Upstate), MAb 6×His (α-His), MAb AC-15 (α-β-actin), and MAb 2A6 (α-E1B-55K). Molecular sizes, in kDa, are indicated on the left, and relevant proteins are on the right.
E1B-55K promotes RNF4-dependent Daxx modification with ubiquitin moieties.
Since our results imply that E1B-55K connects Daxx and the SUMO-dependent ubiquitin ligase RNF4 during infection, we tested whether Daxx PTM and protein stability are also affected. First, cells were transfected with different combinations of Daxx, E1B-55K, and RNF4 expression plasmids. Under proteasome inhibition, immunoprecipitation of Daxx showed significant Daxx modification exclusively when both proteins E1B-55K and RNF4 are present (Fig. 4B, lane 20). These experiments substantiate the possibility that SUMOylated E1B-55K recruits Daxx and connects it to RNF4 to promote Daxx PTM and, most presumably, proteasomal degradation of this anti-HAdV transcription factor.
To further investigate this novel virus-host cross talk, we transfected cells with ubiquitin-His expression constructs and different combinations of Daxx, E1B-55K, and RNF4 plasmids. Cells were not treated with proteasome inhibitors prior to harvesting and lysate preparation. Nickel-nitrilotriacetic acid (Ni-NTA) purification of ubiquitin-His conjugates revealed that the fraction of modified Daxx protein was already reduced by proteasomal degradation when E1B-55K and RNF4 were present (Fig. 4C, lane 12). We also observe that E1B-55K alone reduced the Daxx signal (Fig. 4C, lane 11) compared to Daxx levels in RNF4-expressing cells (Fig. 4C, lane 10). Intriguingly, in cells coexpressing RNF4 together with E1B-55K-SCS, the SUMOylation-deficient variant of the viral protein (Fig. 4C, lane 14), we observed no change in immunoprecipitated Daxx protein fraction. This is consistent with earlier results showing that E1B-55K-SCS does not promote Daxx degradation (2, 10, 30).
RNF4 fosters E1B-55K-mediated Daxx inhibition, enhancing HAdV gene expression.
Daxx is involved in transcriptional regulation and cellular intrinsic antiviral resistance against HAdV, as confirmed by earlier results where knockdown of Daxx using RNA interference (RNAi) techniques significantly increased adenoviral replication, including enhanced viral mRNA synthesis and viral protein expression (39–41). However, early protein E1B-55K counteracts this Daxx restriction imposed upon HAdV growth by binding and degrading Daxx through a proteasome-dependent pathway. To investigate whether RNF4 promotes this E1B-55K-mediated inhibition of Daxx's antiviral capacity, experiments were performed in human cells expressing short hairpin RNAs (shRNAs) depleting RNF4. Reduced RNF4 RNA expression and protein synthesis was confirmed by real-time PCR analysis (Fig. 5A) and immunoblotting (Fig. 5B). Reduction of RNF4 expression did not affect cell proliferation within the time frame of 6 days postinfection (Fig. 5C). To see the effect of RNF4 depletion on the virus life cycle, we assessed viral mRNA synthesis (Fig. 5D). HAdV transcription is promoted by RNF4 expression in infected cells, since viral early E1A mRNA production was lower in RNF4-depleted cells than in control cells (Fig. 5D, left). Similar results were obtained for hexon mRNA expression, suggesting either a positive impact of the host STUbL on early gene products or direct activation of the viral promoter (Fig. 5D, right). To substantiate our results, data from the replication assays suggest that RNF4 expression also supports virus progeny production, since fewer virus particles were synthesized in the RNF4-depleted cell culture system after 48 and 72 h postinfection (Fig. 5E). Interestingly, steady-state concentrations of Daxx protein levels were more efficiently reduced in infected cells expressing RNF4 than shRNF4 cells. Our quantification shows that 48 h postinfection a 3.3-fold difference in Daxx protein levels was observed, which increases up to 40-fold after 72 h. These data support the idea that RNF4 contributes to proteasomal Daxx degradation (Fig. 5F, upper left). As the Daxx antibody detects various protein bands, we additionally tested protein signal in cells stably depleted for Daxx expression. These data show that the antibody also detects an additional signal that is not Daxx specific (Fig. 5F, lower, black asterisks). In sum, these observations substantiate our data showing a delayed Daxx reduction in shRNF4 cells (Fig. 5F, upper). Taken together, these results indicate that Daxx-mediated negative regulation of HAdV replication is counteracted by E1B-55K together with the host STUbL RNF4. Earlier, we reported that HAdV virus progeny production was promoted by loss of Daxx expression in Daxx-depleted cells (20). To further clarify whether RNF4 increases HAdV virus yield by Daxx inhibition, we transiently coexpressed shDaxx constructs in RNF-depleted stable cell lines. We reproduced findings showing that shDaxx expression enhances virus yield almost 2-fold (Fig. 5G, lane 2) compared to that of parental cells (Fig. 5G, lane 1). As seen above, stable depletion of RNF4 reduced efficient virus production (Fig. 5G, lane 3). However, this was restored when the shDaxx plasmid was expressed simultaneously (Fig. 5G, lane 4).
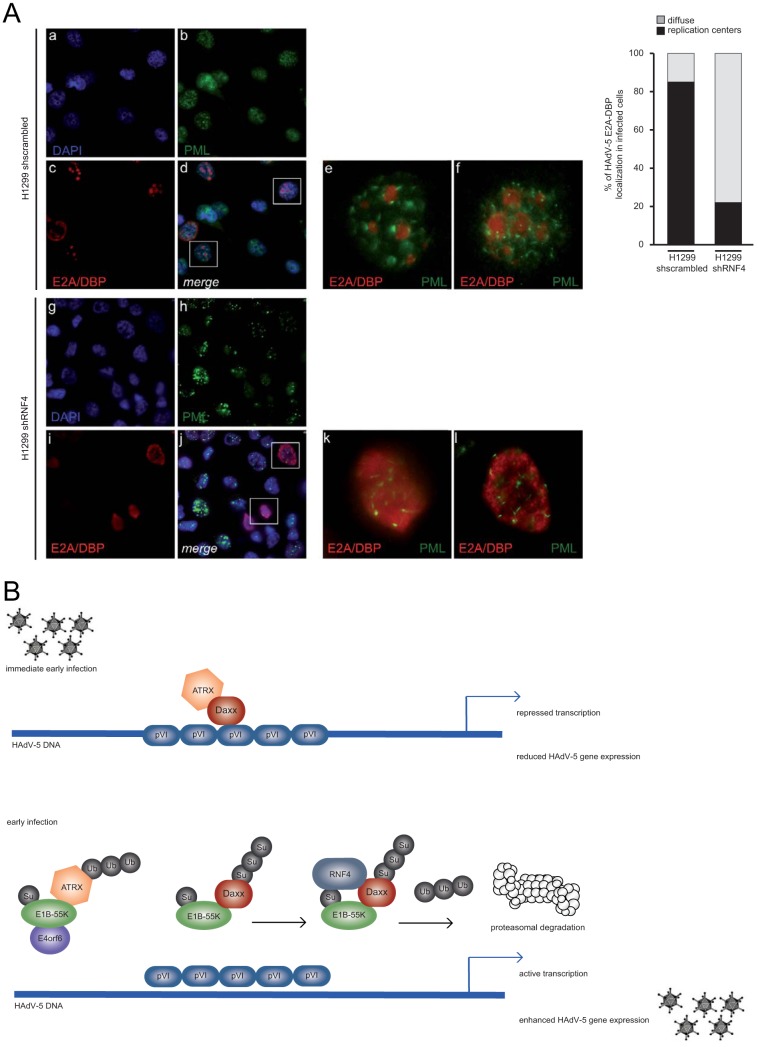
During infection it was shown that PML-NBs are relocalized by viral early proteins into track-like structures juxtaposed with adenoviral replication centers. The question was whether such viral replication centers and PML-NBs are affected in cells depleted for RNF4. Intracellular immunofluorescence analysis revealed no significant difference in PML track formation in the absence of the host STUbL protein (Fig. 6A, panels h, j, k, and l). However, detection of the viral marker protein E2A/DBP (red) intriguingly showed that compared with control cells expressing the scrambled shRNA (Fig. 6A, panels c, e, and f), replication centers are not properly established in cells lacking RNF4 (Fig. 6A, panels i, k, and l). Quantitative analysis showed a more diffuse staining of E2A/DBP within RNF4-depleted cells, whereas in parental cells E2A/DBP staining in replication centers was properly established (Fig. 6A, graph).
FIG 6.
RNF4 affects establishment of viral replication centers. (A) H1299 cells were infected with wt virus (H5pg4100) at a multiplicity of 20 FFU per cell and fixed with methanol after 48 h postinfection. Cells were labeled with α-PML pAb NB 100-59787 (Novus Biologicals, Inc.) and rabbit MAb α-E2A/DBP, detected with Alexa 488 (α-PML; green)- and Cy3 (α-E2A/DBP; red)-conjugated secondary antibody. Nuclei are labeled with DAPI (4,6-diamidino-2-phenylindole). Overlays of images (merge; d and j) and corresponding enlarged overlay (merge; e and f) staining patterns are shown. E2A/DBP staining was quantified and analyzed by counting the ratio between diffuse and replication center localization in infected cells. (B) Model of cross talk between RNF4, Daxx, and E1B-55K. Schematic representation illustrating a proposed model linking E1B-55K-dependent Daxx restriction and modulation by cellular factor RNF4. Upon HAdV infection, RNF4 is recruited to the nucleus in an E1B-55K-dependent manner to promote Daxx PTM prior to proteasomal degradation to counteract the cellular chromatin complex and ensure efficient viral gene expression.
DISCUSSION
Here, we provide evidence that the cellular STUbL RNF4 aids HAdV E1B-55K-dependent Daxx restriction during adenoviral infection. Thus, RNF4 is a novel host factor that significantly promotes HAdV infection by helping to mitigate the Daxx-mediated antiviral host response. We demonstrate that expression of viral E1B-55K promotes relocalization of RNF4 into the nuclear matrix fraction of the cell. These data supports our hypothesis that the host STUbL protein encounters Daxx to inhibit antiviral functions of this transcription factor.
SUMO conjugation of target substrates is a crucial signaling event that regulates diverse processes in the mammalian cell, such as stress response, chromosome segregation, DNA damage response, and meiosis (20). Viruses have also evolved pathways to benefit from these PTM in order to create an efficient replication milieu in the host cell (42). In both scenarios recognition of SUMOylated proteins is mostly mediated through SIMs present on effector proteins (4). The discovery of STUbLs directly links the SUMOylation process to ubiquitinylation and, thus, degradation pathways. Through tandem SIMs, STUbLs recognize poly-SUMOylated proteins and target them for Lys48-linked polyubiquitinylation and degradation through their E3 ubiquitin ligase activities. So far, only two cellular STUbLs have been identified in mammalian cells, such as RNF111/Arkadia (43) and RNF4/SNURF (44). RNF4 is a dimeric STUbL with four functional SIMs in the protein that recognize poly-SUMOylated substrates. The RING domain at the C-terminal part acts together with the SIM domains to facilitate ubiquitinylation of substrates already modified with poly-SUMO chains (21).
HAdV have acquired mechanisms that modulate SUMO- and ubiquitin-mediated regulatory cascades, leading to efficient viral propagation (21, 45). During the course of productive infection, HAdV gene products manipulate destruction pathways to prevent viral clearance or cell death prior to viral genome amplification and release of progeny. We recently demonstrated that chromatin formation by cellular SWI/SNF chromatin remodeling, involving Daxx/ATRX-dependent processes, plays a key role in HAdV transcriptional regulation and virus-mediated transformation (3, 4). Daxx and ATRX are SUMO substrates in the cell and transiently found associated with PML nuclear bodies, large multiprotein complexes representing SUMOylation hotspots in the host cell nucleus. Our recent reports demonstrate the importance of Daxx/ATRX chromatin-remodeling activities for efficient HAdV gene expression; we showed evidence that HAdV promoters are affected by Daxx/ATRX recruitment, leading to a significant block in viral gene expression and progeny production (2, 20, 30).
Early viral gene derepression mediated by incoming capsid protein VI (2, 20, 30) prior to E1B-55K/E4orf6-dependent restriction of Daxx/ATRX functional complexes is necessary for adenoviruses to evade antiviral host cell measures evolved to repress viral gene expression (summarized in Fig. 6B). In detail, based on our reported data, HAdV-mediated protein degradation apparently discriminates between classical E1B-55K/E4orf6-dependent (ATRX) pathways and a novel E1B-55K-dependent (Daxx) degradation route. However, it is still unclear how Daxx degradation works mechanistically and whether this is a kinetic process due to the expression pattern or PTM of the viral protein E1B-55K itself early during HAdV infection.
RNF4 controls protein stability by ubiquitinylation of target substrates, such as PML or the oncogenic fusion protein PML-RAR (1, 46). Thus, the PML-associated factor Daxx, which interacts with the viral E1B-55K protein, might represent a novel STUbL substrate. RNF4 relies on its SIM domains to selectively bind poly-SUMO chains over monomer SUMO. For instance, only poly-SUMOylated PML proteins are recognized by RNF4 (21, 47). Here, we observe that neither RNF4 SIMs nor the ARM region is essential for the cellular STUbL to bind to E1B-55K. Perhaps substrate SUMOylation provides additional binding sites that facilitate protein ubiquitinylation and degradation by the RING domain of RNF4 protein.
Besides the mammalian STUbLs RNF4 and RNF111 (Arkadia), additional viral STUbLs such as varicella zoster virus ORF61, Kaposi's sarcoma-associated herpesvirus K-Rta, and herpes simplex virus (HSV) ICP0 (47) have been described so far. ICP0 functions as a STUbL that preferentially ubiquitinylates poly-SUMOylated PML during HSV infection as a mechanism to inhibit the antiviral activities of PML. Notably, ICP0 represents the first precise viral ortholog of the host STUbL RNF4 to target cellular proteins such as PML and associated factors (48–50).
Although it lacks the canonical RING domain, work by Bridges and coworkers suggest that the adenoviral E4orf3 possess STUbL-like functions or recruit cellular STUbLs to regulate cellular protein stability by SUMO-mediated, ubiquitin-dependent degradation (51). HTLV-1 oncoprotein Tax is also a substrate for RNF4. Upon RNF4-dependent ubiquitinylation, Tax is relocalized into the cytoplasm to activate the NF-κB pathway by direct interaction between Tax and NEMO (52). These findings provide important new insights into STUbL-mediated pathways that regulate the subcellular localization and functional dynamics of viral oncogenes.
In addition, RNF4 blocks Epstein-Bar virus (EBV) infection by ubiquitinylation of the transcription factor Rta, which is required to activate the transcription of EBV lytic genes. Upon ubiquitinylation, Rta is degraded and subsequently EBV lytic replication and virion production are inhibited (53). Here, we find that RNF4 is a positive regulator of HAdV lytic infection, since together with E1B-55K it supports Daxx inhibition.
Chromatin-modifying complexes containing Daxx have also been implicated in human cancer development. Evidence is growing for a correlation between chromatin modifiers and tumor suppression, especially as demonstrated for SWI/SNF complexes, which comprise several subunits displaying tumor suppressor activity. Functional disruption of SWI/SNF complexes may induce a state of epigenetic instability, resulting in altered chromatin structure that affects gene expression and interferes with differentiation processes. These epigenetic changes may be closely linked to genomic instability and predispose organisms to oncogenic transformation (54). Indeed, we recently reported that efficient adenoviral transformation requires E1B-55K-mediated degradation of Daxx (30). In accordance with our current study, we envisage that RNF4 could contribute to cell transformation by modulating Daxx-dependent pathways in cooperation with E1B-55K and consequently through disrupting SWI/SNF chromatin-remodeling functions. This is a particularly interesting concept given the oncogenic capabilities of certain STUbLs, which have been shown to cooperate with either Daxx or associated determinants.
Further elucidating the cross talk between the cellular and viral regulators discussed above will help us better understand the role of chromatin remodeling in HAdV transcriptional regulation and adenoviral transformation of primary cells. Moreover, further investigation of STUbLs during virus infection will help to identify novel therapeutic approaches to modern antiviral therapy and inhibitor development.
MATERIALS AND METHODS
Cell culture and generation of knockdown cell lines.
H1299 (no. CRL-5803; ATCC Global Bioresource Center) and HEK293 cells (no. 85120602-1VL; ECACC [European Collection of Authenticated Cell Cultures]; Sigma-Aldrich) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 U of penicillin, 100 μg of streptomycin per ml in a 5% CO2 atmosphere at 37°C. To generate RNF4 knockdown cell lines, H1299 cells were transduced with lentiviral vectors expressing shRNA targeted to the coding strand sequence 5′-CCGGACGTATATGTGACTACCCATACTCGAGTATGGGTAGTCACATATACGTTTTTTG-3′ (mission RNA no. NM_002938.3-650s21c1; Sigma-Aldrich). Knockdown cell lines were selected and maintained in medium containing puromycin (2 μg/ml). All cell lines are frequently tested for mycoplasma contamination.
Plasmids and transient transfections.
HAdV-C5 proteins examined in this study were expressed from their respective complementary DNAs under the control of cytomegalovirus (CMV) immediate-early promoter, derived from the pcDNA3 vector (Invitrogen) to express E1B-55K and, accordingly, E1B-55K mutants (55). SFB (S tag, Flag epitope tag, and streptavidin-binding peptide tag)-derived wild-type, ΔARM, ΔSIM, and double mutant Δ(ARM+SIM) plasmids expressing RNF4 were kindly provided by Junjie Chen. RNF4 point mutations were introduced by site-directed mutagenesis using oligonucleotides shown in Table 1. pcDNA3-derived pUbiquitin-His plasmid was kindly provided by Ron Hay. pDaxx-HA protein was expressed from pcDNA3-derived vector under the CMV immediate-early promoter. shDaxx was targeted to the coding strand sequence 5′-GGAGTTGGATCTCTCAGAA-3′ located at nucleotides (nt) 626 to 643 in Daxx (29, 56). For transient transfections, subconfluent cells were treated with a mixture of DNA and 25-kDa linear polyethylenimine (PEI) as described recently (20).
TABLE 1.
Overview of oligonucleotides
| Primer no. | Primer description | Primer sequence |
|---|---|---|
| 1371 | 18S rRNA fw | 5′-CGGCTACCACATCCAAGGAA-3′ |
| 1372 | 18S rRNA rev | 5′-GCTGGAATTACCGCGGCT-3′ |
| 1441 | Hexo fw | 5′-CGCTGGACATGACTTTTGAG-3′ |
| 1442 | Hexon rev | 5′-GAACGGTGTGCGCAGGTA-3′ |
| 1686 | E1A fwd | 5′-GTGCCCCATTAAACCAGTTG-3′ |
| 1687 | E1A rev | 5′-GGCGTTTACAGCTCAAGTCC-3′ |
| 3356 | RNF4 fwd | 5′-GGTGGAGCAATAAATTCTAGACAAGC-3′ |
| 3357 | RNF4 rev | 5′-CCACCACAGGCTCTAAAGATTCACAAGTGAGG-3′ |
| 2978 | RNF4 RTR fwd | 5′-CAAGCTCAGAAGGCAGCGGCGGAAGCAACCTCC-3′ |
| 2979 | RNF4 RTR rev | 5′-GGAGGTTGCTTCCGCCGCTGCCTTCTGAGCTTG-3′ |
| 3070 | RNF4 K5R fwd | 5′-GCTCCATGAGTACAGGAAAGCGTCGTGG-3′ |
| 3071 | RNF4 K5R rev | 5′-CCACGACGCTTTCCTGTACTCATGGAGC-3′ |
Viruses.
H5pg4100 served as the wt virus (41). H5pm4149 carries stop codons in the E1B-55K open reading frames to prevent expression of E1B-55K (57, 58). Viruses were propagated and titrated in HEK293 cells. For this, infected cells were harvested after 48 h p.i. and lysed with three cycles of freeze and thaw and reinfected into HEK293 cells. Virus growth was determined by immunofluorescence staining of the adenoviral DNA binding protein E2A/DBP.
Antibodies and protein analysis.
Primary antibodies specific for viral proteins included E1B-55K mouse monoclonal antibody (MAb) 2A6 (38), E4orf6 mouse MAb RSA3 (59), L4-100K rat MAb 6B-10 (60), E2A/DBP mouse MAb B6-8 (61), E1A mouse MAb M73 (62), and HAdV-5 rabbit polyclonal serum L133 (63). Primary antibodies specific for cellular and ectopically expressed proteins included Daxx rabbit polyclonal antibody (pAb) 07-471 (Upstate), RNF4 mouse pAb A01 (Abnova), RNF4 mouse MAb (kindly provided by T. Urano), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) Ab (sc-32233; Santa Cruz), H3 (histone 3) Ab (1326-1; Epitomics), Mre11 rabbit pAB pNB 100-142 (Novus Biologicals, Inc.), α-Flag mouse MAb M2 (Sigma-Aldrich, Inc.), α-HA-tag rat MAb (Roche), α-ubiquitin mouse MAb (FK2; Affinity Research), α-His tag mouse MAb (Clontech), and β-actin mouse MAb AC-15 (Sigma-Aldrich, Inc.). Secondary Ab conjugated to horseradish peroxidase (HRP) for detection of proteins by immunoblotting were α-rabbit IgG, α-mouse IgG, α-mouse light chain IgG, and α-rat IgG (Jackson/Dianova). All protein extracts were prepared in RIPA lysis buffer as described recently (38). For immunoprecipitation, protein A-Sepharose beads (Sigma-Aldrich Inc.) coupled with 1 μg of Ab for 1 h at 4°C were used (3 mg/immunoprecipitation). The Ab-coupled protein A-Sepharose was added to pansorbin-Sepharose (50 μl/lysate; Calbiochem) precleared extracts and rotated for 2 h at 4°C. Proteins bound to the Ab-coupled protein A-Sepharose were precipitated by centrifugation, washed three times, boiled for 5 min at 95°C in 2× Laemmli buffer, and analyzed by immunoblotting. Cell fractionation was performed based on a modified protocol described by Leppard et al. (64), which we reported previously (65). For Ni-NTA pulldown, cells were harvested 48 h after treatment. Twenty percent of cells were pelleted to determine steady-state protein concentrations as described above, whereas the remaining cells were resuspended in 5 ml guanidinium hydrochloride (GuHCl) lysis buffer (0.1 M Na2HPO4, 0.1 M NaH2PO4, 10 mM Tris-HCl, pH 8.0, 20 mM imidazole, and 5 mM β-mercaptoethanol). Lysed cells in GuHCl were sonicated for 30 s (40 pulses, output of 0.6, 0.8 impulses/s) and supplemented with 25 μl Ni-NTA agarose (Qiagen) prewashed with GuHCl. The samples were incubated overnight at 4°C, followed by centrifugation (4,000 rpm, 10 min, 4°C). Sedimented agarose was washed once with buffer A (8 M urea, 0.1 M Na2HPO4, 0.1 M NaH2PO4, 10 mM Tris-HCl, pH 8.0, 20 mM imidazole, and 5 mM β-mercaptoethanol) and two times with buffer B (8 M urea, 0.1 M Na2HPO4, 0.1 M NaH2PO4, 10 mM Tris-HCl, pH 6.3, 20 mM imidazole, and 5 mM β-mercaptoethanol). 6His-ubiquitin conjugates were eluted from the Ni-NTA agarose with 30 μl nickel resin elution buffer (200 mM imidazole, 5% [wt/vol] SDS, 150 mM Tris-HCl [pH 6.7], 30% [vol/vol] glycerol, 720 mM β-mercaptoethanol, 0.01% [wt/vol] bromophenol blue).
After denaturation, proteins were separated by SDS-PAGE, transferred to nitrocellulose blotting membranes (0.45 μm), and visualized by immunoblotting. Autoradiograms were scanned and cropped using Adobe Photoshop CS6, and figures were prepared using Adobe Illustrator CS6 software.
Ubiquitinylation assay.
Transfected cells were treated with 10 μM MG132 and 25 mM N-ethylmaleimide for 4 h before harvesting to inhibit proteasome and protease function. Lysis was performed in 1% SDS lysis buffer (150 mM NaCl, 25 mM HEPES [pH 7.5], 0.2% NP-40, 1 mM glycerol, 10 mM NaF, 8 mM β-glycerophosphate, 1 mM dithiothreitol, 300 μM sodium-vanadate, complete protease inhibitor, and 1% SDS). For immunoprecipitation, lysates were immunoprecipitated with Daxx Ab, followed by immunoblotting with α-Daxx and α-ubiquitin Ab.
Indirect immunofluorescence.
For indirect immunofluorescence, H1299 cells were grown on glass coverslips in 1.5 × 10E5 cells per well. At different times, cells were fixed in 4% paraformaldehyde (PFA) for 20 min at 4°C or with ice-cold ethanol for 10 min at −20°C. Subsequently cells were permeabilized in phosphate-buffered saline (PBS) with 0.5 Triton X-100 for 5 min at room temperature. After 15 min of blocking in Tris-buffered saline-BG (TBS-BG; BG is 5% [wt/vol] bovine serum albumin and 5% [wt/vol] glycine), buffer coverslips were treated for 30 min with the indicated primary antibody diluted in PBS and washed three times in TBS-BG. After 20 min of incubation with the corresponding Alexa 488 (Invitrogen)- or Cy3 (Dianova)-conjugated secondary antibodies, they were washed two times in TBS-BG and one time in PBS. The coverslips were then mounted in Glow medium (Energene), and digital images were acquired with a confocal laser scanning microscope (Nikon). Images were sampled to Nyquist and analyzed using Fiji (64).
ACKNOWLEDGMENTS
We thank Takeshi Urano and Junjien Cheng for kindly providing reagents and Rudolph Reimer for his support with microscopic analyses. We greatly appreciate the critical comments and scientific discussion from Nicole Fischer.
S.M. and S.S. were supported by the Else Kröner-Fresenius-Stiftung. Part of this work was supported by the Deutsche Forschungsgemeinschaft DFG (SFB TRR179), Deutsche Krebshilfe e.V., Dräger Stiftung e.V., and the Manchot Stiftung. The Heinrich Pette Institute, Leibniz Institute for Experimental Virology, is supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit (BMG).
REFERENCES
- 1.Schreiner S, Martinez R, Groitl P, Rayne F, Vaillant R, Wimmer P, Bossis G, Sternsdorf T, Ruszsics Z, Dobner T, Wodrich H. 2012. Transcriptional activation of the adenoviral genome is mediated by capsid protein. PLoS Pathog 8:e1002549. doi: 10.1371/journal.ppat.1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schreiner S, Wimmer P, Sirma H, Everett RD, Blanchette P, Groitl P, Dobner T. 2010. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J Virol 84:7029–7038. doi: 10.1128/JVI.00074-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wimmer P, Schreiner S, Dobner T. 2012. Human pathogens and the host cell SUMOylation system. J Virol 86:642–654. doi: 10.1128/JVI.06227-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wimmer P, Schreiner S. 2015. Viral mimicry to usurp ubiquitin and SUMO host pathways. Viruses 7:4854–4872. doi: 10.3390/v7092849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker A, Rohleder KJ, Hanakahi LA, Ketner G. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J Virol 81:7034–7040. doi: 10.1128/JVI.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dallaire F, Blanchette P, Groitl P, Dobner T, Branton PE. 2009. Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J Virol 83:5329–5338. doi: 10.1128/JVI.00089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, Kaelin WG, Conaway RC, Conaway JW, Branton PE. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev 15:3104–3117. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stracker TH, Carson CT, Weitzman MD. 2002. Adenovirus oncoproteins inactivate the Mre11 Rad50 NBS1 DNA repair complex. Nature 418:348–352. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- 9.Schreiner S, Kinkley S, Bürck C, Mund A, Wimmer P, Schubert T, Groitl P, Will H, Dobner T. 2013. SPOC1-mediated antiviral host cell response is antagonized early in human adenovirus type 5 infection. PLoS Pathog 9:e1003775. doi: 10.1371/journal.ppat.1003775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schreiner S, Wimmer P, Dobner T. 2012. Adenovirus degradation of cellular proteins. Future Microbiol 7(2):211–225. doi: 10.2217/fmb.11.153. [DOI] [PubMed] [Google Scholar]
- 11.Ullman AJ, Hearing P. 2008. Cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein. J Virol 82:7325–7335. doi: 10.1128/JVI.00723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torii S, Egan DA, Evans RA, Reed JC. 1999. Human Daxx regulates Fas-induced apoptosis from nuclear PML oncogenic domains (PODs). EMBO J 18:6037–6049. doi: 10.1093/emboj/18.21.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF III, Maul GG. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol 147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dellaire G, Bazett-Jones DP. 2004. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays 26:963–977. doi: 10.1002/bies.20089. [DOI] [PubMed] [Google Scholar]
- 15.Xu ZX, Zhao RX, Ding T, Tran TT, Zhang W, Pandolfi PP, Chang KS. 2004. Promyelocytic leukemia protein 4 induces apoptosis by inhibition of survivin expression. J Biol Chem 279:1838–1844. doi: 10.1074/jbc.M310987200. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi Y, Lallemand-Breitenbach V, Zhu J, de The H. 2004. PML nuclear bodies and apoptosis. Oncogene 23:2819–2824. doi: 10.1038/sj.onc.1207533. [DOI] [PubMed] [Google Scholar]
- 17.Gostissa M, Morelli M, Mantovani F, Guida E, Piazza S, Collavin L, Brancolini C, Schneider C, Del Sal G. 2004. The transcriptional repressor hDaxx potentiates p53-dependent apoptosis. J Biol Chem 279:48013–48023. doi: 10.1074/jbc.M310801200. [DOI] [PubMed] [Google Scholar]
- 18.Ishov AM, Vladimirova OV, Maul GG. 2004. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J Cell Sci 117:3807–3820. doi: 10.1242/jcs.01230. [DOI] [PubMed] [Google Scholar]
- 19.Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. 2002. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J Cell Sci 115:3319–3330. [DOI] [PubMed] [Google Scholar]
- 20.Schreiner S, Bürck C, Glass M, Groitl P, Wimmer P, Kinkley S, Mund A, Everett RD, Dobner T. 2013. Control of human adenovirus type 5 (Ad5) gene expression by cellular Daxx/ATRX chromatin-associated complexes. Nucleic Acids Res 41:3532–3550. doi: 10.1093/nar/gkt064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 22.Perry JJ, Tainer JA, Boddy MN. 2008. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci 33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. 2012. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev 26:1179–1195. doi: 10.1101/gad.188284.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Plechanovova A, Simpson P, Marchant J, Leidecker O, Kraatz S, Hay RT, Matthews SJ. 2014. Structural insight into SUMO chain recognition and manipulation by the ubiquitin ligase RNF4. Nat Commun 5:4217. doi: 10.1038/ncomms5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Percherancier Y, Germain-Desprez D, Galisson F, Mascle XH, Dianoux L, Estephan P, Chelbi-Alix MK, Aubry M. 2009. Role of SUMO in RNF4-mediated promyelocytic leukemia protein (PML) degradation: sumoylation of PML and phospho-switch control of its SUMO binding domain dissected in living cells. J Biol Chem 284:16595–16608. doi: 10.1074/jbc.M109.006387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orazio NI, Naeger CM, Karlseder J, Weitzman MD. 2011. The adenovirus E1b55K/E4orf6 complex induces degradation of the Bloom helicase during infection. J Virol 85:1887–1892. doi: 10.1128/JVI.02134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta A, Jha S, Engel DA, Ornelles DA, Dutta A. 2013. Tip60 degradation by adenovirus relieves transcriptional repression of viral transcriptional activator EIA. Oncogene 32:5017–5025. doi: 10.1038/onc.2012.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endter C, Hartl B, Spruss T, Hauber J, Dobner T. 2005. Blockage of CRM1-dependent nuclear export of the adenovirus type 5 early region 1B 55-kDa protein augments oncogenic transformation of primary rat cells. Oncogene 24:55–64. doi: 10.1038/sj.onc.1208170. [DOI] [PubMed] [Google Scholar]
- 30.Schreiner S, Wimmer P, Groitl P, Chen SY, Blanchette P, Branton PE, Dobner T. 2011. Adenovirus type 5 early region 1B 55K oncoprotein-dependent degradation of cellular factor Daxx is required for efficient transformation of primary rodent cells. J Virol 85:8752–8765. doi: 10.1128/JVI.00440-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zantema A, Fransen JA, Davis OA, Ramaekers FC, Vooijs GP, DeLeys B, van der Eb AJ. 1985. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology 142:44–58. doi: 10.1016/0042-6822(85)90421-0. [DOI] [PubMed] [Google Scholar]
- 32.Zantema A, Schrier PI, Davis OA, van Laar T, Vaessen RT, van der Eb AJ. 1985. Adenovirus serotype determines association and localization of the large E1B tumor antigen with cellular tumor antigen p53 in transformed cells. Mol Cell Biol 5:3084–3091. doi: 10.1128/MCB.5.11.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodrum FD, Shenk T, Ornelles DA. 1996. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J Virol 70:6323–6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.König C, Roth J, Dobbelstein M. 1999. Adenovirus type 5 E4orf3 protein relieves p53 inhibition by E1B-55-kilodalton protein. J Virol 73:2253–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krätzer F, Rosorius O, Heger P, Hirschmann N, Dobner T, Hauber J, Stauber RH. 2000. The adenovirus type 5 E1B-55k oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene 19:850–857. doi: 10.1038/sj.onc.1203395. [DOI] [PubMed] [Google Scholar]
- 36.Wienzek S, Roth J, Dobbelstein M. 2000. E1B 55-kilodalton oncoproteins of adenovirus types 5 and 12 inactivate and relocalize p53, but not p51 or p73, and cooperate with E4orf6 proteins to destabilize p53. J Virol 74:193–202. doi: 10.1128/JVI.74.1.193-202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanchette P, Wimmer P, Dallaire F, Cheng CY, Branton PE. 2013. Aggresome formation by the adenoviral protein E1B55K is not conserved among adenovirus species and is not required for efficient degradation of nuclear substrates. J Virol 87:4872–4881. doi: 10.1128/JVI.03272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kindsmuller K, Groitl P, Hartl B, Blanchette P, Hauber J, Dobner T. 2007. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc Natl Acad Sci U S A 104:6684–6689. doi: 10.1073/pnas.0702158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pennella MA, Liu Y, Woo JL, Kim CA, Berk AJ. 2010. Adenovirus E1B 55-kilodalton protein is a p53-SUMO1 E3 ligase that represses p53 and stimulates its nuclear export through interactions with promyelocytic leukemia nuclear bodies. J Virol 84:12210–12225. doi: 10.1128/JVI.01442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller S, Dobner T. 2008. The adenovirus E1B-55K oncoprotein induces SUMO modification of p53. Cell Cycle 7:754–758. doi: 10.4161/cc.7.6.5495. [DOI] [PubMed] [Google Scholar]
- 41.Wimmer P, Berscheminski J, Blanchette P, Groitl P, Branton PE, Hay RT, Dobner T, Schreiner S. 2015. PML isoforms IV and V contribute to adenovirus-mediated oncogenic transformation by functional inhibition of the tumor suppressor p53. Oncogene 35:69–82. doi: 10.1038/onc.2015.63. [DOI] [PubMed] [Google Scholar]
- 42.Hay RT. 2013. Decoding the SUMO signal. Biochem Soc Trans 41:463–473. doi: 10.1042/BST20130015. [DOI] [PubMed] [Google Scholar]
- 43.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. 2004. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A 101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poulsen SL, Hansen RK, Wagner SA, van Cuijk L, van Belle GJ, Streicher W, Wikstrom M, Choudhary C, Houtsmuller AB, Marteijn JA, Bekker-Jensen S, Mailand N. 2013. RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J Cell Biol 201:797–807. doi: 10.1083/jcb.201212075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. 2012. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schreiner S, Wodrich H. 2013. Virion factors targeting Daxx to overcome intrinsic immunity. J Virol 87:10412–10422. doi: 10.1128/JVI.00425-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de The H. 2008. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol 10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 48.Sriramachandran AM, Dohmen RJ. 2014. SUMO-targeted ubiquitin ligases. Biochim Biophys Acta 1843:75–85. doi: 10.1016/j.bbamcr.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 49.Izumiya Y, Kobayashi K, Kim KY, Pochampalli M, Izumiya C, Shevchenko B, Wang DH, Huerta SB, Martinez A, Campbell M, Kung HJ. 2013. Kaposi's sarcoma-associated herpesvirus K-Rta exhibits SUMO-targeting ubiquitin ligase (STUbL) like activity and is essential for viral reactivation. PLoS Pathog 9:e1003506. doi: 10.1371/journal.ppat.1003506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boutell C, Cuchet-Lourenco D, Vanni E, Orr A, Glass M, McFarlane S, Everett RD. 2011. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog 7:e1002245. doi: 10.1371/journal.ppat.1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bridges RG, Sohn SY, Wright J, Leppard KN, Hearing P. 2016. The adenovirus E4-ORF3 protein stimulates SUMOylation of general transcription factor TFII-I to direct proteasomal degradation. mBio 7:e02184-15. doi: 10.1128/mBio.02184-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fryrear KA, Guo X, Kerscher O, Semmes JO. 2012. The Sumo-targeted ubiquitin ligase RNF4 regulates the localization and function of the HTLV-1 oncoprotein Tax. Blood 119:1173–1181. doi: 10.1182/blood-2011-06-358564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang YC, Yoshikai Y, Hsu SW, Saitoh H, Chang LK. 2013. Role of RNF4 in the ubiquitination of Rta of Epstein-Barr virus. J Biol Chem 288:12866–12879. doi: 10.1074/jbc.M112.413393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson BG, Roberts CW. 2011. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 55.Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. 2009. Detection of protein SUMOylation in vivo. Nat Protoc 4:1363–1371. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
- 56.Endter C, Kzhyshkowska J, Stauber R, Dobner T. 2001. SUMO-1 modification is required for transformation by adenovirus type 5 early region 1B 55-kDa oncoprotein. Proc Natl Acad Sci U S A 98:11312–11317. doi: 10.1073/pnas.191361798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Groitl P, Dobner T. 2007. Construction of adenovirus type 5 early region 1 and 4 virus mutants. Methods Mol Med 130:29–39. [DOI] [PubMed] [Google Scholar]
- 58.Blanchette P, Kindsmuller K, Groitl P, Dallaire F, Speiseder T, Branton PE, Dobner T. 2008. Control of mRNA export by adenovirus E4orf6 and E1B55K proteins during productive infection requires E4orf6 ubiquitin ligase activity. J Virol 82:2642–2651. doi: 10.1128/JVI.02309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarnow P, Hearing P, Anderson CW, Reich N, Levine AJ. 1982. Identification and characterization of an immunologically conserved adenovirus early region 11,000 Mr protein and its association with the nuclear matrix. J Mol Biol 162:565–583. doi: 10.1016/0022-2836(82)90389-8. [DOI] [PubMed] [Google Scholar]
- 60.Marton MJ, Baim SB, Ornelles DA, Shenk T. 1990. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J Virol 64:2345–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kzhyshkowska J, Kremmer E, Hofmann M, Wolf H, Dobner T. 2004. Protein arginine methylation during lytic adenovirus infection. Biochem J 383:259–265. doi: 10.1042/BJ20040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reich NC, Sarnow P, Duprey E, Levine AJ. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128:480–484. doi: 10.1016/0042-6822(83)90274-X. [DOI] [PubMed] [Google Scholar]
- 63.Harlow E, Franza BR Jr, Schley C. 1985. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J Virol 55:533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wimmer P, Schreiner S, Everett RD, Sirma H, Groitl P, Dobner T. 2010. SUMO modification of E1B-55K oncoprotein regulates isoform-specific binding to the tumour suppressor protein PML. Oncogene 29:5511–5522. doi: 10.1038/onc.2010.284. [DOI] [PubMed] [Google Scholar]
- 65.Leppard KN, Shenk T. 1989. The adenovirus E1B 55 kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J 8:2329–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]