ABSTRACT
An entirely plasmid-based reverse genetics system for rotaviruses was established very recently. We improved the reverse genetics system to generate recombinant rotavirus by transfecting only 11 cDNA plasmids for its 11 gene segments under the condition of increasing the ratio of the cDNA plasmids for NSP2 and NSP5 genes. Utilizing this highly efficient system, we then engineered infectious recombinant rotaviruses expressing bioluminescent (NanoLuc luciferase) and fluorescent (enhanced green fluorescent protein [EGFP] and mCherry) reporters. These recombinant rotaviruses expressing reporters remained genetically stable during serial passages. Our reverse genetics approach and recombinant rotaviruses carrying reporter genes will be great additions to the tool kit for studying the molecular virology of rotavirus and for developing future next-generation vaccines and expression vectors.
IMPORTANCE Rotavirus is one of the most important pathogens causing severe gastroenteritis in young children worldwide. In this paper, we describe a robust and simple reverse genetics system based on only rotavirus cDNAs and its application for engineering infectious recombinant rotaviruses harboring bioluminescent (NanoLuc) and fluorescent (EGFP and mCherry) protein genes. This highly efficient reverse genetics system and recombinant group A rotaviruses expressing reporters could be powerful tools for the study of different aspects of rotavirus replication. Furthermore, they may be useful for next-generation vaccine production for this medically important virus.
KEYWORDS: rotavirus, reverse genetics, NSP2, NSP5, reporters, fluorescent proteins
INTRODUCTION
Group A rotavirus (RVA), a member of the genus Rotavirus within the family Reoviridae, is the leading cause of severe gastroenteritis in young children worldwide. RVA diarrhea accounts for ∼215,000 deaths among children <5 years old annually (1). RVA comprises an 11-segment genome of double-stranded RNA (dsRNA) that encodes 12 proteins (six structural proteins [VP1 to VP4, VP6, and VP7] and six nonstructural proteins [NSP1 to NSP6]) (2).
A reverse genetics technology to engineer infectious viruses from cloned cDNAs is a useful tool for studying different aspects of virus biology. Although reverse genetics systems exist for nearly all major groups of DNA and RNA viruses, development of a technology to engineer an infectious RVA from cDNAs has been more challenging, owing in part to the technical complexity of manipulating its 11-segment genome (3, 4). For RVA, partial plasmid-based reverse genetics systems that require a helper virus have been exploited to engineer novel infectious recombinant RVAs possessing a cDNA-derived gene segment (5–8). However, since these systems require strong selection methods to isolate the recombinant RVA from a helper virus, their use is restricted to certain gene segments. Alternatively, a versatile entirely plasmid-based reverse genetics system was developed recently (9). With this system, recombinant RVA was recovered following transfection of BHK-T7 cells, constitutively expressing T7 RNA polymerase, with 14 plasmids (11 RVA cDNA plasmids for 11 gene segments, flanked by the T7 RNA promoter and hepatitis delta virus [HDV] ribozyme sequences, in combination with three expression plasmids encoding the Nelson Bay orthoreovirus fusion-associated small transmembrane [FAST] protein and the two subunits of the vaccinia virus [VV] capping enzyme). A subsequent study indicated that cotransfection of the two expression plasmids encoding the VV capping enzyme is not essential for virus rescue (10).
Replication-competent recombinant viruses expressing reporters, such as those encoding bioluminescent and fluorescent proteins, have been of great use for studying the molecular biology of viruses and for developing vaccines and antiviral drugs (11, 12). For RVA, many efforts have been made to insert an exogenous gene into its genome. Utilizing a partial plasmid-based reverse genetics system, 24-bp FLAG and hepatitis C virus E2 epitope sequences and the 190-bp internal ribozyme entry site sequence of cricket paralysis virus were successfully inserted out-frame into the 3′-untranslated region (UTR) of the NSP2 segment (13). More recently, the 48-bp split-GFP and 516-bp NanoLuc (Nluc) luciferase genes were successfully inserted in-frame into the NSP1 segment by using a plasmid-only-based reverse genetics system (9). However, the insertion of longer nucleotide sequences, such as those encoding entire fluorescent proteins, has been unsuccessful due in part to the low efficiency of the currently available RVA reverse genetics systems.
In this paper, we first report an improved strategy for generating recombinant RVAs by transfecting just 11 RVA cDNA plasmids for 11 gene segments into BHK-T7 cells. This system is efficient and does not require the use of any other plasmids. This 11-plasmid system was efficient enough for generating recombinant RVAs expressing reporters, including the entire fluorescent proteins enhanced green fluorescent protein (EGFP) and mCherry, that have been impossible to rescue using conventional systems.
RESULTS AND DISCUSSION
Generation of recombinant RVAs by transfecting only 11 RVA T7 plasmids for 11 gene segments.
In the process of modification of the original entirely plasmid-based reverse genetics system, we found that even without the addition of the FAST-expressing plasmid, RVA could be rescued in one of the three wells, although virus rescue with only the 11 RVA plasmids was not expected (Table 1). We could consistently detect appreciable virus generation in repeated experiments (Table 2), indicating reproducible production of RVAs with the 11-plasmid system. We next examined whether the efficiency of virus rescue with the 11-plasmid system could be improved if the amount of a certain T7 plasmid was modified. The original ratios and concentrations of the 11 RVA T7 plasmids were those described in previous reports (9, 10) (0.75 μg of each plasmid) as a guide. Changing the amount of each plasmid carrying the VP1-VP4, VP6, VP7, NSP1, NSP3, or NSP4 gene from 0.75 to 2.25 μg (3-fold) did not affect the efficiency of virus rescue (Table 1). In contrast, a 3-fold increase of the amount of a plasmid carrying the NSP2 or NSP5 gene resulted in more efficient virus generation than that under the original conditions (transfection of 0.75 μg of either plasmid, 1-fold); at 4 days after transfection (at 3 days after starting the coculture with overlaid CV-1 cells), virus was detected in all three individual wells, with virus titers ranging from 4.3 × 104 to 6.8 × 105 PFU/ml (mean, 2.6 × 105 PFU/ml) or 1.0 × 102 to 1.5 × 105 PFU/ml (mean, 5.0 × 104 PFU/ml), respectively, which were higher than the virus titers with the 12-plasmid system, including the FAST-expressing plasmid (Table 1). As a result, we confirmed that 11 RVA T7 plasmids are sufficient for the generation of recombinant RVA (Fig. 1A). Furthermore, it was suggested that the efficiency of virus rescue could be improved by an increase of the amount of a plasmid carrying the NSP2 or NSP5 gene.
TABLE 1.
Screening for determination of the optimal amounts of 11 RVA T7 plasmids required for the generation of recombinant RVAa
| Amt of plasmid (μg) expressingb: |
Efficiency of virus rescuec (no. positive/total no.) | Virus titerd (PFU/ml) for expt: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VP1 | VP2 | VP3 | VP4 | VP6 | VP7 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5 | FAST | 1 | 2 | 3 | |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.2 | 2/3 | <10 | <10 | 0 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0 | 1/3 | <10 | 0 | 0 |
| 2.25 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0 | 0/3 | 0 | 0 | 0 |
| 0.75 | 2.25 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0 | 0/3 | 0 | 0 | 0 |
| 0.75 | 0.75 | 2.25 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0 | 0/3 | 0 | 0 | 0 |
| 0.75 | 0.75 | 0.75 | 2.25 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0 | 0/3 | 0 | 0 | 0 |
| 0.75 | 0.75 | 0.75 | 0.75 | 2.25 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0 | 0/3 | 0 | 0 | 0 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 2.25 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0 | 0/3 | 0 | 0 | 0 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 2.25 | 0.75 | 0.75 | 0.75 | 0.75 | 0 | 1/3 | <10 | 0 | 0 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 2.25 | 0.75 | 0.75 | 0.75 | 0 | 3/3 | 6.8 × 105 | 4.3 × 104 | 5.0 × 104 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 2.25 | 0.75 | 0.75 | 0 | 0/3 | 0 | 0 | 0 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 2.25 | 0.75 | 0 | 0/3 | 0 | 0 | 0 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 2.25 | 0 | 3/3 | 5.8 × 102 | 1.5 × 105 | 1.0 × 102 |
BHK/T7-9 cells in 6-well plates were transfected with the indicated amounts of plasmids. One day later, the transfected cells were washed with incomplete medium and then cultured for 3 days in incomplete medium containing trypsin (0.3 μg/ml) with overlaid CV-1 cells.
Boldface indicates the plasmids whose amounts were 3-fold increased.
Efficiency of virus rescue in three experiments.
Virus titers were determined by plaque assay using the cultures.
TABLE 2.
Determination of the synergistic interaction between the two plasmids encoding the NSP2 and NSP5 genes on the generation of recombinant RVAa
| Amt of rescue T7 plasmid (μg) expressingb: |
Efficiency of virus rescuec (no. positive/total no.) | Virus titerd (PFU/ml) in expt: |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VP1 | VP2 | VP3 | VP4 | VP6 | VP7 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5 | 1 | 2 | 3 | 4 | 5 | 6 | |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 2/6 | <10 | 25 | 0 | 0 | 0 | 0 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 2.25 | 0.75 | 0.75 | 0.75 | 6/6 | 2.0 × 104 | <10 | 3.8 × 104 | 4.8 × 103 | 2.8 × 104 | 5.0 × 104 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 2.25 | 4/6 | 1.5 × 105 | 27.5 | 6.0 × 103 | 15 | 0 | 0 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 2.25 | 0.75 | 0.75 | 2.25 | 6/6 | 3.3 × 106 | 6.5 × 105 | 2.5 × 104 | 3.3 × 105 | 1.0 × 106 | 7.5 × 105 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 3.75 | 0.75 | 0.75 | 0.75 | 6/6 | 2.5 × 104 | 2.8 × 105 | 2.8 × 104 | 4.5 × 104 | 5.0 × 104 | 3.0 × 105 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 3.75 | 5/6 | 45 | 0 | 75 | 4.0 × 104 | 6.0 × 102 | 42.5 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 3.75 | 0.75 | 0.75 | 3.75 | 6/6 | 7.5 × 105 | 8.0 × 104 | 1.2 × 106 | 5.3 × 105 | 6.5 × 104 | 2.8 × 105 |
BHK/T7-9 cells in 6-well plates were transfected with the indicated amounts of plasmids. One day later, the transfected cells were washed with incomplete medium and then cultured for 3 days in incomplete medium containing trypsin (0.3 μg/ml) with overlaid CV-1 cells.
Boldface indicates the plasmids whose amounts were increased (3- or 5-fold).
Efficiency of virus rescue in six experiments.
Virus titers were determined by plaque assay using the cultures.
FIG 1.
Schematic representation of an 11-plasmid system for RVA reverse genetics. The 11 RVA rescue T7 plasmids contain the full-length segment cDNA of an individual gene of SA11-L2 virus, flanked by the T7 RNA promoter (PT7) and the HDV ribozyme (Rib). BHK/T7-9 cells were transfected with the 11 RVA plasmids with/without increased amounts of the two plasmids carrying the NSP2 and NSP5 genes. Recombinant RVAs were rescued from the cultures of the transfected BHK/T7-9 cells by amplification (A) or plaque formation (B) on CV-1 cells.
Because it has been shown that the NSP2 and NSP5 proteins interact with each other to enhance the formation of viroplasm, i.e., the viral factory where RVA replication and assembly take place (14–18), we next tried to determine whether a synergism would be expressed as enhancement of virus generation when the ratio of the plasmids carrying the NSP2 and NSP5 genes was increased (Table 2). For the two doses tested (3- and 5-fold), virus was rescued in all six individual wells, with high virus yields ranging from 2.5 × 104 to 3.3 × 106 PFU/ml (mean, 1.0 × 106 PFU/ml) and 6.5 × 104 to 1.2 × 106 PFU/ml (mean, 5.8 × 105 PFU/ml), respectively. These efficiencies were higher (P value of <0.05, as determined by t test) than those obtained in individual control experiments, in which the amount of each plasmid carrying the NSP2 or NSP5 gene was 3- or 5-fold increased. Thus, synergism was observed between the two genes as improvement of the efficiency of virus rescue.
Robustness of the optimized 11-plasmid system.
Coculture of transfected BHK/T7-9 cells with RVA-susceptible cells (CV-1 or MA104 cells) has been performed to amplify released viruses (9, 10). To confirm the superiority of this optimized 11-plasmid system with increased quantity ratios of plasmids carrying the NSP2 and NSP5 genes, we tried to rescue viruses directly from BHK/T7-9 cells transfected with the 11 RVA plasmids. Under the original conditions (transfection of 0.75 μg of either plasmid, 1-fold), viruses could not be rescued (Table 3). In contrast, the optimized 11-plasmid system successfully rescued viruses directly from transfected BHK/T7-9 cells in one or two of two experiments in which the virus titers reached 1.2 × 102 PFU/ml (Table 3). These results demonstrated the better performance of the optimized 11-plasmid system whatever the conditions used (Fig. 1B).
TABLE 3.
Efficiency of generation of recombinant RVAs from transfected BHK/T7-9 cellsa
| Amt of rescue T7 plasmid (μg) expressingb: |
Efficiency of virus rescuec (no. positive/total no.) | Virus titerd (PFU/ml) in expt: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VP1 | VP2 | VP3 | VP4 | VP6 | VP7 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5 | 1 | 2 | |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0/2 | 0 | 0 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 2.25 | 0.75 | 0.75 | 2.25 | 1/2 | <10 | 0 |
| 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 3.75 | 0.75 | 0.75 | 3.75 | 2/2 | <10 | 1.2 × 102 |
BHK/T7-9 cells in 6-well plates were transfected with the indicated amounts of plasmids. One day later, the transfected cells were washed with incomplete medium and then cultured for 3 days in incomplete medium containing trypsin (0.3 μg/ml).
Boldface indicates the plasmids whose amounts were increased (3- or 5-fold).
Efficiency of virus rescue in two experiments.
Virus titers were determined by plaque assay using the cultures of transfected BHK/T7-9 cells.
Electron microscopy of recombinant RVAs.
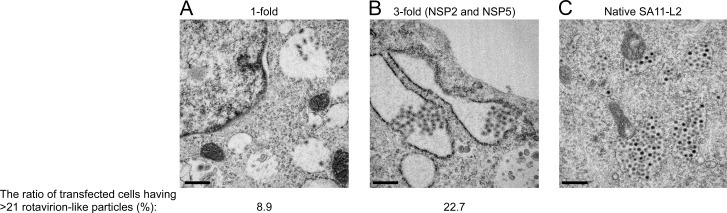
In an attempt to visualize recombinant RVAs entirely from cDNA plasmids, thin sections of BHK/T7-9 cells transfected with the 11 RVA plasmids were examined by electron microscopy (Fig. 2). A few round rotavirion-like particles were observed in the endoplasmic reticulum (ER) of the cells transfected with the 11 plasmids (Fig. 2A and B), but similar particles were not observed in the cells not transfected (data not shown). The structures of these round particles were indistinguishable from those observed in BHK/T7-9 cells infected with native SA11-L2 virus (Fig. 2C). These particles were detected in the proximity of 30% of the cells examined regardless of the quantity ratios of plasmids carrying the NSP2 and NSP5 genes (1- and 3-fold); however, the number of these particles increased when the ratio of the two plasmids was increased to 3-fold (Fig. 2B); at 24 h after transfection, the ratio of transfected cells possessing >21 rotavirion-like particles was 22.7%, compared to only 8.9% for the cells transfected with 0.75 μg of either plasmid (1-fold) (data not shown). These data are compatible with the finding that increasing the amounts of the two plasmids carrying the NSP2 and NSP5 genes improves the efficiency of virus generation.
FIG 2.
Electron microscopy of cells with recombinant RVAs. BHK/T7-9 cells were transfected with 11 RVA plasmids with 0.75 μg of each plasmid (1-fold) (A) or with 11 RVA plasmids with increased amounts of the plasmids carrying the NSP2 and NSP5 genes, from 0.75 to 2.25 μg (3-fold) (B). (C) BHK/T7-9 cells were infected with the native SA11-L2 virus at an MOI of 3. At 24 h after transfection or at 8 h after infection, cells were fixed and stained and then observed under a transmission electron microscope. Bars represent 500 nm.
Because overexpression of the NSP2 and NSP5 proteins in MA104 cells promotes the formation of viroplasm-like structures (15), we predicted that viroplasm formation would be important for a successful RVA reverse genetics system. In contrast to this prediction, we could not identify any electron-dense viroplasms (19, 20) within BHK/T7-9 cells transfected with the 11 plasmids (Fig. 2A and B). This finding was unexpected, but it could be attributed to the fact that viroplasms were only rarely detected even in BHK/T7-9 cells infected with the native SA11-L2 virus at a high multiplicity of infection (MOI) (Fig. 2C). Therefore, BHK/T7-9 and MA104 cells may differ in terms of viroplasm formation in the course of RVA infection, as previous reports documented disparity in protein transport kinetics among cell lines of different origins when infected with RVA (21, 22). Thus, the mechanisms underlying the improvement of the efficiency of virus rescue with increasing quantity ratio of the two plasmids carrying the NSP2 and NSP5 genes are unknown, although the practical implications are obvious.
Generation of a recombinant RVA lacking the NSP1 ORF and expressing the Nluc reporter gene.
After confirming the efficient generation of recombinant RVA using the optimized 11-plasmid system, we attempted to utilize this robust and simplified reverse genetics system to engineer recombinant RVAs expressing the full-length exogenous proteins that were difficult to rescue using the currently available reverse genetics systems. To this end, we made use of a self-cleaving 2A peptide sequence from porcine teschovirus (2A) that drives a cotranslational stop/continue event, with two individual proteins being generated from one open reading frame (ORF) (23, 24).
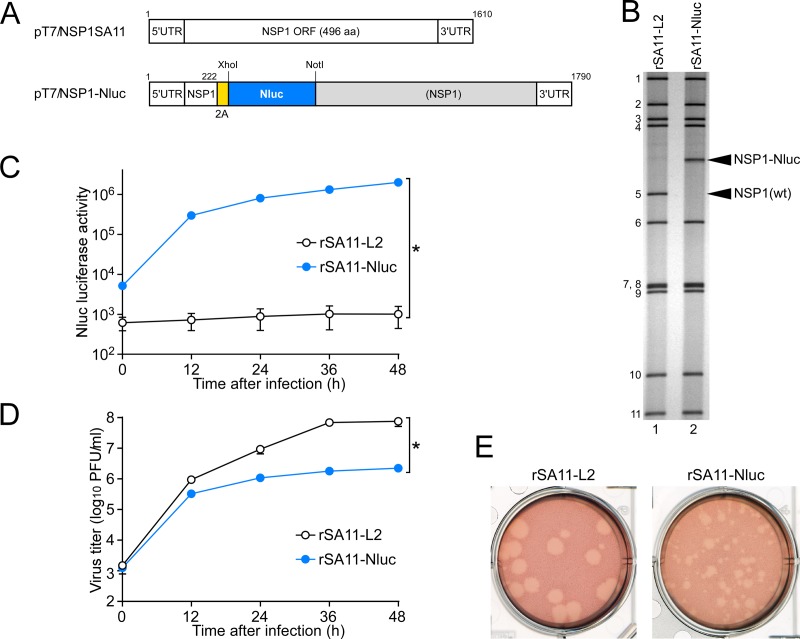
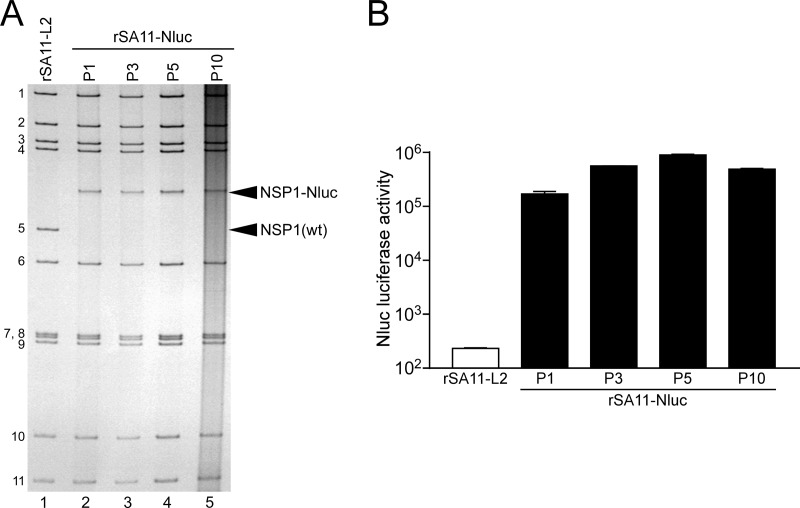
Naturally occurring RVA mutants carrying truncated NSP1 ORFs can replicate in cell culture (25, 26), and RNA interference against NSP1 mRNA (17) has indicated the nonessential role of the NSP1 protein in the RVA replication cycle, suggesting that the NSP1 ORF can be replaced by a foreign gene. To determine whether or not replacement of the NSP1 ORF with a foreign gene could be tolerated by both RVA replication and expression of the exogenous gene, we constructed a T7 plasmid for the NSP1 gene segment, pT7/NSP1-Nluc, carrying the 2A-Nluc gene in the deletion site of the NSP1 gene (Fig. 3A); the deletion site mimicked the corresponding one of the rearranged NSP1 gene of a naturally occurring bovine strain, A5-16 (G8P[1]) (25). Using plasmid pT7/NSP1-Nluc for the NSP1 segment, we successfully generated recombinant rSA11-Nluc virus efficiently in two successive rescue attempts (Fig. 3B). PAGE analysis of viral genomic dsRNAs extracted from the rescued virus showed that the NSP1 segment of rSA11-Nluc migrated slower than the wild-type NSP1 gene segment of the parental rSA11-L2 virus, as expected because of its increased size (1,790 bp) compared to that of the wild-type NSP1 segment (1,610 bp) (Fig. 3B). In MA104 cells infected with rSA11-Nluc, Nluc activity in cell lysates could be detected as early as 12 h after infection and steadily increased with incubation time, whereas the parental rSA11-L2 did not show any Nluc activity (Fig. 3C). To determine whether or not replacement of the N-terminal portion of the NSP1 ORF by the 2A-Nluc gene influenced the RVA infectivity in cell culture, multiple-step growth curves for rSA11-L2 and rSA11-Nluc were obtained after infection of MA104 cells at an MOI of 0.01 PFU/cell. The growth curves demonstrated that the replication of rSA11-Nluc was significantly lower (by about 34-fold) than that of the parental rSA11-L2 (Fig. 3D). We then examined the plaque sizes in CV-1 cells for these viruses by measuring the mean diameter of each of 25 plaques in two different assays (Fig. 3E). rSA11-Nluc formed smaller-sized plaques (diameter, 1.68 ± 0.37 mm) than those of the parental rSA11-L2 (diameter, 3.93 ± 0.40 mm). These results indicated that replacement of the N-terminal portion of the NSP1 ORF with the 2A-Nluc gene did not abolish RVA replication in cell culture. The genetic stability of rSA11-Nluc as to expression of Nluc was indicated to remain unchanged through 10 successive passages by PAGE (Fig. 4A) and Nluc luciferase analyses (Fig. 4B). Based on these results, it was demonstrated that a recombinant RVA can be engineered to express an exogenous gene by replacing the NSP1 ORF with a foreign gene with a simplified method.
FIG 3.
Generation of recombinant rSA11-Nluc virus expressing Nluc luciferase. (A) Schematic presentation of the plasmids carrying the NSP1 genes for rescue of the wild-type (wt) rSA11-L2 and rSA11-Nluc viruses (pT7/NSP1SA11 and pT7/NSP1-Nluc, respectively). To generate plasmid pT7/NSP1-Nluc, nucleotides 223 to 643 in the NSP1 ORF were replaced with the 2A-Nluc gene. (B) PAGE of viral genomic dsRNAs extracted from rSA11-L2 and rSA11-Nluc. Lane 1, dsRNAs from rSA11-L2; lane 2, dsRNAs from rSA11-Nluc. The numbers on the left indicate the order of the genomic dsRNA segments of rSA11-L2. (C) Nluc luciferase activities of rSA11-L2 and rSA11-Nluc. MA104 cells were infected with rSA11-L2 or rSA11-Nluc at an MOI of 0.01 and then incubated for various numbers of hours. Nluc activities in the cultures were determined by luminometry. The data shown are the mean Nluc activities and standard deviations (SDs) for three independent cell cultures. *, P < 0.05 (as determined by t test). (D) Multiple-step growth curves for rSA11-L2 and rSA11-Nluc. MA104 cells were infected with rSA11-L2 or rSA11-Nluc at an MOI of 0.01 and then incubated for various numbers of hours. Virus titers in the cultures were determined by plaque assay. The data shown are the mean viral titers and SDs for three independent cell cultures. *, P < 0.05 (as determined by t test). (E) Plaque formation by rSA11-L2 and rSA11-Nluc. rSA11-L2 or rSA11-Nluc was directly plated onto CV-1 cells for plaque formation. The experiment was repeated three times with similar results, and representative results are shown.
FIG 4.
Genetic stability of the recombinant rSA11-Nluc virus. rSA11-Nluc was consecutively passaged 10 times in MA104 cells. (A) PAGE of viral genomic dsRNAs extracted from passage 1 (P1), 3 (P3), 5 (P5), and 10 (P10) stocks of rSA11-Nluc. Lane 1, dsRNAs from rSA11-L2; lanes 2, 3, 4, and 5, dsRNAs from P1 (lane 2), P3 (lane 3), P5 (lane 4), and P10 (lane 5) stocks of rSA11-Nluc. The numbers on the left indicate the order of the genomic dsRNA segments of rSA11-L2. (B) Nluc activities of P1, P3, P5, and P10 stocks of rSA11-Nluc. MA104 cells were infected with rSA11-L2 or rSA11-Nluc at an MOI of 0.01 and incubated for 12 h, and then Nluc activities in the cultures were determined by luminometry. The data shown are the mean Nluc activities and SDs for three independent cell cultures.
Generation of recombinant RVAs expressing EGFP and mCherry reporter genes.
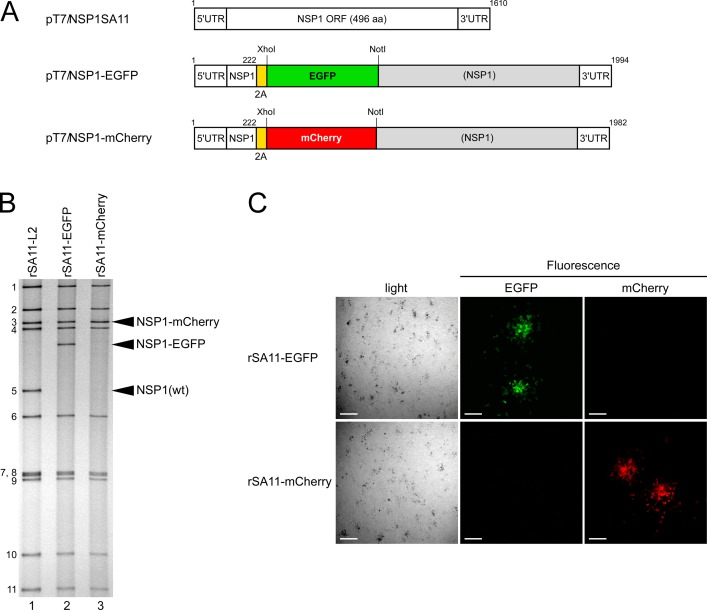
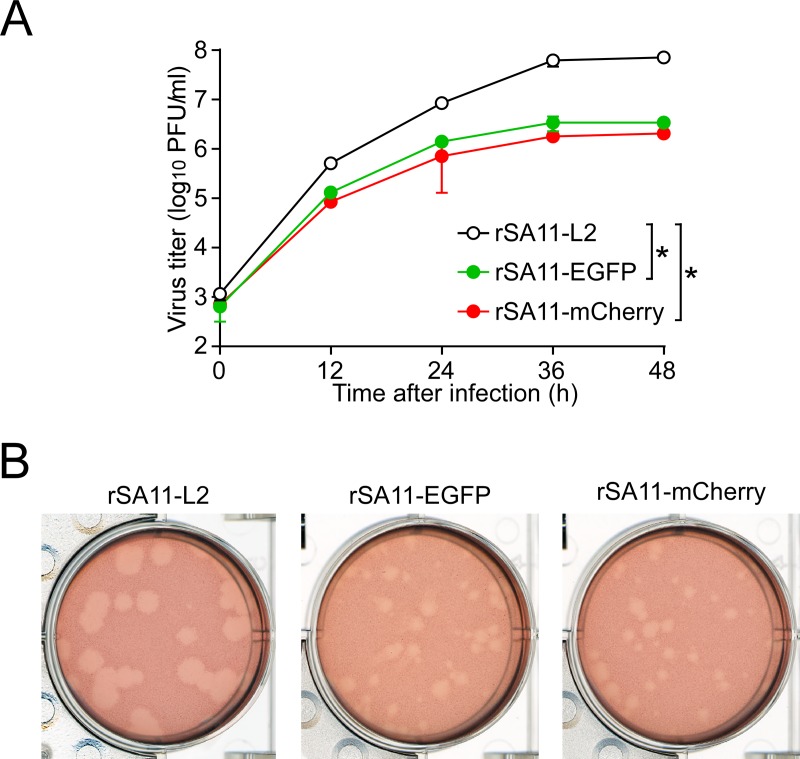
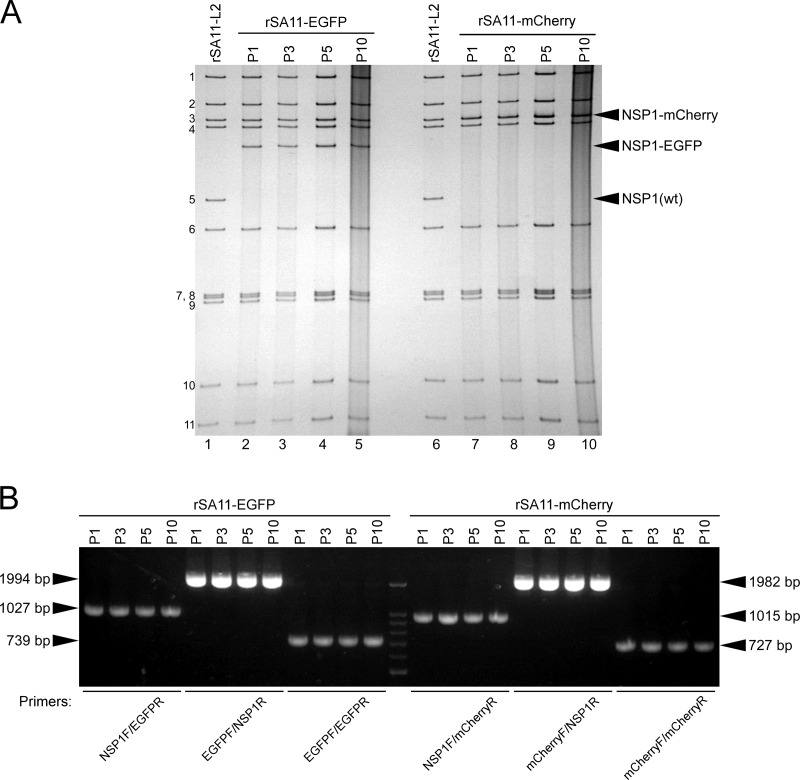
Having succeeded in generating recombinant rSA11-Nluc virus, we next tried to insert longer exogenous genes into the NSP1 segment. The pT7/NSP1-Nluc plasmid was modified to replace the Nluc gene with a longer EGFP or the mCherry gene, using the XhoI-NotI site to yield plasmids pT7/NSP1-EGFP and pT7/NSP1-mCherry, respectively (Fig. 5A). Using these T7 plasmids for the NSP1 segment, viable recombinant RVAs harboring the EGFP and mCherry genes (rSA11-EGFP and rSA11-mCherry virus, respectively) were successfully generated efficiently in two successive rescue attempts (Fig. 5B). PAGE analysis of viral genomic dsRNAs extracted from the rescued viruses demonstrated that the NSP1 segments of rSA11-EGFP and rSA11-mCherry migrated slower than that of the parental rSA11-L2, as expected because of their increased sizes (1,994 bp and 1,982 bp, respectively) compared to that of the wild-type NSP1 segment (1,610 bp) (Fig. 5B). The NSP1 segment of rSA11-mCherry migrated at a much lower rate than rSA11-EGFP, although they have roughly the same number of nucleotides. An explanation for this anomaly may be that the secondary structures of viral dsRNAs in PAGE gels have a significant impact on their migration patterns (27, 28). Expression of fluorescent EGFP and mCherry proteins encoded by the NSP1 segments was detected in RVA foci in CV-1 cells infected with the rSA11-EGFP and rSA11-mCherry viruses, respectively, under a fluorescence microscope (Fig. 5C). To determine whether or not replacement of the N-terminal portion of the NSP1 ORF with an EGFP or mCherry gene influenced RVA infectivity in cell culture, multiple-step growth curves for rSA11-L2, rSA11-EGFP, and rSA11-mCherry were obtained after infection of MA104 cells at a low MOI of 0.01 PFU/cell. The growth curves demonstrated that the replication of rSA11-EGFP and rSA11-mCherry was lower to some extent (by about 26-fold) than that of the parental rSA11-L2 virus (Fig. 6A). We then examined the plaque sizes in CV-1 cells for these viruses by measuring the mean diameter of each of 25 plaques in two different assays (Fig. 6B). rSA11-EGFP and rSA11-mCherry formed smaller-sized plaques (diameters, 1.84 ± 0.34 and 1.77 ± 0.45 mm, respectively) than those of the parental rSA11-L2 (diameter, 3.87 ± 0.32 mm). Thus, these results demonstrated that replacement of the N-terminal portion of the NSP1 ORF with an EGFP or mCherry gene did not abolish RVA replication in cell culture. The genetic stabilities of rSA11-EGFP and rSA11-mCherry were shown to remain unchanged through 10 successive passages by PAGE analysis (Fig. 7A) and reverse transcription-PCR (RT-PCR) using specific primers (Fig. 7B). Expression of EGFP and mCherry was also confirmed through 10 consecutive virus passages under a fluorescence microscope (data not shown). These results demonstrated that recombinant RVAs expressing the entire EGFP and mCherry could be successfully generated using the optimized 11-plasmid system and were stable throughout the course of our study.
FIG 5.
Generation of recombinant rSA11-EGFP and rSA11-mCherry viruses expressing EGFP and mCherry. (A) Schematic presentation of plasmids carrying the NSP1 genes for rescue of wild-type rSA11-L2, rSA11-EGFP, and rSA11-mCherry (pT7/NSP1SA11, pT7/NSP1-EGFP, and pT7/NSP1-mCherry, respectively). To generate plasmids pT7/NSP1-EGFP and pT7/NSP1-mCherry, the Nluc gene in pT7/NSP1-Nluc was replaced by an EGFP or mCherry gene using the XhoI-NotI site (rSA11-EGFP and rSA11-mCherry, respectively). (B) PAGE of viral genomic dsRNAs extracted from rSA11-L2, rSA11-EGFP, and rSA11-mCherry. Lane 1, dsRNAs from rSA11-L2; lanes 2 and 3, dsRNAs from rSA11-EGFP (lane 2) and rSA11-mCherry (lane 3). The numbers on the left indicate the order of the genomic dsRNA segments of rSA11-L2. (C) Expression of EGFP and mCherry from rSA11-EGFP and rSA11-mCherry. CV-1 cells were infected with rSA11-EGFP or rSA11-mCherry at an MOI of 1 and incubated for 34 h, and then expression of EGFP and mCherry in RVA foci, immobilized under 0.7% agar, was assessed under a bright-field and fluorescence microscope using specific filters. Bars represent 200 μm.
FIG 6.
Infectivity of the recombinant rSA11-EGFP and rSA11-mCherry viruses. (A) Multiple-step growth curves for rSA11-L2, rSA11-EGFP, and rSA11-mCherry. MA104 cells were infected with rSA11-L2, rSA11-EGFP, or rSA11-mCherry at an MOI of 0.01 and then incubated for various numbers of hours. Virus titers in the cultures were determined by plaque assay. The data shown are the mean viral titers and SDs for three independent cell cultures. *, P < 0.05 (as determined by t test). (B) Plaque formation by rSA11-L2, rSA11-EGFP, and rSA11-mCherry. rSA11-L2, rSA11-EGFP, or rSA11-mCherry was directly plated onto CV-1 cells for plaque formation. The experiment was repeated three times with similar results, and representative results are shown.
FIG 7.
Genetic stability of the recombinant rSA11-EGFP and rSA11-mCherry viruses. rSA11-EGFP and rSA11-mCherry were consecutively passaged 10 times in MA104 cells. (A) PAGE of viral genomic dsRNAs extracted from P1, P3, P5, and P10 stocks of rSA11-EGFP and rSA11-mCherry. Lanes 1 and 6, dsRNAs from rSA11-L2; lanes 2, 3, 4, and 5, dsRNAs from P1 (lane 2), P3 (lane 3), P5 (lane 4), and P10 (lane 5) stocks of rSA11-EGFP; and lanes 7, 8, 9, and 10, dsRNAs from P1 (lane 7), P3 (lane 8), P5 (lane 9), and P10 (lane 10) stocks of rSA11-mCherry. The numbers on the left indicate the order of the genomic dsRNA segments of rSA11-L2. (B) RT-PCR analysis of engineered NSP1 segments of rSA11-EGFP and rSA11-mCherry. The NSP1 genes were amplified by RT-PCR using viral genomic dsRNAs and specific primers for the NSP1, EGFP, and/or mCherry nucleotide sequences for successive virus passage P1, P3, P5, and P10 stocks of rSA11-EGFP and rSA11-mCherry, followed by separation in a 2% agarose gel. DNA markers are 1,500, 1,000, 900, 800, 700, 600, and 500 bp.
NSP1 protein expression rescues the replication of the recombinant RVAs expressing reporters.
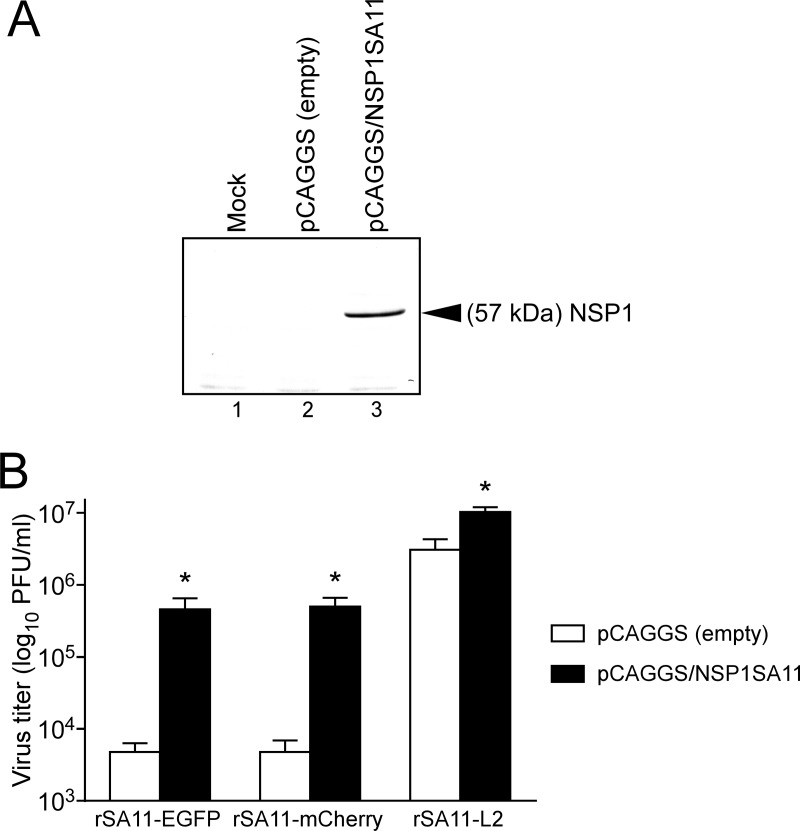
To confirm the significantly impaired replication of recombinant RVAs expressing reporters due to the absence of the full-length NSP1 protein, we assessed viral replication in cells that transiently expressed the authentic NSP1 protein of the SA11-L2 virus (Fig. 8). As in untransduced cells (Fig. 6A), rSA11-EGFP and rSA11-mCherry produced significantly lower yields than the wild-type rSA11-L2 virus at 36 h in MA104 cells transfected with the empty expression plasmid. However, rSA11-EGFP and rSA11-mCherry produced ∼100-fold-higher yields in MA104 cells expressing the NSP1 protein. NSP1 protein expression rescued the replication of rSA11-Nluc in transfected cells in a similar manner (data not shown). These findings indicated that the replication defect for the recombinant RVAs expressing reporters is due to a lack of expression of the full-length NSP1. Furthermore, even the wild-type rSA11-L2 produced slightly higher yields (∼4-fold) in cells overexpressing the NSP1 protein, highlighting the role of NSP1 in optimal RVA replication.
FIG 8.
RVA replication in NSP1-expressing MA104 cells. (A) Expression of NSP1 protein in MA104 cells transfected with the NSP1 expression plasmid, pCAGGS/NSP1SA11. As a control, MA104 cells were transfected with the empty expression plasmid. Whole-cell lysates of transfected cells were analyzed by immunoblotting using anti-NSP1 antiserum. Mock (lane 1), pCAGGS (empty) (lane 2), and pCAGGS/NSP1SA11 (lane 3) plasmid transfection is shown. (B) MA104 cells expressing NSP1 protein were infected with rSA11-EGFP, rSA11-mCherry, or rSA11-L2 at an MOI of 0.01 and then incubated for 36 h. As a control (open columns), cells transfected with the empty expression plasmid were infected with the recombinant RVAs. Virus titers in the cultures were determined by plaque assay. The data shown are the mean viral titers and SDs for three independent cell cultures. *, P < 0.05 (as determined by t test).
In the present study, we described a simple reverse genetics system for RVA, in which only 11 RVA T7 plasmids were transfected into BHK/T7-9 cells. With this system, increasing the amounts of the two plasmids harboring the NSP2 and NSP5 genes drastically improved the rescue efficiency for recombinant RVAs (optimized 11-plasmid system). The recovery efficiency was approximately 1,000 times higher than that of the previously reported 12-plasmid system including the FAST-expressing plasmid (Tables 1 and 2). Although the mechanism underlying the remarkable effect of overexpression of the NSP2 and NSP5 proteins remains unclear, previous studies revealed that overexpression of the viroplasm-forming nonstructural NS2 protein of orbiviruses, in the family Reoviridae, enhances the efficiencies of reverse genetics systems to considerably high levels (29, 30). Thus, the viroplasm-forming nonstructural proteins may play a role in concentration of components of the viral replication complex via formation of viroplasms in transfected cells, resulting in increased virus production in the reverse genetics systems (29).
Taking advantage of this optimized 11-plasmid system, we successfully generated recombinant RVAs expressing bioluminescent (Nluc) and fluorescent (EGFP and mCherry) reporters. This is the first demonstration of the rescue of recombinant RVAs that can express entire fluorescent proteins. We have also succeeded in generating other recombinant RVAs, which have ∼1.5 kbp exogenous genes in their genomes (data not shown), although it remains to be determined how much longer inserts can be packaged into capsids with successful RVA recovery. Our strategy of using a self-cleaving 2A peptide sequence between a truncated NSP1 sequence and a reporter sequence is a simple and effective way to insert a foreign gene into the genome of RVA. Insertion of 2A peptides has been successfully performed to generate replication-competent orthoreoviruses, in the family Reoviridae, with long inserts, such as the 918-bp portion of the simian immunodeficiency virus gag gene (31). These achievements nicely illustrate the potential of engineering dsRNA viruses as expression vectors. The presence of a reporter, such as Nluc, EGFP, and mCherry, in the context of infectious RVA could have several advantages: the expression of these reporters will allow direct measurement of viral infection and replication in RVA-susceptible cells without the use of conventional approaches, which are generally time-consuming and cumbersome.
In conclusion, we report an efficient and simple reverse genetics system to generate recombinant RVA by transfecting only 11 cDNA plasmids for its 11 gene segments. Utilizing this efficient system, we then engineered infectious RVAs carrying bioluminescent and fluorescent reporters. These recombinant viruses carrying reporter genes remained genetically stable during our study. This highly efficient reverse genetics system and recombinant RVAs expressing reporters will be great additions to the tool kit for studying the molecular biology of RVA and developing next-generation vaccine and expression vectors.
MATERIALS AND METHODS
Cells and viruses.
A baby hamster kidney cell line, BHK/T7-9 (32), stably expressing T7 RNA polymerase, was cultured in Dulbecco's modified Eagle medium (DMEM) (Nissui) supplemented with 5% fetal calf serum (FCS) (Gibco) (complete medium) in the presence of 600 ng/ml hygromycin (Invitrogen). Monkey kidney cell lines MA104 and CV-1 were cultured in complete medium. Simian RVA strain SA11-L2 (G3P[2]) (33) was propagated as described previously (6). Briefly, SA11-L2 virus was pretreated with trypsin (type IX, from porcine pancreas) (10 μg/ml; Sigma-Aldrich) and then propagated in MA104 cells in Eagle's minimum essential medium (MEM) (Nissui) without FCS (incomplete medium) but containing trypsin (1 μg/ml).
Construction of rescue T7 plasmids carrying the VP1-VP3, VP6, VP7, and NSP1-NSP5 gene segments of SA11-L2 virus.
In our efforts to improve RVA reverse genetics systems, we newly constructed 10 T7 plasmids expressing the individual mRNAs of 10 gene segments, except for the VP4 segment, of SA11-L2 virus. As vector backbones, we used a pEXR vector (∼2.6 kbp) to minimize the lengths of T7 plasmids carrying medium- and small-sized gene segments (VP6, VP7, and NSP1-NSP5 genes) and a traditional pX8dT vector (∼3.0 kbp) (34) to carry large-sized gene segments (VP1-VP3 genes). To construct T7-driven plasmids, pT7/VP1SA11, pT7/VP2SA11, pT7/VP3SA11, pT7/VP6SA11, pT7/VP7SA11, pT7/NSP1SA11, pT7/NSP2SA11, pT7/NSP3SA11, pT7/NSP4SA11, and pT7/NSP5SA11, each of which encodes the full-length nucleotide sequence of the corresponding gene segment of SA11-L2 virus, cDNAs were amplified by RT-PCR from its genomic dsRNAs with ReverTra Ace reverse transcriptase (Toyobo), PrimeStar HS DNA polymerase (TaKaRa Bio), and specific primers. As described previously (6, 10), the forward primers contain the T7 RNA polymerase promoter sequence and a sequence corresponding to the 5′ terminus of each viral segment. After digestion with restriction enzymes, the cDNAs were ligated into the corresponding restriction enzyme sites of T7 expression plasmids. As T7 expression plasmids, we used a pEX-A2J1-derived pEXR vector that carries HDV ribozyme and T7 RNA polymerase terminator sequences (artificially synthesized by Eurofins Genomics) (for VP6, VP7, and NSP1-NSP5 genes) and a modified pX8dT vector (34) (for VP1-VP3 genes). The constructed T7 plasmids contain the full-length segment cDNAs of individual genes of SA11-L2 virus (GenBank/EMBL/DDBJ accession no. LC333802 to LC333804 and LC333806 to LC333812), flanked by the T7 RNA promoter and HDV ribozyme sequences (5) and followed by the T7 RNA polymerase terminator sequence. To create plasmid pT7/NSP1-Nluc that carries the Nluc ORF in the NSP1 segment, a cDNA fragment was synthesized by Eurofins Genomics in which nucleotides 223 to 643 in the NSP1 ORF were replaced with the 2A-Nluc gene. Thus, the 2A-Nluc gene is flanked by the NSP1 gene sequences (nucleotides 1 to 222 and 644 to 1610). The synthesized cDNA fragment, named 5′NSP1-2A-Nluc-3′NSP1, was inserted into the PstI-SmaI site of the pX8dT vector to create pT7/NSP1-Nluc. The full-length ORFs of EGFP and mCherry were amplified by PCR and then inserted into the XhoI-NotI site of pT7/NSP1-Nluc to replace the Nluc gene with the EGFP or mCherry gene, yielding plasmids pT7/NSP1-EGFP and pT7/NSP1-mCherry, respectively. To create expression plasmid pCAGGS/FAST, which encodes the Nelson Bay orthoreovirus FAST protein, a cDNA fragment encoding FAST (GenBank/EMBL/DDBJ accession no. AB908284) was synthesized by GENEWIZ. The synthesized cDNA fragment was inserted into the EcoRI-XhoI site of the pCAGGS expression vector (35). The NSP1 gene of SA11-L2 virus was amplified by PCR and then inserted into the SacI-XhoI site of pCAGGS to create expression plasmid pCAGGS/NSP1SA11. The nucleotide sequences of all the constructed plasmids were confirmed by direct sequencing. The primer sequences used for plasmid construction will be provided on request.
Generation of recombinant RVAs from only 11 RVA rescue T7 plasmids.
The standard protocol was performed basically as described by Komoto et al. (10). The major modifications were (i) that newly constructed 10 T7 plasmids, pT7/VP1SA11, pT7/VP2SA11, pT7/VP3SA11, pT7/VP6SA11, pT7/VP7SA11, pT7/NSP1SA11, pT7/NSP2SA11, pT7/NSP3SA11, pT7/NSP4SA11, and pT7/NSP5SA11, were employed instead of the pTM1 (36)-based 10 T7 plasmids pT7-VP1SA11, pT7-VP2SA11, pT7-VP3SA11, pT7-VP6SA11, pT7-VP7SA11, pT7-NSP1SA11, pT7-NSP2SA11, pT7-NSP3SA11, pT7-NSP4SA11, and pT7-NSP5SA11 (9), and (ii) that the FAST-expressing plasmid was not included. Briefly, the protocol was as follows. Monolayers of BHK/T7-9 cells in 6-well plates (Falcon) were cotransfected with 11 T7 plasmids, representing the cloned cDNAs of RVA 11 gene segments, using 3 μl of TransIT-LT1 transfection reagent (Mirus) per μg of plasmid DNA. After the optimization experiments, the amounts of the 11 plasmids used were the following: pT7/VP1SA11 (0.75 μg), pT7/VP2SA11 (0.75 μg), pT7/VP3SA11 (0.75 μg), pT7/VP4SA11-ΔPstI (0.75 μg) (6, 10), pT7/VP6SA11 (0.75 μg), pT7/VP7SA11 (0.75 μg), pT7/NSP2SA11 (2.25 μg), pT7/NSP3SA11 (0.75 μg), pT7/NSP4SA11 (0.75 μg), and pT7/NSP5SA11 (2.25 μg), in combination with pT7/NSP1SA11 (0.75 μg), pT7/NSP1-Nluc (0.75 μg), pT7/NSP1-EGFP (0.75 μg), or pT7/NSP1-mCherry (0.75 μg). Following 1 day of incubation, the transfected BHK/T7-9 cells were washed with incomplete medium and then cocultured with overlaid CV-1 cells (5 × 104/well) for 3 days in incomplete medium containing trypsin (0.3 μg/ml). After incubation, the cultures were subjected to two cycles of freezing and thawing and then treated with trypsin (10 μg/ml) for RVA activation, followed by inoculation onto MA104 cells. For the generation of recombinant RVAs expressing reporters, the cultures were passaged in MA104 cells. After 1 or 2 days of incubation, recombinant RVAs were rescued and then plaque purified in CV-1 cells as described previously (37).
Electron microscopy.
Monolayers of BHK/T7-9 cells transfected with 11 rescue T7 plasmids or infected with the native SA11-L2 virus were fixed with 2.5% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 30 min at 4°C and subsequently fixed with 4% PFA and 0.1% glutaraldehyde in PBS for 30 min at 4°C. After washing with PBS, the cells were fixed with 1% osmium tetroxide in PBS for 30 min at room temperature, dehydrated through a graded ethanol series, transferred to n-butyl glycidyl ether, and then embedded in epoxy resin (EPON812; TAAB). Ultrathin sections were cut with an ultramicrotome (Ultracut N; Reichert-Nissei), 100-nm thickness, put on a copper grid (26415; VECO), etched with 5% sodium metaperiodate for 10 min, and then washed with PBS. The sections were stained with uranyl acetate for 7 min and lead citrate for 4 min and then observed under a transmission electron microscope (JEM-1010; JEOL).
Electrophoretic analysis of viral genomic dsRNAs.
Viral genomic dsRNAs were extracted from cell cultures using a QIAamp viral RNA minikit (Qiagen). The extracted viral dsRNAs were loaded onto PAGE gels. The dsRNA species were separated by electrophoresis followed by silver staining, as described previously (6), to visualize viral gene segments.
Nluc luciferase assay.
Monolayers of MA104 cells in 12-well plates (Iwaki) were infected in triplicate with trypsin-pretreated recombinant RVAs at an MOI of 0.01, washed twice with incomplete medium, and then incubated for various numbers of hours in incomplete medium with trypsin (1 μg/ml). Nluc activities in cell cultures were determined using the Nano-Glo luciferase assay system (Promega) with a luminometer (2030 ARVO X5; PerkinElmer).
Multiple-step virus replication.
Monolayers of MA104 cells in 12-well plates (Iwaki) were infected in triplicate with trypsin-pretreated recombinant RVAs at an MOI of 0.01, washed twice with incomplete medium, and then incubated in incomplete medium with trypsin (1 μg/ml) for various numbers of hours. The infected cells were frozen and thawed twice before titration of virus titers by plaque assay.
Plaque assay.
The plaque assays were performed as described previously (38). Briefly, monolayers of CV-1 cells were adsorbed with trypsin-pretreated recombinant RVAs, washed twice with incomplete medium, and then cultured with trypsin (1 μg/ml) in primary overlay medium containing 0.7% agarose. After 2 days of incubation, the cells were stained with secondary overlay medium containing 0.005% neutral red (Sigma-Aldrich) and 0.7% agarose. Plaque sizes were determined by measuring the mean diameters of 25 plaques in two independent assays.
Focus-forming assay.
Monolayers of CV-1 cells in 6-well plates (Falcon) were infected with trypsin-pretreated recombinant RVAs at an MOI of 1, washed twice with incomplete medium, and then incubated with trypsin (1 μg/ml) in overlay medium (0.7% agarose). After 34 h, RVA foci expressing fluorescent EGFP and mCherry proteins were observed under a confocal laser microscope (LSM-710; Carl Zeiss).
Generation of NSP1-specific antiserum.
A polyclonal antibody recognizing NSP1 was generated by Medical and Biological Laboratories. Two RVA-seronegative rabbits were immunized subcutaneously five times with mixed synthetic peptides (corresponding to 216-LPSSKLKQIYFSDFTKE-232 and 234-VIFNTYTKTPGRSIYRN-250 of SA11-L2 NSP1) conjugated to keyhole limpet hemocyanin.
Immunoblotting.
Immunoblotting was carried out as described previously (39, 40). Briefly, monolayers of MA104 cells in 12-well plates (Iwaki) were transfected with 2.5 μg of pCAGGS/NSP1SA11 expression plasmid using FuGENE HD transfection reagent (Promega). Twenty-four hours after transfection, the cells were washed twice with PBS and then lysed in SDS-PAGE sample buffer (63 mM Tris-HCl [pH 6.8], 2% SDS, 5% sucrose, 5% 2-mercaptoethanol, and 0.002% bromophenol blue). Protein expression was assessed by SDS-PAGE (12%) and immunoblotting using anti-NSP1 polyclonal antiserum. The antigen-antibody complex was detected using a 1-step Ultra TMB blotting solution system (Thermo Fisher Scientific).
Transfection/infection assay.
Monolayers of MA104 cells in 12-well plates (Iwaki) were transfected with 2.5 μg of pCAGGS/NSP1SA11 expression plasmid. Twenty-four hours after transfection, the cells were infected with trypsin-pretreated recombinant RVAs, washed twice with incomplete medium, and then incubated for 36 h in incomplete medium with trypsin (1 μg/ml). Virus titers in the cultures were determined by plaque assay.
Statistics.
Virus titers and Nluc luciferase activities were evaluated by means of an unpaired t test. Statistical analyses were performed using GraphPad Prism7 (GraphPad Software). P values of <0.05 were considered statistically significant.
Accession number(s).
The sequences determined in the course of this work have been deposited in GenBank/EMBL/DDBJ under accession numbers LC333802 to LC333812.
ACKNOWLEDGMENTS
We thank Naoe Kotomura, Satoru Ishihara, Naoki Yahata, Akiko Kondo, Kentaro Tsukamoto, and Naoki Yamamoto (Fujita Health University, Aichi) for their technical advice.
This study was supported in part by the MEXT-Supported Program for the Research Program on Emerging and Re-emerging Infectious Diseases of the Japan Agency for Medical Research and Development, AMED (K.T. and S.K.), and JSPS KAKENHI grant number 15K08505 (S.K.).
We have no conflicts of interests to declare.
REFERENCES
- 1.Tate JE, Burton AH, Boschi-Pinto C, Parashar UD, World Health Organization-Coordinated Global Rotavirus Surveillance Network . 2016. Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000-2013. Clin Infect Dis 62(Suppl 2):S96–S105. doi: 10.1093/cid/civ1013. [DOI] [PubMed] [Google Scholar]
- 2.Estes MK, Greenberg HB. 2013. Rotaviruses, p 1347–1401. In Knipe DM, Howley PM (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Komoto S, Taniguchi K. 2013. Genetic engineering of rotaviruses by reverse genetics. Microbiol Immunol 57:479–486. [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi K, Komoto S. 2012. Genetics and reverse genetics of rotavirus. Curr Opin Virol 2:399–407. doi: 10.1016/j.coviro.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Johne R, Reetz J, Kaufer BB, Trojnar E. 2016. Generation of an avian-mammalian rotavirus reassortant by using a helper virus-dependent reverse genetics system. J Virol 90:1439–1443. doi: 10.1128/JVI.02730-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komoto S, Sasaki J, Taniguchi K. 2006. Reverse genetics system for introduction of site-specific mutations into the double-stranded RNA genome of infectious rotavirus. Proc Natl Acad Sci U S A 103:4646–4651. doi: 10.1073/pnas.0509385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trask SD, Taraporewala ZF, Boehme KW, Dermody TS, Patton JT. 2010. Dual selection mechanisms drive efficient single-gene reverse genetics for rotavirus. Proc Natl Acad Sci U S A 107:18652–18657. doi: 10.1073/pnas.1011948107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Troupin C, Dehée A, Schnuriger A, Vende P, Poncet D, Garbarg-Chenon A. 2010. Rearranged genomic RNA segments offer a new approach to the reverse genetics of rotaviruses. J Virol 84:6711–6719. doi: 10.1128/JVI.00547-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanai Y, Komoto S, Kawagishi T, Nouda R, Nagasawa N, Onishi M, Matsuura Y, Taniguchi K, Kobayashi T. 2017. Entirely plasmid-based reverse genetics system for rotaviruses. Proc Natl Acad Sci U S A 114:2349–2354. doi: 10.1073/pnas.1618424114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komoto S, Kanai Y, Fukuda S, Kugita M, Kawagishi T, Ito N, Sugiyama M, Matsuura Y, Kobayashi T, Taniguchi K. 2017. Reverse genetics system demonstrates that rotavirus non-structural protein NSP6 is not essential for viral replication in cell culture. J Virol 91:e00695-17. doi: 10.1128/JVI.00695-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breen M, Nogales A, Baker SF, Martínez-Sobrido L. 2016. Replication-competent influenza A viruses expressing reporter genes. Viruses 8:E179. doi: 10.3390/v8070179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rostad CA, Currier MC, Moore ML. 2016. Fluorescent and bioluminescent reporter myxoviruses. Viruses 8:E214. doi: 10.3390/v8080214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navarro A, Trask SD, Patton JT. 2013. Generation of genetically stable recombinant rotaviruses containing novel genome rearrangements and heterologous sequences by reverse genetics. J Virol 87:6211–6220. doi: 10.1128/JVI.00413-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campagna M, Eichwald C, Vascotto F, Burrone OR. 2005. RNA interference of rotavirus segment 11 mRNA reveals the essential role of NSP5 in the virus replicative cycle. J Gen Virol 86:1481–1487. doi: 10.1099/vir.0.80598-0. [DOI] [PubMed] [Google Scholar]
- 15.Fabbretti E, Afrikanova I, Vascotto F, Burrone OR. 1999. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J Gen Virol 80:333–339. doi: 10.1099/0022-1317-80-2-333. [DOI] [PubMed] [Google Scholar]
- 16.López T, Rojas M, Ayala-Bretón C, López C, Arias CF. 2005. Reduced expression of the rotavirus NSP5 gene has a pleiotropic effect on virus replication. J Gen Virol 86:1609–1617. doi: 10.1099/vir.0.80827-0. [DOI] [PubMed] [Google Scholar]
- 17.Silvestri LS, Taraporewala ZF, Patton JT. 2004. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J Virol 78:7763–7774. doi: 10.1128/JVI.78.14.7763-7774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vascotto F, Campagna M, Visintin M, Cattaneo A, Burrone OR. 2004. Effects of intrabodies specific for rotavirus NSP5 during the virus replicative cycle. J Gen Virol 85:3285–3290. doi: 10.1099/vir.0.80075-0. [DOI] [PubMed] [Google Scholar]
- 19.Petrie BL, Graham DY, Hanssen H, Estes MK. 1982. Localization of rotavirus antigens in infected cells by ultrastructural immunocytochemistry. J Gen Virol 63:457–467. doi: 10.1099/0022-1317-63-2-457. [DOI] [PubMed] [Google Scholar]
- 20.Petrie BL, Greenberg HB, Graham DY, Estes MK. 1984. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res 1:133–152. doi: 10.1016/0168-1702(84)90069-8. [DOI] [PubMed] [Google Scholar]
- 21.Delmas O, Gardet A, Chwetzoff S, Breton M, Cohen J, Colard O, Sapin C, Trugnan G. 2004. Different ways to reach the top of a cell. Analysis of rotavirus assembly and targeting in human intestinal cells reveals an original raft-dependent, Golgi-independent apical targeting pathway. Virology 327:157–161. [DOI] [PubMed] [Google Scholar]
- 22.Gibbons TF, Storey SM, Williams CV, Mclntosh A, Mitchel DM, Parr RD, Schroeder ME, Schroeder F, Ball JM. 2011. Rotavirus NSP4: cell type-dependent transport kinetics to the exofacial plasma membrane and release from intact infected cells. Virol J 8:278. doi: 10.1186/1743-422X-8-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY. 2011. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma P, Yan F, Doronina VA, Escuin-Ordinas H, Ryan MD, Brown JD. 2012. 2A peptides provide distinct solutions to driving stop-carry on translational recoding. Nucleic Acids Res 40:3143–3151. doi: 10.1093/nar/gkr1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taniguchi K, Kojima K, Urasawa S. 1996. Nondefective rotavirus mutants with an NSP1 gene which has a deletion of 500 nucleotides, including a cysteine-rich zinc finger motif-encoding region (nucleotides 156 to 248), or which has a nonsense codon at nucleotides 153 to 155. J Virol 70:4125–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patton JT, Taraporewala Z, Chen D, Chizhikov V, Jones M, Elhelu A, Collins M, Kearney K, Wagner M, Hoshino Y, Gouvea V. 2001. Effect of intragenic rearrangement and changes in the 3′ consensus sequence on NSP1 expression and rotavirus replication. J Virol 75:2076–2086. doi: 10.1128/JVI.75.5.2076-2086.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clarke IN, McCrae MA. 1982. Structural analysis of electrophoretic variation in the genome profiles of rotavirus field isolates. Infect Immun 36:492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roner MR, Joklik WK. 2001. Reovirus reverse genetics: incorporation of the CAT gene into the reovirus genome. Proc Natl Acad Sci U S A 98:8036–8041. doi: 10.1073/pnas.131203198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuo E, Roy P. 2013. Minimum requirements for bluetongue virus primary replication in vivo. J Virol 87:882–889. doi: 10.1128/JVI.02363-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Rijn PA, van de Water SG, Freenstra F, van Gennip RG. 2016. Requirements and comparative analysis of reverse genetics for bluetongue virus (BTV) and African horse sickness virus (AHSV). Virol J 13:119. doi: 10.1186/s12985-016-0574-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demidenko AA, Blattman JN, Blatmann NN, Greenberg PD, Nibert ML. 2013. Engineering recombinant reoviruses with tandem repeats and a tetravirus 2A-like element for exogenous polypeptide expression. Proc Natl Acad Sci U S A 110:E1867–E1876. doi: 10.1073/pnas.1220107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito N, Takayama-Ito M, Yamada K, Hosokawa J, Sugiyama M, Minamoto N. 2003. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiol Immunol 47:613–617. doi: 10.1111/j.1348-0421.2003.tb03424.x. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi K, Nishikawa K, Kobayashi N, Urasawa T, Wu H, Gorziglia M, Urasawa S. 1994. Differences in plaque size and VP4 sequence found in SA11 virus clones having simian authentic VP4. Virology 198:325–330. doi: 10.1006/viro.1994.1035. [DOI] [PubMed] [Google Scholar]
- 34.Schnell MJ, Mebatsion T, Conzelmann KK. 1994. Infectious rabies viruses from cloned cDNA. EMBO J 13:4195–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. doi: 10.1016/0378-1119(91)90434-D. [DOI] [PubMed] [Google Scholar]
- 36.Moss B, Elroy-Stein O, Mizukami T, Alexander WA, Fuerst TR. 1990. Product review. New mammalian expression vectors. Nature 348:91–92. [DOI] [PubMed] [Google Scholar]
- 37.Taniguchi K, Morita Y, Urasawa T, Urasawa S. 1987. Cross-reactive neutralization epitopes on VP3 of human rotavirus: analysis with monoclonal antibodies and antigenic variants. J Virol 61:1726–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urasawa S, Urasawa T, Taniguchi K. 1982. Three human rotavirus serotypes demonstrated by plaque neutralization of isolated strains. Infect Immun 38:781–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komoto S, Kinomoto M, Horikoshi H, Shiraga M, Kurosu T, Mukai T, Auwanit W, Otake T, Oishi I, Ikuta K. 2002. Ability to induce p53 and caspase-mediated apoptosis in primary CD4+ T cells is variable among primary isolates of human immunodeficiency virus type 1. AIDS Res Hum Retrovir 18:435–446. doi: 10.1089/088922202753614209. [DOI] [PubMed] [Google Scholar]
- 40.Komoto S, Wakuda M, Ide T, Niimi G, Maeno Y, Higo-Moriguchi K, Taniguchi K. 2011. Modification of the trypsin cleavage site of rotavirus VP4 to a furin-sensitive form does not enhance replication efficiency. J Gen Virol 92:2914–2921. doi: 10.1099/vir.0.033886-0. [DOI] [PubMed] [Google Scholar]