Abstract
Background and Purpose
TRPM4 is a calcium‐activated non‐selective cation channel expressed in many tissues and implicated in several diseases, and has not yet been validated as a therapeutic target due to the lack of potent and selective inhibitors. We sought to discover a novel series of small‐molecule inhibitors by combining in silico methods and cell‐based screening assay, with sub‐micromolar potency and improved selectivity from previously reported TRPM4 inhibitors.
Experimental Approach
Here, we developed a high throughput screening compatible assay to record TRPM4‐mediated Na+ influx in cells using a Na+‐sensitive dye and used this assay to screen a small set of compounds selected by ligand‐based virtual screening using previously known weakly active and non‐selective TRPM4 inhibitors as seed molecules. Conventional electrophysiological methods were used to validate the potency and selectivity of the hit compounds in HEK293 cells overexpressing TRPM4 and in endogenously expressing prostate cancer cell line LNCaP. Chemical chaperone property of compound 5 was studied using Western blots and electrophysiology experiments.
Key Results
A series of halogenated anthranilic amides were identified with TRPM4 inhibitory properties with sub‐micromolar potency and adequate selectivity. We also showed for the first time that a naturally occurring variant of TRPM4, which displays loss‐of‐expression and function, is rescued by the most promising compound 5 identified in this study.
Conclusions and Implications
The discovery of compound 5, a potent and selective inhibitor of TRPM4 with an additional chemical chaperone feature, revealed new opportunities for studying the role of TRPM4 in human diseases and developing clinical drug candidates.
Abbreviations
- FFA
flufenamic acid
- GABAA
Gamma‐aminobutyric acid receptor subunit alpha
- NMDA
N‐methyl‐D‐aspartate
- LNCaP
Lymph node carcinoma of the prostate
- TRPM
Transient receptor potential melastatin
- TRPV
Transient receptor potential vanilloid
- xLOS
extended Ligand Overlap Score
Introduction
The ion channel http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=496 belongs to the 28‐member TRP (transient receptor potential) channel family. It is activated upon an increase in intracellular Ca2+ and conducts monovalent cations such as Na+, K+ and Cs+, hence modulating the transmembrane electrical potential. TRPM4 is expressed in many cell types and tissues, but its role in physiology is still poorly understood (Launay et al., 2002; Nilius et al., 2003; Fonfria et al., 2006). With regard to its role in human diseases, more than 20 TRPM4 genetic variants have been described in families with cardiac conduction alterations such as progressive conduction block and congenital atrioventricular block (AVB) (Kruse and Pongs, 2014; Syam et al., 2016). A significant number of these pathogenic TRPM4 variants lead to gain‐of‐expression and function, while others to loss‐of‐expression and function (Kruse et al., 2009; Liu et al., 2010; 2013; Syam et al., 2016). Based on studies with experimental autoimmune encephalitis (EAE) mice model, TRPM4 has been suggested to be a key player in the neurodegenerative process of multiple sclerosis (Schattling et al., 2012; Malhotra et al., 2013; Makar et al., 2015). Interestingly, TRPM4‐deficient mice with EAE displayed much milder clinical manifestations than wild‐type (WT) EAE mice (Schattling et al., 2012). Recently, TRPM4 has also been shown to be involved in migration and proliferation of human prostate cancer cells (Ashida et al., 2004; Singh et al., 2006; Suguro et al., 2006; Prevarskaya et al., 2007; Schinke et al., 2014; Holzmann et al., 2015; Berg et al., 2016; Sagredo et al., 2018). Taken together, these recent findings strongly suggest that TRPM4 is a potential pharmacological target for treating neurological and cardiovascular disorders, as well as certain types of cancer.
Most of the characterizations of TRPM4 in different tissues have heavily relied on the patch clamp technique with pharmacological inhibitors that have several issues. Until now, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4114 has been the most commonly used TRPM4 inhibitor to dissect its roles in physiology and diseases (Figure 1) (Grand et al., 2008; Guinamard et al., 2014). However, the low potency and lack of selectivity of 9‐phenanthrol on TRPM4 limit its usefulness in studies using animal models or primary cell lines (Burris et al., 2015; Garland et al., 2015). Other reported TRPM4 inhibitors include http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2447 (Figure 1), acting similarly weakly on TRPM4 but with a longer list of documented off‐target effects that are related to its primary role as a non‐steroidal anti‐inflammatory drug (Gardam et al., 2008). While http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2414, an even weaker TRPM4 inhibitor also inhibits ATP‐dependent K+ channels, which are its primary targets in treatment of type II diabetes (Demion et al., 2007; Alexander et al., 2017d).
Figure 1.
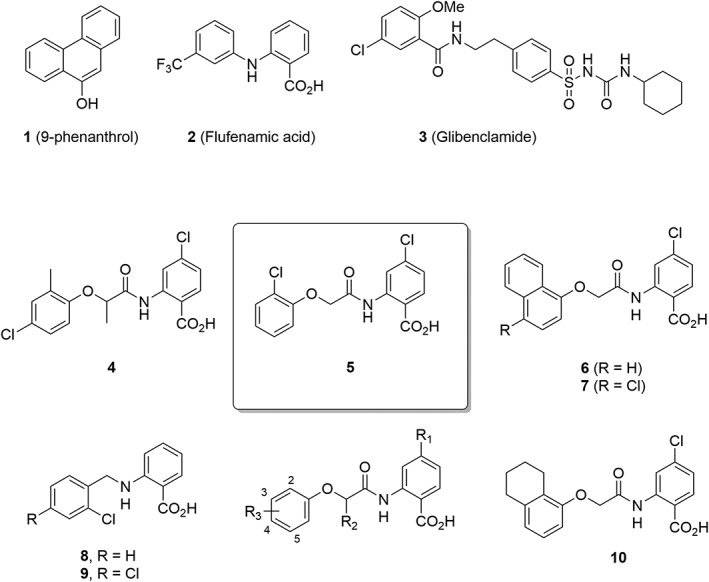
Chemical structures of TRPM4 inhibitors used in this study.
Here, we developed a fluorescence cell‐based screening assay monitoring TRPM4‐induced Na+ influx using an intracellular Na+ specific dye (Asante Natrium Green‐II, ANG‐II). This assay allowed a reliable assessment of TRPM4 activity modulation by small molecules. In the absence of structural information on inhibitor binding sites on TRPM4, we used a ligand‐based virtual screening (LBVS) approach (Scior et al., 2012) to search commercially available compounds for analogues of the abovementioned weak and non‐selective TRPM4 inhibitors (Sterling and Irwin, 2015). We performed LBVS with the 3D‐shape and pharmacophore similarity algorithm xLOS (atom category eXtended Ligand Overlap Score) as a similarity measure. xLOS is a scaffold‐hopping algorithm (Schneider et al., 1999) that recently proved its validity in related searches for selective Ca2+ channel (Simonin et al., 2015) and kinase inhibitors (Kilchmann et al., 2016). Activity screening of a small series of compounds selected by the LBVS procedure allowed us to rapidly identify anthranilic amides 4 and 5, two analogues of FFA with a 10‐fold stronger and more selective inhibition of TRPM4 compared with 9‐phenanthrol (Figure 1). A structure–activity relationship (SAR) study further uncovered 6 as the first selective sub‐micromolar TRPM4 inhibitor. These TRPM4 blockers reversibly inhibited the overexpressed TRPM4 channels in HEK293 cells and blocked TRPM4‐mediated currents in human prostate cancer cells. Their direct and specific interaction with TRPM4 is further exemplified by their activity as a chemical chaperone to restore the expression and activity of a loss‐of‐function TRPM4 variant. These results prepare the way for more potent and specific TRPM4 inhibitors.
Methods
Ligand‐based virtual screening
The first round of virtual screening (VS) was performed in the Princeton database, which contains 900 000 purchasable compounds, using three different reference compounds, namely, 9‐phenanthrol, glibenclamide and flufenamic acid. The VS was carried out with an in‐house developed extended Ligand Overlap Score (xLOS) method, which computes the 3D‐shape and –pharmacophore similarity between any two compounds (typically reference and database compounds) (Simonin et al., 2015). The single lowest energy 3D‐models of reference and database molecules were generated using the CORINA programme available from Molecular Networks Pvt. Ltd. xLOS used these 3D models to calculate a 3D‐similarity score between the reference compound and the database molecules. Afterward, for each of the three reference molecules, 235 molecules were visually selected from the top 1000 xLOS scoring compounds, 214 of those were purchased from Princeton Biomolecular Research. These compounds were dissolved in DMSO solution and tested on TRPM4 activity. Next, three hits from this first round of VS were used for a second round of VS in the Princeton database using xLOS. From this second round, 247 compounds were selected and purchased for biological testing.
Visualization of screening library in 3D
The visualization of screening library was facilitated by an in‐house developed webMolCS web server (http://www.gdb.unibe.ch). WebMolCS takes a user‐defined list of molecules (in SMILES format) as input and produces interactive colour‐coded 3D‐maps using either principal component analysis or similarity mapping of any of the six different molecular fingerprints used for molecular representation. In the present study, similarity mapping (Sim map) and substructures fingerprint (sFP) were used to generate the 3D‐maps. The colours of the 3D‐maps shown in Figure 8 represent the similarity (calculated using sFP fingerprint) of the screened compounds to the best compound 6 from synthetic optimization; from low to high similarity: blue‐cyan‐green‐yellow (Awale and Reymond, 2017).
Figure 8.
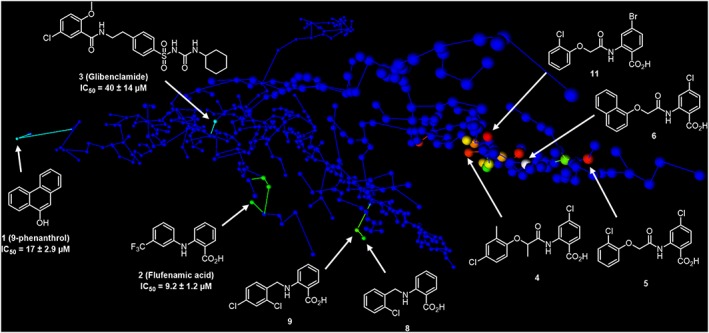
Chemical space analysis. Selected view of an interactive 3D map of substructure fingerprint similarity representing each of the 470 compounds tested as one sphere. The map is colour‐coded by pIC50 value from blue (lowest value) to red (highest value). The interactive map is accessible at http://gdbtools.unibe.ch:8080/webMolCS/yourSIM.html?jobID=1506356578836&fp=Sfp.
Chemical synthesis of compounds
The synthesis and characterization of all compounds are described in the Supporting Information.
Cell culture
HEK293 and tetracycline‐inducible HEK293 Flag‐TRPM4‐expressing cells were used. The HEK293 cells were a gift from Dr R.S. Kass (Columbia University, NY, USA). These cells were given to the group of Dr R.S. Kass by Dr B. Stillman (Cold Spring Harbor Laboratories, Cold Spring, USA). Tetracycline‐inducible HEK293 Flag‐TRPM4 cells were a gift from Dr P. Bouvagnet (University of Lyon, France). These cells were cultured at 37°C in DMEM supplemented with 10% FBS (Invitrogen, CA, USA), 4 mM glutamine (Sigma), 5 μg·mL−1 S‐blasticidin (Invitrogen) and 0.4 mg·mL−1 http://topics.sciencedirect.com/topics/page/Zeocin (Invitrogen). TRPM4 expression was induced by adding 1 μg·mL−1 of tetracycline to the induction medium [DMEM without phenol red (Gibco 31053, Paisley, UK) supplemented with 10% FBS (Invitrogen) and 2% L‐glutamine (Sigma)] 15–20 h before the experiment. For TRPM4 variant studies, HEK293 cells cultured at 37°C in DMEM supplemented with 10% FBS and 4 mM glutamine were transiently transfected with 250 ng of HA‐tagged TRPM4 WT or HA‐tagged TRPM4 p.A432T in a 100 mm dish, mixed with 4 μL of JetPEI (Polyplus transfection, Illkirch, France) and 46 μL of 150 mM NaCl. The cells were incubated for 24 h at 37°C with 5% CO2. All transfections included 100 ng of eGFP as a reporter gene. Cells were used 24 h after transfection. LNCaP cells were purchased from the American Type Cell Culture Collection (ATCC, Rockville, MD, USA) and cultured in RPMI Medium 1640 (Gibco) supplemented with 10% FCS and 1% penicillin/streptomycin (Invitrogen).
The TRPM4 KD cell line was provided by Transposagen Biopharmaceuticals (Lexington, KY, USA) and generated using the CRISPR/Cas9 technique. The cells were transfected with both the Cas9 vector and the corresponding U6 expressed sgRNA vector in a 1:1 molar ratio. Transfection was done with Lipofectamine 2000 (ThermoFischer Scientific, MA, USA) following the manufacturer's protocols. Loss of TRPM4 gene function and protein was then checked in single clones amplified using qPCR and Western blot. The selected clones were cultured and expanded in conditions similar to those used for the HEK293 cells, as mentioned above.
sgRNA targets:
| TRPM4 TS1 | GCCACCTCGCCGCTCTCGC |
| TRPM4 TS2 | GCTCCAGGGGGCCTGCCCG |
Compound cytotoxicity assay
The cytotoxicity of compounds 4, 5 and 6 was studied in RAW 264.7 and HeLa cells for anti‐proliferative effects. The cells were seeded at 2000 cells per well in 100 μL culture media in a 96‐well plate. Following overnight incubation, different concentrations of the compounds were added and incubated together with the solvent control for 72 h at 37°C. After that, the cells were incubated with MTT (3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide, final concentration 0.5 mg·mL−1) for 4 h at 37°C and solubilized with DMSO. The plates were measured at 550 nm (absorbance) and the IC50 value related to cell survival/proliferation was calculated and plotted. Paclitaxel was used as a positive control. Experiments were performed in at least three independent experiments, each in triplicate.
Western blot
To detect TRPM4 protein expression, whole‐cell lysates were prepared by lysing cells in lysis buffer [50 mM HEPES pH 7.4; 150 mM NaCl; 1.5 mM MgCl2, 1 mM EGTA pH 8.0; 10% glycerol; 1% Triton X‐100; 1× complete protease inhibitor cocktail (Roche, Mannheim, Germany)] for 1 h at 4°C. Cell lysates were centrifuged for 15 min at 16 000 × g at 4°C, and protein concentration was evaluated using the Bradford assay. Sixty micrograms of each protein sample was run on 9% polyacrylamide gels, later transferred with the TurboBlot dry blot system (Biorad, Hercules, CA, USA). After transfer, membranes were blocked with 0.1% BSA in PBS and incubated with primary antibodies anti‐TRPM4 (generated by Pineda, Berlin, Germany) and anti‐α‐actin A2066 (Sigma‐Aldrich, St. Louis, MO, USA) using the SNAP i.d. system (Millipore, Billerica, MA, USA), followed by an incubation with secondary antibody, IRDye 800CW, (LI‐COR Biosciences, Lincoln, NE, USA). Membranes were scanned using LiCor Odyssey Infrared imaging system (LI‐COR Biosciences), and protein band intensity was quantified using Image Studio Lite software from LI‐COR Biosciences.
For detecting TRPM4 WT and A432T variant whole‐cell (total) and surface expression after incubation with compound 5, the cells were washed twice with cold 1× PBS and treated with EZlinkTM Sulfo‐NHS‐SS‐Biotin (Thermo Scientific, Waltham, MA, USA) 0.5 mg·mL−1 in cold 1× PBS for 15 min at 4°C. Subsequently, the cells were washed twice with 200 mM glycine in cold 1× PBS and twice with cold 1× PBS to inactivate and to remove the excess biotin respectively. The cells were then lysed with 1× lysis buffer for 1 h at 4°C. Cell lysates were centrifuged at 16 000 × g at 4°C for 15 min. Two milligrams of the supernatant were incubated with 50 μL Streptavidin Sepharose High Performance beads (GE Healthcare, Uppsala, Sweden) for 2 h at 4°C, and the remaining supernatant was kept as the input. The beads were subsequently washed five times with 1× lysis buffer before elution with 50 μL of 2× NuPAGE sample buffer (Invitrogen) plus 100 mM DTT at 37°C for 30 min. These biotinylated fractions were analysed as TRPM4 expressed at the cell surface. The input fractions, analysed as total expression of TRPM4, were resuspended with 4× NuPAGE Sample Buffer plus 100 mM DTT to give a concentration of 1 mg·mL−1 and incubated at 37°C for 30 min.
Na+‐influx screening assay
TRPM4‐expressing cells were plated in 96‐well black‐walled clear bottomed poly‐D‐lysine coated plates (Corning, NY, USA) at a density of 30 000 cells per well in 100 μL of induction medium (described above). After being plated, cells were incubated for 48 h at 37°C in a 5% CO2 incubator. On the day of the assay, the induction medium was replaced with 100 μL per well of the assay buffer [in mM: 140 NMDG‐Cl, 10 KCl, 1 CaCl2, 1 MgCl2, 10 HEPES (pH 7.2) with NMDG‐OH and 290 to 300 mOsM] and incubated for 45 min at room temperature (RT). Later, the assay buffer was replaced with 95 μL per well of dye loading solution prepared in assay buffer supplemented with 0.1% pluronic acid F‐127 (Teflabs, TX, USA) and 5 μM of Na+ sensitive dye, Asante Natrium Green–II (ANGII) (Teflabs). The cells were incubated in the dark at RT for 45 min. After incubation, the dye loading solution was completely drained and replaced with 72 μL assay buffer and incubated in the dark for 20 min, followed by fluorescence readout on FLIPR TETRA® (Molecular Devices, CA, USA).
For evaluation of the effects of TRPM4 inhibitors using the Na+ influx assay, 10 compound plates were prepared in assay buffer in 96‐well polystyrene plates. All stocks were dissolved in DMSO. To initiate Na+ influx through activation of TRPM4, a 5× stimulus buffer plate containing (in mM) 700 NaCl, 4 CaCl2, 4 MgCl2, 40 HEPES, (pH 7.2 with NaOH) along with 50 μM of ionomycin was prepared in a 96‐well polystyrene plate. The background control wells contained stimulus buffer without ionomycin (W/O).
Fluorophores were excited by the 488 nm line of an argon laser. Emission was filtered with a 540 ± 30 nm bandpass filter. Initial baseline fluorescence was measured at 1 Hz from the plate loaded with ANGII dye for 1 min. Followed by addition of 8 μL solution from 10× compound plate, fluorescence was monitored at an interval of 1 Hz for 5 min. Finally, to activate TRPM4, 20 μL of solution from the stimulus buffer plate was added, and fluorescence was acquired at 0.5 Hz intervals during and after addition for 5 min. Data from individual assay wells were normalized to initial baseline fluorescence. The initial rise in fluorescence was measured as area under the curve (AUC), and the counts were normalized using the formula {[(Ionomycin − Compound)/(Ionomycin − W/O Ionomycin)] × 100} to plot concentration–response curves for inhibitor hits. The experimenter for the screening experiments (L.C.O.) was blinded and was not aware of any chemical properties of the compounds until the best hits were validated. The steps are summarized as workflow, presented in Table 1.
Table 1.
Steps for Na+ influx screening assay campaign
| Assay Workflow | |||
|---|---|---|---|
| No. | Step | Value | Detail |
| 1 | Cell plating and induction | 100 μL |
30 000 cells per well Incubation for 48 h |
| Drain the content | |||
| 2 | Na+ depletion | 100 μL |
Assay buffer Incubation for 45 min |
| Drain the content | |||
| 3 | Dye loading | 95 μL |
ANGII dye prepared in assay buffer Incubation in dark for 45 min at RT |
| Drain the content | |||
| 4 | Assay buffer | 72 μL | Store plate in dark at RT |
| 5 | Baseline readout (FLIPR) | 540 nm | Fluorescence readout 1 Hz for 1 min |
| 6 | Test compound | 8 μL |
10× compounds in assay buffer plate Incubation for 5 min FLIPR readout 1 Hz |
| 7 | Stimulus | 20 μL |
5× stimulus buffer plate Incubation for 5 min FLIPR readout at 0.5 Hz |
Electrophysiology
Cells were plated in 35 mm poly‐D‐lysine coated dishes in induction medium [phenol red free DMEM medium (GIBCO‐31053)] supplemented with 10% FBS and 1 μg·mL−1 tetracycline (Invitrogen). After being plated, cells were incubated for 48 h at 37°C in a 5% CO2 incubator. Electrophysiological recordings were performed in the inside‐out patch clamp configuration with patch pipettes (1–2 μm tip opening) pulled from 1.5 mm borosilicate glass capillaries (World Precision Instruments, Inc., FL, USA) using DMZ Universal puller (Zeitz‐Instruments, GmbH, München, Germany). Pipette tips were polished to have a pipette resistance of 2–4 M Ω in the bath solution. The pipette solution contained (in mM) 150 NaCl, 10 HEPES, 2 CaCl2 (pH 7.4 with NaOH). The bath solution contained (in mM) 150 NaCl, 10 HEPES, 2 HEDTA (pH 7.4 with NaOH) as Ca2+‐free solutions. Solutions containing 0.1 to 2 mM Ca2+ were prepared by adding the appropriate concentration of CaCl2 without a Ca2+ chelator to a solution containing (in mM) 150 NaCl, 10 HEPES (pH 7.4 with NaOH) as reported previously (Zhang et al., 2005). Bath solutions with different Ca2+ concentrations were applied to cells by a modified rapid solution exchanger (Perfusion Fast‐Step SF‐77B; Warner Instruments Corp., CT, USA). Inhibitors in DMSO stock were diluted to appropriate concentrations in the pipette solution and applied to the extracellular side of the cells. Membrane currents were recorded with a Multiclamp 700B amplifier (Molecular Devices) controlled by Clampex 10 via a Digidata 1332A (Molecular Devices). Data were low‐pass filtered at 5 kHz and sampled at 10 kHz. Experiments were performed at RT (20–25°C). For I‐V relations, currents were recorded using a stimulation protocol consisting of voltage steps of 200 ms from a holding potential of 0 mV ranging from −80 to +100 mV, followed by a tail voltage at −100 mV. For constructing concentration–response curves, steady‐state currents at +100 mV recorded in 300 μM Ca2+ were normalized to current in absence of any inhibitor. For rescue experiments with compound 5, before performing patch clamp recordings the culture medium was replaced with bath solution without any added inhibitor.
For whole‐cell patch clamp on LNCaP cells, 50 ms voltage ramps spanning −100 to +100 mV were delivered every 2 s from a holding potential of 0 mV. Whole‐cell currents were recorded with a HEKA patch clamp system (EPC‐10, HEKA Elektronik Dr. Schulze GmbH, Lambrecht, Germany). Series resistance (Rs) was monitored in response to a 5 mV voltage step and compensated online up to 80% if needed. Bath solution contained (in mM): 140 NaCl, 0.5 CaCl2, 3 MgCl2, 10 HEPES. Glucose was added to adjust osmolarity to 330 mOsmol·L−1 and pH was adjusted to 7.2 with NaOH. Pipette solution contained (in mM): 140 Cs‐glutamate, 10 HEDTA, 10 HEPES, 8 NaCl, 12.24 MgCl2 and 0.00049 CaCl2. Free Mg2+ of 3 mM and 10 μM Ca2+ was calculated according to http://web.stanford.edu/~cpatton/webmaxcS.htm. Whole‐cell currents at −80 and +80 mV were extracted, normalized to the cell capacity and plotted versus time.
Data analysis
To quantify the Na+ influx after addition of stimulus buffer, the AUC was calculated using the ScreenWorks™ software (Molecular Devices). Data points acquired from the Na+ influx assay from FLIPR or electrophysiology data were exported and analysed using IGOR PRO 6 (Wavematrix, OR, USA). Z` factor was calculated as described previously (Zhang et al., 1999), using the following equation: Z` = 1 − [(3SDp + 3SDn)/(3Meanp − 3Meann)] where SD is standard deviation, and p and n are positive and negative control respectively. Michaelis–Menten constant for Na+ dependence of the signal was deduced using the following equation: V = [Vmax [S]/(K M + [S])], where Vmax is maximal fluorescence signal, S is Na+ concentration and K M is Michaelis–Menten constant. Concentration‐response curves were fitted using the Hill equation fit parameter of IGOR (NC = NCmax [Cmpd]nH/{[Cmpd]nH + EC50 nH}), where NC is normalized current or counts, [Cmpd] is compound concentration and n H is the Hill coefficient. Data are presented as mean ± SEM except for Figure 2A, panels i, ii, where each data point n = 2 and data are presented as mean ± SD. Statistically significant differences between means were determined using Student's t‐test for comparison between two means with n ≥ 5. P values ≤ 0.05 were accepted as significant and represented as * in respective figure panels. The data and statistical analyses comply with the recommendations on experimental design and analysis in pharmacology (Curtis et al., 2015).
Figure 2.
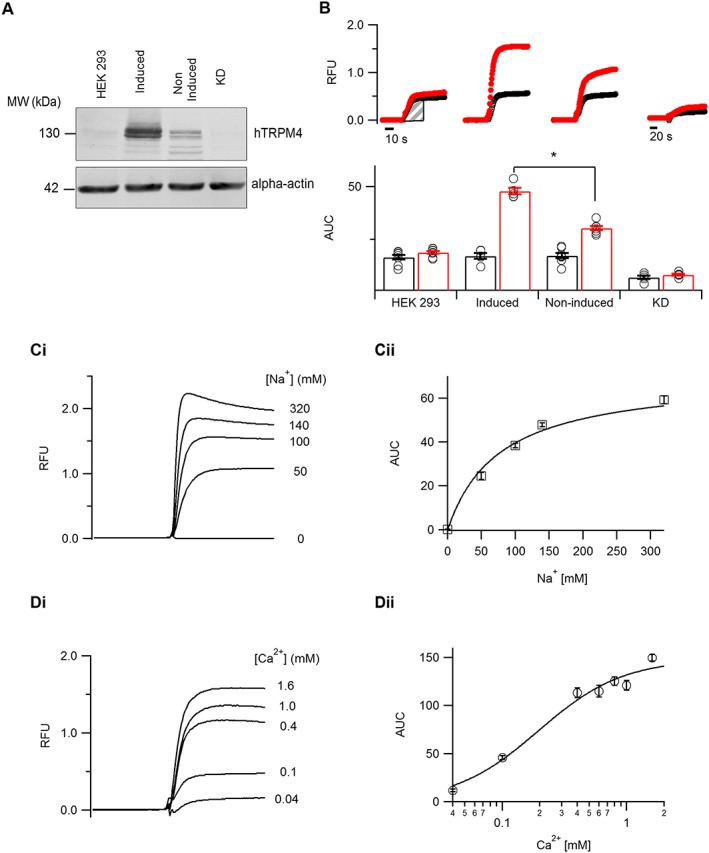
Development and validation of a Na+ influx screening assay. (A) Western blot analysis showing TRPM4 expression as detected by anti‐TRPM4 antibody from whole‐cell extracts prepared from HEK293 cells, TRPM4‐expressing cells either induced with tetracycline or non‐induced and TRPM4 CRISPR‐Cas9 knocked‐down cell line (KD). Alpha‐actin served as a loading control. (B) Averaged traces obtained on the FLIPR with Na+‐sensitive dye ANG‐II in different cellular conditions as mentioned (panel A). The initial rate of fluorescence increase in either the absence (black) or presence (red) of 10 μM ionomycin was deduced by measuring the AUC from the shaded portion for different cellular conditions (n ≥ 5). (C, panel i) Averaged traces obtained on the FLIPR from TRPM4‐expressing cells, with a varying concentration of extracellular Na+. (C, panel ii) The initial rates of fluorescence increase measured as AUC in different Na+ concentrations were fitted using the Michaelis–Menten equation (n ≥ 5). (D, panel i) Averaged traces obtained on the FLIPR from TRPM4‐expressing cells, with varying concentrations of extracellular Ca2+. (D, panel ii) The initial rates of fluorescence increase measured as AUC in different Ca2+ concentrations were fitted using the Hill equation (n = 6). *P ≤ 0.05, Student's t‐test.
Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY(Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 (Alexander et al., 2017a,b,c,d).
Results
Development and validation of a TRPM4 dependent Na+ influx‐screening assay
First, we developed a Na+ influx assay to screen large libraries of chemically diverse small molecules. We used a cell line stably expressing TRPM4 under a tetracycline‐inducible promoter (Amarouch et al., 2013). Functional membrane expression of TRPM4 in the tetracycline‐induced cells was compared with the non‐induced cells and non‐transfected HEK293 cells. Furthermore, endogenous expression of TRPM4 in HEK293 cells as reported in our previous studies (Amarouch et al., 2013) was knocked down by CRISPR/Cas9 technology (KD) (Figure 2A and Supporting Information Figure S1). In order to perform FLIPR (Fluorescent Imaging Plate Reader) based screening of compounds, the cells were pre‐incubated with Na+‐free assay buffer to deplete intracellular Na+, followed by loading the cells with a Na+‐sensitive ANG‐II dye. Reintroducing Na+ along with ionomycin in the stimulus buffer for Ca2+‐dependent activation of TRPM4 resulted in an increase of fluorescence signal over the baseline (Figure 2B). We used the AUC (marked area, Figure 2B) to quantify the signal intensity with or without ionomycin. Upon TRPM4 overexpression, the fluorescence signal increased threefold compared with non‐transfected HEK293 or KD cells (Figure 2B). The increase in fluorescence in non‐transfected HEK293 cells and non‐induced TRPM4 cells could be due to Na+ influx via various endogenous Na+ transporters (Figure 2B); however, the effect is significantly smaller than in TRPM4‐overexpressing cells (Figure 2B). Following the demonstration that the ANG‐II dye could detect Na+ influx, which was largely mediated through TRPM4 channel activation, we further assessed the dependence of the fluorescence signal on the extracellular Na+ and Ca2+ concentration in the stimulus buffer. We determined Na+ dependence with stimulus buffer containing different concentrations of Na+ by replacing the ion with equimolar N‐methyl‐d‐glucamine (NMDG). ANG‐II dye presented an affinity for Na+ ions with a Michaelis–Menten constant K M of 76.6 ± 6.3 mM (Figure 2C). Next, we determined Ca2+ dependence using stimulus buffer containing 140 mM Na+ and varying concentrations of extracellular Ca2+ without any chelator. The increase in fluorescence signal showed a dependence on Ca2+ concentration in the stimulus buffer with an EC50 of 0.2 ± 0.1 mM (Figure 2D). We finally assessed the robustness of the assay by monitoring the statistical Z` factor, which always scored 0.5 or more in all our experiments, while the ratio of AUC in presence and absence of ionomycin remained above 2 (Supporting Information Figure S2A, B ). Finally, the assay developed here tolerated DMSO up to 1%, the highest concentration used to dissolve the test compounds (Supporting Information Figure S2C, D ).
Discovery of small molecule TRPM4 inhibitors
To identify potent TRPM4 inhibitors, we performed a focused screening campaign using LBVS with xLOS (Simonin et al., 2015) to search a catalogue of 900 000 commercially available drug‐like small molecules for analogues of the known but relatively weak TRPM4 inhibitors 9‐phenanthrol, FFA and glibenclamide (Figure 1). We selected 214 compounds among the top scoring virtual hits, purchased 1 mg solid samples that we conditioned as 10 mM stock solution in DMSO and screened these compounds for inhibition of TRPM4 at 10 μM using 9‐phenanthrol (25 μM) as positive control (Figure 3A, panel i). Initial screen revealed three hits for which we purchased and tested an additional 247 analogues selected by LBVS with xLOS as well as other fingerprint similarity search methods (Awale and Reymond, 2014). As a result of these screening efforts, we identified four hits, compounds 4, 5, 8 and 9, repurchased 25 mg solid samples and confirmed their inhibitory activity at 5 μM concentration (Figure 1 and Figure 3A, panel ii). These compounds inhibited the initial rate of TRPM4‐mediated Na+ influx in a concentration‐dependent manner. The best TRPM4 inhibitors were the aryloxyacyl‐anthranilic acids 4 (IC50 = 1.6 ± 0.3 μM) and 5 [IC50 = 1.5 ± 0.1 μM, ligand efficiency (LE) = 0.371], showing a 20 times stronger inhibition than 9‐phenanthrol (IC50 = 29.1 ± 5.8 μM) (Figure 3B–D).
Figure 3.
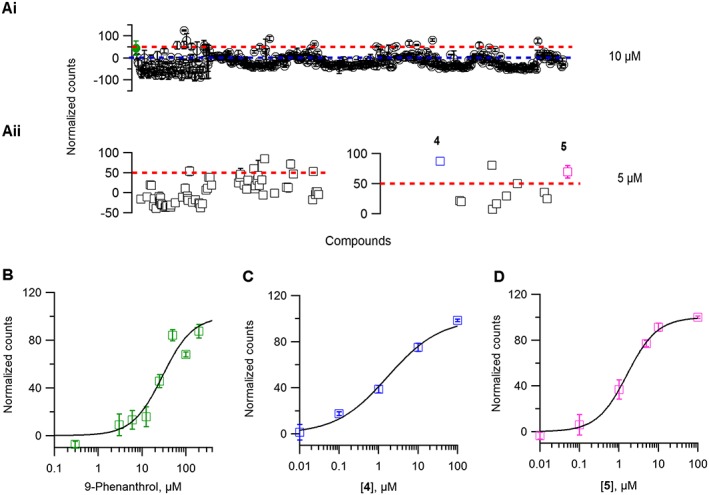
Screening campaign for TRPM4 inhibitor. (A, panel i) Representative scatter plot of normalized counts obtained from 470 compounds evaluated for inhibition of TRPM4‐mediated Na+ influx at 10 μM. Compounds above 0 (blue dotted line) were considered active. (A, panel ii) Compounds with counts 50 or more (red dotted line) were retested twice at 5 μM. Each data point is averaged from two independent 96‐well plates. (B–D) A concentration–response curve was constructed and the values were fitted with the Hill equation to extrapolate IC50 values for 9‐phenanthrol (B) and the newly identified inhibitors 4 (C), and 5 (D) n ≥ 4 for each concentration–response curve.
Structure–activity relationship (SAR) study of TRPM4 inhibitors
For SAR studies, we initially focused on anthranilic inhibitor 5 and devised a synthetic route for varying substituents on both aromatic rings and within the acetyl linker and prepared 49 analogues. (Figure 4A, Table 2 and Supporting Information Table S1–S3). Changing the substituents of the aromatic ring often abolished the activity of the compound. However, 13 anthranilic acid derivatives showed TRPM4 inhibition with IC50 values in the range 1–10 μM, confirming the robustness of the identified scaffold (Table 2, Figure 1). Overall, the results obtained with the different analogues indicated that TRPM4 inhibition required the presence of the anthranilic acid moiety with a halogen substituent at position 4 and a defined substitution pattern on the aryloxy group (Figure 4B). The SAR profile uncovered the 1‐naphthyloxy analogues 6 as the first submicromolar inhibitor in the series with an IC50 = 0.4 ± 0.3 μM (LE = 0.345) (Table 2). However, when considering its LE (Hopkins et al., 2014), compound 6 does not bring additional improvement over compound 5. On the other hand, pure enantiomers (>99% ee) of 4 (52 and 53) were obtained using preparative chiral‐phase HPLC and analysed for purity using the same method. Both enantiomers were similarly active, and chirality of the linker did not significantly influence activity (Supporting Information Table S3), while other modifications of the linker were detrimental to activity ( 54 to 58, Supporting Information Table S3).
Figure 4.
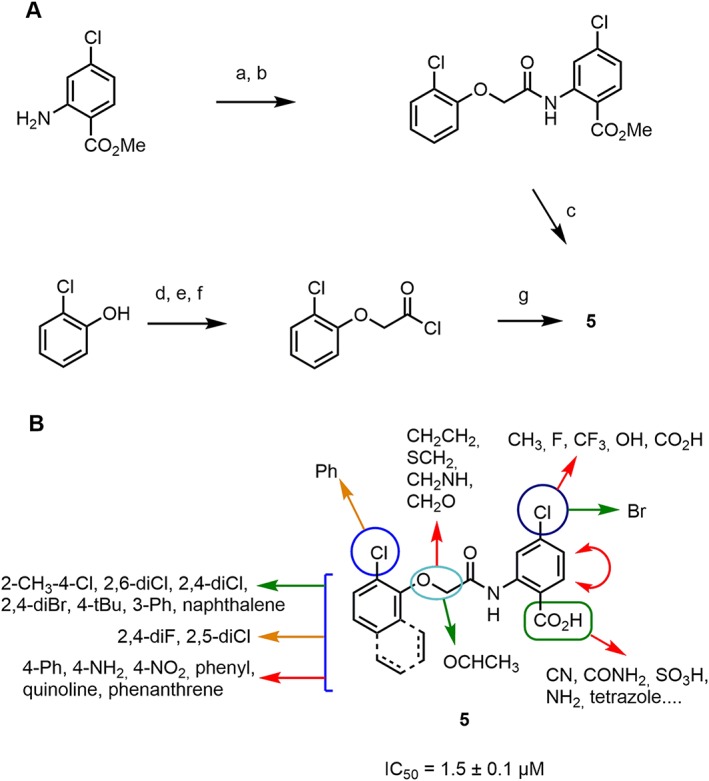
Virtual screening guided discovery and optimization of TRPM4 inhibitors. (A) Synthetic routes used to prepare compound 5 and analogues. Conditions: (a) ClCH2COCl, K2CO3, THF, RT, 4 h. (b) 2‐ClPhOH, K2CO3, DMF, 90°C, 2 h 30. (c) KOH, MeOH/H2O, reflux, 1 h or LiOH, dioxane, RT, 1 h. (d) ClCH2COOt‐Bu, K2CO3, DMF, 80°C, 1 h. (e) TFA, RT, 1 h. (f) SOCl2, DCM, reflux, 4 h. (g) 4‐Cl‐anthranilic acid, K2CO3, THF, RT, 2 h. (B) Overview of the structure–activity relationship uncovered by surveying 49 analogues. Green arrows indicate beneficial modifications and red arrows indicate deleterious ones.
Table 2.
IC50 values of TRPM4 inhibitors tested using the Na+ influx screening assay and electrophysiology recordings
| TRPM4 inhibitors | |||||
|---|---|---|---|---|---|
| ID | Na+ influx IC50 (μM) | Electrophysiology IC50 (μM) | R1 | R2 | R3 |
| 4 | 1.6 ± 0.3 | 1.0 ± 0.2 | Cl | CH3 | 2‐CH3‐4‐Cl |
| 5 | 1.5 ± 0.1 | 1.8 ± 0.1 | Cl | H | 2‐Cl |
| 11 | 1.0 ± 0.2 | 1.6 ± 0.5 | Br | H | 2‐Cl |
| 12 | 1.8 ± 0.2 | – | Cl | CH3 | 2,4‐diCl |
| 13 | 1.8 ± 0.1 | – | Cl | H | 2,4‐diBr |
| 14 | 3.4 ± 1.1 | – | Cl | H | 2,4‐diF |
| 15 | 2.6 ± 0.3 | – | Cl | H | 2‐Cl‐5‐nitro |
| 16 | 2.6 ± 0.4 | – | Cl | H | 3‐phenyl |
| 6 | 0.4 ± 0.3 | 0.2 ± 0.07 | – | – | – |
| 7 | 2.2 ± 0.4 | – | – | – | – |
| 8 | 8.3 ± 0.7 | – | – | – | – |
| 9 | 5.9 ± 0.5 | – | – | – | – |
| 10 | 1.2 ± 0.5 | – | – | – | – |
| 9P | 29.1 ± 5.8 | 17.0 ± 2.8 | – | – | – |
9P, 9 Phenanthrol.
Compound 5 selectively inhibits TRPM4 overexpressed in HEK293 cells
To gain further insight into the three most potent inhibitors found here 4, 5 and 6, we analysed their predicted off‐target effects using the polypharmacology browser (Awale and Reymond, 2017), an online tool that predicts possible biological activity by multi‐fingerprint comparisons with bioactive compounds from the ChEMBL database (Gaulton et al., 2012). While 4 and 5 were essentially free of off‐target predictions, we found that 6 had a reported activity at 10 μM on the http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=249 (glucagon‐like peptide receptor 1) [https://pubchem.ncbi.nlm.nih.gov/bioassay/624417 (accessed July 6, 2017)].
We further confirmed the inhibitory activity of compounds 4 and 5 using classical patch‐clamp electrophysiology recordings on TRPM4 overexpressing cell line. In excised membrane patches, we quantified TRPM4 inhibition using the generic TRPM4 inhibitors 9‐phenanthrol (Figure 5A) and FFA (Figure 5B) and compared it with our newly identified hit compounds 4 (Figure 5C) and 5 (Figure 5D). Maximal current at +100 mV was used to construct concentration–response curves (Average current traces: Supporting Information Figure S3). Interestingly, both compounds inhibited TRPM4 with an IC50 of 1.0 ± 0.2 and 1.8 ± 0.1 μM for 4 and 5 respectively, which are several times more potent than IC50 values of 17.0 ± 2.8 and 9.2 ± 1.2 μM for 9‐phenanthrol and FFA respectively. In addition, we also observed reversible inhibition by 4 and 5 when applied to the cytosolic side of the ruptured membrane patches (Supporting Information Figure S4).
Figure 5.
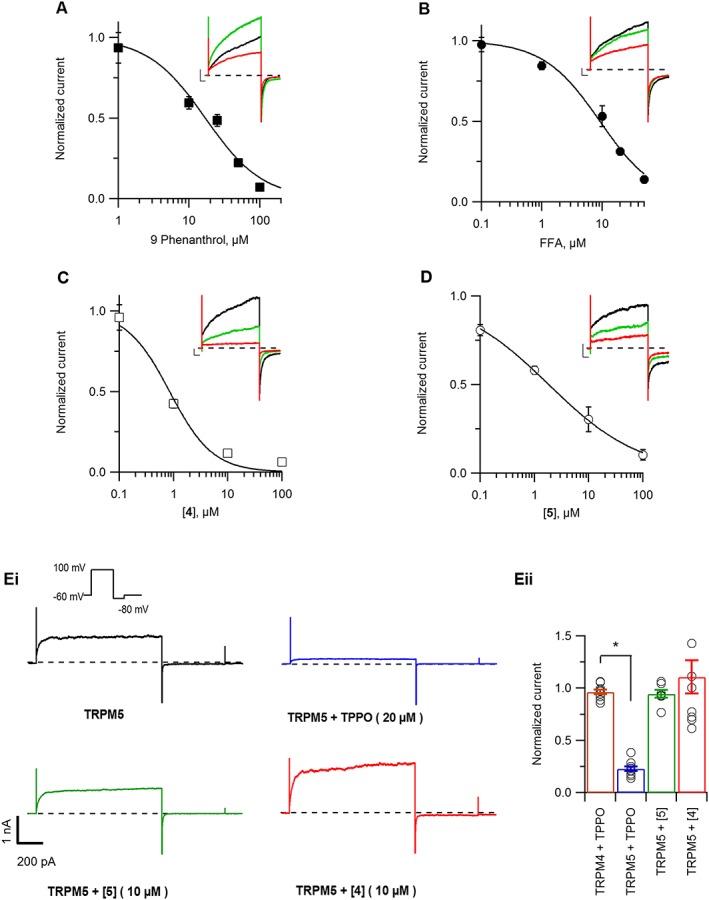
Compound 5 is a potent and selective inhibitor of TRPM4 current. TRPM4 currents were recorded from excised membrane patches exposed to 300 μM free Ca2+ on the cytosolic side of the patches. (A–D) Concentration–response curves were obtained by applying various concentrations of 9‐phenanthrol (A), FFA (B), 4 (C) and 5 (D), dissolved in pipette solution. Currents were normalized to current without any inhibitor and fitted with the Hill equation to extrapolate IC50 values for each inhibitor. n ≥ 4 for each concentration–response curve. Representative current traces were overlapped at 0 μM (black), 1 μM (green) and 10 μM (red) concentration of the inhibitor (scaling: X‐axis 50 ms, Y‐ axis 500 pA). (E) Selectivity of compounds 4 and 5 for mediating inhibition of TRPM5 currents recorded using a voltage‐step protocol (figure inset) from HEK293 cells transiently transfected with TRPM5. (E, panel i) Representative trace of TRPM5 current from excised membrane patches exposed to 50 μM free Ca2+ on the cytosolic side of the patches either inhibited by TPPO (blue), 5 (green) or 4 (red). (E, panel ii) Average currents measured at +100 mV were normalized to current without any inhibitor (n ≥ 8). *P ≤ 0.05, Student's t‐test.
Further, to evaluate the selectivity of the newly identified hits against TRPM5, the closest homologue of TRPM4, we performed excised membrane voltage clamp recordings on HEK293 cells expressing TRPM5. As a positive control, we used triphenylphosphine oxide (TPPO), a known http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=497 inhibitor at 20 μM. (Palmer et al., 2010). TPPO inhibited TRPM5 current but did not affect TRPM4 current (Figure 5E, panels i, ii). However, compound 4 had a variable potentiating effect on TRPM5, while compound 5 had no significant effect on the TRPM5 current (Figure 5E, panel i). In addition, we also tested the selectivity of compound 5 against other TRP family members including http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=499, http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=500, http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=507 and http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=512 using whole‐cell patch clamp recordings. At 10 or 100 μM, compound 5 did not show any inhibition of currents from these targets (Supporting Information Figure S5).
Next, we investigated if compound 5 had any off‐target effects on 17 different ion channel proteins and membrane receptors, including http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=404, http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=75, http://www.guidetopharmacology.org/GRAC/FamilyIntroductionForward?familyId=80 and http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=81, which are targets of the parent drug FFA. At 10 μM, compound 5 did not show any significant effect on these targets and showed only <5% inhibition of dofetilide binding to the cardiac http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=572 (hERG) channel (Supporting Information Table S4).
Compound 5 inhibits endogenous TRPM4 currents in LNCaP cells
We next assessed if compound 5 also inhibits endogenous TRPM4 currents. We recorded TRPM4 whole‐cell currents with 10 μM Ca2+ in the patch pipette in the prostate cancer cell line LNCaP as described previously (Holzmann et al., 2015; Kilch et al., 2016). The currents activated slowly (Figure 6A) with the typical TRPM4 current–voltage relationship (I‐V) shown in Figure 6B. After TRPM4 currents were activated, we applied compound 5 at different concentrations. Compound 5 inhibited TRPM4 currents reversibly in a concentration dependent manner with an IC50 of 1.1 ± 0.3 μM (Figure 6C). This is consistent with our findings in a TRPM4 overexpression system (Figures 3 and 5), where compound 5 also inhibits TRPM4 currents with an IC50 of ~1 μM.
Figure 6.
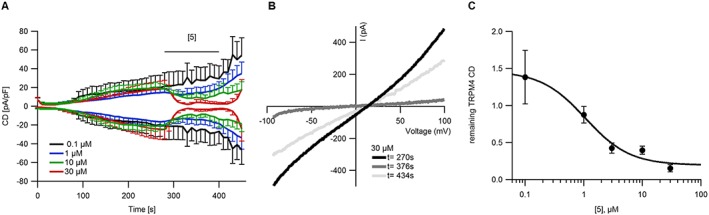
Compound 5 reversibly blocks endogenous TRPM4 currents in LNCaP (prostate cancer) cells. (A) Time course of average TRPM4 current densities (CD) activated in LNCaP cells by intracellular Ca2+ when various concentrations of 5 were applied. n = 5 (0.1 and 1 μM), 6 (10 μM) and 3 (30 μM). (B) Exemplary I‐Vs from cells in (A) when TRPM4 current was developed (t = 270 s) and blocked with 30 μM compound 5 (t = 376 s) and washout (t = 434 s). (C) Dose–response curve for TRPM4 currents when different concentrations of 5 were applied (same cells as in A). CD at 376 s was normalized to CD at 270 s and plotted versus concentration of 5. n ≥ 3 for each concentration of 5. Data were fitted with a Hill equation to extrapolate the IC50.
Compound 5 restores functional expression of A432T, a loss‐of‐expression TRPM4 variant
Based on the TRPM4 selectivity of compound 5, we investigated whether compound 5 can rescue membrane expression of TRPM4 A432T, a loss‐of‐expression variant found in cardiac AVB patients (Syam et al., 2016). It is reported that decreased expression and altered trafficking of ion channel disease causing variants, such as in chloride http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=707 and Kv11.1 channels, can be corrected either by incubation at a lower temperature or by using selective chemical compounds (Denning et al., 1992; Zhou et al., 1999). We followed a similar strategy and observed that the expression of A432T was increased after a 24 h long incubation of the cells at 28°C ( Supporting Information Figure S6A ).
Next, we pre‐incubated cells expressing A432T with 50 μM compound 5 overnight and evaluated any change in A432T total and surface expression using Western blot (Figure 7A, panel i). Compound 5 showed only ~30% cytotoxicity at 100 μM, making it feasible for longer incubation with cells (Supporting Information Figure S7). Densitometric quantification of the upper fully glycosylated (FG) and the lower core glycosylated (CG) bands showed partial rescue of total and surface expression(biotinylated fraction) of A432T by compound 5 (Figure 7A, panel ii). We further evaluated if the restored expression of A432T with pre‐incubation of compound 5 also led to increase in current density in excised membrane patches. As illustrated in Figure 7B, C, A432T showed a significant decrease in the Ca2+ sensitive outward current in comparison with WT. Pre‐incubation with compound 5 at 50 μM overnight and followed by a washout before current recordings, partially restored the functional expression and the Ca2+ sensitive outward current of A432T. Conversely, an inactive congener of compound 5, that is, compound 51, did not restore the expression of A432T at 50 μM (Figure 7D).
Figure 7.
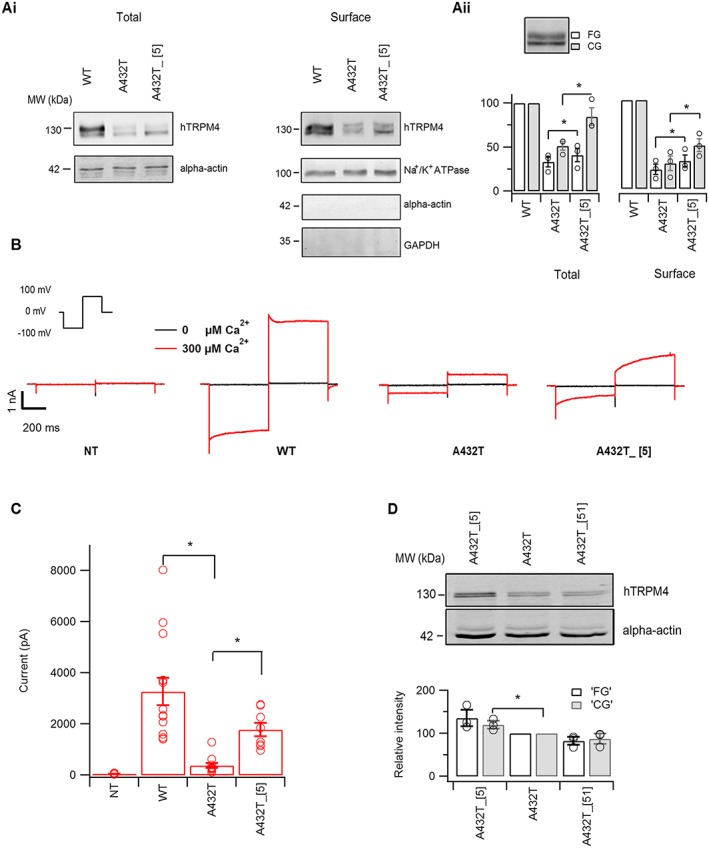
Functional assessment of A432T variant rescue by compound 5. (A, panel i) Western blot analysis shows rescue of A432T variant total and surface expression by 50 μM compound 5, as detected by anti‐TRPM4 antibody. (A, panel ii) Densitometry quantification of blots at total and surface level expression show compound 5 rescued both the core and fully glycosylated form of A432T. The protein band intensity at each concentration was quantified from three individual experiments. (B) Representative inside‐out voltage clamp recordings on membrane patches from non‐transfected (NT) or heterologously expressing TRPM4 WT, A432T or A432T pre‐incubated with 50 μM compound 5 (A432T_[5]), when exposed to nominally either 0 or 300 μM Ca2+ on the cytosolic side. (C) Average current amplitude measured at the end of voltage step to +100 mV (inset, panel B) (n ≥ 5). (D) Western blot analysis showing restoration of membrane expression of A432T variant by active compound 5 and not by inactive congener 51 at 50 μM (n = 3).
Selectivity of compound 5 for rescuing A432T was further evaluated by incubating the loss‐of‐expression of Kv11.1 channel (hERG) variant named Dupl hERG, which was found in a congenital long‐QT syndrome patient (Grilo et al., 2010), both with 50 μM compound 5 and with lower‐temperature (28°C) incubation. As presented in (Supporting Information Figure S6B), Western blot experiments showed that a reduction of incubation temperature rescued the trafficking defect of Dupl hERG, whereas no effect was observed after incubation with compound 5. However, the rescuing property of compound 5 was not limited to A432T but also increased WT TRPM4 expression similar to incubation with lower temperature (Supporting Information Figure S6C, D ).
Discussion
Human mutations in the TRPM4 gene have been implicated in cardiac conduction disorders (Kruse et al., 2009; Liu et al., 2010, 2013; Stallmeyer et al., 2012; Syam et al., 2016). So exploring the role of TRPM4 in cardiac physiology and pathophysiology upholds great clinical relevance. However, most of these physiological characterizations relied upon patch clamp studies with TRPM4 KO mice models combined with pharmacological inhibitors. Notably, the most reported TRPM4 inhibitor, 9‐phenanthrol, did show some selectivity for TRPM4 compared with other TRP channels. However, it is not sufficiently potent, and high concentrations reversibly inhibit delayed outward rectifying K+ and voltage‐gated Ca2+ currents in mouse ventricular myocytes (Simard et al., 2012). A recent study showed that 9‐phenanthrol inhibits http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=130, a Ca2+‐activated Cl− channel in arterial smooth myocytes (Burris et al., 2015). Furthermore, 9‐phenanthrol belongs to the class of polycyclic aromatic hydrocarbons, which contribute to its cellular toxicity (Feng et al., 2012). The non‐steroidal anti‐inflammatory drug FFA and glibenclamide are also used as TRPM4 inhibitors (Ullrich et al., 2005; Demion et al., 2007). Although these compounds inhibit TRPM4 current reversibly, they also have many off‐target effects on other ion channels (Gardam et al., 2008; Alexander et al., 2017d). These findings support the need for more potent and selective TRPM4 inhibitors. In this study, we developed a novel Na+‐sensitive screening assay and performed a focused screening campaign using LBVS with xLOS, which lead to the identification of compounds 4, 5 and 6, as potent TRPM4 inhibitors. In addition, we found that compound 5 possessed a dual function as a TRPM4 inhibitor and also as a chemical chaperon rescuing the membrane expression of a loss‐of‐expression TRPM4 variant.
Compound 5 as a potential inhibitor of TRPM4
Recently, virtual screening methods have emerged as an efficient approach for identifying potent and specific inhibitors for different ion channels (Langer and Krovat, 2003; Fernández‐Ballester et al., 2011; Simonin et al., 2015). In this study, we carried out an initial virtual screening using an in‐house developed pharmacophore similarity search algorithm: xLOS, which compared the spatial overlap between a catalogue of commercially available drug‐like small molecules to the three reference TRPM4 inhibitors, 9‐phenanthrol, glibenclamide and FFA. The purchased compounds selected from the top scoring list were further validated using a new fluorescence based Na+ sensitive screening assay, which measured the activity of TRPM4 in a stably expressing cell line. This screening identified two aryloxyacyl‐anthranilic acid compounds 4 and 5 as the most potent compounds with IC50 in ~1 μM range, which is several fold lower compared with all the reference inhibitors (Abriel et al., 2012). It is noteworthy that compound 5 inhibited TRPM4 current in both an overexpressed cell line and in an endogenously expressed in prostate cancer cell line with similar potency and presented selectivity over other TRP family members including TRPM5, TRPM7, TRPM8, TRPV1, http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=509, TRPV6. Analysis of our in silico and cell‐based results highlighted compound 5 as having a good balance of potency and selectivity that encouraged us for further optimization of this compound through chemical modifications. Although altering substituents on aromatic rings and within the acetyl linker often abolished the inhibitory activity, we identified compound 6 with increased potency of <1 μM IC50 inhibition (Table 2 and Supporting Information Tables S1–S3). However, due to an off‐target effect of compound 6 on the GLP‐1 receptor as predicted by a polypharmacology browser, we did not perform any further experiments with this compound. Future optimization studies on compound 6 could provide a promising tool compound for TRPM4 pharmacology.
Success in finding these inhibitors demonstrates the capability of combining in silico and Na+ sensitive screening assay to explore focused chemical landscapes to identify potent and specific inhibitors of TRPM4. The chemical diversity surveyed here in the virtual screening and subsequent SAR study is summarized in Figure 8 with a colour‐coded map of the chemical space covered by the compounds tested, plotted with our recently reported web‐based tool WebMolCS (Awale et al., 2017) using the principle of similarity mapping (Awale and Reymond, 2015). This 3D‐map positions molecules as individual spheres in a space where distances represent the relative similarities of molecules to one another, as measured by the Tanimoto coefficient of a 1024‐bit binary Daylight type substructure fingerprint, which perceives detailed features of the molecules (Hagadone, 1992). Each sphere is colour‐coded according to the observed TRPM4 activities (inactive = blue, most active = red). This representation illustrates that the anthranilic acid TRPM4 inhibitors occupy a chemical space significantly shifted from the reference inhibitors 9‐phenanthrol, FFA and glibenclamide used for LBVS. Furthermore, the map shows that only very few analogues of 9‐phenanthrol were actually tested due to limited commercial availability.
Compound 5 as a potential chemical chaperone for TRPM4 loss‐of‐expression variant
A number of mutations in the human TRPM4 gene linked with various cardiac pathologies have led to reduced membrane expression and function either due to misfolding or improper assembly of the channel protein (Stallmeyer et al., 2012; Syam et al., 2016). In the past decade, several studies used in silico and in vitro screening to identify small molecules that can partially restore the trafficking defect of many proteins (Naik et al., 2012) including ion channels such as CFTR (Van Goor et al., 2006; Rowe and Verkman, 2013). Most of these chemical chaperones had shown certain selectivity towards the target either by direct interactions or via other accessory proteins. We reasoned that, since compound 5 outlays better potency, selectivity and lower cytotoxicity, it could be a potential candidate to be used as chemical chaperone for A432T‐TRPM4 mutant channel. Indeed compound 5 rescued the total and surface expression of both the core and fully glycosylated forms of the A432T variant channel. This chemical chaperoning effect may be related to a compound‐dependent increase in protein stability. Moreover, the observed increase in A432T‐TRPM4‐mediated current upon compound 5 pre‐incubation, followed by a washout protocol, is consistent with an increased number of channels at the cell surface. The partial functional rescuing may be explained by either mutation‐dependent altered gating or only partial washout of the compound 5. However, it is notable that this rescuing effect was specific to the structure of compound 5, since an inactive congener of 5 with no inhibitory activity, that is, 51, did not show any rescuing effect. It was also channel‐specific since 5 did not rescue a loss‐of‐expression Kv11.1 (hERG) channel variant. These results suggest that this positive effect could be from a direct and selective interaction of compound 5 with the TRPM4 protein and hence reducing its degradation by the endoplasmic reticulum‐associated degradation process. Note that at this stage, there is no evidence that the protein binding of 5 leading to TRPM4 inhibition is identical to the one resulting in its restoration. Nevertheless, to the best of our knowledge, this is the first report of a small molecule compound functionally rescuing the membrane expression of a TRP channel loss‐of‐expression variant.
In conclusion, this study demonstrates that using the newly developed xLOS method linked to a Na+‐sensitive screening assay, a series of halogenated anthranilic amide compounds with potent and selective TRPM4 inhibition properties was discovered. The highly potent compounds 4, 5 and 6 that we identified in this study will open new ways in TRPM4 pharmacology. In the context of recent structural data of TRPM4 (Guo et al., 2017; Winkler et al., 2017; Autzen et al., 2018), future structure–function relationship studies coupled with our selective and potent inhibitor compound 5 will benefit the better understanding of channel function. Moreover, compound 5 demonstrated not only selective inhibition but also a chemical chaperone property, which provide a good starting point for developing clinical drug candidates for rescuing loss‐of‐expression variants.
Author contributions
L.C.O., C.D., B.B., G.N., S.K., M.R., J.G., M.L., C.P., J.L.R. and H.A. designed the experiments. L.C.O. developed and validated the screening assay. L.C.O. and M.R. performed compound screening. L.C.O. and S.K. performed and analysed the patch clamp experiments. B.B. performed western blots for chemical chaperone experiments. U.T. performed additional compound selectivity profiling and D.R‐K. performed biotinylation experiments. C.D., G.N. and C.S. performed virtual screening, purchased compounds. C.D. and G.N. performed chemical synthesis for SAR studies. L.C.O., J.L.R. and H.A. wrote the manuscript with extensive comments from C.D., B.B., G.N., M.R., J.G., S.K., M.L. and C.P.
Conflict of interest
The authors declare no conflicts of interest.
Declaration of transparency and scientific rigour
This http://onlinelibrary.wiley.com/doi/10.1111/bph.13405/abstract acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research recommended by funding agencies, publishers and other organisations engaged with supporting research.
Supporting information
Figure S1 TRPM4 currents in excised membrane patches: Representative current traces from excised membrane patches when exposed to either 0 (black) or 500 μM Ca2+ (blue) from TRPM4 expressing cells either induced with tetracycline or non‐induced and TRPM4 CRISPR‐Cas9 knocked down cell line (KD). Steady state IVs was constructed from currents at the end of 200 ms voltage steps (inset) (n = 3).
Figure S2 Assay validation A) Overlay of traces obtained on the FLIPR from TRPM4‐expressing cells induced with ionomycin (red, signal) or without ionomycin (black, background). B) Z` prime factor and ratio of AUC from signal over background (S/B) calculated over 24 plates on different d of experiment were plotted on left and right Y‐axis respectively. C) Overlay of traces with different extracellular vehicle DMSO concentrations. D) Average AUC after incubation with varying DMSO concentrations (n = 3).
Figure S3 Concentration‐response studies. Average current traces recorded from excised membrane patches in voltage clamp with either vehicle control or inhibitor at different concentrations. n ≥ 4 for each concentrations of different inhibitors.
Figure S4 Cytosolic application of 5 and 4. Representative current traces from excised membrane patches on TRPM4 expressing cells when exposed to either 0 or 300 μM Ca2+. Furthermore, on the same cells, the currents were inhibited by cytosolic application of either 1 μM 5 (A) or 1 μM 4 (B) and subsequently washed out (n ≥ 6).
Figure S5 Compound 5 TRP selectivity. A) Left, representative whole‐cell current density trace at +80 mV from HEK cells stably expressing TRPM7 either induced with tetracyline or non‐induced. After the currents were fully developed, either DMSO or 100 μM compound 5 were applied in bath solution. Right, the currents recorded in presence of DMSO (grey) or compound 5 (red) was normalized to the current before application on the same cell. B) Left, representative whole cell current density trace at –80 mV evoked by 100 μM menthol from HEK cells transiently transfected with TRPM8. The cells were later applied with either DMSO or 10 μM compound 5. Right, the peak currents recorded in presence of DMSO (grey) or compound 5 (red) were normalized to the current before application on the same cell. C) Left,representative whole cell current density trace at – 80mV evoked by 10 μM capsaicin from HEK cells transiently transfected with TRPV1. Right, the peak currents recorded in presence of DMSO (grey) or compound 5 (red) were normalized to the current before application on the same cell. D) Left, representative whole‐cell current density trace at ‐80 mV from HEK cells stably expressing TRPV6. Inward currents were observed with increase of Ca2+ to 10 mM in the bath solution. After the currents were fully developed, either DMSO or 100 μM compound 5 were applied in bath solution. Right, the currents recorded in presence of DMSO (grey) or compound 5 (red) was normalized to the current before application on the same cell.
Figure S6 Specificity of compound 5 in rescuing A432T loss‐of‐expression variant. A) Western blot analysis showing total expression rescue of A432T variant after pre‐incubation at 28°C. B) Western blot analysis shows that compound 5 failed to rescue membrane expression of Dupl hERG variant, whereas the variant was successfully rescued by pre‐incubation at 28°C. C) Western blot analysis showing membrane expression rescue of WT TRPM4 after pre‐incubation at 28°C. D) Western blot analysis showing total expression rescue of WT TRPM4 after pre‐incubation with 50 μM 5. The protein band intensity at each concentration was quantified from 3 individual experiments. * P ≤ 0.05, Student's t‐test.
Figure S7 Cytotoxicity study of the most potent inhibitors. An MTT viability assay was performed for 4, 5 and 6 on HeLa cells. The data represent the means of three values and are normalized against DMSO control.
Table S1 Anthranilic acid variations: from top to bottom, isosteric substitution of the chlorine group and variation of the carboxylic acid's position. a) Racemic mixture. b) Activity on TRPM4 given as Normalized counts of Na+ influx at 5 μM. Values above 0 are active.
Table S2 From top to bottom, substitution variations and bicyclic variations of the phenoxy moiety. a) Racemic mixture. b) Activity on TRPM4 given as normalized counts of Na+ influx at 5 μM. Values above 0 are active.
Table S3 Acetyl linker modifications. a) Racemic mixture. b) Activity on TRPM4 given as Normalized counts of Na+ influx at 5 μM. c) The enantiomerically pure isomers of the racemic 4 were separated by chiral HPLC. d) Activity measured at 10 μM.
Table S4 In vitro pharmacology profiling of 5 on several ion channels. a) Compound 5 was tested on radio ligand binding assay at 10 μM. b) Compound 5 was tested on cellular and nuclear functional assays at 10 μM. The hERG channel inhibition was also tested on automated whole‐cell patch clamp at 10 μM. Inhibition below 50% is considered not significant. The data represent the mean of two experiments. No significant inhibition (NSI).
Table S5 Analytical RP‐UPLC purity. a) The spectra were obtained according to method B (see description in general remarks). b) A partial decomposition of the compound was observed preventing any purity analysis. c) Any UPLC trace could be obtained due to either too few material or precipitation in UPLC solvents. NMR purity was considered (>90%).
Acknowledgements
We thank Jean‐Sebastien Rougier, PhD and Sarah Vermij for helpful discussions and critical proof reading. We thank Prof. Matthias Hediger for the HEK293 TRPV6 cell line and Barbara Hauert for technical assistance. This work was supported by National Centre of Competence in Research (NCCR), TransCure 51NF40‐160620 and a grant from the Schweizerische Herzstiftung to H. A.
Ozhathil, L. C. , Delalande, C. , Bianchi, B. , Nemeth, G. , Kappel, S. , Thomet, U. , Ross‐Kaschitza, D. , Simonin, C. , Rubin, M. , Gertsch, J. , Lochner, M. , Peinelt, C. , Reymond, J.‐L. , and Abriel, H. (2018) Identification of potent and selective small molecule inhibitors of the cation channel TRPM4 . British Journal of Pharmacology, 175: 2504–2519. doi: 10.1111/bph.14220.
Contributor Information
Jean‐Louis Reymond, Email: jean-louis.reymond@dcb.unibe.ch.
Hugues Abriel, Email: hugues.abriel@ibmm.unibe.ch.
References
- Abriel H, Syam N, Sottas V, Amarouch MY, Rougier J‐S (2012). TRPM4 channels in the cardiovascular system: physiology, pathophysiology, and pharmacology. Biochem Pharmacol 84: 873–881. [DOI] [PubMed] [Google Scholar]
- Alexander SPH, Christopoulos A, Davenport AP, Kelly E, Marrion NV, Peters JA et al (2017a). The Concise Guide to PHARMACOLOGY 2017/18: G protein‐coupled receptors. Br J Pharmacol 174: S17–S129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Kelly E, Marrion NV, Peters JA, Faccenda E, Harding SD et al (2017b). The Concise Guide to PHARMACOLOGY 2017/18: Other ion channels. Br J Pharmacol 174: S195–S207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Peters JA, Kelly E, Marrion NV, Faccenda E, Harding SD et al (2017c). The Concise Guide to PHARMACOLOGY 2017/18: Ligand‐gated ion channels. Br J Pharmacol 174: S130–S159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander S, Striessnig J, Kelly E, Marrion NV, Peters JA, Faccenda E et al (2017d). The Concise Guide to PHARMACOLOGY 2017/18: Voltage‐gated ion channels. Br J Pharmacol 174: S160–S194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarouch M‐Y, Syam N, Abriel H (2013). Biochemical, single‐channel, whole‐cell patch clamp, and pharmacological analyses of endogenous TRPM4 channels in HEK293 cells. Neurosci Lett 541: 105–110. [DOI] [PubMed] [Google Scholar]
- Ashida S, Nakagawa H, Katagiri T, Furihata M, Iiizumi M, Anazawa Y et al (2004). Molecular features of the transition from prostatic intraepithelial neoplasia (PIN) to prostate cancer: genome‐wide gene‐expression profiles of prostate cancers and PINs. Cancer Res 64: 5963–5972. [DOI] [PubMed] [Google Scholar]
- Autzen HE, Myasnikov AG, Campbell MG, Asarnow D, Julius D, Cheng Y (2018). Structure of the human TRPM4 ion channel in a lipid nanodisc. Science 359: 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awale M, Probst D, Reymond J‐L (2017). WebMolCS: a web‐based interface for visualizing molecules in three‐dimensional chemical spaces. J Chem Inf Model 57: 643–649. [DOI] [PubMed] [Google Scholar]
- Awale M, Reymond J‐L (2014). A multi‐fingerprint browser for the ZINC database. Nucleic Acids Res 42: W234–W239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awale M, Reymond J‐L (2015). Similarity mapplet: interactive visualization of the directory of useful decoys and chembl in high dimensional chemical spaces. J Chem Inf Model 55: 1509–1516. [DOI] [PubMed] [Google Scholar]
- Awale M, Reymond J‐L (2017). The polypharmacology browser: a web‐based multi‐fingerprint target prediction tool using ChEMBL bioactivity data. J Chem 9: 11 10.1186/s13321-017-0199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg KD, Soldini D, Jung M, Dietrich D, Stephan C, Jung K et al (2016). TRPM4 protein expression in prostate cancer: a novel tissue biomarker associated with risk of biochemical recurrence following radical prostatectomy. Virchows Arch Int J Pathol 468: 345–355. [DOI] [PubMed] [Google Scholar]
- Burris SK, Wang Q, Bulley S, Neeb ZP, Jaggar JH (2015). 9‐Phenanthrol inhibits recombinant and arterial myocyte TMEM16A channels. Br J Pharmacol 172: 2459–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MJ, Bond RA, Spina D, Ahluwalia A, Alexander SPA, Giembycz MA et al (2015). Experimental design and analysis and their reporting: new guidance for publication in BJP. Br J Pharmacol 172: 3461–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demion M, Bois P, Launay P, Guinamard R (2007). TRPM4, a Ca2+−activated nonselective cation channel in mouse sino‐atrial node cells. Cardiovasc Res 73: 531–538. [DOI] [PubMed] [Google Scholar]
- Denning GM, Anderson MP, Amara JF, Marshall J, Smith AE, Welsh MJ (1992). Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature‐sensitive. Nature 358: 761–764. [DOI] [PubMed] [Google Scholar]
- Feng T‐C, Cui C‐Z, Dong F, Feng Y‐Y, Liu Y‐D, Yang X‐M (2012). Phenanthrene biodegradation by halophilic Martelella sp. AD‐3. J Appl Microbiol 113: 779–789. [DOI] [PubMed] [Google Scholar]
- Fernández‐Ballester G, Fernández‐Carvajal A, González‐Ros JM, Ferrer‐Montiel A (2011). Ionic channels as targets for drug design: a review on computational methods. Pharmaceutics 3: 932–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfria E, Murdock PR, Cusdin FS, Benham CD, Kelsell RE, McNulty S (2006). Tissue distribution profiles of the human TRPM cation channel family. J Recept Signal Transduct Res 26: 159–178. [DOI] [PubMed] [Google Scholar]
- Gardam KE, Geiger JE, Hickey CM, Hung AY, Magoski NS (2008). Flufenamic acid affects multiple currents and causes intracellular Ca2+ release in Aplysia bag cell neurons. J Neurophysiol 100: 38–49. [DOI] [PubMed] [Google Scholar]
- Garland CJ, Smirnov SV, Bagher P, Lim CS, Huang CY, Mitchell R et al (2015). TRPM4 inhibitor 9‐phenanthrol activates endothelial cell intermediate conductance calcium‐activated potassium channels in rat isolated mesenteric artery. Br J Pharmacol 172: 1114–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A et al (2012). ChEMBL: a large‐scale bioactivity database for drug discovery. Nucleic Acids Res 40: D1100–D1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grand T, Demion M, Norez C, Mettey Y, Launay P, Becq F et al (2008). 9‐phenanthrol inhibits human TRPM4 but not TRPM5 cationic channels. Br J Pharmacol 153: 1697–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grilo LS, Pruvot E, Grobéty M, Castella V, Fellmann F, Abriel H (2010). Takotsubo cardiomyopathy and congenital long QT syndrome in a patient with a novel duplication in the Per‐Arnt‐Sim (PAS) domain of hERG1. Heart Rhythm 7: 260–265. [DOI] [PubMed] [Google Scholar]
- Guinamard R, Hof T, Del Negro CA (2014). The TRPM4 channel inhibitor 9‐phenanthrol. Br J Pharmacol 171: 1600–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, She J, Zeng W, Chen Q, Bai X‐C, Jiang Y (2017). Structures of the calcium‐activated, non‐selective cation channel TRPM4. Nature 552: 205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagadone TR (1992). Molecular substructure similarity searching: efficient retrieval in two‐dimensional structure databases. J Chem Inf Comput Sci 32: 515–521. [Google Scholar]
- Harding SD, Sharman JL, Faccenda E, Southan C, Pawson AJ, Ireland S et al (2018). The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucl Acids Res 46: D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann C, Kappel S, Kilch T, Jochum MM, Urban SK, Jung V et al (2015). Transient receptor potential melastatin 4 channel contributes to migration of androgen‐insensitive prostate cancer cells. Oncotarget 6: 41783–41793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins AL, Keserü GM, Leeson PD, Rees DC, Reynolds CH (2014). The role of ligand efficiency metrics in drug discovery. Nat Rev Drug Discov 13: 105–121. [DOI] [PubMed] [Google Scholar]
- Kilch T, Kappel S, Peinelt C (2016). Regulation of Ca(2+) signaling in prostate cancer cells. Channels Austin Tex 10: 170–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchmann F, Marcaida MJ, Kotak S, Schick T, Boss SD, Awale M et al (2016). Discovery of a selective aurora a kinase inhibitor by virtual screening. J Med Chem 59: 7188–7211. [DOI] [PubMed] [Google Scholar]
- Kruse M, Pongs O (2014). TRPM4 channels in the cardiovascular system. Curr Opin Pharmacol 15: 68–73. [DOI] [PubMed] [Google Scholar]
- Kruse M, Schulze‐Bahr E, Corfield V, Beckmann A, Stallmeyer B, Kurtbay G et al (2009). Impaired endocytosis of the ion channel TRPM4 is associated with human progressive familial heart block type I. J Clin Invest 119: 2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer T, Krovat EM (2003). Chemical feature‐based pharmacophores and virtual library screening for discovery of new leads. Curr Opin Drug Discov Devel 6: 370–376. [PubMed] [Google Scholar]
- Launay P, Fleig A, Perraud A‐L, Scharenberg AM, Penner R, Kinet J‐P (2002). TRPM4 is a Ca2+‐activated nonselective cation channel mediating cell membrane depolarization. Cell 109: 397–407. [DOI] [PubMed] [Google Scholar]
- Liu H, Chatel S, Simard C, Syam N, Salle L, Probst V et al (2013). Molecular genetics and functional anomalies in a series of 248 Brugada cases with 11 mutations in the TRPM4 channel. PloS One 8: e54131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, El Zein L, Kruse M, Guinamard R, Beckmann A, Bozio A et al (2010). Gain‐of‐function mutations in TRPM4 cause autosomal dominant isolated cardiac conduction disease. Circ Cardiovasc Genet 3: 374–385. [DOI] [PubMed] [Google Scholar]
- Makar TK, Gerzanich V, Nimmagadda VKC, Jain R, Lam K, Mubariz F et al (2015). Silencing of Abcc8 or inhibition of newly upregulated Sur1‐Trpm4 reduce inflammation and disease progression in experimental autoimmune encephalomyelitis. J Neuroinflammation 12: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra S, Castilló J, Negrotto L, Merino‐Zamorano C, Montaner J, Vidal‐Jordana A et al (2013). TRPM4 mRNA expression levels in peripheral blood mononuclear cells from multiple sclerosis patients. J Neuroimmunol 261: 146–148. [DOI] [PubMed] [Google Scholar]
- Naik S, Zhang N, Gao P, Fisher MT (2012). On the design of broad based screening assays to identify potential pharmacological chaperones of protein misfolding diseases. Curr Top Med Chem 12: 2504–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius B, Prenen J, Droogmans G, Voets T, Vennekens R, Freichel M et al (2003). Voltage dependence of the Ca2+−activated cation channel TRPM4. J Biol Chem 278: 30813–30820. [DOI] [PubMed] [Google Scholar]
- Palmer RK, Atwal K, Bakaj I, Carlucci‐Derbyshire S, Buber MT, Cerne R et al (2010). Triphenylphosphine oxide is a potent and selective inhibitor of the transient receptor potential melastatin‐5 ion channel. Assay Drug Dev Technol 8: 703–713. [DOI] [PubMed] [Google Scholar]
- Prevarskaya N, Zhang L, Barritt G (2007). TRP channels in cancer. Biochim Biophys Acta 1772: 937–946. [DOI] [PubMed] [Google Scholar]
- Rowe SM, Verkman AS (2013). Cystic fibrosis transmembrane regulator correctors and potentiators. Cold Spring Harb Perspect Med 3: a009761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagredo AI, Sagredo EA, Cappelli C, Báez P, Rodrigo AM, Blanco C et al (2018). TRPM4 regulates Akt/GSK3‐β activity and enhances β‐catenin signaling and cell proliferation in prostate cancer cells. Mol Oncol 12: 151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattling B, Steinbach K, Thies E, Kruse M, Menigoz A, Ufer F et al (2012). TRPM4 cation channel mediates axonal and neuronal degeneration in experimental autoimmune encephalomyelitis and multiple sclerosis. Nat Med 18: 1805–1811. [DOI] [PubMed] [Google Scholar]
- Schinke EN, Bii V, Nalla A, Rae DT, Tedrick L, Meadows GG et al (2014). A novel approach to identify driver genes involved in androgen‐independent prostate cancer. Mol Cancer 13: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G, Neidhart W, Giller T, Schmid G (1999). “Scaffold‐Hopping” by topological pharmacophore search: a contribution to virtual screening. Angew Chem Int Ed 38: 2894–2896. [PubMed] [Google Scholar]
- Scior T, Bender A, Tresadern G, Medina‐Franco JL, Martínez‐Mayorga K, Langer T et al (2012). Recognizing pitfalls in virtual screening: a critical review. J Chem Inf Model 52: 867–881. [DOI] [PubMed] [Google Scholar]
- Simard C, Sallé L, Rouet R, Guinamard R (2012). Transient receptor potential melastatin 4 inhibitor 9‐phenanthrol abolishes arrhythmias induced by hypoxia and re‐oxygenation in mouse ventricle. Br J Pharmacol 165: 2354–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonin C, Awale M, Brand M, van Deursen R, Schwartz J, Fine M et al (2015). Optimization of TRPV6 calcium channel inhibitors using a 3D ligand‐based virtual screening method. Angew Chem Int Ed Engl 54: 14748–14752. [DOI] [PubMed] [Google Scholar]
- Singh J, Manickam P, Shmoish M, Natik S, Denyer G, Handelsman D et al (2006). Annotation of androgen dependence to human prostate cancer‐associated genes by microarray analysis of mouse prostate. Cancer Lett 237: 298–304. [DOI] [PubMed] [Google Scholar]
- Stallmeyer B, Zumhagen S, Denjoy I, Duthoit G, Hébert J‐L, Ferrer X et al (2012). Mutational spectrum in the Ca(2+)‐activated cation channel gene TRPM4 in patients with cardiac conductance disturbances. Hum Mutat 33: 109–117. [DOI] [PubMed] [Google Scholar]
- Sterling T, Irwin JJ (2015). ZINC 15–ligand discovery for everyone. J Chem Inf Model 55: 2324–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suguro M, Tagawa H, Kagami Y, Okamoto M, Ohshima K, Shiku H et al (2006). Expression profiling analysis of the CD5+ diffuse large B‐cell lymphoma subgroup: development of a CD5 signature. Cancer Sci 97: 868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syam N, Chatel S, Ozhathil LC, Sottas V, Rougier J‐S, Baruteau A et al (2016). Variants of transient receptor potential melastatin member 4 in childhood atrioventricular block. J Am Heart Assoc 5: e001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich ND, Voets T, Prenen J, Vennekens R, Talavera K, Droogmans G et al (2005). Comparison of functional properties of the Ca2+−activated cation channels TRPM4 and TRPM5 from mice. Cell Calcium 37: 267–278. [DOI] [PubMed] [Google Scholar]
- Van Goor F, Straley KS, Cao D, González J, Hadida S, Hazlewood A et al (2006). Rescue of DeltaF508‐CFTR trafficking and gating in human cystic fibrosis airway primary cultures by small molecules. Am J Physiol Lung Cell Mol Physiol 290: L1117–L1130. [DOI] [PubMed] [Google Scholar]
- Winkler PA, Huang Y, Sun W, Du J, Lü W (2017). Electron cryo‐microscopy structure of a human TRPM4 channel. Nature 552: 200–204. [DOI] [PubMed] [Google Scholar]
- Zhang J‐H, Chung TDY, Oldenburg KR (1999). A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4: 67–73. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Okawa H, Wang Y, Liman ER (2005). Phosphatidylinositol 4,5‐bisphosphate rescues TRPM4 channels from desensitization. J Biol Chem 280: 39185–39192. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Gong Q, January CT (1999). Correction of defective protein trafficking of a mutant HERG potassium channel in human long QT syndrome. Pharmacological and temperature effects. J Biol Chem 274: 31123–31126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 TRPM4 currents in excised membrane patches: Representative current traces from excised membrane patches when exposed to either 0 (black) or 500 μM Ca2+ (blue) from TRPM4 expressing cells either induced with tetracycline or non‐induced and TRPM4 CRISPR‐Cas9 knocked down cell line (KD). Steady state IVs was constructed from currents at the end of 200 ms voltage steps (inset) (n = 3).
Figure S2 Assay validation A) Overlay of traces obtained on the FLIPR from TRPM4‐expressing cells induced with ionomycin (red, signal) or without ionomycin (black, background). B) Z` prime factor and ratio of AUC from signal over background (S/B) calculated over 24 plates on different d of experiment were plotted on left and right Y‐axis respectively. C) Overlay of traces with different extracellular vehicle DMSO concentrations. D) Average AUC after incubation with varying DMSO concentrations (n = 3).
Figure S3 Concentration‐response studies. Average current traces recorded from excised membrane patches in voltage clamp with either vehicle control or inhibitor at different concentrations. n ≥ 4 for each concentrations of different inhibitors.
Figure S4 Cytosolic application of 5 and 4. Representative current traces from excised membrane patches on TRPM4 expressing cells when exposed to either 0 or 300 μM Ca2+. Furthermore, on the same cells, the currents were inhibited by cytosolic application of either 1 μM 5 (A) or 1 μM 4 (B) and subsequently washed out (n ≥ 6).
Figure S5 Compound 5 TRP selectivity. A) Left, representative whole‐cell current density trace at +80 mV from HEK cells stably expressing TRPM7 either induced with tetracyline or non‐induced. After the currents were fully developed, either DMSO or 100 μM compound 5 were applied in bath solution. Right, the currents recorded in presence of DMSO (grey) or compound 5 (red) was normalized to the current before application on the same cell. B) Left, representative whole cell current density trace at –80 mV evoked by 100 μM menthol from HEK cells transiently transfected with TRPM8. The cells were later applied with either DMSO or 10 μM compound 5. Right, the peak currents recorded in presence of DMSO (grey) or compound 5 (red) were normalized to the current before application on the same cell. C) Left,representative whole cell current density trace at – 80mV evoked by 10 μM capsaicin from HEK cells transiently transfected with TRPV1. Right, the peak currents recorded in presence of DMSO (grey) or compound 5 (red) were normalized to the current before application on the same cell. D) Left, representative whole‐cell current density trace at ‐80 mV from HEK cells stably expressing TRPV6. Inward currents were observed with increase of Ca2+ to 10 mM in the bath solution. After the currents were fully developed, either DMSO or 100 μM compound 5 were applied in bath solution. Right, the currents recorded in presence of DMSO (grey) or compound 5 (red) was normalized to the current before application on the same cell.
Figure S6 Specificity of compound 5 in rescuing A432T loss‐of‐expression variant. A) Western blot analysis showing total expression rescue of A432T variant after pre‐incubation at 28°C. B) Western blot analysis shows that compound 5 failed to rescue membrane expression of Dupl hERG variant, whereas the variant was successfully rescued by pre‐incubation at 28°C. C) Western blot analysis showing membrane expression rescue of WT TRPM4 after pre‐incubation at 28°C. D) Western blot analysis showing total expression rescue of WT TRPM4 after pre‐incubation with 50 μM 5. The protein band intensity at each concentration was quantified from 3 individual experiments. * P ≤ 0.05, Student's t‐test.
Figure S7 Cytotoxicity study of the most potent inhibitors. An MTT viability assay was performed for 4, 5 and 6 on HeLa cells. The data represent the means of three values and are normalized against DMSO control.
Table S1 Anthranilic acid variations: from top to bottom, isosteric substitution of the chlorine group and variation of the carboxylic acid's position. a) Racemic mixture. b) Activity on TRPM4 given as Normalized counts of Na+ influx at 5 μM. Values above 0 are active.
Table S2 From top to bottom, substitution variations and bicyclic variations of the phenoxy moiety. a) Racemic mixture. b) Activity on TRPM4 given as normalized counts of Na+ influx at 5 μM. Values above 0 are active.
Table S3 Acetyl linker modifications. a) Racemic mixture. b) Activity on TRPM4 given as Normalized counts of Na+ influx at 5 μM. c) The enantiomerically pure isomers of the racemic 4 were separated by chiral HPLC. d) Activity measured at 10 μM.
Table S4 In vitro pharmacology profiling of 5 on several ion channels. a) Compound 5 was tested on radio ligand binding assay at 10 μM. b) Compound 5 was tested on cellular and nuclear functional assays at 10 μM. The hERG channel inhibition was also tested on automated whole‐cell patch clamp at 10 μM. Inhibition below 50% is considered not significant. The data represent the mean of two experiments. No significant inhibition (NSI).
Table S5 Analytical RP‐UPLC purity. a) The spectra were obtained according to method B (see description in general remarks). b) A partial decomposition of the compound was observed preventing any purity analysis. c) Any UPLC trace could be obtained due to either too few material or precipitation in UPLC solvents. NMR purity was considered (>90%).


