Abstract
Polo-like kinase 1 (Plk1) is an essential protein kinase that promotes faithful mitotic progression in eukaryotes. The subcellular localization and substrate interactions of Plk1 are tightly controlled and require its binding to phosphorylated sequences. Here, to identify phosphorylation-dependent interactions within the Plk1 network in human mitotic cells we performed quantitative proteomics on HeLa cells cultured with kinase inhibitors or expressing a Plk1 mutant that was deficient in phosphorylation-dependent substrate binding. We found that many interactions were abolished upon kinase inhibition; however, a subset were protected from phosphatase opposition or were unopposed, resulting in persistent interaction of the substrate with Plk1. This subset includes phosphoprotein phosphatase 6 (PP6), whose activity towards Aurora kinase A (Aurora A) was inhibited by Plk1. Our data suggest that this Plk1-PP6 interaction creates a feedback loop that coordinates and reinforces the activities of Plk1 and Aurora A during mitotic entry and is terminated by the degradation of Plk1 during mitotic exit. Thus, we have identified a mechanism for the previously puzzling observation of Plk1-dependent regulation of Aurora A.
Introduction
In mitosis, cells undergo a dramatic reorganization of their cytoskeleton structure and cellular content to divide into two viable daughter cells with identical genomic content. The division of the cytoplasm, organelles, and chromosomes is spatially and temporally coordinated to ensure fidelity. Errors in these processes are often detrimental to the emerging daughter cells and can lead to aberrant chromosome numbers, a state known as aneuploidy and a hallmark of human cancers and birth defects. Dynamic protein phosphorylation by kinases and phosphatases is one of the main regulatory mechanisms that drives mitotic progression and ensures its fidelity (1–6).
Polo-like kinase 1 (Plk1) is an essential regulator of mitosis (7). Plk1 promotes the activation of the cyclin-dependent kinase 1 (CDK1)/cyclin B complex and thereby mitotic entry, centrosome maturation, and spindle assembly, removal of sister chromatid cohesion, spindle checkpoint signaling, and microtubule-kinetochore attachment (8–10). Plk1 contributes to mitotic exit and cytokinesis by recruiting proteins to the central spindle and the midbody, and by activating the anaphase promoting complex/cyclosome (APC/C) which leads to the destruction of Plk1 itself (8, 9, 11)
Plk1 contains two distinct functional domains: an amino-terminal kinase domain and a carboxyl-terminal polo-box domain (PBD). Activation of Plk1 requires phosphorylation of its activation T-loop on Thr210 by Aurora kinase A (Aurora A) (12–14). Activation of Aurora A is regulated by autophosphorylation of its activation T-loop on Thr288 (15), and inhibition of Aurora A results in a decrease of its own T-loop phosphorylation as well as that of Plk1. Curiously, chemical inhibition of Plk1 activity also reduces the phosphorylation of Thr288 in Aurora A, suggesting a connection between their activities (14, 16, 17), but the mechanism(s) that underlie this relationship are not yet known. In cases where persistent Plk1 substrate targeting is important, the PBD facilitates recognition of and binding to proteins that contain phosphorylated amino acids within the PBD motif (Ser-pSer/pThr-Pro), leading to additional phosphorylation of them (direct substrate phosphorylation) or other nearby proteins (distributive phosphorylation) (18, 19). In the absence of a PBD-substrate interaction, the PBD forms an auto-inhibitory conformation through weak intramolecular interactions with its kinase domain; binding to a phosphorylated PBD motif sequence liberates these inhibitory interactions and increases the specific activity of Plk1 by ~3 fold (20). For many substrates, CDK1 is the priming kinase that phosphorylates the PBD motif (non-self-priming) (19). However, in some cases Plk1 itself can phosphorylate a PBD-binding motif and facilitate its own PBD-dependent target interactions (self-priming), which has been shown to contribute to its subcellular localization (21–24). In prophase, Plk1 localizes to centrosomes and the mitotic spindle; in prometaphase and metaphase, Plk1 can also be found at kinetochores before it translocates to the central spindle in anaphase and the midbody during cytokinesis (7).
In alignment with diverse Plk1 regulatory functions, its activity and substrate interactions are tightly controlled in a spatially and temporally resolved manner. At least in part, Plk1’s dynamic relocalization to subcellular structures during mitotic progression depends not only on phosphorylation, but also dephosphorylation (25). However, the contribution of phosphorylation dynamics to the regulation of Plk1 substrate interactions and signaling is unclear. Here, we determined the extent and dynamics of phosphorylation-dependent priming within the Plk1 interactome network and established the contribution of Plk1 self-priming to substrate targeting. Towards these goals, we conducted global quantitative proteomic analyses of Plk1-interacting proteins in mitotically-arrested HeLa cells in the presence and absence of small molecule kinase inhibitors (26–33) as well as with a mutant version of Plk1 (“Pincer-Plk1”) that abolishes phosphorylation-dependent binding through its PBD (18, 19). Among the insight gleaned from this priming-dependent interactome, we identified proteins with which Plk1 interaction is protected from phosphatase-dependent turnover, leading to a mechanism that explains earlier observations of feedback regulation between Plk1 and Aurora A activities.
Results
Identification of kinase activity-dependent and stable Plk1 interactors by SILAC
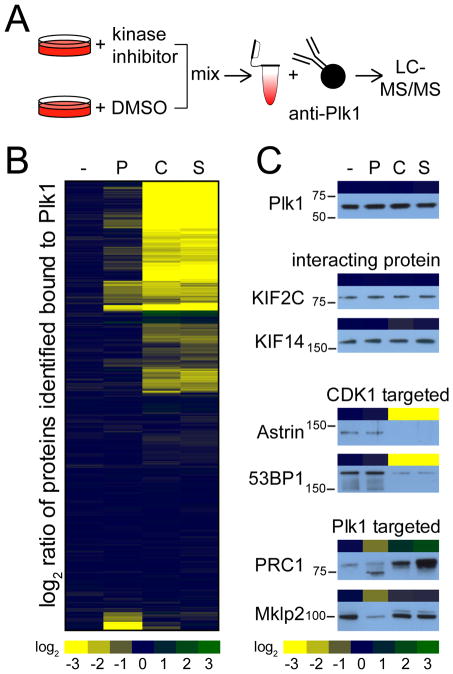
We identified Plk1 interacting proteins in mitosis by performing immunoprecipitations of endogenous Plk1 from mitotically-arrested HeLa cells (Figure 1A). To distinguish kinase activity-dependent from stable interactors and identify the relative contribution of CDK1 and Plk1 kinases to Plk1 substrate priming, we used stable isotope labeling in cell culture (SILAC) (34) to quantitatively compare the Plk1 interactome in the presence and absence of small molecule inhibitors of Plk1 (26), CDK (27), or the general kinase inhibitor staurosporine (31–33) (Figure 1A). Briefly, HeLa cells labeled with either isotopically “heavy” or “light” amino acids were arrested in mitosis using the microtubule stabilizer Taxol (commonly known as paclitaxel). Both heavy- and light-labeled HeLa cells were pre-treated for 30 minutes with the proteasome inhibitor MG132, after which either 100 nM Plk1 inhibitor BI2536, 5 μM CDK inhibitor flavopiridol, or 1 μM staurosporine were added to separate cultures of heavy-labeled HeLa cells for an additional 30 minutes. Afterwards, equal counts of heavy- and light-labeled cells were mixed, lysed, and Plk1 and its interacting proteins were then immunoprecipitated (fig. S1). To control for nonspecific binding, we conducted the same analysis on Taxol-arrested HeLa cells using beads without Plk1 antibody (fig. S1). Plk1 and control immunoprecipitations were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and the degree of confidence for these Plk1 interactors was assessed using a statistical tool developed for this purpose [Significance Analysis of INTeractome (SAINT); (35, 36)]. Using this approach, we identified 437 high-confidence Plk1-interacting proteins (table S1).
Figure 1. Kinase activity-dependent Plk1 interactome.
(A) Experimental design to identify kinase activity-dependent Plk1 interactors by SILAC. Heavy- and light-labeled HeLa cells were arrested in mitosis and treated with MG132. Heavy-labeled cells were treated with a specified kinase inhibitor or DMSO. Heavy- and light-labeled cells were mixed, lysed, and then Plk1 and its interacting proteins were immunoprecipitated from the lysates, resolved by gel electrophoresis and analyzed by LC-MS/MS. (B) Hierarchical clustering of quantitative differences in the binding of Plk1-interacting proteins in HeLa cells arrested with Taxol (-) or treated with Plk1 inhibitor (“P”; BI2536), Cdk inhibitor (“C”; flavopiridol), or staurosporine (S). (C) Comparison between quantitative mass spectrometry data (heatmap) with Western blot analyses of immunoprecipitated Plk1, the two stable interacting proteins KIF2C and KIF14, the two CDK1 kinase activity-dependent Plk1-interacting proteins Astrin and 53BP1, and the two Plk1 kinase activity-dependent Plk1-interacting proteins Prc1 and Mklp2 upon differential kinase inhibitor treatment.
The kinase activity-dependent Plk1 interactome
We clustered these high-confidence interactors based on their Plk1-binding behavior upon kinase inhibition (Figure 1B). Of the 437 proteins, 164 (38%) exhibited reduced interaction with Plk1 by two-fold or more upon CDK inhibition with flavopiridol, and a similar number (and similar set) of proteins exhibited reduced Plk1 binding upon general kinase inhibition with staurosporine (Figure 1B). Staurosporine inhibits many kinases, including the mitotic kinases CDK1, Aurora A and Aurora B, but is less specific for Plk1 (37). With this in mind, we compared the relative changes in binding of all Plk1-interacting proteins upon addition of flavopiridol with those upon addition of staurosporine and found a high correlation (R2 = 0.94) (fig. S2), supporting the concept that in mitosis, CDK activity is responsible for most of the kinase-dependent substrate targeting of Plk1 (37). Inhibition of Plk1 itself regulated the second largest group of activity-dependent interactors. Fifty-one proteins (12%) (Figure 1B) exhibited reduced interaction with Plk1 upon inhibition of Plk1 activity using BI2536. Of these 51 proteins, 33 also exhibited reduced Plk1 binding upon inhibition of CDK, while the rest were reduced only upon addition of the Plk1 inhibitor (table S2). Notably, whereas most proteins that differentially bound to Plk1 upon kinase inhibition exhibited reduced binding, three (Bora, CDCG3, and PSMD6) and 10 (Bora, 6 subunits of the proteasome, PKMYT1, FA83H, and PRC1) proteins exhibited increased binding upon Plk1 and CDK inhibition, respectively. For at least one of these proteins, Bora (which was common to both increased-binding sets), the increase may be easily explained by the loss of negative regulation. Bora protein stability is regulated by Plk1-mediated phosphorylation in a CDK1-dependent manner (38), such that inhibition of either kinase would be expected to reduce β-TRCP-dependent degradation of Bora, resulting in an increased abundance of available protein capable of interacting with Plk1.
To validate these quantitative MS data, we analyzed Plk1 immunoprecipitations under control and kinase inhibitor conditions by Western blotting for select known Plk1 (“Plk1-targeted”) or CDK1 (“CDK1-targeted”) activity-dependent interactors, as well as stable interacting proteins (Figure 1C, table S2). Consistent with the MS results (Fig. 1C, atop each blot; excerpted from Fig. 1B), none of the inhibitor conditions affected the binding of KIF2C or KIF14 to Plk1 by Western blot (Fig. 1C). However, inhibition of CDK1 strongly reduced binding of Astrin and 53BP1 to Plk1 (39, 40). The group of Plk1 targeted proteins contained the known Plk1 self-priming substrates Mklp2 (22) and PRC1 (25), both of which exhibited reduced Plk1 binding upon inhibition of Plk1 activity, as assessed by MS and Western blotting (Figure 1C). PRC1 is phosphorylated by CDK1 early in mitosis, and this phosphorylation inhibits Plk1’s binding to it (25). Indeed, this behavior is recapitulated in our analyses. Upon inhibition of Plk1, binding of PRC1 to Plk1 was reduced; conversely, inhibition of CDK1 increased PRC1 binding to Plk1 (Figure 1C) (25). In addition to Mklp2 and PRC1, the group of Plk1 activity-dependent interactors also included the known Plk1 self-priming substrates CenpU/Pbip1 (21) and JIP4 (24). Based on our survey of the published literature, our analysis appears to have identified all previously reported Plk1 self-priming substrates, which provided confidence in the data set.
We then grouped the identified Plk1-interacting proteins into three categories: (i) proteins that bind to Plk1 only in a Plk1 activity-dependent manner (“self-priming targets”, Plk1-targeted), (ii) proteins that bind to Plk1 in a CDK1 activity-dependent manner (“non-self-priming targets”, CDK1-targeted), and (iii) stable interacting proteins (“stable”). At a threshold of 2-fold or greater difference in binding to Plk1 upon inhibitor addition, we identified 18 proteins as Plk1-targeted, 164 proteins as CDK1-targeted, and 255 proteins as stable interactors (table S2).
Biological functions of Plk1 interactors
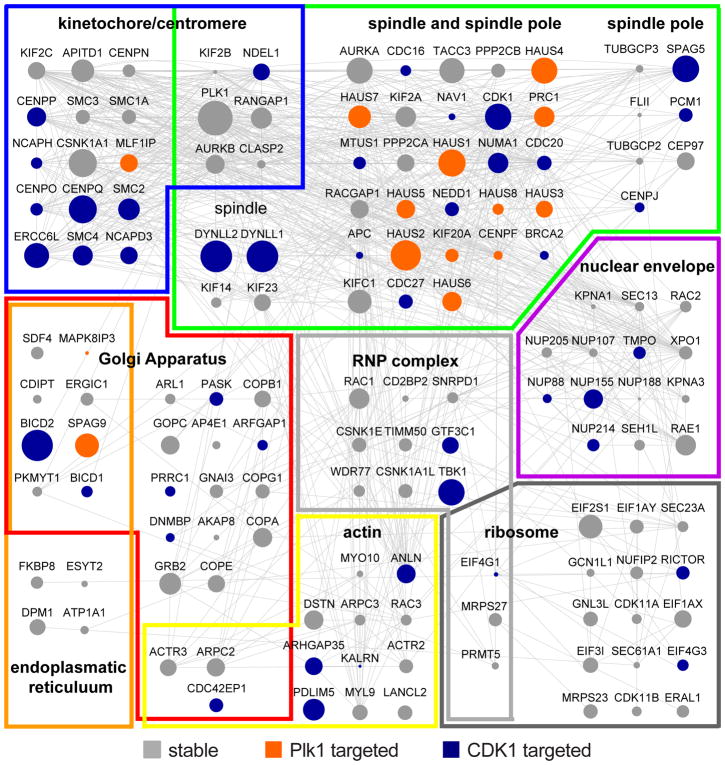
Plk1 is essential for mitotic progression by functionally regulating a plethora of biological processes. Gene ontology (GO) analysis (41–44) of our Plk1 interactome revealed a strong enrichment in aspects of chromosome segregation, organization, condensation, and cohesion; microtubule cytoskeleton, microtubule organization center and centrosome organization as well as spindle assembly and cytokinesis (fig. S3A). A regulatory function for Plk1 in these processes is supported by the dynamic subcellular localization of Plk1 to kinetochores, spindle poles, the mitotic and central spindle, as well as the midbody. We also observe a strong enrichment of Plk1 interactors at these subcellular structures (Figure 2 and fig. S3B) resulting in highly connected and spatially organized Plk1-interacting protein networks that are consistent with the regulation of biological processes essential for mitotic progression. We also discovered many multimeric protein complexes in the Plk1 interactome, including proteins involved in the control of the actin cytoskeleton (Arp2/3 complex), spindle (HAUS complex) and chromosome organization (cohesin, condensin, and synaptonemal complexes), and in the regulation of protein ubiquitination (anaphase-promoting complex, other ubiquitin ligase complexes, and the proteasome) (fig. S4, A and B). Unexpectedly, proteins involved in Golgi vesicle trafficking as well as rRNA and tRNA transcription were significantly enriched in the interactome (fig. S3A), which may be interesting to explore in the future.
Figure 2. Subcellular localization map of Plk1 interactors.
Subcellular localization map of Plk1 interactors to kinetochores/centromeres, microtubule cytoskeleton and spindle poles, Golgi apparatus, nuclear envelope, endoplasmic reticulum, actin cytoskeleton, ribonucleoprotein (RNP) complexes, and ribosomes.
Stable Plk1 interactors and dynamic phosphatase opposition of PBD-binding motifs
We were intrigued by the observation that the majority of Plk1-interacting proteins remain stably bound to Plk1 upon the addition of kinase inhibitor(s). One caveat of this experimental strategy is that upon cessation of kinase activity, the activity of an opposing protein phosphatase is necessary to reduce phosphorylation site occupancy and therefore interaction with Plk1. In other words, in the absence of a counteracting phosphatase, a bone-fide phosphorylation-dependent Plk1 interaction would not change in abundance in the presence of inhibitors targeting the responsible kinase, once the interaction was already established.
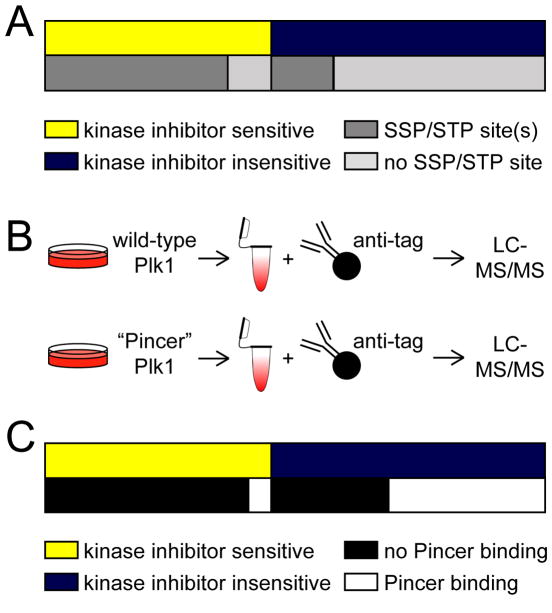
To assess the degree to which lack of or protection from phosphatase opposition could contribute to the observation of stable Plk1-protein interactions, we determined the prevalence of Ser-Ser/Thr-Pro or Ser/Thr-Pro sequences that could be phosphorylated to generate a PBD motif (Ser-pSer/pThr-Pro) in Plk1 interactors. We found that the majority of Plk1 interactors whose binding was sensitive to kinase inhibition contained a Ser-Ser/Thr-Pro motif (Figure 3A). Of the 39 proteins that did not contain a Ser-Ser/Thr-Pro motif, 10 were only sensitive to the Plk1 but not the CDK inhibitor (table S2), and 25 had a least one first-degree neighbor identified in our analysis that did contain a candidate PBD motif (fig. S5). Thus, it is plausible that kinase activity-dependent binding of these 25 proteins is mediated indirectly, through a binding partner that is phosphorylated by CDK1. For the four proteins that lacked a PBD-binding motif and for which we did not identify a CDK1-dependent first-degree neighbor, it is possible that they have binding partners not identified in our analyses that are CDK1-dependent, or that CDK1-mediated phosphorylation of Ser/Thr-Pro or other sites is sufficient in rare cases to promote Plk1 interactions. Notably, 22% of proteins that stably interacted with Plk1 upon kinase inhibition contained a Ser-Ser/Thr-Pro motif that has previously been observed to be phosphorylated in the PhosphoSitePlus database (Figure 3A), an enrichment that was significantly greater than random chance (2,550 Ser-(phos-Ser/Thr)-Pro proteins in PhosphoSitePlus & 20,225 proteins in UniProt (ratio: 0.126082); cumulative binomial probability p=0.000005), making them candidates for phosphorylation-dependent Plk1 interactions whose phosphorylation of the PBD-binding motif is not dynamically turned over by a protein phosphatase under our experimental conditions (such as cell cycle phase and duration of inhibitor treatment).
Figure 3. Stable interactors of Plk1 include phosphorylation-dependent interactions.
(A) Diagram depicting the distribution of kinase inhibitor-sensitive (yellow) and inhibitor-insensitive (blue) Plk1 interactors and, below, their relative enrichment in candidate PBD motifs (SSP/STP) (dark grey) or not (light grey). (B) Experimental design to identify kinase activity-dependent Plk1 interactors using wild-type and “Pincer” mutant Plk1. HeLa cells expressing with myc- or Flag-tagged wild-type or “Pincer” mutant Plk1 were arrested in mitosis and treated with MG132. Plk1 and its interacting proteins were immunoprecipitated, resolved by gel electrophoresis and analyzed by LC-MS/MS. (C) Diagram depicting the distribution of kinase inhibitor sensitive (yellow) and insensitive (blue) Plk1 interactors and their ability to bind (white) or not bind (black) to “Pincer” mutant Plk1.
To experimentally determine whether the lack of phosphorylation site turnover contributes to the observed stable interaction of some candidate Plk1 substrates upon kinase inhibition, we introduced mutations (“Pincer” mutations; H538A and K540M) into the linker between the two polo-boxes in the PBD of Plk1, which abolishes priming phosphorylation-dependent interactions (18, 19). We reasoned that the differential interaction behavior of stable interactors between wild-type and “Pincer” mutant Plk1 would provide greater evidence of candidate kinase activity-dependent interactors that are protected from or unopposed by a phosphatase and help us distinguish them from activity-independent interactors. A similar strategy was reported previously using recombinant, purified wild-type or “Pincer” mutant PBDs alone as baits for assessing candidate phosphorylation-dependent interactions of the Plk1 PBD in mitotic U2OS lysates (45). We established HeLa cell lines stably expressing all combinations of Myc- or Flag-tagged, wild-type or “Pincer” Plk1 to account for potential epitope tag-specific interaction effects, synchronized them in mitosis, immunoprecipitated wild-type and “Pincer” Plk1, and determined their respective interactomes by LC-MS/MS (Figure 3B). Using this approach, we identified 426 of the aforementioned 437 Plk1-interacting proteins, including 247 of the 255 stable interactors (table S3), and conclusively sorted most of them (354 proteins) into two categories based on their binding behavior to “Pincer” Plk1 and wild-type Plk1: proteins that bound to both (“Pincer bound”), and proteins that bound to wild-type Plk1 but not “Pincer” Plk1 (“Pincer lost”) (table S3, last column). Finally, we queried the number of kinase activity-dependent and stable Plk1 interactors for their “Pincer” mutant-binding behavior. Similar to the results of candidate PBD motif analysis, the majority (88%) of kinase activity-dependent interactors bound to wild-type but not “Pincer” Plk1 (Figure 3C). Importantly, we found that 34% of stable Plk1-interacting proteins were lost with “Pincer” but not wild-type Plk1 (Figure 3C), supporting our hypothesis that a subset of stable Plk1 interactors may bind in a phosphorylation-dependent manner but that their PBD-binding motifs could be protected from or not opposed by phosphatases; alternatively, a kinase not inhibited by any of these inhibitors, including staurosporine, may be responsible for a phosphorylation-priming event.
Plk1 regulatory interaction with phosphoprotein phosphatase 6 (PP6)
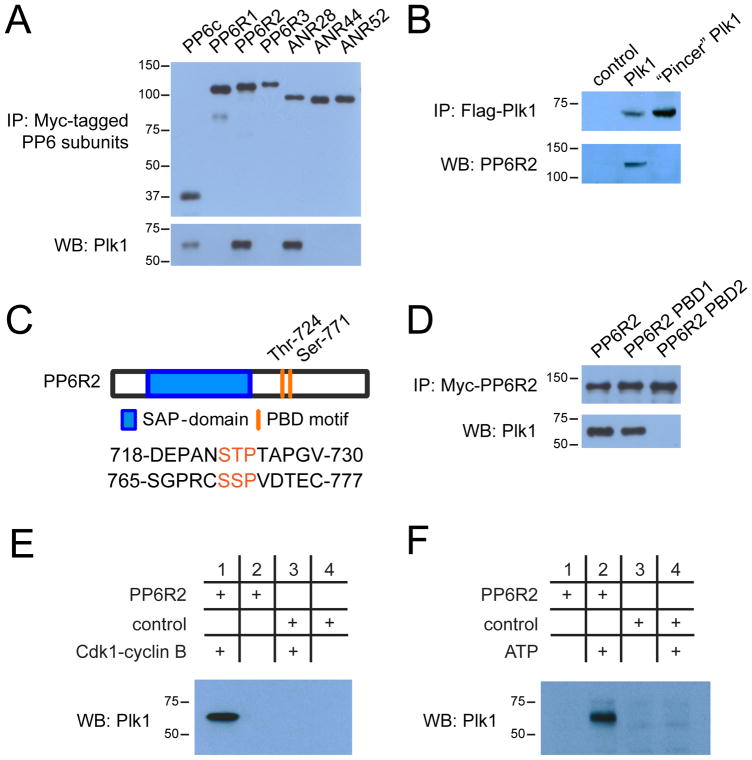
One of the stable Plk1 interactors that showed differential binding to wild-type and “Pincer” Plk1 was the phosphoprotein phosphatase 6 (PP6) holoenzyme complex. PP6 is a heterotrimeric holoenzyme complex that consists of a catalytic subunit (PP6c), a SAPS-domain containing subunit (PP6R1-3), and an ankyrin-repeat domain containing subunit (ANR28, ANR44, ANR52) (46, 47), all of which were detected to varying degrees in the Plk1 immunoprecipitations by mass spectrometry (fig. S6, A and B). To determine whether Plk1 preferentially interacts with a specific PP6 holoenzyme, we compared the median and average total peptide counts of the different PP6 subunits that we identified in Plk1 pulldowns (fig. S6B and table S1). PP6R2 and ANR28 were represented by roughly four times as many peptides than the other two PP6R and ANR subunits, suggesting that Plk1 preferentially binds to a PP6 holoenzyme consisting of PP6c, PP6R2, and ANR28. To validate the PP6-Plk1 interaction, we stably expressed all seven Myc-tagged PP6 subunits in HeLa cells and performed individual immunoprecipitations from mitotically-arrested cells. Although we precipitated comparable amounts of PP6 subunits, Plk1 co-precipitated in higher amounts with PP6c, PP6R2, and ANR28 than the others (Figure 4A), supporting the semi-quantitative mass spectrometry data that Plk1 preferentially interacts with the PP6c/PP6R2/ANR28 holoenzyme.
Figure 4. Regulatory interaction of PP6 and Plk1.
(A) Western blot analysis of Myc-tagged PP6 subunit binding to endogenous Plk1 in immunoprecipitates (IP) from lysates of HeLa cells expressing the indicated Myc-tagged PP6 subunit. (B) Immunoprecipitation of Flag-tagged wild-type and “Pincer” mutant Plk1 from transfected HeLa cells and Western blot analysis of co-precipitated endogenous PP6R2. (C) Diagram depicting PP6R2 domain structure and location of PBD motifs. (D) Immunoprecipitations of wild-type, PBD1-, and PBD2-mutant PP6R2 and Western blot analysis of co-precipitated Plk1. (E) Immunoprecipitation assessing the dependence of the PP6R2-Plk1 interaction on the activity of CDK1-cyclin B complex. Bacterially expressed and purified PP6R2 was immobilized on activated Sepharose and incubated with ATP and CDK1-cyclin B complex or not, followed by addition to mitotically-arrested (Taxol) HeLa cell lysate, isolation of PP6R2-containing complexes and Western blotting for Plk1. (F) As described in (E), except that immobilized PP6R2 was incubated directly with mitotic HeLa cell lysates with or without additional ATP prior to isolation of PP6R2 complexes and Western blotting for Plk1. BSA-Sepharose was used as a control (E and F).
PP6 protein-protein interactions can be established through the regulatory SAPS-domain containing subunits (PP6R) (48, 49). To support our hypothesis that the PP6 holoenzyme might interact with Plk1 in a phosphorylation-dependent manner that could be protected from or unopposed by phosphatases under mitotic arrest, we performed pull-downs for wild-type and “Pincer” Plk1 and analyzed them by western blotting for the presence of PP6R2 (Figure 4B). Indeed, while wild-type Plk1 readily bound PP6R2, PP6R2 binding was not detected in “Pincer”-Plk1 pulldowns. PP6R2 contains two candidate PBD-binding motifs surrounding Thr724 (Ser-Thr-Pro; PBD1) and Ser771 (Ser-Ser-Pro; PBD2), the latter of which has been previously observed to be phosphorylated in cells, including in mitotic HeLa cells [www.phosphosite.org; (17)] (Figure 4C). Notably, alignment of these two candidate PBD-binding motifs across chordates (50, 51) indicates a high degree of sequence conservation of PBD2 but not PBD1 (fig. S7). To determine if either PBD motif is required for the Plk1-PP6 interaction, we separately mutated Thr724 and Ser771 to alanine and compared the extent to which wild-type and mutant PP6R2s could interact with Plk1 in mitotic HeLa cells. We found that Plk1 readily bound to wild-type and PBD1-mutant PP6R2, but that the interaction with Plk1 was abolished in PBD2-mutant PP6R2 pulldowns (Figure 4D). To determine whether phosphorylation is required for this interaction, PP6R2 was expressed and purified from bacteria, followed by covalent immobilization on Sepharose beads and incubation in vitro in the presence or absence of CDK1/cyclin B complex and ATP. These PP6R2 beads were then incubated with mitotic cell lysate, followed by re-isolation of PP6R2 and Western blotting of the pulldowns for Plk1. Furthermore, control and in vitro phosphorylated PP6R2 were analyzed by LC-MS/MS to identify phosphorylation sites. We confirmed that the CDK1/cyclin B complex phosphorylates Ser771 and that this phosphopeptide was not detectable in control reactions (table S4). Only after phosphorylation by Cdk1 was PP6R2 capable of co-precipitating Plk1 from lysates (Figure 4E). Finally, we incubated unphosphorylated PP6R2 covalently immobilized on Sepharose with mitotic HeLa cell lysates treated with ATP or not, followed by isolation of PP6R2 complexes and Western blotting for Plk1 (Figure 4F). In this experiment, Plk1 bound to PP6R2 only when additional ATP was present in lysates.
These data demonstrate that Plk1 interacts with PP6 through the PP6R2 subunit in a phosphorylation-dependent manner that appears to be protected from or not opposed by a phosphatase in Taxol-arrested HeLa cells. Many phosphatases are inhibited early in mitosis and are reactivated at the metaphase-to-anaphase transition (2). To determine whether a phosphatase capable of dephosphorylating PP6R2 Ser771 is only active in later phases of mitosis, we investigated the Plk1-PP6 interaction during mitotic exit. We arrested HeLa cells in mitosis using low doses of the microtubule depolymerizing drug nocodazole, which can be washed out to allow for a synchronous population of cells to exit mitosis. We collected cells at 0, 30, 60, 90, and 180 minutes after nocodazole wash-out and performed immunoprecipitations of endogenous PP6R2. When we probed the immunoprecipitates for PP6R2 and Plk1, we found that the while the amount of PP6R2 remained constant as HeLa cells progress into later phases of mitosis, the amount of co-precipitated Plk1 decreased (fig. S8A) . This decrease in Plk1-PP6 interaction could be suggestive of reactivation of a putative PP6R2 Ser771 phosphatase; however, it was previously reported that Plk1 is degraded in an anaphase-promoting complex/cyclosome-dependent manner, beginning in anaphase and continuing throughout cytokinesis, to allow for proper exit from mitosis (52). To assess whether the kinetics of Plk1 degradation mirrored the loss of Plk1-PP6 interaction, we measured total Plk1 and PP6R2 protein abundance during nocodazole washout by Western blotting. Indeed, while the abundance of total PP6R2 stayed constant, total Plk1 abundance was markedly reduced by 60 minutes after nocodazole washout suggesting that the loss of the Plk1-PP6 interaction is likely due to protein degradation, rather than PP6R2 Ser771 dephosphorylation (fig. S8B). To directly test this, we immunoprecipitated Plk1 from HeLa cells as they exited mitosis, probed these precipitates for Plk1 and PP6R2, and found that the ratio of Plk1 to PP6R2 remained constant as Plk1 is degraded during exit (fig. S8C). Together, these results indicate that the Plk1-PP6 interaction is likely protected from or not opposed by a phosphatase, and is eventually terminated by the degradation of Plk1.
Plk1-dependent activation of Aurora A through inhibition of PP6
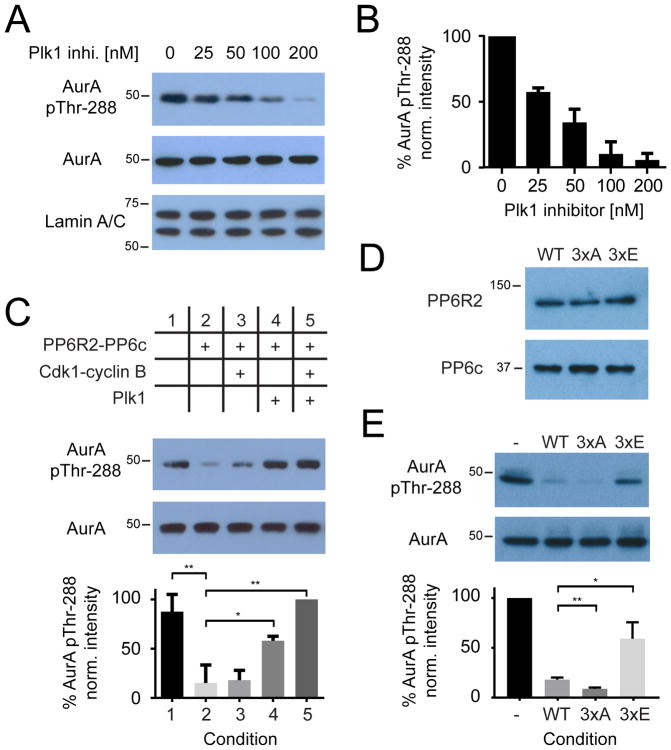
Although early reports demonstrated protein phosphatase 1 (PP1c) as capable of dephosphorylating the T-loop of Aurora A in vitro (53, 54), more recent studies support a PP6 holoenzyme as the primary T-loop phosphatase for Aurora A in cells (55, 56). As introduced above, Aurora A is the activating kinase for Plk1 (13, 14). Interestingly, we and others have observed that inhibition of Plk1 results in a reduction of Aurora A T-loop phosphorylation at Thr288 and thereby Aurora A activity (14, 16, 17) (Figure 5, A and B). Although this effect is thought to be indirect, as Aurora A autophosphorylates its T-loop on a basic Aurora A consensus motif (RRXp[S/T]) that is distinct from the Plk1 consensus motif ([D/E/N]Xp[S/T][Φ], Φ = hydrophobic) (57), the precise mechanism underlying this observation is not known. Thus, we hypothesized that Plk1 might negatively regulate PP6 activity, thereby explaining prior observations of the dependency of Aurora A activity on Plk1. To test this hypothesis, we reconstituted all components of the proposed regulatory circuit including Plk1 and the Aurora A-TPX2 (microtubule nucleation factor), PP6R2-PP6c, and CDK1-cyclin B complexes in vitro. We preincubated PP6R2-PP6c with buffer alone (Figure 5C, column 2), with the CDK1-cyclin B complex (Figure 5C, column 3), with Plk1 (Figure 5C, column 4), or with both the CDK1-cyclin B complex and Plk1 (Figure 5C, column 5) in the presence of ATP to phosphorylate PP6R2, while a pool of the PP6 substrate Aurora A-TPX2 was separately preincubated with staurosporine to inhibit Aurora A autophosphorylation activity (37). After the preincubation step, Aurora A-TPX2 was added to each condition to assess PP6R2-PP6c activity (Figure 5C). Reactions were analyzed by Western blotting for Aurora A Thr288 phosphorylation (pThr288) relative to total Aurora A protein as a loading control and compared to normalized Aurora A pThr288 abundance in an untreated control sample (Figure 5C, column 1). As expected, incubation of Aurora A-TPX2 with PP6R2-PP6c alone resulted in significant dephosphorylation of Thr288 compared to control. Pre-incubation of PP6R2-PP6c with CDK1 alone did not significantly affect PP6R2-PP6c activity, whereas pre-incubation with Plk1 alone significantly decreased PP6R2-PP6c–mediated Aurora A Thr288 dephosphorylation; this effect was further increased upon preincubation with both CDK1 and Plk1. Plk1 does not strictly require priming by CDK1 for substrate recognition and phosphorylation in vitro (57); thus, incubation with Plk1 alone could be capable of phosphorylating the PP6 holoenzyme and, thus, could result in some inhibition of PP6R2-PP6. However, the efficiency of Plk1 phosphorylation is increased by CDK1-dependent priming, resulting in greater inhibition of PP6R2-PP6c activity.
Figure 5. Regulation of PP6 phosphatase activity by Plk1 in vitro.
(A and B) Western blotting (A) and quantification (B) of Aurora A phosphorylation at Thr288 in mitotically arrested HeLa cells treated with increasing amounts of Plk1 inhibitor, BI2536. Lamin A/C was a loading control. (C) followed by Western blotting for total and phosphorylated (Thr288) Aurora A after preincubation with PP6R2-PP6c and CDK1/cyclinB, Plk1, or both in in vitro kinase/phosphatase assays as indicated (top). (D) Western blot analysis of wild-type and Plk1 phosphorylation-site mutants of PP6R2 holoenzymes containing PP6c, in vitro. (E) Representative Western blots of total and phosphorylated (Thr288) Aurora A (E) in basal conditions in vitro (-) or upon incubation with wild-type (WT) or mutant PP6R2 holoenzyme described in (D). Blots (A–E) are representative (n=3), and quantified data (B, C, and E) are mean ± S.D. of 3 biological replicates; * p < 0.05, ** p < 0.01).
To identify the specific sites on PP6R2 phosphorylated by Plk1, we performed SILAC experiments in triplicate in which mitotically arrested HeLa cells stably expressing Myc-PP6R2 were treated with the Plk1 inhibitor BI2536 (heavy-labeled condition) or vehicle (light-labeled condition), followed by collecting and combining these “heavy” and “light” cells, immunoprecipitation using antibodies against the Myc tag, and gel electrophoresis to isolate the PP6R2 protein. Subsequent tryptic digestion and quantitative analysis by LC-MS/MS identified multiple phosphopeptides from PP6R2 but only three that significantly decreased in the presence of Plk1 inhibitor: one containing pThr10, one containing pSer289, and one containing a poorly localized phosphorylation site within a cluster of Ser residues spanning Ser822 to Ser825 (table S5). Both the pThr10 and pSer289 peptides were localized within a Plk1 consensus motif; Ser823 is also within a Plk1 consensus sequence and so was considered the most likely Plk1 site within this cluster.
We generated multi-site PP6R2 mutants that contained all three Plk1 sites mutated to either alanine (PP6R2-3xA) or glutamic acid (PP6R2-3xE) to ablate or mimic Plk1 phosphorylation respectively, purified the corresponding PP6c holoenzymes and compared their relative activities towards Aurora A-TPX2 in in vitro dephosphorylation assays against wild-type PP6R2-PP6c holoenzymes. Whereas all three PP6R2 holoenzymes contained comparable amounts of catalytic PP6c (Figure 5D), PP6R2-3xE was significantly less active than either wild-type or 3xA-mutant PP6R2 (Figure 5E); indeed, the non-phosphorylatable PP6R2-3xA mutant exhibited the highest activity towards Aurora A-TPX2 pThr288 in this assay. Thus, we conclude that the effect of multi-site Plk1 phosphorylation on inhibiting PP6R2-PP6c holoenzymes towards Aurora A is by a mechanism other than regulation of catalytic subunit binding, perhaps by negatively regulating substrate recognition and interactions.
To further support the physiological role of PP6 as the phosphatase responsible for Aurora A T-loop dephosphorylation in cells, we separately incubated purified PP1c and ATP with Plk1 or not and used these preparations to test whether Plk1 activity could reduce the capacity for PP1c to dephosphorylate the Aurora A T-loop. Notably, PP1c was equally capable of dephosphorylating Aurora A pThr288 in the presence or absence of Plk1 activity (fig. S9), contrary to our and others’ (14–16) observations of the dependence of Aurora A T-loop phosphorylation on Plk1 activity in cells or with PP6R2/PP6c complexes in vitro (Figures 5, A and C).
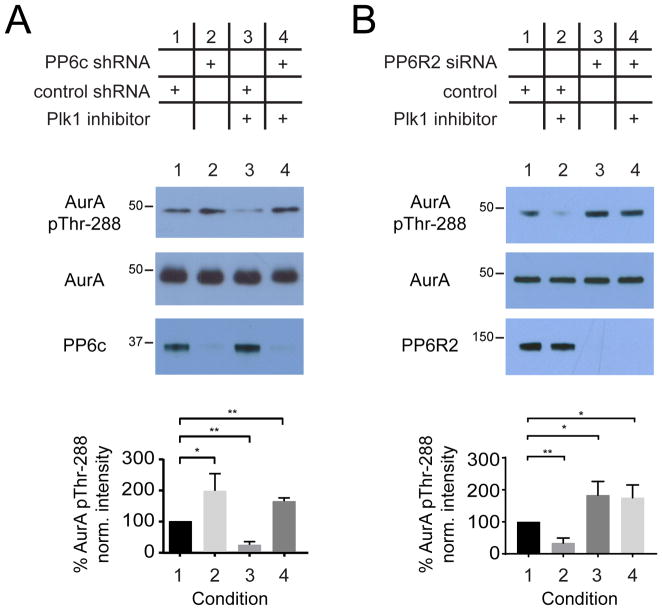
Next, we tested our hypothesis in mitotically arrested HeLa cells transduced with pooled short hairpin RNAs (shRNAs)-containing baculoviruses previously shown to efficiently reduce PP6c protein abundance and increase Aurora A Thr288 phosphorylation site occupancy (56). HeLa cells were infected with either control or PP6c shRNA baculoviruses and synchronized in mitosis using Taxol. As previously shown (55, 56), Aurora A Thr288 phosphorylation significantly increased upon reduction of PP6c protein abundance (Figure 6A). Conversely, Aurora A Thr288 phosphorylation significantly decreased upon treatment with Plk1 inhibitor BI2536 (Figure 6A, column 3) and was abolished in the absence of PP6c, which suggests that Plk1’s effect on Aurora A Thr288 phosphorylation is mediated by PP6c. To determine whether this effect is specific to a PP6R2-containing holoenzyme, we repeated this analysis with small interfering RNA (siRNA) to reduce PP6R2 abundance and saw similar effects on Aurora A Thr288 phosphorylation (Figure 6B). As with PP6c, this PP6R2-dependent increase in Aurora A Thr288 phosphorylation was insensitive to Plk1 inhibition (Figure 6B).
Figure 6. Regulation of PP6 phosphatase activity by Plk1 in cells.
(A and B) Representative Western blot and analysis of total and phosphorylated (Thr288) Aurora A upon depletion of PP6c (A) or PP6R2 (B), Plk1 inhibition with BI2536, or both. Data are mean ± S.D. of 3 biological replicates; * p < 0.05, ** p < 0.01).
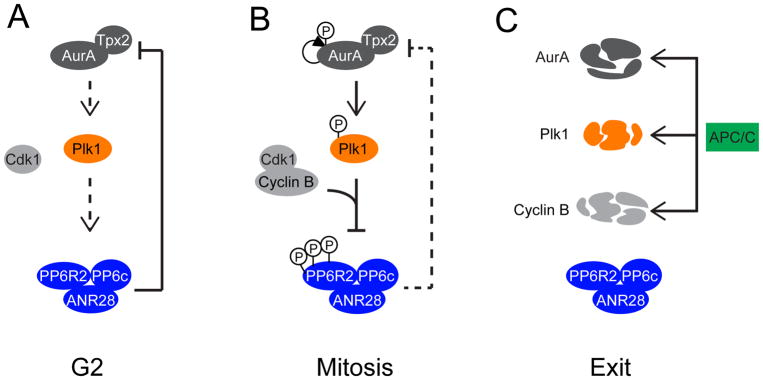
These results suggest a model in which, in pre-mitotic cells, active PP6R2-PP6c keeps Aurora A activity in check by T-loop dephosphorylation (Figure 7A). As cells advance towards and into mitosis, increased activation of the CDK1-cyclin B complex generates a Plk1 binding site on the PP6R2-PP6 complex, thereby allowing for Plk1-mediated phosphorylation of the holoenzyme and reducing PP6 activity towards Aurora A (and potentially other substrates) (Figure 7B). This has the effect of positively reinforcing Plk1 activity, as decreased PP6 activity leads to increased Aurora A activity, the Plk1 T-loop kinase. Starting in anaphase, however, Plk1 decreases in abundance by protein degradation, which reduces the Plk1-PP6R2 interaction and resets the potential for reactivation of PP6R2-PP6c activity in subsequent cell cycles (Figure 7C).
Figure 7. Model of regulatory feedback governing Aurora A, Plk1 and PP6.
(A) In G2, Aurora A and Plk1 activities are low and PP6R2-PP6c activity is high, serving as a check on premature Aurora A activation. (B) As cells enter mitosis, Aurora A is activated and PP6R2 is primed by the CDK1-cyclin B complex, promoting Plk1 activation and phosphorylation-dependent inhibition of PP6. This results in a net increase in Aurora A phosphorylation and activity. (C) At the onset of anaphase, protein turnover of cyclin B, Plk1 and Aurora A breaks this feedback loop and resets the PP6R2-PP6c complex for the subsequent cell cycle.
Discussion
Plk1 is an essential regulator of mitotic progression from entry to exit, and of many associated cellular processes including centrosome maturation, mitotic spindle assembly, chromosome segregation, and cytokinesis. Plk1 conducts this diverse set of functions through phosphorylation of a diverse array of substrates. Plk1 identifies these substrates by phosphorylation-dependent targeting via its PBD (18, 19). It was previously shown that either Plk1 itself (self-priming) (21–24) or other kinases (non-self-priming) (18, 19) can generate the phospho-binding motif on Plk1 substrates. Both priming-dependent behaviors are considered essential mechanisms that regulate Plk1 subcellular localization.
Several groups, including ours, have conducted proteomics analyses to identify Plk1 substrates using small-molecule Plk1 kinase inhibitors, RNA interference gene silencing, ATP analog-sensitive alleles, and interactome studies (17, 45, 58–60). While these analyses have buttressed our knowledge of Plk1 substrates, the broader mechanisms that underlie Plk1 substrate targeting, including regulatory inputs by other kinases and phosphatases, have remained less well understood.
Here, we sought to address these open questions by studying the Plk1 interactome in a kinase activity-dependent manner. Using quantitative proteomics, we identified 437 high confidence Plk1 interactors and determined their differential Plk1 binding behavior upon inhibition of Plk1 itself or of CDK1, as well as upon conditions of general, unselective kinase inhibition. We found that CDK1 is indeed the primary upstream kinase responsible for PBD-binding motif phosphorylation of Plk1 substrates and the comparison with general kinase inhibition suggest little effects from other kinases. In these analyses, we were also able to identify many known as well as new CDK1-dependent Plk1 interactors as well as Plk1 self-priming substrates. As noted earlier, a previous study used recombinant wild-type and “Pincer” PBD fragments of Plk1 linked to a solid support as bait to fish out candidate phosphorylation-dependent PBD interactors from U2OS cell lysates (45). Although the overlap between our and their datasets is low, likely due to the unique methods and approaches used in each study, we note that they include CDK1 activity-dependent, Plk1 activity-dependent, and stable interactors among them. Thus, the present work adds additional context to our still incomplete understanding of Plk1 substrate recognition and biology.
Consistent with Plk1 localization to spindle poles, kinetochores, central spindle and the midbody, we identified many Plk1 interactors with known localization to these structures. But we also observed interactions of Plk1 with proteins known to localize to the Golgi apparatus and the endoplasmic reticulum. Plk1 phosphorylation was previously shown to inhibit GRASP65 function in Golgi tethering (61, 62). Our analysis identified 25 proteins thought to localize to the Golgi apparatus, which potentially expands the regulatory role of Plk1 for this organelle in mitosis. Furthermore, we found proteins that are part of the nuclear pore and envelope, both of which undergo phosphorylation-dependent disassembly and reassembly in prophase and telophase, respectively. Finally, proteins were identified as Plk1 interactors that constitute the ribosome, which could contribute to the halting of transcription and translation in mitosis. Thus, our analyses reveal new entry points into the investigation of Plk1 function(s) in organelle organization and distribution in mitosis.
When we grouped Plk1-interacting proteins by their binding behavior into kinase activity-dependent and stable interactors, we were surprised to find such a large complement of stable binders in the Plk1 interactome. Further bioinformatics interrogation of these proteins revealed that these stable interactors were enriched in PBD sequence motifs previously reported to be phosphorylated, suggesting that their Plk1 binding might be phosphorylation-dependent but not dynamically turned over by a phosphatase. When kinases are inhibited in cells, reduction in their substrate phosphorylation requires either substrate protein turnover or dephosphorylation by a protein phosphatase, as cessation of kinase activity under static conditions does not by itself change the abundance of substrate phosphorylation. Here, in the absence of phosphatase activity, such a phosphorylation site and thus its phosphorylation-dependent protein-protein interaction would be transparent to our approach. This prompted us to further dissect the stability of interactor binding using “Pincer” mutant Plk1. While not conclusively demonstrative of phosphorylation-dependent interactions, the combined use of wild-type and conservative “Pincer” mutations in Plk1 suggest the potential for differential phosphatase recognition and/or opposition of the PBD motif of Plk1 substrates, which would represent an additional layer of regulation of Plk1 signaling for further investigation.
One of these stable substrates is the PP6c-PP6R2-ANR28 holoenzyme complex. PP6 was previously shown to be the T-loop phosphatase that opposes Aurora A (55), the kinase that activates Plk1 (12–14). We found that while the Plk1-PP6 interaction requires CDK1 phosphorylation of a PBD motif in PP6R2, it is not dynamically turned over. Degradation of Plk1 during mitotic exit abolished the PP6R2 interaction, enabling the potential reactivation of PP6 in subsequent cell cycle phases. We and others have observed that inhibition of Plk1 indirectly reduces Aurora A T-loop phosphorylation and thereby activity (14, 16, 17); our study reveals that PP6 is the missing piece in this regulatory feedback loop, that the prolonged interaction of Plk1 with PP6 inhibits PP6-mediated inhibition of Aurora A, and thus dually reinforces Plk1 and Aurora A activity.
Our findings provide a mechanistic explanation for the coordination of these three enzyme activities and their temporal regulation during mitotic progression. In G2 phase of the cell cycle, the activities of Aurora A and Plk1 are low and that of PP6R2-PP6c is high, serving as a check on premature Aurora A activation (Figure 7A). As cells enter mitosis, Aurora A is activated by CDK1-cyclin B complexes (63), increasing phosphorylation occupancy of its T-loop as the balance of Aurora A autophosphorylation and PP6-dependent dephosphorylation shifts. Furthermore, our findings suggest that CDK1-cyclin B–mediated phosphorylation of PP6R2 primes it for Plk1-mediated phosphorylation without inhibiting its activity. In our model, increased Aurora A activity promotes Plk1 activation through phosphorylation of its T-loop, which in turn leads to phosphorylation and inhibition of PP6 (Figure 7B). This results in a net increase in Aurora A phosphorylation and reduction in T-loop dephosphorylation as cells enter mitosis. Finally, protein turnover of cyclin-B, Plk1 and Aurora A at the onset of anaphase break this feedback loop (Figure 7C). Although beyond the scope of the present work, it is possible that an as-yet unidentified phosphatase activated in or unique to G1 serves to further reset the PP6R2-PP6c complex for the subsequent cell cycle. Future investigations of other Plk1 interacting proteins, their regulatory inputs by phosphatases, and their biological function will further enrich our understanding of Plk1 function.
Materials and Methods
Small molecule inhibitors, antibodies, and siRNAs
BI2536 was synthesized in house. MG132, Taxol, nocodazole and staurosporine were purchased from Tocris Bioscience, and flavopiridol was purchased from Sigma-Aldrich. Stock solutions were prepared in DMSO (Sigma-Aldrich).
Antibodies to Plk1 (cat. no. P5998) and Aurora A (cat. no. A1321) were purchased from Sigma-Aldrich; antibodies to 53BP1 (cat. no. 4937), Prc1 (cat. no. 3639), and phosphorylated Aurora A (Thr288) (cat. no. 3079) were purchased from Cell Signaling Technology; antibodies to Mklp2 (cat. no. 172620) and PP6R2 (cat. no. 72032) were purchased from Abcam; antibody to Myc (clone 9E10) was purchased from Bio-Xcell; and antibody to KIF14 (cat. no. GTX103196) was purchased from GeneTex. Antibodies to KIF2C and Astrin were gifts from Duane Compton, Dartmouth. Antibody to lamin A/C was a gift from Dr. Frank McKeon, Harvard. Secondary antibodies (Alexa-fluor conjugated 488/568 goat anti-rabbit/mouse) were obtained from Molecular Probes (Invitrogen) and (goat-anti rabbit/mouse-HRP) Jackson ImmunoResearch.
Single siRNA against PP6R2 was obtained from GE Dharmacon. ShRNAs against PP6c and control shRNA were previously described (56).
Cloning, mutagenesis and transfection
Human Plk1 was amplified from HeLa cDNA and cloned into the p3xFlag-CMV10 (Sigma) or pBMN-6Myc-Neo vector and confirmed by DNA sequencing. “Pincer” (H538A and K540M) and kinase-deficient (KD1: K82M; KD2: D176N) mutations were introduced into the Plk1-coding sequence by site direct mutagenesis the QuickChange II® Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to manufacturer’s instructions and confirmed by DNA sequencing. HeLa cells were transfected with p3xFlag-CMV10-Plk1 or p3xFlag-CMV10-“Pincer”-Plk1 using Lipofectamine2000 (Invitrogen) according to the manufacturer’s instructions and selected with G418. pBMN-6Myc-Plk1, pBMN-6Myc-“Pincer”-Plk1, pBMN-6Myc-Plk1-KD1, pBMN-6Myc-Plk1-KD2 vectors were packaged in 293T A’ cells, viral supernatants were used for HeLa infections, and cells were selected with G418. Human PP6R1, PP6R2, PP2R3, ANR28, ANR44, and ANR52 were amplified from HeLa cDNA and cloned into the pBMN-6Myc-neo vector and confirmed by DNA sequencing. PBD motif mutations T724A (PBD1) and S771A (PBD2), and Plk1 substrate site mutations T10A or T10E, S289A or S289E, and S823A or S823E were introduced into the PP6R2 coding sequence by site direct mutagenesis the QuickChange II® Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to manufacturer’s instructions and confirmed by DNA sequencing. Vectors were packaged in 293T A’ cells, viral supernatants were used for HeLa infections, and cells were selected with G418.
Cell culture
HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) with 10% fetal bovine serum (Hyclone, South Logan, Utah) and penicillin-streptomycin (100U/mL and 100μg/mL, Invitrogen), at 37°C in a humidified atmosphere with 5% CO2. For SILAC experiments, HeLa cells were grown in arginine- and lysine-free DMEM with 10% dialyzed fetal bovine serum (Hyclone) supplemented with either 100mg/L 13C615N2-lysine and 100mg/L 13C615N4-arginine (Cambridge Isotope Laboratories, Inc., Andover, MA) (“heavy”) or identical concentrations of isotopically-normal lysine and arginine (“light”) for at least six cell doublings.
Small molecule inhibitor treatment of SILAC HeLa cells
Heavy- and light-labeled HeLa cells were synchronized by a double thymidine block; three hours after washout of the second thymidine block, 100 nM Taxol was added to the cells for 10 hours. Cells were treated with 10 μM MG132 for 30 minutes, followed by addition of 100 nM BI2536, 5 μM flavopiridol, or 1 μM staurosporine to heavy-labeled HeLa cells for an additional 30 minutes; a total of 4 control (no Plk1 antibody), 2 Taxol-only (vehicle), 4 BI2536, 3 flavopiridol and 2 staurosporine-treated SILAC samples were prepared. After inhibitor treatment, HeLa cells were collected by mitotic shake-off and counted. Equal counts of heavy- and light-labeled HeLa cells were mixed, washed twice in PBS, and lysed under non-denaturing conditions with a mild detergent for immunoprecipitation.
SDS-PAGE and Western blotting
SDS-PAGE was performed using NOVEX (Life Technologies) precast 4–12% Bis-Tris gels. Samples were prepared by heating to 95C in 4x LDS sample buffer containing 5mM DTT (Life Technologies) for 5 minutes, followed by electrophoresis at 150V for 90 minutes in MOPS running buffer. Western blotting was performed using the BioRad transfer system to nitrocellulose in 20% methanol transfer buffer at 100V for 1 hour in ice. Membranes were blocked for 1 hour in 3% milk / TBS-T, followed by incubation with primary antibody overnight, washing, and secondary antibody for 1 hour. Antibodies to Plk1 and Aurora A were used at 1:2000, antibodies to 53BP1, Prc1, anti-Myc clone 9E10, Kif14, and phosphorylated Aurora A (Thr288) were used at 1:500, antibodies to Mklp2, Kif2c, Astrin and PP6R2 were used at 1:1000, and lamin A/C was used at 1:10,000. All secondary antibodies were used at 1:3000.
Immunoprecipitation
For Plk1 immunoprecipitation, antibody to Plk1 (Sigma Aldrich) was loaded on protein G-agarose beads (Roche) at 1mg/ml antibody to beads and cross-linked. HeLa cells were lysed in lysis buffer (50 mM Tris-HCl, pH7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM EDTA, 0.5% Triton X-100, 1 mM beta-glycerophosphate, 1mM sodium molybdate, 1 mM sodium fluoride, 1 mM sodium tartrate, and protease inhibitors). The lysate was clarified by centrifugation at 12,000 x g for 30 minutes at 4°C. The supernatant was transferred to a new tube and incubated with 15 micrograms antibody for 2 hours at 4°C while rotating. Anti-Myc and anti-Flag immunoprecipitations were carried out as described above with the exception that for anti-Myc immunoprecipitations, antibody clone 9E10 was cross-linked to protein G-agarose beads (Roche) with 10mM dimethylpimelimidate (Sigma) and used at 15 micrograms antibody per IP, and for anti-Flag immunoprecipitations 10 microliters M2-Flag-agarose suspension (Sigma) was used per IP. Each Myc-Plk1, Myc-Pincer, Flag-Plk1 and Flag-Pincer immunoprecipitation was performed in duplicate. Afterwards beads were washed three times with lysis buffer and 3 times with PBS, eluted with SDS-sample buffer at 70°C for 5 minutes, reduced with 5 mM DTT at 55°C for 15 min, cooled to room temperature, and alkylated with 15 mM iodoacetamide (Sigma-Aldrich) in the dark for one hour, and resolved by SDS-PAGE. Gel bands were excised, destained, trypsin-digested, and analyzed by LC-MS/MS (described below).
Mass spectrometry
Plk1 immunoprecipitations were analyzed by nanoscale microcapillary LC-MS/MS on a LTQ-Orbitrap (ThermoFisher Scientific) as described in (17). Anti-Myc and anti-Flag immunoprecipitations (described above) were analyzed on an Orbitrap Fusion (Thermo Electron) or Q Exactive Plus (ThermoFisher Scientific) equipped with an Easy-nLC 1000 (ThermoFisher Scientific) and nanospray source. Peptides were resuspended in 5% methanol / 1% formic acid and loaded on to a trap column [1 cm length, 100 μm inner diameter, ReproSil, C18 AQ 5 μm 120 Å pore (Dr. Maisch HPLC GMBH (Ammerbuch, Germany))], vented to waste via a micro-tee and eluted across a fritless analytical resolving column (35 cm length, 100 μm inner diameter, ReproSil, C18 AQ 3 μm 120 Å pore) pulled in-house (Sutter P-2000, Sutter Instruments, San Francisco, CA) with a 60 minute gradient of 5–30% LC-MS buffer B (LC-MS buffer A: 0.0625% formic acid, 3% ACN; LC-MS buffer B: 0.0625% formic acid, 95% ACN). The Q-Exactive Plus was set to perform an Orbitrap MS1 scan (R=70K; AGC target = 3e6) from 350 – 1500 Thomson, followed by high collision energy dissociation (HCD) MS2 spectra on the 10 most abundant precursor ions detected by Orbitrap scanning (R=17.5K; AGC target = 1e5; max ion time = 75 ms) before repeating the cycle. Precursor ions were isolated for HCD by quadrupole isolation at width = 0.8 Thomson and HCD fragmentation at 26 normalized collision energy (NCE). Charge state 2, 3 and 4 ions were selected for MS2. Precursor ions were added to a dynamic exclusion list +/− 20ppm for 20 seconds. The resulting data files were searched using Comet software (64) against a target-decoy (reversed) (65) version of the human proteome sequence database (UniProt; downloaded 2/2013) with a precursor mass tolerance of +/− 1 Da and requiring fully tryptic peptides with up to three miscleavages. Carbamidomethylcysteine was enabled as a fixed modification. Oxidized Met, isotopically heavy-labeled Arg and Lys were enabled as variable modifications. The resulting peptide spectral matches were filtered to < 1% false discovery rate (FDR) for peptides, based on reverse-hit counting (mass measurement accuracy cutoffs (MMA) within −/+ 2.5ppm, a delta-XCorr (dCn) of greater than 0.08, and appropriate XCorr values). Protein ratio quantification was performed using MassChroQ (66). Log2 H/L ratios were median-adjusted for mixing errors. Label-free protein quantification for Myc- and Flag-tagged Plk1 and “Pincer”-Plk1 interacting proteins was carried out by intensity-based absolute quantification (iBAQ) (67).
MS data analysis
All total peptide counts were input into the computational tool SAINT (36) using the Contaminant Repository for Affinity Purification (CRAPome) interface (35). For a protein to be considered for further analysis, we required the interaction to have an average probability in individual replicates (AvgP) score in the SAINT analysis of 0.9 or above in at least three of the six conditions. Furthermore, we required that proteins were identified in at least one of the replicates for all conditions, be highly enriched above the control immunoprecipitations (1 peptide or 20-fold increase in peptide count compared to sample) and have a mean peptide count of at least 2. Agglomerative hierarchical clustering was carried out using the Euclidian distance similarity metric and centroid linkage in Cluster 3.0 software (68). Significance of differences in binding upon kinase inhibitor treated was determined by an unpaired, two-tailed Student’s t-test. To control for multiple comparisons, Benjamini-Hochberg correction was employed. Cut-offs were chosen based on known Plk1-activity-dependent interactors. The output was visualized by Java Treeview 1.1.1 (69). Protein-protein interactions between identified Plk1 interacting proteins were determined using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database and analyzed in Cytoscape (70, 71). Edges represent protein-protein interactions based on the STRING database. GO annotations were performed in Cytoscape using the Biological Networks Gene Ontology (BiNGO) Java tool to test for ontology enrichment. To determine significance of enrichment of terms a Bonferroni corrected P-value cutoff of 0.05 was used.
Quantitative comparisons of protein abundance by Western blot or immunofluorescence microscopy were performed on biological replicates (n >= 3) using two-tailed Student’s T-test in Prism 5 (GraphPad, La Jolla, CA). Sequence alignment for PP6R2 was performed using the OrthoMCL-DB for chordates as described (50, 51).
In vitro kinase and phosphatase reactions
Aurora A-TPX2 and Plk1 were purified from Sf9 insect cells as previously described (17), including addition of okadaic acid (100nM) to Sf9 cells 2 hours prior to collection to maximize T-loop phosphorylation site occupancy. Cyclin B was amplified from HeLa cDNA and cloned into pFastBac1-GST (Gibco) vector. Cdk1 was cloned into empty pFastBac1 (Gibco). Constructs were sequenced confirmed, transformed into DH10Bac E. coli (Gibco), recombinant bacmids DNA was isolated and recombination was confirmed by PCR. Bacmids were transfected into Sf9 cells using Cellfectin (Gibco) according to the manufacturer’s instructions to generate the P1 virus stock. For insect cell protein expression, SF9 cells were infected with virus stocks and 72 hours after infection cells were harvested, lysed in GST lysis buffer containing: PBS, 0.5% Triton X-100, 1 mM EDTA, 0.5 mM DTT, and protease inhibitors. Lysates was clarified at 8000xg at 4°C for 30 min, Glutathione-Sepharose was added, incubated for 1 hr with rotation at 4°C. Sepharose was washed, eluted with reduced glutathione, and dialyzed overnight against 50 mM Hepes pH 7.5, 10 mM MgCl2, 5 mM MnCl2, 1 mM DTT, and 10% glycerol.
For bacterial cell protein expression, full length SAPS2 was subcloned into pET16better vector containing a N-terminal 10-His tag using traditional cloning methods. Plasmid was transformed into E. coli (BL21-DE3-Rosettas) and grown to an OD600 ~ 1.0 and induced with 0.5mM IPTG for 18 hours at 18°C. Cells were centrifuged at 8,000 x g at 4°C for 10 minutes and stored at −80C. For purification, cells were resuspended buffer A (50 mM Tris-HCl pH 7.5, 300 mM NaCl, 5% glycerol, 5 mM β-mercaptoethanol, and a protease inhibitor tablet) and sonicated. Lysates were then cleared at 20,000 x g at 4°C for 20 minutes and passed through a .22um filter. Purification was then performed by passing lysate through nickel-NTA column and eluted with 100% buffer B (Buffer A with the addition of 500 mM imidazole). Coupling to CNBr-Sepharose was performed per manufacturer’s protocol.
PP6R2 was amplified from HeLa cDNA and cloned into p3xFlag-CMV10. PP6c was amplified from HeLa cDNA and cloned into pcDNA3.1. PP6R2 and PP6c were co-transfected into 293T cells and purified using M2-Flag agarose (Sigma). For in vitro kinase and phosphatase assays, 10 ng of PP6R2-PP6c was incubated with either 5 ng of Plk1, 5ng Cdk1-cyclin B, or both in 20 mM Hepes pH 7.5, 10 mM MgCl2, 5 mM MnCl2, 1 mM DTT, 0.1 mg/ml BSA, and 200 μM ATP for 30 min at 30˚C. 100 ng of AuroraA-TPX2 was incubated with 1 μM staurosporine in 20 mM Hepes pH 7.5, 10 mM MgCl2, 5 mM MnCl22, 1 mM DTT, and 0.1 mg/ml BSA for 30 min at 30˚C. After30 min, 100 ng of AuroraA-TPX2 were added to the respective PP6R2-PP6c condition and further incubated for 30 min at 30˚C. Reactions were quenched by the addition of 2X SDS-PAGE sample buffer and analyzed by Western blotting (described below) for total and pThr288 Aurora A.
Supplementary Material
Figure S1: SDS-PAGE gels of Plk1 and control immunoprecipitations.
Figure S2: Comparison of effects of flavopiridol and staurosporine on Plk1 interactions.
Figure S3: Gene ontology analyses.
Figure S4: Protein complexes in the Plk1 interactome.
Figure S5: First-degree neighbors of CDK1-targeted Plk1 interactors.
Figure S6: Regulatory interaction of PP6 and Plk1.
Figure S7: Conservation of candidate PBD-binding motifs in PP6R2.
Figure S8: Termination of the Plk1-PP6 interaction.
Figure S9: Dependence of PP1c on Plk1 activity in dephosphorylating the Aurora A-TPX2 complex.
Table S1: SILAC-based identification of kinase inhibitor-responsive Plk1 protein-protein interactions.
Table S2: Number of SP/TP and SSP/STP motifs in Plk1 interactors.
Table S3: Wild-type and “Pincer” mutant Plk1 protein-protein interactions.
Table S4: SILAC-based identification of Plk1 inhibitor-responsive sites on PP6R2.
Table S5: In vitro confirmation of Cdk1-cyclin B phosphorylation occupancy of PP6R2.
Acknowledgments
The authors would like to thank members of the Kettenbach and Gerber labs for helpful discussions. We thank D. Compton (Dartmouth Medical School) and F. McKeon (Harvard Medical School) for gifting us antibodies.
Funding: Research reported here was funded by NIH/NIGMS (R35GM119455, P20GM113132) and The V Foundation for Cancer Research (V2016-022) to ANK and by NIH/NCI/NIGMS (R01CA155260, R01GM122846) to SAG. The Orbitrap Fusion Tribrid mass spectrometer was acquired with support from the National Institutes of Health (S10-OD016212).
Footnotes
Competing interests: The authors declare no conflict of interest.
Data and materials availability: The raw proteomics data have been deposited to the PRIDE/ProteomeXchange database (http://www.proteomexchange.org/), accession identifier PXD009168. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
Author Contributions: ANK and SAG designed the study and wrote the paper. ANK performed all of the experiments not otherwise specified, IN generated SAPS2-Sepharose, SPL cloned, expressed and purified bacterial SAPS2, and KAS generated Myc-SAPS and Myc-ANR constructs and cell lines and SAPS2-PBD phosphorylation site mutants. ANK, SAG and MEA analyzed the data. JG supported the statistical data analysis. MEA supported the proteomics pipeline, generated software, and conducted data analysis.
References and Notes
- 1.Barr FA, Elliott PR, Gruneberg U. Protein phosphatases and the regulation of mitosis. J Cell Sci. 2011 Jul 15;124:2323. doi: 10.1242/jcs.087106. [DOI] [PubMed] [Google Scholar]
- 2.Wurzenberger C, Gerlich DW. Phosphatases: providing safe passage through mitotic exit. Nature reviews Molecular cell biology. 2011 Aug;12:469. doi: 10.1038/nrm3149. [DOI] [PubMed] [Google Scholar]
- 3.Bruinsma W, Raaijmakers JA, Medema RH. Switching Polo-like kinase-1 on and off in time and space. Trends in biochemical sciences. 2012 Dec;37:534. doi: 10.1016/j.tibs.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Domingo-Sananes MR, Kapuy O, Hunt T, Novak B. Switches and latches: a biochemical tug-of-war between the kinases and phosphatases that control mitosis. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2011 Dec 27;366:3584. doi: 10.1098/rstb.2011.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medema RH, Lindqvist A. Boosting and suppressing mitotic phosphorylation. Trends Biochem Sci. 2011 Nov;36:578. doi: 10.1016/j.tibs.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Ma HT, Poon RY. How protein kinases co-ordinate mitosis in animal cells. Biochem J. 2011 Apr 1;435:17. doi: 10.1042/BJ20100284. [DOI] [PubMed] [Google Scholar]
- 7.Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nature Reviews Molecular Cell Biology. 2004;5:429. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 8.Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nature Reviews Molecular Cell Biology. 2009;10:265. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- 9.Petronczki M, Lenart P, Peters JM. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Developmental Cell. 2008;14:646. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 10.Hood EA, Kettenbach AN, Gerber SA, Compton DA. Plk1 regulates the kinesin-13 protein Kif2b to promote faithful chromosome segregation. Mol Biol Cell. 2012 Jun;23:2264. doi: 10.1091/mbc.E11-12-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petronczki M, Glotzer M, Kraut N, Peters JM. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev Cell. 2007 May;12:713. doi: 10.1016/j.devcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Jang YJ, Ma S, Terada Y, Erikson RL. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J Biol Chem. 2002 Nov 15;277:44115. doi: 10.1074/jbc.M202172200. [DOI] [PubMed] [Google Scholar]
- 13.Macurek L, et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008 Sep 4;455:119. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- 14.Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008 Jun 20;320:1655. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eyers PA, Erikson E, Chen LG, Maller JL. A novel mechanism for activation of the protein kinase Aurora A. Curr Biol. 2003 Apr 15;13:691. doi: 10.1016/s0960-9822(03)00166-0. [DOI] [PubMed] [Google Scholar]
- 16.Scutt PJ, et al. Discovery and exploitation of inhibitor-resistant aurora and polo kinase mutants for the analysis of mitotic networks. J Biol Chem. 2009 Jun 5;284:15880. doi: 10.1074/jbc.M109.005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kettenbach AN, et al. Quantitative phosphoproteomics identifies substrates and functional modules of aurora and polo-like kinase activities in mitotic cells. Sci Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elia AE, et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003 Oct 3;115:83. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 19.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003 Feb 21;299:1228. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 20.Elia AEH, et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003 Oct 3;115:83. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 21.Kang YH, et al. Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Molecular cell. 2006 Nov 3;24:409. doi: 10.1016/j.molcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 22.Neef R, et al. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J Cell Biol. 2003 Sep 1;162:863. doi: 10.1083/jcb.200306009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neef R, Klein UR, Kopajtich R, Barr FA. Cooperation between mitotic kinesins controls the late stages of cytokinesis. Curr Biol. 2006 Feb 7;16:301. doi: 10.1016/j.cub.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 24.Pinder A, et al. JIP4 is a PLK1 binding protein that regulates p38MAPK activity in G2 phase. Cell Signal. 2015 Nov;27:2296. doi: 10.1016/j.cellsig.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Neef R, et al. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol. 2007 Apr;9:436. doi: 10.1038/ncb1557. [DOI] [PubMed] [Google Scholar]
- 26.Steegmaier M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007 Feb 20;17:316. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro GI. Preclinical and clinical development of the cyclin-dependent kinase inhibitor flavopiridol. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004 Jun 15;10:4270s. doi: 10.1158/1078-0432.CCR-040020. [DOI] [PubMed] [Google Scholar]
- 28.Manfredi MG, et al. Antitumor activity of MLN8054, an orally active small-molecule inhibitor of Aurora A kinase. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4106. doi: 10.1073/pnas.0608798104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson RW, et al. AZD1152, a selective inhibitor of Aurora B kinase, inhibits human tumor xenograft growth by inducing apoptosis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007 Jun 15;13:3682. doi: 10.1158/1078-0432.CCR-06-2979. [DOI] [PubMed] [Google Scholar]
- 30.Yang J, et al. AZD1152, a novel and selective aurora B kinase inhibitor, induces growth arrest, apoptosis, and sensitization for tubulin depolymerizing agent or topoisomerase II inhibitor in human acute leukemia cells in vitro and in vivo. Blood. 2007 Sep 15;110:2034. doi: 10.1182/blood-2007-02-073700. [DOI] [PubMed] [Google Scholar]
- 31.Omura S, et al. A new alkaloid AM-2282 OF Streptomyces origin. Taxonomy, fermentation, isolation and preliminary characterization. J Antibiot (Tokyo) 1977 Apr;30:275. doi: 10.7164/antibiotics.30.275. [DOI] [PubMed] [Google Scholar]
- 32.Tamaoki T, et al. Staurosporine, a potent inhibitor of phospholipid/Ca++dependent protein kinase. Biochem Biophys Res Commun. 1986 Mar 13;135:397. doi: 10.1016/0006-291x(86)90008-2. [DOI] [PubMed] [Google Scholar]
- 33.Nakano H, et al. Staurosporine inhibits tyrosine-specific protein kinase activity of Rous sarcoma virus transforming protein p60. J Antibiot (Tokyo) 1987 May;40:706. doi: 10.7164/antibiotics.40.706. [DOI] [PubMed] [Google Scholar]
- 34.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular & Cellular Proteomics. 2002;1:376. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 35.Mellacheruvu D, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013 Aug;10:730. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi H, et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods. 2011 Jan;8:70. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011 Nov;29:1039. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan EH, Santamaria A, Sillje HH, Nigg EA. Plk1 regulates mitotic Aurora A function through betaTrCP-dependent degradation of hBora. Chromosoma. 2008 Oct;117:457. doi: 10.1007/s00412-008-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan J, et al. Astrin regulates meiotic spindle organization, spindle pole tethering and cell cycle progression in mouse oocytes. Cell Cycle. 2009 Oct 15;8:3384. doi: 10.4161/cc.8.20.9885. [DOI] [PubMed] [Google Scholar]
- 40.van Vugt MA, et al. A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G(2)/M DNA damage checkpoint. PLoS Biol. 2010 Jan;8:e1000287. doi: 10.1371/journal.pbio.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camon E, et al. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004 Jan 1;32:D262. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris MA, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004 Jan 1;32:D258. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camon E, Barrell D, Lee V, Dimmer E, Apweiler R. The Gene Ontology Annotation (GOA) Database--an integrated resource of GO annotations to the UniProt Knowledgebase. In silico biology. 2004;4:5. [PubMed] [Google Scholar]
- 44.Barrell D, et al. The GOA database in 2009--an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009 Jan;37:D396. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lowery DM, et al. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. Embo J. 2007 May 2;26:2262. doi: 10.1038/sj.emboj.7601683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guergnon J, Derewenda U, Edelson JR, Brautigan DL. Mapping of protein phosphatase-6 association with its SAPS domain regulatory subunit using a model of helical repeats. BMC Biochem. 2009;10:24. doi: 10.1186/1471-2091-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stefansson B, Ohama T, Daugherty AE, Brautigan DL. Protein phosphatase 6 regulatory subunits composed of ankyrin repeat domains. Biochemistry. 2008 Feb 5;47:1442. doi: 10.1021/bi7022877. [DOI] [PubMed] [Google Scholar]
- 48.Hosing AS, Valerie NC, Dziegielewski J, Brautigan DL, Larner JM. PP6 regulatory subunit R1 is bidentate anchor for targeting protein phosphatase-6 to DNA-dependent protein kinase. The Journal of biological chemistry. 2012 Mar 16;287:9230. doi: 10.1074/jbc.M111.333708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Couzens AL, et al. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Science signaling. 2013 Nov 19;6:rs15. doi: 10.1126/scisignal.2004712. [DOI] [PubMed] [Google Scholar]
- 50.Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic acids research. 2006;34:D363. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome research. 2003;13:2178. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindon C, Pines J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J Cell Biol. 2004 Jan 19;164:233. doi: 10.1083/jcb.200309035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walter AO, Peng ZY, Cartwright CA. The Shp-2 tyrosine phosphatase activates the Src tyrosine kinase by a non-enzymatic mechanism. Oncogene. 1999 Mar 18;18:1911. doi: 10.1038/sj.onc.1202513. [DOI] [PubMed] [Google Scholar]
- 54.Satinover DL, Leach CA, Stukenberg PT, Brautigan DL. Activation of Aurora-A kinase by protein phosphatase inhibitor-2, a bifunctional signaling protein. Proc Natl Acad Sci U S A. 2004 Jun 8;101:8625. doi: 10.1073/pnas.0402966101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng K, Bastos RN, Barr FA, Gruneberg U. Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2. J Cell Biol. 2010 Dec 27;191:1315. doi: 10.1083/jcb.201008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rusin SF, Schlosser KA, Adamo ME, Kettenbach AN. Quantitative phosphoproteomics reveals new roles for the protein phosphatase PP6 in mitotic cells. Science signaling. 2015 Oct 13;8:rs12. doi: 10.1126/scisignal.aab3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kettenbach AN, et al. Rapid determination of multiple linear kinase substrate motifs by mass spectrometry. Chem Biol. 2012 May 25;19:608. doi: 10.1016/j.chembiol.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grosstessner-Hain K, et al. Quantitative phospho-proteomics to investigate the polo-like kinase 1-dependent phospho-proteome. Mol Cell Proteomics. 2011 Nov;10:M111 008540. doi: 10.1074/mcp.M111.008540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santamaria A, et al. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteomics. 2011 Jan;10:M110 004457. doi: 10.1074/mcp.M110.004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oppermann FS, et al. Combination of chemical genetics and phosphoproteomics for kinase signaling analysis enables confident identification of cellular downstream targets. Mol Cell Proteomics. 2012 Apr;11:O111 012351. doi: 10.1074/mcp.O111.012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Preisinger C, et al. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. Embo J. 2005 Feb 23;24:753. doi: 10.1038/sj.emboj.7600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sengupta D, Linstedt AD. Mitotic inhibition of GRASP65 organelle tethering involves Polo-like kinase 1 (PLK1) phosphorylation proximate to an internal PDZ ligand. J Biol Chem. 2010 Dec 17;285:39994. doi: 10.1074/jbc.M110.189449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Horn RD, et al. Cdk1 activity is required for mitotic activation of aurora A during G2/M transition of human cells. The Journal of biological chemistry. 2010 Jul 09;285:21849. doi: 10.1074/jbc.M110.141010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013 Jan;13:22. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 65.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007 Mar;4:207. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 66.Valot B, Langella O, Nano E, Zivy M. MassChroQ: a versatile tool for mass spectrometry quantification. Proteomics. 2011 Sep;11:3572. doi: 10.1002/pmic.201100120. [DOI] [PubMed] [Google Scholar]
- 67.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011 May 19;473:337. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 68.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004 Jun 12;20:1453. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 69.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004 Nov 22;20:3246. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 70.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003 Nov;13:2498. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saito R, et al. A travel guide to Cytoscape plugins. Nat Methods. 2012 Nov;9:1069. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: SDS-PAGE gels of Plk1 and control immunoprecipitations.
Figure S2: Comparison of effects of flavopiridol and staurosporine on Plk1 interactions.
Figure S3: Gene ontology analyses.
Figure S4: Protein complexes in the Plk1 interactome.
Figure S5: First-degree neighbors of CDK1-targeted Plk1 interactors.
Figure S6: Regulatory interaction of PP6 and Plk1.
Figure S7: Conservation of candidate PBD-binding motifs in PP6R2.
Figure S8: Termination of the Plk1-PP6 interaction.
Figure S9: Dependence of PP1c on Plk1 activity in dephosphorylating the Aurora A-TPX2 complex.
Table S1: SILAC-based identification of kinase inhibitor-responsive Plk1 protein-protein interactions.
Table S2: Number of SP/TP and SSP/STP motifs in Plk1 interactors.
Table S3: Wild-type and “Pincer” mutant Plk1 protein-protein interactions.
Table S4: SILAC-based identification of Plk1 inhibitor-responsive sites on PP6R2.
Table S5: In vitro confirmation of Cdk1-cyclin B phosphorylation occupancy of PP6R2.