Abstract
Eukaryotic centromeres are defined by the presence of nucleosomes containing the histone H3 variant, centromere protein A (CENP-A). Once incorporated at centromeres, CENP-A nucleosomes are remarkably stable, exhibiting no detectable loss or exchange over many cell cycles. It is currently unclear whether this stability is an intrinsic property of CENP-A containing chromatin or whether it arises from proteins that specifically associate with CENP-A chromatin. Two proteins, CENP-C and CENP-N, are known to bind CENP-A human nucleosomes directly. Here we test the hypothesis that CENP-C or CENP-N stabilize CENP-A nucleosomes in vitro and in living cells. We show that CENP-N stabilizes CENP-A nucleosomes alone and additively with CENP-C in vitro. However, removal of CENP-C and CENP-N from cells, or mutating CENP-A so that it no longer interacts with CENP-C or CENP-N, had no effect on centromeric CENP-A stability in vivo. Thus, the stability of CENP-A nucleosomes in chromatin does not arise solely from its interactions with CENP-C or CENP-N.
INTRODUCTION
During mitosis, vertebrate cells assemble one kinetochore on each chromosome to connect chromosomes to spindle microtubules, monitor chromosome alignment on the spindle, and move chromosomes to poles during anaphase. The assembly site for the kinetochore is the centromere, a specialized chromatin domain that is epigenetically specified by the replacement of histone H3 in nucleosomes with the centromere-specific histone variant centromere protein A (CENP-A) (McKinley and Cheeseman, 2016). Unlike histones H3.1 and H3.2, CENP-A nucleosome assembly is uncoupled from replication and occurs only after mitotic exit in G1 (Jansen et al., 2007). To maintain centromere identity, CENP-A nucleosomes must remain at centromeres outside of the assembly period (Hoffmann et al., 2016). This occurs because CENP-A nucleosomes are stable once incorporated into chromatin, showing loss only by dilution through replication (Bodor et al., 2013). CENP-A appears to be stably maintained at centromeres over days in dividing cells and for months or even years in postmitotic cells (Smoak et al., 2016).
Currently, it is unclear whether the stability of CENP-A nucleosomes in chromatin is an intrinsic property of CENP-A containing nucleosomes or results from other factors that bind to and stabilize CENP-A nucleosomes. On a single nucleosome level, purified or reconstituted CENP-A nucleosomes are more prone to disassembly than H3 nucleosomes when challenged with the destabilizing effects of heat, heparin, or nucleosome assembly protein 1 (NAP-1) (Conde e Silva et al., 2007; Arimura et al., 2014). In dividing cells, CENP-A at centromeres exhibit slower turnover rates than most H3 and are as stable as the most stable H3 (Bodor et al., 2013). While much of H3 turnover is mediate by active processes like chromatin remodeling and transcription (Henikoff, 2008), it is surprising that these processes do not also act on CENP-A given that there is transcription at active centromeres (McNulty et al., 2017). Therefore, we hypothesize that factors that distinguish between CENP-A and H3 nucleosomes can help stabilize CENP-A nucleosomes either through direct binding or exclusion of these active processes.
Two essential proteins, CENP-C and CENP-N, have been found to preferentially interact with CENP-A nucleosomes at the centromere. Previous studies have suggested that CENP-C stabilizes CENP-A nucleosomes (Falk et al., 2015, 2016). Another study showed that replacing the loop1 and α2-helix region on H3 with the CENP-A targeting domain (CATD) region of CENP-A, the region that binds CENP-N (Carroll et al., 2009), is sufficient to confer stability to CENP-A nucleosomes in vivo (Bodor et al., 2013). In a recent report, it was proposed that centromeres are maintained by CENP-N through fastening of CENP-A to DNA (Guo et al., 2017). We tested the direct effects of CENP-C and CENP-N on CENP-A nucleosomes maintenance in vitro and in cells. Here we show that in vitro, CENP-N protects CENP-A mono-nucleosomes against the destabilizing effects of salt, dilution, or plunge-freezing on an EM-grid, and this stabilization is further increased by CENP-C. In contrast, rapid in vivo degradation of either CENP-C, CENP-N, or both, had no effect on CENP-A stability at centromeres within a single cell cycle. Similarly, mutating residues R80 and G81 in the context of CENP-A, or a chimera of H3 containing just the CENP-A CATD, had no effect on the maintenance of these proteins at centromeres. Our results indicate that although CENP-C and CENP-N provide added stability to the CENP-A nucleosome in vitro, this effect does not account for any additional stability of the CENP-A nucleosome in vivo.
RESULTS
CENP-A nucleosome DNA ends are less accessible to micrococcal nuclease when CENP-N interacts with the nucleosome
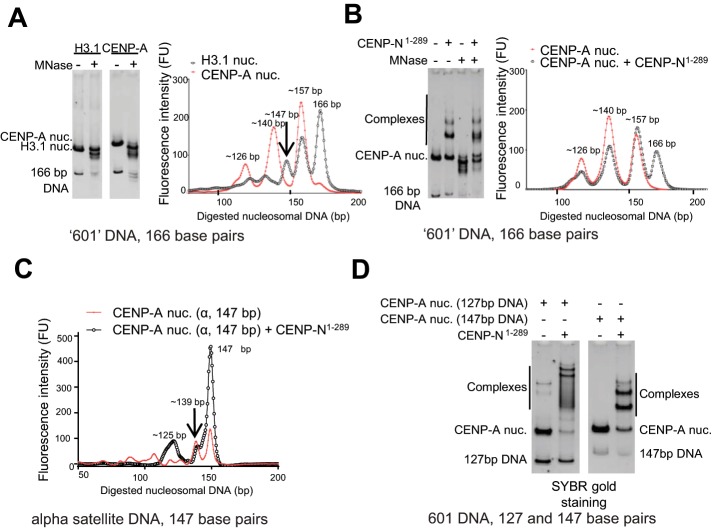
The CENP-A nucleosome coordinates fewer base pairs of DNA near the DNA entry and exit sites than does the histone H3 nucleosome (Yoda et al., 2000; Dechassa et al., 2011; Kingston et al., 2011; Tachiwana et al., 2011). This reduced wrapping of DNA around the histone core promotes the loss of H2A/H2B and is thought to be one key feature that causes or reflects the instability of centromeric nucleosomes (Voltz et al., 2012). Here we performed limited micrococcal nuclease digestion (MNase) with both CENP-A nucleosome and H3 nucleosomes to test whether this affects MNase accessibility. Nucleosomes were reconstituted with a 166–base pair DNA fragment derived from the 601 nucleosome positioning sequence identified by Widom and colleagues (Lowary and Widom, 1998) (sequence listed under Materials and Methods). While the majority of DNA in the H3 nucleosome sample remained unaffected by MNase (leaving an intact 166–base pair DNA fragment; Figure 1A), nearly all DNA in the CENP-A nucleosome was degraded to smaller fragments (157, 140, and 126 base pairs; Figure 1A). This indicates differences in the way in which DNA ends are organized by the histone core, with H3 conferring more stable wrapping than CENP-A and thus less access for MNase.
FIGURE 1:
CENP-N protects CENP-A nucleosomes from micrococcal nuclease digestion. (A) Comparative MNase digestion analysis of H3.1- and CENP-A nucleosomes, reconstituted with a 166–base pair DNA fragment derived from the 601 sequence. After digestion (quenched with 50 mM EDTA), samples were analyzed in a gel shift assay prior to DNA extraction (left panel) and after DNA extraction (right panel). The length of the DNA fragments was determined according to the standard DNA ladder curve obtained from the Bioanalyzer (Agilent). (B) MNase digestion analysis of CENP-A nucleosome (with the same DNA sequence as in A) in the absence and presence of CENP-N1-289. The reactions were performed and analyzed as in A. (C) MNase digestion analysis of CENP-A nucleosome (147–base pair DNA derived from α satellite DNA) in the absence and presence of CENP-N1-289. (D) The DNA ends of the CENP-A nucleosome are important for the proper orientation of CENP-N1-289 on the CENP-A nucleosome. CENP-A nucleosomes were reconstituted with either 147 or 127 base pairs of 601 nucleosome-positioning DNA. CENP-N1-289 was mixed with CENP-A nucleosome at a 3:1 ratio. After a 5-min incubation at 37°C, samples were analyzed by 5% native PAGE.
We then asked whether the nucleosome-binding domain (1-289) of CENP-N would confer increased stability on DNA ends when bound to CENP-A nucleosomes by performing MNase assays in presence of CENP-N. CENP-N1-289 elutes as a monomer from an S200 gel filtration column (Supplemental Figure S1A). Native PAGE suggests that more than one CENP-N1-289 can interact with a CENP-A nucleosome, as expected from the presence of two equivalent binding sites on the nucleosome (Supplemental Figure S1B). To determine the effect of CENP-N binding on DNA accessibility, we repeated the MNase treatment with a 1:2 complex of the CENP-A-nucleosome with CENP-N1-289. While the patterns of generated DNA fragments were the same for the CENP-A nucleosome in the presence or absence of CENP-N, more of the 166–base pair undigested DNA fragment was extracted from the sample with CENP-N present (Figure 1B). We repeated this with nucleosomes reconstituted with either a 147– or 207–base pair DNA fragment with the same 601 core sequence, both of which showed similar results (unpublished data). To exclude that this effect was a result of the rather unique properties of the 601 sequence, we repeated the experiment with the palindromic DNA derived from α-satellite DNA, which closely resembles the native DNA template for CENP-A at the centromere. Consistent with the results described above, CENP-N conferred increasing resistance toward MNase digestion in CENP-A nucleosomes reconstituted with α-satellite DNA (Figure 1C). Therefore, we conclude that the interaction between CENP-N and CENP-A nucleosome stabilizes the DNA ends in CENP-A nucleosomes and protects them from digestion by MNase. When we tested a CENP-A nucleosome reconstituted with a 127–base pair DNA fragment (where the penultimate 10 base pairs were removed from the 147–base pair 601 DNA fragment used in the other experiments), these CENP-A nucleosomes formed less-defined complexes with CENP-N as judged by native PAGE, where complexes formed a smear rather than two discrete shifted bands (Figure 1D). This implies that the DNA ends are essential for the formation of a well-defined complex between the CENP-A nucleosome and CENP-N.
CENP-N increases the stability of the CENP-A nucleosome in vitro
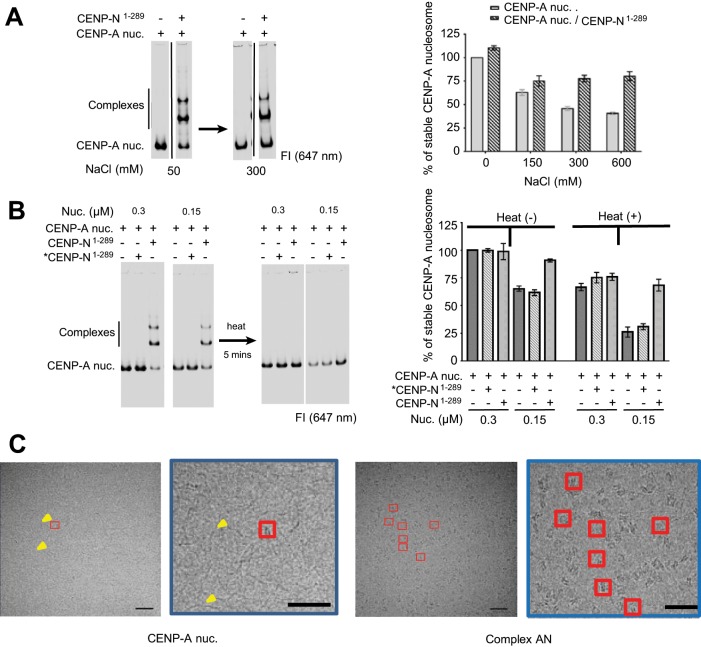
To further explore the effect of CENP-N on CENP-A nucleosome stability, we used an electrophoretic mobility shift assay (EMSA) to quantify the amount of stable nucleosome remaining under various destabilizing conditions. First, we incubated nucleosomes in the absence and presence of CENP-N1-289 at increasing ionic strength. CENP-A nucleosomes dissociated in increased ionic strength such that less than half of the input nucleosome was present at 300 and 600 mM salt. This dissociation was reduced in the presence of CENP-N1-289 (Figure 2A), such that 75% of the nucleosomes remained intact throughout the salt titration series (Figure 2A, left panel). CENP-N also stabilized CENP-A nucleosomes against dissociation in response to dilution and heat (Figure 2B). Heating CENP-N for 5 min at 55ºC was sufficient to denature CENP-N and prevented it from binding to the CENP-A nucleosome, and this served as a control in these experiments. In the presence of CENP-N, 75% of CENP-A nucleosomes remained intact after twofold dilution and heat treatment, while only 25–30% of nucleosomes remained intact in the absence of CENP-N or with CENP-N that had been denatured before addition to CENP-A nucleosomes. Finally, application of CENP-A nucleosomes onto a cryo-electron microscopy (cryoEM) grid (C-Flat gold) followed by plunge-freezing yielded very few (fewer than 10 per image) intact nucleosome particles, while the very same preparations, in the presence of a two- to threefold excess of CENP-N, yielded a large number of well-formed particles (more than 700 nucleosome-like particles per image; Figure 2C and Table 1). Negative-staining electron microscopy (EM) was also used to test the stability of the CENP-A nucleosome. Interestingly, the particle sizes varied drastically when no CENP-N was present in solution, while the presence of CENP-N dramatically increased not only the total number but also the quality of particles (Supplemental Figure S1C and Table 1). Together, these data indicate a direct role for CENP-N in CENP-A nucleosome stabilization in vitro. It should be noted that we and others have successfully prepared EM grids with H3 nucleosomes (Chua et al., 2016), and thus the increased fragility of CENP-A containing nucleosomes that is alleviated by CENP-N is due to CENP-A. A recent report showed that CENP-N fastens the nucleosome by simultaneously binding to the CATD on CENP-A and to nucleosomal DNA around CATD, which could stabilize the CENP-A nucleosome (Guo et al., 2017). This was recently confirmed by cryo-EM studies by us and others (Chittori et al., 2017; Pentakota et al., 2017). Our data provided direct evidence of the stabilizing effect of CENP-N on CENP-A nucleosome in vitro.
FIGURE 2:
CENP-N increases the stability of CENP-A nucleosomes against dissociation in vitro. (A) CENP-N1-289 increases the stability of CENP-A nucleosomes at increased ionic strength. CENP-A nucleosome (100 nM) was mixed with either buffer or 200 nM CENP-N1-289 at the indicated NaCl concentration. Nucleosome stability was quantified by measuring the intensity of all bands, including shifted bands (right panel; normalized as described under Materials and Methods), n = 3. All assays were performed in a final buffer containing 20 mM Tris HCl, pH 7.5, the indicated NaCl concentration, 5% glycerol, 0.5 mM EDTA (no detergent). The ∼25% of nucleosomes that are not protected in the presence of CENP-N likely reflects the amount of unbound CENP-A nucleosome (right panel). (B) CENP-N1-289 stabilizes CENP-A nucleosomes against the combined effects of dilution and heat treatment. Left panel: native PAGE. CENP-N1-289 was mixed with CENP-A nucleosomes at a molar ratio of 3:1. As a control, *CENP-N1-289 indicates CENP-N1-289 that was denatured by heating at 55°C for 5 min before mixing with CENP-A nucleosomes. The same amount of sample was loaded immediately after treatment. All bands including shifted bands were quantified to determine the percentage of remaining nucleosome, and error bars are derived from three independent gels (n = 3). Intensity at 647 nm was measured. (C) CENP-A mono-nucleosomes on an EM grid are greatly stabilized in the presence of CENP-N1-289. The CENP-A nucleosome sample (2.5 µM) was mixed with 7.5 µM CENP-N1-289, and the control was adjusted with buffer. Red boxes indicate nucleosome-shaped particles. Yellow arrows show free DNA. Scale bar = 50 nm. The blue box highlights the area from the left micrograph. The intact particles were counted, and the numbers are listed in Table 1.
TABLE 1:
Particle analysis from electron micrographs.
| Particle number (per image) | ||
|---|---|---|
| Sample | Cryo-EM grid | Negative stain |
| CENP-A nucleosome | <10 | 290 |
| Complex AN | 700 | 460 |
| Complex ANC | 800 | N/A |
Particles on a cryo-EM micrograph were picked and counted by using nucleosome-like two-dimensional classes as reference in Relion. Particles on images obtained from negative-stained grids were picked and counted by “dogPicker,” where nucleosomes were defined by a diameter of ∼10 nm. No negative staining EM grids were obtained for complex ANC.
CENP-N and CENP-C additively stabilize CENP-A nucleosomes in vitro
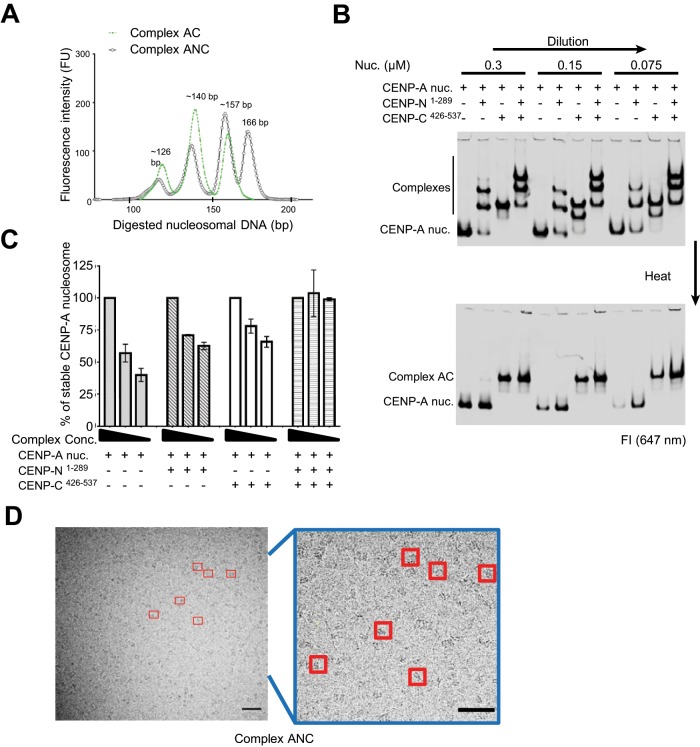
A second CENP-A nucleosome-specific binding protein, CENP-C, has also been shown to stabilize CENP-A nucleosomes and is thought to bind CENP-A nucleosomes through a binding site that differs from that of CENP-N (Carroll et al., 2010). We confirmed that the nucleosome-binding domains of CENP-C and CENP-N (CENP-N1-289 and CENP-C426-537) form distinct complexes when incubated with CENP-A nucleosomes individually (Supplemental Figure S2A). When CENP-N1-289 is added to a preformed complex of CENP-C426-537 with the CENP-A nucleosome (complex AC), we observe a distinct pattern by EMSA that is consistent with independent binding events. Using pull-down assays (Supplemental Figure S2B), we show that indeed CENP-N and CENP-C bind to the same nucleosome (complex ANC), in agreement with previous reports (Carroll et al., 2010; Guo et al., 2017). To test whether simultaneous binding of CENP-C and CENP-N provides added stability to nucleosomes, we repeated the MNase protection experiment with the complex AC and with complex ANC. The complex AC exhibited the same sensitivity toward MNase digestion as CENP-A nucleosomes alone, while the complex ANC exhibited the same level of protection from MNase as the AN complex (Figure 3A). We conclude that CENP-C does not contribute to the protection of DNA ends in CENP-A nucleosomes. Thus, the coordinated binding of CENP-N and CENP-C to CENP-A nucleosomes could further increase the overall stability of the nucleosome. Using EMSA and cryoEM, we found that CENP-C426-537 and CENP-N1-289 showed similar stabilizing effects on CENP-A nucleosomes, but the combination of CENP-C426-537 and CENP-N1-289 increased the stability of the CENP-A nucleosome more than CENP-C or CENP-N alone (Figure 3, B–D; Table 1). Thus, CENP-N and CENP-C independently protect CENP-A nucleosomes against dissociation in vitro.
FIGURE 3:
The CENP-A nucleosome-binding domains of CENP-N and CENP-C have additive effects on stabilizing CENP-A nucleosomes in vitro. (A) MNase digestion analysis for complex AC and complex ANC. CENP-C426-537 was mixed with CENP-A nucleosome at a 2:1 ratio to form the complex AC. CENP-N1-289 was mixed with preformed AC complex at 2:1 ratio to form the ANC complex. DNA fragments after digestion were analyzed as in Figure 1A. (B) CENP-N1-289 and CENP-C426-537 showed additive effects in stabilizing the CENP-A nucleosome against dilution and heat. CENP-N1-289 or CENP-C426-537 was mixed with CENP-A nucleosome at molar ratios of 3:1. The buffer was adjusted to a final 20 mM Tris HCl, pH 7.5, 150 mM NaCl, 2% glycerol, 0.5 mM EDTA. Dilution and heat treatments were done as described in under Materials and Methods. Samples were kept at 4°C for 2 h before analysis by native PAGE. (C) Quantification of the percentage of stable nucleosomes under different nucleosome concentrations without heat (from the upper panel in B, n = 2. The raw signal (intensity of fluorescence signal at 647 nm) of nucleosome or complex after dilution (150 and 75 nM) was normalized to the signal from samples before dilution (at 300 mM NaCl). (D) The stabilizing effect of CENP-N and CENP-C on CENP-A nucleosome, as demonstrated by cryo-EM. Both CENP-N1-289 and CENP-C426-537 were mixed with CENP-A nucleosome at molar ratio 3:1 to form the ANC complex. Buffer was adjusted to the same condition for both samples. Concentration of both samples was 2.5 µM. Red boxes indicate nucleosome-shaped particles. Scale bar = 50 nm. Blue box highlights the area from the left micrograph. The intact particles were counted, and the numbers are listed in Table 1.
Loss of CENP-C and CENP-N does not alter CENP-A nucleosome levels in chromatin
We tested whether the stability of CENP-A nucleosomes in vivo results from the nucleosome-stabilizing effects of CENP-N and/or CENP-C observed in vitro. To this end, we generated cells expressing conditionally degradable CENP-C and/or CENP-N by fusing the auxin-inducible degron (AID) tag to the C-terminus of the endogenous CENP-C and CENP-N genes in cells expressing the F-box protein, Tir1 (Nishimura et al., 2009; Holland et al., 2012; McKinley et al., 2015). We also tagged CENP-N with superfolder GFP (sfGFP) and CENP-C with mRuby2 and a 3xFLAG epitope. We validated that we had modified both alleles of the endogenous genes by PCR or Western blotting (Supplemental Figure S3, C and D). On addition of 1 mM indole-3-acetic acid (IAA), either CENP-N alone or CENP-N and C were degraded to background levels within 30 min (Supplemental Figure S3).
We validated the specificity of CENP-N degradation by stably introducing mRuby2-3xFLAG-tagged full-length CENP-N, mRuby2-3xFLAG-tagged CENP-N truncations, or the mRuby2-3xFLAG tag alone as transgenes into our AID tagged CENP-N cells. The mRuby2-3xFLAG tag expressed alone was not stable in cells (Supplemental Figure S4A). However, when fused to full-length or truncated CENP-N, the fusion proteins were stable and expressed at similar levels (Supplemental Figure S4A). On IAA addition to degrade the endogenously tagged CENP-N, cells expressing only the tag or N-terminal truncations of CENP-N showed signs of chromosome segregation defects on CENP-N loss, indicated by the appearance of micronuclei, as previously described (McKinley et al., 2015). No significant increase in micronuclei was detected in cells expressing the full-length CENP-N, indicating that these defects result specifically from CENP-N degradation and that the full-length CENP-N transgene complements the IAA-induced loss of CENP-N (Supplemental Figure S4, B and C).
We tested whether the loss of CENP-C, CENP-N, or both had an impact on the levels of CENP-A at centromeres by degrading the proteins and measuring the levels of centromere associated CENP-A using an anti-CENP-A antibody. To avoid assaying secondary effects from chromosome segregation defects, we degraded the proteins during drug-induced cell-cycle arrests. In all cases, cells were treated with 0.5 mM thymidine to induce arrest at the G1–S boundary. Cells were then released from this arrest and allowed to go through mitosis before the addition of 1 mM IAA. Cells were cultured in the presence of IAA during G1 ending in a second thymidine arrest at the G1–S boundary (Figure 4A); through a release from the second thymidine arrest into a roscovitine arrest, roughly corresponding to G1–S–G2; or starting from the release from the second thymidine arrest into a roscovitine arrest, roughly corresponding to early S-phase through G2 (Figure 4B). Despite near complete degradation of these two centromere proteins (Supplemental Figure S5, A–C), we observed no significant differences in total CENP-A levels as assayed by immunofluorescence (Figure 4, A and B). To distinguish between the maintenance of CENP-A in chromatin and the new assembly of CENP-A-nucleosomes, we made a stable cell line that constitutively overexpressed SNAP-tagged CENP-A at ∼8× the endogenous level. We then used fluorescent-pulse labeling of the SNAP-tag with TMR-Star to measure the levels of preexisting CENP-A in chromatin, newly assembled CENP-A, or total SNAP-tagged CENP-A (Figure 4C). We observed a significant decrease in centromeric TMR-Star signal when we labeled the total SNAP-tagged CENP-A or the newly assembled SNAP-tagged CENP-A but, in contrast to published observations (Guo et al., 2017), we saw no difference in the SNAP-tagged CENP-A previously incorporated into the centromere. We were unable to reproduce a loss of previously incorporated CENP-A in either our cell lines or the published cell lines using the published methods (Supplemental Figure S6 and Discussion). Our data demonstrating that CENP-A assembly depends on CENP-N is consistent with our previous observations (Carroll et al., 2009) but also demonstrates that the loss of CENP-N has no effect on the maintenance of preexisting CENP-A at centromeres.
FIGURE 4:
CENP-C and CENP-N degradation has no significant effect on centromeric CENP-A maintenance. (A, B) Cell lines containing AID tagged CENP-N, CENP-C, or both were treated with IAA to degrade the indicated proteins. Blue bars represent cells not treated with IAA. Cells were maintained in IAA beginning just after mitosis (red bars) and harvested in an early S-phase thymidine (thym) arrest (A) or after mitosis (mustard bars) or in early S phase (green bars) and harvested in a G2-phase roscovitine (rosco) arrest (B). Centromeric CENP-A immunofluorescence signal was normalized to the no-IAA signal. (C) Degradation of CENP-N inhibits new CENP-A assembly but not preexisting CENP-A in chromatin. The CENP-N AID-sfGFP cell line containing a stably integrated SNAP-tagged CENP-A was either fluorescently labeled or quenched according to the schematic (left panel). Green bars represent the time of synthesis of the fluorescent population of SNAP-tagged CENP-A. IAA was added as in A. Centromeric TMR-Star intensity represents the fluorescent population of SNAP-tagged CENP-A. Data are presented as mean ± SEM for three independent replicates. *p < 0.05.
Loss of CENP-C and/or CENP-N does not alter the salt extraction of CENP-A from centromeric chromatin
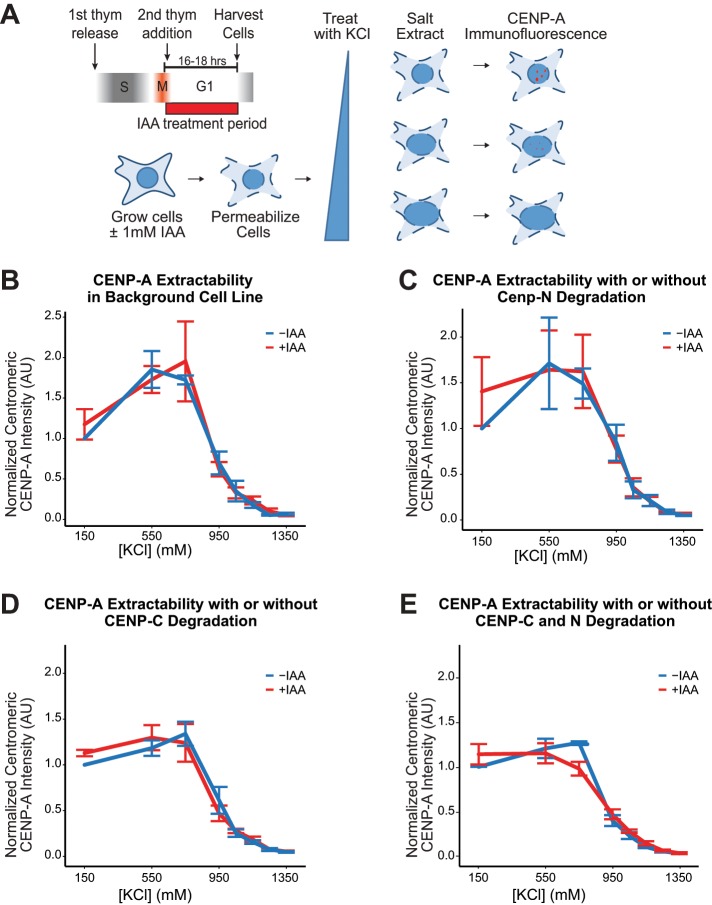
Although we saw no change in CENP-A levels in chromatin after degradation of CENP-C and/or CENP-N, this does not directly assay nucleosome stability in chromatin. We therefore measured the ease with which centromeric CENP-A could be extracted with salt in the presence or absence of CENP-C and/or CENP-N. The difference in CENP-A nucleosome stability that we see in vitro predicts that we would extract CENP-A from chromatin at lower salt concentrations in the absence of CENP-C and/or CENP-N. We permeabilized cells and treated them with increasing concentrations of KCl as previously described (Moree et al., 2011), removed the salt, and localized CENP-A using immunofluorescence (Figure 5A). IAA treatment did not have a significant effect on CENP-A salt extraction in cells with no AID tagged proteins (Figure 5B). Degrading either CENP-C or CENP-N for 16–18 h during G1 also resulted in no difference in salt extraction of CENP-A (Figure 5, C and D). This lack of difference was not due to loss of CENP-C or CENP-N during the salt extraction because both proteins remained localized at centromeres at all tested KCl concentration without IAA treatment, independent of CENP-A levels (Supplemental Figure S5, D–F). When both CENP-C and CENP-N were degraded, we saw a slight effect on CENP-A extractability at 600 mM KCl (p value = 0.076) but no significant difference at higher or lower salt concentrations (Figure 5E). Our data suggest that the presence of CENP-N and CENP-C at centromeres does not have a strong stabilizing effect on CENP-A nucleosomes in vivo.
FIGURE 5:
CENP-C or CENP-N alone does not affect the salt-extractability of centromeric CENP-A. (A) Schematic of experimental workflow. Cells were treated with different concentrations of KCl to extract CENP-A from chromatin. Cells were then fixed and stained for CENP-A before imaging. (B) IAA alone has no effect on CENP-A extractability. CENP-A extraction with increasing salt concentration in the parental OsTIR1 expressing cell line with no AID tagged proteins. (C–E) Salt extractability curves for three different cell lines where CENP-C, CENP-N, or both were tagged with AID. Cells were left untreated or treated with IAA to degrade AID-CENP-N (C), AID-CENP-C (D), or both (E). Centromeric CENP-A immunofluorescence signal was normalized to the no-IAA, 150 mM KCl signal. Data are presented as mean ± SEM for three independent replicates.
Mutation of the CENP-A L1 loop to reduce CENP-N affinity does not affect CENP-A or CATD chimera maintenance at centromeres
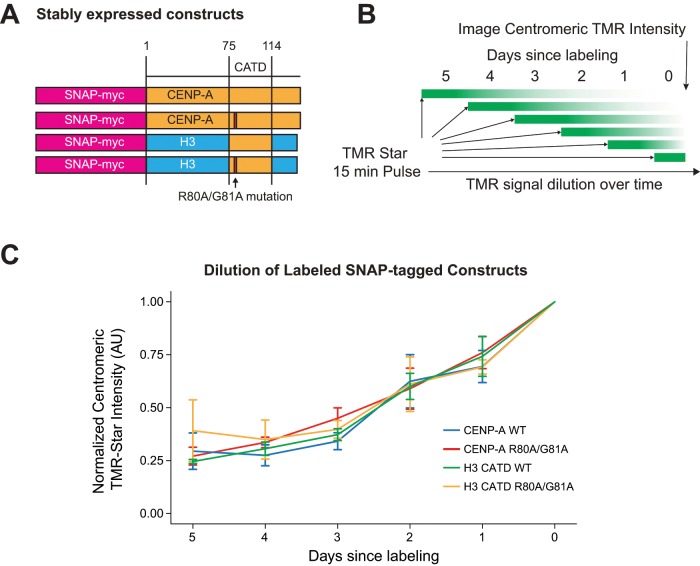
CENP-C and CENP-N are both essential proteins, making it impossible to test whether long-term loss of these factors alters CENP-A nucleosome stability in dividing cells. To circumvent this issue, we engineered stable cell lines that constitutively express four different mutated forms of CENP-A to 4–8 × the endogenous level: wild-type CENP-A, a chimeric CENP-A with the CENP-A CATD but the corresponding N and C-terminus from H3.1, or these two species where residues R80 and G81 on CENP-A were mutated to alanine (Figure 6A). The CATD chimera should engage neither CENP-B nor CENP-C (Carroll et al., 2010; Guse et al., 2011; Fachinetti et al., 2013, 2015; Fujita et al., 2015), and the CENP-A-R80A,G81A mutation reduces CENP-N’s affinity for CENP-A nucleosomes (Fang et al., 2015; Pentakota et al., 2017). These mutations make it possible to compare the long-term maintenance of these four CENP-A mutants as a readout for CENP-A stability when CENP-N and CENP-C binding are perturbed.
FIGURE 6:
R80A G81A mutation does not affect CENP-A or CATD chimera maintenance at centromeres. (A) Schematic of constructs stably integrated into cells for this experiment. Mustard-colored bars represent CENP-A sequences. Blue-colored bars represent H3.1 sequences. (B) Schematic of labeling scheme used for C. Cells were seeded on coverslips at the same density 5 d before harvest. Cells were labeled with TMR-Star for 15 min on different days so that the fluorescent population of CENP-A is diluted to different degrees when cells are harvested at day 0. (C) Centromeric TMR-Star intensity was determined by microscopy. Signals across the different cell lines were normalized to the intensity at 0 d since labeling. Intensities were not background subtracted because of variable nuclear background. There are no significant differences between the four curves. Data are presented as mean ± SEM for three independent replicates.
We measured the CENP-A transgene levels at centromeres by fluorescently labeling cells with a short pulse of SNAP-Cell TMR-Star each day over the course of five days (Figure 6B). The kinetics of loss were equivalent between all four species until day 5 after pulse labeling when we could no longer reliably detect a centromeric signal (Figure 6C). Our results recapitulated published data showing that the CATD chimera was maintained similarly to normal CENP-A at centromeres (Bodor et al., 2013). Our findings are consistent with our conclusion that CENP-C and CENP-N binding to CENP-A nucleosomes does not have a strong stabilizing effect on CENP-A in nuclear chromatin.
DISCUSSION
CENP-A nucleosomes were previously shown to be more persistent in chromatin than most H3 nucleosomes (Bodor et al., 2013). However, what precisely causes stabilization of these nucleosomes at the molecular level is unknown. We hypothesized that direct interactors of the CENP-A nucleosome, CENP-C and CENP-N, could stabilize the CENP-A nucleosome resulting in more stable retention. To test this hypothesis, we examined the functions of CENP-C and CENP-N in stabilizing CENP-A mono-nucleosomes in vitro and CENP-A chromatin at centromeres.
We observed that CENP-N (additively with CENP-C) stabilizes the penultimate 10 base pairs of DNA in a single CENP-A nucleosome, thereby protecting it from disassembly caused by dilution, increased ionic strength, or adsorption onto various EM grids. A recent report pointed out that CENP-N could stabilize the CENP-A nucleosome by fastening CENP-A and DNA (Guo et al., 2017). This was shown very recently by cryo-EM studies of a CENP-A nucleosome in complex with CENP-N (Chittori et al., 2017; Pentakota et al., 2017). While no direct interaction with DNA ends was observed in these structures, CENP-N has a large (15–base pair) DNA interaction interface near the CENP-A L1 loop, with which it also interacts extensively. Possibly, breathing of DNA ends is prevented by the contacts between CENP-N and nucleosomal DNA further into the particle. Our in vitro data are consistent with the structures in that CENP-A mononucleosomes form much more stable particles in the presence of CENP-C and/or CENP-N.
In contrast, in cells, we do not observe a significant change in centromeric CENP-A levels in the absence of CENP-C, CENP-N, or both. Our results are inconsistent with a recently published report (Guo et al., 2017) that shows a loss of CENP-A in chromatin when CENP-N is removed. In attempts to resolve this difference, we obtained the CENP-N-AID-GFP cell line with SNAP-tagged CENP-A from Guo et al. (2017). We directly compared their cell line with the ones we used in this study and replicated their experimental protocol. Despite this, we found no significant difference between the IAA-treated and control cells in any of the cell lines (Supplemental Figure S6E). Notably, when the data were analyzed as in Guo et al. (2017), the cell line obtained used in that study exhibited the largest difference, a 23% ± 20% decrease on treatment with IAA-treated, when both centromeric TMR-Star signal (Supplemental Figure S6E, top) and changes in cell number (Supplemental Figure S6E, middle) were factored in. However, this observed difference (p value = 0.3485) might be largely due to nonspecific, IAA-dependent changes in cell number (Supplemental Figure S6E, middle) as a cell line expressing no AID-tagged proteins showed a 16 ± 19% decrease when treated with IAA, when changes in cell number were considered. Without cell number normalization, this value would be 3 ± 8%. Though there are modest differences in SNAP-CENP-A and TIR1 expression levels between the cell lines (Supplemental Figure S6B), these differences cannot account for the lack of destabilization of CENP-A nucleosomes in our hands. Thus, while we observe a stabilization of the CENP-A nucleosome by CENP-C and CENP-N in vitro, removing CENP-C and CENP-N in vivo does not result in significant destabilization of CENP-A in chromatin. We found it surprising that we did not observe any loss of retention of CENP-A nucleosomes without CENP-C and/or CENP-N in cells given the stabilizing effect of these proteins in vitro. We propose three possible explanations for why the loss of CENP-C and CENP-N has relatively little effect on CENP-A nucleosome stability in vivo, despite the demonstrated stabilizing effect on the CENP-A nucleosome by CENP-C and CENP-N in vitro. One possibility is that CENP-A nucleosomes interact with other factors at the centromere that promote their stability. For example, CENP-B interacts with the N-terminal tail of CENP-A and also binds to DNA and thus may compensate for the loss of CENP-C or CENP-N. Currently only CENP-N, CENP-C, and CENP-B have been identified as centromere-specific CENP-A nucleosome binding proteins. It was recently shown that M18BP1/KNL2 can also bind directly to CENP-A nucleosomes in frogs (French et al., 2017), birds (Hori et al., 2017), and plants (Sandmann et al., 2017), suggesting that other factors may also exist in human cells that bind and stabilize CENP-A nucleosomes.
A second interpretation of our findings is that despite the requirement for CENP-N and CENP-C in centromere assembly, the interaction with CENP-A may be less critical once the CCAN has properly assembled. We found that overexpression of full-length Ruby-tagged CENP-N displaced GFP-tagged CENP-N expressed at endogenous levels, while the overexpression of truncated versions of CENP-N that lacked the CENP-L interaction domain did not displace GFP-tagged CENP-N, despite being able to localize at centromeres (Supplemental Figure S4, D and E). The fact that their localization does not displace endogenous CENP-N suggests two possible models: 1) that the number of CENP-A binding sites exceeds the number of full-length CENP-N molecules at the centromere or 2) that once CENP-N localizes to the centromere by interacting with CENP-A, it no longer requires CENP-A interaction to remain at the centromere and instead depends on its interactions with other members of the CCAN. This second model is supported by our cell-based salt-wash assays in which CENP-A levels decreased with increasing salt yet leaving CENP-C and CENP-N localization to the centromere unaffected (Supplemental Figure S5, D–F). A recent study demonstrated that complete removal of CENP-A from the centromere did not result in a significant decrease in CENP-N that was already bound at the centromere (Hoffmann et al., 2016). A portion of centromeric CENP-C is also resistant to either degradation or replicative dilution of CENP-A (Fachinetti et al., 2013; Hoffmann et al., 2016). If the second model were correct, then we cannot assume that CENP-C or CENP-N constitutively bind CENP-A directly, and in the absence of such an interaction we would expect no difference in CENP-A stability without CENP-C or CENP-N.
Third, the environment surrounding CENP-A nucleosomes differ substantially in vitro and in cells. In vitro, we observed changes in the stability of CENP-A nucleosomes in response to CENP-N binding at intermediate salt concentrations. While these measurements represent real differences in the stability of the CENP-A nucleosome, this stability change may not translate to the cellular environment where there is substantial crowding and regulated levels of monovalent and divalent cations. In vitro, a substantial stability difference with and without CENP-N was observed only above 150 mM NaCl. Similarly, in the dilution assay, the presence of CENP-N did not result in additional stabilization at 300 nM nucleosome concentration, while there was a reproducible threefold effect at 150 nM. While it is difficult to assess the local concentration of CENP-A in the cell, we observed that CENP-A appears as a diffraction limited spot in microscopy. Using the estimated number of 200 CENP-A nucleosomes per centromere (Bodor et al., 2014), and assuming that the centromere occupies a sphere with a 300-nm diffraction limited radius, then the concentration of CENP-A nucleosomes would be ∼3 µM, and thus beyond the “critical concentration.” The actual concentration of all nucleosomes if we include H3 nucleosomes would be even higher.
Since we did not observe a change in the retention of CENP-A nucleosomes in cells, the nature of this retention remains elusive. Rather than relying on CCAN interactions, CENP-A nucleosomes could be locked in place by interaction with other nearby nucleosomes brought into proximity by the three-dimensional organization of the chromatin fiber. Several other factors such as posttranslational modifications, RNA interactions, or yet-to-be-discovered CENP-A interacting proteins might also contribute to CENP-A nucleosome stability. It may be that all nucleosomes meet the basic level of stability to demonstrate this retention in cells but CENP-A nucleosomes may be protected from destabilizing processes that displace nucleosomes from chromatin. Several studies demonstrate that a fraction of H3 nucleosomes persist on chromatin in cycling cells (Kimura and Cook, 2001; Radman-Livaja et al., 2011).
Also, while we focused on rapidly dividing cells, Smoak et al. (2016) recently showed that there is long-term retention of CENP-A at centromeres in mouse oocytes for greater than a year with no detectable assembly. However, this may not be unique, as H3 is one of the longest-lived proteins in postmitotic tissues (Commerford et al., 1982; Waterborg, 1993; Toyama et al., 2013). Unfortunately, experiments in cultured cells can only access much shorter timescales, and thus the stabilization of CENP-A by CENP-C and CENP-N that we see in vitro may be relevant when observed over the lifetime of an oocyte or egg.
MATERIALS AND METHODS
Protein purification
Cenp-N1-289 with a (His)6 tag at the C-terminus was constructed in plasmid pACEbac1 and expressed in Sf21 insect cells. Infected cells were collected and suspended in lysis buffer (50 mM sodium phosphate buffer, pH 7.5, 300 mM NaCl, 10% glycerol). Cells were sonicated in presence of benzonase nuclease (2 µl per 40 ml). Nickel beads, equilibrated with lysis buffer, were added, and the mixture was incubated overnight at 4°C. Beads were washed and protein was eluted with a 40 to 500 mM imidazole buffer in lysis buffer. Size exclusion chromatography over a GE S200 column was performed in 20 mM Tris HCl, pH 7.5, 300 mM NaCl, 10% glycerol, 1 mM dithiothreitol (DTT), and 0.5 mM EDTA. CENP-A-H4 was coexpressed from a bicistronic plasmid in Escherichia coli and purified as described (Guse et al., 2012).
Nucleosome reconstitution
DNA, (CENP-A - H4)2 tetramer, and H2A-H2B were mixed at a 1:1.1:2.2 M ratio in buffer containing 20 mM Tris HCl, pH 7.5, 2 M NaCl, 1 mM DTT, and 1 mM EDTA. Reconstitutions were performed either by stepwise dilution or by dialysis against TEN buffer (20 mM Tris HCl, pH 7.5, 1 mM EDTA, 10 mM NaCl, and 1 mM DTT) as described (Muthurajan et al., 2016). Some preparations of nucleosome were also reconstituted with atto 647N dye labeled H2B (T112C) (D’Arcy et al., 2013).
The following DNA templates were used in this study:
Widom 601 sequence DNA (147 base pairs; Lowary and Widom, 1998):
ATCTGAGAATCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCGAT
Widom 601 sequence DNA (127 base pairs): CCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATA
Widom 601 sequence DNA (166 base pairs):
ATCCCTATACGCGGCCGCCCTGGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCGAT
DNA derived from α satellite DNA (147 base pairs):
ATCAATATCCACCTGCAGATACTACCAAAAGTGTATTTGGAAACTGCTCCATCAAAAGGCATGTTCAGCTGGATT
CCAGCTGAACATGCCTTTTGATGGAGCAGTTTCCAAATACACTTTTGGTAGTATCTGCAGGTGGATATTGAT
Binding assay
Nucleosomes or DNA were mixed with the indicated amount of Cenp-N1-289 or CENP-C426-537 in buffer (20 mM Tris HCl, pH 7.5, 1 mM EDTA, 50–150 mM NaCl, and 1 mM DTT) for 30 min at room temperature and then analyzed by 5% native PAGE. The gel image was captured at 488 nm (for SYBR Gold staining) or 647 nm (for labeled 18–base pair DNA) using a Typhoon imager.
Micrococcal nuclease digestion
Nucleosome (166–base pair DNA, 300 nM, 20 µl) was incubated with 1U/100 ng MNase in 20 mM Tris HCl, pH 7.5, 1 mM EDTA, 50 mM NaCl, and 1 mM DTT at 37°C for 5 min. Reactions were quenched by addition of 50 mM EDTA. Native PAGE was used to analyze the digestion without DNA extraction. In parallel, DNA from the treated sample was extracted with the Minelute DNA extraction kit, followed by quantification on a bioanalyzer (Agilent). The length of DNA fragment was calculated from the standard DNA ladder. For CENP-A nucleosome containing 147–base pair α satellite DNA, 2 U/100 ng MNase was added into the reaction. The reaction was quenched after 10 min’ incubation at 37°C under same buffer condition above.
Nucleosome stability test assay (gel based)
Assembled nucleosomes were incubated in buffer (20 mM Tris HCl, pH 7.5, 1 mM EDTA, and 1 mM DTT) containing the indicated concentration of NaCl. Alternatively, nucleosome samples were diluted with buffer at room temperature or 55°C. Nucleosome stability was defined by dividing the intensity of the remaining nucleosome intensity by that of the input nucleosome on native PAGE. All raw signals were detected using a Typhoon at 647 nm (H2B in nucleosomes was labeled on residue T122C with atto 647N).
Cryo-EM grid preparation and microscopy for testing stability of nucleosome
Fresh preparations of nucleosomes were divided into two equal aliquots. One was mixed with a two- to threefold excess of either CENP-N1-289 or CENP-C426-537/CENP-N1-289, while the other was mixed with the same amount of buffer to adjust buffer components and sample concentration. Of each sample (final concentration of 2.5 µM), 4 µl was loaded on to C-Flat(Au) grid and blotted for 4 s in a FEI Vitrobot before plunge freezing in liquid ethane. Samples were imaged at magnification 62K× by a FEI F30 electron microscope with a Gatan K2 Summit direct detector device. MotionCor2 (Zheng et al., 2017) was used to align 45 frames for each image. Relion2.1 (Scheres, 2016) was used to pick the nucleosome-like particles. Particles were extracted and quantified in Relion.
Negative grid preparation and microscopy for testing nucleosome stability
Samples was prepared as above. Final concentration was ∼30 nM. Of each sample, 4 µl was loaded onto holey carbon grids for 30 s. Grids were rinsed twice with water. Uranyl acetate (1%) was used to fix and stain the particle. Images were acquired on a FEI F20 microscope equipped with a charge-coupled device (CCD) detector. Particles were picked and quantified by using dogpick in Appion (Lander et al., 2009). Nucleosome particles were defined by the diameter around 10–16 nm.
Cell culture
All cell lines were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin/0.1 mg/ml streptomycin, 2 g/l sodium bicarbonate, and 2 µg/ml puromycin. Degradation during G1 was achieved by treating with 0.5 mM thymidine for 16–18 h, release into 17 µM deoxycytidine for 3 h, normal media for 6 h, followed by a second 0.5 mM thymidine arrest and the addition of 1 mM IAA. Degradation during G1 and S phases were done by adding 1 mM IAA during an additional 3 h release into 17 µM deoxycytidine followed by 20–22 h in 0.1 mM roscovitine. All drugs were added directly to the media. Expression of transgenes were induced using 1 µg/ml doxycycline.
Generation of cell lines
CRISPR-Cas9 based genome engineering was used to tag CENP-N with AID-sfGFP at the C-terminus (guide RNA target sequence: GTGCATGTGCAATATCAAGA, 500–base pair homology arms flanking the stop codon were used in a pUC18 backbone donor construct) in osTir1 Flp-In TRex-DLD1 cells (a gift from the Don W. Cleveland Lab, University of California, San Diego [UCSD]). CENP-C was tagged with AID-mRuby2-3xFlag at the C-terminus (guide RNA target sequence: GAATGAGTAGACATATTAATC, 1292– and 1251–base pair homology arms before and after the stop codon, respectively, in a pUC18 backbone donor construct) in the CENP-N-AID-sfGFP background to generate double AID cell lines. Cells were cotransfected with 0.5 μg of plasmid expressing Cas9-GFP (pX458-Addgene 48138) (for CENP-C tagging) or Cas9-mCherry (modified pX458 with GFP replaced with mCherry, a gift from the Rajat Rohatgi lab, Stanford University) (for CENP-N tagging) and guide RNA targeting sequence and 0.5 µg of homology arm donor constructs using Promega Fugene HD in a six-well plate well. Cells were sorted using a Sony SH800Z Cell sorter for Cas9 expression 2 d posttransfection. Sorted cells were outgrown to confluency and then sorted for single cells that expressed the endogenous fusion tags and counter selected for Cas9 expression. CENP-C-AID-YFP cell lines were a gift from Dani Fachinetti and Don Cleveland (UCSD). CENP-N rescue constructs and SNAP-CENP-A were induced using FRT/Flp-mediated recombination of pcDNA5/FRT/TO- and pEF5/FRT-based vectors, respectively, and selected with 100 µg/ml Hygromycin B. CENP-A R80A/G81A mutants were generated with the following oligonucleotides: ATTTACCGCTGCCGTGGACTTCAACTGG, AGTCCACGGCAGCGGTAAATTTCACGCAGATTTC. Full plasmid sequences are available from the Stanford Digital Repository (https://purl.stanford.edu/gz478gq3828). Cloning was done using DH5alpha strains of E. coli bacteria. Cells were tested for mycoplasma contamination at the start of cell line generation by PCR and conditions were monitored by the absence of cytoplasmic DNA staining by microscopy.
CENP-A labeling with TMR-Star
Cell labeling or blocking was done by incubating cells with 2 µM SNAP-Cell TMR-Star (New England BioLabs) or 6.7 µM SNAP-Cell Block (New England Biolabs) diluted with media for 15 min, washed with phosphate-buffered saline (PBS) + 1 mM MgCl2 and CaCl2, incubated in media for 1 h, washed with PBS + 1 mM MgCl2 and CaCl2, and put back in media until ready to harvest.
Immunofluorescence and microscopy
For all but the salt extraction experiments, cells were grown on glass coverslips and washed with PBS + 1 mM MgCl2 and CaCl2, permeabilized with PBKCl (139.7 mM KCl, 11.8 mM KH2PO4) with 0.5% Triton X-100 for 5 min and fixed in PBKCl/0.5% Triton X-100/3.7% formaldehyde for 10 min, and blocked in antibody dilution buffer (20 mM Tris HCl, pH 7.4, 150 mM NaCl, 0.1% Triton X-100, 2% bovine serum albumin, and 0.1% sodium azide). For salt extraction experiments, cells were permeabilized for 10 min followed by treatment with salt solutions (80 mM K-PIPES, pH 6.8, 1 mM MgCl2, and 1 mM EGTA, 30% glycerol, and 0.5% Triton X-100) with additional KCl added as indicated for 15 min. Fixation was done following staining. Proteins were detected with the following primary antibodies:
| α-Centromere derived from human CREST patient serum (Antibodies, Inc., 15-234-0001) | 1:100 |
| Custom rabbit α-CENP-A | 2 µg/ml |
| α-Flag M2 (Sigma, F3165) | 2 µg/ml for CENP-C-mRuby2-3xFlag |
| 5 µg/ml for CENP-N-mRuby2-3xFlag truncations | |
| Llama nanobody α-GFP directly conjugated with Alexa Fluor 488 | 1 µg/ml for salt-wash experiments |
Primary antibodies were detected by either 568 or 647 directly conjugated goat secondary antibodies (Molecular Probes). DNA was stained with 10 µg/ml Hoechst 33258. Those not stained with the nanobody were imaged using endogenous GFP or YFP fluorescence. Images were acquired on a DeltaVision Core deconvolution microscope equipped with a CoolSnap HQ CCD camera (Photometrics). Z-sections were acquired at 0.2 µm steps using a 60× 1.4 NA objective. Image analysis was done using a centromere finder as in Moree et al. (2011) (http://cjfuller.github.io/imageanalysistools/). Average background signal was subtracted from centromeric signal. All quantification of microscopy experiments covered three independent experiments with at least 30 cells per condition per experiment.
Immunoblotting
Cells were harvested using 0.25% Trypsin-EDTA and quenched with media. Cells were spun down and washed with PBS + 1 mM MgCl2 and CaCl2 and washed twice with PBKCl + 0.5% Triton X-100 with 1 mM PMSF and 1 mM Benzamidine HCl, spinning down at 10,000 × g for 5 min at 4°C between washes. Cells were resuspended in 100 µl denaturing protein lysis buffer (20 mM Tris HCl, pH 7.4, 15 mM NaCl, 10 mM EDTA, 0.5% Igepal CA630, 0.1% Triton X-100, 0.1% SDS, 0.1% NaDeoxycholate) and sonicated before being separated by SDS–PAGE and transferred onto polyvinylidene fluoride membrane (Bio-Rad Laboratories). Samples were transferred in CAPS transfer buffer (10 mM 3-[cyclohexylamino]-1-propanesulfonic acid, pH 11.3, 0.1% SDS, and 20% methanol) for 3 h at 4°C. Rabbit anti-GFP antibodies (custom), rabbit anti-CENP-C antibodies (custom), and mouse anti-Flag M2 antibodies (F1804; Sigma-Aldrich) were used at 5 µg/ml. Mouse anti-RNA PolII (ab5408; Abcam) was used at 0.25 µg/ml. These were detected using donkey anti-rabbit conjugated to IRDye800 used at 1:10,000 (LiCor) or goat anti-mouse directly conjugated to Alexa Fluor 647 (Invitrogen).
Supplementary Material
Acknowledgments
We thank the Luger and Straight Lab members for helpful discussions and comments on this study. We thank Dani Fachinetti and Don Cleveland for the Tir1 DLD1 and CENP-C AID YFP cell lines. We thank Ben Black and Lucie Guo for exchange of cell lines and methods toward understanding the differences in our studies. This work was supported by Stanford Bio-X and a Stanford interdisciplinary graduate fellowship. Research reported in this publication was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health under award numbers T32GM007276 and GM074728 to A.F.S. and by GM067777 to K.L., who is also supported by the Howard Hughes Medical Institute. Some of this work was performed at the National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the National Institutes of Health NIGMS (GM103310) and the Simons Foundation (349247).
Abbreviations used:
- AID
auxin-inducible degron
- CATD
CENP-A targeting domain
- CENP-A
centromere protein A
- CENP-C
centromere protein C
- CENP-N
centromere protein N
- EM
electron microscopy
- EMSA
electrophoretic mobility shift assay
- IAA
indole-3-acetic acid
- MNase
micrococcal nuclease
- TMR
tetramethylrhodamine.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-10-0596) on January 17, 2018.
REFERENCES
- Arimura Y, Shirayama K, Horikoshi N, Fujita R, Taguchi H, Kagawa W, Fukagawa T, Almouzni G, Kurumizaka H. (2014). Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. Sci Rep , 7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE. (2014). The quantitative architecture of centromeric chromatin. Elife , e02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor DL, Valente LP, Mata JF, Black BE, Jansen LET. (2013). Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol Biol Cell , 923–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF. (2010). Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol , 1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Silva MCC, Godek KM, Jansen LET, Straight AF. (2009). Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol , 896–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittori S, Hong J, Saunders H, Feng H, Ghirlando R, Kelly AE, Bai Y, Subramaniam S. (2017). Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science , 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua EYD, Vogirala VK, Inian O, Wong ASW, Nordenskiöld L, Plitzko JM, Danev R, Sandin S. (2016). 3.9 Å structure of the nucleosome core particle determined by phase-plate cryo-EM. Nucleic Acids Res , 8013–8019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commerford SL, Carsten AL, Cronkite EP. (1982). Histone turnover within nonproliferating cells. Proc Natl Acad Sci USA , 1163–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde e Silva N, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A. (2007). CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J Mol Biol , 555–573 [DOI] [PubMed] [Google Scholar]
- D’Arcy S, Martin K, Panchenko T, Chen X, Bergeron S, Stargell L, Black B, Luger K. (2013). Chaperone Nap1 shields histone surfaces used in a nucleosome and can put H2A-H2B in an unconventional tetrameric form. Mol Cell , 662–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa ML, Wyns K, Li M, Hall MA, Wang MD. (2011). Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun , 310–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LET, Cleveland DW. (2013). A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol , 1056–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW. (2015). DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function. Dev Cell , 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LET, Van Duyne GD, Vinogradov SA, et al. (2015). CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science , 699–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk SJ, Lee J, Sekulic N, Sennett MA, Lee T-H, Black BE. (2016). CENP-C directs a structural transition of CENP-A nucleosomes mainly through sliding of DNA gyres. Nat Struct Mol Biol , 204–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Liu Y, Wei Y, Deng W, Yu Z, Huang L, Teng Y, Yao T, You Q, Ruan H, et al. (2015). Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N. Genes Dev , 1058–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French BT, Westhorpe FG, Limouse C, Straight AF. (2017). Xenopus laevis M18BP1 directly binds existing CENP-A nucleosomes to promote centromeric chromatin assembly. Dev Cell , 190–199.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita R, Otake K, Arimura Y, Horikoshi N, Miya Y, Shiga T, Osakabe A, Tachiwana H, Ohzeki J, Larinov V, et al. (2015). Stable complex formation of CENP-B with the CENP-A nucleosome. Nucleic Acids Res , 4909–4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LY, Allu PK, Zandarashvili L, McKinley KL, Sekulic N, Dawicki-McKenna JM, Fachinetti D, Logsdon GA, Jamiolkowski RM, Cleveland DW, et al. (2017). Centromere maintenance is achieved by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome structural transition. Nat Commun , 15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. (2011). In vitro centromere and kinetochore assembly on defined chromatin templates. Nature , 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A, Fuller CJ, Straight AF. (2012). A cell-free system for functional centromere and kinetochore assembly. Nat Protoc , 1847–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. (2008). Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet , 15–26 [DOI] [PubMed] [Google Scholar]
- Hoffmann S, Dumont M, Barra V, Ly P, Nechemia-Arbely Y, McMahon MA, Hervé S, Cleveland DW, Fachinetti D. (2016). CENP-A is dispensable for mitotic centromere function after initial centromere/kinetochore assembly. Cell Rep , 2394–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Han JS, Cleveland DW. (2012). Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc Natl Acad Sci USA , E3350–E3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Shang W-H, Hara M, Ariyoshi M, Arimura Y, Fujita R, Kurumizaka H, Fukagawa T. (2017). Association of M18BP1/KNL2 with CENP-A nucleosome is essential for centromere formation in non-mammalian vertebrates. Dev Cell , 181–189.e3 [DOI] [PubMed] [Google Scholar]
- Jansen LET, Black BE, Foltz DR, Cleveland DW. (2007). Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol , 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Cook PR. (2001). Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2b. J Cell Biol , 1341–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston IJ, Yung JSY, Singleton MR. (2011). Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem , 4021–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. (2009). Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol , 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary P, Widom J. (1998). New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol , 19–42 [DOI] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM. (2016). The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol , 16–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Sekulic N, Guo LY, Tsinman T, Black BE, Cheeseman IM. (2015). The CENP-L-N complex forms a critical node in an integrated meshwork of interactions at the centromere-kinetochore interface. Mol Cell , 886–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty SM, Sullivan LL, Sullivan BA. (2017). Human centromeres produce chromosome-specific and array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Dev Cell , 226–240.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. (2011). CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol , 855–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthurajan U, Mattiroli F, Bergeron S, Zhou K, Gu Y, Chakravarthy S, Dyer P, Irving T, Luger K. (2016). In vitro chromatin assembly: strategies and quality control. Methods Enzymol , 3–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. (2009). An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods , 917–923 [DOI] [PubMed] [Google Scholar]
- Pentakota S, Zhou K, Smith C, Maffini S, Petrovic A, Morgan GP, Weir JR, Vetter IR, Musacchio A, Luger K. (2017). Decoding the centromeric nucleosome through CENP-N. Elife , e33442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M, Verzijlbergen KF, Weiner A, van Welsem T, Friedman N, Rando OJ, van Leeuwen F. (2011). Patterns and mechanisms of Ancestral Histone protein inheritance in Budding yeast. PLoS Biol , e1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann M, Talbert P, Demidov D, Kuhlmann M, Rutten T, Conrad U, Lermontova I. (2017). Targeting of A. thaliana KNL2 to centromeres depends on the conserved CENPC-k motif in its C-terminus. Plant Cell , 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. (2016). Processing of structurally heterogeneous Cryo-EM data in RELION. Methods Enzymol , 125–157 [DOI] [PubMed] [Google Scholar]
- Smoak EM, Stein P, Schultz RM, Lampson MA, Black BE. (2016). Long-term retention of CENP-A nucleosomes in mammalian oocytes underpins transgenerational inheritance of centromere identity. Curr Biol , 1110–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park S, et al. (2011). Crystal structure of the human centromeric nucleosome containing CENP-A. Nature , 232–235 [DOI] [PubMed] [Google Scholar]
- Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Iii JRY, Hetzer MW. (2013). Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell , 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voltz K, Trylska J, Calimet N, Smith JC, Langowski J. (2012). Unwrapping of nucleosomal DNA ends: a multiscale molecular dynamics study. Biophys J , 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterborg JH. (1993). Histone synthesis and turnover in alfalfa. Fast loss of highly acetylated replacement histone variant H3.2. J Biol Chem , 4912–4917 [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K. (2000). Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. 2–7. [DOI] [PMC free article] [PubMed]
- Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods , 331–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.