Significance
Bacteria and other organisms make extensive use of signaling molecules that are derived from ribonucleotides or their derivatives. Previously, five riboswitch classes had been discovered that sense the four RNA-derived signaling molecules: c-di-GMP, c-di-AMP, c-AMP-GMP, and ZTP. We now report the discovery and biochemical validation of bacterial riboswitches for the widespread alarmone guanosine tetraphosphate (ppGpp), which signals metabolic and physiological adaptations to starvation. These findings expand the number of natural partnerships between riboswitches and ribonucleotide-like signaling molecules, and provide RNA-based sensors for detecting ppGpp production in cells.
Keywords: branched chain amino acid, guanosine tetraphosphate, logic gate, stringent response, ykkC motif RNA
Abstract
Riboswitches are noncoding portions of certain mRNAs that bind metabolite, coenzyme, signaling molecule, or inorganic ion ligands and regulate gene expression. Most known riboswitches sense derivatives of RNA monomers. This bias in ligand chemical composition is consistent with the hypothesis that widespread riboswitch classes first emerged during the RNA World, which is proposed to have existed before proteins were present. Here we report the discovery and biochemical validation of a natural riboswitch class that selectively binds guanosine tetraphosphate (ppGpp), a widespread signaling molecule and bacterial “alarmone” derived from the ribonucleotide GTP. Riboswitches for ppGpp are predicted to regulate genes involved in branched-chain amino acid biosynthesis and transport, as well as other gene classes that previously had not been implicated to be part of its signaling network. This newfound riboswitch–alarmone partnership supports the hypothesis that prominent RNA World signaling pathways have been retained by modern cells to control key biological processes.
Among the 38 distinct riboswitch (1–3) classes validated previously (4), five respond to RNA-derived signaling molecules that might have emerged in an RNA World, but have been retained to trigger regulatory processes in modern cells. We have proposed (5, 6) that riboswitches for the RNA-derived molecules c-di-GMP (7, 8), c-di-AMP (9), ZTP (10), and c-AMP-GMP (11, 12) represent modern versions of primordial signaling processes built entirely from RNA.
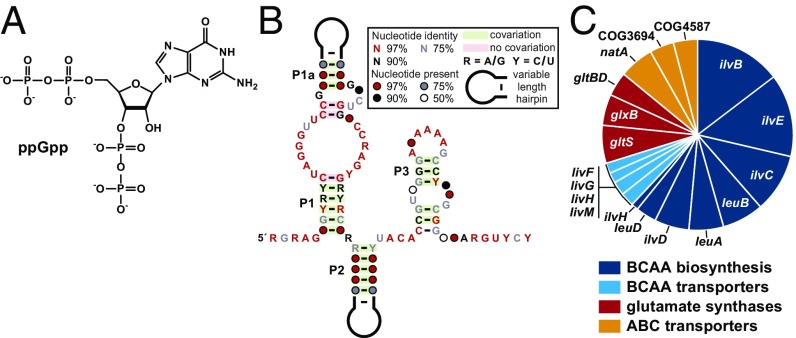
A widely distributed RNA signaling molecule that is essential for many bacteria is ppGpp (Fig. 1A), a guanosine tetraphosphate alarmone originally called magic spot (13). This compound, along with its natural precursor pppGpp, is widely implicated in regulating RNA polymerase and other protein factors to control metabolic and physiological responses to various stresses (14–16), including the long-studied stringent response. The stringent response is a major starvation-induced adaptation that can be triggered by amino acid depletion (17–19). In Proteobacteria, this adaptation is accomplished at least in part via the direct inhibition of RNA polymerase by ppGpp binding (16).
Fig. 1.
ppGpp riboswitches and their regulatory network. (A) The chemical structure of ppGpp (guanosine-3′,5′-bisdiphosphate). (B) Consensus sequence and secondary structure model for ppGpp riboswitches derived from 115 unique representatives. (C) Genes predicted to be regulated by ppGpp riboswitches.
In Firmicutes, transcription regulation by ppGpp is known to act indirectly. GTP levels drop as a result of (p)ppGpp synthesis to specifically down-regulate transcription of genes that initiate with a G nucleotide, which affects the production of certain mRNAs and also rRNAs. GTP depletion still permits transcription of genes that initiate with an A nucleotide, such as those for branched-chain amino acid (BCAA) biosynthesis (20, 21). Specific gene activation also occurs by the dissociation of the CodY transcriptional repressor protein when GTP levels drop as a result of (p)ppGpp synthesis (22). This protein-based system thus allows ppGpp to indirectly activate CodY-controlled genes. Herein we describe another mechanism for ppGpp-triggered regulation of gene expression in Firmicutes mediated by numerous examples of selective ppGpp-sensing riboswitches.
Results and Discussion
Identification of ppGpp Riboswitch Candidates.
The widespread distribution of ppGpp in biology, its role in regulating fundamental biological processes, and its construction from an RNA mononucleotide are consistent with an ancient RNA World origin (5). Given these characteristics, we previously speculated that ppGpp would be an ideal ligand candidate for a riboswitch class that had yet to be discovered (5, 6, 10, 12). Thus, we were well positioned to test ppGpp as a candidate ligand when we recently (23) identified a riboswitch subtype (Fig. 1B) of unknown function that commonly associates with genes for BCAA metabolism (Fig. 1C). The ppGpp riboswitch candidates were recognized as “subtype 2a” variants of what we originally called a ykkC motif RNAs (24).
The predominant members (subtype 1) of the ykkC motif collection were recently determined to function as guanidine-sensing RNAs, which are collectively called guanidine-I riboswitches (23). In this same study, it was discovered that certain ykkC motif RNAs (subtype 2) carry distinctive mutations at nucleotide positions found directly in the binding pocket (25, 26). Members of the ykkC subtype 2 collection do not bind guanidine, confirming that the binding site differences indeed alter riboswitch specificity.
Although ppGpp had previously been tested for binding to ykkC RNAs (27), it was determined in retrospect that the specific RNA sequences tested belong to the guanidine-I riboswitch class. After further separation of the subtypes into four additional classes, called subtype 2a through 2d (based on distinctive gene associations and aptamer sequence variations), ppGpp became a likely candidate ligand because of the association of many subtype 2a representatives with genes for BCAA biosynthesis. Furthermore, BCAA biosynthesis is activated during the stringent response in Firmicutes, which are the predominant organisms that contain subtype 2a ykkC motif variants.
Subtype 2a ykkC Motif RNAs Selectively Bind ppGpp.
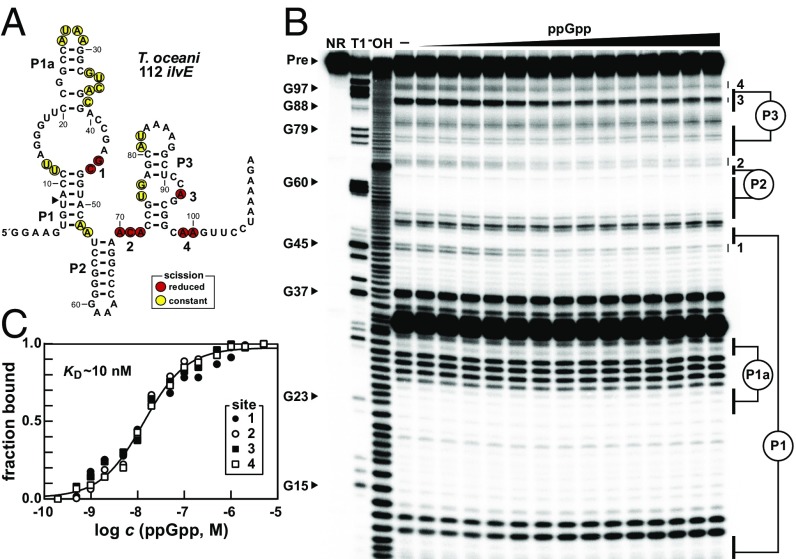
To test for ppGpp riboswitch function, we first arbitrarily chose to examine a BCAA-associated candidate RNA construct from the ilvE gene of Thermosediminibacter oceani (Fig. 2A) by using in-line probing. This method exploits the differential instability of RNA phosphoester bonds in unstructured versus structured regions, to provide evidence that ligand docking alters the folding state of the ligand-binding aptamer (28, 29). We observed a pattern of ppGpp-dependent RNA structural modulation with this construct (Fig. 2B) to yield a binding curve consistent with 1:1 complex formation and an apparent dissociation constant (KD) of ∼10 nM (Fig. 2C). Nucleotides that exhibit structural modulation are located in regions that are known to form the binding pocket for guanidine-I riboswitches (25, 26). Remarkably, these two riboswitch classes utilize the same general architecture but have altered key nucleotides to create binding pockets capable of recognizing completely different ligands.
Fig. 2.
Ligand binding by a ppGpp riboswitch aptamer. (A) Sequence and secondary structure of the 112 nucleotide RNA derived from the ilvE gene of T. oceani. Data collected in B were used to determine regions of constant or reduced scission upon the addition of ppGpp to in-line probing reactions. (B) PAGE analysis of the products of in-line probing reactions with 5′-32P–labeled 112 ilvE RNA in the presence of increasing ppGpp concentrations. NR, T1, and −OH represent RNA undergoing no reaction, partial digest with RNase T1 (cleaves after guanosine nucleotides), or partial digest under alkaline conditions (cleaves after every nucleotide), respectively. Selected bands corresponding to RNA strand scission after G residues are annotated. In-line probing reactions contained either no ligand (–) or ppGpp ranging from 200 pM to 5 µM. Regions 1–4 of the RNA exhibit structural stabilization in a ppGpp-dependent manner. (C) Plot of the fraction of RNA bound to ppGpp, as determined by the normalized fraction of RNA scission at regions 1–4 as a function of the logarithm of ppGpp concentration (c). The dissociation constant (KD) was estimated by a sigmoidal fit of the average of the data points from all regions of RNA structural modulation using a procedure described in Materials and Methods. Data shown are representative of at least three separate in-line probing experiments.
Individual mutation of six highly conserved nucleotides (M1 through M6) within the T. oceani RNA (SI Appendix, Fig. S1A), predicted to be directly involved in ligand binding, each cause at least a three order-of-magnitude loss in ppGpp binding affinity (SI Appendix, Fig. S1B). We speculate that these nucleotide positions might form the binding pocket for ppGpp because their equivalent positions are directly involved in contacting the ligand in the highly similar members of the guanidine-I riboswitch class. Reversing the nucleotide identity of a conserved base pair (M7) similarly reduces ligand affinity by about one order-of-magnitude. These findings demonstrate that conserved nucleotides are critical for optimal binding by the aptamer.
Riboswitch binding specificity was assessed via in-line probing by using a collection of ppGpp analogs comprised of guanosine nucleotide derivatives. In Bacillus subtilis, the most abundant natural version of the signaling molecule is the pentaphosphate species (pppGpp), which is directly synthesized when GTP is used as the biosynthetic precursor. The riboswitch displays equal affinities for ppGpp (Fig. 2C) and pppGpp (SI Appendix, Fig. S2B), indicating they are both likely to trigger gene expression in cells. At higher concentrations, similar compounds, such as pGp, GTP, and GDP, modulate the RNA structure in the same regions as pppGpp and ppGpp (SI Appendix, Fig. S3A). Both GDP and GTP require >1,000-fold higher concentration than ppGpp to half-maximally saturate the riboswitch aptamer (SI Appendix, Fig. S3B), whereas other nucleotides have no effect regardless of the concentrations tested (SI Appendix, Fig. S3C).
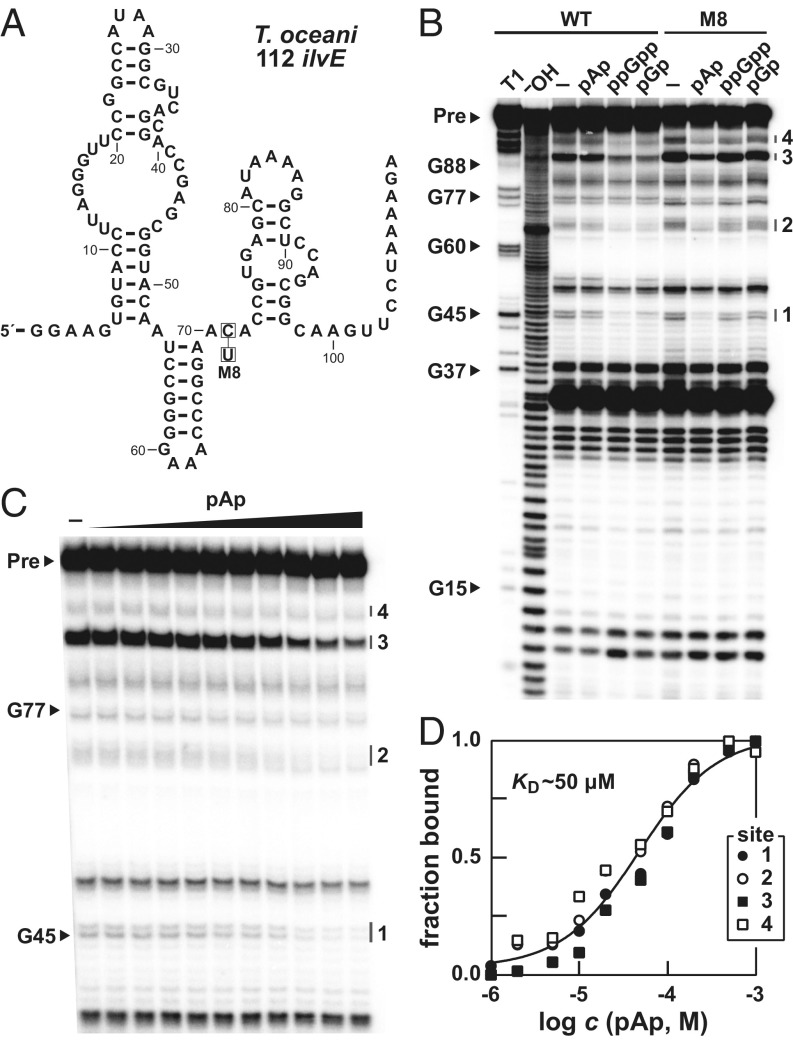
We hypothesized that the ppGpp aptamer RNA might recognize the guanine base of its ligand through Watson–Crick pairing and sought to identify a conserved cytosine residue that might perform this function. The conserved cytosine between the P2 and P3 stems was identified as a likely candidate due to its role in the guanidine-I riboswitch structure (25, 26). Specifically, this C forms a long-range base pair with a conserved G in the internal loop of the P3 stem, which appears to be absent in ppGpp riboswitches because the corresponding nucleotide is no longer a conserved G nucleotide. It seemed plausible that the C nucleotide (C71 in the T. oceani 112 ilvE RNA), which is in a region of reduced scission upon ligand binding in-line probing reactions (Fig. 2A), might be available to base pair to the nucleobase of ppGpp. Indeed, introducing a C to U mutation at this position (M8) (Fig. 3A) switches the specificity of the aptamer from a G-containing ligand to an A-containing ligand, as demonstrated by ability of the mutant RNA to preferentially recognize pAp over pGp or ppGpp (Fig. 3B).
Fig. 3.
Mutation of a conserved nucleotide changes ligand specificity. (A) Sequence and secondary structure of the WT 112 ilvE RNA from T. oceani. The boxed nucleotide identifies the M8 mutation. (B) PAGE analysis of in-line probing assays with WT and M8 112 ilvE RNA in the absence (–) or presence of 1 mM pAp, ppGpp, or pGp. Additional annotations are as described in the legend to Fig. 2B. (C) PAGE analysis of in-line probing assays of M8 112 ilvE RNA with various concentrations of pAp ranging from 0 (–) to 1 mM. (D) Plot of the fraction of RNA bound to pAp. Additional annotations are as described in the legend to Fig. 2C.
In-line probing was also used to estimate the KD of pAp binding by the M8 RNA construct (Fig. 3C). The binding curve generated from these data (Fig. 3D) reveals a KD of ∼50 μM for one-to-one binding of pAp by M8 RNA. This KD is similar to that measured for pGp binding to the WT version of this same RNA (SI Appendix, Fig. S3B). These data suggest that this C nucleotide at position 71 of the 112 ilvE aptamer is the primary determinant of nucleobase recognition for the riboswitch ligand. Currently, we have no evidence for a naturally occurring aptamer for ppApp (or a similar adenosine-containing molecule) because this position of the aptamer is conserved as a C nucleotide across all natural examples of the sequences resembling ppGpp riboswitches examined to date.
Transcription Termination Regulated by a ppGpp Riboswitch.
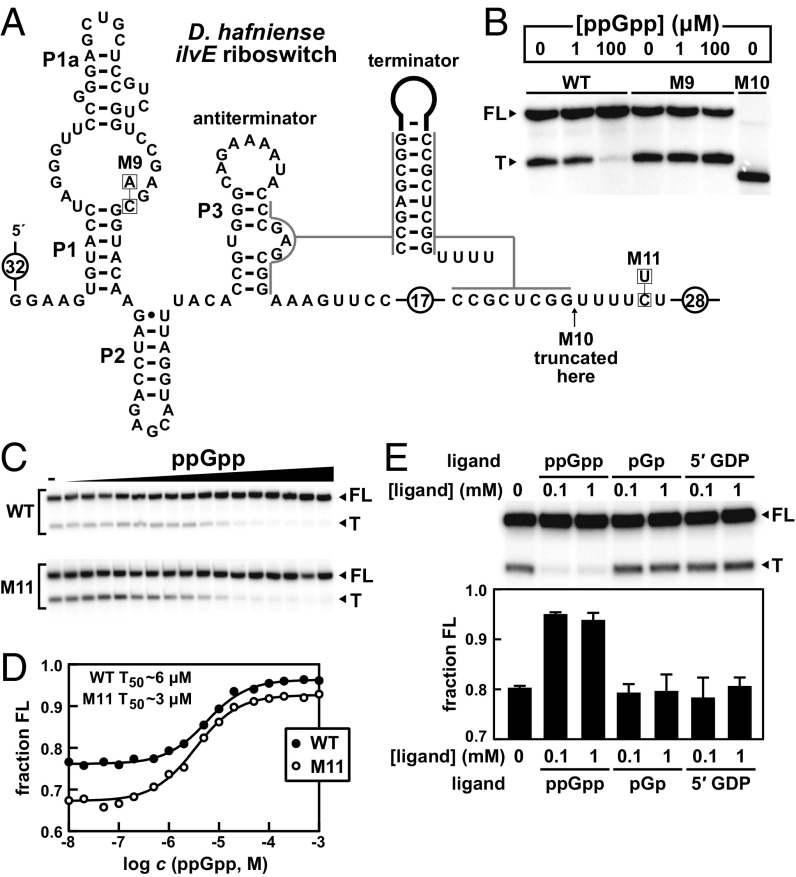
To determine whether ppGpp binding by a member of this riboswitch class directly affects gene expression, single turnover in vitro transcription termination assays (30) were performed as previously described (31) with a representative from the 5′UTR of the ilvE gene of Desulfitobacterium hafniense (Fig. 4A). This construct was chosen because the expression platform exhibited an architecture that suggested the D. hafniense riboswitch uses a transcription termination mechanism, and this construct was confirmed to operate with RNA polymerase from Escherichia coli. Specifically, we predicted that the P3 helix of the aptamer acts as an antiterminator stem, and so transcription of the full-length mRNA should increase when ppGpp is bound. As expected, the WT riboswitch permits RNA polymerase to proceed through the stop site of the adjoining intrinsic terminator stem with greater efficiency only when sufficient ppGpp is added to the reaction (Fig. 4B). An RNA (M9) with a single mutation at a nucleotide position that disrupts the binding pocket terminates with the same frequency regardless of the addition of ppGpp. This finding demonstrates that transcription read-through requires ppGpp binding by the riboswitch, rather than the binding of this signaling molecule to the RNA polymerase protein complex.
Fig. 4.
ppGpp regulates riboswitch-dependent transcript termination. (A) Sequence and secondary structure of the ppGpp riboswitch derived from the ilvE gene of D. hafniense. An alternative “terminator” RNA structure, which only forms in the absence of ppGpp, is depicted wherein a hairpin forms followed by a U-rich tract. (B) PAGE analysis of single-round transcription termination assays with WT, M9, and M10 D. hafniense ilvE riboswitches conducted in the absence or presence of ppGpp, as indicated. Construct sequences are as depicted in A. FL and T denote full-length and terminated transcripts, respectively. Construct M10 provides a size marker for a transcript that terminates several nucleotides before the product generated when the intrinsic terminator forms. (C) PAGE analysis of single-round transcription termination assays of the WT and M11 D. hafniense ilvE RNAs with either no ligand (–) or ppGpp ranging from 10 nM to 1 mM. (D) Plot of the fraction of FL WT or M11 D. hafniense ilvE riboswitch transcripts contributing to the total number of transcripts (FL plus T) as a function of the logarithm of ppGpp molar concentration. The concentration of ppGpp required to cause half-maximal change in termination (T50) was determined by a sigmoidal curve fit. (E, Upper) PAGE analysis of single-round transcription termination assays of the WT D. hafniense ilvE RNA with either no ligand (–) or each ligand as indicated at a concentration of 0.1 or 1 mM. Data are representative of multiple trials. (Lower) Plot of the fraction of FL WT D. hafniense ilvE riboswitch transcripts contributing to the total number of transcripts (FL plus T) in the absence of ligand (–) or with each ligand as indicated. Values reported are the average and SD of three separate trials, including the PAGE image depicted above.
The concentration of ppGpp needed to achieve half-maximal termination (T50) is 6 µM for the D. hafniense ilvE RNA (Fig. 4 C and D). This is nearly 1,000-fold higher than the KD value measured for direct binding of ppGpp to the T. oceani 112 ilvE RNA construct (Fig. 2C). This difference might be partly due to the distinct sequences of the two ppGpp riboswitches, or to the different conditions in which the two types of assays are conducted. However, it is well established (31–34) that some riboswitches operate under kinetic control (31) (likely experienced during transcription termination assays), rather than reaching thermodynamic equilibrium (as attained during in-line probing assays). Riboswitches operating under kinetic control have been shown to require higher concentrations of ligand to trigger gene control compared with KD values of the same constructs measured under thermodynamic equilibrium. In other words, KD values measured in a test tube do not reflect the concentration of ligand naturally required to trigger large changes in gene expression because each riboswitch aptamer does not have time to reach thermodynamic equilibrium before RNA polymerase transcribes beyond the intrinsic terminator stem.
The amount of full-length transcript is quite high even when no ppGpp is present and the terminator is expected to form (Fig. 4B). This is again likely due to kinetic aspects, such as the speed of RNA polymerase, which can influence the amount of ligand needed to regulate riboswitch-mediated termination, and the absence of the transcription factor NusA, which can increase the dynamic range of transcription termination yields (31, 35). Also, the modest dynamic range of the D. hafniense ilvE riboswitch in vitro might be due in part to the fact that the run of U residues traditionally following a terminator stem is interrupted with a C nucleotide. A construct wherein this C residue is altered to a U (M11) exhibits a modestly improved dynamic range (Fig. 4 C and D). Regardless of the factors affecting the dynamic range of the riboswitch when measured in vitro, similar dynamic ranges for other riboswitch classes examined by in vitro transcription assays have been observed previously (36–38).
To further assess ppGpp as the natural ligand for this riboswitch class, we tested other guanine nucleotide compounds using in vitro transcription termination assays. Although pGp and GDP cause modulation of the RNA under in-line probing conditions, albeit with decreased affinity compared with ppGpp, neither is able to cause a shift from terminated to full-length product even at a concentration of 1 mM (Fig. 4E). GTP was not tested because it is already present at 150 µM in the transcription reaction. These results demonstrate that these RNAs selectively regulate gene expression in response to ppGpp or pppGpp, but not any other guanine nucleotide derivative.
Gene Associations of ppGpp Riboswitches.
The ligand binding and in vitro regulatory function of ppGpp riboswitch constructs described above are entirely consistent with the predominant gene associations observed for members of this riboswitch class. The link between BCAA metabolism and ppGpp signaling is well established (13–18), and our observations that ppGpp and pppGpp are bound by members of this riboswitch class are consistent with this natural gene association.
However, ppGpp riboswitches are also commonly associated with members of two other gene classes, which are connections that were not well established previously. The first includes genes encoding various glutamate synthase domains, including gltS, gltB, gltD, and glxB. The second includes operons containing three ATP-binding cassette (ABC) transporters, one of which is predicted to be natA while the other two (COG3694 and COG4587) have unknown functions. Representative RNAs associated with glutamate synthase (SI Appendix, Fig. S4) and natA (SI Appendix, Fig. S5) gene classes bind ppGpp with dissociation constants similar to that observed for the T. oceani 112 ilvE riboswitch aptamer (Fig. 2).
Organisms that use ppGpp riboswitches to control glutamate synthase genes typically also use ppGpp riboswitches to regulate BCAA biosynthesis-related gene expression, suggesting that these two metabolic processes are linked. Glutamate levels are known to be elevated during the stringent response (39), and this riboswitch discovery provides a direct mechanism for up-regulation of glutamate biosynthesis during this stress response. Moreover, the known connection between these biochemical responses reinforces our hypothesis that the natural ligand for some ykkC subtype 2 RNAs indeed is ppGpp or its immediate biosynthetic precursor pppGpp.
There was no prior established link between ppGpp signaling and the ABC transporters associated with ppGpp riboswitches. Organisms with ppGpp riboswitches upstream of these transporter genes are mutually exclusive to organisms with ppGpp riboswitches upstream of BCAA and glutamate synthesis genes. A possible explanation for this gene-association pattern comes from the fact that one of these transporters shares homology with NatA, which has a predicted cation transport function and is known to be expressed under membrane stress conditions (40). Various cell envelope stresses are also known to activate the relP ppGpp synthase gene in B. subtilis (41–43). Therefore, it seems logical for these transporters, if they are indeed NatA homologs, to be expressed when ppGpp levels are high, although in this case ppGpp production is not directly related to the stringent response.
Some ppGpp Riboswitches Reside in Tandem with T Box RNAs.
In ∼40% of the instances when the ppGpp riboswitch is found in the 5′UTR of BCAA biosynthesis genes, a leucine T box RNA is found either directly upstream or downstream of the riboswitch in the same UTR. T box RNAs are regulatory domains that control gene expression in response to binding certain uncharged tRNAs, thereby indirectly detecting a deficiency of an amino acid (44). These two RNA elements presumably function independently because each has its own expression platform. When found in tandem, they function as a two-input Boolean AND logic gate, which would be similar to riboswitch logic gate architectures reported previously (45). Specifically, both uncharged tRNALeu and ppGpp would be necessary for transcription of the downstream protein coding regions.
This tandem arrangement likely exploits two required inputs to ensure that the expression of associated genes is reduced when nutrients are plentiful. Additionally, because RelA is allosterically regulated by any uncharged tRNA, the ppGpp riboswitch will turn on gene expression when any amino acid is limiting, whereas each T box RNA indirectly detects a limitation only of its cognate amino acid. At the promoter level, BCAA biosynthesis genes are regulated by the CodY transcriptional repressor, which uses GTP and isoleucine as regulatory inputs (22, 46). It is unclear whether all three of these mechanisms are utilized simultaneously because, while organisms with ppGpp riboswitches typically have the codY gene, the canonical CodY binding site consensus sequence (46) was not found upstream of ppGpp riboswitch sequences.
Bioinformatic and Biochemical Data Reveal Additional Riboswitch Classes.
All other RNAs examined that were grouped with ykkC subtype 2 fail to bind either ppGpp or guanidine, and therefore represent distinct riboswitch classes. Subtypes 2a through 2d were identified by sorting RNA representatives based on their distinct gene associations, and then by comparative sequence analysis (23), as was conducted to identify ykkC subtype 2a RNAs and to verify that they function as ppGpp riboswitches (Fig. 1). We recently determined that some of these RNAs (subtype 2b) bind phosphoribosyl pyrophosphate, a precursor of nucleotide biosynthesis (47). However, we speculate that at least two additional candidate riboswitch classes (subtypes 2c and 2d) whose ligands remain undiscovered are represented among the remaining ykkC RNA variants.
Conclusions
A riboswitch class for ppGpp has long been anticipated (5, 6, 10, 12), but representatives of this particular class were challenging to identify due to their striking similarity to riboswitches for guanidine (23). At one point early in evolutionary history, ppGpp riboswitches might have been extremely widespread, but it appears that their function has been replaced by the CodY repressor protein factor in many organisms.
Notably, the discovery of ppGpp riboswitches in tandem with T box regulators serves as an additional demonstration of how complex molecular devices can be formed purely from RNA. The Boolean AND gate architecture described herein involves a tRNA sensed by a T box RNA and a riboswitch RNA that senses the RNA-derived alarmone ppGpp. This all-RNA system is similar to that for the two-input NOR logic gate formed by tandem riboswitches for S-adenosylmethionine and adenosylcobalamin (45). Similarly, it has previously been demonstrated that a riboswitch located in tandem with a group I self-splicing ribozyme functions as a two-input AND gate that requires both the signaling molecule c-di-GMP and the nucleotide GTP to properly splice and express a messenger RNA from Clostridium difficile (8, 48). Such complex all-RNA systems suggest that riboswitch-mediated responses to nucleotide-like signaling molecules such as ppGpp could be of ancient RNA World origin.
Materials and Methods
Chemicals and Reagents.
Chemicals were purchased from Sigma-Aldrich with the exceptions of guanosine 5′,3′-bisdiphosphate (ppGpp), guanosine pentaphosphate (pppGpp) (Jena Bioscience), and guanosine 5′,3′-bisphosphate (pGp) (TriLink Biotech). [γ-32P]-ATP was purchased from Perkin-Elmer and used within 2 wk of delivery. All bulk chemicals were purchased from J. T. Baker and enzymes were purchased from New England Biolabs, unless otherwise noted. All solutions were prepared using deionized water (dH2O) and were either autoclaved or filter-sterilized (using 0.22-µm filters; Millipore) before use. DNA oligonucleotides used in this study (SI Appendix, Table S1) were purchased from Sigma-Aldrich and Integrated DNA Technologies. Strains of B. subtilis were obtained from the Bacillus Genetic Stock Center at The Ohio State University.
Bioinformatics Analyses.
Additional examples of ykkC motif RNAs were identified using Infernal 1.1 (49) to search RefSeq v76 plus additional environmental microbial databases (50). Iterative searches for new sequences were performed based on the previously published alignment of ykkC subtype 2 RNAs (23). These sequences were sorted by nucleotide identity at a specific position as previously indicated (23) to exclude guanidine-I riboswitches, then further manually sorted by downstream gene association. This analysis identified 105 examples of subtype 2a RNAs (ppGpp riboswitches), of which 78 are upstream of BCAA metabolism genes, 15 are upstream of various glutamate synthase genes, and 12 are upstream of natA and other ABC transporter genes. It should be noted that this is an underestimate of the true number of representatives of this riboswitch class in this dataset, as examples with incomplete gene contexts were not evaluated. The consensus sequence and secondary structure model of these 105 ppGpp riboswitch RNAs was constructed using R2R software (51).
RNA Oligonucleotide Preparation.
Preparation of RNAs for use in in-line probing experiments was performed as previously described (23, 52) with the following modifications. All double-stranded DNA (dsDNA) templates used for RNA transcription were assembled via extension of overlapping synthetic DNAs (SI Appendix, Table S1) using SuperScript II reverse transcriptase (Thermo Fisher Scientific). Forward primers contain the T7 RNA polymerase (T7 RNAP) promoter followed by guanosine residues (when not present in the WT sequence) to enable transcription. Thereafter, RNA constructs were prepared from these dsDNAs by in vitro transcription, purified, and subsequently 5′ 32P-labeled as previously described (23, 52).
RNA In-Line Probing Analyses.
In-line probing assays were performed as previously described (28, 29). The values for the amount of RNA aptamer bound to ligand or “fraction bound” were estimated based on the ligand-mediate changes in the band intensities from in-line probing assays also as reported previously (23, 52). Briefly, quantified changes in band intensities at designated sites reflect the fraction of RNAs that are bound to ligand, such that half-maximal increases or decreases in band intensities represent the KD for the RNA–ligand interaction.
Single-Round Transcription Termination Assays.
Template dsDNA constructs for single-round in vitro transcription were designed to include the aptamer and expression platform of the D. hafniense riboswitch starting at the predicted natural transcription start site and ending 28 nucleotides beyond the terminator stem. The promoter used to enable transcription was derived from the B. subtilis lysC gene, which is compatible with E. coli RNA polymerase. The described WT and mutant templates were purchased as synthetic dsDNAs and subsequently amplified by PCR. Approximately 2 pmol of the resulting, purified DNA template was added to a transcription initiation mixture [20 mM Tris⋅HCl (pH 7.5 at 23 °C), 75 mM KCl, 5 mM MgCl2, 1 mM DTT, 10 µg mL−1 BSA, 130 µM ApA dinucleotide, 1% glycerol, 0.04 U µL−1 E. coli RNA polymerase holoenzyme, 2.5 µM GTP, 2.5 µM ATP, and 1 µM UTP]. Approximately 1 µCi [α-32P]-UTP was added per 8-µL reaction and transcription was allowed to proceed at 37 °C for 10 min, leading to formation of a stalled polymerase complex at the first cytidine nucleotide of each transcript. For each 8-µL transcription reaction, 1 µL of 10× elongation buffer [20 mM Tris⋅HCl (pH 7.5 at 23 °C), 75 mM KCl, 5 mM MgCl2, 1 mM DTT, 2 mg mL−1 heparin, 1.5 mM ATP, 1.5 mM GTP, 1.5 mM CTP, and 250 µM UTP] as well as 1 µL of a 10× solution of the ligand of interest were added sequentially. Because ppGpp is a known inhibitor of E. coli RNA polymerase initiation (16), the holoenzyme was first allowed to assemble into a stalled, stable ternary complex before ppGpp was added concurrently with the elongation mixture. Transcription reactions were then incubated at 37 °C for an additional 60 min.
The transcription products were subsequently analyzed via denaturing 10% PAGE and visualized using a phosphorimager (GE Healthcare Life Sciences). Fraction full-length (FL) values were calculated by varying the ligand concentration in separate reactions and quantifying the changes in band intensity of both FL and terminated (T) transcription products using the formula (FL)/(FL + T). The T50 values were determined by plotting the fraction FL as a function of the logarithm of ligand concentration and using a sigmoidal four parameter logistic fit in GraphPad Prism 7.
Supplementary Material
Acknowledgments
We thank Shira Stav, Adam Roth, Kimberly Harris, and other members of the R.R.B. laboratory for helpful discussions. This work was supported by NIH Grants GM022778 and DE022340 (to R.R.B.). M.E.S. was supported by NIH Cellular and Molecular Biology Training Grant T32GM007223. R.R.B. is also supported by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720406115/-/DCSupplemental.
References
- 1.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherwood AV, Henkin TM. Riboswitch-mediated gene regulation: Novel RNA architectures dictate gene expression responses. Annu Rev Microbiol. 2016;70:361–374. doi: 10.1146/annurev-micro-091014-104306. [DOI] [PubMed] [Google Scholar]
- 4.McCown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR. Riboswitch diversity and distribution. RNA. 2017;23:995–1011. doi: 10.1261/rna.061234.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson JW, Breaker RR. The lost language of the RNA World. Sci Signal. 2017;10:1–10. doi: 10.1126/scisignal.aam8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breaker RR. Riboswitches: From ancient gene control systems to modern drug targets. Future Microbiol. 2009;4:771–773. doi: 10.2217/fmb.09.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudarsan N, et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson JW, et al. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol. 2013;9:834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim PB, Nelson JW, Breaker RR. An ancient riboswitch class in bacteria regulates purine biosynthesis and one-carbon metabolism. Mol Cell. 2015;57:317–328. doi: 10.1016/j.molcel.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kellenberger CA, et al. GEMM-I riboswitches from Geobacter sense the bacterial second messenger cyclic AMP-GMP. Proc Natl Acad Sci USA. 2015;112:5383–5388. doi: 10.1073/pnas.1419328112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson JW, et al. Control of bacterial exoelectrogenesis by c-AMP-GMP. Proc Natl Acad Sci USA. 2015;112:5389–5394. doi: 10.1073/pnas.1419264112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cashel M. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J Biol Chem. 1969;244:3133–3141. [PubMed] [Google Scholar]
- 14.Potrykus K, Cashel M. (p)ppGpp: Still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 15.Dalebroux ZD, Swanson MS. ppGpp: Magic beyond RNA polymerase. Nat Rev Microbiol. 2012;10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 16.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cashel M, Rudd K. The stringent response. In: Neidhardt FC, editor. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology; Washington, DC: 1987. pp. 1410–1438. [Google Scholar]
- 18.Traxler MF, et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shivers RP, Sonenshein AL. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Microbiol. 2004;53:599–611. doi: 10.1111/j.1365-2958.2004.04135.x. [DOI] [PubMed] [Google Scholar]
- 20.Krásný L, Tiserová H, Jonák J, Rejman D, Sanderová H. The identity of the transcription +1 position is crucial for changes in gene expression in response to amino acid starvation in Bacillus subtilis. Mol Microbiol. 2008;69:42–54. doi: 10.1111/j.1365-2958.2008.06256.x. [DOI] [PubMed] [Google Scholar]
- 21.Wolz C, Geiger T, Goerke C. The synthesis and function of the alarmone (p)ppGpp in firmicutes. Int J Med Microbiol. 2010;300:142–147. doi: 10.1016/j.ijmm.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Sonenshein AL. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol. 2005;8:203–207. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Nelson JW, Atilho RM, Sherlock ME, Stockbridge RB, Breaker RR. Metabolism of free guanidine in bacteria is regulated by a widespread riboswitch class. Mol Cell. 2017;65:220–230. doi: 10.1016/j.molcel.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrick JE, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reiss CW, Xiong Y, Strobel SA. Structural basis for ligand binding to the guanidine-I riboswitch. Structure. 2017;25:195–202. doi: 10.1016/j.str.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Battaglia RA, Price IR, Ke A. Structural basis for guanidine sensing by the ykkC family of riboswitches. RNA. 2017;23:578–585. doi: 10.1261/rna.060186.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer MM, et al. Challenges of ligand identification for riboswitch candidates. RNA Biol. 2011;8:5–10. doi: 10.4161/rna.8.1.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 30.Landick R, Wang D, Chan CL. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: His leader pause site as paradigm. Methods Enzymol. 1996;274:334–353. doi: 10.1016/s0076-6879(96)74029-6. [DOI] [PubMed] [Google Scholar]
- 31.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 32.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 33.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J Mol Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Haller A, Soulière MF, Micura R. The dynamic nature of RNA as key to understanding riboswitch mechanisms. Acc Chem Res. 2011;44:1339–1348. doi: 10.1021/ar200035g. [DOI] [PubMed] [Google Scholar]
- 35.Lemay J-F, et al. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDaniel BAM, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: Direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci USA. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 39.Imaizumi A, Kojima H, Matsui K. The effect of intracellular ppGpp levels on glutamate and lysine overproduction in Escherichia coli. J Biotechnol. 2006;125:328–337. doi: 10.1016/j.jbiotec.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 40.Cheng J, Guffanti AA, Krulwich TA. A two-gene ABC-type transport system that extrudes Na+ in Bacillus subtilis is induced by ethanol or protonophore. Mol Microbiol. 1997;23:1107–1120. doi: 10.1046/j.1365-2958.1997.2951656.x. [DOI] [PubMed] [Google Scholar]
- 41.Nanamiya H, et al. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol. 2008;67:291–304. doi: 10.1111/j.1365-2958.2007.06018.x. [DOI] [PubMed] [Google Scholar]
- 42.Geiger T, Wolz C. Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria. Int J Med Microbiol. 2014;304:150–155. doi: 10.1016/j.ijmm.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 43.Geiger T, Kästle B, Gratani FL, Goerke C, Wolz C. Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J Bacteriol. 2014;196:894–902. doi: 10.1128/JB.01201-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010;584:318–324. doi: 10.1016/j.febslet.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sudarsan N, et al. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 46.den Hengst CD, et al. The Lactococcus lactis CodY regulon: Identification of a conserved cis-regulatory element. J Biol Chem. 2005;280:34332–34342. doi: 10.1074/jbc.M502349200. [DOI] [PubMed] [Google Scholar]
- 47.Sherlock ME, Sudarsan N, Stav S, Breaker RR. Tandem riboswitches form a natural Boolean logic gate to control purine metabolism in bacteria. eLife. 2018;7:e33908. doi: 10.7554/eLife.33908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen AG, Sudarsan N, Breaker RR. Mechanism for gene control by a natural allosteric group I ribozyme. RNA. 2011;17:1967–1972. doi: 10.1261/rna.2757311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinberg Z, et al. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol. 2015;11:606–610. doi: 10.1038/nchembio.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinberg Z, Breaker RR. R2R—Software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics. 2011;12:3. doi: 10.1186/1471-2105-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherlock ME, Malkowski SN, Breaker RR. Biochemical validation of a second guanidine riboswitch class. Biochemistry. 2017;56:352–358. doi: 10.1021/acs.biochem.6b01270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.