Significance
Circadian misalignment (i.e., behavioral processes such as food intake or sleep occurring at inappropriate endogenous circadian times) commonly occurs during shift work and is associated with health problems. Identifying mechanisms underlying health problems associated with circadian misalignment will help develop precision medicine countermeasures. Thus, we investigated the impact of circadian misalignment on the human plasma proteome using a simulated nightshift protocol in healthy volunteers. We demonstrate that circadian and/or behavioral wake–sleep/food intake–fasting cycles regulate 24-h time-of-day patterns of the human plasma proteome. Further, we show that proteins altered during circadian misalignment are associated with biological pathways involved in immune function, metabolism, and cancer and with altered glucose and energy metabolism, identifying potential mechanisms contributing to metabolic dysregulation.
Keywords: circadian rhythm, eating at night, shift work, peripheral clocks, personalized medicine
Abstract
Proteomics holds great promise for understanding human physiology, developing health biomarkers, and precision medicine. However, how much the plasma proteome varies with time of day and is regulated by the master circadian suprachiasmatic nucleus brain clock, assessed here by the melatonin rhythm, is largely unknown. Here, we assessed 24-h time-of-day patterns of human plasma proteins in six healthy men during daytime food intake and nighttime sleep in phase with the endogenous circadian clock (i.e., circadian alignment) versus daytime sleep and nighttime food intake out of phase with the endogenous circadian clock (i.e., circadian misalignment induced by simulated nightshift work). We identified 24-h time-of-day patterns in 573 of 1,129 proteins analyzed, with 30 proteins showing strong regulation by the circadian cycle. Relative to circadian alignment, the average abundance and/or 24-h time-of-day patterns of 127 proteins were altered during circadian misalignment. Altered proteins were associated with biological pathways involved in immune function, metabolism, and cancer. Of the 30 circadian-regulated proteins, the majority peaked between 1400 hours and 2100 hours, and these 30 proteins were associated with basic pathways involved in extracellular matrix organization, tyrosine kinase signaling, and signaling by receptor tyrosine-protein kinase erbB-2. Furthermore, circadian misalignment altered multiple proteins known to regulate glucose homeostasis and/or energy metabolism, with implications for altered metabolic physiology. Our findings demonstrate the circadian clock, the behavioral wake–sleep/food intake–fasting cycle, and interactions between these processes regulate 24-h time-of-day patterns of human plasma proteins and help identify mechanisms of circadian misalignment that may contribute to metabolic dysregulation.
The circadian system regulates ∼24-h physiological cycles. At the cellular level, core clock genes generate circadian rhythms through self-sustained transcriptional translational feedback loops (1, 2), and self-sustained metabolic oscillations are also reported to contribute to circadian rhythmicity (3, 4). In mammals, the environmental light–dark cycle is the primary synchronizer of the master clock located in the suprachiasmatic nucleus (SCN) (2, 5), and this SCN clock drives rhythms in hormones, body temperature, and food intake, all of which in turn can synchronize peripheral clocks in tissues such as the liver, muscle, adipose tissue, pancreas, and brain regions outside the SCN (6). In humans, the circadian system promotes wakefulness, physical activity, and food intake during the biological day and sleep, physical inactivity, and fasting during the biological night (7). Circadian misalignment in humans often occurs when 24-h behavioral cycles (e.g., sleep–wake/fasting–food intake) are misaligned with the timing of the endogenous master circadian SCN clock. Thus, by definition, during circadian misalignment behavioral processes such as food intake or sleep occur at inappropriate endogenous circadian times. Worldwide, ∼20% of people in the workforce are shift workers and therefore work and eat during hours typically reserved for sleep. Circadian misalignment is prevalent among shift workers as they undergo abrupt changes in their behavioral cycles (8, 9). In nocturnal rodents, findings show such behaviorally induced circadian misalignment results in metabolic disease (10–12). Furthermore, epidemiological findings show shift work is associated with elevated metabolic disease risk (13, 14), and findings from controlled laboratory studies indicate circadian misalignment contributes to metabolic dysregulation (15), including reduced insulin sensitivity (16–18), reduced glucose tolerance (19), and risk of weight gain (20).
Findings from studies in mice suggest the circadian system regulates thousands of protein-coding genes (21, 22), and up to half of these proteins are regulated in an organ-specific manner (23–25). Furthermore, findings show many phosphoproteins (26) and nuclear proteins (27) in mouse liver exhibit diurnal rhythms. The rhythmicity of the human whole-blood transcriptome was investigated in two studies, and findings indicated ∼6–9% of transcripts assessed had ∼24-h rhythmicity under circadian-aligned conditions at the 5% false discovery rate (FDR) level (28, 29). However, during circadian misalignment, 97% of these transcripts were arrhythmic (28) using a 5% FDR, suggesting many rhythmic transcripts under circadian alignment are regulated, in part, by the behavioral cycle. Moreover, many rate-limiting steps in metabolic pathways implicated in metabolic disease are under circadian regulation (22, 28), further implicating circadian misalignment in metabolic dysregulation. Whether such changes in gene expression result in changes in protein levels remains to be determined. Here we studied 24-h time-of-day patterns of the plasma proteome, which includes proteins derived from circulating cells, the endothelium, and most peripheral tissues and therefore reflects a broad range of physiological processes and states (30, 31). As most studies to date have examined the human plasma proteome at only a few or one time of day, a primary aim of the current effort was to examine changes in the proteome across the 24-h day and assess the contribution of the circadian versus the behavioral cycle in 24-h time-of-day patterns in protein levels. We also aimed to examine alterations in protein levels during circadian misalignment induced by simulated nightshift work (20) that may contribute to altered physiological function (Fig. 1A).
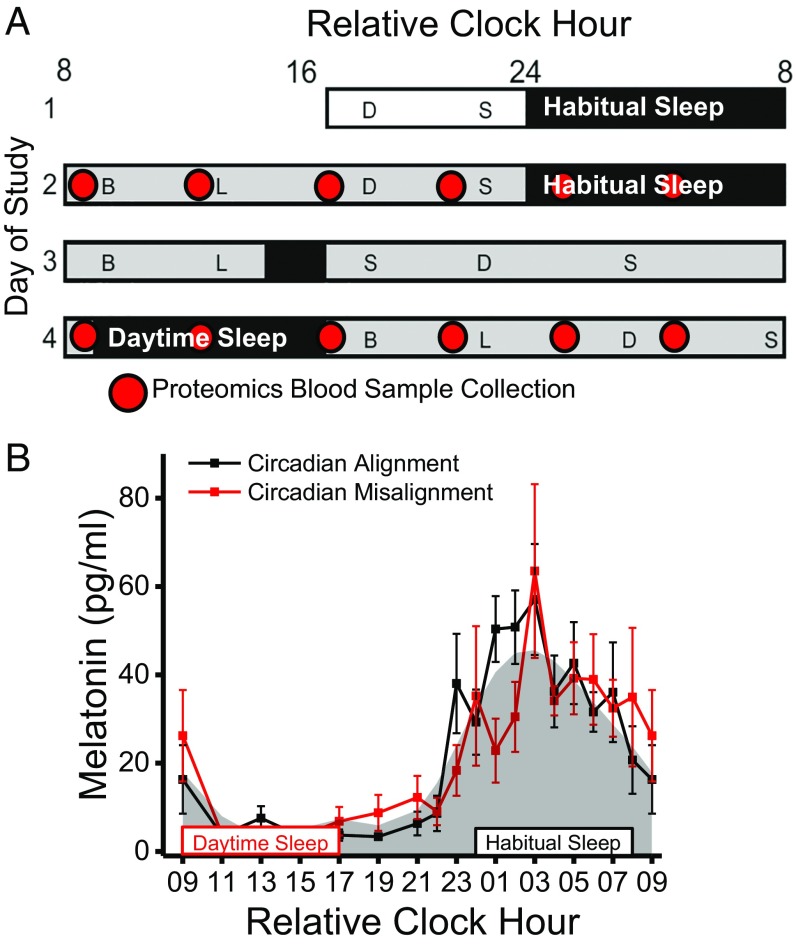
Fig. 1.
Protocol and circadian melatonin timing (n = 6). (A) In-laboratory protocol with time of day plotted as relative clock hour with scheduled waketime arbitrarily assigned a value of 0800 h and all other times referenced to this. The open rectangle represents room light (<40 lx), black rectangles represent scheduled sleep, and gray rectangles represent scheduled wakefulness in dim light (<1 lx). On study day 2 subjects were circadian aligned, and on study day 4 subjects were circadian misaligned. Red circles represent the timing of blood collection for proteomics analyses. B, breakfast; D, dinner; L, lunch; S, snack. (B) Melatonin levels during circadian alignment (black line) and misalignment (red line). Gray shading represents the average fitted melatonin values across circadian alignment and misalignment conditions. Black- and red-outlined rectangles represent scheduled sleep opportunities during circadian alignment (habitual sleep) and circadian misalignment (daytime sleep), respectively. Data are mean ± SEM.
Results
Circadian Timing and Total Sleep Time.
In humans, the biological day and night are defined by low and high melatonin levels, respectively (32). As reported previously in a larger sample from this study (20), timing of the biological day and night as measured by the endogenous melatonin rhythm was similar (Fig. 1B) between circadian alignment (study day 2; daytime wakefulness and food intake with nighttime sleep) and circadian misalignment (study day 4; nighttime wakefulness and food intake with daytime sleep). Furthermore, total sleep time was lower (P < 0.05) during circadian misalignment (6 h 38 min ± 13 min) versus circadian alignment (7 h 19 min ± 10 min) in the current subsample.
Proteins That Increased or Decreased During Circadian Misalignment.
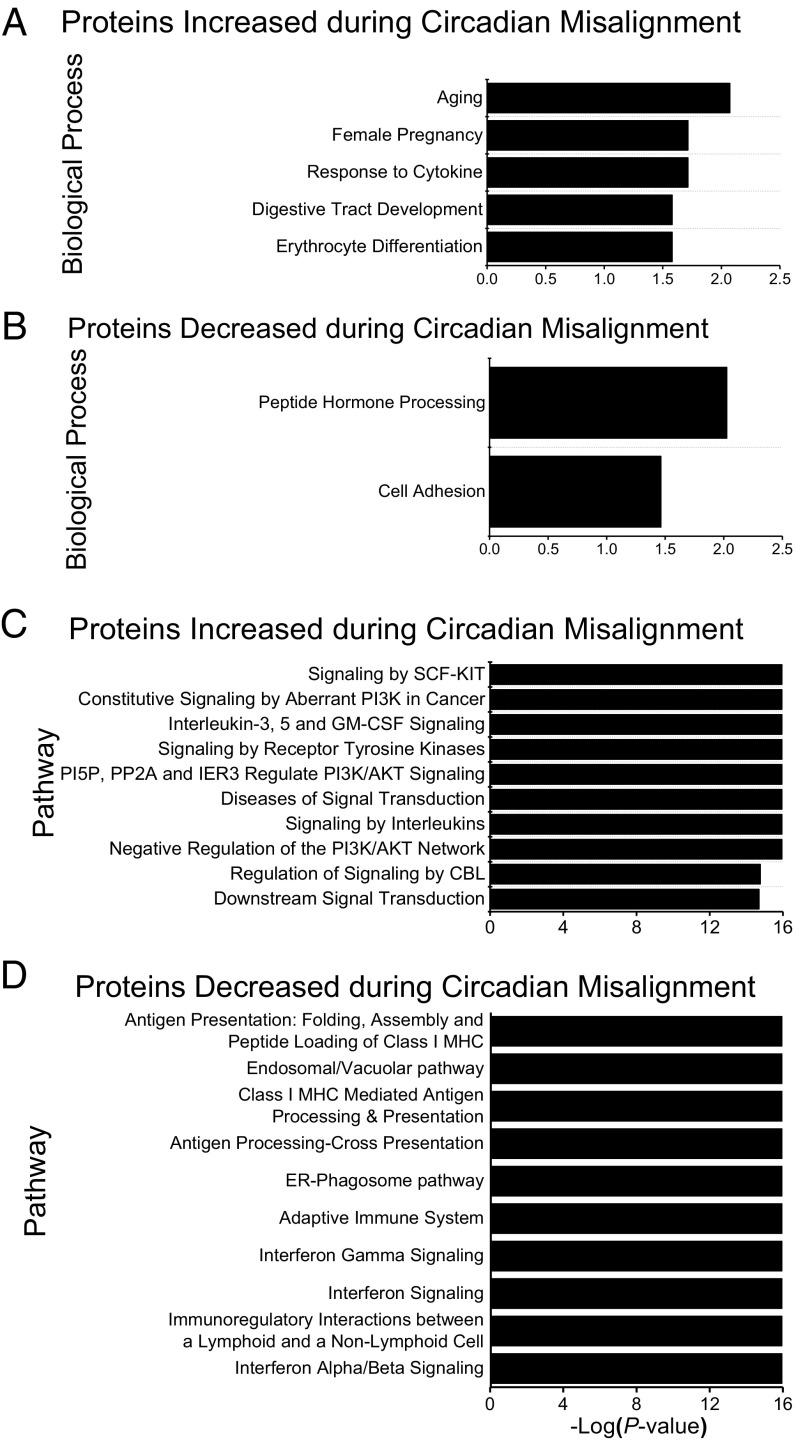
Using Significance Analysis of Microarray (SAM) (33), we found that 62 of the 1,129 proteins analyzed had increased or decreased average 24-h plasma levels during circadian misalignment versus alignment at the 10% FDR level. Of these 62 proteins, the levels of 38 were increased, and the levels of 24 were decreased during circadian misalignment versus alignment. Gene Ontology (GO; DAVID v6.8) (34, 35) analyses identified multiple biological processes, including response to cytokines, erythrocyte differentiation, peptide hormone processing, and cell adhesion, associated with these increased or decreased proteins (Fig. 2 A and B). Pathways associated with proteins that increased during circadian misalignment include multiple pathways linked to PI3K subunit alpha isoform signaling, signaling by receptor tyrosine kinases, and signaling by interleukins (Fig. 2C). PI3K signaling is a key component of the insulin-signaling pathway, and aberrant PI3K signaling is reported to be implicated in cancer. Pathways associated with proteins that decreased during circadian misalignment include multiple pathways linked to antigen presentation and processing and IFN signaling (Fig. 2D). As IFNs are known to increase antigen presentation (36), these findings suggest the adaptive immune system may be altered during circadian misalignment. Follow-up studies are required to specifically investigate the impact of circadian misalignment on PI3K, interleukin signaling, and the adaptive immune system in humans.
Fig. 2.
Proteins increased and decreased during circadian misalignment (n = 6) as identified with two-way ANOVA. (A) GO biological processes associated with proteins with increased protein levels when circadian misaligned versus circadian aligned. (B) GO biological processes associated with proteins with decreased protein levels when circadian misaligned versus aligned. (C) Pathways associated with proteins with increased protein levels when circadian misaligned versus aligned. (D) Pathways associated with proteins with decreased protein levels when circadian misaligned versus aligned. Biological processes and pathways are ranked by P value. Enriched biological processes and pathways were nominally statistically significant at the P < 0.05 level.
Proteins with 24-H Time-of-Day Patterns and with Alterations During Circadian Misalignment.
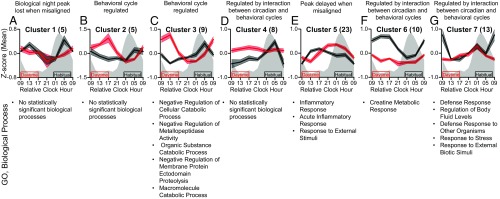
Proteins were assessed for 24-h time-of-day patterns when circadian aligned and misaligned using the R package MetaCycle (37) and by fitting mixed-effects models with a cubic time component (SI Appendix). Resulting P values from these analyses for the circadian alignment and misalignment days were combined using the minP method (38) and were considered statistically significant at P < 0.05 and 10% FDR level. We used multiple approaches for this analysis, as biological rhythms are not expected to all fit one single model of rhythmicity (39). Supporting this concept, the results of these two approaches overlapped for just 40 of the 573 proteins that met the statistical significance criteria on at least one of the circadian alignment or misalignment study days (Dataset S1). This indicates that the majority of proteins with a significant 24-h time-of-day pattern were detected by only one of these algorithms. The mixed-effects cubic model detected more proteins with 24-h time-of-day patterns when circadian aligned (391 proteins) versus misaligned (107 proteins), whereas MetaCycle detected fewer proteins with 24-h time-of-day patterns when aligned (24 proteins) versus misaligned (202 proteins) at the P < 0.05 level (SI Appendix, Figs. S8 and S9). These 573 proteins were further analyzed for differential 24-h time-of-day patterns between circadian alignment and misalignment using the Extraction of Differential Gene Expression (EDGE) (40) and Detection of Differential Rhythmicity (DODR) R packages (41). Resulting DODR and EDGE P values were corrected for multiple comparisons (SI Appendix); and at the corrected P < 0.05 significance level EDGE detected 27 proteins and DODR detected 55 proteins with significantly different 24-h time-of-day patterns during circadian misalignment versus alignment. As six proteins overlapped between EDGE and DODR outcomes, we identified 76 total proteins with significantly different 24-h time-of-day patterns between circadian misalignment and alignment (Fig. 3 and Dataset S1). Hierarchical cluster analyses were used to assess 24-h time-of-day patterns for these 76 proteins. At the P < 0.05 level, we identified seven statistically significant clusters (Fig. 3 and SI Appendix, Fig. S1). GO analyses identified biological processes associated with each cluster (Fig. 3). Cluster 1, consisting of five proteins, peaked in the biological night during scheduled sleep when circadian aligned; in general, this peak was lost when circadian misaligned (Fig. 3A). Cluster 2, consisting of five proteins, peaked during scheduled sleep when both circadian aligned and misaligned (Fig. 3B). While clusters 1 and 2 were not associated with any specific biological processes, insulin-like growth factor-binding proteins (IGFBPs) 4 and 7 were in clusters 1 and 2, respectively; a detailed discussion of the IGFBPs as they relate to metabolism is shown in SI Appendix, Fig. S2. Similar to the pattern for cluster 2, cluster 3, consisting of nine proteins, was lowest during scheduled wakefulness and peaked during scheduled sleep when both circadian aligned and misaligned (Fig. 3C). Cluster 3 was associated with multiple biological processes related to cellular catabolic process. The specific definition of the term with the lowest P value, negative regulation of cellular catabolic process, is any process that stops, prevents, or reduces the frequency, rate, or extent of the chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells. Our findings that clusters 2 and 3 peaked during scheduled sleep when both circadian aligned and misaligned suggest that the behavioral sleep–wake/fasting–food intake cycle strongly regulates these proteins and related biological processes, including reducing cellular catabolism. Cluster 4, consisting of eight proteins, peaked in the biological night during scheduled sleep when circadian aligned, and levels were high over the 24 h when circadian misaligned (Fig. 3D). Cluster 4 was not associated with any biological processes. Cluster 5, consisting of 23 proteins, peaked during the middle of scheduled wakefulness when circadian aligned, and this peak was delayed when circadian misaligned (Fig. 3E). Cluster 5 was associated with biological processes related to inflammatory response and response to external stimuli. The term response to external stimuli is a broad GO term defined as any process that results in a change in state or activity of a cell or an organism as a result of an external stimulus. As such, proteins that respond to the external environment may be expected to change with the altered behavioral cycle in our protocol. These findings also suggest some inflammatory processes, notably acute inflammation, may be under regulation by the behavioral sleep–wake/fasting–food intake cycle. Cluster 6, consisting of 10 proteins, peaked during scheduled wakefulness when circadian aligned, and this peak was lost when circadian misaligned (Fig. 3F). Cluster 6 was associated with the biological process creatine metabolic response. Cluster 7, consisting of 13 proteins, peaked during the biological daytime following scheduled sleep when circadian aligned, and this peak was lost when circadian misaligned (Fig. 3G). Cluster 7 was associated with biological processes related to defense response and responses to external biotic stimuli/stress.
Fig. 3.
Plasma proteins with altered 24-h time-of-day patterns between circadian misalignment and alignment. A–G show statistically significant (P < 0.05) clusters derived from the 76 proteins with different 24-h time-of-day patterns when circadian misaligned versus aligned as identified by EDGE and DODR analyses. (Upper) Represent mean z-score (±SEM) data based on all proteins within each cluster. The number of proteins in each cluster is indicated in parentheses. Gray shading and black and red boxes and lines are as in Fig. 1. Data for relative clock hour 0900 are replotted for visual purposes only and were not used for any analyses. (Lower) The top five GO biological processes (ranked by P value) associated with the set of proteins within each cluster. Enriched biological processes were nominally statistically significant at the P < 0.05 level.
Overall, proteins that gained or lost 24-h time-of-day patterns when circadian misaligned, such as those in clusters 4, 6, and 7, are likely regulated by an interaction between circadian and behavioral sleep–wake/fasting–food intake cycles. For example, thyroid-stimulating hormone (TSH) from cluster 4 has well-established regulation by sleep and circadian rhythms (42). Specifically, TSH is circadian regulated and peaks during the biological night, whereas sleep has an inhibitory effect that reduces TSH levels (42). Consistent with previous findings (42), here, TSH levels peaked in the biological night during both circadian alignment and misalignment (SI Appendix, Fig. S3A). During circadian alignment, when sleep occurred during the biological night and therefore was expected to blunt the circadian-driven TSH peak, raw TSH levels in relative florescence units (RFU) were ∼10.0% lower versus circadian misalignment with daytime sleep, although this reduction was not statistically significant (P = 0.25). Additionally, the reduction in total sleep time that occurs during circadian misalignment may have also contributed to the gain or loss of 24-h time-of-day patterns and to changes in average 24-h levels.
Proteins Primarily Regulated by the Circadian Clock.
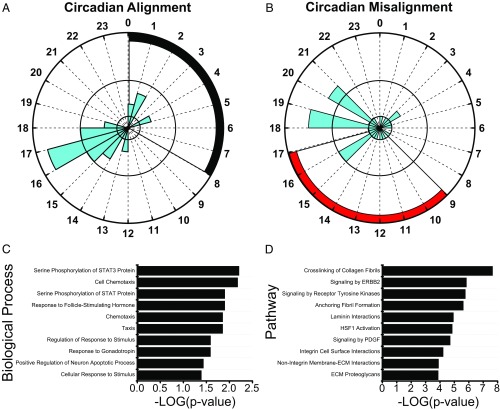
As the master human circadian clock did not adapt rapidly to the simulated nightshift schedule in dim light, strongly circadian-regulated proteins remain, by definition, synchronized to the endogenous master circadian SCN clock. To identify strongly circadian-regulated proteins, we first selected proteins with statistically significant (P < 0.05, 15% FDR level) 24-h time-of-day patterns during both circadian alignment and misalignment days based on the MetaCycle and the mixed-effects models noted above. Fifty-seven proteins met these criteria. Next, we used MetaCycle to estimate the phase of these 57 proteins when circadian aligned and misaligned (SI Appendix). As strongly circadian-regulated proteins remain synchronized to the master circadian SCN clock, given our limited sampling frequency of 4 h, only proteins with less than a 4-h difference in phase estimate (peak abundance) when circadian aligned versus misaligned were defined here as strongly circadian regulated. The difference in phase estimates during circadian alignment versus misalignment is <0.5 h for three proteins, <1.0 h for seven proteins, <2.0 h for 12 proteins, <3.0 h for 21 proteins, and <4.0 h for 30 proteins (Dataset S1). Depending on the difference in phase-estimate cutoff used, up to 30 proteins meet our criteria (Fig. 4 and SI Appendix, Fig. S4) for being strongly circadian regulated. The average phase difference (±SEM) between circadian alignment and misalignment of these 30 strongly circadian-regulated proteins is 2.2 h ± 0.2 h. Of these 30 circadian-regulated proteins, 23 have not previously been identified as being circadian regulated (Dataset S1), and previous findings suggest two of the circadian-regulated proteins, Lamin-B1 (43) and cyclin-dependent kinase-5 (44), interact with the molecular circadian clock and more specifically that cyclin-dependent kinase-5 can phosphorylate the CLOCK protein (44). When circadian aligned, 20 of these 30 circadian-regulated proteins peaked between 1400 h and 1900 h (relative clock hour), and, similarly, when circadian misaligned, 19 of these 30 circadian-regulated proteins peaked between 1400 h and 2100 h (Fig. 4 A and B). However, the phase of these 30 circadian-regulated proteins was more spread out across the day when circadian misaligned than when aligned (Fig. 4 A and B). As noted, the latter analysis is limited by our sampling rate, and future protocols with more frequent sampling rates are needed to verify the circadian regulation of these proteins. GO analyses using the 30 circadian-regulated proteins identified several associated biological processes, including serine phosphorylation of STAT3 protein, taxis, cell chemotaxis, response to follicle-stimulating hormone, regulation of response to stimulus, and response to gonadotropin (Fig. 4C). In general, the biological processes associated with these circadian-regulated proteins do not overlap with specific biological processes associated with proteins that showed either increased or decreased levels (Fig. 2 A and B) or with the clusters of proteins with altered 24-h time-of-day patterns (Fig. 3). The biological process terms positive chemotaxis, macrophage chemotaxis, neutrophil chemotaxis, and positive regulation of monocyte chemotaxis were all associated with proteins whose levels were decreased when circadian misaligned versus aligned (Fig. 2B), and these biological processes are child terms of taxis, chemotaxis, and cell chemotaxis that are associated with the 30 circadian-regulated proteins (Fig. 4C). While these biological processes are related, the terms associated with proteins whose levels decreased are more specific to inflammatory cells or positive chemotaxis, whereas the terms associated with the circadian-regulated proteins are broad taxis terms. Similarly, the biological process terms response to external stimuli and response to external biotic stimuli are associated with proteins in clusters 5 and 7, respectively (Fig. 3 E and G), and these are child terms of cellular response to stimulus that is associated with the 30 circadian-regulated proteins (Fig. 4C). These findings support the concept that some biological processes are likely regulated by an interaction between the circadian and behavioral cycles. Fig. 4D presents the top 10 pathways (ranked by P value) associated with the 30 circadian-regulated proteins. Similar to the biological processes, only one pathway associated with circadian-regulated proteins, signaling by receptor tyrosine kinases, overlapped with pathways associated with altered proteins when circadian misaligned versus aligned. Six of the top 10 pathways are related to the extracellular matrix (ECM): crosslinking of collagen fibrils, anchoring fibril formation, laminin interactions, integrin cell-surface interactions, nonintegrin membrane ECM interactions, and ECM proteoglycans; thus our findings suggest a potential role of the circadian clock in regulating the ECM. Previous findings derived from mouse macrophages also suggest a role of the circadian clock in regulating ECM homeostasis (45), representing a potential link between the circadian clock and the immune system. Outside the top 10 pathways ranked by P value, the 14th pathway associated with circadian-regulated proteins is G2/M DNA damage checkpoint (P = 0.0002), and this is consistent with prior findings suggesting the cell cycle and circadian cycle are coupled (46). Overall, desynchronization between these strongly circadian-regulated proteins and the behavioral cycle may contribute to physiological dysregulation associated with circadian misalignment.
Fig. 4.
Plasma proteins with circadian-regulated 24-h time-of-day patterns as identified with the MetaCycle and mixed-effects models. (A and B) All 30 proteins identified with circadian-regulated time-of-day patterns are represented for both circadian alignment (A) and circadian misalignment (B) days. Black and red shading represents scheduled sleep during circadian alignment and misalignment days, respectively. Data (blue shading) represent the number of proteins with an estimated peak level of abundance for each relative clock hour as determined by MetaCycle analyses. Radial axes range from 0 to 8 proteins with one and four indicated by black concentric circles. (C and D) Biological processes (C) and pathways (D) associated with proteins with strongly circadian-regulated 24-h time-of-day patterns. All 30 proteins identified with strongly circadian-regulated 24-h time-of-day patterns and their established protein interactions were used for pathway analyses. The top 10 biological processes and pathways (ranked by P value) are represented. Enriched biological processes and pathways were nominally statistically significant at the P < 0.05 level.
Targeted Data Analyses on Proteins and Physiological Outcomes Implicated in Metabolic Function.
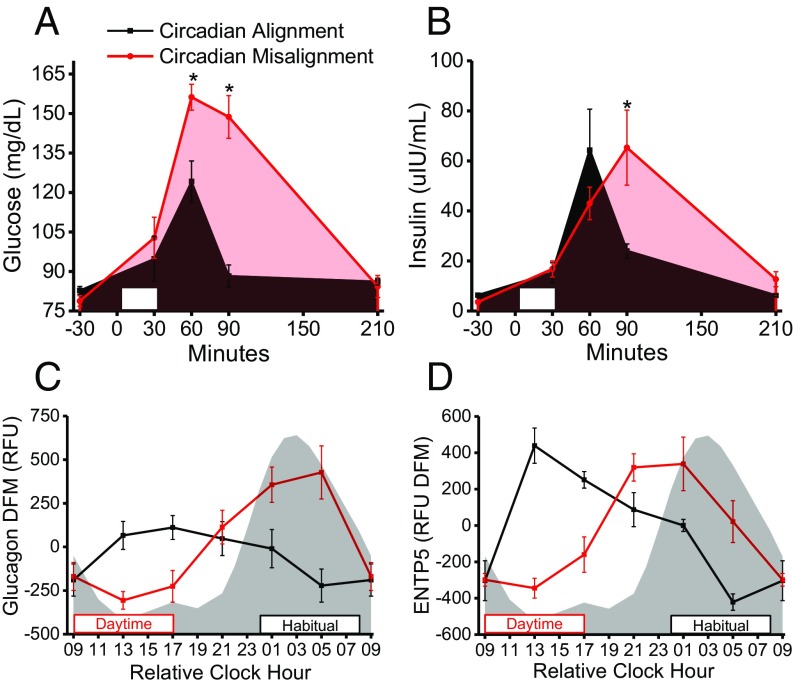
Subjects consumed identical breakfast meals 1.5 h after the scheduled waketime following >10-h fasts on study days 2 and 4 (Fig. 1A). Based on findings from our group (18) and others (19), we hypothesized the postprandial glucose and insulin responses to the breakfast would be elevated during circadian misalignment versus alignment. Before breakfast, fasting plasma glucose and insulin were similar during circadian misalignment versus alignment (Fig. 5 A and B), whereas glucose levels at 60 min and 90 min postbreakfast were ∼26% and ∼68% higher, respectively, (all P < 0.05) when circadian misaligned versus aligned (Fig. 5A). Furthermore, insulin levels were ∼172% higher (P < 0.05) at 90 min postbreakfast during circadian misalignment versus alignment (Fig. 5B). Collectively, these changes suggest that circadian misalignment results in lower glucose tolerance and impaired insulin sensitivity, as is consistent with prior findings of disrupted glucose metabolism during circadian misalignment (16, 47). No statistically significant differences in blood glucose and insulin responses to lunch and dinner meals were detected (SI Appendix, Fig. S5 A–D). Follow-up studies using more sensitive metabolic tests (e.g., glucose tolerance tests and hyperinsulinemic/euglycemic clamps) are needed to examine insulin sensitivity during circadian misalignment.
Fig. 5.
Effects of circadian misalignment on glucose homeostasis and associated proteins (n = 6). (A and B) Postprandial plasma glucose (study day, P = 0.01; study day × time, P = 0.00003; mixed-model ANOVA) (A) and postprandial plasma insulin (study day × time, P = 0.02; mixed-model ANOVA) (B) following fixed breakfast meals. White rectangles represent the time (30 min) allocated to ingest the breakfast meal. (C) Plasma glucagon measured from the proteomics platform. (D) Plasma ENTP5 measured from the proteomics platform. Gray shading and black and red boxes and lines are as in Fig. 1. *P < 0.05 versus circadian alignment at same time point for glucose and insulin (one-tailed t test). Glucagon (EDGE; FDR = 0.002) and ENTP5 (EDGE; FDR = 0.002) have significantly different 24-h time-of-day patterns when circadian misaligned versus aligned. Data are mean ± SEM. The difference from mean (DFM) was calculated for each subject individually using the grand mean of all samples within each individual subject. Data for relative clock hour 0900 are as in Fig. 3.
We next examined proteins involved in glucose metabolism: insulin (SI Appendix, Fig. S5F), glucagon (Fig. 5C), ectonucleoside triphosphate diphosphohydrolase 5 (ENTP5) (Fig. 5D), lipocalin-2 (SI Appendix, Fig. S6A), kallikrein-7 (SI Appendix, Fig. S6B), adiponectin (SI Appendix, Fig. S6C), phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (SI Appendix, Fig. S6D), erythropoietin (SI Appendix, Fig. S6E), FGF receptor 4 (FGFR4) (SI Appendix, Fig. S6F), hepcidin (SI Appendix, Fig. S6G), insulin-like growth factor-1 (SI Appendix, Fig. S2A), and IGFBPs 1–7 (SI Appendix, Fig. S2 B–H).
In response to low blood glucose, pancreatic alpha cells normally secrete glucagon, leading to increased hepatic glucose production. However, excess glucagon is a key factor contributing to diabetes risk (48) and therefore must be tightly regulated. Findings from studies of nocturnal rodents show a day/night rhythm of plasma glucagon with regulation by the master SCN clock and the behavioral food-intake cycle (49). Here, plasma glucagon had significantly different 24-h time-of-day patterns when circadian misaligned versus aligned (Fig. 5C), showing plasma glucagon is strongly regulated by the behavioral cycle in humans. Furthermore, glucagon levels were increased when food intake occurred during the biological night. Related, previous findings show human pancreatic alpha cells express melatonin receptor MT1, and exogenous melatonin stimulates glucagon secretion in perfused human islets (50). Thus, food intake during the biological night when melatonin levels are elevated may contribute to higher plasma glucagon levels and dysregulated glucose metabolism in humans. In addition to directly regulating glucose metabolism, findings from animal studies suggest glucagon regulates hepatic circadian clock timing (10, 51, 52). Thus, in humans the elevated plasma glucagon when eating during the biological night may contribute to misalignment between the master SCN clock and peripheral liver clocks. Collectively, these findings indicate future studies should investigate the impact of circadian misalignment on glucagon regulation in chronic shift workers. Metformin, the drug most widely used to manage type 2 diabetes, targets glucagon regulation of hepatic glucose output (53), and thus if our findings translate to chronic shift workers, future trials investigating the use of metformin in chronic shift workers with risk factors for diabetes may be warranted.
ENTP5 is an endoplasmic reticulum enzyme reportedly up-regulated by protein kinase B (AKT) activation and is a key component of an ATP hydrolysis cycle that elevates AMP levels (54). Previous findings show elevated ENTP5 promotes glycolysis, likely through AMP allosteric activation of the glycolytic enzyme phosphofructokinase (54). Our findings show plasma ENTP5 had significantly different 24-h time-of-day patterns when circadian misaligned versus aligned (Fig. 5D), indicating ENTP5 is strongly regulated by the behavioral cycle. Thus, changes in the 24-h time-of-day pattern of ENTP5 may contribute to altered glucose metabolism during circadian misalignment; investigation in future studies is warranted.
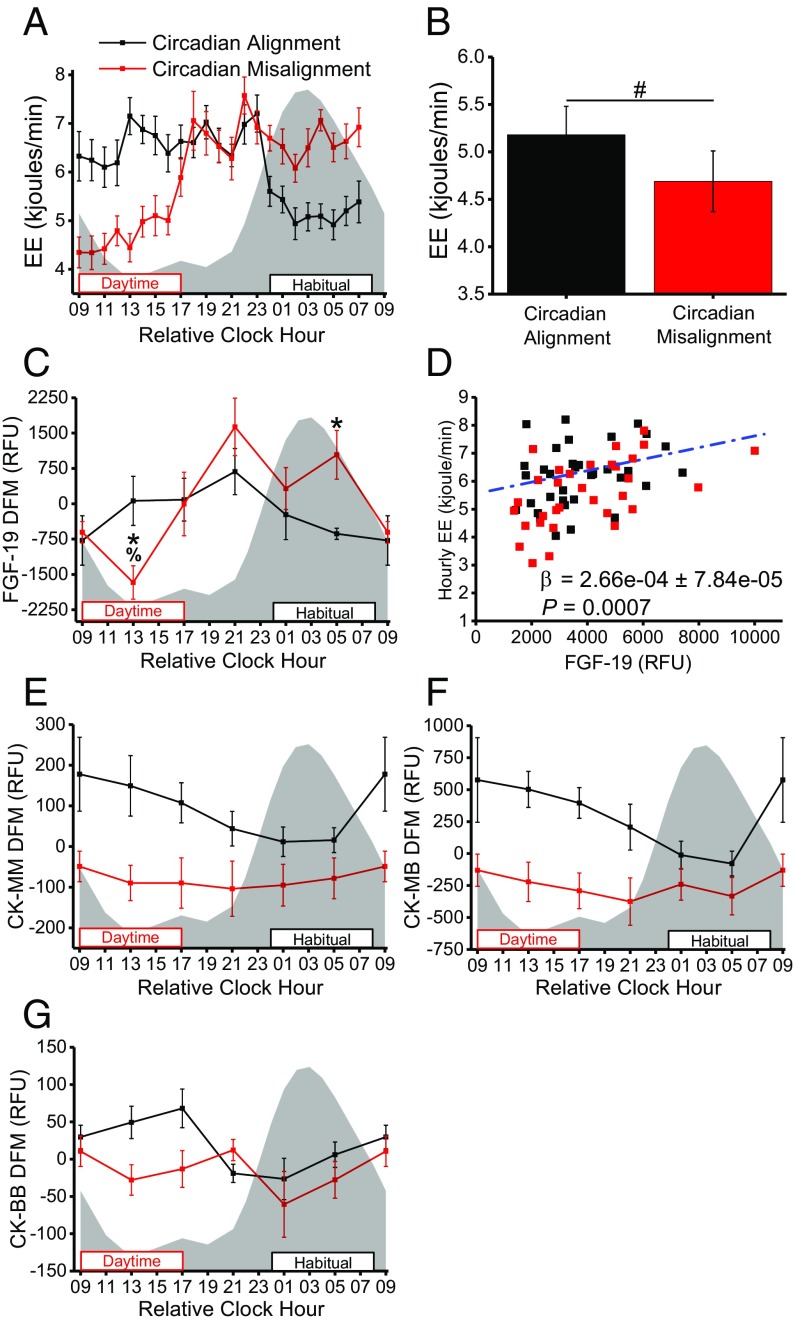
We next examined proteins involved in energy expenditure, as total daily energy expenditure was ∼185 ± 94.9 kJ lower (Fig. 6A) and per-minute sleeping energy expenditure was 10% lower (Fig. 6B) (all P < 0.05) when circadian misaligned versus aligned, as is consistent with our previously reported findings from the larger sample from this study (20). FGF-19 (Fig. 6C) and creatine kinase (CK) M-type (CK-MM) (Fig. 6E), the CK M-B heterodimer (CK-MB) (Fig. 6F), and CK B-type (CK-BB) (Fig. 6G) levels were significantly altered when circadian misaligned. Transgenic mice overexpressing FGF-15, the mouse ortholog of human FGF-19, have elevated energy expenditure, weigh less than their wild-type littermates, and remain lean when fed high-fat diets (55). Furthermore, administration of recombinant FGF-19 increases energy expenditure and protects mice from high-fat diet–induced weight gain (56). We hypothesized that FGF-19 levels would therefore be lower in the current study during sleep versus wakefulness and when sleep occurred during the biological day versus the biological night. Plasma FGF-19 showed significantly different 24-h time-of-day patterns when circadian misaligned versus aligned (Fig. 6C), such that, in parallel to lower daytime sleeping energy expenditure when circadian misaligned (Fig. 6 A and B), plasma FGF-19 was ∼34% lower (P < 0.05) during the approximate midpoint of sleep during circadian misalignment (daytime sleep) versus alignment (nighttime sleep) (Fig. 6C). Additionally, across both circadian alignment and misalignment, higher FGF-19 protein levels were associated (P < 0.001) with higher hourly energy expenditure (Fig. 6D). FGFR4, the primary FGF-19 receptor, levels were similar during circadian misalignment and alignment (SI Appendix, Fig. S6F), suggesting changes in FGF-19 signaling are not due to altered FGFR4 levels. Collectively, these findings suggest altered FGF-19 protein levels may contribute to lower sleeping and total daily energy expenditure during circadian misalignment, elevating the risk of weight gain (20).
Fig. 6.
Altered energy metabolism during circadian misalignment (n = 6). (A) Average per-minute energy expenditure by hour (time, P < 0.00001; study day × time, P < 0.00001; mixed-model ANOVA). (B) Average per-minute energy expenditure during scheduled 8-h sleep opportunities. (C) Plasma FGF-19 measured from the proteomics platform. (D) Association between average hourly energy expenditure (EE) and FGF-19 protein abundance at all proteomics time points across all subjects (linear mixed model). (E) Plasma CK-MM measured from the proteomics platform. (F) Plasma CK-MB measured from the proteomics platform. (G) Plasma CK-BB measured from the proteomics platform. Gray shading and black and red boxes and lines are as in Fig. 1. *P < 0.05 versus circadian alignment at same relative clock hour (one-tailed t test). #P < 0.05 for circadian misalignment versus circadian alignment (two-tailed). %P < 0.05 versus circadian alignment at relative clock hour 0500 (one-tailed t test). FGF-19 (DODR; P = 0.02), CK-MM (DODR; P = 0.004), and CK-MB (DODR; P = 0.04) have significantly different 24-h time-of-day patterns when circadian misaligned versus aligned. Data are mean ± SEM. The DFM was calculated for each subject individually using the grand mean of all samples within each individual subject. Data for relative clock hour 0900 are as in Fig. 3. kjoules, kilojoules.
CK is a critical enzymatic component of the CK/phosphocreatine system responsible for connecting subcellular sites of ATP production (mitochondria and glycolysis) with sites of ATP utilization (ATPases) (57). Our findings show CK-MM and CK-MB had significantly different 24-h time-of-day patterns when circadian misaligned versus aligned (Fig. 6 E and F). Based on results from individual mixed-model ANOVAs for each CK isoform, only CK-BB had a statistically significant (P < 0.05) effect of study day, with a small effect size (η2G = 0.10) (Fig. 6G). Further highlighting the changes in CK during circadian misalignment, creatine metabolic response is the biological process associated with cluster 6 (Fig. 3F). As such, altered levels of the CK isoforms during circadian misalignment may contribute to our finding of reduced energy expenditure. However, unlike FGF-19, across all time points, levels of all CK isoforms analyzed were not associated (all P ≥ 0.60) with hourly energy expenditure. Findings for additional metabolic proteins are provided in SI Appendix, Fig. S6.
Discussion
Our findings identify human plasma proteins that are regulated across the 24-h day by the endogenous circadian SCN clock, by the behavioral sleep–wake/fasting–food intake cycle, and by interactions between the circadian and behavioral cycles. Understanding such regulation is critical for the development of health and disease biomarkers, precision medicine, and biomarkers of circadian misalignment. Our findings also show that acute circadian misalignment, induced by simulated nightshift work, can have rapid and wide-reaching impacts on 24-h time-of-day protein patterns and protein levels. Overall, 62 proteins had altered 24-h average levels, and 76 had altered 24-h time-of-day patterns with 11 proteins overlapping between these two categories, resulting in 127 total proteins being altered during circadian misalignment. Disease processes associated with these altered proteins, such as heart disease and cancer, are commonly found in shift workers (SI Appendix, Fig. S7). Our protocol was designed to simulate shift work and the associated circadian misalignment. Similar to shift workers in the real world, our subjects had less total sleep time during circadian misalignment versus circadian alignment, and thus reduced total sleep time may contribute to our findings. Additionally, as the timing of food intake and the sleep–wake cycle were altered in parallel, we cannot separate the effects of mistimed food intake versus mistimed sleep or interactions between the two on our outcomes.
Circadian misalignment impacted proteins implicated in glucose and energy metabolism and in immune and other basic physiological functions. Previous findings consistently show glucose tolerance decreases across the 24-h day and is under circadian regulation (47, 58). Here, since the endogenous circadian SCN clock did not change across study days, subjects consumed breakfast at a later circadian time during circadian misalignment versus alignment. The majority of cycling metabolic proteins in our analyses showed regulation by the behavioral sleep–wake/fasting–food intake cycle. From an evolutionary perspective, humans developed under conditions of intermittent food availability, and under such conditions it is likely advantageous for our metabolic physiology to adapt rapidly to periods of food intake and fasting (59), as is consistent with our findings showing behavioral cycle regulation of many key metabolic proteins such as glucagon and peptide YY (SI Appendix, Fig. S3B). Alternatively, many of the proteins we identify as strongly circadian regulated are involved in immune and blood coagulation functions. Our findings are consistent with prior human transcriptomics findings suggesting transcripts that are robustly rhythmic and are unaffected by circadian misalignment are related to intrinsic blood-specific functions (28). Importantly, our current findings identify some immune- and inflammatory-related proteins that appear to be regulated by the circadian cycle and some that appear to be regulated by the behavioral cycle. Thus, circadian misalignment disrupts the normal phase relationship in these two groups of immune- and inflammatory-related proteins. Findings are also consistent with the idea that both sleep and circadian rhythms influence immune function (60–62).
Since messenger RNA expression does not always predict protein abundance, our proteomics assessment in humans more closely reflects functional physiological changes during circadian misalignment, building on previous transcriptomics findings (21, 28, 29) and highlighting the need for cross-omics studies. In general, we cannot identify the specific tissue of origin for the plasma proteins we analyzed, as most of the proteins are expressed in multiple tissues. In some cases, such as glucagon being produced in pancreatic alpha cells, we can identify the tissue of origin. It is well established that peripheral tissues have local circadian clocks, and 24-h time-of-day patterns are likely regulated locally within tissues by tissue-specific clocks to varying degrees. Findings from nocturnal rodent models show clocks in peripheral tissues have different rates of re-entrainment to phase shifts (63, 64), with mice requiring 8 d to fully re-entrain to a 6-h phase advance (63). Different rates of re-entrainment between peripheral tissues disrupt the normal phase relationships between tissues during circadian misalignment. Potentially, some of the differential changes in 24-h time-of-day patterns reported here represent desynchrony between clocks in peripheral tissues. Since the endogenous SCN circadian clock did not change during circadian misalignment in our study, strongly behaviorally driven metabolic protein hormones were out of phase with the master SCN clock and potentially were out of phase with proteins regulated by clocks in other peripheral tissues. During shift-work schedules, chronic recurrent internal desynchronization between peripheral tissue clocks and misalignment between the circadian and behavioral cycle likely contribute to the risk of disease.
Our findings have implications for studying the plasma proteome, indicating that future human proteomics studies should account for circadian- and behavioral cycle-regulated time-of-day changes in protein levels. Such knowledge is especially important for developing biomarkers of overall sleep health and circadian misalignment and alignment and for identifying therapeutic targets in the era of precision medicine, especially for shift workers, patients with circadian rhythm sleep–wake disorders, and others with circadian disruption. More specifically, our findings suggest studies sampling at one or a few times per day may miss the peak timing of many proteins and important mechanistic changes underlying physiology, disease risk, or treatment responses. A key strength of our study design is that we utilized a sample of extensively screened and phenotyped healthy subjects in a highly controlled laboratory trial. However, follow-up studies investigating populations with chronic circadian misalignment, including shift workers, those with circadian rhythm sleep–wake disorders, and those with chronic sleep loss (18), targeting a larger number of known proteins as the technology develops, and including men and women, are needed, as the current study included only a small sample of healthy men. A related issue is that our sample size likely results in detecting the largest effects, and thus we may underestimate the number of altered proteins or proteins with 24-h time-of-day patterns. Future trials using protocols such as the constant routine with more frequent sampling rates will be needed to precisely determine the circadian amplitude and phase of the detected circadian-driven proteins reported here. The human circadian system regulates the timing of behavioral and physiological events across the 24-h day. Our findings highlight that the human plasma proteome is impacted by the behavioral and circadian cycles and by interactions between these two processes. These findings suggest that under circadian-entrained conditions when the circadian clock promotes the occurrence of behaviors such as food intake, physical activity, and sleep at optimal internal biological times, the composition of the human plasma proteome promotes healthy physiological function. Overall, greater precision and knowledge focused on the timing of assessing omics health biomarkers is likely to improve our understanding of healthy physiology and disease processes and the specificity of personalized medicine.
Materials and Methods
Subjects.
The current analyses utilized all the male subjects from a larger dataset in which subjects completed a 6-d in-laboratory protocol simulating nightshift work (20). In the larger dataset of 14 subjects, females (n = 8) were studied under different menstrual cycle phases, and thus we chose to study males (n = 6) for this initial analysis. Plasma from six healthy young males aged 26.2 ± 5.6 y (mean ± SD), with a normal body mass index (BMI) (22.5 ± 1.6), percent body fat (21.2 ± 5.1), and fasting blood glucose (82.8 ± 3.7 mg/dL) was analyzed using proteomics. Procedures were approved by the scientific and advisory review committee of the Colorado Clinical and Translational Sciences Institute, the Colorado Multiple Institutional Review Board (IRB), and the University of Colorado Boulder IRB. After providing written informed consent, subjects underwent health screening consisting of medical, psychological, and sleep histories, a semistructured clinical psychiatric interview, a physical examination, a complete blood cell count and a comprehensive metabolic panel, urine toxicology, a 12-lead electrocardiogram, and a polysomnographic sleep disorders screen (20). Based on these tests, subjects were deemed free of medical and psychological disorders. Inclusion criteria were age 18–39 y; BMI 18.5–24.9; habitual nightly sleep duration >7 h and <9.25 h; low to moderate caffeine use (<500 mg/d); average alcohol use (fewer than two drinks per day and fewer than five drinks per week), and nonsmokers (20). Exclusion criteria were current or chronic medical/psychiatric conditions; working a shift-work schedule, dwelling below the Denver altitude (1,600 m above sea level) in the year before study, travel across more than one time zone in the 3 wk before study, recent self-reported weight loss, and a positive urine toxicology screen (20).
Protocol.
Subjects were minimally physically active before the in-laboratory segment of the study to reduce the effects of detraining. For 1 wk before admission to the University of Colorado Hospital Clinical Translational Research Center (CTRC), subjects discontinued use of caffeine, alcohol, nicotine, and over-the-counter medication and maintained a consistent ∼8 h per night sleep schedule based on habitual sleep and circadian timing (20). Sleep timing was verified via wrist actigraphy with light-exposure monitoring (Actiwatch-L; MiniMitter/Respironics), sleep logs, and call-ins to a time-stamped voice recorder to report sleep and wakefulness times. Urine toxicology screen and breath alcohol assessments (model FC10; Lifeloc Technologies) verified the drug- and alcohol-free status of subjects upon CTRC admission. Three days before CTRC admission, exercise was proscribed, and subjects were provided a 3-d outpatient diet designed to meet individual daily caloric needs determined from the resting metabolic rate with a 1.5 activity factor (20), ensuring subjects were in energy balance upon entering the CTRC. All protocol events were scheduled relative to each subject’s habitual sleep and wake times based on the week of prestudy monitoring. The in-laboratory segment for the current analyses included study days 1–4 from the protocol (Fig. 1A) (20). Subjects were provided an 8-h sleep opportunity at habitual bedtime on day 1 for polysomnographic screening to ensure subjects were free from sleep disorders (20). Following scheduled sleep on day 1, subjects were awakened at habitual waketime and maintained a constant posture protocol with brief breaks: seated semirecumbent posture in a hospital bed with the head raised to ∼35°, room temperature maintained at 22–24 °C, and dim lighting (<1 lx in the angle of gaze, <5 lx maximum) during scheduled wakefulness and 0 lx during scheduled sleep. Day 2 served as circadian alignment, with an 8-h nighttime sleep opportunity at habitual bedtime. Day 3 served as transition to nightshift work with subjects awakened at habitual waketime and then provided a 2-h afternoon nap opportunity leading into the simulated nightshift work. Study day 4 served as circadian misalignment with subjects provided an 8-h daytime sleep opportunity beginning 1 h after habitual waketime followed by a second simulated nightshift on day 4. Blood samples were analyzed for proteomic and glucose assessments every 4 h for 24 h on study days 2 (circadian alignment) and 4 (circadian misalignment) with the first blood collection occurring 1 h after habitual waketime (represented as relative clock hour 0800 h in all figures). All blood samples were immediately centrifuged (2,000–3,000 × g); then plasma was collected and subsequently stored at −80 °C until assayed.
Subjects were provided scheduled meals (percent daily caloric intake: 30% breakfast, 30% lunch, 30% dinner, 10% snack) at 1.5, 5.5, 10.5, and 14.5 h postawakening on study days 2 and 4 (20). Meals were identical (total caloric and macronutrient composition) across study days (e.g., the food served for breakfast was the same each day), and subjects were required to consume all food provided.
Circadian Phase Assessment.
Plasma melatonin was initially assessed every 2 h during scheduled wakefulness and every 1 h starting at 13 h after the habitual waketime as a marker of internal circadian timing (20).
Energy Expenditure.
Whole-room indirect calorimetry was used to assess daily energy expenditure on all study days as previously described (20).
Proteomics Platform.
Proteomics were performed by SomaLogic, Inc. utilizing a modified aptamer-based proteomics platform as previously described (65–67). Briefly, each of the 1,129 proteins analyzed has its own binding reagent made from chemically modified ssDNA, termed “modified aptamers” (65). The modified aptamers are fluorescently labeled, allowing quantification. Plasma samples were incubated with the modified aptamers generating protein–aptamer complexes. Unbound aptamers, proteins, and nonspecifically bound proteins were washed away using two bead-based immobilization steps, leaving only aptamer–protein complexes. Aptamers were then eluted from target proteins and directly quantified on an Agilent hybridization array (Agilent Technologies). Calibrators were included so the degree of fluorescence quantitatively represented the relative protein abundance of each protein corresponding to the specific aptamer.
Plasma Glucose.
Plasma glucose was assessed with a YSI 2900 autoanalyzer (Yellow Springs Instrument Co.) at the same time points used for proteomics assessments.
Glucose and Insulin Responses to Test Meals.
On study days 2 (circadian alignment) and 4 (circadian misalignment), subjects consumed identical breakfast, lunch, and dinner meals (total caloric and macronutrient composition). Each meal was ∼55% carbohydrate, 30% fat, and 15% protein. Subjects were provided 30 min to consume the meals (minutes 0–30). To assess glucose (Beckman Coulter) and insulin (Beckman Coulter) responses to test meals, blood was collected 30 min before (−30 min) and at 30 min, 60 min, 90 min, and 210 min following the start of food intake (minute 0). Due to other protocol events, baseline glucose and insulin levels were analyzed 90 min before (−90 min) the start of food intake for the dinner meal on study day 2 (circadian alignment).
Statistics.
The glucose and insulin meal responses and targeted data analyses of specific proteins were analyzed by mixed-model ANOVAs with study day and relative clock hour as fixed factors and subject as a random factor using Statistica version 10.0 (Statsoft). One-tailed a priori directional planned comparison-dependent t tests examined the hypothesized changes in glucose and insulin responses to test meals, FGF-19, and adiponectin. The hypothesized changes in these outcomes were based on both preliminary data and previously published findings as noted in Results. Two-tailed comparisons for other measures without directional hypotheses were calculated. Trapezoidal area under the curve (AUC) analyses were performed using OriginPro version 92E (OriginLab Corporation).
For the proteomic data, all samples were normalized and calibrated using standard hybridization and median normalization procedures defined in the good laboratory practice quality system of SomaLogic, Inc. (68). Hybridization normalization was performed using elution probes on a per-sample basis. Hybridization scale factors are expected to be within the range 0.4–2.5, and all samples passed. Median normalization was applied on a per-sample basis, within each dilution, to control for bulk differences in protein concentration and other potential sources of variation such as pipetting errors. Median normalization scale factors are derived from the overall magnitude of the median signal from each sample and can be indicative of study bias. We also examined median normalization scale factors for bias introduced as a function of time. Based on these analyses, we did not detect any indications of study bias. Scale factors are expected to be within the range of 0.4–2.5, and only one sample failed these criteria. This sample was excluded from all data analyses.
To detect proteins with significantly different average 24-h levels during circadian misalignment versus alignment, we used the nonparametric SAM method using the R package SAMR v2.0 (33) with statistical significance set at the 10% FDR level. See SI Appendix for detailed parameters.
Because not all physiologically derived rhythmicity or time-of-day patterns are expected to fit one single model, we used two independent approaches to analyze proteins for ∼24-h patterns. A linear mixed effects (LME) model was used to determine which proteins have a time-dependent component. LME models are extensions of linear regression models, where the fixed effects terms are the conventional linear regression terms and the random effects are associated with subject-specific variation from individuals drawn at random from a population. LME models were fit for each protein target, treating study day 2 (circadian alignment) and 4 (circadian misalignment) data separately. Each subject was allowed a random intercept to account for subject-specific baseline differences in RFU levels. Mixed models assume the random intercept term follows a Gaussian distribution. To control for violation of this assumption by large baseline differences in RFU signal, the time series for each subject was individually mean-centered using the grand mean (i.e., the entire time series across both days for an individual subject) of all samples from each study day. Change in RFUs as a function of time was modeled using a cubic fit (y = t + t2 + t3), constraining the fit to a maximum of two critical points. Second, we used the meta3d function within the MetaCycle package (37) for R (v3.3.1) to detect rhythmicity in large-scale time-series datasets. Within MetaCycle, we utilized individually mean-centered data and the Lomb–Scargle, JTK, and Arser approaches to detect proteins with significant ∼24-h patterns. P values from the Lomb–Scargle, JTK, and Arser analyses were combined using Fisher’s method (69).
To detect proteins with significantly different 24-h time-of-day patterns between study day 2 (circadian alignment) and 4 (circadian misalignment), we used EDGE and DODR packages for R (v3.3.1) (40, 41). The EDGE analysis incorporates data from each subject individually and amounts to fitting a B-spline to mean-centered data (treating each data point as independent) for each day separately and determining if this is a better fit to the data than a single B-spline fit for both days (see SI Appendix for details). P values are calculated from a simulated null distribution using a novel bootstrapping technique developed and described by Storey et al. (70, 71). Alternatively, the DODR analysis detects differences in phase, amplitude, and signal-to-noise ratios between two datasets. Here, we applied the DODR package to identify proteins with different 24-h time-of-day patterns when circadian misaligned versus aligned (see SI Appendix for details). Since EDGE and DODR analyses use different approaches to detect differential rhythmicity and are likely to identify proteins with different patterns of differential rhythmicity, we applied both approaches to our analyses. Resulting P values from EDGE and DODR analyses were combined using the minP method (38).
For GO disease association and biological process analyses, UNIPROT accession numbers were submitted to DAVID (https://david.ncifcrf.gov/summary.jsp) (34, 35). The background gene set consisted of all 1,129 proteins in our proteomics platform, and default settings within DAVID were used. Pathway analyses were conducted using Reactome (v64) (72, 73). For each pathway analysis, protein interactions with the set of proteins for each analysis were identified using the IntAct database (v4.2.10) (74). Default settings for IntAct were used, and spoke-expanded protein interactions were excluded. All pathway analyses included the set of proteins identified for each analysis and the protein interactors as identified by IntAct. The human proteome was set as the background for all pathway analyses.
Cluster analyses to identify common profile patterns among the 76 proteins with significantly different 24-h time-of-day patterns between circadian alignment and misalignment (Fig. 3 and SI Appendix, Fig. S1) were performed in R (v3.3.1) using Ward hierarchical clustering (see SI Appendix for details) on mean z-scored data. For the cluster analyses, RFU data for each subject and protein were z-scored using the grand mean and SD from all time points on study day 2 (circadian alignment) and study day 4 (circadian misalignment) together. Statistically significant clusters were identified using a multiscale bootstrap resampling approach in the R package pvclust (75). Within pvclust we used 10,000 bootstrap replications to calculate the approximately unbiased P values for all clusters. P values < 0.05 were considered statistically significant. Clusters meeting statistical significance criteria are presented in Fig. 3 as the mean z-score of all proteins within a given cluster at each time-point on each study day.
Linear mixed model analysis using R (v.3.3.1) was used to test the association between protein levels (FGF-19, CK-MM, CK-MB, and CK-BB) and hourly energy expenditure. Hourly energy expenditure was derived from the corresponding hour for each of the proteomics blood-collection time points. Protein levels as RFUs, study day, and time were entered as fixed factors, and subject was entered as a random factor in each model.
Supplementary Material
Acknowledgments
We thank the participants; the University Colorado Boulder Clinical Translational Research Center staff; B. Birks, B. Smith, B. Brainard, B. Griffin, T. Dear, S. Morton, J. Broussard, and G. Wright for study assistance; and Thomas H. Hraha, M.S. for performing mixed-effects models with the cubic time component. This research was supported by NIH Grants DK092624, HL132150, DK111161, TR001082, and DK048520; Sleep Research Society Foundation Grant 011-JP-16; and SomaLogic, Inc. The contents do not represent the views of the US Department of Veterans Affairs or the United States Government.
Footnotes
Conflict of interest statement: E.L.M. has received grants/research support from Philips Inc. and Somalogics, Inc. K.P.W. has received grants/research support from CurAegis Technologies (formerly known as Torvec, Inc.), Philips Inc., and Somalogics, Inc. K.P.W. has received consulting fees or has served as a paid member of the scientific advisory boards for CurAegis Technologies, the NIH, and Circadian Therapeutics. K.P.W. has received speaker honoraria from the American Academy of Sleep Medicine, the American College of Chest Physicians, the American Diabetes Association, The Obesity Society, and Philips, Inc. K.P.W. holds stock options for CurAegis Technologies.
This article is a PNAS Direct Submission.
Data deposition: Raw protein abundance data expressed as relative florescence units have been deposited in Figshare (https://figshare.com/) and are publicly available at the following DOI: 10.6084/m9.figshare.5752650.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714813115/-/DCSupplemental.
References
- 1.Shearman LP, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 2.Lowrey PL, Takahashi JS. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menaker M. Circadian rhythms. Circadian photoreception. Science. 2003;299:213–214. doi: 10.1126/science.1081112. [DOI] [PubMed] [Google Scholar]
- 6.Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sack RL, Blood ML, Lewy AJ. Melatonin rhythms in night shift workers. Sleep. 1992;15:434–441. doi: 10.1093/sleep/15.5.434. [DOI] [PubMed] [Google Scholar]
- 9.Roden M, Koller M, Pirich K, Vierhapper H, Waldhauser F. The circadian melatonin and cortisol secretion pattern in permanent night shift workers. Am J Physiol. 1993;265:R261–R267. doi: 10.1152/ajpregu.1993.265.1.R261. [DOI] [PubMed] [Google Scholar]
- 10.Hatori M, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 2009;17:2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turek FW, et al. Obesity and metabolic syndrome in circadian clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F, et al. Meta-analysis on night shift work and risk of metabolic syndrome. Obes Rev. 2014;15:709–720. doi: 10.1111/obr.12194. [DOI] [PubMed] [Google Scholar]
- 14.Knutsson A. Health disorders of shift workers. Occup Med (Lond) 2003;53:103–108. doi: 10.1093/occmed/kqg048. [DOI] [PubMed] [Google Scholar]
- 15.Depner CM, Stothard ER, Wright KP., Jr Metabolic consequences of sleep and circadian disorders. Curr Diab Rep. 2014;14:507. doi: 10.1007/s11892-014-0507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leproult R, Holmbäck U, Van Cauter E. Circadian misalignment augments markers of insulin resistance and inflammation, independently of sleep loss. Diabetes. 2014;63:1860–1869. doi: 10.2337/db13-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckel RH, et al. Morning circadian misalignment during short sleep duration impacts insulin sensitivity. Curr Biol. 2015;25:3004–3010. doi: 10.1016/j.cub.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 19.Morris CJ, et al. Endogenous circadian system and circadian misalignment impact glucose tolerance via separate mechanisms in humans. Proc Natl Acad Sci USA. 2015;112:E2225–E2234. doi: 10.1073/pnas.1418955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McHill AW, et al. Impact of circadian misalignment on energy metabolism during simulated nightshift work. Proc Natl Acad Sci USA. 2014;111:17302–17307. doi: 10.1073/pnas.1412021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci USA. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 23.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 24.Møller M, Sparre T, Bache N, Roepstorff P, Vorum H. Proteomic analysis of day-night variations in protein levels in the rat pineal gland. Proteomics. 2007;7:2009–2018. doi: 10.1002/pmic.200600963. [DOI] [PubMed] [Google Scholar]
- 25.Deery MJ, et al. Proteomic analysis reveals the role of synaptic vesicle cycling in sustaining the suprachiasmatic circadian clock. Curr Biol. 2009;19:2031–2036. doi: 10.1016/j.cub.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 26.Robles MS, Humphrey SJ, Mann M. Phosphorylation is a central mechanism for circadian control of metabolism and physiology. Cell Metab. 2017;25:118–127. doi: 10.1016/j.cmet.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, et al. Nuclear proteomics uncovers diurnal regulatory landscapes in mouse liver. Cell Metab. 2017;25:102–117. doi: 10.1016/j.cmet.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Archer SN, et al. Mistimed sleep disrupts circadian regulation of the human transcriptome. Proc Natl Acad Sci USA. 2014;111:E682–E691. doi: 10.1073/pnas.1316335111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Möller-Levet CS, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci USA. 2013;110:E1132–E1141. doi: 10.1073/pnas.1217154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malmström E, et al. Large-scale inference of protein tissue origin in gram-positive sepsis plasma using quantitative targeted proteomics. Nat Commun. 2016;7:10261. doi: 10.1038/ncomms10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farrah T, et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol Cell Proteomics. 2011;10:M110.006353. doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright KP, Jr, et al. Entrainment of the human circadian clock to the natural light-dark cycle. Curr Biol. 2013;23:1554–1558. doi: 10.1016/j.cub.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Parkin J, Cohen B. An overview of the immune system. Lancet. 2001;357:1777–1789. doi: 10.1016/S0140-6736(00)04904-7. [DOI] [PubMed] [Google Scholar]
- 37.Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB. MetaCycle: An integrated R package to evaluate periodicity in large scale data. Bioinformatics. 2016;32:3351–3353. doi: 10.1093/bioinformatics/btw405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tseng GC, Ghosh D, Feingold E. Comprehensive literature review and statistical considerations for microarray meta-analysis. Nucleic Acids Res. 2012;40:3785–3799. doi: 10.1093/nar/gkr1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deckard A, Anafi RC, Hogenesch JB, Haase SB, Harer J. Design and analysis of large-scale biological rhythm studies: A comparison of algorithms for detecting periodic signals in biological data. Bioinformatics. 2013;29:3174–3180. doi: 10.1093/bioinformatics/btt541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci USA. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thaben PF, Westermark PO. Differential rhythmicity: Detecting altered rhythmicity in biological data. Bioinformatics. 2016;32:2800–2808. doi: 10.1093/bioinformatics/btw309. [DOI] [PubMed] [Google Scholar]
- 42.Allan JS, Czeisler CA. Persistence of the circadian thyrotropin rhythm under constant conditions and after light-induced shifts of circadian phase. J Clin Endocrinol Metab. 1994;79:508–512. doi: 10.1210/jcem.79.2.8045970. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, Ptáčk LJ, Fu YH. Nuclear envelope regulates the circadian clock. Nucleus. 2015;6:114–117. doi: 10.1080/19491034.2015.1010949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwak Y, et al. Cyclin-dependent kinase 5 (Cdk5) regulates the function of CLOCK protein by direct phosphorylation. J Biol Chem. 2013;288:36878–36889. doi: 10.1074/jbc.M113.494856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keller M, et al. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci USA. 2009;106:21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masri S, Cervantes M, Sassone-Corsi P. The circadian clock and cell cycle: Interconnected biological circuits. Curr Opin Cell Biol. 2013;25:730–734. doi: 10.1016/j.ceb.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Cauter E, et al. Modulation of glucose regulation and insulin secretion by circadian rhythmicity and sleep. J Clin Invest. 1991;88:934–942. doi: 10.1172/JCI115396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee YH, Wang MY, Yu XX, Unger RH. Glucagon is the key factor in the development of diabetes. Diabetologia. 2016;59:1372–1375. doi: 10.1007/s00125-016-3965-9. [DOI] [PubMed] [Google Scholar]
- 49.Ruiter M, et al. The daily rhythm in plasma glucagon concentrations in the rat is modulated by the biological clock and by feeding behavior. Diabetes. 2003;52:1709–1715. doi: 10.2337/diabetes.52.7.1709. [DOI] [PubMed] [Google Scholar]
- 50.Ramracheya RD, et al. Function and expression of melatonin receptors on human pancreatic islets. J Pineal Res. 2008;44:273–279. doi: 10.1111/j.1600-079X.2007.00523.x. [DOI] [PubMed] [Google Scholar]
- 51.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukherji A, Kobiita A, Chambon P. Shifting the feeding of mice to the rest phase creates metabolic alterations, which, on their own, shift the peripheral circadian clocks by 12 hours. Proc Natl Acad Sci USA. 2015;112:E6683–E6690. doi: 10.1073/pnas.1519735112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foretz M, Guigas B, Bertrand L, Pollak M, Viollet B. Metformin: From mechanisms of action to therapies. Cell Metab. 2014;20:953–966. doi: 10.1016/j.cmet.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 54.Fang M, et al. The ER UDPase ENTPD5 promotes protein N-glycosylation, the Warburg effect, and proliferation in the PTEN pathway. Cell. 2010;143:711–724. doi: 10.1016/j.cell.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 55.Tomlinson E, et al. Transgenic mice expressing human fibroblast growth factor-19 display increased metabolic rate and decreased adiposity. Endocrinology. 2002;143:1741–1747. doi: 10.1210/endo.143.5.8850. [DOI] [PubMed] [Google Scholar]
- 56.Fu L, et al. Fibroblast growth factor 19 increases metabolic rate and reverses dietary and leptin-deficient diabetes. Endocrinology. 2004;145:2594–2603. doi: 10.1210/en.2003-1671. [DOI] [PubMed] [Google Scholar]
- 57.Wallimann T, Tokarska-Schlattner M, Schlattner U. The creatine kinase system and pleiotropic effects of creatine. Amino Acids. 2011;40:1271–1296. doi: 10.1007/s00726-011-0877-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee A, Ader M, Bray GA, Bergman RN. Diurnal variation in glucose tolerance. Cyclic suppression of insulin action and insulin secretion in normal-weight, but not obese, subjects. Diabetes. 1992;41:750–759. doi: 10.2337/diab.41.6.750. [DOI] [PubMed] [Google Scholar]
- 59.Mattson MP, et al. Meal frequency and timing in health and disease. Proc Natl Acad Sci USA. 2014;111:16647–16653. doi: 10.1073/pnas.1413965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Irwin MR, Opp MR. Sleep health: Reciprocal regulation of sleep and innate immunity. Neuropsychopharmacology. 2017;42:129–155. doi: 10.1038/npp.2016.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ringgold KM, Barf RP, George A, Sutton BC, Opp MR. Prolonged sleep fragmentation of mice exacerbates febrile responses to lipopolysaccharide. J Neurosci Methods. 2013;219:104–112. doi: 10.1016/j.jneumeth.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edgar RS, et al. Cell autonomous regulation of herpes and influenza virus infection by the circadian clock. Proc Natl Acad Sci USA. 2016;113:10085–10090. doi: 10.1073/pnas.1601895113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davidson AJ, Castanon-Cervantes O, Leise TL, Molyneux PC, Harrington ME. Visualizing jet lag in the mouse suprachiasmatic nucleus and peripheral circadian timing system. Eur J Neurosci. 2009;29:171–180. doi: 10.1111/j.1460-9568.2008.06534.x. [DOI] [PubMed] [Google Scholar]
- 64.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gold L, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vaught JD, et al. Expanding the chemistry of DNA for in vitro selection. J Am Chem Soc. 2010;132:4141–4151. doi: 10.1021/ja908035g. [DOI] [PubMed] [Google Scholar]
- 67.Brody E, et al. Life’s simple measures: Unlocking the proteome. J Mol Biol. 2012;422:595–606. doi: 10.1016/j.jmb.2012.06.021. [DOI] [PubMed] [Google Scholar]
- 68.SomaLogic Inc 2016 SOMAscan proteomic assay technical white paper. Available at: somalogic.com/wp-content/uploads/2017/06/SSM-002-Technical-White-Paper_010916_LSM1.pdf. Accessed February 1, 2018.
- 69.Fisher R. Statistical Methods for Research Workers. Oliver and Boyd; Edinburgh: 1925. [Google Scholar]
- 70.Storey JD, Dai JY, Leek JT. The optimal discovery procedure for large-scale significance testing, with applications to comparative microarray experiments. Biostatistics. 2007;8:414–432. doi: 10.1093/biostatistics/kxl019. [DOI] [PubMed] [Google Scholar]
- 71.Woo S, Leek JT, Storey JD. A computationally efficient modular optimal discovery procedure. Bioinformatics. 2011;27:509–515. doi: 10.1093/bioinformatics/btq701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Milacic M, et al. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers (Basel) 2012;4:1180–1211. doi: 10.3390/cancers4041180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orchard S, et al. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suzuki R, Shimodaira H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.