Significance
The Bromo- and Extra-Terminal (BET) family proteins regulate transcription of several oncogenes. For this reason, targeting BET protein may be a promising strategy for cancer therapy. Checkpoint inhibitors, such as anti–CTLA-4, sustain cytotoxic T cells in their anticancer activity. This paper develops a mathematical model to determine the efficacy of combination therapy with BET and CTLA-4 inhibitors. Simulations of the model are in agreement with experimental results. It is shown that the two drugs are positively correlated in reducing the tumor volume. The model also shows, under different combinations of doses that lead to the same tumor volume reduction, in what proportion to choose the two drugs to minimize the increased level of the proinflammatory TNF-.
Keywords: BET inhibitor, anti–PD-L1, combination therapy, mathematical model
Abstract
CTLA-4 is an immune checkpoint expressed on active anticancer T cells. When it combines with its ligand B7 on dendritic cells, it inhibits the activity of the T cells. The Bromo- and Extra-Terminal (BET) protein family includes proteins that regulate the expression of key oncogenes and antiapoptotic proteins. BET inhibitor (BETi) has been shown to reduce the expression of MYC by suppressing its transcription factors and to down-regulate the hypoxic transcriptome response to VEGF-A. This paper develops a mathematical model of the treatment of cancer by combination therapy of BETi and CTLA-4 inhibitor. The model shows that the two drugs are positively correlated in the sense that the tumor volume decreases as the dose of each of the drugs is increased. The model also considers the effect of the combined therapy on levels of myeloid-derived suppressor cells (MDSCs) and the overexpression of TNF-, which may predict gastrointestinal side effects of the combination.
Lysine (lys) acetylation is a widespread regulatory posttranslational modification of proteins. In particular, it is involved in regulating transcription and cell signaling (1). Bromodomains (BRDs) are protein–protein interaction modules that selectively recognize and bind to acetylated lys residues on histones and transcription factors and thereby have important roles in the regulation of gene expression. BRD-containing proteins are frequently dysregulated in cancer. Many of these proteins are oncogenes, and mutations in BRD proteins have been shown to be present in malignant cells (2).
The Bromo- and Extra-Terminal (BET) family proteins include proteins that share the feature of containing two BRDs and an extraterminal domain. The BET family proteins perform transcriptional regulatory functions under normal conditions, but in cancer they regulate transcription of several oncogenes, including c-Myc and Bcl-2 (3). For this reason, targeting BET proteins may be a promising strategy for cancer therapy. Indeed, there are currently several clinical trials with BET inhibitors (BETis) (4, 5). The mechanism underlying the therapeutic process of BETi was explained in ref. 6 and, with emphasis on breast cancer, in ref. 7. Recent experiments show that BETi (e.g., JQ1) may also have a beneficial effect in treating T-cell–mediated inflammatory diseases (8, 9).
In the context of cancer, BETi (e.g., JQ1) has been shown to (i) reduce the expression of MYC by suppressing its transcription factors (3, 10), (ii) significantly down-regulate the hypoxic transcriptome response to VEGF-A (7, 11), and (iii) reduce production of cytokine TNF-, which impairs macrophage inflammatory response (12).
BETi acts as an anticancer agent by blocking VEGF production and by down-regulating MYC expression (and hence cancer cell proliferation). Although BETi down-regulates production of TNF- by M1 macrophages, the total effect of BETi is anticancer. It suppresses tumor growth in breast cancer cell lines (13, 14), in acute myeloid leukemia (15), in myeloma (16, 17), in pancreatic ductal adenocarcinoma (18), and in nuclear protein of the testis (NUT) midline carcinoma (19).
Immune checkpoints are regulatory pathways in the immune system that inhibit its active response against specific targets. Under normal physiological conditions, immune checkpoints are crucial for maintenance of self-tolerance (e.g., prevention of autoimmune diseases) and for protecting tissue from damage following immune system response to pathogenic infection (20). Engagement of the immune checkpoint on antitumor T cells leads to dampening of anticancer immune response and results in the “exhausted” T-cell phenotype. CTLA-4 is an immune inhibitor surface protein expressed on activated T cells; when it combines with its receptor B7 on dendritic cells, the complex CTLA-4–B7 acts as a checkpoint on antitumor T cells (21, 22). There has been much progress in recent years in developing checkpoint inhibitors, primarily anti–CTLA-4 (23, 24), anti–PD-1, and anti–PD-L1 neutralizing antibodies (25).
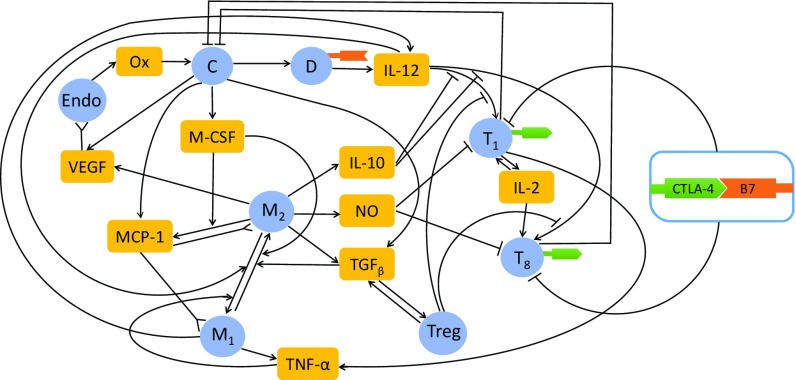
It was recently shown that BETi inhibits expression of the immune checkpoint ligand PD-L1 and that combining anti–PD-1 with BETi (JQ1) leads to higher antitumor responses compared with each drug given alone (2). In this paper, we use a mathematical model to address the efficacy of combination of BETi (e.g., JQ1) and checkpoint inhibitor. We focus on anti–CTLA-4 (e.g., ipilimumab), but the same method, with some changes, can be applied to anti–PD-1 and anti–PD-L1 (see SI Text). The model includes several types of T cells, macrophages and dendritic cells, endothelial cells and cancer cells, as well as signaling molecules involved in the cross-talk among these cells. The interactions among these cells and molecules are schematically described in Fig. 1, as explained below.
Fig. 1.
Interaction of immune cells with cancer cells. Sharp arrows indicate proliferation/activation, blocked arrows indicate killing/blocking, and inverted sharp arrows indicate recruitment/chemoattraction. C, cancer cells; D, dentritic cells; Endo, endothelial cells; , M1 macrophages; , M2 macrophages and MDSCs; NO, Nitric Oxide; Ox, oxygen from the blood; , CD Th1 cells; , CD T cells; Treg, T regulatory cells. and T cells express CTLA-4; dendritic cells express B7.
Dendritic cell-derived IL-12 activates effector T cells (CD Th1 and CD lymphocytes cells) (26, 27). Th1 produces IL-2, which further promotes proliferation of effector T cells. Effector T cells kill cancer cells, while cancer cells suppress the functions of effector T cells by producing immunosuppressive cytokines TGF- and IL-10 (28). IL-10 inhibits the activation of CD Th1 and CD T cells (28). TGF- activates T regulatory (Treg) cells, which inhibit Th1 and CD T cells (29, 30), thus promoting tumor growth. Treg cells produce TGF- (29). Cancer cells also produce monocyte chemotactic protein 1 (MCP-1), macrophage colony-stimulating factor 1 (M-CSF), and vascular endothelial growth factor (VEGF). VEGF recruits endothelial cells to promote angiogenesis by forming new blood vessels in the tumor microenvironment that provide oxygen to the cancer cells (31, 32). MCP-1 and M-CSF recruit macrophages into the tumor (30, 33–36). Myeloid-derived suppressor cells (MDSCs) are also attracted into the tumor microenvironment (33, 37). Macrophages are polarized into two phenotypes, M1 and M2 (38). M1 macrophages change to the M2 phenotype under the influence of TGF- and M-CSF, while M2 macrophages change to M1 phenotypes under the influence of IL-12 and TNF- (32, 34). We consider here only the monocytic subsets of MDSCs (excluding the granulocytic MDSCs, which have different immunosuppressive function). Since the phenotype of these MDSCs is similar to that of M2 macrophages, we shall combine both types of cells and view them as M2 cells. M1 macrophages as well as Th1 cells produce TNF- (12, 39, 40). M2 macrophages produce TGF-, IL-10, nitric oxide (NO) and arginase 1, and VEGF (37, 41). NO and arginase 1 suppress the function of Th1 and CD T cells (37, 41).
One of the checkpoints on T cells is the membrane protein CTLA-4. Its receptor B7 is expressed on activated dendritic cells (42). The complex CTLA-4–B7 inhibits the function of effector T cells (43). CTLA-4 is also constitutionally expressed on Treg cells, but it plays a different role than in Th1 cells and CD T cells (44, 45). CTLA-4 on Treg cells promotes cancer by forming a complex with B7 on dendritic cells, thereby decreasing the source of B7 available for modulation of Th1 cells and CD T cells.
The mathematical model developed in the present paper is based on Fig. 1. We simulate the model in the control case (no drugs) and in treatment with anti–CTLA-4 and BETi. It is shown that the two drugs are positively correlated in the sense that tumor volume decreases as each of the drugs is increased. We use the model also to determine one negative side effect of the combined therapy—namely, gastrointestinal disorder—which we represent by the increase in the level of TNF-. It is shown that CTLA-4 inhibition increases the level of TNF- at a larger rate than BETi.
Mathematical models on cancer vaccine combined with another agent were introduced in refs. 46–48 and, more recently, with checkpoint inhibitor in ref. 49. These papers considered, as a second drug in the combination, chemotherapy (47), anti–TGF- (48), anti–PD-1 (49), or a nonspecific immunotherapy (46). Refs. 46–48 used ordinary differential equation models, while ref. 49 used a partial differential equation model. In the present paper, we consider the efficacy of combination therapy with BETi and anti–CTLA-4 by a partial differential equation model, focusing on the cross-talk among these anticancer agents, cancer cells and the immune cells, and signaling molecules in the tumor microenvironment.
Mathematical Model
The mathematical model is based on the network shown in Fig. 1 and includes also BET and CTLA-4 inhibitors. Since the only function of NO and arginase in the mathematical model is in the suppression of Th1 and CD T cells, for simplicity we shall use NO to represent both NO and arginase.
We assume that the total density of cells within the tumor remains constant in space and time:
| [1] |
Equation for DCs ().
The dynamics of DCs is given by
| [2] |
where the first term on the right-hand side (RHS) of the equation represents indirect activation by necrotic cancer cells (), and is the diffusion coefficient (see SI Text for more details).
Equations for Th1 Cells () and CD T Cells ().
Naive CD T cells differentiate into active Th1 cells () under the IL-12 () environment (26, 27), while IL-10 () and Treg cells () inhibit the differentiation of naive CD T cells into Th1 cells (28, 29). The proliferation of activated Th1 cells is enhanced by IL-2 (). Both processes of activation and proliferation of Th1 cells are assumed to be inhibited by the complex CTLA-4–B7 () by a factor and enhanced by BETi () since BET inhibition promotes Th1 cell activation by suppressing PD-L1 (2, 50). Hence, Th1 satisfies the following equation:
| [3] |
where is the density of the naive CD T cells. Similarly,
| [4] |
where is the density of the inactive CD T cells.
Equation for Activated Treg Cells ().
The production of is induced by TGF- () (29, 30). BETi attenuates Treg cells’ suppressive function (51). Hence,
| [5] |
Equations for M1 Macrophages () and M2 Macrophages ().
The equation for M1 macrophages takes the following form:
| [6] |
where we used the notation if , and if ; the first term on the RHS represents a source of macrophages differentiated from monocytes that are activated by MPC-1 () (34); the third and fourth terms on the RHS represent phenotype changes from M1 to M2 under the influence of TGF- and M-CSF (), and M2 to M1 under the influence of IL-12 and TNF- () (32, 34). Similarly,
| [7] |
Equation for Endothelial Cells ().
Endothelial cells are chemoattracted by VEGF, and their proliferation is increased by VEGF (31, 32). Hence,
| [8] |
where is a threshold below which the proliferation of does not occur (52).
Equation for Tumor Cells ().
We assume a logistic growth for cancer cells with carrying capacity () to account for competition for space among these cells. The proliferation rate depends on the density of oxygen () (32). BETi suppresses MYC (10), which reduces proliferation of tumor cells. Cancer cells are killed by Th1 and CD T cells, and the killing rate is resisted by NO (and arginase 1) (37, 41). The equation for takes the form:
| [9] |
where and and are the killing rates of cancer cells by Th1 and CD T cells, is the natural death rate of cancer cells, and and represent the resistance by BETi and NO, respectively.
The equations for all cytokines are given in SI Text.
Equation for Anti–CTLA-4 ().
Anti–CTLA-4 () is injected intraperitoneally twice weekly for 20 d in mouse experiments (see SI Text). For simplicity, we assume that the level of the drug is constant, so that it provides a constant source of anti–CTLA-4, and we denote by the effective level of the source. The drug is degraded at a rate and is depleted in the process of blocking CTLA-4 (). Hence,
| [10] |
Equation for BETi ().
BETi is administered by oral gavage once daily for 20 d as in mouse experiments (see SI Text), providing a constant source of BETi. We denote by the effective level of the source, BETi, absorbed by the cancer cells and M1 macrophages (12). We represent these absorption rates by and and hence obtain the following equation for :
| [11] |
To simplify the computations, we assume that the tumor is spherical and denote its radius by . We also assume that all of the densities and concentrations are radially symmetric—that is, functions of , where . In particular, , where is the unit radial vector.
Equation for Free Boundary.
We assume that the free boundary moves with the velocity of cells, so that
| [12] |
Results
The simulations of the model were performed by Matlab based on the moving mesh method for solving partial differential equations with free boundary (53). The computational method, parameter estimation, and sensitivity analysis are given in SI Text.
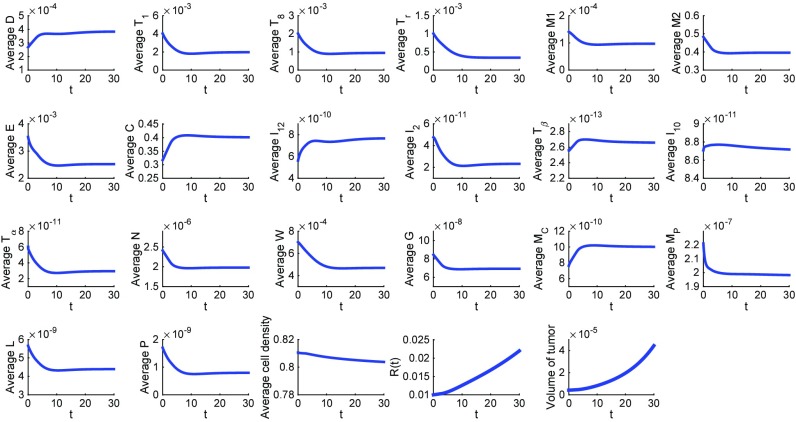
Fig. 2 is a simulation of the model with no drugs (the control case) for the first 30 d; the average densities and concentrations of all species are their total mass in the tumor divided by the tumor volume. The simulations show consistency in the choice of the model parameters. Indeed, as can be quickly checked, the steady states of all of the cytokines and cells are approximately equal to the values that were assumed in estimating the parameters of the model in SI Text.
Fig. 2.
Average densities/concentrations of all of the variables in the model in the control case (no drugs). All parameter values are the same as in Tables S2–S4.
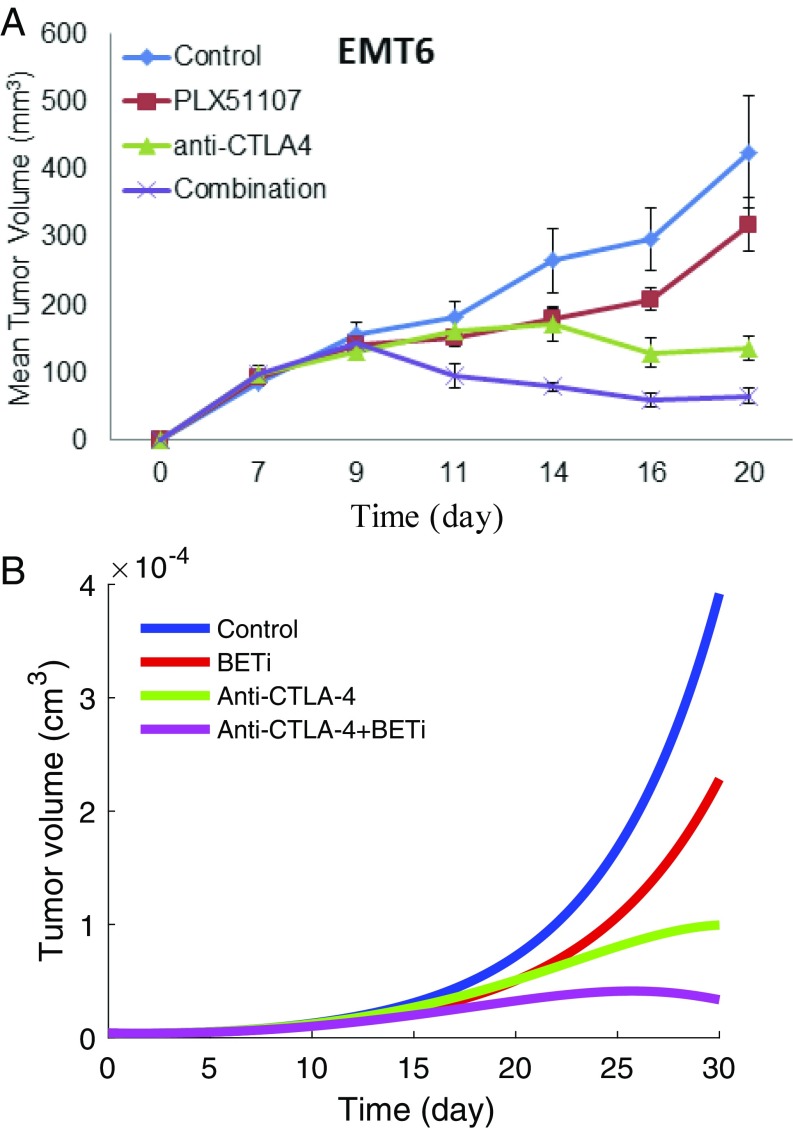
Fig. 3A shows the tumor volume growth in mice experiments, where BETi is administered by oral gavage at 20 mg/kg once daily and CTLA-4 antibody is injected at 100 g twice weekly; the details of the experiment are given in SI Text. The mouse experiment was approved by the Institutional Review Board of Ohio State University. In the mathematical model, the administration of BETi () and anti–CTLA-4 () should be proportional to the amount of drugs in the mice experiments; however, the proportionality coefficients are not known. We determined the order of magnitude of and so that the growth or decrease of the tumor volume in the simulations will be in qualitative agreement with the mice experiments. Fig. 3B, with and , shows that BETi and anti–CTLA-4 as single agents reduce tumor volume, and in combination, the reduction increases to more than 75% at day 30, in agreement with our experimental results in Fig. 3A.
Fig. 3.
The growth of tumor volume during the administration of anti–CTLA-4 antibody and BETi. (A) Mouse experiment results (see SI Text for details). (B) Numerical simulation result. Anti–CTLA-4 is administered at the rate , and BETi is administered at the rate . All other parameter values are the same as in Tables S2–S4.
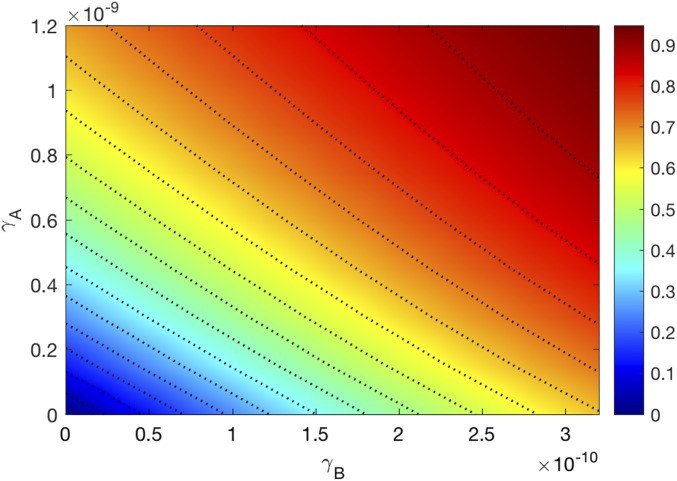
We next consider the efficacy of the combination therapy for a range of values of BETi and anti–CTLA-4. We define the efficacy of a combination therapy, with , by the formula
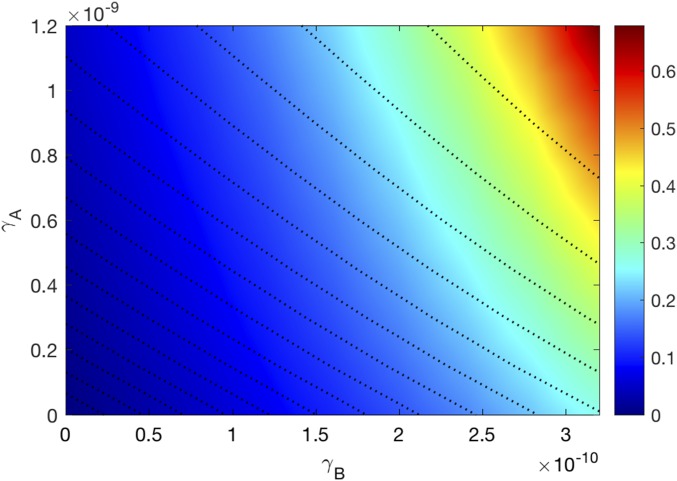
where the tumor volume is computed at day 30; is the tumor volume at day 30 in the control case. The efficacy values lie between 0 and 1 (or between 0% and 100%). Fig. 4 is the efficacy map of the combined therapy, with in the range of and in the range of . The color column shows the efficacy for any pair of ; the maximum efficacy is 0.98 (98%). We see that the two drugs are positively correlated in the sense that tumor volume decreases as each of the drugs is increased.
Fig. 4.
Drug efficacy map. The color column shows the efficacy when varies and varies . All other parameter values are the same as in Tables S2–S4.
From Fig. 4 we can draw, for any given level of efficacy, a curve in the plane consisting of the pairs , which yield the same efficacy level . We consider the following question: Which is the best choice of , on the curve , given a specific additional aim of the treatment. One such aim could be the reduction in the inflammation associated with TNF-. Proceeding with this example, we denote by the average concentration of TNF- at day 30 under combined therapy with anti–CTLA-4 () and BETi () and introduce the function
measures the relative overexpression of the TNF- at day 30: the color column in Fig. 5 shows the relative overexpression of TNF- for any pair . We see that is a decreasing function of . Hence, given the aim of minimizing TNF-, the best choice of for a given efficacy is the pair with the smallest .
Fig. 5.
Average density of TNF-. The color column shows the adverse effect function when varies and varies . All other parameter values are the same as in Tables S2–S4.
Few potential negative side effects of BETi have been identified: memory lost in mice (54), and suppression of chondrocyte differentiation and restrained bone growth (55). However, because of the multitude of the BRD proteins, many more adverse effects may turn out in clinical trials (5, 56).
Checkpoint inhibitors cause a whole range of autoimmune effects, including pneumonitis, rashes, vitiligo, hepatitis, and adrenal insufficiency (57). They also cause gastrointestinal disorder. For instance, among patients receiving anti–CTLA-4, 30% of patients develop diarrhea (54, 57), and 10% develop severe diarrhea due to autoimmune colitis. High level of TNF- is associated with colitis, and anti–TNF- blockade is a strategy used to suppress the disease (58). We may view a high level of TNF- as representing the adverse gastrointestinal reaction to treatment with checkpoint inhibitors.
Conclusion
Under normal healthy conditions, the BET family proteins perform transcriptional regulatory functions, but in cancer they regulate transcription of several oncogenes, including c-Myc (3). BETi has been shown to reduce the expression of MYC by suppressing its transcription factors (3, 10). BETi acts as an anticancer agent also by down-regulating the hypoxic transcriptome response of VEGF-A (7, 11) and by suppressing PD-L1 expression (2, 50). BET also participates in transcription of NF-B target genes, which often encode inhibitory cytokines, and BETi could decrease expression of these genes as well (59, 60). For these reasons, it was suggested that targeting BET proteins may be a promising strategy for cancer therapy. Indeed, several BETis are currently in clinical development for several cancer types (61). We have recently shown in preclinical in vivo experiments using immunocompetent tumor-bearing mouse models that combining anti–CTLA-4 with BETi increases the anticancer responses greater than either drug alone (see Fig. 3A). We have also shown theoretically that BETi as a single agent decreased the ratio of MDSC to CD cells and experimentally that anti–CTLA-4 as a single agent also decreased the ratio of MDSC to CD (see Figs. S1 and S2). The last result is in agreement with mice experiments (62) that show that anti-CTLA decreased the ratio of MDSC to CD T cells.
In the present paper, we developed a mathematical model to compute the efficacy of combination of BETi (e.g., JQ1) and anti–CTLA-4 (e.g., ipilimumab). The model is represented by a system of partial differential equations within the tumor tissue. The model includes immune cells [Th1 and CD T cells, Treg cells, M1 and M2 macrophages (or monocytic MDSCs), and dendritic cells] and endothelial cells, cytokines (IL-12, IL-2, IL-10, TGF-, and TNF-), NO, oxygen, VEGF, M-CSF, MCP-1, CTLA-4, B7, and the complex CTLA-4–B7. We simulated the model with a combination of drugs, BETi at the “level” and anti–CTLA-4 antibody at the level , and computed the tumor volume at day 30. We showed that the model simulations are in agreement with our mice experiments, both in terms of the tumor volume (see Fig. 3) and the ratio of MDSC to CD cells (see SI Text). We introduced the efficacy function, , an expression that quantifies the relative reduction in tumor volume compared with the control case (no drugs).
The efficacy map in Fig. 4 shows that for any levels of and , the two drugs are positively correlated in the sense that the tumor volume decreases as each of the drugs is increased.
Given any efficacy value , one can mark a curve in the plane of Fig. 5 such that each pair on yields the same efficacy . We can therefore try to choose a specific pair on that satisfies an additional aim of the treatment. If one could quantify all of the negative side effects collectively by a single function , then the aim of achieving efficacy with the least negative side effects would be to choose , on the equi-efficacy curve , for which attains its minimum. The construction of such a function is beyond the scope of present paper, but this concept was illustrated by considering the function as representing one of the adverse effects that one could aim to minimize. The adverse effect map in Fig. 5 shows that the pair on with the smallest is the optimal choice.
In this paper, we did not consider the imperfect specificity/selectivity of small-molecule inhibitors and how this may affect signaling of and the interaction between the two therapies in the combination. These issues may limit the validity of the results. Furthermore, since the model verification is based on experiments in the murine tumor model, its application to human cancer therapy will need further explanation, both preclinically and clinically.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants P01 CA95426 and K24 CA93670 (to W.E.C.), T32 CA90338-27, P30 CA016058; Ohio State University Comprehensive Cancer Center Translational Therapeutics Award TT142; National Natural Science Foundation of China Grant 11501568; Renmin University of China; and the Mathematical Biosciences Institute, Ohio State University.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721559115/-/DCSupplemental.
References
- 1.Fujisawa T, Filippakopoulos P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat Rev Mol Cell Biol. 2017;18:246–262. doi: 10.1038/nrm.2016.143. [DOI] [PubMed] [Google Scholar]
- 2.Hogg SJ, et al. Bet-bromodomain inhibitors engage the host immune system and regulate expression of the immune checkpoint ligand pd-l1. Cell Rep. 2017;18:2162–2174. doi: 10.1016/j.celrep.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu LL, et al. Inhibition of bet bromodomains as a therapeutic strategy for cancer drug discovery. Oncotarget. 2015;6:5501–5516. doi: 10.18632/oncotarget.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kagoya Y, et al. Bet bromodomain inhibition enhances t cell persistence and function in adoptive immunotherapy models. J Clin Invest. 2016;126:3479–3494. doi: 10.1172/JCI86437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrieu G, Belkina AC, Denis GV. Clinical trials for bet inhibitors run ahead of the science. Drug Discov Today Tech. 2016;19:45–50. doi: 10.1016/j.ddtec.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Junwei S, Vakoc CR. The mechanisms behind the therapeutic activity of bet bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shu S, Polyak K. Bet bromodomain proteins as cancer therapeutic targets. Cold Spring Harb Symp Quant Biol. 2017;81:123–129. doi: 10.1101/sqb.2016.81.030908. [DOI] [PubMed] [Google Scholar]
- 8.Toniolo PA, et al. Inhibiting stat5 by the bet bromodomain inhibitor jq1 disrupts human dendritic cell maturation. J Immunol. 2015;194:3180–3190. doi: 10.4049/jimmunol.1401635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schilderink R, et al. Bet bromodomain inhibition reduces maturation and enhances tolerogenic properties of human and mouse dendritic cells. Mol Immunol. 2016;79:66–76. doi: 10.1016/j.molimm.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 10.Wadhwa E, Nicolaides T. Bromodomain inhibitor review: Bromodomain and extra-terminal family protein inhibitors as a potential new therapy in central nervous system tumors. Cureus. 2016;8:e620. doi: 10.7759/cureus.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Motta LL, et al. The bet inhibitor JQ1 selectively impairs tumour response to hypoxia and downregulates CA9 and angiogenesis in triple negative breast cancer. Oncogene. 2017;36:122–132. doi: 10.1038/onc.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belkina AC, Nikolajczyk BS, Denis GV. Bet protein function is required for inflammation: Brd2 genetic disruption and bet inhibitor jq1 impair mouse macrophage inflammatory responses. J Immunol. 2013;190:3670–3678. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi J, et al. Disrupting the interaction of brd4 with diacetylated twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;2:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai L, et al. Targeted degradation of bet proteins in triple-negative breast cancer. Cancer Res. 2017;77:2476–2487. doi: 10.1158/0008-5472.CAN-16-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mertz JA, et al. Targeting myc dependence in cancer by inhibiting bet bromodomains. Proc Natl Acad Sci USA. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aird F, Kandela I, Mantis C, Biology RPC. Replication study: Bet bromodomain inhibition as a therapeutic strategy to target c-myc. eLife. 2017;6:e21253. doi: 10.7554/eLife.21253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delmore JE, et al. Bet bromodomain inhibition as a therapeutic strategy to target c-myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bian B, et al. Gene expression profiling of patient-derived pancreatic cancer xenografts predicts sensitivity to the bet bromodomain inhibitor jq1: Implications for individualized medicine efforts. EMBO Mol Med. 2017;9:482–497. doi: 10.15252/emmm.201606975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.French CA. Nut midline carcinoma. Cancer Genet Cytogenet. 2010;203:16–20. doi: 10.1016/j.cancergencyto.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchbinder EI, Desai A. Ctla-4 and pd-1 pathways: Similarities, differences, and implications of their inhibition. Am J Clin Oncol. 2016;39:98–106. doi: 10.1097/COC.0000000000000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol. 2015;33:1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blank CU, Enk A. Therapeutic use of anti-CTLA-4 antibodies. Int Immunol. 2015;27:3–10. doi: 10.1093/intimm/dxu076. [DOI] [PubMed] [Google Scholar]
- 24.Funt SA, Page DB, Wolchok JD, Postow MA. Ctla-4 antibodies: New directions, new combinations. Oncol (Williston Park) Suppl. 2014;3:6–14. [PubMed] [Google Scholar]
- 25.He J, Hu Y, Hu M, Li B. Development of pd-1/pd-l1 pathway in tumor immune microenvironment and treatment for non-small cell lung cancer. SciRep. 2015;5:13110. doi: 10.1038/srep13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janco JMT, Lamichhane P, Karyampudi L, Knutson KL. Tumor-infiltrating dendritic cells in cancer pathogenesis. J Immunol. Apr 2015;194:2985–2991. doi: 10.4049/jimmunol.1403134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y, Shurin1 GV, Peiyuan Z, Shurin MR. Dendritic cells in the cancer microenvironment. J Cancer. 2013;4:36–44. doi: 10.7150/jca.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perrot CY, Javelaud D, Mauviel A. Insights into the transforming growth factor-beta signaling pathway in cutaneous melanoma. Ann Dermatol. 2013;25:135–144. doi: 10.5021/ad.2013.25.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiteside TL. The role of regulatory t cells in cancer immunology. Immunotargets Ther. 2015;4:159–171. doi: 10.2147/ITT.S55415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umansky V, Blattner C, Gebhardt C, Utikal J. The role of myeloid-derived suppressor cells (mdsc) in cancer progression. Vaccines. 2016;4:36. doi: 10.3390/vaccines4040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szomolay B, Eubank TD, Roberts RD, Marsh CB, Friedman A. Modeling the inhibition of breast cancer growth by gm-csf. J Theor Biol. 2012;303:141–151. doi: 10.1016/j.jtbi.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers. 2014;6:1670–1690. doi: 10.3390/cancers6031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markowitz J, Wesolowski R, Papenfuss T, Brooks TR, Carson WE. Myeloid-derived suppressor cells in breast cancer. Breast Cancer Res Treat. 2013;140:13–21. doi: 10.1007/s10549-013-2618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obeid E, Nanda R, Fu YX, Olopade OI. The role of tumor-associated macrophages in breast cancer progression (review) Int J Oncol. 2013;43:5–12. doi: 10.3892/ijo.2013.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourette RP, Rohrschneider LR. Early events in m-csf receptor signaling. Growth Factors. 2000;17:155–166. doi: 10.3109/08977190009001065. [DOI] [PubMed] [Google Scholar]
- 36.FJ FJP, Stanley ER. Csf-1 regulation of the wandering macrophage: Complexity in action. Trends Cell Biol. 2004;14:628–638. doi: 10.1016/j.tcb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 37.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–74. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sousa S, et al. Human breast cancer cells educate macrophages toward the m2 activation status. Breast Cancer Res. 2015;17:1–14. doi: 10.1186/s13058-015-0621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Patel MB, et al. Tumor necrosis factor-alpha produced in the kidney contributes to angiotensin II-dependent hypertension. Hypertension. 2014;64:1275–1281. doi: 10.1161/HYPERTENSIONAHA.114.03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X, Oppenheim JJ. Contrasting effects of tnf and anti-tnf on the activation of effector t cells and regulatory t cells in autoimmunity. FEBS Lett. 2011;585:3611–3618. doi: 10.1016/j.febslet.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng X, et al. Structure and interactions of the human programmed cell death 1 receptor. J Biol Chem. 2013;288:11771–11785. doi: 10.1074/jbc.M112.448126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng X, et al. 2017 Human pd-l1/b7–h1/cd274 protein. Sino Biological Inc. Available at www.sinobiological.com/PD-L1-B7-H1-CD274-Protein-g-533.html.
- 44.Jain N, Nguyen H, Chambers C, Kang J. Dual function of ctla-4 in regulatory t cells and conventional t cells to prevent multiorgan autoimmunity. Proc Natl Acad Sci USA. 2010;107:1524–1528. doi: 10.1073/pnas.0910341107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker LS. Treg and ctla-4: Two intertwining pathways to immune tolerance. J Autoimmun. 2013;45:49–57. doi: 10.1016/j.jaut.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joshi B, et al. On immunotherapies and cancer vaccination protocols: A mathematical modelling approach. J Theor Biol. 2009;259:820–827. doi: 10.1016/j.jtbi.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 47.de Pillis LG, Gu W, Radunskaya AE. Mixed immunotherapy and chemotherapy of tumors: Modeling, applications and biological interpretations. J Theor Biol. 2006;238:841–862. doi: 10.1016/j.jtbi.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 48.Wilson S, Levy D. A mathematical model of the enhancement of tumor vaccine efficacy by immunotherapy. Bull Math Biol. 2012;74:1485–1500. doi: 10.1007/s11538-012-9722-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai X, Friedman A. Combination therapy of cancer with cancer vaccine and immune checkpoint inhibitor: A mathematical model. PLoS One. 2017;12:e0178479. doi: 10.1371/journal.pone.0178479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu H, et al. Bet bromodomain inhibition promotes anti-tumor immunity by suppressing pd-l1 expression. Cell Rep. 2017;16:2829–2873. doi: 10.1016/j.celrep.2016.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adeegbe D, et al. Synergistic immunostimulatory effects and therapeutic benefit of combined histone deacetylase and bromodomain inhibition in non-small cell lung cancer. Cancer Discov. 2017;7:852–867. doi: 10.1158/2159-8290.CD-16-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hao W, Friedman A. Serum upar as biomarker in breast cancer recurrence: A mathematical model. PLoS One. 2016;11:e0153508. doi: 10.1371/journal.pone.0153508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D’Acunto B. 2004. Computational Methods for PDE in mechanics, Series on Advances in Mathematics for Applied Sciences (Word Scientific, Singapore), Vol 67.
- 54.Michot J, et al. Immune-related adverse events with immune checkpoint blockade: A comprehensive review. Eur J Cancer. 2016;54:139–148. doi: 10.1016/j.ejca.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 55.Niu N, Shao R, Yan G, Zou W. Bromodomain and extra-terminal (bet) protein inhibitors suppress chondrocyte differentiation and restrain bone growth. J Biol Chem. 2016;291:26647–26657. doi: 10.1074/jbc.M116.749697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Picaud S, et al. Rvx-208, an inhibitor of bet transcriptional regulators with selectivity for the second bromodomain. Proc Natl Acad Sci USA. 2013;110:19754–19759. doi: 10.1073/pnas.1310658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Linardou H, Gogas H. Toxicity management of immunotherapy for patients with metastatic melanoma. Ann Transl Med. 2016;4:272. doi: 10.21037/atm.2016.07.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lo WC, Arsenescu V, Arsenescu RI, Friedman A. Inflammatory bowel disease: How effective is tnf-alpha suppression? PLoS One. 2016;11:e0165782. doi: 10.1371/journal.pone.0165782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of nf-kb via specific binding to acetylated rela. Mol Cell Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang DJ, Ratnam NM, Byrd JC, Guttridge DC. Nf-kb functions in tumor initiation by suppressing the surveillance of both innate and adaptive immune cells. Cell Rep. 2014;9:90–103. doi: 10.1016/j.celrep.2014.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Filippakopoulos P, Knapp S. Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 62.Curran MA, Montalvo W, Yagita H, Allison JP. Pd-1 and ctla-4 combination blockade expands infiltrating t cells and reduces regulatory t and myeloid cells within b16 melanoma tumors. Proc Natl Acad Sci USA. 2010;107:4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.