Significance
Reliable proteome-wide quantification in large biological cohorts is highly valuable for clinical and pharmaceutical research yet remains extremely challenging despite recent technical advancements. Specifically, elevated missing data levels and compromised quantitative quality are common issues for prevalent methods. Here, we describe an IonStar technique taking advantage of sensitive and selective MS1 ion current-base quantification via innovations in effective and reproducible quantitative feature generation. Compared with several label-free strategies, IonStar showed superior performance in large-cohort analysis, manifested by excellent accuracy/precision, extremely low missing data, and confident discovery of subtle protein changes. In a proof-of-concept study, we demonstrated that IonStar quantified >7,000 unique proteins in 100 brain samples with no missing data and excellent quantitative quality, which has not been achievable by existing methods.
Keywords: quantitative proteomics, label-free quantification, MS1 ion current-based methods, large-cohort analysis, missing data
Abstract
Reproducible quantification of large biological cohorts is critical for clinical/pharmaceutical proteomics yet remains challenging because most prevalent methods suffer from drastically declined commonly quantified proteins and substantially deteriorated quantitative quality as cohort size expands. MS2-based data-independent acquisition approaches represent tremendous advancements in reproducible protein measurement, but often with limited depth. We developed IonStar, an MS1-based quantitative approach enabling in-depth, high-quality quantification of large cohorts by combining efficient/reproducible experimental procedures with unique data-processing components, such as efficient 3D chromatographic alignment, sensitive and selective direct ion current extraction, and stringent postfeature generation quality control. Compared with several popular label-free methods, IonStar exhibited far lower missing data (0.1%), superior quantitative accuracy/precision [∼5% intragroup coefficient of variation (CV)], the widest protein abundance range, and the highest sensitivity/specificity for identifying protein changes (<5% false altered-protein discovery) in a benchmark sample set (n = 20). We demonstrated the usage of IonStar by a large-scale investigation of traumatic injuries and pharmacological treatments in rat brains (n = 100), quantifying >7,000 unique protein groups (>99.8% without missing data across the 100 samples) with a low false discovery rate (FDR), two or more unique peptides per protein, and high quantitative precision. IonStar represents a reliable and robust solution for precise and reproducible protein measurement in large cohorts.
For clinical and pharmaceutical proteomics studies, analysis of large biological cohorts is necessary to alleviate the impacts of high biological variability of animal/human subjects and enhance the reliability of quantification (1–3). Label-free approaches thus appear to be an attractive option because of a theoretically unlimited number of samples quantifiable in one batch, as well as more flexible and cost-effective sample preparation procedures (4–7). In reality, however, label-free quantification of large cohorts remains challenging despite the recent advancements in liquid chromatography (LC)-MS instrumentation and informatics efforts. Common issues include compromised quantitative accuracy and precision because of the typically remarkable experimental variations without using internal calibration measures (4, 8, 9), as well as snowballing missing data levels as the sample number increases (10, 11). These two problems severely undermine the quality of protein quantification and elevate false-positive discovery of significant protein changes, impeding appropriate interpretation of biological relevance (12, 13).
One primary source for diminished quantitative quality is the stochasticity of data-dependent acquisition (DDA) (14), which leads to the undersampling of ions low in abundance. Commonly practiced DDA features, such as dynamic exclusion, devised to enhance identification depth, can also compromise MS2 spectra quality (15, 16). Consequently, most MS2-based quantitative approaches [e.g., spectral counting (SpC), MS2 ion currents (ICs) (10, 15, 17, 18)] suffer from suboptimal reproducibility for lower abundance peptide species, resulting in a high missing data rate (e.g., 20–50% of proteins have missing data in sample sets with sizes from 6 to 20 replicates, and a markedly worse rate as cohort size enlarges) when analyzing a large number of samples (13, 19). MS2-based data-independent acquisition (MS2-DIA) approaches, with sequential window acquisition of all theoretical fragment ion spectra (SWATH)-MS as the most prominent example, profoundly alleviate missing data (<10%) in large-scale proteomics experiments by triggering MS2 scans in a mass-to-charge ratio (m/z) window-based manner (10, 20, 21). Although MS2-DIA strategies represent a tremendous advancement in reproducible protein measurement, problems include difficulties in interpreting MS2 spectra typically containing multiple cofragmented precursors (22) and limited quantitative depth using spectral library-based peptide identification (23). Methods such as spectral library-free identification approaches [e.g., PECAN, DIA-Umpire, DirectDIA in Spectronaut Pulsar (24, 25)] have been developed to promote MS2-DIA performance, but the data quality for large-cohort analysis has yet to be examined.
An alternative strategy to improve quantitative quality is MS1 IC-based methods, which acquire peptide precursor ICs as quantitative features, completely independent of MS2. Frequently, an MS1 quantitative feature is identified by the retention time (RT) and high-resolution m/z of a precursor ion, and is acquired across all samples (26), while MS2-DDA is merely employed to assign a peptide identification (ID) to extracted MS1 features. Hence, a peptide can be quantified in the entire dataset even with only one successful MS2 identification [i.e., peptide-spectrum match (PSM)] across all runs; this feature, when combined with the use of high-resolution MS, provides much improved sensitivity compared with MS2-based methods (27). MS1-based methods thereby hold high potential for sensitive and reproducible peptide measurement in larger sample sets (28–30). Quintessential examples of MS1-based quantitative packages are MaxQuant and OpenMS, which are excellent pipelines and have been widely employed in label-free quantitative proteomics studies. Other examples include Proteome Discoverer, Skyline, Platform for Experimental Proteomics Pattern Recognition (PEPPeR), Census, and Superhirn (31). However, it turns out that missing data rates returned from these MS1-based methods still range from 10 to 20% with 10–20 replicates and become dramatically higher with larger sample numbers (10, 17). More recently, several MS1-based quantitative packages reported a low rate of missing data for quantitative proteomics, such as Progenesis QI (nonlinear dynamics) (32), DeMix-Q (11), and the FeatureFinderIdentification (FFId) add-on in OpenMS (33). Nonetheless, to date, these packages either have yet to show the capacity for analyzing larger sample cohorts (e.g., ≥20 samples) or have exhibited inferior data quality (e.g., suboptimal accuracy/precision) (32). Furthermore, the ability to robustly determine subtle changes (e.g., <50%) is crucial for clinical and pharmaceutical studies, especially in biological systems with mild systematic proteomic changes ( e.g., brain) (34–37). Consequently, high analytical precision (e.g., <10% median intragroup variability on the protein level for technical replicates) is essential; however, to our knowledge, none of the existing methods has demonstrated such capability for large-cohort studies.
Here we describe IonStar, a novel MS1-based approach to address the current limits in large-scale quantitative proteomics. IonStar incorporates (i) unique experimental procedures for efficient and reproducible sample preparation and liquid chromatography-mass spectrometry (LC-MS) analysis, which enable sensitive and robust data acquisition for many biological replicates [as described previously (30)], and (ii) a data processing pipeline for reliable large-cohort analysis. The key components of this pipeline include 3D chromatographic alignment, extensive and sensitive feature detection/propagation, and stringent postfeature generation quality control. These unique components in IonStar enabled efficient and reproducible procurement of quantitative features from large-cohort datasets, which, in turn, resulted in superior quantitative accuracy/precision and much lower missing data levels compared with existing methods, as well as allowing confident identification of changed proteins, especially subtle ones that usually elude detection. The applicability of IonStar in large biological sample sets was demonstrated by an investigation of 100 brain samples subjected to different severities of traumatic brain injury (TBI) and subsequent pharmacological treatments, which yielded >7,000 unique protein groups with two or more unique peptides per protein and >99.8% of these proteins without missing data in any of the 100 samples. To our knowledge, a proteomics analysis with such a high sampling number in one analytical batch, deep proteome coverage, and low missing data level has not been achieved previously.
Results
Development and Optimization of IonStar Data Processing Pipeline.
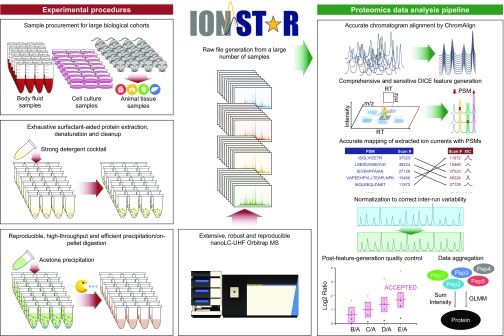
The IonStar workflow encompasses a series of experimental and data processing components as illustrated in Fig. 1. The experimental components for efficient and reproducible sample treatment and LC-MS analysis were reported by Shen et al. (30), while this work focuses on the IonStar data processing components.
Fig. 1.
Overview of IonStar quantitative workflow. IonStar comprises experimental procedures and a proteomics data analysis pipeline. The experimental procedures include an efficient and exhaustive sample preparation protocol plus a sensitive and reproducible LC-MS setup to allow high-quality conversion of large biological sample cohorts to LC-MS raw files. The data analysis pipeline, taking full advantages of high-resolution (120,000 FWHM) MS1 and consistent LC separation from the experimental procedures, allows low-missing data quantification in large sample cohorts with superior quantitative quality. XIC, extracted ion chromatogram.
Effective chromatographic alignment with ChromAlign.
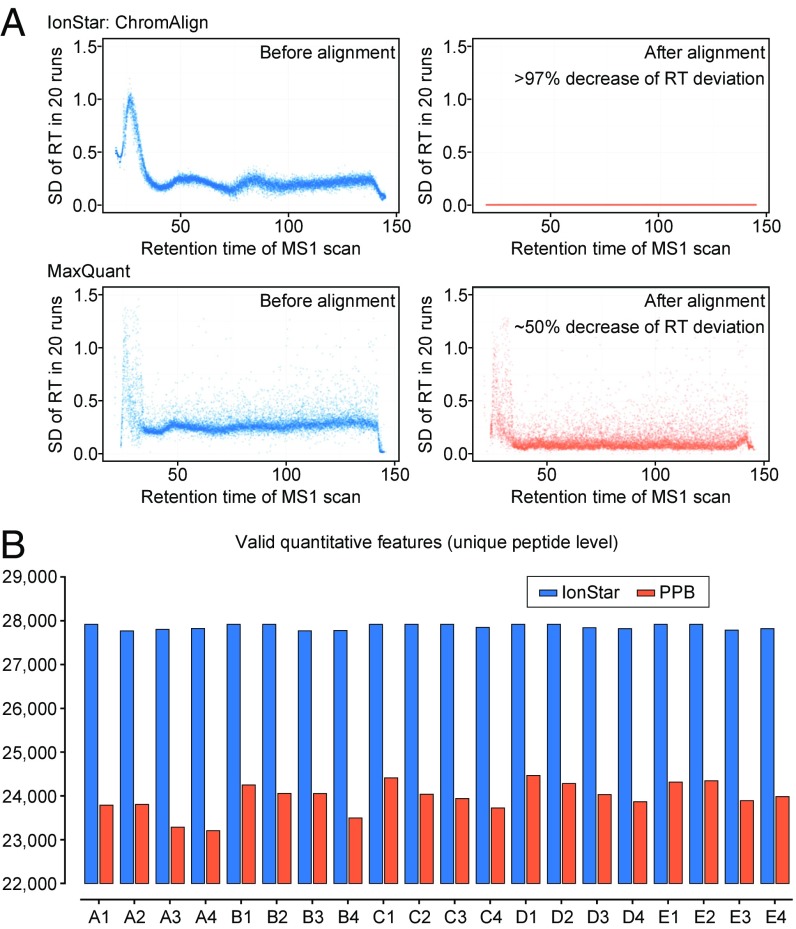
First and foremost, efforts to improve data quality (i.e., higher quantitative precision and lower missing data) of MS1-based quantification in large sample cohorts should emphasize the accurate matching of ICs of the same peptide, which is highly critical. This is usually accomplished via the reference of m/z and RT of ICs across the whole dataset [e.g., accurate mass and time tag strategy (AMT) (38)]. Because of the inevitable fluctuations of LC conditions and slight matrix differences among samples, global correction of peptide RT deviation via chromatographic alignment is essential to ensure the sensitivity and specificity of the IC matching step (31). This also applies to IonStar, despite the fact that the LC-MS setup of IonStar provides considerable reproducible analysis with minor RT variation (30) (compare Fig. 2A).
Fig. 2.
Much improved chromatographic alignment and more comprehensive/reproducible feature generation by IonStar. (A) Performance of alignment algorithms used in IonStar (ChromAlign) and MaxQuant. SDs of peptide RT in 20 benchmark LC-MS runs were plotted as a function of RT before and after alignment. IonStar decreased RT variations >97% compared with <50% by MaxQuant. (B) Comparison of valid quantitative feature numbers generated by IonStar vs. a representative PPB method.
Most prevalent MS1-based methods exploit 2D alignment algorithms, which rely on m/z and RT, while a reduced isotopic envelope is considered by a few for RT correction. For example, OpenMS employs identification-based RT adjustment via establishing reference points across LC-MS runs, and the median RT shift of a peptide in one run is corrected by either the reference RT or median RT of all runs (19). MaxQuant applies pairwise alignment of identified peptides in a time-dependent and nonlinear way via 2D Gaussian kernel smoothing to achieve RT correction (4). Here, we demonstrated the use of a ChromAlign algorithm optimized from a previously published technique (39) and now incorporated in the SIEVE package, for effective dataset-wide RT correction. ChromAlign employs a two-step, 3D algorithm (RT, MS1 peaks in full scan, and variations in MS1 relative abundances) considering all representative peak features before peptide identification, as well as a correlation-optimized, time warping-like algorithm to achieve significantly improved RT alignment compared with the above-mentioned 2D alignment algorithms. Examples comparing ChromAlign and the algorithm used by MaxQuant are shown in Fig. 2A. It should be noted that a strict peak-to-peak comparison of the two algorithms is not feasible, as features being aligned are considerably different: ChromAlign in IonStar aligns the RT of a large number of abundant MS1 peaks in each run against a reference run (identified by comparison of alignment scores among multiple candidates) regardless of MS2 identification, and the algorithm in MaxQuant only aligns RT of peaks with valid peptide identification in each run without using a reference run. These distinct strategies result in profoundly different data distribution (Fig. 2A), while it is evident that ChromAlign performs more effectively in RT correction: For the 20-run benchmark dataset (described in the following sections), the algorithm in MaxQuant decreased RT deviations by ∼50%, while ChromAlign decreases RT deviations by an average of ∼97%. This substantially contributes to reduced missing data and enhanced reproducibility in combination with our unique feature generation method (discussed below). By working with ThermoFisher Scientific, the ChromAlign process has been incorporated into the IonStar pipeline.
Comprehensive and sensitive quantitative feature generation.
In MS1-based quantification, the most prevalent strategy for feature generation, termed the peak property-based (PPB) method (SI Appendix, Fig. S1), extracts peaks by wavelet-based techniques and then passes the peaks through a series of quality check thresholds (e.g., intensities, peak occurrence with the same m/z, peak shapes, isotopic patterns) to ascertain their validity as features. Peptide identities are then propagated from runs with valid PSMs to those without PSMs using AMT or similar algorithms. Examples of MS1-based packages using the PPB method include OpenMS (19), MaxQuant (4, 40), Census (41), and Superhirn (42). The stringent quality control during feature generation ensures that the PPB method only selects signals with excellent quality for quantification; on the other hand, the PPB method may compromise sensitivity for feature detection when analyzing complex biological samples, where peak characteristics (e.g., peak shapes, isotopic patterns) of lower abundance peptides are profoundly affected by coeluted perplexing matrix interferences, and thereby are difficult to model. Consequently, a large portion of peaks from lower abundance peptides may fail to pass the preset thresholds and are excluded from quantification. This gives rise to elevated levels of missing data as well as impaired quantitative accuracy/precision because of fewer usable quantitative features per protein, which is likely exacerbated as sample size increases.
An alternative type of strategy for feature generation, direct ion-current extraction (DICE), has been developed more recently to improve sensitivity and lower missing data. DICE methods generate quantitative features by directly extracting the ICs of a precursor in the aligned dataset using a predefined window of RT and high-resolution MS1 m/z, often with no or few requirements for peak properties. DICE-type methods are illustrated in SI Appendix, Fig. S1. While a number of DICE methods have been developed and employed in packages such as Skyline (7), DeMix-Q (11), and FFId (33), to our knowledge, none of them has been demonstrated to be functional in large-cohort analysis. Here, we developed a unique DICE-type approach in IonStar for large-scale proteomic quantification, taking advantage of the high-resolution (120,000 FWHM) MS1 signals and consistent chromatographic separation from IonStar (30) experimental steps, as well as accurately aligned peak clusters by ChromAlign. The ICs of precursors with triggered MS2 scans are extracted stringently using narrow MS1 m/z-RT windows from all LC-MS runs, and the set of ICs unambiguously linked to the same precursor ion is mapped together to form a “quantitative frame.” Peptide identity of a frame is then retrieved by matching the MS2 scan number linked to individual features with database searching results, while frames with ambiguous PSM assignment are discarded to improve data quality (43). This approach significantly enhances reliability and sensitivity for feature generation and markedly reduces mismatching. Examples of peptide ICs extracted by DICE are shown in SI Appendix, Fig. S2. In practice, the combination of ChromAlign and the unique feature generation method in IonStar enables more consistent and sensitive feature generation. On average, 16.3% more quantitative features were generated with much improved intra-run consistency in a 20-sample human-Escherichia coli spike-in dataset (described below) (Fig. 2B), which greatly contributed to the better sensitivity, accuracy, and precision, as well as the lower rate of missing data, by IonStar, as discussed below.
Postfeature generation quality control based on peptide quantitative data.
Although the DICE method may generally confer significantly enhanced sensitivity in feature generation, low-quality features from coeluted interferences, which severely deteriorate data quality, are inevitably introduced owing to the lack of quality control during the procedure. Postfeature generation quality control is thus a necessary measure to reinforce the reliability of quantification (e.g., target-decoy scoring of quantitative features by DeMix-Q) (11). In IonStar, a mean and variation modeling approach is adopted for postfeature generation quality control, which examines the consistency of intergroup quantitative ratios obtained independently from each of the multiple peptides belonging to the same protein. Peptides with outlier ratios, likely attributed to low-quality features, are excluded to ensure reliability of quantitative results. Based on our evaluation results, we selected principal component-based outlier detection (PCOut) (44) because of its superior performance for detecting outliers, especially from multiple-group data generated from large-cohort analysis. A classic Grubb’s test is also included in IonStar for outlier rejection in the two-group comparison. Using the benchmark sample set, it has been shown that PCOut prominently improved quantitative accuracy by decreasing the median deviation between observed and theoretical protein ratios from 0.09 to 0.02 (log2 scale), while quantitative precision was unaffected (SI Appendix, Fig. S3 and Dataset S4), implying that chemical/instrumental noises and coeluted peaks, rather than quantitative variability, were mainly responsible for the emergence of low-quality features.
Data normalization and aggregation to protein level.
Previously, we tested a number of normalization strategies for MS1-based quantification and found that several approaches significantly improved quantitative precision (26). In one previous study, we evaluated the performance of different normalization methods on proteomics data and found that normalization by quantiles achieved the best results ubiquitously, followed by total IC (26). In IonStar, both methods are made available for users to select from based on characteristics of datasets; in general, whichever achieves the lowest intragroup variation for technical replicates is preferred.
An optimal method to aggregate quantitative data from the frame or peptide level to the protein level is also essential for reliable protein quantification. Former evaluation by our group and others suggested that summing up the peak areas of ICs from all peptides could provide straightforward and accurate measurement of proteins (9, 45). Here, we found that aggregation using an in-house–developed generalized linear mixed model (GLMM) also yielded high accuracy comparable to or better than sum of intensity, and appeared to perform markedly better in sample sets with relatively high intragroup variation (SI Appendix, Fig. S4). The GLMM model differs from the traditional linear mixed model in that relationships of mean and variance are modeled to avoid the introduction of biases during data transformation. Also, a Bayesian approach was taken for parameter estimation and statistical significance inference. In IonStar, both GLMM and sum of intensity are available for data aggregation and GLMM is recommended.
Comparative Evaluation of IonStar with Prevalent Label-Free Methods.
The performance of IonStar was compared with several prevalently used software packages for MS1-based quantification, including OpenMS, MaxQuant, and Proteome Discoverer 2.1, as well as MS2-based SpC, a robust and popularly used method for label-free quantification (15, 18). DeMix-Q and Skyline were not included in the study as they were not capable of processing our dataset despite our strenuous attempts. It is likely that IonStar provides superior performance (discussed below). A multigroup benchmark sample set containing 20 concocted samples was prepared by spiking small portions (<10% total protein amount) of E. coli lysates at five levels (n = 4 per level) into a constant background of human cell lysates and then analyzed using the IonStar experimental procedures (n = 20 in total; SI Appendix, Fig. S5). Using our most recent experimental setup involving selective trapping/delivery and Orbitrap Fusion Lumos MS, we demonstrated the quantification of >6,000 unique protein groups (two or more unique peptides per protein) without missing data after removal of shared peptides between the two species (30). However, as the most recent versions of OpenMS and MaxQuant do not yet process such large datasets, we employed an older dataset by Orbitrap Fusion MS [Proteomics Identifications (PRIDE) identifier PXD003881], where >3,800 protein groups were identified with the same criteria. To ensure unbiased and comprehensive comparison, a set of rules was applied to all packages; for example, each package was optimized individually to achieve the best performance and uniform cutoffs for peptide and protein identification [both peptide and protein false discovery rates (FDRs) < 1%, and at least two unique peptides per protein for reliable quantification, as justified in SI Appendix, Fig. S6]. No missing data imputation was adopted, and proteins with missing data were excluded from evaluation. Details are provided in Methods.
Extremely low missing data and wide protein intensity range by IonStar.
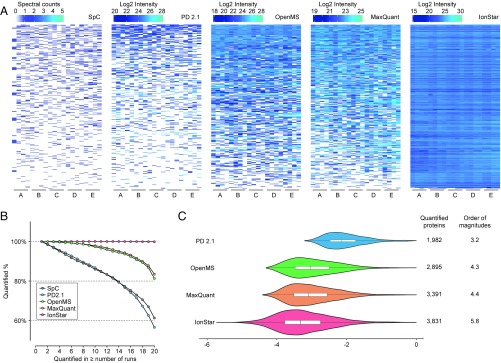
Missing data levels of the five methods, defined as the percentage of proteins with missing quantitative values in all proteins quantified, were first compared. As shown in Fig. 3A, for proteins with the lowest 10% of abundance, SpC exhibited a very high missing data rate, rendering reliable quantification of these proteins impractical; OpenMS and MaxQuant remarkably alleviated the missing data problem but still with significant missing values, while IonStar showed excellent coverage and extremely low missing data. From another perspective (Fig. 3B), proteins quantified without missing data decreased drastically to ∼60% of total quantified proteins at 20 runs for SpC and PD 2.1, while the numbers increased to ∼81% and 83%, respectively, for OpenMS and MaxQuant. In comparison, ∼99.9% of all proteins quantified by IonStar were missing data-free in 20 samples (i.e., 0.1% dataset-wide missing data rate). Although DeMix-Q was not included in the comparison, the reported missing data rate (2.8%) using the iPRG-2015 dataset was higher than IonStar with a lower sample number (n = 12 vs. n = 20 here) (11). As a result, the numbers of quantifiable proteins (i.e., proteins without missing data) were 3,834 for IonStar, much higher than 3,391 for MaxQuant, 2,895 for OpenMS, 1,982 for PD 2.1, and 2,484 for SpC.
Fig. 3.
Missing data levels and quantitative intensity ranges among quantitative approaches compared in this study. (A) Abundance heat maps of proteins with the lowest 10% abundances in the benchmark sample set (n = 20 in total). White areas indicate missing data. (B) Percentage of proteins without missing data as a function of run (sample) number by different quantitative approaches. (C) Distribution of quantitative intensities (in log10 scale) for proteins without missing data by the four MS1-based quantitative approaches. The area of each violin plot is proportional to the number of quantifiable proteins (i.e., without missing data). The orders of magnitude of protein intensity ranges by all methods are labeled to the right of the plots.
SpC suffered from a severe missing data problem as a result of the inherent defects of MS2 DDA. PD 2.1 also produced high missing data owing to the lack of an effective approach to infer the peptide ID of MS1 peaks missing identification from other runs with valid PSMs. MaxQuant and OpenMS showed similar missing data levels likely because they use similar alignment and feature generation algorithms, as discussed above. The extremely low levels of missing data by IonStar can be attributed to the unique alignment and feature generation methods, permitting comprehensive and accurate feature clustering/procurement/matching. The missing data levels of IonStar even compared favorably with MS2-based DIA methods such as SWATH-MS, where 1–10% protein-level missing data rates were reported (10, 21, 25). This unique characteristic of IonStar enables highly reproducible protein measurement in large sample cohorts (as shown in the following application) and improves analytical quality, especially for low-abundance proteins. The superior performance by IonStar also extended the intensity range of quantifiable proteins (Fig. 3C) to 5.8 orders of magnitude in the benchmark dataset, prominently wider than those by other methods (3.2–4.4).
Superior quantitative reproducibility, accuracy, and precision by IonStar.
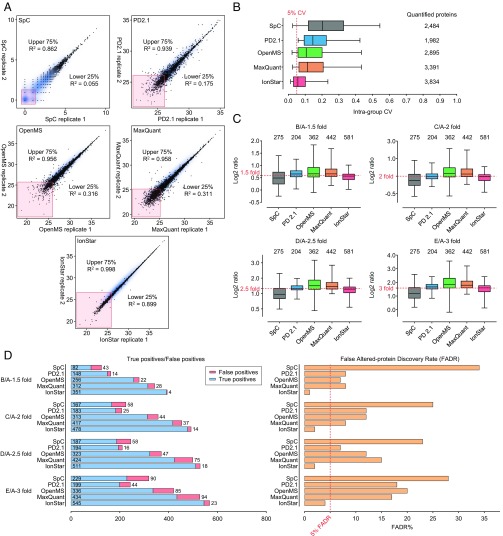
Quantitative accuracy and precision were then evaluated. Here, we define accuracy as the closeness between the true and observed protein ratios, and we define precision as the intragroup coefficient of variation (CV) for protein quantification (i.e., among technical replicates). Since reproducibility of protein measurement profoundly affects both accuracy and precision for relative quantification, run-to-run reproducibility of the five methods was first evaluated by correlating quantitative intensities of proteins from two replicate LC-MS runs of the same pooled sample (Fig. 4A). It was observed that proteins with the upper 75% abundance (i.e., high-abundance proteins) exhibited good reproducibility across all methods evaluated, with R2 ranging from 0.862 to 0.998, while IonStar achieved the best reproducibility. Nonetheless, reproducibility for proteins with the lower 25% abundance (i.e., low-abundance proteins) showed striking differences among the methods, as evidenced by the R2 values for each method: 0.055 by SpC, 0.175 by PD 2.1, 0.316 by OpenMS, and 0.311 by MaxQuant, while much higher by IonStar (0.899). The excellent quantitative reproducibility by IonStar led to exceptional quantitative precision by IonStar, as shown in Fig. 4B. IonStar achieved the lowest intragroup CV of ∼5% for all quantified proteins across the five groups, compared with ∼19%, 14%, 10%, and 11% by SpC, PD 2.1, OpenMS, and MaxQuant, respectively, roughly consistent with previous literature (46). The precision of IonStar also appeared to be superior to that of other published methods (not experimentally compared here because of infeasibility of comparison). For example, Skyline showed an ∼20% median CV in the 12-replicate iPRG-2015 sample set and DeMix-Q showed an ∼7% protein-level CV in the same sample set (11). Quantitative precision by IonStar also appears to surpass MS2-DIA methods, which typically report an ∼10% or higher median CV among technical replicates (10, 21, 22). The excellent quantitative precision of IonStar is critical for robust detection of subtle abundance differences between biological samples, as well as for accurate determination of intergroup ratios. For this reason, IonStar showed superior accuracy in quantifying both E. coli (i.e., true positive) and human proteins (i.e., true negative) from the benchmark sample set. As shown in Fig. 4C, IonStar quantified by far the largest number of E. coli proteins (581 vs. 204–442 by other methods) without missing data, and yet provided the most accurate measurement in every spike-in group. PD 2.1 also showed relatively accurate estimation of protein ratios, but only 204 E. coli proteins were quantifiable due to high missing data levels; OpenMS and MaxQuant tended to overestimate protein ratios in all groups, in line with previous observations (19, 26). IonStar also achieved the highest quantitative accuracy for human proteins, especially for low-abundance ones (SI Appendix, Fig. S7).
Fig. 4.
Comparison of quantitative quality among different approaches. (A) Reproducibility examined by Pearson correlation of two replicate runs from the same sample. The R2 values were separately calculated for proteins in the upper 75% and lower 25% abundance percentiles. Proteins with missing data were removed from comparison. (B) Quantitative precision of proteins quantified by each method, indicated by intragroup CV levels. (C) Quantitative accuracy of E. coli proteins quantified under all spike-in levels by each method. The true ratios are highlighted by red dotted lines. (D) Numbers of true and false positives (Left) in significantly changed proteins and FADR (Right) by each method. All spike-in levels are included.
Again, these results demonstrated the much improved quantitative performance by IonStar, especially for low-abundance proteins, likely attributed to the optimal RT correction and feature generation approaches. Moreover, the postfeature generation quality control in IonStar effectively removes low-quality quantitative data that may otherwise severely diminish quantitative accuracy and precision, further substantiating the quality of quantification (SI Appendix, Fig. S8).
The lowest levels of false discovery of altered proteins by IonStar.
One ultimate goal of quantitative proteomics is to discover significant protein changes, often by applying cutoff thresholds for both fold of change and significance tests (9, 47). False positives represent a prominent problem leading to incorrect biological clues and misuse of resources in downstream analysis and validation. Here, we examine the false altered-protein discovery rate (FADR) by each method. In the benchmark dataset, a cutoff threshold of ≥1.4-fold protein change and <0.05 P value was used to determine significant protein changes, optimized by employing an experimental null method (9). In the benchmark dataset, FADR can be easily calculated, as all E. coli proteins are true positives, while all human proteins are true negatives (i.e., changed human proteins are false positives). IonStar identified the highest number of true positives and the lowest number of false positives, resulting in a 0.5–4.5% FADR across the four comparisons (Fig. 4D), while the other MS1-based methods generated a FADR ranging from 8 to 20%. SpC exhibited a >20% FADR at all spike-in levels, mostly arising from the suboptimal quantification of low-abundance proteins. FADRs under other cutoffs were also calculated (SI Appendix, Fig. S9), where IonStar also substantially outperformed other methods. The high sensitivity and low false-positive rates in altered protein discovery by IonStar are likely a result of its excellent quantitative accuracy and precision.
Application of IonStar in Large-Scale Quantitative Proteomics (N = 100).
As a proof of concept, IonStar was applied to a large-scale neuroproteomics study of TBI encompassing 100 biological samples. TBI is a debilitating disease triggered by damage to the brain from an external force, which accounts for 2.2 million emergency room visits annually in the United States alone (48). While mild TBI is usually temporary and self-healing, severe TBI causes various levels of neurological, cognitive, and physical disability or even mortality. To develop an effective therapy for treating severe TBI, it is essential to understand the highly complex and heterogeneous mechanisms of the disease (49). Recently, phenoxybenzamine (PBZ) and methamphetamine (METH) have been identified as potential neuroprotective agents against TBI in a rodent lateral fluid percussion (LFP) injury model, substantiated by significant alleviation of behavioral and cognitive deficits as well as regulation of several critical gene targets (50, 51). Nonetheless, dysregulations in protein signaling networks after PBZ or METH administration remain unknown, impeding the elucidation of mechanisms underlying the neuroprotective effects of the two drugs.
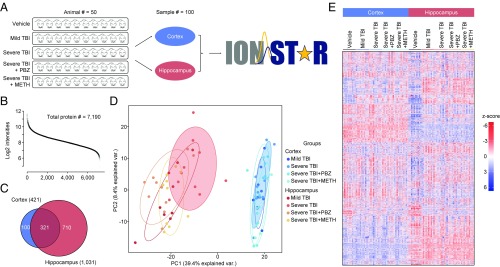
In the current study, the early-stage alteration of proteome patterns in two brain regions (i.e., cortex, hippocampus) from a rodent LFP model with varied TBI severities and pharmacological treatments was investigated using IonStar. Briefly, 50 male Wistar rats were assigned to five groups (n = 10 per group) and were subjected to different TBI procedures and treatments, including uninjured (+PBS), mild TBI (+PBS), severe TBI (+PBS), severe TBI with PBZ treatment, and severe TBI with METH treatment. The cortex and hippocampus were separately procured 32 h postinjury (24 h posttreatment), and then analyzed by the IonStar workflow (Fig. 5A). Missing data-free quantification of 7,190 unique protein groups (i.e., 99.8% of all quantified proteins) was achieved in the 100 samples analyzed (two or more peptides per protein, and protein FDR < 1%). Intensities of quantified proteins spanned 6.8 orders of magnitude (Fig. 5B), indicating excellent sensitivity that allowed reliable quantification of low-abundance proteins. Median intragroup CVs of protein quantitative values were ∼8% for technical replicates and 9.3–15.9% for biological replicates across the entire sample set (SI Appendix, Fig. S10). The abundance heat map for the 7,190 proteins quantified without missing data is shown in Fig. 5E. Quantitative results for proteins and peptides are provided in Datasets S5 and S6 separately.
Fig. 5.
Application of IonStar in a large-scale quantitative investigation to explore the mechanism of TBI and pharmaceutical recovery (PBZ or METH). (A) Overview of study design. The cortex and hippocampus from 50 LFP rats with varied TBI severities and pharmacological treatments were included in the study (n = 100 in total). (B) Intensity distribution of the 7,190 proteins quantified without missing data. (C) Significantly changed proteins in the two brain regions. (D) PC analysis of significantly changed proteins. The untreated severe TBI groups are highlighted with shading. var., variation. (E) Abundance heat map of the 7,190 proteins quantified from the 100 brain samples.
Using an experimental null method reported previously (9), 421 and 1,031 proteins were determined to be differentially expressed across all groups in the cortex and hippocampus, respectively, under cutoff thresholds of >1.4-fold protein change and <0.05 P value (Fig. 5C). Low FADRs ranging from 1.91 to 7.86% were achieved across groups, indicating highly confident discovery of protein changes. Clearly, the two brain regions showed distinct biological responses to injury and drug treatment, as manifested by the different altered proteins in Fig. 5C. Furthermore, PC analysis of the significantly changed proteins showed the complete segregation of the cortex and hippocampus, with PC1 and PC2 summarizing ∼50% of the variability (Fig. 5D). More interestingly, severe TBI (highlighted in shades) exhibited more dissimilarity compared with other conditions in both brain regions (Fig. 5D), suggesting PBZ and METH treatment of severe TBI altered the proteome patterns to be more similar to mild TBI. This notion is further supported by Pearson correlation of quantitative values of the 7,190 quantified proteins in the five groups (SI Appendix, Fig. S11). Gene ontology and pathway analysis of the changed proteins revealed a number of potential upstream molecules that might regulate the neuroprotective effects of PBZ and METH, such as TGF-β, Rho GTPase, FMR1, BDNF, and TNF (SI Appendix, Fig. S12).
Discussion
In-depth, reproducible, and robust quantification of proteins in large sample sets is critically important for pharmaceutical and clinical proteomics studies. MS1-based label-free quantitative methods confer great potential for this purpose, considering no theoretical limit of quantifiable samples and that acquisition of quantitative values is independent of MS2. Nonetheless, this potential has not been fully exploited by most current quantitative approaches. Here, we achieved robust large-cohort proteomics analysis using the IonStar approach, which features effective chromatographic alignment and sensitive feature generation/assignment with efficient postgeneration quality control, substantially improving quantitative quality. Using a premixed benchmarking sample set, it was observed that IonStar offered much lower missing data levels (0.1% on protein level), superior accuracy (10.8% median error) and precision (<5% CV), and the lowest false-positive rate (<5% FADR) in identifying protein changes, compared with prevalent quantitative approaches that were feasible to evaluate in this study. We demonstrated excellent quantitative performance of IonStar in large-scale investigations by a TBI proteomics study, which quantified 100 biological samples in a single batch. Under stringent cutoffs, including two or more unique peptides per protein and <1% protein FDR, ∼7,000 protein groups were quantified without missing data in any of the 100 samples, with excellent quantitative quality (∼0.2% missing data and ∼8% median CV among technical replicates). The results unveiled viable targets for subsequent interrogation of the detailed mechanisms underlying the injury and pharmaceutical treatment. To our knowledge, label-free quantification with so many samples with such depth, quantitative precision, and missing data has not been achieved previously. It appears that the performance of IonStar compares favorably with MS2-DIA approaches in terms of proteome coverage, quantitative precision, and missing data levels, and thereby may potentially serve as a promising alternative to MS2-DIA methods for reproducible and reliable protein measurement in large-scale studies.
Finally, IonStar is quite straightforward and can be easily implemented in large-scale quantitative proteomics studies. Future work on IonStar will feature the development of an MS1-peptide ID library matching strategy to retrieve peptide IDs to unassigned quantitative frames, thus further enhancing proteome coverage of large-cohort sample sets.
Methods
Data and Program Sharing Plan.
Raw files generated from the benchmark spike-in sample set will be available upon publication on ProteomeXchange Consortium via the PRIDE partner repository (identifier PXD003881) (52). In the present form, the IonStar pipeline can be used by combining the SIEVE (v2.2 SP2, ThermoFisher Scientific) package and the R package IonStarStat. IonStarStat, relevant R scripts (IonStar_FrameGen.R and IonStar_Run.R), and the user manual for IonStar (SI Appendix, File S2) are available for downloading at https://github.com/shichens1989/IonStarStat.
Experimental Design and Statistical Rationales.
Multigroup benchmark spike-in sample set.
To benchmark the quantitative performance of different label-free proteomics methods, a set of concocted samples was prepared by spiking small and variable amounts of DH5α E. coli digest [mimicking changed proteins (true positive)] into a large and constant background of human PANC-1 cell digest [mimicking unchanged proteins (true negative)]. Protein extraction and digestion procedures are specified below. A total of five E. coli spike-in groups were included (wt/wt, percentage of E. coli in total proteins): 3% (onefold, A), 4.5% (1.5-fold, B), 6% (twofold, C), 7.5% (2.5-fold, D), and 9% (threefold, E), each containing four replicates (n = 20 in total). Samples were analyzed by LC-MS in an alternating sequence to minimize carryover. Group A was used as the control group for intergroup ratio calculation, and P values of each comparison pair were determined by a two-sample equal variance Student’s t test. Grouping and replicate information is denoted in the file names by A–E and 1–4.
Technical replicates and mimic biological replicate sample sets.
To evaluate different data aggregation strategies, we concocted an additional two sample sets: (i) technical replicates and (ii) mimic biological replicates. These sample sets were previously described elsewhere (9). Details are provided in SI Appendix, Fig. S4A.
Large-scale TBI rat brain sample set.
All animal experiment procedures were conducted with the approval of the Institutional Animal Care and Use Committee, University at Buffalo, The State University of New York. Male Wistar rats (350–500 g) obtained from Charles River Laboratories were used in the study, and TBI was induced by an LFP procedure. A neurological severity scoring (NSS) test was performed 8 h after TBI to examine the severity of TBI in individual rats. In total, 50 rats were selected for the study, including 10 with a craniotomy but not receiving TBI (i.e., sham/uninjured), 10 with mild TBI (NSS ≤ 5), and 30 with severe TBI (NSS ≥ 10, divided into three groups each containing 10 animals). At 8 h after injury, PBZ at was injected i.p. at a dose of 1 mg/kg in a 10% ethanol/saline solution. A separate group of 10 rats with severe TBI was injected with an i.v. bolus of METH at a dose of 0.425 mg/kg, dissolved in sterile saline, followed by continuous i.v. infusion of METH at 0.5 mg⋅kg⋅h−1 for 24 h. Rats were killed 32 h after TBI, and brains were procured. The cortex (ipsilateral to the injury) and hippocampus were then separated and snap-frozen for sample preparation. Details about the LFP procedure, NSS testing, and sample procurement can be found in papers by Rau et al. (50, 51)
Protein Extraction, Digestion, and Nano-Flow LC-MS/MS Analysis.
Samples collected were treated with a cold lysis buffer [50 mM Tris-formic acid (FA), 150 mM NaCl, 0.5% sodium deoxycholate, 2% SDS, 2% IGEPAL CA-630 (pH 8.4)] supplemented with cOmplete protease inhibitor tablets and PhosSTOP phosphatase inhibitor tablets (Roche Applied Science). Homogenization was first performed using a Polytron PT2100 homogenizer (Kinematica AG) to conduct a homogenization-cooling cycle (15,000 rpm), which was performed five to 10 times, followed by approximately three to five sonication-cooling cycles (20 s each). The sample mixture was then centrifuged at 20,000 × g at 4 °C for 30 min, and the supernatant was transferred to new Eppendorf tubes. Protein concentrations for all samples were determined by means of a bicinchoninic acid assay kit (Pierce Biotechnology, Inc.).
For each sample, 100 μg of total proteins was used for digestion. Proteins were first reduced by 5 mM DTT for 30 min and alkylated by 20 mM iodoacetamide for 30 min (in darkness). Both the reduction and alkylation steps were performed at 37 °C with rigorous oscillation in an Eppendorf Thermomixer (Eppendorf). Protein precipitation was then conducted by addition of 6 vol of chilled acetone with constant vortexing, and the mixture was incubated at −20 °C for 3 h. After centrifugation (20,000 × g at 4 °C for 30 min), the supernatant was removed and the pelleted proteins were washed with 500 μL of methanol and left to dry. Pellet proteins were wetted by addition of 80 μL of 50 mM Tris-FA. A total volume of 20 μL of trypsin (Sigma–Aldrich) dissolved in 50 mM Tris-FA (0.25 μg/μL) was added to protein pellets at an enzyme-to-substrate ratio of 1:20 (wt/wt), and tryptic digestion was performed at 37 °C for 6 h with rigorous oscillation in an Eppendorf Thermomixer. Digestion was terminated by addition of 1% FA. The samples were centrifuged at 20,000 × g at 4 °C for 30 min, and supernatant was carefully transferred to LC vials.
For each sample, peptides derived from 4 μg of proteins were separated and analyzed by a nano LC-MS system consisting of a Spark Endurance autosampler, an ultra-high-pressure Eksigent Nano-2D Ultra capillary/nano LC system, and an Orbitrap Fusion mass spectrometer (ThermoFisher Scientific) for the benchmarking sample set, or a Dionex Ultimate 3000 nano LC system, a Dionex Ultimate 3000 gradient micro LC system with a WPS-3000 autosampler, and an Orbitrap Fusion Lumos mass spectrometer for the TBI proteomics sample set. A large-ID trapping column (300-μm i.d. × 5 mm) was implemented before the nano LC column (75-μm i.d. × 100 cm, packed with 3 μm of Pepmap C18) for large-capacity sample loading, hydrophilic/hydrophobic garbage removal, and selective peptide delivery. Mobile phases A and B were 0.1% FA in 2% acetonitrile and 0.1% FA in 88% acetonitrile. The 180-min LC gradient profile was as follows: 4–13% B for 15 min, 13–28% B for 110 min, 28–44% B for 5 min, 44–60% B for 5 min, 60–97% B for 1 min, and isocratic at 97% B for 17 min. The mass spectrometer was operated under DDA mode, with a maximal duty cycle time of 3 s. MS1 spectra were acquired in the m/z range of 400–1,500 under 120,000 resolution with dynamic exclusion settings (60 s ± 10 ppm). Precursor ions were filtered by quadrupole using a 1-Thomson (Th)-wide window and fragmented by high-energy collision dissociation at a normalized collision energy of 35%. MS2 spectra were acquired under 15,000 resolution in an Orbitrap. More detailed information about the LC-MS system can be found in study by Shen et al. (30).
Protein Identification.
For the E. coli-human spike-in dataset and the TBI rat brain dataset, LC-MS raw files (.raw) were matched with the following protein sequence databases concatenated with reversed sequences: (i) human-E. coli SwissProt database for the benchmark spike-in sample set (version 201407; 33,232 entries) and (ii) rat SwissProt/TrEMBL database for the TBI rat brain sample set (version 201608; 35,953 entries), using the MS-GF+ search engine (version 10089, released on July 16, 2013) (43). The search parameters were as follows: (i) precursor ion mass tolerance: 20 ppm, (ii) instrument type: Q-Exactive, (iii) matches per spectrum: 1, (iv) fixed modification: carbamidomethylation of cysteine, (v) dynamic modification: oxidation of methionine and acetylation of N-terminal lysine, and (vi) maximal missed cleavages per peptide: 2. PSM filtering, protein inference/grouping, and global FDR control were accomplished using Scaffold (v4.3.2; Proteome Software, Inc.) (53) or IDPicker (54). Both protein and peptide FDRs were controlled at ≤1%, while a minimum of two unique peptides per protein was required. Proteins with no unique peptides are grouped into one protein group. Peptide and protein identification information for the benchmark dataset is provided in Dataset S1.
Protein Quantification.
Quantification in IonStar involves SIEVE and our R package IonStarStat. Chromatographic alignment and ion intensity-based MS1 feature detection/extraction were performed using functions in SIEVE v2.2 (working collaboratively with ThermoFisher Scientific) (55). The main processes in SIEVE include the following:
-
i)
Chromatographic alignment with ChromAlign for inter-run RT adjustment (39). Quality control as well as the selection of the optimal reference run for the alignment step were achieved by monitoring alignment scores (>0.8 as qualified) and base-peak intensity.
-
ii)
Data-independent MS1 feature generation using the DICE method. Characteristics of the both steps are discussed in Results. DICE in IonStar generates quantitative features for all precursor ions with corresponding MS2 scans by extracting ICs in the aligned dataset with a defined m/z-RT window. We optimized the width of the extraction window using different combinations of m/z and RT (i.e., 5 to 20 ppm, 1 to 2 min), and it turned out that 10 ppm (i.e., m/z ± 5 ppm) and 1 min (i.e., RT ± 0.5 min) returned the best outcomes for the benchmark dataset.
-
iii)
A Structured Query Language database containing the list of quantitative features (i.e., defined as “frames”) as well as corresponding intensities in each LC-MS run was exported and matched to the filtered PSM list by MS2 scan number using a customized R script, IonStar_FrameGen.R. Frames with valid PSMs were then subjected to dataset-wide normalization, multivariate outlier detection/removal, and aggregation to protein level using IonStarStat. Details are enclosed in the user manual for IonStar.
Comparison of IonStar with Other Label-Free Quantitative Packages.
A comparison of quantitative performance was performed between IonStar and several popular label-free quantitative packages, including OpenMS (56), MaxQuant (40), Proteome Discoverer 2.1, and SpC. Due to various technical issues experienced when processing such a large-scale dataset (i.e., many LC-MS files using UHF-Orbitrap with a 1-m-long nano LC column), it was not feasible to compare Skyline and DeMix-Q in this evaluation. For the benchmark dataset, an eight-core, 32-GB, random-access memory personal computer was used for data processing; it took IonStar 2 d to accomplish the evaluation set compared with 3 d for MaxQuant and 2 wk for OpenMS.
To ensure an unbiased and comprehensive comparison, the following rules were applied to all packages:
-
i)
Based on literature guidance and experimental evaluations, the parameters for each package were individually optimized to achieve the best performance. Specifically, optimization of OpenMS was performed according to Weisser et al. (19), and processing procedures included chromatogram alignment and peptide identity transfer. For MaxQuant, optimization was based on configurations used by Cox et al. (4) (e.g., the use of proper instrumental parameters and the “match between runs” feature, with a 20-min alignment window and a 1-min matching tolerance). For PD 2.1, the “Precursor Ion Area Detection” module was used under the guidance of our colleagues at ThermoFisher. For SpC, the SpC function in MaxQuant was employed. Detailed parameters for each software program are provided in SI Appendix, File S1.
-
ii)
Database searching was conducted against the same human-E. coli concatenated database, with both peptide and protein FDR < 1%, and at least two unique peptides per protein. The two-unique-peptide rule was adopted to ensure confident quantification by all packages, and we found that using the two-peptide rule obtained notably better quantitative outcomes than using one peptide per protein (SI Appendix, Fig. S6).
-
iii)
Proteins with missing data were excluded from comparison of reproducibility, precision, accuracy, and FADR, which is a common practice used in many other comparative studies (11, 33, 54, 57). Missing data imputation was not employed, as it was not the focus of this study, although it appears to be complicated and widely debated (13, 57, 58).
-
iv)
Optimal normalization methods were applied for individual packages to correct analytical variance and biases. Raw and processed quantitative results are provided in Datasets S2 and S3.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.R.Y. is a guest editor invited by the Editorial Board.
Data deposition: The benchmark spike-in sample set has been deposited on ProteomeXchange Consortium via the Proteomics Identifications (PRIDE) partner repository (PRIDE identifier PXD003881). IonStarStat, relevant R scripts (IonStar_FrameGen.R and IonStar_Run.R), and the user manual for IonStar have been deposited on GitHub (https://github.com/shichens1989/IonStarStat).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800541115/-/DCSupplemental.
References
- 1.Mischak H, et al. Clinical proteomics: A need to define the field and to begin to set adequate standards. Proteomics Clin Appl. 2007;1:148–156. doi: 10.1002/prca.200600771. [DOI] [PubMed] [Google Scholar]
- 2.Qian W-J, Jacobs JM, Liu T, Camp DG, 2nd, Smith RD. Advances and challenges in liquid chromatography-mass spectrometry-based proteomic profiling for clinical applications. Mol Cell Proteomics. 2006;5:1727–1744. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: The long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 4.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higgs RE, Knierman MD, Gelfanova V, Butler JP, Hale JE. Label-free LC-MS method for the identification of biomarkers. Methods Mol Biol. 2008;428:209–230. doi: 10.1007/978-1-59745-117-8_12. [DOI] [PubMed] [Google Scholar]
- 6.Merl J, Ueffing M, Hauck SM, von Toerne C. Direct comparison of MS-based label-free and SILAC quantitative proteome profiling strategies in primary retinal Müller cells. Proteomics. 2012;12:1902–1911. doi: 10.1002/pmic.201100549. [DOI] [PubMed] [Google Scholar]
- 7.Schilling B, et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: Application to protein acetylation and phosphorylation. Mol Cell Proteomics. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nahnsen S, Bielow C, Reinert K, Kohlbacher O. Tools for label-free peptide quantification. Mol Cell Proteomics. 2013;12:549–556. doi: 10.1074/mcp.R112.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen X, Hu Q, Li J, Wang J, Qu J. Experimental null method to guide the development of technical procedures and to control false-positive discovery in quantitative proteomics. J Proteome Res. 2015;14:4147–4157. doi: 10.1021/acs.jproteome.5b00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruderer R, et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol Cell Proteomics. 2015;14:1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang B, Käll L, Zubarev RA. DeMix-Q: Quantification-centered data processing workflow. Mol Cell Proteomics. 2016;15:1467–1478. doi: 10.1074/mcp.O115.055475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domon B, Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nat Biotechnol. 2010;28:710–721. doi: 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- 13.Webb-Robertson BJ, et al. Review, evaluation, and discussion of the challenges of missing value imputation for mass spectrometry-based label-free global proteomics. J Proteome Res. 2015;14:1993–2001. doi: 10.1021/pr501138h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mann M, Hendrickson RC, Pandey A. Analysis of proteins and proteomes by mass spectrometry. Annu Rev Biochem. 2001;70:437–473. doi: 10.1146/annurev.biochem.70.1.437. [DOI] [PubMed] [Google Scholar]
- 15.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 16.Michalski A, Cox J, Mann M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J Proteome Res. 2011;10:1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 17.Chen YY, et al. IDPQuantify: Combining precursor intensity with spectral counts for protein and peptide quantification. J Proteome Res. 2013;12:4111–4121. doi: 10.1021/pr400438q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 19.Weisser H, et al. An automated pipeline for high-throughput label-free quantitative proteomics. J Proteome Res. 2013;12:1628–1644. doi: 10.1021/pr300992u. [DOI] [PubMed] [Google Scholar]
- 20.Gillet LC, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selevsek N, et al. Reproducible and consistent quantification of the Saccharomyces cerevisiae proteome by SWATH-mass spectrometry. Mol Cell Proteomics. 2015;14:739–749. doi: 10.1074/mcp.M113.035550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Röst HL, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 23.Guo T, et al. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nat Med. 2015;21:407–413. doi: 10.1038/nm.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ting YS, et al. PECAN: Library-free peptide detection for data-independent acquisition tandem mass spectrometry data. Nat Methods. 2017;14:903–908. doi: 10.1038/nmeth.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsou C-C, et al. DIA-umpire: Comprehensive computational framework for data-independent acquisition proteomics. Nat Methods. 2015;12:258–264, 7, 264. doi: 10.1038/nmeth.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu C, et al. ICan: An optimized ion-current-based quantification procedure with enhanced quantitative accuracy and sensitivity in biomarker discovery. J Proteome Res. 2014;13:5888–5897. doi: 10.1021/pr5008224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu C, Li J, Sheng Q, Zhang M, Qu J. Systematic assessment of survey scan and MS2-based abundance strategies for label-free quantitative proteomics using high-resolution MS data. J Proteome Res. 2014;13:2069–2079. doi: 10.1021/pr401206m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lott K, et al. Global proteomic analysis in trypanosomes reveals unique proteins and conserved cellular processes impacted by arginine methylation. J Proteomics. 2013;91:210–225. doi: 10.1016/j.jprot.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nouri-Nigjeh E, et al. Highly multiplexed and reproducible ion-current-based strategy for large-scale quantitative proteomics and the application to protein expression dynamics induced by methylprednisolone in 60 rats. Anal Chem. 2014;86:8149–8157. doi: 10.1021/ac501380s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen X, et al. An ionStar experimental strategy for MS1 ion current-based quantification using ultrahigh-field orbitrap: Reproducible, in-depth, and accurate protein measurement in large cohorts. J Proteome Res. 2017;16:2445–2456. doi: 10.1021/acs.jproteome.7b00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandin M, Teleman J, Malmström J, Levander F. Data processing methods and quality control strategies for label-free LC-MS protein quantification. Biochim Biophys Acta. 2014;1844:29–41. doi: 10.1016/j.bbapap.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 32.Chawade A, Sandin M, Teleman J, Malmström J, Levander F. Data processing has major impact on the outcome of quantitative label-free LC-MS analysis. J Proteome Res. 2015;14:676–687. doi: 10.1021/pr500665j. [DOI] [PubMed] [Google Scholar]
- 33.Weisser H, Choudhary JS. Targeted feature detection for data-dependent shotgun proteomics. J Proteome Res. 2017;16:2964–2974. doi: 10.1021/acs.jproteome.7b00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen LN, et al. Proteomic analyses for the global S-Nitrosylated proteins in the brain tissues of different human prion diseases. Mol Neurobiol. 2016;53:5079–5096. doi: 10.1007/s12035-015-9440-7. [DOI] [PubMed] [Google Scholar]
- 35.Licker V, et al. Proteomic analysis of human substantia nigra identifies novel candidates involved in Parkinson’s disease pathogenesis. Proteomics. 2014;14:784–794. doi: 10.1002/pmic.201300342. [DOI] [PubMed] [Google Scholar]
- 36.Shen S, et al. Large-scale, ion-current-based proteomic investigation of the rat striatal proteome in a model of short- and long-term cocaine withdrawal. J Proteome Res. 2016;15:1702–1716. doi: 10.1021/acs.jproteome.6b00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musunuri S, et al. Quantification of the brain proteome in Alzheimer’s disease using multiplexed mass spectrometry. J Proteome Res. 2014;13:2056–2068. doi: 10.1021/pr401202d. [DOI] [PubMed] [Google Scholar]
- 38.Smith RD, et al. An accurate mass tag strategy for quantitative and high-throughput proteome measurements. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 39.Sadygov RG, Maroto FM, Hühmer AF. ChromAlign: A two-step algorithmic procedure for time alignment of three-dimensional LC-MS chromatographic surfaces. Anal Chem. 2006;78:8207–8217. doi: 10.1021/ac060923y. [DOI] [PubMed] [Google Scholar]
- 40.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 41.Park SK, Venable JD, Xu T, Yates JR., 3rd A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 2008;5:319–322. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mueller LN, et al. SuperHirn–A novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics. 2007;7:3470–3480. doi: 10.1002/pmic.200700057. [DOI] [PubMed] [Google Scholar]
- 43.Kim S, Pevzner PA. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filzmoser P, Maronna R, Werner M. Outlier identification in high dimensions. Comput Stat Data Anal. 2008;52:1694–1711. [Google Scholar]
- 45.Carrillo B, Yanofsky C, Laboissiere S, Nadon R, Kearney RE. Methods for combining peptide intensities to estimate relative protein abundance. Bioinformatics. 2010;26:98–103. doi: 10.1093/bioinformatics/btp610. [DOI] [PubMed] [Google Scholar]
- 46.Tabb DL, et al. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res. 2010;9:761–776. doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Margolin AA, et al. Empirical Bayes analysis of quantitative proteomics experiments. PLoS One. 2009;4:e7454. doi: 10.1371/journal.pone.0007454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Control CD. Prevention, Report to Congress on Traumatic Brain Injury in the United States: Epidemiology and Rehabilitation. National Center for Injury Prevention and Control; Atlanta: 2014. pp. 1–72. [Google Scholar]
- 49.McConeghy KW, Hatton J, Hughes L, Cook AM. A review of neuroprotection pharmacology and therapies in patients with acute traumatic brain injury. CNS Drugs. 2012;26:613–636. doi: 10.2165/11634020-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 50.Rau TF, Kothiwal A, Rova A, Rhoderick JF, Poulsen DJ. Phenoxybenzamine is neuroprotective in a rat model of severe traumatic brain injury. Int J Mol Sci. 2014;15:1402–1417. doi: 10.3390/ijms15011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rau TF, et al. Administration of low dose methamphetamine 12 h after a severe traumatic brain injury prevents neurological dysfunction and cognitive impairment in rats. Exp Neurol. 2014;253:31–40. doi: 10.1016/j.expneurol.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 52.Vizcaíno JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Searle BC. Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10:1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 54.Ma ZQ, et al. IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J Proteome Res. 2009;8:3872–3881. doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez MF, et al. Mass spectrometric discovery and selective reaction monitoring (SRM) of putative protein biomarker candidates in first trimester Trisomy 21 maternal serum. J Proteome Res. 2011;10:133–142. doi: 10.1021/pr100153j. [DOI] [PubMed] [Google Scholar]
- 56.Sturm M, et al. OpenMS–An open-source software framework for mass spectrometry. BMC Bioinformatics. 2008;9:163. doi: 10.1186/1471-2105-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karpievitch YV, Dabney AR, Smith RD. Normalization and missing value imputation for label-free LC-MS analysis. BMC Bioinformatics. 2012;13(Suppl 16):S5. doi: 10.1186/1471-2105-13-S16-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lazar C, Gatto L, Ferro M, Bruley C, Burger T. Accounting for the multiple natures of missing values in label-free quantitative proteomics data sets to compare imputation strategies. J Proteome Res. 2016;15:1116–1125. doi: 10.1021/acs.jproteome.5b00981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.